Open Access Article
Open Access ArticleCharacterization of a LanC-free pathway for the formation of an LL-MeLan residue and an alloAviMeCys residue in the newly identified class V lanthipeptide triantimycins†
Weizhong
Ding‡
a,
Xiaofeng
Wang‡
ab,
Yu
Yin
c,
Jiang
Tao
*d,
Yanqing
Xue
 *a and
Wen
Liu
*a
*a and
Wen
Liu
*a
aState Key Laboratory of Chemical Biology, Shanghai Institute of Organic Chemistry, University of Chinese Academy of Sciences, Chinese Academy of Sciences, 345 Lingling Road, Shanghai 200032, China. E-mail: yanqing@illinois.edu; wliu@sioc.ac.cn
bSchool of Chemistry and Materials Science, Hangzhou Institute for Advanced Study, University of Chinese Academy of Sciences, 1 Sublane Xiangshan, Hangzhou 310024, China
cShanghai Jiao Tong University, Shanghai 200240, China
dDepartment of General Dentistry, Shanghai Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine, College of Stomatology, Shanghai Jiao Tong University, National Center for Stomatology, National Clinical Research Center for Oral Diseases, Shanghai Key Laboratory of Stomatology, Shanghai Research Institute of Stomatology, Research Unit of Oral and Maxillofacial Regenerative Medicine, Chinese Academy of Medical Science, No. 639 Zhizaoju Road, Shanghai 200011, China. E-mail: taojiang_doctor@hotmail.com
First published on 15th May 2024
Abstract
The thioether-connected bis-amino acid lanthionine (Lan) residues are class-defining residues of lanthipeptides. Typically, the cyclization step of lanthionine formation, which relies on the addition of a cysteine to an unsaturated dehydroamino acid, is directed either by a standalone cyclase LanC (class I) or by a cyclase domain (class II–IV). However, the pathways of characterized class V members often lack a known cyclase (domain), raising a question on the mechanism by which their multi-macrocycle systems are formed. Herein, we report a new RiPP gene cluster in Streptomyces TN 58, where it encodes the biosynthesis of 3 distinct class V lanthipeptides—termed triantimycins (TAMs). TAM A1∼A3 share an N-terminal LL-MeLan residue, and only TAM A1 contains an additional internal LL-Lan residue. TAM A1 also has a C-terminal (2S, 3R)-S-((Z)-2-aminovinyl)-3-methyl-D-cysteine (alloAviMeCys) residue, which is distinct from the previously reported (2S, 3S)-AviMeCys residue in other RiPPs. Gene deletion, heterologous coexpression, and structural elucidation demonstrated that the cyclization for an LL-MeLan formation occurs spontaneously and is independent of any known lanthionine cyclase. This study provides a new paradigm for lanthionine formation and facilitates genome mining and engineering efforts on RiPPs containing (Me)Lan and (allo)Avi(Me)Cys residues.
Introduction
Lanthipeptides represent one of the largest classes of ribosomally synthesized and post-translationally modified peptides (RiPPs),1 with a continual surge in new members emerging from diverse bacterial phyla.2 Recent genome mining and activity-based screening efforts collectively contributed to the identification of class V lanthipeptides.3 Based on their structural ring pattern, the identified class V lanthipeptides could be divided into 2 subgroups (Fig. 1A). Class Va subgroup members include cacaoidin,3b,4 lexapeptide,3c and pristinins,3a as well as recently reported massatides, sistertides, and kebanetides (Fig. 1A and S1†).5 Besides the N-terminal and/or internal lanthionine (Lan) and/or methyl-lanthionine (MeLan) residue(s), they also contain a C-terminal (2S, 3S)-S-((Z)-2-aminovinyl)-3-methyl-D-cysteine ((2S, 3S)-AviMeCys) residue which is believed to be critical for bioactivity.6 mSmoAC is the parent and the sole member to date of the class Vb subgroup.7 It is constrained in a stapled configuration by 2 overlapping (Me)Lan residues and a C-terminal Lan residue (Fig. 1A). | ||
| Fig. 1 Bioinformatical exploration of class Va lanthipeptides. (A) Structural classification of known class Va and Vb lanthipeptides. The D-amino acids are colored in blue, and other residues that underwent PTMs are in bold. (B) SSN analysis of class Va candidates. Precursors containing 0–3 Cys residue(s) are colored in blue, light blue, gray, and orange, respectively. Precursors of cacaoidin, lexapeptide, and pristinins (that is CaoA, LxmA, and SprA1-A3, respectively) are shaped in diamond. (C) Core sequence analysis of selected subgroups. For details, see Fig. S2 and Table S1.† | ||
The lanthipeptide class-defining (Me)Lan residues are typically synthesized by distinct sets of post-translational modification (PTM) enzymes known as lanthipeptide synthetases in class I–IV lanthipeptides.2e,8 A pathway-dedicated cyclase LanC (class I) or a cyclase domain (class II–IV) fused to a megasynthetase directs the thia-Michael addition of a cysteine to an intramolecular dehydroalanine (Dha) or dehydrobutyrine (Dhb) residue for the formation of Lan or MeLan residue.8a For class V lanthipeptides, the split kinase-like LanK and effector-like protein LanY act as the primary PTM enzymes for the formation of Dhx (Dha or Dhb) residues,9 which are presumed to be substrates for D-amino acid(s), (Me)Lan residue(s), and the (2S, 3S)-AviMeCys residue (Fig. S1†).4 The cyclization of (Me)Lan residues in class Vb involves a dedicated LanC cyclase that catalyzes the otherwise energetically unfavored cyclization of the overlapping (Me)Lan residues.7 However, the class Va members lack a known cyclase in their encoding gene clusters (Fig. S1†).3,5 While a non-enzymatic macrocyclization hypothesis has been proposed,7 there is a lack of experimental support. Available biosynthetic insights into the cyclization of (Me)Lan and AviMeCys residues are primarily derived from lexapeptide.3c The formation of Lan and AviMeCys residues in lexapeptide involves 4 PTM enzymes: a LanK (LxmK), a LanY (LxmY), and 2 PTM enzymes (LxmD and LxmX) that have limited homology with the minimum AviMeCys synthase in thioviridamides.10 Omitting any one of these four genes failed to produce lexapeptide or any pathway-related metabolites.
