Open Access Article
Open Access ArticleNanocomplexation between curcumin and proteins by charge-switch method for improved physicochemical and biological properties of curcumin†
Anyu Zhanga,
Manish Mahotraa,
Hong Yua,
Tianqi Zhua and
Say Chye Joachim Loo *abc
*abc
aSchool of Materials Science and Engineering, Nanyang Technological University, 50 Nanyang Avenue, 639798, Singapore. E-mail: joachimloo@ntu.edu.sg
bLee Kong Chian School of Medicine, Nanyang Technological University, 59 Nanyang Drive, 636921, Singapore
cSingapore Centre for Environmental Life Sciences Engineering, Nanyang Technological University, 60 Nanyang Drive, 637551, Singapore
First published on 27th March 2025
Abstract
Curcumin, a natural polyphenol, has various biological functions including antioxidant and antimicrobial properties, but the functions are limited by its low aqueous solubility. To address this, two proteins, bovine serum albumin (BSA) and gelatin were used to form curcumin–protein nanocomplexes using a charge-switch method, by mixing oppositely charged curcumin and proteins. Complexation with BSA and gelatin increased the curcumin solubility to 391.77 (±15.70) μg mL−1 and 143.64 (±2.29) μg mL−1 respectively, and the loading amount (LA) of curcumin to 21.36% and 15.57%, respectively. Moreover, enhanced bioaccessibility, and antioxidant and antimicrobial properties were observed after complexation. After complexation, the minimum inhibitory concentration (MIC) of curcumin decreased by eightfold against Vibrio parahaemolyticus, more than eightfold against Vibrio harveyi, and twofold against Bacillus cereus and Streptococcus iniae, which are food and aquaculture related pathogens. The curcumin–protein nanocomplexes presented in this work could serve as non-antibiotic additives for preventing and managing microbial diseases in agri-food applications.
1. Introduction
Curcumin is a polyphenol isolated from the rhizome of turmeric (Curcuma longa).1 Commercially, it is used as a spice and food-coloring agent,2 and is reported to provide many health benefits, including antioxidant, antimicrobial and anti-inflammatory properties.1–3 Besides its use as a functional food ingredient, curcumin has also been explored for agri-food applications (i.e. aquaculture), where it has been reported to exhibit antimicrobial effects against pathogens responsible for disease outbreaks.4The use of curcumin, however, does have its challenges: the key challenge is its extremely low aqueous solubility (11 ng mL−1) and consequently low bioavailability.5 To improve its bioavailability, lipid-based delivery systems had been developed, because lipids can be transformed into micelles to solubilize curcumin for enhanced digestion. However, the lipid composition and the use of surfactants in such systems may raise safety concerns, and the toxicity assessments of these have yet to be sufficiently investigated.6 Complexation with proteins, on the other hand, is a safe, simple and energy-saving method to improve the aqueous solubility, physicochemical and biological properties of curcumin.7 Previous studies on curcumin–protein complexation mainly focused on two fabrication techniques: (1) solvent-based method: directly adding curcumin (dissolved in organic solvents) into an aqueous protein solution;8 and (2) pH-shift method: dissolving both curcumin and proteins at alkali pH values, and shifting the pH of the binary solution to neutral to form a curcumin–protein complex. Compared to the solvent-based method, the pH-shift method is free of organic solvent, and can further improve the aqueous solubility of curcumin. However, the main limitation of the pH-shift method is its low curcumin loading amount (LA), as large amounts of protein is required during fabrication.9–11
In this work, a charge-switch nanocomplexation method was developed to promote the solubility, bioaccessibility, antioxidant properties, and anti-microbial properties of curcumin, while achieving a higher curcumin loading amount. Curcumin has three pKa values (8.31, 9.88 and 10.51), above which it can carry net negative charges;12 and proteins can carry net positive charges below their isoelectric points.13,14 By introducing opposite charges to curcumin and protein, the complexation process, arising from an electrostatic interaction, can be completed in a single mixing step. Two proteins, bovine serum albumin (BSA) and gelatin were chosen for complexation with curcumin, because of their non-toxicity, biodegradability, and previously reported applications in complexation with curcumin.15,16 BSA, in this work, was applied as a model protein for complexation, and gelatin, which is widely applied in food and pharmaceutical industries,16 was applied as a more cost-effective alternative protein. In contrast to previous studies for curcumin–BSA and curcumin–gelatin complexes,15,16 this present method is organic solvent free. Additionally, the electrostatic interaction during fabrication was hypothesized to achieve a high curcumin loading amount. The improved solubility of curcumin after complexation was expected to contribute to its antimicrobial properties against pathogens such as Vibrio parahaemolyticus, Vibrio harveyi, Bacillus cereus, and Streptococcus iniae: when present in food, Vibrio parahaemolyticus can cause acute gastroenteritis, sepsis, and other diseases,17 and Bacillus cereus can cause diarrheal and emetic syndromes;18 Vibrio harveyi and Streptococcus iniae are two common fish pathogens that can cause various fish diseases and also affect human health.19,20 To overcome these infections, antibiotics are usually employed but these often comes with negative impacts, including the emergence of antibiotic-resistant bacterial strains.4 Given the improved antimicrobial properties of curcumin against these pathogens after complexation, the nanocomplex could therefore potentially be used as a non-antibiotic functional additive for human food and in aquaculture feeds.
