Open Access Article
Open Access ArticleKinetic–thermodynamic correlation of conformational changes in ammonium complexes of a flexible naphthocage†
Shan
He
ab,
Mao
Quan
bc,
Liu-Pan
Yang
 b,
Ho Yu
Au-Yeung
b,
Ho Yu
Au-Yeung
 *a and
Wei
Jiang‡
*a and
Wei
Jiang‡
 b
b
aDepartment of Chemistry and State Key Laboratory of Synthetic Chemistry, The University of Hong Kong, Hong Kong, China. E-mail: hoyuay@hku.hk
bDepartment of Chemistry and Guangdong Provincial Key Laboratory of Catalysis, Shenzhen Grubbs Institute, Southern University of Science and Technology, Shenzhen 518055, China
cShenzhen Key Laboratory of New Information Display and Storage Materials, College of Materials Science and Engineering, Shenzhen University, Shenzhen 518055, China
First published on 30th August 2024
Abstract
Conformational changes in non-covalent complexes are of fundamental importance to many chemical and biological processes. Yet, these low-energy structural changes are usually fast and difficult to monitor, which poses challenges in their detailed kinetic understanding. The correlation between kinetics and thermodynamics of the conformational change of a model supramolecular system featuring a flexible naphthocage and quaternary ammonium guests is described in this work. Guest binding initially locks the host in two major conformations, which then equilibrates over time to the more stable conformer. The overall rate of the system to attain conformational equilibrium is found to inversely correlate with the thermodynamic stability of the host–guest complexes, and hence not only can the kinetic parameters of the conformational exchange be predicted from the easily obtainable thermodynamic data, but the kinetic profile can also be rationalized by using the structural properties of the different guests.
Introduction
Conformational changes in non-covalent complexes, and their underlying thermodynamics and kinetics, are of fundamental importance in many chemical and biological processes such as catalysis, transport, protein–drug interactions, etc.1–4 For example, many neurodegenerative disorders including Alzheimer's and Parkinson's diseases are due to protein misfolding that can be considered as large-scale conformational changes.5 Understanding the relationship between the stability and dynamics of the various conformational states of the proteins is not only essential to the disease pathology, but also valuable to the development of new therapeutic strategies that can intervene in the onset and progression of the diseases.6–8While information on thermodynamics is generally more easily available, studies of fast, low-energy conformational changes to obtain rate and other kinetic parameters are however more challenging.9,10 Indeed, reported studies on conformationally flexible host–guest systems are largely focusing on structures and binding thermodynamics, whereas their kinetic studies remain rare and poorly understood despite also being important to the binding mechanism,11 dynamics,12 guest selectivity,13 and further downstream events.14 In this regard, appropriate supramolecular systems with systematically varying host/guest structures will be essential to investigate and establish a structure–thermodynamic–kinetic correlation behind the conformational exchange of non-covalent complexes. Kinetic parameters could also be predicted from readily accessible thermodynamics and structural data from such a correlation.
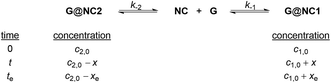
Similar to the linear free energy relationship between reaction rates and equilibrium constants for chemical reactions as described by Hammett and Brønsted plots,15 we report herein a study on correlating the binding thermodynamics and conformational kinetics of a host–guest system which consists of the conformationally flexible naphthocage NC and a series of quaternary ammonium guests. Upon guest binding, two conformers of the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 host–guest complexes are initially obtained, and the thermodynamically less stable conformer is found to convert to the more stable one over time whose kinetics can be followed by 1H NMR. Kinetic analysis showed that the rate of the conformational conversion is correlated with the stability of the two conformers, and hence the observed rate and kinetic parameters of the conformational change can be rationalized and predicted by the overall and local structural features of the guests (Fig. 1).
1 host–guest complexes are initially obtained, and the thermodynamically less stable conformer is found to convert to the more stable one over time whose kinetics can be followed by 1H NMR. Kinetic analysis showed that the rate of the conformational conversion is correlated with the stability of the two conformers, and hence the observed rate and kinetic parameters of the conformational change can be rationalized and predicted by the overall and local structural features of the guests (Fig. 1).
