Open Access Article
Open Access ArticleSingle-cell parallel plate mechanics by side-view optical microscopy-assisted atomic force microscopy†
Mingyang
Yang
ab,
Yanqi
Yang
ab,
Lianqing
Liu
 a and
Mi
Li
a and
Mi
Li
 *a
*a
aState Key Laboratory of Robotics, Shenyang Institute of Automation, Chinese Academy of Sciences, Shenyang 110016, China. E-mail: limi@sia.cn
bUniversity of Chinese Academy of Sciences, Beijing 100049, China
First published on 19th March 2025
Abstract
The mechanical forces of individual cells are crucial for physiological and pathological processes, and particularly atomic force microscopy (AFM)-based force spectroscopy has become an important and widely used tool for single-cell mechanical measurements. Here, we present a single-cell parallel plate mechanical measurement method based on side-view optical microscopy-assisted AFM by using a wedged probe, which enables visualization of the uniaxial AFM force spectroscopy process of the cell in real time from the vertical cross-sectional view. With this method, AFM whole-cell compression assays with uniaxial loading forces were performed to quantify the Young's modulus of the entire cell, and the effects of experimental parameters (ramp rate of the AFM probe) and drugs (actin inhibitors) on the measured cell Young's modulus were examined. Additionally, by attaching a living cell to the parallel plate of the wedged probe, AFM-based uniaxial single-cell force spectroscopy (SCFS) assays were performed to measure the adhesion forces of individual cells. The study illustrates a promising approach for single-cell mechanical analysis, which will benefit the application of AFM-based force spectroscopy in the field of mechanobiology to reveal additional insights into the regulatory role of mechanical cues in life activities.
1. Introduction
Mechanical forces are essential for cellular functions, and single-cell mechanical measurements are gaining increasing attention. Cells are both biochemical and mechanical systems, and the cytoskeleton network is responsible for mechanically and biochemically connecting cells to the external environment.1 Cells are constantly exposed to various mechanical and physical stimuli during their life activities, which regulate fundamental subcellular and cellular processes.2 There are many different types of sensor biomolecules on the cell surface, such as integrins and ion channels, through which cells are able to feel the mechanical signals in the extracellular microenvironment.3 Cells can not only respond to these mechanical forces by altering cell structures and functions, but also actively generate forces to reshape their microenvironmental mechanics to meet the needs of cellular physiological/pathological processes, and such a bidirectional crosstalk is termed dynamic reciprocity.4–6 For microbes, they have been found to exploit the physical cues to optimize their functions and ultimately improve microbial fitness.7 For eukaryotic cells, mechanical cues have been shown to regulate cell fate and behavior and direct developmental processes8 and disease progression.9 Therefore, mechanobiology, a branch at the interdisciplinary field of biology and physics that focuses on how cell/tissue mechanics and physical forces affect cell behavior, cell and tissue morphogenesis, and diseases associated with these processes,10 is expected to help unveil the mysteries of life from an additional viewpoint complementary to traditional biochemistry. Various tools have been developed to quantify the physical forces generated by single cells,11 among which atomic force microscopy (AFM)-based force spectroscopy is an important and unique one due to its exceptional signal-to-noise ratio at (sub-)nanometer resolution and no need of sample pretreatment.12–15 Particularly, due to the ability to exert uniaxial forces and inhibit cell sliding during AFM force spectroscopy experiments, wedged cantilevers have been used to sense the rounding forces generated by single mitotic cells.16,17 Besides, providing a capability to image the deformation of the target cell in real time along the axis of loading18 during AFM force measurements is beneficial for enhancing the application of AFM-based force spectroscopy in life sciences. We have previously developed a detachable side-view optical microscopy module for visualizing the thickness of the specimen being measured in an AFM indentation assay.19 Here we demonstrate the use of side-view optical microscopy-assisted AFM to achieve single-cell parallel plate mechanics with uniaxial loading forces (including whole-cell compression to measure the Young's modulus of the entire cell and single-cell force spectroscopy to measure the intercellular adhesion force) by using a wedged AFM probe.2. Materials and methods