2. Experimental
2.1 Chemicals
ABTS was bought from Roche Diagnostics GmbH (Germany). Acetic acid was purchased from Aladdin Industrial Corporation (China). Acetonitrile was bought from Tedia (USA). BSA, ciprofloxacin, curcumin, dimethyl sulfoxide, gelatin from cold water fish skin, L-ascorbic acid, ox-bile, pancreatin from porcine pancreas, pepsin from porcine gastric mucosa, phosphate buffered saline (PBS), sodium acetate and sodium persulfate were purchased from Sigma Aldrich (USA). Nutrient Broth (NB) and Wilkins-Chalgren Anaerobe (WCA) media were purchased from Thermo Fisher Scientific, USA. Sodium chloride was purchased from Nice Chemicals (India). Sodium hydroxide was bought from Schedelco (Singapore).2.2 Methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 30 min, to separate out the insoluble curcumin. Similarly, same concentrations of curcumin (dissolved in 0.1 M NaOH) were mixed with same concentrations of gelatin (dissolved in 0.6% v/v acetic acid) and centrifuged. All the resulting curcumin–protein complexes were then used to test the solubility, and some groups were used for subsequent characterizations.
000g for 30 min, to separate out the insoluble curcumin. Similarly, same concentrations of curcumin (dissolved in 0.1 M NaOH) were mixed with same concentrations of gelatin (dissolved in 0.6% v/v acetic acid) and centrifuged. All the resulting curcumin–protein complexes were then used to test the solubility, and some groups were used for subsequent characterizations.Fluorescence spectroscopy was performed to investigate curcumin–protein interactions using an Infinite M200 (TECAN) microplate reader at room temperature. 0.1 mg mL−1 BSA (or gelatin) in 0.6% v/v acetic acid was treated with equal volume of curcumin in 0.1 M NaOH with the concentrations of 0 (control), 2, 4, 6, 8, and 10 μg mL−1 respectively. Both curcumin–BSA and curcumin–gelatin samples were illuminated using an excitation wavelength of 270 nm, and the emission spectra were measured in the 300-320 nm range.16
The fluorescence quenching process is described by the Stern–Volmer equation:
| F0/F = 1 + kqτ0[Q] = 1 + KSV[Q] | (1) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 v/v) with deionized water, and added to cuvettes with 1 cm optical pathway (Kartell, Italy). The refractive index and absorption coefficient were fixed at 1.42 and 0.01 respectively.22 Pure water was used as the dispersant (viscosity = 0.8872 cP, refractive index = 1.330). The zeta potential values of curcumin–BSA complex and curcumin–gelatin complex were measured with the same Malvern Nanosizer (Malvern Instruments, U.K.). The samples were filtered (0.22 μm) but not diluted before zeta potential measurement.
100 v/v) with deionized water, and added to cuvettes with 1 cm optical pathway (Kartell, Italy). The refractive index and absorption coefficient were fixed at 1.42 and 0.01 respectively.22 Pure water was used as the dispersant (viscosity = 0.8872 cP, refractive index = 1.330). The zeta potential values of curcumin–BSA complex and curcumin–gelatin complex were measured with the same Malvern Nanosizer (Malvern Instruments, U.K.). The samples were filtered (0.22 μm) but not diluted before zeta potential measurement.The loading amount (LA) of curcumin after complexation was also determined, as defined in eqn (2):
| Loading amount (LA) (%) = (curcumin complexed in protein)/(total amount of curcumin and protein) | (2) |
| ABTS scavenging activity (%) = (absorbancecontrol − absorbancesample)/(absorbancecontrol) × 100 | (3) |
2.2.8.1 Growth and preparation of pathogens. The bacterial strains used in this work were all purchased from American type culture collection (ATCC). Frozen stock culture of the pathogens was streaked on respective agar plates and incubated for 24 h, and single colonies were inoculated into 6 mL fresh broth and incubated for 24 h, following the growth conditions shown in Table 1.
| Pathogens | G+/G− | Media | Incubation temperature (°C) |
|---|---|---|---|
| Vibrio parahaemolyticus ATCC®17802 | − | Nutrient Agar/Broth (with 3% NaCl) | 37 |
| Vibrio harveyi ATCC®14126 | − | WCA Agar/Broth | 30 |
| Bacillus cereus ATCC®11778 | + | WCA Agar/Broth | 30 |
| Streptococcus iniae ATCC 29178 | + | WCA Agar/Broth | 37 |
2.2.8.2 Agar well diffusion assay. The ability of free curcumin, curcumin–BSA complex, and curcumin–gelatin complex to inhibit the growth of the pathogens was assessed using the agar well diffusion assay.3 A high concentration “stock solution” was prepared by dissolving curcumin in dimethyl sulfoxide (DMSO) at 50 mg mL−1, and the stock solution was added into respective broth (Table 1) to yield curcumin concentration of 2 mg mL−1 and DMSO concentration of 4%. Curcumin and proteins (BSA/gelatin) were dissolved in autoclaved 0.1 M NaOH and 0.6% v/v acetic acid at 4 mg mL−1 and 10 mg mL−1 respectively, and equal volume of both solutions were mixed to prepare the curcumin–protein complex solutions with curcumin concentration of 2 mg mL−1. 100 μL of each pathogen that was diluted in the media to 108 CFU mL−1 was spread on an agar plate, and three 6 mm diameter wells were formed in each agar plate. 50 μL of free curcumin, curcumin–BSA complex, and curcumin–gelatin complex were added into each well, and the plates were incubated according to the growth conditions shown in Table 1 for 24 h. DMSO (4%), BSA (5 mg mL−1) and gelatin (5 mg mL−1) at corresponding concentrations were applied as negative controls. After incubation, the diameter of the inhibition zones around the wells were recorded.