Results and discussion
Structures and thermodynamics of NC complexes
NC is a molecular cage having two 1,3,5-triethylbenzene covers and three (2,6-dibutoxy)-1,5-naphthyl side walls connected via ether linkages (Fig. 2).16 Quaternary ammonium cations can bind strongly to NC, and a series of quaternary ammoniums of different sizes and functional groups (G1–G24) were studied in the present work. Broad peaks were observed for the naphthyl protons in the 1H NMR spectrum of free NC, showing that the host exists as multiple exchanging conformations due to the flipping of the flexible naphthyl side walls with an exchange rate similar to that of the NMR time-scale (i.e. ∼102 s−1). Occupying the cage cavity by encapsulating an ammonium guest hindered the flipping motions and gave two major conformers, in which the naphthyl walls are either all in the same relative orientation (NC1), or one of the naphthyl walls orients differently than the other two (NC2).Binding of ammonium guests was followed by 1H NMR spectroscopy. Addition of the guest to a 1 mM solution of NC in CD2Cl2/CD3CN (v/v = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) resulted in two sets of sharp signals assignable to G@NC1 and G@NC2 that are in slow exchange. Concentration of G@NC1 was found to increase at the expense of G@NC2 over time except for G8, G10, G11, G14, G19 and G20. At equilibrium, 85% to 98% of the host–guest complexes were found to be G@NC1, suggesting that the symmetrical conformer is more stable. Depending on the structure of the guests and the overall symmetry of the inclusion complex, different 1H NMR spectral features were observed. For guests that can be fully encapsulated inside the cage, two doublets were observed for the naphthyl protons of G@NC1 with an overall D3 symmetry, and six naphthyl doublets were observed for the G@NC2 conformer with an overall C2 symmetry (“2 + 6” type). For guests with longer substituents that extend to the outside of the cage, the host–guest complexes are less symmetrical, in which G@NC1 will have an overall C2 symmetry with six naphthyl doublets, and the G@NC2 conformer will have a C1 symmetry with twelve doublets for the naphthyl protons (“6 + 12” type). Larger ammonium ions such as nPr4N+ and nBu4N+ showed no binding to NC, presumably because the cavity and/or the openings of the cage are not large enough for these larger ions.
1) resulted in two sets of sharp signals assignable to G@NC1 and G@NC2 that are in slow exchange. Concentration of G@NC1 was found to increase at the expense of G@NC2 over time except for G8, G10, G11, G14, G19 and G20. At equilibrium, 85% to 98% of the host–guest complexes were found to be G@NC1, suggesting that the symmetrical conformer is more stable. Depending on the structure of the guests and the overall symmetry of the inclusion complex, different 1H NMR spectral features were observed. For guests that can be fully encapsulated inside the cage, two doublets were observed for the naphthyl protons of G@NC1 with an overall D3 symmetry, and six naphthyl doublets were observed for the G@NC2 conformer with an overall C2 symmetry (“2 + 6” type). For guests with longer substituents that extend to the outside of the cage, the host–guest complexes are less symmetrical, in which G@NC1 will have an overall C2 symmetry with six naphthyl doublets, and the G@NC2 conformer will have a C1 symmetry with twelve doublets for the naphthyl protons (“6 + 12” type). Larger ammonium ions such as nPr4N+ and nBu4N+ showed no binding to NC, presumably because the cavity and/or the openings of the cage are not large enough for these larger ions.
The binding constant of these guests to NC was determined by the NMR competitive experiment and the data are summarized in Table 1.17 Binding constants (Ka1) of the more stable G@NC1 complexes are found to range from 2.4 × 103 M−1 (for G8) to 2.4 × 107 M−1 (for G9), and are generally related to the structure and bulkiness of the ammonium guests. For example, comparing G2 with G9 to G14, a smaller binding constant is found when more of the methyl groups in the ammonium guests are replaced by the relatively bulkier ethyl and propyl substituents. A similar observation can also be found when comparing the binding constants of G18 and G19, and that of G21 and G22. Also, binding of G17–G24 that feature either a phenyl, cyclohexyl or naphthyl substituent group is also generally weaker than that of other tetraalkylammoniums. G@NC2 is less stable and Ka2 is generally an order of magnitude smaller than Ka1. A plot of ln(Ka1) against ln(Ka2) shows a good positive correlation (Fig. 3), suggesting that there is a similar extent of differential stabilities for the two conformers of these host–guest complexes.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). Guests highlighted in purple and red are those that showed no conformational evolution and do not conform to the kinetic model (vide infra) respectively