2.1 Cell culture and sample preparation
The cell lines used in this study include Raji (human B lymphoblastoid cells) and C2C12 (mouse myoblasts), both of which were purchased from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). The C2C12 cells were cultured in flasks in the high-glucose Dulbecco's Modified Eagle Medium (DMEM) (DearyTech, Shanghai, China) supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, Waltham, MA, USA) and 1% penicillin–streptomycin (Solarbio, Beijing, China) at 37 °C (5% CO2), and the Raji cells were cultured in flasks in the RPMI-1640 medium (DearyTech, Shanghai, China) supplemented with 10% FBS and 1% penicillin–streptomycin at 37 °C (5% CO2). To ensure the tight attachment of cells, coverslips (10 mm × 10 mm) used in the experiments were coated with poly-L-lysine (PLL) (ZSGB-BIO, Beijing, China) in advance. After cleaning and drying the coverslips, the PLL solution was dropped onto the coverslips and incubated for 10 min. Subsequently, the excess PLL solution was removed and the coverslips were air-dried at room temperature overnight.For preparing the cell sample for AFM experiments, 20 mL of Leibovitz's L-15 medium (Thermo Fisher Scientific, Waltham, MA, USA) was added into the square Petri dish of the side-view optical microscopy-assisted AFM, and a PLL-modified coverslip was then placed in the dish (Fig. 1). The cultured C2C12 cells were harvested by using trypsin (Thermo Fisher Scientific, Waltham, MA, USA) and resuspended in 2 mL of the fresh culture medium to prepare a homogeneous cell suspension. For Raji cells, they were directly harvested by centrifugation and resuspended in 2 mL of the fresh culture medium. The prepared cell suspension (C2C12 or Raji) was then added to the coverslip and the cells were allowed to settle for 10 min to facilitate cell attachment. During AFM measurements, the heating plate below the square dish (Fig. 1) was used to provide the 37 °C temperature required for cell growth and metabolism.
To examine the effect of cytochalasin B on the mechanical properties of Raji cells, the cytochalasin B solution (Solarbio, Beijing, China) was added to the Raji cell suspension. First, 0.2 μL of the cytochalasin B solution (the concentration is 1 mg mL−1) was added to the 2 mL of the Raji cell suspension and incubated at 37 °C (5% CO2) for 2 h. After the incubation, the cells were washed by phosphate-buffered saline (PBS) (DearyTech, Shanghai, China) to remove residual cytochalasin B and then centrifuged and resuspended in 2 mL of the fresh culture medium. The resuspended cell solution was then used for subsequent AFM experiments to analyze the effect of cytochalasin B on cellular mechanical properties.
2.2 Preparation of wedged parallel plate probe
The first step is to fabricate the wedge-shaped plate structures, which was done using two-photon lithography here. The wedge-shaped plate was used to compensate the tilt of the tipless silicon nitride AFM cantilever (NP-O10-B, Bruker, Santa Barbara, CA, USA) used here so that the bottom surface of the probe was parallel to the substrate during experiments. According to this requirement, the wedge-shaped plate was designed to be 40 μm in length and width, with its thickness gradually decreasing from one end to the other (the maximum thickness was designed to be 20 μm and the minimum thickness was designed to be 14 μm), forming a trapezoidal side-view profile. This trapezoidal design ensures that the bottom surface of the AFM probe remains parallel to the substrate during loading. The wedge-shaped plate structure was designed by the SolidWorks (Dassault Systemes, Velizy-Villacoublay, France), and the generated STL file was then imported into the two-photon lithography system (Photonic Professional GT2, Nanoscribe Company, Germany) to perform 3D microprinting of the microstructures on a fused silica substrate (Nanoscribe Company, Germany) using resin as the printing material. After printing, the substrate was treated with propylene glycol methyl ether acetate (PGMEA) (Aladdin Company, Shanghai, China) for 20 min and then treated with isopropanol (Aladdin Company, Shanghai, China) for 30 s to remove the residual resin and form the wedge-shaped plate structures.The fabricated wedge-shaped plate structure was then glued to the tipless AFM cantilever by using a 3D digital optical microscope (Hirox Company, Tokyo, Japan). To this end, the cantilever was mounted onto the probe arm of a 3D micropositioner (Everbeing, Beijing, China). Next, a small amount of UV-curable adhesive (Gunasen-AA-3504, Gunasen, Foshan, China) was dropped onto a fresh coverslip, and the coverslip was placed on the sample stage of the 3D digital optical microscope. The micropositioner has three knobs for precise