2.2.8.3 Determination of minimum inhibitory concentration (MIC). The susceptibility of the pathogens to curcumin was also evaluated using MIC assays, and the experiments were all done for three times independently. To determine the MIC of free curcumin, 50 mg mL−1 curcumin in DMSO was added into broth to yield curcumin concentration of 2 mg mL−1 and DMSO concentration of 4%. 100 μL of this solution was added to each well of 96-well plate, and was two-fold serially diluted using the broth. 50 μL of the bacteria suspension with the working concentration of 106 CFU mL−1 was added into each well to obtain the highest curcumin concentration of 1000 μg mL−1 (in 2% DMSO) to lowest curcumin concentration of 7.81 μg mL−1 (in 0.016% DMSO). A negative control was included in each plate by adding 100 μL broth only. The 96-well plate was incubated for 18 h, and the curcumin concentration that yielded no visible bacterial growth (turbidity increase) was determined as MIC.26
To measure the MIC of curcumin–protein complex against the pathogens, curcumin–protein complex solutions prepared from 4.0 mg mL−1 curcumin (in 0.1 M NaOH) and 10.0 mg mL−1 proteins (in 0.6% v/v acetic acid) were mixed with respective broth prepared at double the intended concentration.26 100 μL of this solution (with 1000 μg mL−1 curcumin) was added to each well of 96-well plate, and was two-fold serially diluted using the broth. Similarly, 50 μL of pathogens with the working concentration of 106 CFU mL−1 was added into each well to obtain the highest curcumin concentration of 500 μg mL−1 to lowest curcumin concentration of 3.91 μg mL−1. A negative control was included in each plate by adding 100 μL broth only. The 96-well plate was incubated for 18 h. The curcumin concentration that yielded no visible bacterial growth (turbidity increase) was determined as MIC.
As control, the same bacteria suspension was added into same concentrations of DMSO, BSA and gelatin in the broth, to ensure that the BSA and gelatin did not affect the viability of the pathogens. 0.1 mg mL−1 penicillin and 0.1 mg mL−1 ciprofloxacin (dissolved in designated broth) were used for Gram-positive (Bacillus cereus and Streptococcus iniae) and Gram-negative pathogens (Vibrio parahaemolyticus and Vibrio harveyi) respectively, as positive control.
3. Results and discussion
3.1 Characterizations of curcumin–protein complex
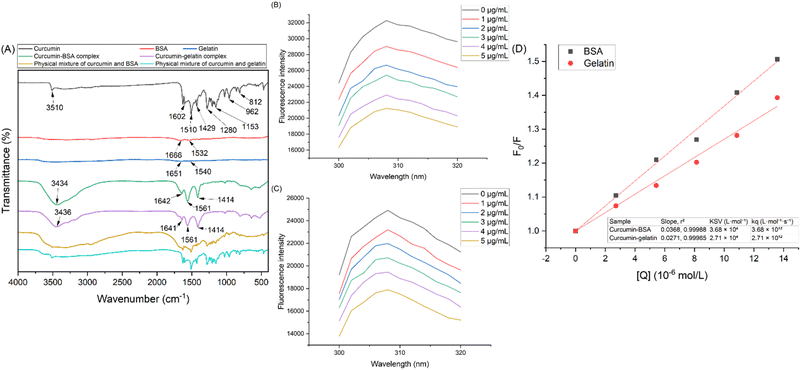
The charge-switch nanocomplexation method utilizes the net charges of curcumin and protein. When curcumin (dissolved in 0.1 M NaOH) and protein (dissolved in 0.6% v/v acetic acid) are mixed, neutralization occurs and the attraction of opposite charges from the two molecules can lead to the formation of the water-soluble curcumin–protein complex. The FTIR spectrums of curcumin, BSA, gelatin, curcumin–BSA complex, curcumin–gelatin complex, physical mixture of curcumin and BSA, and physical mixture of curcumin and gelatin, are shown in Fig. 1A, and the interpretation of respective adsorption peaks are summarized in ESI 1.†12,15,16,27 Characteristic peaks of curcumin and proteins (BSA and gelatin) were both observed in the spectrum of curcumin–protein complexes, indicating that both curcumin and proteins existed in the curcumin–protein complexes. The difference in the spectrum of curcumin–protein physical mixtures and curcumin–protein complexes indicated that the stretching and bending vibrations in curcumin molecule were limited after complexation.11 Moreover, in the spectrum of curcumin, the peak indicating the –OH group at 3510 cm−1 shifted to lower wavenumbers at 3434 and 3436 cm−1 for the curcumin–protein complexes, demonstrating the formation of hydrogen bonds between curcumin and the two proteins.27Fluorescence spectroscopy was used to further investigate the interactions between curcumin and the proteins, and the fluorescence emission spectra of BSA and gelatin (both at 0.1 mg mL−1) with the addition of different concentrations of curcumin are shown in Fig. 1B and C. Curcumin, as a quencher, interacts with proteins and reduces their fluorescence emission. Emission spectra of BSA and gelatin were recorded from 300 to 320 nm with an excitation wavelength of 270 nm. For both curcumin–protein complexes, an intense emission band centered at 308 nm was observed, which was due to tyrosine residues.28 With increasing curcumin concentration, the emission peak did not shift, while the intensity decreased, indicating that curcumin, as quencher, did not change the molecular conformation of BSA and gelatin.16,29
The Stern–Volmer plots for curcumin–BSA and curcumin–gelatin complexes were generated and shown in Fig. 1D, and the corresponding parameters are shown in the table inset. It could be derived from the slopes of the two curves that the KSV values of curcumin–BSA and curcumin–gelatin were 3.68 × 104 and 2.71 × 104 L mol−1 respectively. The Stern–Volmer constant (KSV) of curcumin–BSA complex was higher than that of curcumin–gelatin complex, which indicated that the interaction and affinity between curcumin–BSA was stronger than curcumin–gelatin.