1). Guests highlighted in purple and red are those that showed no conformational evolution and do not conform to the kinetic model (vide infra) respectively
| K a1/M−1 | K a2/M−1 | Type | K a1/M−1 | K a2/M−1 | Type | ||
|---|---|---|---|---|---|---|---|
| G1 | 1.9 × 106 | 3.9 × 104 | 2 + 6 | G13 | 5.6 × 105 | 9.1 × 104 | 2 + 6 |
| G2 | 2.0 × 107 | 8.3 × 105 | 2 + 6 | G14 | 1.1 × 105 | 5.3 × 104 | 2 + 6 |
| G3 | 6.2 × 106 | 6.9 × 105 | 2 + 6 | G15 | 1.0 × 107 | 7.7 × 105 | 2 + 6 |
| G4 | 2.5 × 106 | 1.6 × 105 | 2 + 6 | G16 | 1.4 × 107 | 2.9 × 105 | 2 + 6 |
| G5 | 8.6 × 105 | 7.5 × 104 | 6 + 12 | G17 | 1.4 × 105 | 2.1 × 104 | 2 + 6 |
| G6 | 4.2 × 106 | 4.1 × 105 | 2 + 6 | G18 | 2.1 × 105 | 2.9 × 104 | 6 + 12 |
| G7 | 1.6 × 106 | 1.6 × 105 | 2 + 6 | G19 | 6.2 × 104 | 5.5 × 103 | 6 + 12 |
| G8 | 2.4 × 103 | 3.4 × 102 | 2 + 6 | G20 | 3.0 × 103 | 3.0 × 102 | 6 + 12 |
| G9 | 2.4 × 107 | 1.3 × 106 | 2 + 6 | G21 | 3.8 × 104 | 4.2 × 103 | 6 + 12 |
| G10 | 1.2 × 106 | 4.6 × 106 | 2 + 6 | G22 | 1.9 × 104 | 1.4 × 103 | 6 + 12 |
| G11 | 5.9 × 105 | 2.7 × 105 | 2 + 6 | G23 | 1.1 × 105 | 7.0 × 103 | 6 + 12 |
| G12 | 3.4 × 106 | 7.5 × 104 | 2 + 6 | G24 | 1.6 × 105 | 6.9 × 103 | 6 + 12 |
Kinetics of the conformational changes
Kinetics of conformational evolution was studied by monitoring changes in the concentrations of G@NC1 and G@NC2 over time. During the conformational conversion, only resonances corresponding to the two complex conformers were observed, and no free host was detected, suggesting that the guest association was fast relative to the NMR timescale. The time required for the complexes to re-configure their conformations and reach equilibrium varies and ranges from hours to days. In particular, the time to reach conformational equilibrium is extremely slow for G12, and changes in the concentration of the two complex conformers were still observed even after the complex mixture has been analysed for over 8 days. Taking the complex of G1 (a “2 + 6” type) as an example (Fig. 4), two and six doublets in a ratio of 0.38 to 0.62, assignable to G1@NC1 and G1@NC2 respectively, were observed after 5 minutes of addition of the guest to NC (total concentration = 1 mM). Concentration of G1@NC1 gradually increased over time at the expense of G1@NC2, and equilibrium was reached in about four hours with the final concentration of G1@NC1 at 0.98 mM. The absence of observable conformational changes for G8, G10, G11, G14, G19 and G20 could either be due to a very fast or extremely slow conformational change that the timescale of the equilibrium is out of the range of that of the NMR.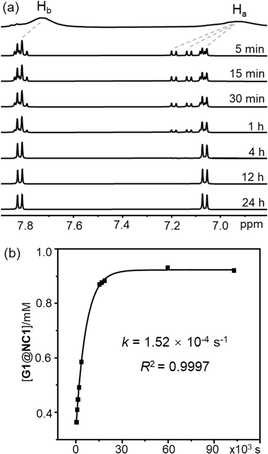 | ||
Fig. 4 (a) Partial 1H NMR spectra (500 MHz, CD2Cl2/CD3CN (v/v = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), 298 K) of NC after addition of G1 at different times, and (b) time-dependent changes of G1@NC1 concentration. 1), 298 K) of NC after addition of G1 at different times, and (b) time-dependent changes of G1@NC1 concentration. | ||
The relatively slow conformational change that allows convenient monitoring is unusual for flexible hosts like NC, which also provides a good opportunity for following the conformational exchange by 1H NMR spectroscopy that is convenient and sensitive for monitoring subtle structural changes. While UV-vis and other spectroscopic methods with a faster timescale are generally more suitable for studying fast conformational exchanges, it is also necessary for the different conformers of the host, guest, or host–guest complex to possess unique spectroscopic features for efficient differentiation and monitoring, which renders the kinetic study of host–guest systems generally challenging. Nevertheless, since it is less plausible for G@NC2 to convert directly to G@NC1 when the host cavity is occupied by the guest and flipping of the naphthyl walls is inhibited, a “guest dissociation-host conformational change-guest re-association” mechanism involving the unbound NC as a conformationally flexible intermediate is proposed. Although both the conformational exchange of free NC and guest association are fast, the substantial stability of both complex conformers implies that the slower guest dissociation would be the rate-determining step of the observed conformational evolution. As a result, kinetics of the conformational change can be described similarly by using the kinetic rate equation of reversible reactions, and the rate of the net increase in the concentration of G@NC1 is hence the difference between the dissociation rates of the two conformers under a steady-state approximation.18,19
 | (1) |