To elucidate the cyclization process in class Va lanthipeptides, we started to bioinformatically analyze the gene clusters encoding the co-occurrence of LanK and LanY counterparts but without a cyclase (domain) homologous to those for class I–IV members. Our efforts led to the discovery of triantimycins (TAMs), 3 new class V lanthipeptides with synergistic activity against several Gram-positive pathogens. TAMs share an N-terminal LL-MeLan residue, while TAM A1 also occupies an extra internal LL-Lan and a C-terminal (2S, 3R)-S-((Z)-2-aminovinyl)-3-methyl-D-cysteine (alloAviMeCys). To our knowledge, the alloAviMeCys residue is stereochemically distinct from the previously reported (2S, 3S)-AviMeCys residue in other RiPPs,6 indicating a different forming process. Gene deletion, heterologous co-expression, and extensive structural elucidation efforts collectively supported that the cyclization step for the LL-MeLan formation occurs spontaneously. Our work showcases a site- and stereochemistry-controlled cyclization paradigm that is independent of a LanC cyclase or a cyclase domain.
Results and discussion
Bioinformatic exploration of class Va lanthipeptides
We investigated Streptomyces species (taxonomy ID: 1883) in the NCBI reference Sequence database for class Va lanthipeptide candidates using the cblaster tool,11 with the sequences of CaoK/CaoY (cases of LanK/LanY in cacaoidin) as the input.9 To filter out false positives in which LanC cyclases or other lanthipeptide synthetases are co-existent,7,12 the co-occurrence of CaoD/CaoX (cases of LanD/LanX in cacaoidin) was tentatively utilized as a secondary confirmation. This approach was based on the observation that all known class Va lanthipeptides exhibit the co-occurrence of LanK/LanY/LanD/LanX (Fig. S1†). A total of 194 promising hits were identified from 188 Streptomyces strains, which encode 129 non-redundant precursor peptides (Table S1†). All hits lack a LanC cyclase (domain) and share a highly similar gene set encoding PTM enzymes found in known class Va lanthipeptides, except for N-methyltransferase. Sequence similarity network (SSN) analysis of the 129 precursors revealed that the class Va candidates could be further classified into more than 9 subgroups (Fig. 1B). Among the 129 precursors, 43 lack an N-methyltransferase within their putative gene clusters. Most of them are concentrated within subgroups 2 (12/18), 5 (9/9), 6 (5/7), and 7 (7/7). Precursors in subgroups 1–4 and 9 have 2 Cys residues (n = 2), while members in subgroups 5–8 only contain 1 Cys residue (n = 1). Over half of the singletons contain 3 Cys residues (n = 3), as demonstrated by SprA3 (precursor for pristinin A3). Other singletons contain varying numbers of Cys residues (n = 1, 2), except for 1 sequence from S. sp. L2, which does not have any Cys residue (n = 0) (Accession: WP_164992318, Table S1†). The core sequence analysis suggested the presence of 2 remarkable sequence motifs: (S/T)2X2C and (S/T)X4C (Fig. 1C and S2†). These 2 motifs strongly correlate with the ring pattern of mature products. Both the N-terminal and internal (S/T)2X2C motif contribute to (Me)Lan residues, while the C-terminal (S/T)X4C motif leads to an Avi(Me)Cys residue. For example, pristinin A2,3a a mature product from subgroup 1, has an N-terminal MeLan residue derived from the T2X2C motif and a C-terminal AviMeCys residue from the TX4C motif (Fig. 1). Lexapeptide from subgroup 3 is another exemplar. Members in subgroup 8 with an N-terminal T2X2C motif drew our attention because they likely form a single MeLan residue. This makes them an ideal model for studying the cyclization mechanism of class Va lanthipeptides. In subgroup 8, 2 out of 6 precursors encoded in the stn cluster from S. sp. TN58 are termed StnA2 and StnA3. StnA2 shares a highly conserved N-terminal 34-amino acid (aa) leader peptide (LP) and a nearly identical C-terminal 22-aa core peptide (CP) with StnA3 (Fig. 2A). In addition to StnA2/StnA3, the stn cluster also contains a longer precursor peptide termed StnA1, which consists of a 32-aa LP and a 36-aa CP. Besides the 3 precursors (StnA1, StnA2, and StnA3) and the 4 bait PTM enzymes (StnK, StnY, StnD, and StnX), the stn cluster likely encodes an N-methyltransferase (StnM),3b a LanJC family reductase (StnJ),3c an insulinase family protease (StnP), 3 ABC transporters (StnT1, StnT2, and StnT3), a luxR family regulator (StnR), and 2 hypothetic orfs (termed StnU1 and StnU2, Fig. 2A and Table S2,† the nomenclature for each gene was derived from the corresponding gene in lexapeptide).Heterologous expression of the stn cluster resulted in compounds 1, 2, and 3