Dynamic light scattering (DLS) is an optical technique to measure the particle size and surface charge. The effect of curcumin–protein mass ratios on the particle size and zeta potential is shown in Fig. 2A and B. The size of the curcumin–BSA complex decreased from 141.57 (±19.28) nm to 67.30 (±6.56) nm as the curcumin–BSA mass ratio decreased from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, indicating that the nanoparticles became more compact with increasing BSA concentration.12 For curcumin–gelatin complex, the effect of curcumin–gelatin mass ratio on particle size was insignificant. While the curcumin–protein mass ratio decreased from 1
5, indicating that the nanoparticles became more compact with increasing BSA concentration.12 For curcumin–gelatin complex, the effect of curcumin–gelatin mass ratio on particle size was insignificant. While the curcumin–protein mass ratio decreased from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, the zeta potential of curcumin–BSA complex decreased from −7.80 (±0.79) mV to −10.08 (±1.95) mV, and the zeta potential of curcumin–gelatin complex increased from 1.97 (±0.44) mV to 2.67 (±0.05) mV. The isoelectric points of BSA and gelatin are 4.6 and 7.79 respectively,13,14 and the pH of the solution after neutralization was measured as 5.99 ± 0.01 in this work, which was above the isoelectric point of BSA but below the isoelectric point of gelatin. From the negative and positive zeta potentials of curcumin–BSA and curcumin–gelatin complexes respectively, it could be concluded that the proteins were likely to be on the surface of the curcumin–protein complex particles.
5, the zeta potential of curcumin–BSA complex decreased from −7.80 (±0.79) mV to −10.08 (±1.95) mV, and the zeta potential of curcumin–gelatin complex increased from 1.97 (±0.44) mV to 2.67 (±0.05) mV. The isoelectric points of BSA and gelatin are 4.6 and 7.79 respectively,13,14 and the pH of the solution after neutralization was measured as 5.99 ± 0.01 in this work, which was above the isoelectric point of BSA but below the isoelectric point of gelatin. From the negative and positive zeta potentials of curcumin–BSA and curcumin–gelatin complexes respectively, it could be concluded that the proteins were likely to be on the surface of the curcumin–protein complex particles.
 | ||
| Fig. 2 Particle size and zeta potential of the curcumin–BSA (A) and curcumin–gelatin (B) complexes. DSC curves of curcumin, BSA, gelatin, curcumin–BSA complex, and curcumin–gelatin complex (C). | ||
The thermal properties of curcumin, BSA, gelatin, curcumin–BSA complex, and curcumin–gelatin complex were investigated using DSC analysis, as shown in Fig. 2C. The smooth DSC curves of BSA and gelatin indicates their amorphous structures and thermal stability up to 200 °C, and the endothermic peak at 180 °C in the DSC curve of curcumin was attributed to the melting of curcumin crystals.25 The peak disappeared in the curves of both curcumin–BSA complex and curcumin–gelatin complex, indicating that curcumin was in an amorphous form after complexation with proteins. The amorphous form of hydrophobic bioactives is known to offer higher bioavailability and better absorption rates than the crystalline state, because the molecules in amorphous state need less energy to break down the solid structure; while in the crystalline state, the strong bonds between the molecules in the lattice will hinder the dissolution process.30
3.2 Solubility of curcumin after complexation
The solubility images of free curcumin, curcumin–BSA complex, and curcumin–gelatin complex in water is shown in Fig. 3A, and the aqueous solubility values of curcumin after complexation with BSA and gelatin is shown in Fig. 3B and C. The solubility was determined as the amount of curcumin in the supernatant after centrifugation and before extraction. Complexation with BSA and gelatin could increase curcumin solubility up to 391.77 (±15.70) μg mL−1 (Fig. 3B) and 143.64 (±2.29) μg mL−1 (Fig. 3C) respectively. The maximum solubility of curcumin–BSA complex was achieved when 2 mg mL−1 of curcumin was mixed with 5 mg mL−1 BSA, and the maximum solubility of curcumin–gelatin complex was achieved when 0.5 mg mL−1 curcumin was mixed with 5 mg mL−1 gelatin. Compared to the aqueous solubility of free curcumin, which was 11 ng mL−1,5 complexation with BSA and gelatin increased the solubility of curcumin by over 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and 13
000 and 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 times. The solubility improvement could be attributed to the decrease in size and the amorphous form of curcumin after complexation, as verified in DLS and DSC results. When protein concentration was 0 (control), curcumin could not be solubilized in water; when protein concentration increased, the solubility of curcumin also increased accordingly. It could also be concluded that curcumin can only be significantly solubilized when the mass of proteins is higher than curcumin. After complexation, the aqueous solubility of curcumin–BSA complex was generally higher than that of curcumin–gelatin complex, which was consistent with the result showed by fluorescence spectroscopy that the interaction and affinity between curcumin–BSA was stronger than curcumin–gelatin. The possible mechanism of curcumin–protein complex formation using the charge-switch method is shown in Fig. 3D. BSA has a higher percentage of polar amino acids than gelatin (44.7% vs. 10.9%).31 Among these, the basic amino acids are lysine, arginine, and histidine, with side chain pKa of 10.53, 12.48, and 6.04.32 These basic amino acids can be protonated and carry positive charges at low pH. It is postulated that BSA, with a higher percentage of these amino acids than gelatin, can provide more cationic sites for interacting electrostatically with the negatively charged curcumin, which is one possible reason that curcumin–BSA interaction is stronger than curcumin–gelatin, and curcumin–BSA is more soluble than curcumin–gelatin.