By integrating eqn (1),
 | (2) |
 | (3) |
at equilibrium,
 | (4) |
 | (5) |
| x = −xee−kt + xe | (6) |
Fitting the concentration changes of G@NC1 determined by 1H NMR at different times to the above equation gives the dissociation rate constants k−1 and k−2, and the initial concentrations c1,0 and c2,0. The results are summarized in Table 2, and all the guests that showed conformational evolution were found to conform to the above equation. Several analyses can be made using the obtained data. First, consistent with the proposed mechanism, it is found that k−2 has a major contribution to the overall rate k (∼90% or above), showing that the observed rate of conformational evolution is largely dependent on the dissociation of the less stable G@NC2. Second, the relationship between the measured binding constants and dissociation rate constants obtained from the fitting is analysed. Since the binding constant can be expressed as the ratio between the association rate constant and dissociation rate constant,
 | (7) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1)) experiment and kinetic equation fitting. The total concentration of the host is 1 mM
1)) experiment and kinetic equation fitting. The total concentration of the host is 1 mM
| From the experiment | From fitting | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| c 1,5min/mM | c 1,e/mM | c 1,0/mM | c 2,0/mM | c 1,e/mM | c 2,e/mM | k/s−1 | k −1/s−1 | k −2/s−1 | R 2 | |
| a No conformational change was observed. b The conformational equilibrium was not reached after 8 days and the equilibrium concentration was therefore not measured. | ||||||||||
| G1 | 0.38 | 0.98 | 0.33 | 0.67 | 0.98 | 0.02 | 3.0 × 10−4 | 4.7 × 10−6 | 2.9 × 10−4 | 0.9999 |
| G2 | 0.36 | 0.95 | 0.36 | 0.64 | 0.96 | 0.04 | 2.2 × 10−5 | 9.3 × 10−7 | 2.1 × 10−5 | 0.9998 |
| G3 | 0.34 | 0.90 | 0.34 | 0.66 | 0.90 | 0.10 | 1.9 × 10−5 | 1.9 × 10−6 | 1.8 × 10−5 | 0.9999 |
| G4 | 0.33 | 0.94 | 0.32 | 0.68 | 0.94 | 0.06 | 4.7 × 10−5 | 2.8 × 10−6 | 4.4 × 10−5 | 0.9999 |
| G5 | 0.35 | 0.90 | 0.34 | 0.66 | 0.92 | 0.08 | 1.1 × 10−4 | 8.9 × 10−6 | 1.1 × 10−4 | 0.9996 |
| G6 | 0.36 | 0.90 | 0.36 | 0.64 | 0.91 | 0.09 | 1.5 × 10−5 | 1.3 × 10−6 | 1.3 × 10−5 | 0.9991 |
| G7 | 0.35 | 0.91 | 0.34 | 0.66 | 0.91 | 0.09 | 5.3 × 10−5 | 4.7 × 10−6 | 4.8 × 10−5 | 0.9999 |
| G8 | 0.90 | 0.90 | — | — | — | — | — | — | — | — |
| G9 | 0.41 | 0.93 | 0.41 | 0.59 | 0.95 | 0.05 | 2.6 × 10−6 | 1.4 × 10−7 | 2.4 × 10−6 | 0.9999 |
| G10 | 0.73 | 0.73 | — | — | — | — | — | — | — | — |
| G11 | 0.69 | 0.69 | — | — | — | — | — | — | — | — |
| G12 | 0.67 | 0.67 | 0.33 | 0.98 | 0.02 | 6.0 × 10−7 | 1.3 × 10−8 | 5.9 × 10−7 | 0.9979 | |
| G13 | 0.65 | 0.85 | 0.65 | 0.35 | 0.86 | 0.14 | 6.3 × 10−6 | 8.7 × 10−7 | 5.4 × 10−6 | 0.9992 |
| G14 | 0.68 | 0.68 | — | — | — | — | — | — | — | — |
| G15 | 0.31 | 0.92 | 0.31 | 0.69 | 0.93 | 0.07 | 1.0 × 10−5 | 7.1 × 10−7 | 9.6 × 10−6 | 0.9981 |
| G16 | 0.31 | 0.96 | 0.31 | 0.69 | 0.98 | 0.02 | 1.3 × 10−5 | 3.1 × 10−7 | 1.3 × 10−5 | 0.9980 |
| G17 | 0.47 | 0.84 | 0.28 | 0.72 | 0.87 | 0.13 | 8.5 × 10−4 | 1.1 × 10−4 | 7.4 × 10−4 | 0.9965 |
| G18 | 0.67 | 0.88 | 0.67 | 0.33 | 0.88 | 0.12 | 5.4 × 10−5 | 6.7 × 10−6 | 4.8 × 10−5 | 0.9996 |
| G19 | 0.92 | 0.92 | — | — | — | — | — | — | — | — |
| G20 | 0.89 | 0.89 | — | — | — | — | — | — | — | — |
| G21 | 0.68 | 0.90 | 0.46 | 0.54 | 0.90 | 0.10 | 2.4 × 10−3 | 2.3 × 10−4 | 2.1 × 10−3 | 0.9980 |
| G22 | 0.78 | 0.93 | 0.78 | 0.22 | 0.93 | 0.07 | 1.9 × 10−4 | 1.4 × 10−5 | 1.8 × 10−4 | 0.9836 |
| G23 | 0.64 | 0.94 | 0.50 | 0.50 | 0.94 | 0.06 | 1.4 × 10−3 | 8.4 × 10−5 | 1.3 × 10−3 | 0.9960 |
| G24 | 0.54 | 0.95 | 0.35 | 0.65 | 0.96 | 0.04 | 1.2 × 10−3 | 5.3 × 10−5 | 1.1 × 10−3 | 0.9975 |
hence,
ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ka1 = ln Ka1 = ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) k1 − ln k1 − ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) k−1 k−1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) | (8) |
ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ka2 = ln Ka2 = ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) k2 − ln k2 − ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) k−2 k−2 | (9) |