adjustment of the probe's position in the X, Y, and Z directions to ensure accurate alignment. Based on the micropositioner manipulations, the bottom surface of the AFM cantilever was controlled to gently contact with the adhesive, after which the cantilever was immediately retracted. Subsequently, the fused silica substrate with wedge-shaped plate structures was placed on the sample stage of the 3D digital optical microscope. Notably, to avoid misalignment when gluing the wedge-shaped plate to the tipless cantilever, a rectangular marker was added to the higher side of the top surface of the wedge during design to facilitate visual confirmation of the orientation of the wedge. The wedge was then precisely bonded to the free end of the cantilever using micropositioner manipulations, after which a UV lamp was turned on to harden the adhesive for 5 min. After curing, the wedge-shaped plate structure was firmly attached to the cantilever, and the prepared wedged parallel plate probe was stored for AFM experiments. The prepared wedged parallel plate probe was characterized by a scanning electron microscope (SEM) (Thermo Fisher Scientific, Waltham, MA, USA). Before SEM imaging, a thin layer of gold was sputtered on the probe.
2.3 Functionalization of wedged parallel plate probe for single-cell force spectroscopy (SCFS)
For SCFS experiments, the AFM wedged parallel plate probe was functionalized with concanavalin A (ConA) molecules according to a previous protocol.19,20 The functionalization reagents, including biotinylated bovine serum albumin (biotin-BSA), streptavidin, and biotinylated ConA (biotin-ConA), were purchased from Solarbio (Beijing, China). First, the wedged parallel plate probe was sterilized with UV light for 45 min. After sterilization, the probe was immersed in 50 μL of biotin-BSA solution and incubated overnight at 37 °C in a cell culture incubator (Thermo Fisher Scientific, Waltham, MA, USA) to facilitate molecular binding on the probe surface. After incubation, the probe was rinsed three times with PBS to remove any loosely bound molecules. Next, the probe was immersed in 50 μL of streptavidin solution for 30 min at room temperature and then washed three times with PBS. Finally, the probe was immersed in 50 μL of biotin-ConA solution for 30 min at room temperature and washed three times with PBS. The ConA-functionalized wedged parallel plate probe was stored in PBS at 4 °C before SCFS experiments.2.4 AFM experiments
AFM experiments (whole-cell compression and SCFS) were performed with the commercial Dimension Icon AFM (Bruker, Santa Barbara, CA, USA) integrated with a side-view optical microscopy module19 at 37 °C in the CO2-independent L-15 medium. Before experiments, the probe was pressed against a glass substrate to obtain force–distance curves, and then the spring constant of the probe was calibrated by using the AFM's thermal tune module. Whole-cell compression experiments (i.e. conventional AFM indentation assay using the wedged parallel plate probe) were performed on Raji cells. Force–distance curves were acquired with a ramp size of 4 μm (512 sampling points) at different ramp rates (1 Hz, 2 Hz, 3 Hz, 4 Hz, and 5 Hz). The experimental parameters were kept the same when performing whole-cell compression experiments on cytochalasin B-treated Raji cells. SCFS experiments were performed on C2C12 cells. Under the guidance of the AFM's optical microscope, the ConA-functionalized wedged parallel plate probe was positioned above a living C2C12 cell. The probe was controlled to touch the cell for 30 s with a loading force of 4 nN. The probe was then retracted and maintained in the medium for 20 min to allow firm binding formation between the cell and the probe. For obtaining force–distance curves of SCFS experiments, the ramp size was 12 μm, the ramp rate was 1 Hz, the sampling point number was 512, and the dwell time was 1 s. During AFM experiments, after obtaining side-view optical images of cells, an image dehazing method was utilized to improve the image quality,19,21 and the cell thickness was calculated by using the measurement tool in the open-source software ImageView.2.5 Data analysis
For the whole-cell compression experiments, the obtained force–distance curves were analyzed by a modified Hertz model (in the case of compressing a round cell with a plate larger than the cell diameter)22 to obtain the cellular Young's modulus: | (1) |
3. Results and discussion