000 times. The solubility improvement could be attributed to the decrease in size and the amorphous form of curcumin after complexation, as verified in DLS and DSC results. When protein concentration was 0 (control), curcumin could not be solubilized in water; when protein concentration increased, the solubility of curcumin also increased accordingly. It could also be concluded that curcumin can only be significantly solubilized when the mass of proteins is higher than curcumin. After complexation, the aqueous solubility of curcumin–BSA complex was generally higher than that of curcumin–gelatin complex, which was consistent with the result showed by fluorescence spectroscopy that the interaction and affinity between curcumin–BSA was stronger than curcumin–gelatin. The possible mechanism of curcumin–protein complex formation using the charge-switch method is shown in Fig. 3D. BSA has a higher percentage of polar amino acids than gelatin (44.7% vs. 10.9%).31 Among these, the basic amino acids are lysine, arginine, and histidine, with side chain pKa of 10.53, 12.48, and 6.04.32 These basic amino acids can be protonated and carry positive charges at low pH. It is postulated that BSA, with a higher percentage of these amino acids than gelatin, can provide more cationic sites for interacting electrostatically with the negatively charged curcumin, which is one possible reason that curcumin–BSA interaction is stronger than curcumin–gelatin, and curcumin–BSA is more soluble than curcumin–gelatin.
In this work, the maximum solubility of curcumin after complexation with BSA [391.77 (±15.70) μg mL−1] was achieved by mixing equal volumes of 2 mg mL−1 curcumin (negatively charged) with 5 mg mL−1 BSA (positively charged), which was one of the highest curcumin solubilities compared to literature.9–11 At this concentration, the loading amount (LA) of curcumin was 13.55%, which was significantly higher than the previous studies. Similarly, the maximum solubility of curcumin after complexation with gelatin in this work was 143.64 (±2.29) μg mL−1, which was achieved by mixing equal volumes of 0.5 mg mL−1 curcumin (negatively charged) with 5 mg mL−1 gelatin (positively charged), and the LA of curcumin at this concentration was 5.27%. Moreover, curcumin–BSA complex prepared from 0.5 mg mL−1 curcumin and 1 mg mL−1 BSA showed 135.83 (±5.69) μg mL−1 solubility but a much higher LA at 21.36%, and curcumin–gelatin complex prepared from 0.25 mg mL−1 curcumin and 1 mg mL−1 gelatin showed 92.22 (±0.63) μg mL−1 solubility and a much higher LA at 15.57%. In summary, complexation with BSA and gelatin could increase curcumin solubility up to 391.77 (±15.70) μg mL−1 and 143.64 (±2.29) μg mL−1, and the curcumin loading amount was up to 21.36% and 15.57%, which was one of the highest reported LA for water-soluble curcumin–protein complex, compared those reported in literature, as mentioned above.9–11 There were mainly two reported methods to make the curcumin–protein complex, i.e. “solvent-based method” and “pH-shift method”. Using the “solvent-based method”, the solubility of curcumin has been improved up to 78.26 μg mL−1 (curcumin–soy protein isolate)33 and 81.68 μg mL−1 (curcumin–ovalbumin complex).34 Using the “pH-shift method”, the solubility of curcumin has been increased to 95 μg mL−1 (curcumin–ovalbumin complex),9 160.2 μg mL−1 (curcumin–egg white protein isolate complex),10 and 319.2 μg mL−1 (curcumin–whey protein isolate complex),11 but the corresponding loading amount (LA) of curcumin was only 4.53%, 3.10%, and 1.57%, respectively. In the curcumin molecule, the hydrophobic phenyl groups contribute to its hydrophobic interactions, and the phenolic hydroxyl and methoxy groups can participate in hydrogen-bonding interactions.5 In addition to these, the charge-switch method made use of the electrostatic interactions between negatively charged curcumin and positively charged proteins, which was one possible reason of higher curcumin loading.