Except for G12, G13, G18 and G22, a good inverse linear relationship was found in the ln(Ka1) − ln(k−1) and ln(Ka2) − ln(k−2) plots for all other guests (Fig. 5). Such an inverse linear relationship shows that these guests have a very similar association rate (i.e. k1 and k2) when forming the two complex conformers, implying that these guests bind to NCvia a similar process. In fact, these guests that conform to the inverse linear relationship are mostly trimethylammonium ions. While G9 is only slightly larger with two methyl and two ethyl groups, the 5- and 6-membered rings in G15 and G16 would also be less bulky than comparable linear/branched alkyls. On the other hand, both G12 and G13 contain at least three ethyl groups on the ammonium nitrogen, and G18 and G22 also have one ethyl and a benzyl substituent, making these four guests larger in size than the trimethylammonium ones. Furthermore, for G8, G10, G11, G14, G19 and G20 that showed no conformational evolution, for which the corresponding rate will also not conform to the above linear relationship, their steric bulkiness is even greater due to the presence of the bulky neopentyl (G8), 2-methylbenzyl (G20), two propyl (G10 and G11) or one propyl plus one ethyl/benzyl (G14 and G19) groups on the ammonium nitrogen. Consistent with the structure of these inclusion complexes in which the cationic ammonium portion of the guest is always encapsulated, this steric dependence may suggest that the guest first approaches and enters the host cavity via its ammonium portion during binding. The corresponding structural change of the host (e.g. flipping of the naphthyl walls) and activation barrier for accommodating the guest would hence be similar if the bulkiness around the ammonium nitrogen is similar. Intuitively, while the entry of a guest with a trimethylammonium head (or smaller) may involve a certain extent of naphthyl wall rearrangement, a guest with a larger ammonium head would need a larger opening and a larger extent of naphthyl wall rearrangement when entering the cavity, which would hence result in a higher activation barrier (Fig. 6). In fact, in the ln(Ka1) − ln(k−1) and ln(Ka2) − ln(k−2) plots, the data points for G12, G13, G18 and G22 are all found to be below the fitted line obtained from other trimethylammonium guests, showing that the corresponding association rate constants (i.e. k1 and k2) of these four larger ammonium guests are indeed smaller.
 | ||
| Fig. 5 Plot of (a) ln(k−1) against ln(Ka1) and (b) ln(k−2) against ln(Ka2). Data points for G12, G13, G18 and G22 are excluded in the fitting of the trend line. | ||
The similar binding mechanisms for most of the studied guests can also be reflected by the similar ratio of the initial concentration of G@NC1 and G@NC2. Statistically, the ratio of c1,0/c2,0 would be 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 if the activation barrier for forming both conformers are the same. Except for G9, G12, G13, G18, G21, G22 and G23, a similar c1,0/c2,0 ratio of ∼1
3 if the activation barrier for forming both conformers are the same. Except for G9, G12, G13, G18, G21, G22 and G23, a similar c1,0/c2,0 ratio of ∼1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 was found for all other guests. While this similar ratio is again suggestive of a similar binding process, the higher c1,0/c2,0 ratio than that statistically expected may indicate a relatively lower activation barrier for forming the symmetrical conformer than that of the unsymmetrical one, which is also consistent with the slightly larger ln(k1) than ln(k2) obtained from the ln(Ka1) − ln(k−1) and the ln(Ka2) − ln(k−2) plots (i.e. the y-intercepts). For the sterically bulkier G12, G13, G18 and G22 that do not conform to the ln(Ka1) − ln(k−1) and ln(Ka2) − ln(k−2) plots, the corresponding c1,0/c2,0 ratio of 1
2 was found for all other guests. While this similar ratio is again suggestive of a similar binding process, the higher c1,0/c2,0 ratio than that statistically expected may indicate a relatively lower activation barrier for forming the symmetrical conformer than that of the unsymmetrical one, which is also consistent with the slightly larger ln(k1) than ln(k2) obtained from the ln(Ka1) − ln(k−1) and the ln(Ka2) − ln(k−2) plots (i.e. the y-intercepts). For the sterically bulkier G12, G13, G18 and G22 that do not conform to the ln(Ka1) − ln(k−1) and ln(Ka2) − ln(k−2) plots, the corresponding c1,0/c2,0 ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.3 to 1
0.3 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 is also significantly different than that of the other guests. While for the slightly larger G9 (with two methyl and two ethyl) and G21 (with three methyl and one 3-methylbenzyl), the c1,0/c2,0 ratio of 1
0.5 is also significantly different than that of the other guests. While for the slightly larger G9 (with two methyl and two ethyl) and G21 (with three methyl and one 3-methylbenzyl), the c1,0/c2,0 ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.4 and 1
1.4 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.2, respectively, are closer to the 1
1.2, respectively, are closer to the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 observed for most other guests, further supporting that the local substituents around the ammonium are playing a more important role in the initial guest association.