Fig. 1 shows the established experimental platform of single-cell parallel plate mechanics based on side-view optical microscopy-assisted AFM by using a wedged cantilever. Previously, we have developed a detachable side-view optical microscopy module for AFM-based nanomechanical analysis.19 Herein, by using a wedged cantilever, we demonstrate the utility of side-view optical microscopy-assisted AFM for parallel plate mechanical measurements of individual living cells under uniaxial loading forces (Fig. 1A). Detailed descriptions of the configuration of the side-view optical microscopy-assisted AFM are shown in our previous work.19 The side-view optical microscopy module (I in Fig. 1B) was customized according to the commercial AFM used here, which involves the use of a square Petri dish (II in Fig. 1B). The square dish was placed on a heating plate to provide the temperature environment (37 °C) required for cell growth. Since the CO2-independent L-15 medium was used here, the CO2 environment is not required. Compared with the side-view optical microscopy module we built previously, here we use the dielectric reflectors (LBTEK Company, Shenzhen, China) (II in Fig. 1B) instead of ordinary mirrors to improve the side-view imaging quality. We also established the procedure of fabricating the wedged parallel plate probe based on two-photon lithography and micromanipulations (Fig. 1E), which is described in detail in the Materials and methods section. SEM (Fig. 1C) images show the details of the wedge on the cantilever, and the side-view optical imaging (Fig. 1D) results clearly show that the bottom surface of the wedged probe is parallel to the substrate in the AFM experimental state.The established experimental platform allows performing AFM whole-cell compression experiments on single living cells. Raji cells were used as an example. We firstly performed experiments on regular living Raji cells. One can see that side-view optical imaging clearly visualizes the dynamic compression process of the wedged parallel plate (denoted by the white arrow in Fig. 2A(I)) on the target cell (denoted by the yellow arrow in Fig. 2A(I)), including approaching (I and II in Fig. 2A) and pressing (III in Fig. 2A), which is inaccessible by the AFM's own optical microscope (the inset in Fig. 2A(I)). The real-time whole-cell compression process is shown in ESI Movie 1.† Force–distance curves were recorded during the compression process. In order to analyze the influence of the approach speed of the wedged cantilever on the measurement results, force–distance curves were obtained at different ramp rates of the probe. We can see that (Fig. 2C(I)) increasing the probe approach speed results in an increase in the slope of the contact portion of the acquired force–distance curve (the complete force–distance curves are shown in Fig. S1†), and the fitting results show that increasing the probe loading speed leads to an increase in the measured cell Young's modulus (Fig. 2C(II) and S1†), which is consistent with the results obtained from the indentation experiments using conventional AFM probes.23 In addition, one can see that the modified flat plate Hertz model fits the indentation curve well (Fig. 2C(II) and S1†), suggesting the effectiveness of the model. For control, we then performed experiments on Raji cells treated with cytochalasin B (a drug commonly used to inhibit actin polymerization and soften cells24). Side-view optical microscopy clearly shows the real-time process of the measurement on the cytochalasin B-treated cell (Fig. 2B), including approaching (I in Fig. 2B), touching (II in Fig. 2B) and pressing (III in Fig. 2B), and force–distance curves were obtained at different ramp rates (Fig. 2D and S2†). We measured 7 regular Raji cells and 7 drug-treated Raji cells, and the statistical results are shown in Fig. 3. It can be seen that the measured cell Young's modulus is independent of the cell diameter (Fig. 3A and B). This is because that Young's modulus is a measure of intrinsic material stiffness and is independent of structure.25 We can see that Raji cells became significantly softer after treatment with cytochalasin B, which is consistent with the previous results,26 indicating the effectiveness of the proposed measurement method. Besides, the statistical results clearly show that the probe loading rate (Fig. 3C and D) affects the measurement results. Notably, experiments were performed under the L-15 medium at 37 °C here to meet the requirements of cellular physiological activities. Studies have shown that the growth medium composition (e.g., serum concentration and additional supplements) can affect the elasticity of cells in experiments characterizing the mechanical properties of cells using AFM,27 and the medium temperature also influences the elastic modulus of cells.28 Therefore, one need to ensure the identical experimental conditions (including medium composition and temperature as well as the AFM instrumental parameters such as probe loading rate) when performing comparative studies on cell mechanical properties.