3.3 Bioaccessibility of curcumin–protein complex
The application of curcumin is significantly limited by its low aqueous solubility and fast degradation under physiological conditions.35 Therefore, the in vitro bioaccessibility of free curcumin, curcumin–BSA and curcumin–gelatin complex, were evaluated under sequential simulated gastric and intestinal digestion (Fig. 4). “Bioaccessibility” is defined here as the amount of curcumin present in the aqueous phase of the digests after simulated gastric and intestinal digestion.7 As seen in Fig. 4, at the end of simulated gastric phase, 2.22 (±0.45)%, 8.42 (±2.33)%, and 9.43 (±1.14)% of curcumin was released from free curcumin, curcumin–BSA complex, and curcumin–gelatin complex respectively. While at the end of the full in vitro digestion process, the total amount of curcumin released was 13.33 (±1.46)% for free curcumin, 74.11 (±1.01)% for curcumin–BSA complex, and 74.48 (±0.13)% for curcumin–gelatin complex. For free curcumin, the bioaccessibility after simulated intestinal digestion was higher than after simulated gastric digestion, which was attributed to the bile salts that forms micelles to solubilize curcumin.35 During gastric digestion, the curcumin–protein complex has been reported to be resistant to pepsin,1,35 which may have contributed to the lower release of curcumin from the complex, as observed in Fig. 4. However, upon exposure to intestinal digestion, pancreatin likely facilitated the breakdown of the complex, as reported previously.1,35 This breakdown released curcumin, which was subsequently solubilized by bile salts, leading to increased bioaccessibility.35 Overall, the amount of soluble curcumin (i.e., curcumin potentially available for absorption) from the curcumin–BSA and curcumin–gelatin complexes after in vitro digestion were significantly higher than free curcumin (p < 0.05). These results indicated that complexation with BSA and gelatin both increased curcumin bioaccessibility, which could be attributed to the increased aqueous solubility, decreased size, and amorphous state of curcumin after complexation.3.4 Antioxidant properties of curcumin–protein complex
ABTS radical scavenging is an important method to test the antioxidant property. In ABTS radical scavenging assay, 2,2′-azino-bis-3-ethylbenzthiazoline-6-sulphonic acid (ABTS) can react with oxidizers and generate ABTS cation radicals (ABTS˙+) which has a characteristic absorption peak at 734 nm. Antioxidants can suppress the production of ABTS cation radicals and thereby decrease the absorbance. The ABTS scavenging activity of all the samples are shown in Fig. 5. When directly dispersed in water, curcumin showed low radical scavenging activity at 12.85 (±0.20)%, which was attributed to its low solubility in water. BSA and gelatin also showed mild antioxidant property, which were 25.90 (±0.80)% and 31.11 (±0.39)% respectively, and it was because of the presence of cysteine in the two proteins.36 curcumin–BSA complex and curcumin–gelatin complex both showed high radical scavenging activity (around 100%), similar to the antioxidant vitamin C as positive control, which showed that complexation with BSA and gelatin both significantly improved the antioxidant activity of curcumin (p < 0.05). The complexation contributed to both increased aqueous solubility of curcumin and the antioxidant properties of BSA and gelatin. The result was similar to previous study that curcumin–mung bean protein complex showed improved aqueous solubility and therefore higher ABTS radical scavenging activity.25 With significantly improved antioxidant activity, the curcumin–BSA complex and the curcumin–gelatin complex could be used for strengthening the antioxidant status, and various applications that require antioxidant property.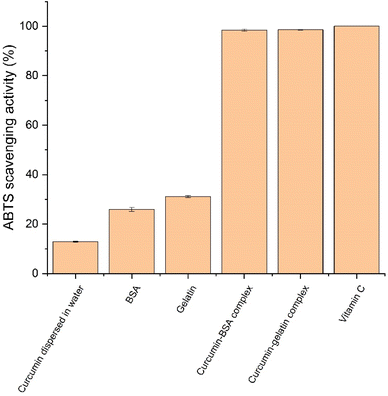 | ||
| Fig. 5 ABTS scavenging activity of curcumin dispersed in water, BSA, gelatin, curcumin–BSA complex, curcumin–gelatin complex, and vitamin C (positive control). | ||
3.5 Antimicrobial properties of curcumin–protein complex
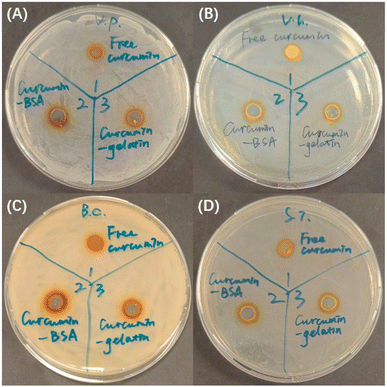
Agar well diffusion assay was used to evaluate the inhibitory effect of curcumin–protein complex on Vibrio parahaemolyticus, Vibrio harveyi, Bacillus cereus, and Streptococcus iniae. Inhibition zones are clear sites with no visible bacteria growth, and the antimicrobial activity was evaluated by measuring the diameter of inhibition zone against tested pathogens,3 as shown in Table 2 and Fig. 6. For all the four pathogens studied, larger inhibition zone was observed for curcumin–protein complex, compared to free curcumin (p < 0.05). DMSO (4%), BSA (5 mg mL−1), and gelatin (5 mg mL−1) at corresponding concentrations, as negative control, did now show inhibition effect on any of the pathogens. Therefore, it can be concluded that complexation with proteins improved the antimicrobial effect of curcumin for certain pathogens, which was attributed to the improved solubility and decreased size. For all the four pathogens investigated, the inhibition zones of curcumin–BSA complex were larger than that of curcumin–gelatin complex, which was attributed to higher aqueous solubility of curcumin–BSA than curcumin–gelatin.| Pathogen | Diameter of inhibition zone of free curcumin (mm) | Diameter of inhibition zone of curcumin–BSA complex (mm) | Diameter of inhibition zone of curcumin–gelatin complex (mm) |
|---|---|---|---|
| Vibrio parahaemolyticus | 8.3 ± 0.2 | 10.3 ± 0.6 | 9.8 ± 0.6 |
| Vibrio harveyi | 7.5 ± 0.0 | 10.7 ± 0.2 | 9.2 ± 0.2 |
| Bacillus cereus | 7.5 ± 0.0 | 9.7 ± 0.2 | 9.2 ± 0.2 |
| Streptococcus iniae | 7.7 ± 0.2 | 10.3 ± 0.5 | 9.7 ± 0.2 |
To further evaluate the inhibitory effect of curcumin–protein complex on Vibrio parahaemolyticus, Vibrio harveyi, Bacillus cereus, and Streptococcus iniae, minimum inhibitory concentration (MIC) of curcumin (before and after complexation) against these four pathogens was determined through broth micro-dilution techniques. For Vibrio parahaemolyticus, the MIC of free curcumin was 250 μg mL−1, but the MICs of both curcumin–protein complexes decreased to 31.25 μg mL−1. For Vibrio harveyi, the MIC of free curcumin was not observed in the range of concentrations studied (1000 to 7.81 μg mL−1), but the MICs of both curcumin–protein complexes decreased to 125 μg mL−1. For Bacillus cereus, the MIC of free curcumin was 62.5 μg mL−1, while the MICs of both curcumin–protein complexes decreased to 31.25 μg mL−1. For Streptococcus iniae, the MIC of free curcumin was 125 μg mL−1, but the MICs of both curcumin–protein complexes decreased to 62.5 μg mL−1. As control, DMSO, BSA and gelatin alone, at corresponding concentrations, did not inhibit the growth of any of these pathogens.