2 observed for most other guests, further supporting that the local substituents around the ammonium are playing a more important role in the initial guest association.
Guest dissociation, on the other hand, is correlated with the thermodynamic stability of the complex conformers. Since the guest dissociation barrier is the sum of the guest association barrier and the free energy of guest binding, both the local structure (i.e. related to the association barrier) and the overall structure (i.e. related to the thermodynamic stability) of the guests will be contributing factors in guest dissociation. As mentioned above, both G@NC1 and G@NC2 are conformationally rigid and the observed conformational evolution likely involves the conformationally flexible unbound NC; the dissociation of the less stable G@NC2 would be the rate-determining step. Since guests with similar local structural features bind to NCvia a similar mechanism with a similar association barrier, and Ka1 and Ka2 are positively correlated due to the comparable differential stability of the two complex conformers, for a guest that forms a more stable G@NC1 complex, the corresponding G@NC2 conformer will also be more stable, and therefore the dissociation barrier for G@NC2 will be proportionally higher with a slower guest dissociation rate (Fig. S106†). The overall rate of conformational evolution will therefore be slower which explains the observed time required for the system to reach the conformational equilibrium. In other words, these results show that sufficient time is essential for an evolving, dynamic system to search for the thermodynamically favored product (e.g.G@NC1), especially if it is a highly stable one in which the possible existence of other kinetic products (e.g.G@NC2) of significant stability with slow reverse kinetics cannot be excluded.
Predicting the time required for attaining conformational equilibrium
By applying the established structure–thermodynamic–kinetic correlation for NC and the model ammonium guests, we also sought to predict the conformational equilibration time for the NC complexes of trimethylammonium guests derived from biogenic amines including acetylcholine, glycine, γ-aminobutyric acid, dopamine and tryptamine (i.e.A1–A7). These biogenic amines are important neurotransmitters and their host–guest complexes could find applications in sensing and triggered-release. In addition, a triethylammonium guest (i.e.A8) was also selected as a control. The binding constant of these guests was first determined by a competitive NMR experiment, and the results are summarized in Table 3. These guests were found to bind to NC with ln(Ka1) ranging from 10 to 15, suggesting that their dissociation rate constants would also span across several orders of magnitude.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1)
1)
With the measured Ka1 and Ka2 for A1 to A7, values of k−1 and k−2 can be obtained from the ln(k−1) − ln(Ka1) and ln(k−2) − ln(Ka2) plots (Fig. 5), from which the time required for the complexes to reach conformational equilibrium (te) can be predicted (see ESI† for details). The conformational changes in these host–guest complexes were also independently studied by 1H NMR, and the corresponding dissociation rate constants were determined as previously described (Table 4). A good agreement was found between the predicted and experimentally obtained values of k−1, k−2 and te, demonstrating that the conformational kinetics of the host–guest system can be satisfactorily predicted from the more easily available binding constants. On the other hand, NC complexes of A8 were found to display no conformational exchange.