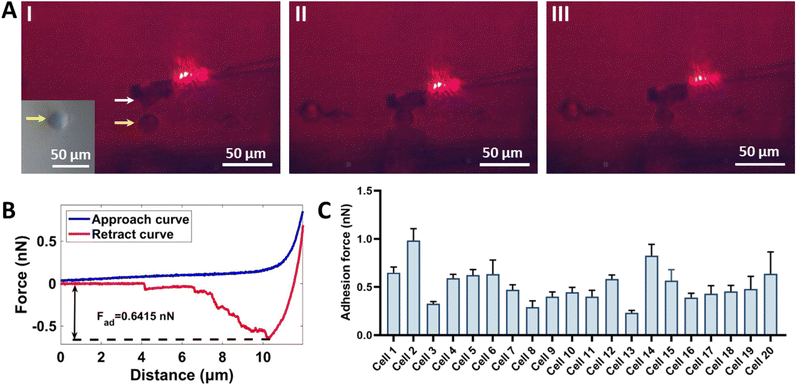
The established experimental platform is also able to perform SCFS experiments, and the results are shown in Fig. 4. Here we used C2C12 cells as an example and the adhesion forces between C2C12 cells were measured. We can clearly see the SCFS experimental process by the side-view optical microscopy (Fig. 4A), including approaching (Fig. 4A(I)), contacting and compressing (Fig. 4A(II)), and retracting (Fig. 4A(III)). The real-time SCFS process is shown in ESI Movie 2.† Representative SCFS force–distance curves with specific unbinding events (including membrane tethers and force steps)20 were obtained (Fig. 4B). We performed SCFS measurements on 20 cells (Fig. 4C) and the experimental results demonstrate the reliability of the proposed SCFS based on side-view optical microscopy-assisted AFM using the single-cell wedged parallel plate probe. Compared with the conventional SCFS method which directly attaches a cell to a tipless cantilever,29 the advantage the SCFS by using the wedged cantilever with a parallel plate to carry a cell is that it can apply uniaxial loading forces and can avoid cell sliding during measurement, which thus helps to improve the stability of the measurement. Notably, functionalizing the AFM cantilever with appropriate linker molecules to prepare single-cell probe is crucial for SCFS experiments. For cantilever functionalization, the cell-cantilever binding strength should be larger than that between the single-cell probe and the substrate (another cell or a functionalized surface), so that the bond formed between the single-cell probe and the substrate is broken during the measurement. Since ConA molecules are widely used to coat AFM cantilevers for SCFS experiments,29,30 we here coated the AFM wedged probe with ConA molecules to prepare the single-cell probe. In addition to ConA, some other biomolecules have also been used to attach single cell to the AFM cantilever, such as polydopamine31 and Matrigel,32 which offer additional options for cantilever functionalization.