S. sp. TN58 is a prolific producer of a series of antimicrobial molecules.13 However, no peptidyl metabolites related to the stn cluster were reported, possibly due to transcriptional silence under routine laboratory cultures. To obtain the products of the stn cluster, we employed our recently developed heterologous expression strategy.14 In short, 3 structural genes (stnA1/A2/A3) and 7 PTM genes (stnK/Y/D/E/M/J/P) for RiPP maturation were cloned into the resulting plasmid pXYDuet-stn (Fig. 2B), while the regulatory and transporter genes were omitted. This construct was then transferred into and overexpressed in a S. coelicolor M1146 chassis.15 After a 5 day fermentation, the crude methanol extract of the mycelium was analyzed by high-performance liquid chromatography (HPLC) coupled with high-resolution mass spectrometry (HRMS). 2 new ion peaks were observed with the molecular weights over 2000 Da. HRMS of the first peak revealed a triple-protonated ion [M+3H]3+ = 1020.8387, while the second peak was dominated by 2 double-protonated ions [M+2H]2+ = 1059.0021 and 1043.0165 (Fig. 2C). These data indicated that the heterologous expression of the stn cluster led to 3 new compounds, designated as 1, 2, and 3, respectively (Fig. 2D). To further corroborate the relationship between the compounds 1∼3 and the structural genes stnA1∼stnA3, we conducted 3 in-frame double knockouts. The resulting ΔstnA1A2, ΔstnA1A3 and ΔstnA2A3 strains abolished the production of compounds 1 and 2, compounds 1 and 3, and compounds 2 and 3, respectively (Fig. S3†). Compounds 1, 2, and 3 were confirmed as the mature metabolites of structural genes stnA1, stnA2, and stnA3, respectively.Planar structure of compounds 1, 2, and 3
With the genomic sequence of the structural genes (stnA1∼A3) in mind (Fig. 2A), HR-MS/MS analysis was conducted to map PTMs occurring on their core sequences during maturation. As expected, the fragments of 1 revealed a 3-ring system, including an N-terminal 5-aa MeLan residue (A ring) formed by Thr1/Cys5, an internal 5-aa Lan residue (B ring) formed by Ser15/Cys19, and a C-terminal (C ring) AviMeCys residue formed by Thr31/Cys36 (Fig. 2D and S4†). The in-source fragment ion at [M+2H]2+ = 1014.4880 (y24 ion of 1) was fragmented in a pseudo-MS3 approach (Fig. S5†), which further supported the location of the B and C ring. The fragments of 2 and 3 were nearly identical, which indicated that similar PTMs occurred on their CPs (Fig. 2D, and S6, S7†). The noncanonical residues, such as Dhx, and aminobutyric acid (Abu), were also found in 1, 2 and 3. Taken together, compounds 1, 2, and 3 exhibit PTMs commonly observed in other known class Va lanthipeptides, establishing them as 3 new members.To avoid the mixing of compounds 2 and 3 during isolation due to their structural similarity, we engineered the stnA2 gene in the heterologous expression system to produce a precursor peptide in which the Ala8 and Met12 residues were replaced with Val8 and Ala12 residues, respectively. The recombinant stnA2:stnA3 strain abolished the production of compound 2, while the production of compound 1 remained unaffected, and the production of compound 3 was enhanced. To confirm their planar structures, a scaled-up batch fermentation was conducted. We purified compounds 1 (∼10 mg) and 3 (∼10 mg) through a 120 L fermentation. NMR-based sequences correlated well with the genomic and tandem MS data for both 1 and 3 (Fig. S8 and S9†). All the Dhb residues in compounds 1 and 3 were assigned as Z-geometry based on the NOE correlations of the NH with the γ-Me group (Fig. S8 and S9†).