| From the experiment | From the prediction | |||||
|---|---|---|---|---|---|---|
| k −1/s−1 | k −2/s−1 | t e/min | k −1/s−1 | k −2/s−1 | t e/min | |
| A1 | 2.9 × 10−6 | 5.2 × 10−5 | 920 | 2.6 × 10−6 | 4.6 × 10−5 | 1020 |
| A2 | 6.7 × 10−6 | 7.5 × 10−5 | 610 | 6.4 × 10−6 | 6.9 × 10−5 | 660 |
| A3 | 1.0 × 10−5 | 5.5 × 10−5 | 770 | 1.1 × 10−5 | 5.1 × 10−5 | 810 |
| A4 | 5.0 × 10−4 | 3.7 × 10−3 | 12 | 3.7 × 10−4 | 2.5 × 10−3 | 17 |
| A5 | 2.7 × 10−5 | 3.1 × 10−4 | 150 | 2.5 × 10−5 | 2.8 × 10−4 | 160 |
| A6 | 1.1 × 10−5 | 9.9 × 10−5 | 450 | 1.2 × 10−5 | 1.1 × 10−4 | 420 |
| A7 | 1.0 × 10−4 | 2.1 × 10−3 | 22 | 7.6 × 10−5 | 1.6 × 10−3 | 29 |
Conclusions
In summary, a supramolecular system consisting of the flexible molecular cage NC and a series of quaternary ammonium guests is discovered to be a suitable model for studying conformational kinetics in host–guest binding. Binding of quaternary ammonium guests resulted in two conformers whose stability is positively correlated. After the initial binding, the less stable conformer converts to the more stable one via a “guest dissociation-host conformational change-guest re-association” mechanism, and as such the guest dissociation from the less stable conformer to form the conformationally flexible, unbound host is the rate determining step of the observed conformational evolution, which in turn correlates with the overall thermodynamics of the complexes. As a result, the rate of the fast conformational exchange of the free NC is significantly slowed down from ∼102 s−1 to 10−3 s−1 to 10−7 s−1via binding to ammonium guests with association constants from 104 M−1 to 107 M−1. Applying the established thermodynamic–kinetic correlation also allows the successful prediction of the kinetic parameters from thermodynamic data for another series of guests derived from biogenic amines. Further analysis of the rate and binding data shows that while the complex stability is related to the overall structural features of the guests, kinetics of the guest association is governed by the local steric properties around the ammonium head that first approaches the host during the binding.The present study demonstrates for the first time the free energy analysis and correlation of the kinetics and thermodynamics of conformational changes in non-covalent complexes. Similar to Hammett analysis that explains and predicts the behaviours and outcomes of chemical reactions from reactant structures, understanding of such a thermodynamics–kinetics relationship of conformational changes will have broad implications in host–guest binding, structural adaption, dynamic assembly and induced motions in different supramolecular systems.
Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
S. He: conceptualization, methodology, formal analysis, investigation, writing – original draft, writing – review & editing; M. Quan: methodology, formal analysis; L.-P. Yang: methodology; H. Y. Au-Yeung: formal analysis, writing – original draft, writing – review & editing, supervision, funding acquisition; W. Jiang: conceptualization, resources, supervision, funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was financially supported by the National Natural Science Foundation of China (No. 22125105 and 22101125), the Shenzhen Science and Technology Innovation Committee (JCYJ20180504165810828), the CAS-Croucher Funding Scheme for Joint Laboratories and the Collaborative Research Fund (CRF) (C7075-21G) from the Research Grant Council of Hong Kong. We are grateful for the technical support from SUSTech-Core Research Facilities.Notes and references
- K. Dunker, C. J. Brown, J. D. Lawson, L. M. Iakoucheva and Z. Obradović, Biochemistry, 2022, 41, 6573 CrossRef PubMed.
- (a) W. L. Jorgensen, Science, 1991, 254, 954 CrossRef CAS PubMed; (b) M. I. Zavodszky and L. A. Kuhn, Protein Sci., 2005, 14, 1104 CrossRef CAS PubMed; (c) L. Cai and H. X. Zhou, J. Chem. Phys., 2011, 134, 105101 CrossRef PubMed; (d) Y. Wang, J. M. Martins and K. L. Larsen, Chem. Sci., 2017, 8, 6466 RSC.
- (a) B. H. Havsteen and G. P. Hess, J. Am. Chem. Soc., 1962, 84, 491 CrossRef CAS; (b) G. G. Hammes, Biochemistry, 2002, 41, 8221 CrossRef CAS PubMed; (c) R. Callender and R. B. Dyer, Acc. Chem. Res., 2015, 48, 407 CrossRef CAS PubMed.
- (a) D. R. Madden, Nat. Rev. Neurosci., 2002, 3, 91 CrossRef CAS PubMed; (b) S. S. Ramaswamy, D. M. MacLean, A. A. Gorfe and V. Jayaraman, J. Bio. Chem., 2013, 288, 35896 CrossRef CAS PubMed; (c) S. Talwar and J. W. Lynch, Neuropharmacology, 2015, 98, 3 CrossRef CAS PubMed.
- P. J. Tummino and R. A. Copeland, Biochemistry, 2008, 47, 5481 CrossRef CAS PubMed.
- (a) S. B. Prusiner, Science, 1991, 252, 1515 CrossRef CAS PubMed; (b) S. B. Prusiner, Science, 1997, 278, 245 CrossRef CAS PubMed.
- (a) E. H. Corder, A. M. Saunders and W. J. Strittmatter, Science, 1993, 261, 921 CrossRef CAS PubMed; (b) J. Poirier, J. Davignon, D. Bouthillier, S. Kogan, P. Bertrand and S. Gauthier, Lancet, 1993, 342, 697 CrossRef CAS PubMed; (c) A. M. Saunders, W. J. Strittmatter and D. Schmechel, Neurology, 1993, 43, 1467 CrossRef CAS PubMed.
- (a) Y. Mizuno, N. Hattori and H. Matsumine, J. Neurochem., 1998, 71, 893 CrossRef CAS PubMed; (b) H. Shimura, N. Hattori, S. Kubo, Y. Mizuno, S. Asakawa and S. Minoshima, Nat. Genet., 2020, 25, 302 CrossRef PubMed.