The study expands the capabilities of AFM-based single-cell mechanical analysis and will benefit the field of AFM-based mechanobiology. Traditionally, AFM-based force spectroscopy has been mainly used to characterize the local mechanical properties of single cells because the size of the AFM probe tip is much smaller than that of the measured cell.33 Here, we demonstrate that side-view optical microscopy-assisted AFM can perform whole-cell compression on single cells to measure the mechanical properties of the entire cell by using a wedged cantilever, which complements the traditional AFM-based local mechanics of single cells and is conducive to the comprehensive characterization of cellular mechanical properties at different scales. We anticipate that one application of the proposed method will be to detect the mechanical properties of circulating tumor cells (CTCs) (CTCs typically have a round morphology34), which will provide novel possibilities for AFM-based nanomechanical analysis of cancer liquid biopsies.35 Besides, by attaching a cell to the bottom flat surface of the wedged cantilever, side-view optical microscopy-assisted AFM allows performing SCFS experiments under uniaxial loading forces to steadily characterize the adhesion forces of single cells, thus facilitating the studies of SCFS experiments. Particularly, the whole-cell compression and SCFS experimental process can be directly displayed in real time by the side-view optical imaging system, which enhances the visualization of AFM-based force spectroscopy and helps to understand the intermediate details of the force spectroscopy experiments. In current experiments utilizing AFM to characterize cell mechanics, cells are commonly assumed to be incompressible materials and so the Poisson ratio of cells is simply assumed to be 0.5, but a recent study has shown that the Poisson ratio of the cell is frequency dependent.36 Therefore, accurately measuring the Poisson ratio of cells during AFM force spectroscopy experiments to better characterize the mechanical behaviors of cells will benefit the field of mechanobiology. In addition to measuring the thickness of the cell for mechanical analysis, the side-view optical microscopy module may also be used to simultaneously monitor the transversal and longitudinal deformations of the cell during AFM-based compression assay to directly calculate the Poisson ratio of the cell. To this end, the resolution of the side-view optical microscopy system needs to be further improved. Furthermore, since the established methodology does not require any modification of the AFM system itself, it can be conveniently and directly applied to more cell types and biological systems. Altogether, the study adds additional tools to the toolbox of AFM-based force spectroscopy, which will facilitate the application of AFM in mechanobiology.
4. Conclusions
The study has demonstrated the ability of side-view optical microscopy-assisted AFM combined with a wedged cantilever to realize single-cell parallel plate mechanics. The experimental process of single-cell parallel plate mechanical measurement can be optically visualized in real time from the sectional viewpoint. The method proposed in this work can not only perform whole-cell compression under uniaxial loading forces to characterize the Young's modulus of the entire cell, but also perform uniaxial SCFS assay to measure the adhesion force of individual cells. Future studies, such as further improving the resolution of side-view optical imaging and applying the established methodology to address more mechanical issues in life sciences, will contribute to the application of AFM-based force spectroscopy in the field of single-cell mechanobiology.Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
Conceptualization (M. L.); funding acquisition (M. L. and L. L.); investigation (M. Y. and Y. Y.); formal analysis (M. Y.); methodology (M. Y., Y. Y. and M. L.); resources (M. L. and L. L.); project administration (M. L.); supervision (M. L.); writing-original draft (M. L. and M. Y.); writing-review & editing (M. L. and M. Y.).Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 62273330), the Natural Science Foundation of Liaoning Province (Grant No. 2024JH3/50100021) and the New Cornerstone Science Foundation through the XPLORER PRIZE.References
- D. A. Fletcher and R. D. Mullins, Cell mechanics and the cytoskeleton, Nature, 2010, 463(7280), 485–492 CAS.
- V. M. Weaver, Cell and tissue mechanics: the new cell biology of frontier, Mol. Biol. Cell, 2017, 28(14), 1815–1818 CrossRef CAS PubMed.
- B. D. Hoffman, C. Grashoff and M. A. Schwartz, Dynamic molecular processes mediate cellular mechanotransduction, Nature, 2011, 475(7356), 316–323 CrossRef CAS.