The extensively collected NMR correlations were then utilized for the regio- and geometry-assignments of the thioether linkages. For compound 3, HMBC correlations between β-CH2 of former Cys5 and β-CH of former Thr1 confirmed the 5-aa MeLan-type linkage (Fig. S9†). A strong NOE correlation between NMe2 and γ-Me of former Thr1 revealed a cis geometry (Fig. 3A), which supports either an LL-MeLan or a DL-MeLan residue and precludes the possibility of other allo isomers.16 For compound 1, as expected, the correlation signals of the A ring were similar to those observed in compound 3 (Fig. 3A and S8†), which also supports a 5-aa MeLan residue. HMBC correlations from β-CH2 of former Cys19 to β-C of former Ser15 evidenced a 5-aa Lan-type ring (B ring, Fig. S8†). Correlation signals between β-CH of former Thr31 and β-CH of former Cys36 validated the 6-aa AviMeCys-type C ring (Fig. S8†). A Z-geometry double bond of the AviMeCys was determined based on the 3JH,H coupling value (8.5 Hz, Table S3†). Upon careful analysis of the rotating frame Overhauser effect spectroscopy (ROESY) of compound 1, we observed correlations between α-H and β-H, α-H and γ-Me, and α-NH and β-H, but not with γ-Me (Fig. S10†). These observations, coupled with the S configuration for α-C obtained by Marfey's analysis (vide infra), align well with a conformation of the (2S, 3R) configuration according to Newman projection analysis (Fig. 3B), inspired by the pioneering work of Ren et al.18
Absolute configuration of the residues in compounds 1 and 3
We examined the absolute configurations of the residues in 1 and 3 using the advanced Marfey's method.3b,c,14 For compound 1, the acid hydrolysate was reacted with excessive Marfey's reagent, 1-fluoro-2, 4-dinitrophenyl-5-L-alanine amide (L-FDAA) and/or its enantiomer D-FDAA, and then compared with the standard amino acids using HPLC-MS. This analysis revealed the presence of both L- and D-Ala residues, with a peak area ratio of 4 L-Ala residues to 1 D-Ala residue (Fig. S11†). Considering there are 12 genetically encoded L-Ala residues, the prorated 3 D-Ala residues were possibly transformed from 3 former Ser residues. Furthermore, the Abu residue was observed in the D configuration (Fig. S11†). For compound 3, the chiral analysis of its hydrolysate also revealed a mixture of L- and D-Ala, with a peak area ratio of 3 to 2. This is consistent with the scenario in which the proteinogenic 3 L-Ala residues remained unmodified, and the newly generated 2 Ala residues were both in the D-configuration (Fig. S12†). Except for the achiral Gly residue, all the other canonical residues in 1 and 3 are in the L configuration (Fig. S11 and S12†). Collectively, these data supported that the enzymatic hydrogenation of Dhx residues results in D-amino acids3c.Next, we focused on the absolute stereochemistry of (Me)Lan residues. The L-FDAA derivative of NMe2-MeLan residue (m/z 727.17) was detected in the hydrolysates of both 1 and 3. However, NMe2-MeLan standards are resistant to routine synthesis. Based on the knowledge from cypemycin,14,17b the N,N-dimethylation catalyzed by the methyltransferase would not change the chirality. To obtain the desmethylated metabolites, we deleted the stnM gene on the ΔstnA1A2 strain, which ultimately led to the compound 3-1 (Fig. S13†). HR-MS/MS analysis supported that 3-1 is the desmethylated congener of 3 (Fig. S13†). We purified 3-1 (∼0.5 mg) for acidic hydrolysis and chemically synthesized DL- and LL-MeLan as the standards (see ESI†).17 The L-FDAA-MeLan from 3-1 coeluted with LL-MeLan, which confirmed that the N-terminal MeLan residue in 3-1 is in LL-stereochemistry (Fig. 3C). This observation somewhat contradicts a previous report on cacaoidin,3b in which the configuration of its N-terminal Lan residue was empirically assigned as DL, despite being in the same (T/S)2X2C motif. We suspected that the (T/S)2X2C motif leads to an LL-(Me)Lan residue in the metabolites of the stn cluster. To support this, we accordingly compared the internal Lan residue in compound 1 with the chemically synthesized LL-Lan and DL-Lan standards (see ESI†).17 HPLC-MS revealed that the Lan from 1 coeluted with the LL-Lan standard, which confirmed that the internal Lan residue in 1 is indeed an LL-Lan residue (Fig. 3D).