For all the four pathogens, the MIC of curcumin decreased after complexation with the proteins (eight times for Vibrio parahaemolyticus, more than eight times for Vibrio harveyi, two times for Bacillus cereus, and two times for Streptococcus iniae), which was attributed to both increased solubility and decreased size of curcumin after complexation.3 Currently, there are only a few studies on the antimicrobial properties of curcumin–protein complex, and the antimicrobial properties of curcumin–protein complex have been found to be equal or better than free curcumin.5 Based on the result of this study, the curcumin–protein complex, with improved solubility, bioaccessibility, antioxidant and antimicrobial properties, could effectively decrease the dosage of curcumin in controlling and preventing diseases that are relevant to these pathogens.
4. Conclusions
The charge-switch nanocomplexation method presented in this work is an effective strategy to improve the physicochemical and biological properties of curcumin. The complexation process can be carried out by introducing opposite net charges to curcumin and protein respectively and simple mixing. The interactions between curcumin and protein were verified by FTIR and fluorescence quenching studies. Complexation with BSA and gelatin increased curcumin's aqueous solubility to 391.77 (±15.70) μg mL−1 and 143.64 (±2.29) μg mL−1 respectively, and the loading amount (LA) of curcumin was up to 21.36% and 15.57% respectively. While curcumin–BSA complex generally showed better solubility than curcumin–gelatin complex, the lower cost of proteins like gelatin can be more suitable for forming curcumin–protein complexes for food and aquaculture applications. In vitro digestion showed increased bioaccessibility of curcumin after complexation as more curcumin was accessible from curcumin–BSA complex and curcumin–gelatin complex. The curcumin–protein nanocomplexes developed in this work also showed improved antioxidant properties and antimicrobial properties against four food and aquaculture related pathogens. Therefore, the improved physiochemical and biological properties of the curcumin–protein nanocomplexes shows its potential as functional food additives in agri-food applications. Future studies should focus on large scale production of curcumin–protein complexes, incorporation into food matrices, and in vivo bioavailability studies of the curcumin–protein complex to study the possible advantages on absorption and bioavailability after oral administration.Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.Author contributions
Anyu Zhang: conceptualization, methodology, data curation, formal analysis, writing – original draft. Manish Mahotra: conceptualization, formal analysis, methodology, writing – review & editing. Hong Yu: methodology, formal analysis, writing – review & editing. Tianqi Zhu: methodology, data curation. Say Chye Joachim Loo: conceptualization, visualization, resources, funding acquisition, supervision, writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to acknowledge the financial support from the Ministry of Education (RG79/22, RG29/24, MOE-T2EP30223-0001, NGF-2023-14-005), and the Singapore Food Agency (SFS_RND_SUFP_001_06).Notes and references
- X. Du, H. Jing, L. Wang, X. Huang, L. Mo, X. Bai and H. Wang, LWT - Food Sci. Technol., 2022, 154, 112753 CAS.
- V. P. Menon and A. R. Sudheer, The Molecular Targets and Therapeutic Uses of Curcumin in Health and Disease, 2007, pp. 105–125 Search PubMed.
- Bhawana, R. K. Basniwal, H. S. Buttar, V. K. Jain and N. Jain, J. Agric. Food Chem., 2011, 59, 2056–2061 CAS.
- M. Alagawany, M. R. Farag, S. A. Abdelnour, M. A. Dawood, S. S. Elnesr and K. Dhama, Aquaculture, 2021, 532, 736030 CAS.
- M. Mohammadian, M. Salami, E. Assadpour and S. M. Jafari, Trends Food Sci. Technol., 2024, 104372 CrossRef CAS.
- V. Andonova and P. Peneva, Curr. Pharm. Des., 2017, 23, 6630–6642 CAS.
- C. H. Tang, Food Hydrocolloids, 2020, 109, 106106 CrossRef CAS.
- M. Esmaili, S. M. Ghaffari, Z. Moosavi-Movahedi, M. S. Atri, A. Sharifizadeh, M. Farhadi, R. Yousefi, J. Chobert, T. Haertlé and A. A. Moosavi-Movahedi, LWT - Food Sci. Technol., 2011, 44, 2166–2172 CrossRef CAS.