- D. E. Discher, P. Janmey and Y. L. Wang, Tissue cells feel and respond to the stiffness of their substrate, Science, 2005, 310(5751), 1139–1143 CrossRef CAS PubMed.
- S. van Helvert, C. Storm and P. Friedl, Mechanoreciprocity in cell migration, Nat. Cell Biol., 2018, 20(1), 8–20 CrossRef CAS PubMed.
- A. Diz-Munoz, O. D. Weiner and D. A. Fletcher, In pursuit of the mechanics that shape cell surfaces, Nat. Phys., 2018, 14(7), 648–652 Search PubMed.
- Y. F. Dufrene and A. Persat, Mechanomicrobiology: how bacteria sense and respond to forces, Nat. Rev. Microbiol., 2020, 18(4), 227–240 Search PubMed.
- K. H. Vining and D. J. Mooney, Mechanical forces direct stem cell behaviour in development and regeneration, Nat. Rev. Mol. Cell Biol., 2017, 18(12), 728–742 CrossRef CAS PubMed.
- H. T. Nia, L. L. Munn and R. K. Jain, Physical traits of cancer, Science, 2020, 370(6516), eaaz0868 CAS.
- K. A. Jansen, D. M. Donato, H. E. Balcioglu, T. Schmidt, E. H. J. Danen and G. H. Koenderink, A guide to mechanobiology: where biology and physics meet, Biochim. Biophys. Acta, 2015, 1853, 3043–3052 CAS.
- P. Roca-Cusachs, V. Conte and X. Trepat, Quantifying forces in cell biology, Nat. Cell Biol., 2017, 19(7), 742–751 CAS.
- Y. F. Dufrene, T. Ando, R. Garcia, D. Alsteens, D. Martinez-Martin, A. Engel, C. Gerber and D. J. Muller, Imaging modes of atomic force microscopy for application in molecular and cell biology, Nat. Nanotechnol., 2017, 12(4), 295–307 CrossRef CAS PubMed.
- M. Krieg, G. Flaschner, D. Alsteens, B. M. Gaub, W. H. Roos, G. J. L. Wuite, H. E. Gaub, C. Gerber, Y. F. Dufrene and D. J. Muller, Atomic force microscopy-based mechanobiology, Nat. Rev. Phys., 2020, 1(1), 41–57 Search PubMed.
- R. Garcia, Nanomechanical mapping of soft materials with the atomic force microscope: methods, theory and applications, Chem. Soc. Rev., 2020, 49(16), 5850–5884 CAS.
- A. Viljoen, M. Mathelie-Guinlet, A. Ray, N. Strohmeyer, Y. J. Oh, P. Hinterdorfer, D. J. Muller, D. Alsteens and Y. F. Dufrene, Force spectroscopy of single cells using atomic force microscopy, Nat. Rev. Methods Primers, 2021, 1, 63 CAS.
- M. P. Stewart, A. W. Hodel, A. Spielhofer, C. J. Cattin, D. J. Muller and J. Helenius, Wedged AFM-cantilevers for parallel plate mechanics, Methods, 2013, 60(2), 186–194 CAS.
- C. J. Cattin, M. Duggelin, D. Martinez-Martin, C. Gerber, D. J. Muller and M. P. Stewart, Mechanical control of mitotic progression in single animal cells, Proc. Natl. Acad. Sci. U. S. A., 2015, 112(36), 11258–11263 CAS.
- O. Chaudhuri, S. H. Parekh, W. A. Lam and D. A. Fletcher, Combined atomic force microscopy and side-view optical imaging for mechanical studies of cells, Nat. Methods, 2009, 6(5), 383–387 CAS.
- Y. Yang and M. Li, Side-view optical microscopy-assisted atomic force microscopy for thickness-dependent nanobiomechanics, Nanoscale Adv., 2024, 6(13), 3306–3319 RSC.