Finally, we focused on the absolute stereochemistry of the alloAviMeCys residue in 1. One classic reductive cleavage method for assigning the stereochemistry of AviMeCys residue was not suitable for compound 1,3b as it would also reduce the Dhb2 residue to a racemic Abu residue. The AviCys residue of cypemycin was reported to decompose during acidic hydrolysis, leading to an Ala residue with retained stereochemistry.17b We hypothesized that the acidic hydrolysis of the alloAviMeCys-31 residue would similarly result in a chiral-retained Abu residue. Given that the hydrolysate of 1 only contained D-Abu (Fig. S11†), the stereochemistry of α-C of the alloAviMeCys-31 could be designated as “S”. To further support this designation, we conducted a T8S mutation on StnA1, which led to the production of 1-T8S (Fig. S14†). The tandem MS of 1-T8S supported that it is a 1-like compound but lacking the Abu residue (Fig. S14†). Marfey's analysis of 1-T8S again revealed an overwhelming presence of D-Abu residue (Fig. S15†), which strongly supported the existence of a 2S-AviMeCys residue. Combined with the allo relative stereochemistry determined by NMR, a non-canonical (2S, 3R)-S-[(Z)-2-aminovinyl]-3-methyl-D-cysteine (alloAviMeCys) in 1 was assigned (Fig. 4). To our knowledge, the alloAviMeCys residue is stereochemically diverse from the (2S, 3S)-AviMeCys residue reported in other RiPPs to date.6,18 This observation contradicts the long-assumed ‘net’ anti-addition of an enethiol nucleophile to a Z-Dhb residue, which would leave the α-H and β-CH in a cis relative position to produce (2S, 3S)-AviMeCys.3b
Formation of an alloAviMeCys residue requires the enzymatic activities of StnK/Y/D/X
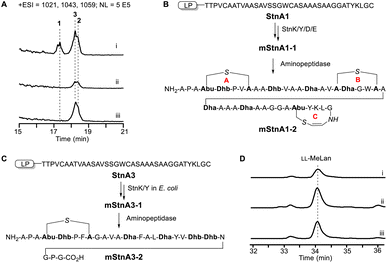
Considering the homology between StnK/Y/D/X and their counterparts in lexapeptide (LxmK/Y/D/X, respectively), it is likely that these 4 PTM enzymes are responsible for the formation of the alloAviMeCys residue in 1. To confirm this, we conducted 4 in-frame single knockout of the 4 genes (stnK/Y/D/X) in the wild-type construct. The ΔstnK or ΔstnY abolished the production of 3 compounds. Interestingly, both the ΔstnD and ΔstnX mutants abolished the production of compound 1, while still producing compounds 2 and 3 without a significant yield decline (Fig. 5A). These data indicated that the formation of the alloAviMeCys residue requires all 4 genes of stnK/Y/D/X.To confirm that 4 genes stnK/Y/D/X are sufficient for the formation of an alloAviMeCys residue, we coexpressed stnA with stnK/Y/D/X in E. coli. To enhance the dehydratase activity of stnK/Y, we engineered a fused dehydratase gene stnYK, by adopting a strategy successfully used in cacaoidin.9b These proteins can be expressed individually (StnK/Y/D) or as a fusion protein (StnYK) in the cytoplasm of E. coli (Fig. S16†). This coexpression led to the production of a highly modified polypeptide mStnA1-1 (Fig. S17†). The labeling of free Cys residues of mStnA1-1 with iodoacetamide (IAA) did not result in IAA adducts (Fig. S18†), which indicated that the 3 rings are formed in mStnA1-1. After treatment with aminopeptidase, the MS analysis of the remaining peptidyl fragment mStnA1-2 supported that all three rings (A, B, and C) were established in the E. coli coexpression system (Fig. 5B and S19†). These observations are reminiscent of the enzymatic synthesis of AviMeCys residue in thioviridamides,10a and we parallelly designated StnD and StnX as the minimum alloAviMeCys synthase.