- H. Li, M. Zhao, S. Zhou, H. Zhang, J. Wang, N. Xia, Y. Liu, S. Hua and G. Tan, Food Hydrocolloids, 2024, 149, 109623 CrossRef CAS.
- Y. Wang, L. Zhang, P. Wang, X. Xu and G. Zhou, Food Res. Int., 2020, 137, 109366 CrossRef CAS PubMed.
- H. T. Kevij, M. Mohammadian and M. Salami, J. Food Process. Preserv., 2019, 43, e14227 Search PubMed.
- Y. Ma, S. Chen, W. Liao, L. Zhang, J. Liu and Y. Gao, J. Agric. Food Chem., 2020, 68, 7103–7111 CrossRef CAS PubMed.
- S. H. Brewer, W. R. Glomm, M. C. Johnson, M. K. Knag and S. Franzen, Langmuir, 2005, 21, 9303–9307 CrossRef CAS PubMed.
- L. C. Sow, N. Z. Y. Toh, C. W. Wong and H. Yang, Food Hydrocolloids, 2019, 94, 459–467 CrossRef CAS.
- M. Salehiabar, H. Nosrati, E. Javani, F. Aliakbarzadeh, H. K. Manjili, S. Davaran and H. Danafar, Int. J. Biol. Macromol., 2018, 115, 83–89 CrossRef CAS PubMed.
- T. Yang, H. Yang, Y. Fan, B. Li and H. Hou, Int. J. Biol. Macromol., 2018, 118, 124–131 CrossRef CAS PubMed.
- Y. Zhai, X. Meng, L. Li, Y. Liu, K. Xu, C. Zhao, J. Wang, X. Song, J. Li and M. Jin, RSC Adv., 2021, 11, 38638–38647 RSC.
- S. Gong, C. Jiao, B. Liu, W. Qu, L. Guo and Y. Jiang, J. Funct. Foods, 2023, 105, 105571 CrossRef CAS.
- X. H. Zhang, X. He and B. Austin, Mar. Life Sci. & Technol., 2020, 2, 231–245 Search PubMed.
- W. Agnew and A. C. Barnes, Vet. Microbiol., 2007, 122, 1–15 CrossRef CAS PubMed.
- H. Yu, M. H. Nguyen, W. S. Cheow and K. Hadinoto, Mater. Sci. Eng., C, 2017, 75, 25–33 CrossRef CAS PubMed.
- A. Nayak, C. Genot, A. Meynier, A. Dorlando and I. Capron, LWT - Food Sci. Technol., 2022, 153, 112421 CrossRef CAS.
- F. P. Chen, B. S. Li and C. H. Tang, J. Agric. Food Chem., 2015, 63, 3559–3569 CrossRef CAS PubMed.
- S. Kharel, A. Gautam, M. Mahotra, N. M. Theniko and S. C. J. Loo, J. Funct. Foods, 2021, 87, 104749 CrossRef CAS PubMed.
- M. Mohammadian, M. Salami, M. Moghadam, A. Amirsalehi and Z. Emam-Djomeh, J. Drug Delivery Sci. Technol., 2021, 61, 102148 CrossRef CAS.
- I. Wiegand, K. Hilpert and R. E. Hancock, Nat. Protoc., 2008, 3, 163–175 CrossRef CAS PubMed.
- S. Solghi, Z. Emam-Djomeh, M. Fathi and F. Farahani, J. Food Process Eng., 2020, 43, e13403 CrossRef CAS.
- S. Gorinstein, I. Goshev, S. Moncheva, M. Zemser, M. Weisz, A. Caspi, I. Libman, H. T. Lerner, S. Trakhtenberg and O. Martín-Belloso, J. Protein Chem., 2000, 19, 637–642 Search PubMed.
- A. Papadopoulou, R. J. Green and R. A. Frazier, J. Agric. Food Chem., 2005, 53, 158–163 CrossRef CAS PubMed.
- Y. Hu, C. Qiu, D. J. McClements, Y. Qin, J. Long, A. Jiao, X. Li, J. Wang and Z. Jin, Food Chem., 2022, 376, 131869 Search PubMed.
- P. Khramtsov, T. Kalashnikova, M. Bochkova, M. Kropaneva, V. Timganova, S. Zamorina and M. Rayev, Int. J. Pharm., 2021, 599, 120422 Search PubMed.
- M. S. Alsalhi, P. G. Royall and K. L. A. Chan, RSC Adv., 2022, 12, 19040–19053 Search PubMed.
- F. Ji, Z. Wang, X. Bai, Y. Zhao, X. Zhong, S. Luo, Y. Shen, S. Jiang and Z. Zheng, Food Hydrocolloids, 2023, 143, 108929 CrossRef CAS.
- H. Zhang, T. Wang, F. He and G. Chen, Int. J. Biol. Macromol., 2021, 168, 686–694 CrossRef CAS PubMed.
- O. D. Okagu, O. Verma, D. J. McClements and C. C. Udenigwe, Int. J. Biol. Macromol., 2020, 151, 333–343 CrossRef CAS PubMed.
- M. Roche, P. Rondeau, N. R. Singh, E. Tarnus and E. Bourdon, FEBS Lett., 2008, 582, 1783–1787 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Characteristic peaks and corresponding functional groups in the FTIR spectrums. See DOI: https://doi.org/10.1039/d4ra07613c |
| This journal is © The Royal Society of Chemistry 2025 |