- J. Friedrichs, J. Helenius and D. J. Muller, Quantifying cellular adhesion to extracellular matrix components by single-cell force spectroscopy, Nat. Protoc., 2010, 5(7), 1353–1361 CrossRef CAS PubMed.
- K. He, J. Sun and X. Tang, Single image haze removal using dark channel prior, IEEE Trans. Pattern Anal. Mach. Intell., 2011, 33(12), 2341–2353 Search PubMed.
- D. Chang, T. Hirate, C. Uehara, H. Maruyama, N. Uozumi and F. Arai, Evaluating Young's modulus of single yeast cells based on compression using an atomic force microscope with a flat tip, Microsc. Microanal., 2021, 27(2), 392–399 CrossRef CAS PubMed.
- M. Lekka, Discrimination between normal and cancerous cells using AFM, Bionanoscience, 2016, 6, 65–80 CrossRef.
- R. Xiao, Y. Zhang and M. Li, Automated high-throughput atomic force microscopy single-cell nanomechanical assay enabled by deep learning-based optical image recognition, Nano Lett., 2024, 24(39), 12323–12332 CrossRef CAS PubMed.
- C. F. Guimaraes, L. Gasperini, A. P. Marques and R. L. Reis, The stiffness of living tissues and its implications for tissue engineering, Nat. Rev. Mater., 2020, 5(5), 351–370 Search PubMed.
- C. Rotsch and M. Radmacher, Drug-induced changes of cytoskeletal structure and mechanics in fibroblasts: an atomic force microscopy study, Biophys. J., 2000, 78(1), 520–535 CAS.
- M. Nikkhah, J. S. Strobl, E. M. Schmelz and M. Agah, Evaluation of the influence of growth medium composition on cell elasticity, J. Biomech., 2011, 44(4), 762–766 Search PubMed.
- E. Spedden, D. L. Kaplan and C. Staii, Temperature response of the neuronal cytoskeleton mapped via atomic force and fluorescence microscopy, Phys. Biol., 2013, 10(5), 056002 CAS.
- J. Helenius, C. P. Heisenberg, H. E. Gaub and D. J. Muller, Single-cell force spectroscopy, J. Cell Sci., 2008, 121(11), 1785–1791 CAS.
- P. M. Spoerri, N. Strohmeyer, Z. Sun, R. Fassler and D. J. Muller, Protease-activated receptor signalling initiates α5β1-integrin-mediated adhesion in non-haematopoietic cells, Nat. Mater., 2020, 19(2), 218–226 CAS.
- A. Beaussart, S. El-Kirat-Chatel, P. Herman, D. Alsteens, J. Mahillon, P. Hols and Y. F. Dufrene, Single-cell force spectroscopy of probiotic bacteria, Biophys. J., 2013, 104(9), 1886–1892 CAS.
- M. Huber, J. Casares-Arias, R. Fassler, D. J. Muller and N. Strohmeyer, In mitosis integrins reduce adhesion to extracellular matrix and strengthen adhesion to adjacent cells, Nat. Commun., 2023, 14, 2143 CAS.
- N. Gavara and R. S. Chadwick, Determination of the elastic moduli of thin samples and adherent cells using conical atomic force microscope tips, Nat. Nanotechnol., 2012, 7(11), 733–736 CAS.
- S. H. Cross, Y. S. Jin, J. Rao and J. K. Gimzewski, Nanomechanical analysis of cells from cancer patients, Nat. Nanotechnol., 2007, 2(12), 780–783 CrossRef CAS PubMed.
- M. Li, Atomic force microscopy as a nanomechanical tool for cancer liquid biopsy, Biochem. Biophys. Res. Commun., 2024, 734, 150637 CrossRef CAS PubMed.
- M. Mokbel, K. Hosseini, S. Aland and E. Fischer-Friedrich, The Poisson ratio of the cellular actin cortex is frequency dependent, Biophys. J., 2020, 118(8), 1968–1976 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5na00012b |
| This journal is © The Royal Society of Chemistry 2025 |