Cyclization of the LL-MeLan residue occurs spontaneously
The ΔstnD and ΔstnX mutants did not affect the production of compounds 2 and 3, which indicated that the formation of the MeLan residue in these compounds does not require the activities of stnD and stnX, but only necessitates the dehydratase activity of stnK and stnY. To exclude the involvement of other PTM genes within the stn cluster and other possible cross-cluster lanthipeptide synthetases,17b such as LanC (accession: CAB55532 and CAD55393) and LanKC (Accession: CAA19960) in S. coelicolor M1146, we coexpressed the stnA3 gene with the fused stnKY genes in E. coli. This resulted in the production of an overwhelming 6-dehydrated product mStnA3-1 (Fig. S20†), which was resistant to reaction with IAA (Fig. S21†), indicating the lack of free Cys residue. Subsequent treatment of mStnA3-1 with aminopeptidase produced mStnA3-2, which exhibited better fragmentation in tandem MS and revealed the 5-aa MeLan-type ring similarly formed by Thr1 and Cys5 (Fig. 5C and S22†). To analyse its absolute configuration, we scaled up a 10 L fermentation of the recombinant E. coli. After protein purification, ∼1 mg pure mStnA3-1 solid was used for further Marfey's analysis. The retention time of L-FDAA-derivatized MeLan residue from mStnA3-1 aligned well with that from compound 3-1 (Fig. 5D). The coelution further confirmed the LL geometry (Fig. 5D). The Dhx2X2C motif in the substrate, which is transformed from the (T/X)2X2C motif in the core peptide, demonstrates a robust correlation with the final formation of a 5-aa (Me)Lan-type ring. This correlation is exemplified by cacaoidin,3b pristinins,3a cytolysin L and S,19 carnolysins, and haloduracin β,20 excluding lexapeptide (4-aa Lan instead).3c Pioneering mechanistic studies on cytolysin S revealed that the Dhx2X2C motif forms a rigid helix through bifurcated intra-ring hydrogen bonds between residues i and i + 3 (i = 1, 2), which facilitate a 5-aa regioselective cyclization.21 The secondary structure of the motif also potentially determines the stereoselectivity for a LL-MeLan residue and renders the thia-Michael addition slightly exergonic.21 The formation of a 5-aa LL-MeLan residue was demonstrated to occur spontaneously in vitro with the prerequisite Dhx2X2C motif in cytolysin S, albeit the pathway-dedicated LanM enzyme also contributed to the stereoselective cyclization step.22 We tentatively designated that the site- and stereoselective cyclization of the (Me)Lan residue in the stn cluster occurs spontaneously, independent of any other PTM genes within the stn cluster or any other cross-cluster lanthipeptide synthetases.Synergistic antimicrobial activity against Gram-positive pathogens
Class Va members always exhibit potent antimicrobial activities.3b,c,5 We also conducted antimicrobial activity tests for the isolated compounds 1 and 3. Compound 1 showed moderate antimicrobial activity against several Gram-positive pathogens, while compound 3 exhibited no detectable antimicrobial activity when tested individually (Table 1). However, a mixture of compounds 1 and 3, in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 or 1
1 or 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 mass ratio, demonstrated a 4-fold increase in potency against Enterococcus strains (Table 1). Additionally, the mixture of compounds 1 and 3 led to a reduction in the usage of 1 for inhibiting S. aureus growth. These observations suggested that compounds 1 and 3 exhibit synergistic antimicrobial activity. We designated compounds 1∼3 as triantimycin (TAM) A1∼A3, respectively. The optical mixture ratio, synergistic mechanism, and mode of action await further exploration.
2 mass ratio, demonstrated a 4-fold increase in potency against Enterococcus strains (Table 1). Additionally, the mixture of compounds 1 and 3 led to a reduction in the usage of 1 for inhibiting S. aureus growth. These observations suggested that compounds 1 and 3 exhibit synergistic antimicrobial activity. We designated compounds 1∼3 as triantimycin (TAM) A1∼A3, respectively. The optical mixture ratio, synergistic mechanism, and mode of action await further exploration.
| Antibiotics | MSSAa | MRSAb | VSEc | VREd | Pseudomonas aeruginosa |
|---|---|---|---|---|---|
| a Methicillin-sensitive Staphylococcus aureus. b Methicillin-resistant Staphylococcus aureus. c Vancomycin-sensitive Enterococcus. d Vancomycin-resistant Enterococcus. n.d., not detected. | |||||
| Vancomycin | 0.5 | 1 | 4 | 128 | n.d. |
| 1 | 16 | 16 | 64 | 64 | 256 |
| 3 | >256 | >256 | >256 | >256 | 256 |
1+3, in mass ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
16 | 16 | 16 | 16 | >256 |
1+3, in mass ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
16 | 16 | 16 | 16 | >256 |
Conclusions
In summary, we characterized a LanC-free pathway for the production of TAMs—3 new class Va lanthipeptides from S. sp. TN58. Through careful structural elucidation, we identified an LL-MeLan residue, an LL-Lan residue, and a stereochemically diverse alloAviMeCys residue in TAMs. We evidenced that the site- and stereochemistry-selective formation of an LL-(Me)Lan residue occurred spontaneously, which provides a supplementary cyclization mechanism that is otherwise catalyzed by a pathway-dedicated LanC cyclase (domain).8 Our work provided useful biosynthetic insights into the establishment of a complicated macrocycle system of class Va lanthipeptides, which would benefit the exploration and engineering of RiPPs containing (Me)Lan and (allo)Avi(Me)Cys residues.5Data availability
All experimental and characterization data including NMR and MS spectra are available in the ESI.†Author contributions
W. D. and X. W.: investigation, data curation, validation. Y. Y.: investigation. J. T.: formal analysis, investigation, resources. Y.X.: conceptualization, formal analysis, visualization, writing – original draft. W. L.: supervision, formal analysis, funding acquisition, resources, project administration. All the authors contribute to writing – review & editing. W.D. and X.W. contributed equally to this work.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported in part by grants from the National Natural Science Foundation of China (32030002, 22193070, and 81974495).Notes and references
- M. Montalban-Lopez, T. A. Scott, S. Ramesh, I. R. Rahman, A. J. van Heel, J. H. Viel, V. Bandarian, E. Dittmann, O. Genilloud, Y. Goto, M. J. Grande Burgos, C. Hill, S. Kim, J. Koehnke, J. A. Latham, A. J. Link, B. Martinez, S. K. Nair, Y. Nicolet, S. Rebuffat, H. G. Sahl, D. Sareen, E. W. Schmidt, L. Schmitt, K. Severinov, R. D. Sussmuth, A. W. Truman, H. Wang, J. K. Weng, G. P. van Wezel, Q. Zhang, J. Zhong, J. Piel, D. A. Mitchell, O. P. Kuipers and W. A. van der Donk, Nat. Prod. Rep., 2021, 38, 130–239 RSC.