- (a) G. S. Hammond, J. Am. Chem. Soc., 1955, 77, 334 CrossRef CAS; (b) A. R. Hughes, M. Liu, S. Paul, A. I. Cooper and F. Blanc, J. Phys. Chem. C, 2021, 125, 13370 CrossRef CAS PubMed; (c) G. Schreiber, G. Haran and H.-X. Zhou, Chem. Rev., 2009, 109, 839 CrossRef CAS PubMed.
- For examples of reviews on common supramolecular hosts that include discussion on their binding kinetics, see: (a) S. Fa, T. Kakuta, T. Yamagishi and T. Ogoshi, Chem. Lett., 2019, 48, 1278 CrossRef CAS; (b) E. Masson, X. Ling, R. Joseph, L. Kyeremeh-Mensah and X. Lu, RSC Adv., 2012, 2, 1213 RSC; (c) T. Ogoshi, T.-A. Yamagishi and Y. Nakamoto, Chem. Rev., 2016, 116, 7937 CrossRef CAS PubMed; (d) M. Xue, Y. Yang, X. Chi, Z. Zhang and F. Huang, Acc. Chem. Res., 2012, 45, 1294 CrossRef CAS PubMed; (e) K. Hermann, Y. Ruan, A. M. Hardin, C. M. Hadad and J. D. Badjić, Chem. Soc. Rev., 2015, 44, 500 RSC.
- (a) G. G. Hammes, Y. C. Chang and T. G. Oas, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 13737 CrossRef CAS PubMed; (b) C. M. Hong, D. M. Kaphan, R. G. Bergman, K. N. Raymond and F. D. Toste, J. Am. Chem. Soc., 2017, 139, 8013 CrossRef CAS PubMed; (c) L. P. Yang, L. Zhang, M. Quan, J. S. Ward, Y. L. Ma, H. Zhou, K. Rissanen and W. Jiang, Nat. Commun., 2020, 11, 2740 CrossRef CAS PubMed; (d) Z. Y. Tang and C. A. Chang, J. Chem. Theory Comput., 2018, 14, 303 CrossRef CAS PubMed.
- (a) H. Tang, D. Fuentealba, Y. H. Ko, N. Selvapalam, K. Kim and C. Bohne, J. Am. Chem. Soc., 2011, 133, 20623–20633 CrossRef CAS PubMed; (b) C. Bohne, Chem. Soc. Rev., 2014, 43, 4037 RSC; (c) C. Bohne, Langmuir, 2006, 22, 9100 CrossRef CAS PubMed.
- S. Rieth, X. G. Bao, B. Y. Wang, C. M. Hadad and J. D. Badjić, J. Am. Chem. Soc., 2010, 132, 773 CrossRef CAS PubMed.
- (a) H. Zheng, L. Fu, R. Wang, J. Jiao, Y. Song, C. Shi, Y. Chen, J. Jiang, C. Lin, J. Ma and L. Wang, Nat. Commun., 2023, 14, 590 CrossRef CAS PubMed; (b) T. Zhao, W. Wu and C. Yang, Chem. Commun., 2023, 59, 11469 RSC.
- L. P. Hammett, Chem. Rev., 1935, 17, 125 CrossRef CAS.
- (a) F. Jia, H. V. Schröder, L.-P. Yang, C. von Essen, S. Sobottka, B. Sarkar, K. Rissanen, W. Jiang and C. A. Schalley, J. Am. Chem. Soc., 2020, 142, 3306 CrossRef CAS PubMed; (b) F. Jia, H. Hupatz, L.-P. Yang, H. V. Schröder, D.-H. Li, S. Xin, D. Lentz, F. Witte, X. J. Xie, B. Paulus, C. A. Schalley and W. Jiang, J. Am. Chem. Soc., 2019, 141, 4468 CrossRef CAS PubMed; (c) S. B. Lu, H. X. Chai, J. S. Ward, M. Quan, J. Zhang, K. Rissanen, R. Luo, L. P. Yang and W. Jiang, Chem. Commun., 2020, 56, 888 RSC.
- L. P. Cao, M. Sekutor, P. Y. Zavalij, K. M. Majerski, R. Claser and L. Isaacs, Angew. Chem., Int. Ed., 2014, 53, 988 CrossRef CAS PubMed.
- J. F. Bunnett, Investigation of Rates and Mechanisms of Reactions, John Wiley & Sons, New York, 1986 Search PubMed.
- A. Prabodh, S. Sinn, L. Grimm, Z. Miskolczy, M. Megyesi, L. Biczók, S. Bräse and F. Biedermann, Chem. Commun., 2020, 56, 12327 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc02831g |
| ‡ Deceased 25 December 2022. |
| This journal is © The Royal Society of Chemistry 2024 |