- (a) A. Barbour, L. Smith, M. Oveisi, M. Williams, R. C. Huang, C. Marks, N. Fine, C. Sun, F. Younesi, S. Zargaran, R. Orugunty, T. D. Horvath, S. J. Haidacher, A. M. Haag, A. Sabharwal, B. Hinz and M. Glogauer, Proc. Natl. Acad. Sci. U.S.A., 2023, 120, e2219392120 CrossRef CAS PubMed; (b) Y. He, A. Fan, M. Han, H. Li, M. Li, H. Fan, X. An, L. Song, S. Zhu and Y. Tong, Biochemistry, 2023, 62, 462–475 CrossRef CAS PubMed; (c) L. Li, J. Zhang, L. Zhou, H. Shi, H. Mai, J. Su, X. Ma and J. Zhong, Appl. Environ. Microbiol., 2023, 89, e0212322 CrossRef PubMed; (d) Y. Li, Y. Ma, Y. Xia, T. Zhang, S. Sun, J. Gao, H. Yao and H. Wang, Nat. Commun., 2023, 14, 2944 CrossRef CAS PubMed; (e) A. Sigurdsson, B. M. Martins, S. A. Duttmann, M. Jasyk, B. Dimos-Rohl, F. Schopf, M. Gemander, C. H. Knittel, R. Schnegotzki, B. Schmid, S. Kosol, L. Pommerening, M. Gonzales-Viegaz, M. Seidel, M. Hugelland, S. Leimkuhler, H. Dobbek, A. Mainz and R. D. Sussmuth, Angew. Chem., Int. Ed., 2023, 62, e202302490 CrossRef CAS PubMed; (f) S. S. Singh, D. Sharma, C. Singh, S. Kumar, P. Singh, A. Sharma, D. K. Das, A. K. Pinnaka, K. G. Thakur, R. P. Ringe and S. Korpole, J. Appl. Microbiol., 2023, 134, lxad054 CrossRef PubMed; (g) X. Wang, X. Chen, Z. J. Wang, M. Zhuang, L. Zhong, C. Fu, R. Garcia, R. Muller, Y. Zhang, J. Yan, D. Wu and L. Huo, J. Am. Chem. Soc., 2023, 145, 16924–16937 CrossRef CAS PubMed; (h) X. Wang, Z. Wang, Z. Dong, Y. Yan, Y. Zhang and L. Huo, ACS Chem. Biol., 2023, 18, 1218–1227 CrossRef CAS PubMed.
- (a) A. M. Kloosterman, P. Cimermancic, S. S. Elsayed, C. Du, M. Hadjithomas, M. S. Donia, M. A. Fischbach, G. P. van Wezel and M. H. Medema, PLoS Biol., 2020, 18, e3001026 CrossRef CAS PubMed; (b) F. J. Ortiz-Lopez, D. Carretero-Molina, M. Sanchez-Hidalgo, J. Martin, I. Gonzalez, F. Roman-Hurtado, M. de la Cruz, S. Garcia-Fernandez, F. Reyes, J. P. Deisinger, A. Muller, T. Schneider and O. Genilloud, Angew. Chem., Int. Ed., 2020, 59, 12654–12658 CrossRef CAS PubMed; (c) M. Xu, F. Zhang, Z. Cheng, G. Bashiri, J. Wang, J. Hong, Y. Wang, L. Xu, X. Chen, S. X. Huang, S. Lin, Z. Deng and M. Tao, Angew. Chem., Int. Ed., 2020, 59, 18029–18035 CrossRef CAS PubMed.
- F. Roman-Hurtado, M. Sanchez-Hidalgo, J. Martin, F. J. Ortiz-Lopez and O. Genilloud, Antibiotics, 2021, 10, 403 CrossRef CAS PubMed.
- C. Zhuo, H. Bei-Bei, L. Kangfan, G. Ying, S. Yuqi, Z. Zheng, L. Hongyan, L. Runze, Z. Haili, W. Song, Z. Wenxuan, T. Xiaoyu, L. Yong-Xin, bioRxiv, 2023, DOI:10.1101/2023.10.26.563470.
- (a) E. S. Grant-Mackie, E. T. Williams, P. W. R. Harris and M. A. Brimble, JACS Au, 2021, 1, 1527–1540 CrossRef CAS PubMed; (b) B. Cheng, Y. Xue, Y. Duan and W. Liu, ChemPlusChem, 2024, e202400047 CrossRef PubMed.
- Z. F. Pei, L. Zhu, R. Sarksian, W. A. van der Donk and S. K. Nair, J. Am. Chem. Soc., 2022, 144, 17549–17557 CrossRef CAS PubMed.
- (a) L. M. Repka, J. R. Chekan, S. K. Nair and W. A. van der Donk, Chem. Rev., 2017, 117, 5457–5520 CrossRef CAS PubMed; (b) A. Hernandez Garcia and S. K. Nair, ACS Cent. Sci., 2023, 9, 1944–1956 CrossRef CAS PubMed; (c) C. Ongpipattanakul, S. Liu, Y. Luo, S. K. Nair and W. A. van der Donk, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2217523120 CrossRef CAS PubMed.
- (a) H. Liang, I. J. Lopez, M. Sanchez-Hidalgo, O. Genilloud and W. A. van der Donk, ACS Chem. Biol., 2022, 17, 2519–2527 CrossRef CAS PubMed; (b) Y. Xue, M. Li, L. Hu, J. Liu, L. Pan and W. Liu, Chin. J. Chem., 2023, 41, 3579–3586 CrossRef CAS.
- (a) Y. Qiu, J. Liu, Y. Li, Y. Xue and W. Liu, Cell Chem. Biol., 2021, 28, 675–685 CrossRef CAS PubMed; (b) A. Sikandar, M. Lopatniuk, A. Luzhetskyy, R. Muller and J. Koehnke, J. Am. Chem. Soc., 2022, 144, 5136–5144 CrossRef CAS PubMed; (c) J. Xiong, S. Luo, C. X. Qin, J. J. Cui, Y. X. Ma, M. X. Guo, S. S. Zhang, Y. Li, K. Gao and S. H. Dong, Org. Lett., 2022, 24, 1518–1523 CrossRef CAS PubMed.
- C. L. M. Gilchrist, T. J. Booth, B. van Wersch, L. van Grieken, M. H. Medema, Y.-H. Chooi and A. Ouangraoua, Bioinform. Adv., 2021, 1, vbab016 CrossRef PubMed.
- T. H. Eyles, N. M. Vior, R. Lacret and A. W. Truman, Chem. Sci., 2021, 12, 7138–7150 RSC.
- (a) L. Mellouli, I. Karray-Rebai, S. Sioud, R. B. Ameur-Mehdi, B. Naili and S. Bejar, Curr. Microbiol., 2004, 49, 400–406 CrossRef CAS PubMed; (b) R. Ben Ameur Mehdi, K. A. Shaaban, I. K. Rebai, S. Smaoui, S. Bejar and L. Mellouli, Nat. Prod. Res., 2009, 23, 1095–1107 CrossRef CAS PubMed.
- Y. Xue, X. Wang and W. Liu, J. Am. Chem. Soc., 2023, 145, 7040–7047 CrossRef CAS PubMed.
- J. P. Gomez-Escribano and M. J. Bibb, Microb. Biotechnol., 2011, 4, 207–215 CrossRef CAS PubMed.
- R. Sarksian, J. D. Hegemann, M. A. Simon, J. Z. Acedo and W. A. van der Donk, J. Am. Chem. Soc., 2022, 144, 6373–6382 CrossRef CAS PubMed.
- (a) T. Denoel, A. Zervosen, T. Gerards, C. Lemaire, B. Joris, D. Blanot and A. Luxen, Bioorg. Med. Chem., 2014, 22, 4621–4628 CrossRef CAS PubMed; (b) L. Chu, J. Cheng, C. Zhou, T. Mo, X. Ji, T. Zhu, J. Chen, S. Ma, J. Gao and Q. Zhang, ACS Chem. Biol., 2022, 17, 3198–3206 CrossRef CAS PubMed.
- H. Ren, C. Huang, Y. Pan, S. R. Dommaraju, H. Cui, M. Li, M. G. Gadgil, D. A. Mitchell and H. Zhao, Nat. Chem., 2024 DOI:10.1038/s41557-024-01491-3.
- D. Van Tyne, M. J. Martin and M. S. Gilmore, Toxins, 2013, 5, 895–911 CrossRef CAS PubMed.
- A. L. McClerren, L. E. Cooper, C. Quan, P. M. Thomas, N. L. Kelleher and W. A. van der Donk, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 17243–17248 CrossRef CAS PubMed.
- W. Tang, G. Jiménez-Osés, K. N. Houk and W. A. van der Donk, Nat. Chem., 2014, 7, 57–64 CrossRef PubMed.
- W. Tang, G. N. Thibodeaux and W. A. van der Donk, ACS Chem. Biol., 2016, 11, 2438–2446 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc02302a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |