 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Assessing the ecotoxicity of multicomponent nanomaterials using a classification SAR approach†
G. P.
Gakis
 *,
I. G.
Aviziotis
and
C. A.
Charitidis
*,
I. G.
Aviziotis
and
C. A.
Charitidis

Research Lab of Advanced, Composite, Nano-Materials and Nanotechnology, Materials Science and Engineering Department, School of Chemical Engineering, National Technical University of Athens, 9 Heroon Polytechneiou Street, Zografos, Athens 15780, Greece. E-mail: gakisg@chemeng.ntua.gr; Tel: +30 2107723296
First published on 15th April 2025
Abstract
The emerging applications of nanotechnology have led to the synthesis, production and use of a continuously increasing number of nanomaterials. In recent years, the focus is being shifted to multicomponent nanomaterials (MCNMs), due to the control over their functional properties. At the same time, the increasing exposure of ecosystems to such materials has raised concerns over their environmental hazard, with several in vivo and in vitro studies used to assess the ecotoxicity of MCNMs. The demanding nature of such methods has also led to the increasing development of in silico methods, such as structure–activity relationship (SAR) models. Although such approaches have been developed for single component nanomaterials, models for the ecotoxicity of MCNMs are still sparse in scientific literature. In this paper, we address the case of MCNM ecotoxicity by developing an in silico classification SAR computational framework. The models are built over a dataset of 652 ecotoxicity measurements for 214 metal and metal oxide MCNMs, towards bacteria, eukaryotes, fish, plants and crustaceans. This dataset is, to the best of the authors' knowledge, the largest dataset used for MCNM ecotoxicity. It is found that two descriptors can adequately classify different MCNMs based on their ecotoxicity over the whole heterogeneous dataset. These descriptors are the hydration enthalpy of the metal ion and the energy difference between the MCNM conduction band and the redox potential in biological media. Although the classification does not allow a quantitative ecotoxicity assessment, the heterogeneous nature of the dataset can reveal key MCNM features that induce toxic action, allowing a more holistic understanding of MCNM ecotoxicity, as well as the nature of interaction between the different MCNM components.
Environmental significanceIn recent years, nanotechnology research has focused on multi-component nanomaterials (MCNMs), which allow increased control over nanomaterial properties. However, concern has been raised regarding the safety of extended ecosystem exposure to such materials. The demanding nature of in vitro and in vivo methods has led to emerging in silico techniques, such as structure–activity relationship (SAR) models, for nanomaterial safety assessment. Nevertheless, such approaches for the case of MCNM ecotoxicity are limited, and are built using limited datasets. In this work, a classification approach is presented for MCNM ecotoxicity towards bacteria, eukaryotes, fish, plants, and crustaceans. The use of the heterogeneous datasets allows a more holistic understanding of MCNM toxicity, assisting the synthesis of MCNMs that are safe-by-design towards ecosystems. |
1. Introduction
The wide spectrum of engineered nanomaterial (ENM) applications, ranging from catalysis1 and energy2 to electronics3 and nanomedicine4 has rendered the synthesis of ENMs of varying composition and forms5–10 a key part of nanotechnology research. This increasing use of ENMs has led to the demand for control and tunability of ENM properties, depending on the targeted application.11,12 These demanding requirements have led to the synthesis of multi-component nanomaterials (MCNMs),13,14 such as bimetallic nanomaterials15 or alloys,16 surface functionalized nanomaterials,17 coated nanoparticles,18 as well as doped nanomaterials,19 among others.The emergence of ENM and MCNM applications has also raised concern regarding the safety of extended exposure to such materials. While such exposure can be intentional, with the use of these materials as drug carriers,20,21 non-intentional exposure to ENMs and MCNMs can occur in varying environments.22–25 Ecosystems can also be exposed to ENMs and MCNMs, throughout their manufacturing process, application and disposal stages, via different exposure routes.26–28
The safety of ENMs and MCNMs is usually assessed by measuring their toxicity. In this context, in vitro toxicity methods are used for a faster and cost effective initial toxicity assessment,29 while in vivo methods, which are the most reliable, are used at later stages of regulatory risk assessment,30 as they are time consuming and are characterized by higher cost and ethical concerns regarding animal testing. However, both types of toxicity assessment methods cannot keep up with the innovation, synthesis, and application of novel nanomaterials.31 For this reason, in silico methods have emerged to assess the safety of nanomaterials, with computational models developed for biodistribution32,33 and toxicity,31,34,35 toward different cells and organisms.
During the last decades, the increasing applications of nanotechnology have led to a wide number of research studies regarding nanomaterial toxicity assessment using in vitro36–38 and in vivo39–41 methods, as well as comparative reviews between the different toxicity assessment methods.42,43 In a similar way, interest has shifted towards in silico methods, and in particular structure–activity relationship (SAR) models44,45 based on the correlation of structural characteristics of the materials under study, known as descriptors, to biological activity endpoint data.46 Both quantitative (QSAR) and qualitative (classification SAR)47 models have been developed, especially for the case of metal oxide nanoparticles (NPs),47–49 but also for other types of ENMs.35,50In silico models for such materials have been comprehensively reviewed by Buglak et al.,44 and Li et al.51 However, most models are developed using a limited dataset,47,48,52,53 using a lower number of toxicity data. This limitation can severely hinder the extension of the use of SAR approaches, as models developed using a limited and homogeneous dataset apply only to a range of data similar to their training datasets. This means that the model precision may be severely decreased when comparing with measurements with a slight difference in the experimental design, or the NP properties, such as size and shape, thus damaging the model reproducibility. Furthermore, when larger and more heterogeneous datasets are used for the model development,49,54–57 the extraction of mechanistic information from SAR models challenging due to the complexity of the descriptors used. Complex descriptors can be difficult to be computed for novel NPs or more complex nanomaterial structures. Furthermore, the lack of mechanistic understanding does not allow to extend the use of QSAR models as decision supporting tools for the design and synthesis of safe-by-design nanomaterials.58 Nevertheless, theoretical frameworks have been developed to provide a more cohesive, consistent and mechanistic understanding of metal oxide toxicity, while also leading to predictive SAR models for toxicity.31,53,59–63 Based on a similar framework, we recently used an extensive and heterogeneous dataset of toxicity measurements towards a wide range of cell lines and organisms, to develop a classification SAR model.64
Although in silico methods have been developed for pure metal oxides, the case of MCNMs is not yet sufficiently covered. In particular, the case of metal-loaded TiO2 MCNMs has been studied by means of QSAR modelling,65–69 while only the case of ZnO-based NPs has been covered besides the TiO2 MCNMs.70 Nevertheless, the limited number of MCNMs and toxicity data used to develop these models does not allow a more global understanding of MCNM toxicity mechanisms. In a recent work, we presented a classification SAR model using an extensive dataset for the case of cytotoxicity and antibacterial activity of metal and metal oxide MCNMs,71 which allowed a more mechanistic insight on the dominant MCNM toxicity pathways. Regarding the case of ecotoxicity, however, although there has been an increasing interest using in vitro and in vivo assessment methods,72–75in silico studies with the development of SAR models are still missing for MCNMs.
In this work, a classification SAR approach for the prediction of MCNM ecotoxicity is presented. The model is developed using an extensive dataset of 652 half-maximal concentration measurements for the ecotoxicity of metal and metal oxide MCNMs. The MCNMs considered in the present work consist of doped metal oxides, composite metal oxides, bimetallic NPs, as well as surface-loaded metal oxide NPs. Different subsets of data were used to build different models, based on the target organisms. In particular, models were developed for MCNM ecotoxicity towards E. coli, S. aureus, D. rerio, D. magna and C. albicans. Furthermore, the approach was extended to more heterogeneous datasets, consisting of MCNM ecotoxicity measurements towards different organisms' groups, such as bacteria, eukaryotes, fish, crustaceans and plants. Finally, the complete heterogeneous dataset was used for the development of a SAR model, showing that the approach can offer a more general insight regarding MCNM ecotoxicity.
The novelty of the approach lies in the size and nature of the dataset used for the development of such a model. The present study is, to the best of the author's knowledge, the largest dataset of MCNM ecotoxicity measurements used for the development of a SAR model. Furthermore, the dataset is heterogeneous, consisting of ecotoxicity measurements towards bacteria, eukaryotes, fish, plants and crustaceans. Such a heterogeneous SAR approach has not been applied before for the case of MCNM ecotoxicity while the used approach to compute MCNM descriptors for surface-loaded MCNMs is also novel for ecotoxicity models. Finally, the aim of the present work is not restricted to the development of a predictive classification model, but also to unravel the key characteristics of MCNMs that induce ecotoxicity. The size and heterogeneous nature of the dataset used for the model development will also assist towards a more holistic and mechanistic understanding of the ecotoxic action of MCNMs and the interaction between their components.
2. Methods
2.1. Toxicity endpoints, classification and datasets
The dataset for the model development consists of MCNM ecotoxicity measurements, retrieved from 102 papers published in scientific literature. In particular, the ecotoxicity endpoints consist of reported half maximal effective (EC50), inhibitory (IC50), and lethal (LC50) concentration measurements. For clarity, the different half-maximal concentration measurements are onwards referred to as EC50, as in the work of Simeone and Costa.31 The different EC50 values were derived from dose–response studies towards different fish, plants, and crustaceans. Furthermore, as bacteria and eukaryotic organisms such as yeast, fungi and common pathogens can survive in ecosystems,76–78 they are also included in the ecotoxicity dataset. Although the common practice is to develop SAR models based on a specific endpoint and target cell line or organism, heterogeneous datasets using multiple endpoints and cells/organisms have been previously used for the development of SAR models for ENM cytotoxicity,55,71 ecotoxicity,54 as well as combined cytotoxic and ecotoxic predictions.49,56,64Regarding the classification scheme, a measurement is characterized as toxic or non-toxic based on the criteria reported by Simeone and Costa,31 following the scheme presented in our previous works.64,71 Briefly, the measurement is classified as toxic if the logarithm of the concentration endpoint in molar units (mol L−1) is lower than −2.5 (log(EC50) ≤ −2.5). However, in some works, EC50 concentrations are reported as being higher than the range of experimentally tested concentrations (EC50 > Cmax,tested). In such cases, if the maximum concentration yielded an effect less than 50% and log(Cmax) > −2.5, then the measurement was classified as non-toxic. On the other hand, if log(Cmax) ≤ −2.5, then the following scheme is applied: If Cmax is more than 50% of the threshold concentration (Cthres), the measurement is classified as non-toxic. If, however, Cmax is less than 50% of the threshold value, the measurement is omitted from the dataset. The value of 50% was arbitrarily chosen, so that a significant amount of MCNM has been exposed so that the measurement is classified as non-toxic, and to reduce the number of data omitted from the dataset.
In a similar way, if the concentration tested was lower than the threshold value (log(Cmax) ≤ −2.5), and the effect was higher than 50%, then the EC50 value is set to the concentration tested and classified as toxic. Otherwise, for measurements with an effect higher than 50% where the concentration tested was higher than the threshold value (log(Cmax) > −2.5), the measurement is removed. The classification scheme is summarized in Table 1.
| Reported concentration | Condition | Classification |
|---|---|---|
| EC50 value reported | log(EC50) ≤ −2.5 | Toxic |
| log(EC50) > −2.5 | Non-toxic | |
| C max yields effect >50% | log(Cmax) ≤ −2.5 | Toxic |
| log(Cmax) > −2.5 | Omitted | |
| C max yields effect <50% | log(Cmax) > −2.5 | Non-toxic |
| log(Cmax) ≤ −2.5, Cmax ≥ 0.5·Cthreshold | Non-toxic | |
| log(Cmax) ≤ −2.5, Cmax < 0.5·Cthreshold | Omitted |
Based on the above classification scheme, a final dataset of 652 MCNM ecotoxicity measurements is developed, presented in the ESI† of the paper. The dataset consists of ecotoxicity measurements of bimetallic NPs, metal-doped metal oxide NPs, surface loaded metal oxide NPs, and composite metal oxide NPs. The total dataset is divided to different subsets of data, corresponding to the individual organism that was exposed to the MCNMs. Datasets corresponding to the different organism groups are also created. The dataset size (minimum of 30 measurements) and the number of toxic and non-toxic measurements (minimum of 20% of both data classes) served as the criteria for the creation of a data subset. Based on the above scheme, 5 data subsets are created for individual organisms, as well as 5 datasets for organism groups. The complete dataset is also used for the model development. The final datasets are presented in Table 2.
| Cell type/cell type | No of measurements | No of NPs | % of toxic measurements | % of non-toxic measurements |
|---|---|---|---|---|
| Individual organisms | ||||
| E. coli | 92 | 65 | 50 | 50 |
| S. aureus | 70 | 45 | 50 | 50 |
| D. rerio | 88 | 36 | 21.6 | 78.4 |
| D. magna | 37 | 26 | 64.9 | 35.1 |
| C. albicans | 36 | 31 | 66.7 | 33.3 |
| Organism groups | ||||
|---|---|---|---|---|
| Bacteria | 252 | 82 | 49.2 | 50.8 |
| Eukaryotes | 123 | 69 | 57.7 | 42.3 |
| Fish | 101 | 42 | 26.7 | 73.3 |
| Crustaceans | 74 | 58 | 64.9 | 35.1 |
| Plants | 102 | 40 | 30.4 | 69.6 |
| Complete dataset | 652 | 214 | 46.2 | 53.8 |
2.2. Descriptors
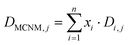 | (1) |
A different computational scheme is used for surface-loaded MCNMs, as presented in our previous work.71 Briefly, the composition of the MCNM surface, which is the area of interaction with the biological media, is computed. This is done by computing the total mass and molecular amount (qi, in moles) for each of the particle components in a single particle, using the nominal densities (25 °C) of each component (from online handbooks/databases) and mass/molar fractions, as well as the particle volume:
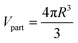 | (2) |
The surface components are all assumed to be situated on the particle surface (qs,i = qi). The amount of the core component on the particle surface (qs,core) is computed based on the core component's unit cell:
 | (3) |
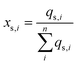 | (4) |
2.3. Model implementation, training and validation
As shown in Table 2, five datasets consist of MCNM ecotoxicity measurements towards individual organisms, while five different datasets were created for organism groups, namely bacteria, eukaryotes, fish, plants and crustaceans. Finally, the total set of data, consisting of all the MCNM ecotoxicity endpoints, was also used for SAR model development.For datasets with a lower number of measurements (n < 100), a five-fold cross validation was used. For larger datasets (n ≥ 100), a hold-out validation is used with 80% of the data as a training set and 20% as a validation set. In the hold-out validation scheme, the model is trained using a five-fold cross validation on the training set, while the validation is performed by comparing the trained model predictions to the toxic class assigned to the measurements in the validation set. The data splitting is random, and performed using MATLAB®. The model training is performed using the classification learner toolkit, by implementing Support Vector Machines (SVM), k-Nearest-Neighbors (kNN) and Random Forests (RF). The optimal models were identified with the use of different statistical metrics based on the resulting confusion matrix, namely accuracy, precision, sensitivity (or recall), and specificity (or selectivity), computed as in ref. 64 and 71. The receiver operating characteristic (ROC) curve was also used as a metric to identify the optimal models, while the models were deemed acceptable when the accuracy exceeded 80%.
2.4. Applicability domain
Besides the descriptor selection and the model development and validation, the definition of the applicability domain is also crucial for the development of SAR models. As the OECD standards state, the applicability domain should be clearly defined in order to determine a descriptor space, where the developed model can be applied.90 In the present work, the methods used for the applicability domain definition are the bounding box PCA, convex hull and Euclidean distance from centroid methods,91,92 which were implemented using the Applicability Domain toolbox developed by the Milano Chemometrics and QSAR Research Group.91,923. Results and discussion
3.1. Descriptor selection
As mentioned in section 2.2.3, the intercorrelation analysis of the individual component descriptors is the first step of the descriptor selection process. The results of this analysis for different metal and metal oxide descriptors are presented in Fig. 1, below. The Pearson coefficient has been selected to analyze the intercorrelation between the different descriptors, while the threshold value to consider the descriptors intercorrelated was set to 0.95.93 Only one of the intercorrelated descriptors is kept, while the other descriptors are discarded. From the remaining features, descriptors that were non-continuous over the dataset, as well as redundant descriptors with similar physical meaning, were subsequently removed from the descriptor matrix.As a second step, the feature selection methods, namely ReliefF and chi-square, are used to rank the remaining descriptors based on their relevance to the response variable (ecotoxic class), as described in section 2.2.3. The four highest ranked descriptors are kept, following the feature selection analysis. Representative results for the four highest ranked descriptors, as derived from the two methods, using the complete dataset, are shown in Fig. 2. The descriptors rank using the two feature selection methods, for the different data subsets, is presented in Table 3. It is mentioned that in Table 3, only the descriptors that were ranked among the four most relevant descriptors for at least one dataset are shown.
 | ||
| Fig. 2 The four most relevant descriptors, for the complete dataset, as derived from a) ReliefF and b) chi-square methods. | ||
| Relief F | Chi-square | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HE | Dbio | S_IP | IP | X_M | VWR | Xion | HE | Dbio | S_IP | IP | X_M | VWR | Xion | |
| E. coli | 3 | — | 2 | — | — | 1 | 4 | 2 | 1 | — | 3 | — | 4 | — |
| S. aureus | 3 | — | 1 | 2 | 4 | — | — | 4 | 1 | — | — | 3 | 2 | — |
| D. rerio | — | 2 | — | 4 | — | 1 | 3 | 1 | — | 3 | — | 2 | 4 | — |
| D. magna | 2 | 4 | — | — | — | 1 | 3 | 1 | — | 2 | 3 | — | 4 | — |
| C. albicans | 3 | 1 | 2 | — | — | — | 4 | 2 | 1 | 3 | 4 | — | — | — |
| Bacteria | 2 | 4 | 1 | — | — | — | 3 | 2 | — | — | 4 | 1 | 3 | — |
| Eukaryotes | 3 | 1 | 4 | 2 | — | — | — | 2 | 1 | — | — | 3 | — | 4 |
| Fish | — | 1 | — | 3 | 2 | 4 | — | 2 | 1 | — | — | — | 4 | 3 |
| Crustaceans | 1 | 3 | — | 4 | 2 | — | — | 1 | 2 | 3 | 4 | — | — | — |
| Plants | 4 | 2 | — | — | 3 | 1 | — | 2 | — | 3 | 1 | — | — | 4 |
| Complete dataset | 4 | 1 | — | — | — | 2 | 3 | 1 | 3 | — | 2 | 4 | — | — |
The descriptor ranking presented in Fig. 2 and Table 3 shows that seven descriptors are ranked within the four highest ranked descriptors at least once, for all the data subsets, using the two feature selection methods. Interestingly, HE (hydration enthalpy61,63,64,71), Dbio (energy difference between the conduction band of the MCNM and redox potential of biological media64,71), and IP (ionic potential of the metal component81) are ranked amongst the four highest ranked descriptors for all 11 datasets, using at least one feature selection method. Based on the above observations, the three abovementioned descriptors are kept for the model development, with different descriptor combinations being tested. The results are presented in the following sections, for the models developed for the different datasets.
3.2. MCNM ecotoxicity models for individual organisms
Following the descriptor selection step, the analysis continues with the development of SAR models for the datasets consisting of MCNM ecotoxicity measurements towards individual organisms. Different combinations of the three descriptors identified in the descriptor selection step (HE, Dbio, IP) are tested, and the optimal model for each dataset is kept. The inclusion of a descriptor is accepted when the model accuracy is significantly improved. The accuracy of the models built using the different descriptor combinations, for the individual organism datasets, is presented in Table 4.| Dataset | Descriptor combination | ||||||
|---|---|---|---|---|---|---|---|
| HE, Dbio, IP | HE, Dbio | HE, IP | Dbio, IP | HE | Dbio | IP | |
| E. coli | 95.2 | 94.6 | 85.9 | 90.2 | 70.7 | 53.3 | 78.3 |
| S. aureus | 97.1 | 97.1 | 81.4 | 95.7 | 84.3 | 51.4 | 77.1 |
| D. rerio | 89.8 | 93.2 | 87.5 | 81.8 | 83.0 | 71.6 | 78.4 |
| D. magna | 86.5 | 89.2 | 89.2 | 86.5 | 89.2 | 78.4 | 83.8 |
| C. albicans | 91.7 | 94.4 | 88.9 | 88.9 | 86.1 | 83.3 | 80.6 |
As seen from the results of Table 4, amongst the single descriptor models, HE is the most predictive descriptor, for most datasets, with the exception of the model developed for the measurements towards E. coli where IP is the most predictive descriptor. The inclusion of a second descriptor improved the model accuracy, with the combination of HE and Dbio being the most predictive set of descriptors, except from the case of the D. magna dataset, where the accuracy did not improve with the addition of Dbio as a descriptor. Finally, the inclusion of all three descriptors produced a non-significant or lower accuracy than the combination of HE and Dbio. Hence, in order to reduce the complexity of the model and enhance the interpretability of the results, the minimum number of descriptors that produce the highest accuracy are kept. This means that the two-descriptor combination of HE and Dbio is kept for the different models, except from the case of the D. magna model, where the single descriptor of HE is kept.
The statistical metrics for the optimal models developed using the ecotoxicity measurements within the individual organism datasets are presented in Table 5.
| Organism | Descriptors | Validation scheme | Acc (%) | Prec (%) | Sens (%) | Sel (%) |
|---|---|---|---|---|---|---|
| E. coli | HE, Dbio | 5-Fold cross validation | 94.6 | 97.7 | 91.3 | 97.8 |
| S. aureus | HE, Dbio | 5-Fold cross validation | 97.1 | 94.6 | 100 | 94.3 |
| D. rerio | HE, Dbio | 5-Fold cross validation | 93.2 | 100 | 68.4 | 100 |
| D. magna | HE | 5-Fold cross validation | 89.2 | 100 | 83.3 | 100 |
| C. albicans | HE, Dbio | 5-Fold cross validation | 94.4 | 95.8 | 95.8 | 91.7 |
The statistical metrics presented in Table 5 show that the models developed have acceptable values for accuracy, towards all the individual model datasets. The lowest accuracy is obtained by the D. magna model, which, however, has an acceptable value of 89.2%. The developed models also show high values for precision, sensitivity and selectivity, with the exception of the D. rerio model, which shows a sensitivity of 68.4%. This lower sensitivity could be assigned to the more imbalanced nature of the dataset, as only 21.6% of the measurements are classified as toxic within the D. rerio ecotoxicity dataset. The very high values of precision and selectivity could also be assigned to this imbalance, for the D. rerio dataset. In any case, the different models were able to classify the ecotoxicity measurements in the different datasets successfully, using the same descriptor combination of HE and Dbio, except from the D. magna model which used the HE descriptor. However, as seen in the results of Table 4, the addition of Dbio did not decrease or increase the accuracy.
The results presented in Tables 4 and 5 show that the classification SAR approach presented can predict the MCNM ecotoxicity towards the different organisms with acceptable accuracy, using similar descriptors. This could hint towards similar underlying mechanisms being responsible for the ecotoxicity of the different MCNMs towards the different organisms. Furthermore, the same results show that the additive mixture approach, as suggested by Mikolajczyk et al.,65 and the approach used for surface-loaded MCNMs previously presented,71 is able to produce predictive descriptors for MCNM ecotoxicity. This can assist towards the understanding of the interaction between the various MCNM components.
The small number of descriptors used for the classification SAR model development, together with the fact that similar descriptors are used for all the individual organism datasets, allows the mapping of the classification results over the descriptor space. The predicted ecotoxic class for the different MCNMs, towards the different organisms, is presented in Fig. 3, as a function of the two descriptors, HE and Dbio. It is noted that the results for the D. magna model are also presented over the same descriptor space for comparison, even though the model is developed using only HE as a descriptor.
 | ||
| Fig. 3 SAR model predictions of the ecotoxic class of the different MCNMs, towards the different datasets, as a function of the two descriptors (HE, Dbio). | ||
Results of Fig. 3 show that similar results are obtained for the ecotoxic class prediction over the descriptor space, for all the different datasets. Specifically, the ecotoxic MCNMs are characterized by a less negative HE and a lower Dbio value, for all the different classification SAR models developed. Specifically, a threshold value of HE close to −50 eV and a Dbio value close to 1 eV separate the two classes, with the exception of the C. albicans model, where ecotoxic classification is obtained for higher Dbio values. This similar behavior may hint towards similar dominating mechanisms for ecotoxic action among the MCNMs of the different datasets, towards different organisms. Such results are consistent with previous findings for the case of pure metal oxides,61–64 and may assist towards a more global understanding of ecotoxic action of MCNMs. Further discussion regarding the ecotoxic mechanisms and the interpretation of model results will be presented in a subsequent section of the present paper.
3.3. MCNM ecotoxicity models for heterogeneous datasets
Based on the results of the previous section, where similar descriptors were able to predict the ecotoxic class of the different MCNMs in the individual organism datasets, the approach is extended for more heterogeneous datasets in terms of the target organism. For this reason, the datasets consisting of ecotoxicity measurements towards similar organism groups are used for the SAR model development. The complete dataset is also used to develop a SAR model, in order to test whether more holistic ecotoxicity predictions can be obtained with the present approach.As in the previous section, the different combinations of the descriptors identified in the selection step (HE, Dbio, IP) are used for SAR model development, in order to identify the optimal descriptor combination. The accuracy of the models built using the different descriptor combinations, for the organism groups and the complete dataset, is presented in Table 6. It is noted that for the descriptor combination tests, all the models were developed using five-fold cross validation.
| Dataset | Descriptor combination | ||||||
|---|---|---|---|---|---|---|---|
| HE, Dbio, IP | HE, Dbio | HE, IP | Dbio, IP | HE | Dbio | IP | |
| Bacteria | 92.9 | 92.9 | 85.3 | 90.9 | 75.4 | 75.0 | 84.9 |
| Eukaryotes | 93.5 | 92.7 | 81.3 | 87.0 | 71.5 | 85.4 | 72.4 |
| Fish | 87.1 | 88.1 | 78.2 | 84.2 | 76.2 | 74.3 | 72.3 |
| Crustaceans | 93.2 | 93.2 | 93.2 | 82.4 | 93.2 | 78.4 | 74.3 |
| Plants | 87.3 | 87.3 | 85.3 | 83.3 | 80.4 | 69.6 | 85.3 |
| Complete dataset | 89.9 | 89.6 | 75.6 | 80.8 | 76.4 | 64.0 | 72.9 |
The results of Table 6 show that among the single descriptor models, HE is the most predictive descriptor for the fish and crustaceans models, as well as for the complete dataset model. IP is the most predictive descriptor for the bacteria and plants models, while Dbio is the most predictive for the eukaryotes model. As for the case of individual organism models, the addition of a second descriptor improved the accuracy in all the heterogeneous models, except from the crustacean model, where the addition of a second descriptor to HE did not affect the model accuracy. Interestingly, the most predictive descriptor pair was found to be the HE and Dbio combination, as in the individual organism models. Finally, the addition of IP to the HE and Dbio combination led to non or insignificant increase of the model accuracy. From the above results, it is concluded that the optimal descriptor set is the combination of HE and Dbio, with the exception of the crustacean model, where the single descriptor model developed using HE was found to be optimal.
Using the abovementioned descriptor combinations, the optimal models are developed. The resulting statistical metrics for the optimal models using the ecotoxicity measurements within the heterogeneous datasets of organism groups and the complete dataset are presented in Table 7. It is mentioned that the models presented in Table 7 display different accuracy than in Table 6 (except from the crustaceans model), due to the fact that they are developed using a hold-out validation scheme, contrary to the models of Table 6, where a five-fold cross validation scheme was used for all models.
| Organism | Descriptors | Validation scheme | Acc (%) | Prec (%) | Sens (%) | Sel (%) |
|---|---|---|---|---|---|---|
| Bacteria | HE, Dbio | Train. (n = 202) | 94.0 | 90.5 | 95.0 | 93.3 |
| Val. (n = 50) | ||||||
| Eukaryotes | HE, Dbio | Train. (n = 99) | 91.7 | 92.9 | 92.9 | 90.0 |
| Val. (n = 24) | ||||||
| Fish | HE, Dbio | Train. (n = 81) | 85.0 | 80.0 | 66.7 | 92.3 |
| Val. (n = 20) | ||||||
| Crustaceans | HE | 5-Fold cross validation | 93.2 | 97.8 | 91.7 | 96.2 |
| Plants | HE, Dbio | Train. (n = 82) | 90.0 | 100 | 75.0 | 100 |
| Val. (n = 20) | ||||||
| Complete dataset | HE, Dbio | Train. (n = 522) | 89.2 | 83.3 | 92.6 | 86.9 |
| Val. (n = 130) |
Results of Table 7 show that acceptable accuracy is obtained for all developed models using the heterogeneous datasets for the organism groups, as well as the complete dataset. Furthermore, acceptable values are obtained for all statistical metrics along the different models, except from the sensitivity obtained by the models developed using the fish and plants datasets. This lower model sensitivity could be assigned to the more imbalanced nature of these datasets (26.7% and 30.4% of toxic measurements, respectively). Nevertheless, the fact that the developed models can predict the ecotoxic class of the various MCNMs in the different validation sets of the different organism group and complete datasets, using similar descriptors, enhances the notion that similar mechanisms dominate the MCNM ecotoxic action. As in the case of the individual organism models, HE and Dbio are found to be the most predictive set of descriptors, except from the case of the crustacean model, which was developed using only the HE descriptor. Moreover, the additive mixture approach used to compute the MCNM descriptors was again able to produce predictive descriptors for the different classification SAR models, allowing a more global understanding of the nature of the interaction between MCNM component.
The ecotoxic class predicted by the different classification SAR models, mapped over the space defined by the two model descriptors, is presented in Fig. 4. This mapping allows more clear understanding of the effect of the different descriptors on the model results, and is possible due to the low number of descriptors used during the model development.
 | ||
| Fig. 4 SAR model predictions of the ecotoxic class of the different MCNMs, towards the different organism groups and complete datasets, as a function of the two descriptors (HE, Dbio). | ||
As in the case of the models developed for the individual organism datasets (Fig. 3), the results of Fig. 4 present a similar ecotoxic class prediction over the descriptor space, for all models developed for the organism group and complete datasets. Ecotoxic class is predicted for MCNMs that exhibit a less negative HE and a lower Dbio value. For all the different classification SAR models developed. A similar trend is observed across all the SAR models, both for the case individual organisms (Fig. 3) and organism groups (Fig. 4). This behavior enhances the notion of similar mechanisms being dominant for the ecotoxic action of MCNMs towards different organisms. Such results may lead to a more holistic understanding of ecotoxic action of MCNMs, which in turn can assist in the development of MCNMs with properties that increase or decrease the interaction of MCNMs with the abovementioned organism, according to the desired application.
3.4. Applicability domain
Based on the guidelines for (Q)SAR model development, as set by the OECD, a validated SAR model should have a defined applicability domain, which is one of the five principles of SAR validation.90 In this way, the descriptor space within which SAR model can be applied is clearly defined. For this reason, the applicability domain identification methods usually consist of geometric, range-based, and distance-based methods.92 In the present work, the bounding box PCA, convex hull and centroid distance methods are employed to identify the applicability domain of the developed classification SAR models, for the different datasets. The number of measurements that fell outside the applicability domain of the model, using the different methods, is presented in Table 8. It is noted that the applicability domain is defined using the training set of each dataset.| Dataset | Bounding box PCA | Convex hull | Centroid distance |
|---|---|---|---|
| E. coli | — | — | 2 |
| S. aureus | — | — | 3 |
| D. rerio | — | — | 2 |
| D. magna | — | — | 2 |
| C. albicans | — | — | 2 |
| Bacteria | — | — | 12 |
| Eukaryotes | — | — | 6 |
| Fish | — | 2 | 2 |
| Crustaceans | — | — | 4 |
| Plants | — | — | 5 |
| Complete dataset | — | 1 | 27 |
The results of Table 8 show that the Bounding box PCA method included all the MCNM measurements within the applicability domain of all the developed classification SAR models. The convex hull method also includes the total set of MCNM measurements within the applicability domain of the models developed for most datasets, with the exception of the fish dataset, where two measurements were deemed to be outside the applicability domain. A single measurement was also deemed to be outside the applicability domain for the complete dataset model, by the convex hull method. On the other hand, the centroid distance method defined narrower applicability domains, which did not include a number of MCNM measurements, for all the different ecotoxicity datasets. Such results show the dependency of the applicability domain on the method used for its definition. This dependency has been identified and discussed in scientific literature.92 The analysis of the applicability domain results reproduces the findings of previous SAR model development, where metal oxides with similar descriptor values are outside the applicability domain of the models.62,64
3.5. Overall predictions and mechanistic information
The results presented in the previous sections showed that the classification SAR models developed for the different datasets consisting of MCNM ecotoxicity measurements towards individual organisms, as well as for heterogeneous datasets for organism groups, were developed using similar sets of descriptors as the optimal combination (HE and Dbio descriptors). Moreover, the optimal model developed for the complete dataset also used the same combination of two descriptors. This similar descriptor combination allows the mechanistic interpretation of results over the whole set of MCNM ecotoxicity measurements, assisted by an overall ecotoxicity prediction by the SAR model.The complete dataset model is used to predict the ecotoxic class of the different MCNMs, and the results are presented in Fig. 5a, over the space defined by the two descriptors used for the model development. In order to compare the model predictions to the actual experimental measurements, Fig. 5b is used to present the corresponding percentage of toxic measurements for each MCNM (as several MCNMs have multiple measurements within the dataset).
 | ||
| Fig. 5 a) Ecotoxic class prediction for the different MCNMs by the complete dataset model, b) ecotoxic measurements percentage for the different MCNMs in the complete dataset. | ||
The results of Fig. 5a show that the ecotoxic MCNMs are characterized by a lower Dbio value, as well as a less negative HE value. Hence, using the physical interpretation of the two descriptors, MCNMs that have a conduction band energy close to the redox potential of biological pairs (lower Dbio value), together with a less exothermic hydration of their respective metal cations (less negative hydration enthalpy, HE), are more probable to be ecotoxic. Similar conclusions have been drawn from previous experimental and modelling works for metal oxide NPs.61–63 In the same direction with those works, our previous works for the case of pure metal oxides toxicity,64 as well as metal oxide MCNMs cytotoxicity,71 have also identified these descriptors as predictive. The measurements presented in Fig. 5b show that the percentage of ecotoxic experimental measurements for the different MCNMs follows the same trend over the descriptor space. Besides the misclassification of some MCNMs, the experimental classification follows the model predictions, as the vast majority of ecotoxic MCNMs are situated in the space bounded by low Dbio and less negative HE values. Similarly, MCNMs that are characterized by a high Dbio or a highly negative HE value exhibit less ecotoxic measurements. The good agreement between the model classification and the measurements in the dataset does not only show the accuracy and predictive ability of the classification model, but also enhances the notion that similar MCNM characteristics may induce ecotoxic action towards the different organisms taken into account in the dataset. These key characteristics can be adequately quantified by the two descriptors taken into account for the classification SAR model development.
It should be noted however, that the exact boundary values of descriptors that define whether a MCNM is ecotoxic is not possible using the present approach, as it does not aim towards a quantitative ecotoxicity prediction, but rather towards a qualitative assessment. Hence, the model results are sensitive towards the classification scheme used. Furthermore, the models developed for heterogeneous datasets (Table 7 and Fig. 4) in terms of the tested organisms do not take into account the varying cell morphology of the different target organisms. Although the cell morphology has an influence on the ecotoxic action of the different MCNMs, all the different models were developed using similar descriptors, with a good accuracy towards the ecotoxicity measurements, which may hint that the MCNMs have similar ecotoxic modes of action. However, these results do not mean that the toxicity mechanisms are unaffected by the morphology of the target cells, but rather that the dominating toxicity mechanisms are of similar nature. Specifically, the cell morphology could severely affect the ecotoxic action in a more quantitative way, with higher or lower uptake and MCNM-cell interaction rates. Due to the qualitative nature of the model presented in this work, the extraction of quantitative information is not possible by the present approach. However, similar mechanisms for the toxic action of metal oxides, towards different morphologies of target cells or organisms have been concluded to occur in previous nanotoxicology studies,61,63 where the conduction band of nanoparticles has been found to induce electron transfer and toxicity towards BEAS-2B and RAW264.7 cells,63 as well as E. coli.61 On the basis of such observations, classification SAR approaches for nanomaterial toxicity using heterogeneous morphology datasets have been developed,49,54–57,64,71 assessing the toxicity of nanomaterials over a wide range of cell morphologies. In a similar way, the general trends seen in Fig. 5 are reproduced over a large and populous dataset of heterogeneous MCNM measurements, which encourages a more general understanding of MCNM ecotoxicity mechanisms.
As previously mentioned, Dbio expresses the energy difference between the conduction band and the mean redox potential of pairs in biological media.60,61,63,94,95 Hence, a low Dbio value means that electron transfer between the MCNM and the cell is more probable. This electron transfer, previously identified as a toxicity mechanism for the case of metal oxides, can increase the oxidative stress on the cells, by unbalancing its reducing capacity.59,60 Furthermore, overlapping metal oxide conduction bands and biological redox potentials have been correlated with the production of reactive oxygen species (ROS).96 Such ROS can include hydrogen peroxides and OH radicals.63,97–103 Similar quantities have been used for the development of QSAR,53 classification SAR61–64,71 models, as well as for nanomaterial toxicity grouping,31 towards cells and organisms of different kind, showing a more holistic metal oxide toxicity pathway.
The hydration enthalpy (HE) denotes the energy released during the hydration of a respective metal ion, that is released from the MCNM. In this way, the descriptor expresses the affinity of these metal ions to water molecules.61 A more negative HE value means that water molecules will be more strongly attracted to the metal ions, leading an increase of the ion's hydration shell. In turn, a larger hydration cell hinders the ion's permeability through the cell membrane.104–108 As in previous works, HE is computed using Latimer's equation:61
 | (5) |
The release of metal ions has been concluded to be a dominant initial pathway of metal and metal oxide nanoparticle toxicity.97 Several underlying mechanisms regarding the toxic interaction between these ions and different cells have been identified, such as enzyme inactivation,111,112 cell membrane damage63,113 and the increase of oxidative stress.111,112,114–116 DNA damage has also been attributed to the interaction with certain metal ions.111,117,118
The predictive capability of the two descriptors towards the various and heterogeneous datasets could hint that the abovementioned mechanisms could be the dominant pathways towards the metal and metal oxide MCNM ecotoxicity. However, it cannot be concluded whether one of the two pathways prevails over the other. As seen from the results of sections 3.2 and 3.3, single descriptor models were less accurate than models developed using the combination of the two descriptors, except from the D. magna and crustaceans models. For the rest of the models, the ecotoxic MCNMs exhibit a lower Dbio value and a less negative HE. This could mean that both electron exchange between the MCNM and the organism cells, as well as metal ion release that permeate the cell must take place for the MCNM to induce ecotoxic action, as defined with the present classification scheme.
Regarding the nature of interaction between the different components of the MCNMs in the different datasets, the additive mixture approach introduced by Mikolajczyk et al. for the case of MCNMs,65 was able to calculate descriptors that were predictive towards the whole dataset range. The additive approach assumes that the different MCNM components have similar modes of action. While other approaches have been used for the descriptor calculation for mixture of chemicals,119,120 QSAR model development for nanomaterial mixtures has mainly employed the additive mixture approach.82,83,121 Works that have developed QSARs for smaller MCNMs datasets have also used similar approaches for the descriptors, showing high predictivity towards toxicity endpoints.65–68,70 In our previous work, we used the additive mixture approach, together with a novel approach to calculate descriptors for surface loaded MCNMs, to calculate predictive descriptors for the cytotoxicity classification over a large MCNM dataset, leading to the development of high accuracy models.71
With the results of the present work, it is seen that the exact same approach can be used to calculate descriptors that are also able to classify MCNMs based on their ecotoxicity, towards a wide range of organisms, such as bacteria, eukaryotes, fish, plants, and crustaceans. As the additive mixture approach assumes, this could hint towards similar ecotoxic pathways of the different MCNM components. The components in the present work are either metals or metal oxides, which have previously been reported to have similar modes of action.51,64 However, the qualitative nature of the classification approach presented in this work does not allow the quantitative interpretation of the individual component impact in certainty. Nevertheless, quantitative methods have deemed that the compositional ratio of MCNM components dictates their respective impact, showing an additive effect to toxicity.66,67 The large and heterogeneous nature of the MCNM ecotoxicity dataset used in the present work, along with the high accuracy of the developed models, enhances the notion that this additive effect also occurs for MCNM ecotoxicity. In any case, this should be tested by developing quantitative models for more homogeneous datasets, under similar experimental conditions, which will be a subject of future work.
To summarize, the results presented in this study can show the potential of data-based models, such as classification SAR approaches, to be used not only as predictive models, but also as tools for a more general understanding of toxicity mechanisms and modes of action. With their use as an inductive tool to extract scientific information from large and heterogeneous datasets of toxicity measurements, such models can potentially be used to assist the synthesis of safe-by-design nanomaterials. A potential example of such an approach is presented in the work of Feng et al.,58 where based on the results of previous research regarding the dependence of metal oxide toxicity on the conduction band energy, the authors managed to synthesize MCNMs with control over their biological activity, by adequately tuning the conduction band energy of the produced nanomaterials, which is consistent with the findings of the present work.
Conclusions
In the present paper, a dataset of 652 measurements is used to develop classification SAR models for MCNM ecotoxicity. This dataset is, to the best of the author's knowledge, the largest dataset of MCNM ecotoxicity measurements used for the development of such a model. Furthermore, the dataset is heterogeneous, consisting of ecotoxicity measurements towards bacteria, eukaryotes, fish, plants and crustaceans. The models developed for the ecotoxicity towards a specific organism classified the MCNMs in the dataset with accuracy exceeding 89%, based on their ecotoxicity towards the different organisms and organism groups. Models were also able to classify the ecotoxic MCNMs towards different organism groups with an accuracy exceeding 90% towards bacteria, eukaryotes, plants and crustaceans, while the accuracy for fish was 85%. Finally, the complete heterogeneous dataset was used to build a classification model for the MCNM ecotoxicity, with an accuracy of 89.2%. The same descriptor set was used for the classification of the different subsets of data, allowing a more holistic understanding of the underlying mechanisms that dictate MCNM ecotoxicity.In particular, the electron transfer between the MCNM and the biological pairs, as well as the release and transport of metal ions from the MCNMs, were deemed to be the dictating ecotoxic pathways for the different MCNMs. These findings are consistent with previous works for the cytotoxicity of metal oxide nanoparticles and MCNMs. However, the identification of these descriptors for the case of ecotoxicity is novel for the case of MCNM. The two descriptors that expressed the abovementioned mechanisms were computed based on the additive mixture approach, shedding light on the nature of the interaction between the MCNM components. These findings are consistent with previous works regarding the QSAR modelling of MCNM toxicity, where the additive mixture approach was found to produce predictive descriptors.
The novelty of the present study lies in the development of a SAR approach using a large, heterogeneous dataset for the prediction of MCNM ecotoxicity. Such an approach allows a more holistic understanding of the ecotoxic action, upon ecosystem exposure of different MCNMs and their constituting components towards various organisms. The mechanistic information extracted by the present approach regarding MCNM ecotoxicity and the interaction of the multiple components of MCNMs, can thus assist towards a more knowledge-driven MCNM ecotoxicity assessment, as well as the synthesis of safe-by-design MCNMs for various applications.
Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was funded by the EU H2020 Project ‘Safe and sUstainable by desigN Strategies for HIgh performance multi-component NanomatErials’ “SUNSHINE” (grant number 952924). The publication of the article in OA mode was financially supported by HEAL-Link.References
- S. Mitchell, R. Qin, N. Zheng and J. Pérez-Ramírez, Nanoscale engineering of catalytic materials for sustainable technologies, Nat. Nanotechnol., 2021, 16, 129–139 CrossRef CAS PubMed.
- F. Christian, E. Selly, D. Adityawarman and A. Indarto, Application of nanotechnologies in the energy sector: A brief and short review, Front. Energy, 2013, 7, 6–18 CrossRef.
- C.-Z. Ning, L. Dou and P. Yang, Bandgap engineering in semiconductor alloy nanomaterials with widely tunable compositions, Nat. Rev. Mater., 2017, 2, 17070 CrossRef CAS.
- X. Hu, Y. Zhang, T. Ding, J. Liu and H. Zhao, Multifunctional Gold Nanoparticles: A Novel Nanomaterial for Various Medical Applications and Biological Activities, Front. Bioeng. Biotechnol., 2020, 8, 990 CrossRef PubMed.
- G. P. Gakis, H. Vergnes, F. Cristiano, Y. Tison, C. Vahlas, B. Caussat, A. G. Boudouvis and E. Scheid, In situ N2-NH3 plasma pre-treatment of silicon substrate enhances the initial growth and restricts the substrate oxidation during alumina ALD, J. Appl. Phys., 2019, 126, 125305 CrossRef.
- G. P. Gakis, C. Vahlas, H. Vergnes, S. Dourdain, Y. Tison, H. Martinez, J. Bour, D. Ruch, A. G. Boudouvis, B. Caussat and E. Scheid, Investigation of the initial deposition steps and the interfacial layer of Atomic Layer Deposited (ALD) Al2O3 on Si, Appl. Surf. Sci., 2019, 492, 245–254 CrossRef CAS.
- G. P. Gakis, H. Vergnes, E. Scheid, C. Vahlas, A. G. Boudouvis and B. Caussat, Detailed investigation of the surface mechanisms and their interplay with transport phenomena in alumina atomic layer deposition from TMA and water, Chem. Eng. Sci., 2019, 195, 399–412 CrossRef CAS.
- R. Sharma, Newer Methods of Nanoparticle Synthesis: Nitroimidazole properties with Nanometal oxides in Polymer Cages as Drug-Biomarker Monitors, Nat. Preced., 2009 DOI:10.1038/npre.2009.3952.1.
- G. P. Gakis, S. Termine, A.-F. A. Trompeta, I. G. Aviziotis and C. A. Charitidis, Unraveling the mechanisms of carbon nanotube growth by chemical vapor deposition, Chem. Eng. J., 2022, 445, 136807 CrossRef CAS.
- G. P. Gakis, E. N. Skountzos, I. G. Aviziotis and C. A. Charitidis, Multi-parametric analysis of the CVD of CNTs: Effect of reaction temperature, pressure and acetylene flow rate, Chem. Eng. Sci., 2023, 267, 118374 CrossRef CAS.
- Q. Tang, Z. Zhou and Z. Chen, Graphene-related nanomaterials: tuning properties by functionalization, Nanoscale, 2013, 5, 4541–4583 RSC.
- X. Y. Wong, A. Sena-Torralba, R. Álvarez-Diduk, K. Muthoosamy and A. Merkoçi, Nanomaterials for Nanotheranostics: Tuning Their Properties According to Disease Needs, ACS Nano, 2020, 14, 2585–2627 CrossRef CAS PubMed.
- Q. Yao, Z. Yu, L. Li and X. Huang, Strain and Surface Engineering of Multicomponent Metallic Nanomaterials with Unconventional Phases, Chem. Rev., 2023, 123, 9676–9717 CrossRef CAS PubMed.
- G. Yang, L. Sun and Q. Zhang, Multicomponent chiral plasmonic hybrid nanomaterials: recent advances in synthesis and applications, Nanoscale Adv., 2024, 6, 318–336 RSC.
- O. Vasylkiv and Y. Sakka, Nanoexplosion Synthesis of Multimetal Oxide Ceramic Nanopowders, Nano Lett., 2005, 5, 2598–2604 CrossRef CAS PubMed.
- Y. Zhang and S.-J. Park, Stabilizing CuPd bimetallic alloy nanoparticles deposited on holey carbon nitride for selective hydroxylation of benzene to phenol, J. Catal., 2019, 379, 154–163 CrossRef CAS.
- S. Alex and A. Tiwari, Functionalized Gold Nanoparticles: Synthesis, Properties and Applications; A Review, J. Nanosci. Nanotechnol., 2015, 15, 1869–1894 CrossRef CAS PubMed.
- S. Patil, R. Tandon and N. Tandon, A current research on silica coated ferrite nanoparticle and their application: Review, Curr. Res. Green Sustainable Chem., 2021, 4, 100063 CrossRef CAS.
- R. Medhi, M. D. Marquez and T. R. Lee, Visible-Light-Active Doped Metal Oxide Nanoparticles: Review of their Synthesis, Properties, and Applications, ACS Appl. Nano Mater., 2020, 3, 6156–6185 CrossRef CAS.
- M. A. Hamzawy, A. M. Abo-youssef, H. F. Salem and S. A. Mohammed, Antitumor activity of intratracheal inhalation of temozolomide (TMZ) loaded into gold nanoparticles and/or liposomes against urethane-induced lung cancer in BALB/c mice, Drug Delivery, 2017, 24, 599–607 CrossRef CAS PubMed.
- A. K. Verma and S. Perlman, Lipid nanoparticle-mRNA: another step in the fight against COVID-19, Cell Res., 2022, 32, 421–422 CrossRef CAS PubMed.
- M. Manigrasso, C. Protano, M. L. Astolfi, L. Massimi, P. Avino, M. Vitali and S. Canepari, Evidences of copper nanoparticle exposure in indoor environments: Long-term assessment, high-resolution field emission scanning electron microscopy evaluation, in silico respiratory dosimetry study and possible health implications, Sci. Total Environ., 2019, 653, 1192–1203 CrossRef CAS PubMed.
- M. J. Bessa, F. Brandão, M. Viana, J. F. Gomes, E. Monfort, F. R. Cassee, S. Fraga and J. P. Teixeira, Nanoparticle exposure and hazard in the ceramic industry: an overview of potential sources, toxicity and health effects, Environ. Res., 2020, 184, 109297 CrossRef CAS PubMed.
- S. Sonwani, S. Madaan, J. Arora, S. Suryanarayan, D. Rangra, N. Mongia, T. Vats and P. Saxena, Inhalation Exposure to Atmospheric Nanoparticles and Its Associated Impacts on Human Health: A Review, Front. Sustain. Cities, 2021, 3, 690444 CrossRef.
- Y. Nazarenko, T. W. Han, P. J. Lioy and G. Mainelis, Potential for exposure to engineered nanoparticles from nanotechnology-based consumer spray products, J. Exposure Sci. Environ. Epidemiol., 2011, 21, 515–528 CrossRef CAS PubMed.
- M. Mortimer and P. A. Holden, in Exposure to Engineered Nanomaterials in the Environment, ed. N. Marmiroli, J. C. White and J. Song, Elsevier, 2019, pp. 61–103, DOI:10.1016/B978-0-12-814835-8.00003-0.
- G. V. Lowry, E. M. Hotze, E. S. Bernhardt, D. D. Dionysiou, J. A. Pedersen, M. R. Wiesner and B. Xing, Environmental Occurrences, Behavior, Fate, and Ecological Effects of Nanomaterials: An Introduction to the Special Series, J. Environ. Qual., 2010, 39, 1867–1874 CrossRef CAS PubMed.
- J. Blasco, I. Corsi and V. Matranga, Particles in the oceans: Implication for a safe marine environment, Mar. Environ. Res., 2015, 111, 1–4 CrossRef CAS PubMed.
- V. Kumar, N. Sharma and S. S. Maitra, In vitro and in vivo toxicity assessment of nanoparticles, Int. Nano Lett., 2017, 7, 243–256 CrossRef CAS.
- D. T. Savage, J. Z. Hilt and T. D. Dziubla, in Nanotoxicity: Methods and Protocols, ed. Q. Zhang, Springer New York, New York, NY, 2019, pp. 1–29, DOI:10.1007/978-1-4939-8916-4_1.
- F. C. Simeone and A. L. Costa, Assessment of cytotoxicity of metal oxide nanoparticles on the basis of fundamental physical–chemical parameters: a robust approach to grouping, Environ. Sci.: Nano, 2019, 6, 3102–3112 RSC.
- G. P. Gakis, A. Krikas, P. Neofytou, L. Tran and C. Charitidis, Modelling the biodistribution of inhaled gold nanoparticles in rats with interspecies extrapolation to humans, Toxicol. Appl. Pharmacol., 2022, 457, 116322 CrossRef CAS PubMed.
- A. Krikas, P. Neofytou, G. P. Gakis, I. Xiarchos, C. Charitidis and L. Tran, Modeling of clearance, retention, and translocation of inhaled gold nanoparticles in rats, Inhalation Toxicol., 2022, 34, 361–379 CrossRef CAS PubMed.
- I. Xiarchos, A. K. Morozinis, P. Kavouras and C. A. Charitidis, Nanocharacterization, Materials Modeling, and Research Integrity as Enablers of Sound Risk Assessment: Designing Responsible Nanotechnology, Small, 2020, 16, 2001590 CrossRef CAS PubMed.
- M. Kotzabasaki, I. Sotiropoulos, C. Charitidis and H. Sarimveis, Machine learning methods for multi-walled carbon nanotubes (MWCNT) genotoxicity prediction, Nanoscale Adv., 2021, 3, 3167–3176 RSC.
- C. F. Jones and D. W. Grainger, In vitro assessments of nanomaterial toxicity, Adv. Drug Delivery Rev., 2009, 61, 438–456 CrossRef CAS PubMed.
- S. G. Ciappellano, E. Tedesco, M. Venturini and F. Benetti, In vitro toxicity assessment of oral nanocarriers, Adv. Drug Delivery Rev., 2016, 106, 381–401 CrossRef CAS PubMed.
- C. Hirsch, M. Roesslein, H. F. Krug and P. Wick, Nanomaterial cell interactions: are current in vitro tests reliable?, Nanomedicine, 2011, 6, 837–847 CrossRef CAS PubMed.
- K. L. Aillon, Y. Xie, N. El-Gendy, C. J. Berkland and M. L. Forrest, Effects of nanomaterial physicochemical properties on in vivo toxicity, Adv. Drug Delivery Rev., 2009, 61, 457–466 CrossRef CAS PubMed.
- S. T. Stern, P. P. Adiseshaiah and R. M. Crist, Autophagy and lysosomal dysfunction as emerging mechanisms of nanomaterial toxicity, Part. Fibre Toxicol., 2012, 9, 20 CrossRef CAS PubMed.
- A. Kermanizadeh, D. Balharry, H. Wallin, S. Loft and P. Møller, Nanomaterial translocation–the biokinetics, tissue accumulation, toxicity and fate of materials in secondary organs–a review, Crit. Rev. Toxicol., 2015, 45, 837–872 CrossRef CAS PubMed.
- A. Rhazouani, H. Gamrani, M. El Achaby, K. Aziz, L. Gebrati, M. S. Uddin and F. Aziz, Synthesis and Toxicity of Graphene Oxide Nanoparticles: A Literature Review of In Vitro and In Vivo Studies, BioMed Res. Int., 2021, 2021, 5518999 CrossRef PubMed.
- R. Landsiedel, U. G. Sauer, L. Ma-Hock, J. Schnekenburger and M. Wiemann, Pulmonary toxicity of nanomaterials: a critical comparison of published in vitro assays and in vivo inhalation or instillation studies, Nanomedicine, 2014, 9, 2557–2585 CrossRef CAS PubMed.
- A. A. Buglak, A. V. Zherdev and B. B. Dzantiev, Nano-(Q)SAR for Cytotoxicity Prediction of Engineered Nanomaterials, Molecules, 2019, 24(24), 4537 CrossRef CAS PubMed.
- C. Oksel, C. Y. Ma, J. J. Liu, T. Wilkins and X. Z. Wang, (Q)SAR modelling of nanomaterial toxicity: A critical review, Particuology, 2015, 21, 1–19 CrossRef CAS.
- L. Lamon, D. Asturiol, A. Vilchez, R. Ruperez-Illescas, J. Cabellos, A. Richarz and A. Worth, Computational models for the assessment of manufactured nanomaterials: Development of model reporting standards and mapping of the model landscape, Comput. Toxicol., 2019, 9, 143–151 CrossRef CAS PubMed.
- T. Puzyn, B. Rasulev, A. Gajewicz, X. Hu, T. P. Dasari, A. Michalkova, H. M. Hwang, A. Toropov, D. Leszczynska and J. Leszczynski, Using nano-QSAR to predict the cytotoxicity of metal oxide nanoparticles, Nat. Nanotechnol., 2011, 6, 175–178 CrossRef CAS PubMed.
- S. Kar, K. Pathakoti, P. B. Tchounwou, D. Leszczynska and J. Leszczynski, Evaluating the cytotoxicity of a large pool of metal oxide nanoparticles to Escherichia coli: Mechanistic understanding through In Vitro and In Silico studies, Chemosphere, 2021, 264, 128428 CrossRef CAS PubMed.
- V. V. Kleandrova, F. Luan, H. González-Díaz, J. M. Ruso, A. Speck-Planche and M. N. D. S. Cordeiro, Computational Tool for Risk Assessment of Nanomaterials: Novel QSTR-Perturbation Model for Simultaneous Prediction of Ecotoxicity and Cytotoxicity of Uncoated and Coated Nanoparticles under Multiple Experimental Conditions, Environ. Sci. Technol., 2014, 48, 14686–14694 CrossRef CAS PubMed.
- A. A. Toropov and A. P. Toropova, Quasi-QSAR for mutagenic potential of multi-walled carbon-nanotubes, Chemosphere, 2015, 124, 40–46 CrossRef CAS PubMed.
- J. Li, C. Wang, L. Yue, F. Chen, X. Cao and Z. Wang, Nano-QSAR modeling for predicting the cytotoxicity of metallic and metal oxide nanoparticles: A review, Ecotoxicol. Environ. Saf., 2022, 243, 113955 CrossRef CAS PubMed.
- J. Cao, Y. Pan, Y. Jiang, R. Qi, B. Yuan, Z. Jia, J. Jiang and Q. Wang, Computer-aided nanotoxicology: risk assessment of metal oxide nanoparticles via nano-QSAR, Green Chem., 2020, 22, 3512–3521 RSC.
- A. Gajewicz, N. Schaeublin, B. Rasulev, S. Hussain, D. Leszczynska, T. Puzyn and J. Leszczynski, Towards understanding mechanisms governing cytotoxicity of metal oxides nanoparticles: Hints from nano-QSAR studies, Nanotoxicology, 2015, 9, 313–325 CrossRef CAS PubMed.
- V. V. Kleandrova, F. Luan, H. González-Díaz, J. M. Ruso, A. Melo, A. Speck-Planche and M. N. D. S. Cordeiro, Computational ecotoxicology: Simultaneous prediction of ecotoxic effects of nanoparticles under different experimental conditions, Environ. Int., 2014, 73, 288–294 CrossRef CAS PubMed.
- F. Luan, V. V. Kleandrova, H. González-Díaz, J. M. Ruso, A. Melo, A. Speck-Planche and M. N. D. S. Cordeiro, Computer-aided nanotoxicology: assessing cytotoxicity of nanoparticles under diverse experimental conditions by using a novel QSTR-perturbation approach, Nanoscale, 2014, 6, 10623–10630 RSC.
- R. Concu, V. V. Kleandrova, A. Speck-Planche and M. N. D. S. Cordeiro, Probing the toxicity of nanoparticles: a unified in silico machine learning model based on perturbation theory, Nanotoxicology, 2017, 11, 891–906 CrossRef CAS PubMed.
- M. K. Ha, T. X. Trinh, J. S. Choi, D. Maulina, H. G. Byun and T. H. Yoon, Toxicity Classification of Oxide Nanomaterials: Effects of Data Gap Filling and PChem Score-based Screening Approaches, Sci. Rep., 2018, 8, 3141 CrossRef PubMed.
- Y. Feng, Y. Chang, K. Xu, R. Zheng, X. Wu, Y. Cheng and H. Zhang, Safety-by-Design of Metal Oxide Nanoparticles Based on the Regulation of their Energy Edges, Small, 2020, 16, 1907643 CrossRef CAS PubMed.
- E. Burello and A. Worth, in Towards Efficient Designing of Safe Nanomaterials: Innovative Merge of Computational Approaches and Experimental Techniques, The Royal Society of Chemistry, 2012, pp. 257–283, 10.1039/9781849735476-00257.
- E. Burello and A. P. Worth, A theoretical framework for predicting the oxidative stress potential of oxide nanoparticles, Nanotoxicology, 2011, 5, 228–235 CrossRef CAS PubMed.
- C. Kaweeteerawat, A. Ivask, R. Liu, H. Zhang, C. H. Chang, C. Low-Kam, H. Fischer, Z. Ji, S. Pokhrel, Y. Cohen, D. Telesca, J. Zink, L. Madler, P. A. Holden, A. Nel and H. Godwin, Toxicity of metal oxide nanoparticles in Escherichia coli correlates with conduction band and hydration energies, Environ. Sci. Technol., 2015, 49, 1105–1112 CrossRef CAS PubMed.
- R. Liu, H. Y. Zhang, Z. X. Ji, R. Rallo, T. Xia, C. H. Chang, A. Nel and Y. Cohen, Development of structure–activity relationship for metal oxide nanoparticles, Nanoscale, 2013, 5, 5644–5653 RSC.
- H. Zhang, Z. Ji, T. Xia, H. Meng, C. Low-Kam, R. Liu, S. Pokhrel, S. Lin, X. Wang, Y.-P. Liao, M. Wang, L. Li, R. Rallo, R. Damoiseaux, D. Telesca, L. Mädler, Y. Cohen, J. I. Zink and A. E. Nel, Use of Metal Oxide Nanoparticle Band Gap To Develop a Predictive Paradigm for Oxidative Stress and Acute Pulmonary Inflammation, ACS Nano, 2012, 6, 4349–4368 CrossRef CAS PubMed.
- G. P. Gakis, I. G. Aviziotis and C. A. Charitidis, Metal and metal oxide nanoparticle toxicity: moving towards a more holistic structure–activity approach, Environ. Sci.: Nano, 2023, 10, 761–780 RSC.
- A. Mikolajczyk, N. Sizochenko, E. Mulkiewicz, A. Malankowska, M. Nischk, P. Jurczak, S. Hirano, G. Nowaczyk, A. Zaleska-Medynska, J. Leszczynski, A. Gajewicz and T. Puzyn, Evaluating the toxicity of TiO2-based nanoparticles to Chinese hamster ovary cells and Escherichia coli: a complementary experimental and computational approach, Beilstein J. Nanotechnol., 2017, 8, 2171–2180 CrossRef CAS PubMed.
- A. Mikolajczyk, A. Gajewicz, E. Mulkiewicz, B. Rasulev, M. Marchelek, M. Diak, S. Hirano, A. Zaleska-Medynska and T. Puzyn, Nano-QSAR modeling for ecosafe design of heterogeneous TiO2-based nano-photocatalysts, Environ. Sci.: Nano, 2018, 5, 1150–1160 RSC.
- A. Mikolajczyk, N. Sizochenko, E. Mulkiewicz, A. Malankowska, B. Rasulev and T. Puzyn, A chemoinformatics approach for the characterization of hybrid nanomaterials: safer and efficient design perspective, Nanoscale, 2019, 11, 11808–11818 RSC.
- F. Stoliński, A. Rybińska-Fryca, M. Gromelski, A. Mikolajczyk and T. Puzyn, NanoMixHamster: a web-based tool for predicting cytotoxicity of TiO2-based multicomponent nanomaterials toward Chinese hamster ovary (CHO-K1) cells, Nanotoxicology, 2022, 16, 276–289 CrossRef PubMed.
- A. Rybińska-Fryca, A. Mikolajczyk and T. Puzyn, Structure–activity prediction networks (SAPNets): a step beyond Nano-QSAR for effective implementation of the safe-by-design concept, Nanoscale, 2020, 12, 20669–20676 RSC.
- T. C. Le, H. Yin, R. Chen, Y. Chen, L. Zhao, P. S. Casey, C. Chen and D. A. Winkler, An Experimental and Computational Approach to the Development of ZnO Nanoparticles that are Safe by Design, Small, 2016, 12, 3568–3577 CrossRef CAS PubMed.
- G. P. Gakis, I. G. Aviziotis and C. A. Charitidis, A structure–activity approach towards the toxicity assessment of multicomponent metal oxide nanomaterials, Nanoscale, 2023, 15, 16432–16446 RSC.
- F. Zhang, Z. Wang, W. J. G. M. Peijnenburg and M. G. Vijver, Review and Prospects on the Ecotoxicity of Mixtures of Nanoparticles and Hybrid Nanomaterials, Environ. Sci. Technol., 2022, 56, 15238–15250 CrossRef CAS PubMed.
- J. Bozich, M. Hang, R. Hamers and R. Klaper, Core chemistry influences the toxicity of multicomponent metal oxide nanomaterials, lithium nickel manganese cobalt oxide, and lithium cobalt oxide to Daphnia magna, Environ. Toxicol. Chem., 2017, 36, 2493–2502 CrossRef CAS PubMed.
- A. M. Z. de Medeiros, L. U. Khan, G. H. da Silva, C. A. Ospina, O. L. Alves, V. L. de Castro and D. S. T. Martinez, Graphene oxide-silver nanoparticle hybrid material: an integrated nanosafety study in zebrafish embryos, Ecotoxicol. Environ. Saf., 2021, 209, 111776 CrossRef CAS PubMed.
- T. X. Trinh and J. Kim, Status Quo in Data Availability and Predictive Models of Nano-Mixture Toxicity, Nanomaterials, 2021, 11(1), 124 CrossRef CAS PubMed.
- J. Jang, H. G. Hur, M. J. Sadowsky, M. N. Byappanahalli, T. Yan and S. Ishii, Environmental Escherichia coli: ecology and public health implications—a review, J. Appl. Microbiol., 2017, 123, 570–581 CrossRef CAS PubMed.
- M. Sautour, J.-P. Lemaître, L. Ranjard, C. Truntzer, L. Basmaciyan, G. Depret, A. Hartmann and F. Dalle, Detection and survival of Candida albicans in soils, Environ. DNA, 2021, 3, 1093–1101 CrossRef.
- H. K. Abbas, J. R. Wilkinson, R. M. Zablotowicz, C. Accinelli, C. A. Abel, H. A. Bruns and M. A. Weaver, Ecology of Aspergillus flavus, regulation of aflatoxin production, and management strategies to reduce aflatoxin contamination of corn, Toxin Rev., 2009, 28, 142–153 CrossRef CAS.
- J. Portier, G. Campet, J. Etourneau and B. Tanguy, A simple model for the estimation of electronegativities of cations in different electronic states and coordinations, J. Alloys Compd., 1994, 209, 285–289 CrossRef CAS.
- J. Portier, H. S. Hilal, I. Saadeddin, S. J. Hwang, M. A. Subramanian and G. Campet, Thermodynamic correlations and band gap calculations in metal oxides, Prog. Solid State Chem., 2004, 32, 207–217 CrossRef CAS.
- P. Ambure, R. B. Aher, A. Gajewicz, T. Puzyn and K. Roy, “NanoBRIDGES” software: Open access tools to perform QSAR and nano-QSAR modeling, Chemom. Intell. Lab. Syst., 2015, 147, 1–13 CrossRef CAS.
- B. Yuan, P. Wang, L. Sang, J. Gong, Y. Pan and Y. Hu, QNAR modeling of cytotoxicity of mixing nano-TiO2 and heavy metals, Ecotoxicol. Environ. Saf., 2021, 208, 111634 CrossRef CAS PubMed.
- L. Sang, Y. Wang, C. Zong, P. Wang, H. Zhang, D. Guo, B. Yuan and Y. Pan, Machine Learning for Evaluating the Cytotoxicity of Mixtures of Nano-TiO2 and Heavy Metals: QSAR Model Apply Random Forest Algorithm after Clustering Analysis, Molecules, 2022, 27(18), 6125 CrossRef CAS PubMed.
- F. Zhang, Z. Wang, W. J. G. M. Peijnenburg and M. G. Vijver, Machine learning-driven QSAR models for predicting the mixture toxicity of nanoparticles, Environ. Int., 2023, 177, 108025 CrossRef CAS PubMed.
- T. X. Trinh, M. Seo, T. H. Yoon and J. Kim, Developing random forest based QSAR models for predicting the mixture toxicity of TiO2 based nano-mixtures to Daphnia magna, NanoImpact, 2022, 25, 100383 CrossRef CAS PubMed.
- T. X. Trinh and J. Kim, Status Quo in Data Availability and Predictive Models of Nano-Mixture Toxicity, Nanomaterials, 2021, 11, 124 CrossRef CAS PubMed.
- I. Guyon and A. Elisseeff, An introduction to variable and feature selection, J. Mach. Learn. Res., 2003, 3, 1157–1182 Search PubMed.
- M. Eklund, U. Norinder, S. Boyer and L. Carlsson, Choosing Feature Selection and Learning Algorithms in QSAR, J. Chem. Inf. Model., 2014, 54, 837–843 CrossRef CAS PubMed.
- Y. Li, Z. Dai, D. Cao, F. Luo, Y. Chen and Z. Yuan, Chi-MIC-share: a new feature selection algorithm for quantitative structure–activity relationship models, RSC Adv., 2020, 10, 19852–19860 RSC.
- OECD, Guidance Document on the Validation of (Quantitative) Structure-Activity Relationship [(Q)SAR] Models, 2014.
- F. Sahigara, D. Ballabio, R. Todeschini and V. Consonni, Assessing the Validity of QSARs for Ready Biodegradability of Chemicals: An Applicability Domain Perspective, Curr. Comput.-Aided Drug Des., 2014, 10, 137–147 CrossRef CAS PubMed.
- F. Sahigara, K. Mansouri, D. Ballabio, A. Mauri, V. Consonni and R. Todeschini, Comparison of Different Approaches to Define the Applicability Domain of QSAR Models, Molecules, 2012, 17, 4791–4810 CrossRef CAS PubMed.
- A. Rácz, D. Bajusz and K. Héberger, Intercorrelation Limits in Molecular Descriptor Preselection for QSAR/QSPR, Mol. Inf., 2019, 38, 1800154 CrossRef PubMed.
- M. Auffan, J. Rose, M. R. Wiesner and J. Y. Bottero, Chemical stability of metallic nanoparticles: a parameter controlling their potential cellular toxicity in vitro, Environ. Pollut., 2009, 157, 1127–1133 CrossRef CAS PubMed.
- G. S. Plumlee, S. A. Morman and T. L. Ziegler, The Toxicological Geochemistry of Earth Materials: An Overview of Processes and the Interdisciplinary Methods Used to Understand Them, Rev. Mineral. Geochem., 2006, 64, 5–57 CrossRef CAS.
- Y. Li, W. Zhang, J. Niu and Y. Chen, Mechanism of Photogenerated Reactive Oxygen Species and Correlation with the Antibacterial Properties of Engineered Metal-Oxide Nanoparticles, ACS Nano, 2012, 6, 5164–5173 CrossRef CAS PubMed.
- A. B. Djurisic, Y. H. Leung, A. M. Ng, X. Y. Xu, P. K. Lee, N. Degger and R. S. Wu, Toxicity of metal oxide nanoparticles: mechanisms, characterization, and avoiding experimental artefacts, Small, 2015, 11, 26–44 CrossRef CAS PubMed.
- M. Premanathan, K. Karthikeyan, K. Jeyasubramanian and G. Manivannan, Selective toxicity of ZnO nanoparticles toward Gram-positive bacteria and cancer cells by apoptosis through lipid peroxidation, Nanomed.: Nanotechnol. Biol. Med., 2011, 7, 184–192 CrossRef CAS PubMed.
- H. L. Karlsson, P. Cronholm, J. Gustafsson and L. Möller, Copper Oxide Nanoparticles Are Highly Toxic: A Comparison between Metal Oxide Nanoparticles and Carbon Nanotubes, Chem. Res. Toxicol., 2008, 21, 1726–1732 Search PubMed.
- G. Applerot, A. Lipovsky, R. Dror, N. Perkas, Y. Nitzan, R. Lubart and A. Gedanken, Enhanced Antibacterial Activity of Nanocrystalline ZnO Due to Increased ROS-Mediated Cell Injury, Adv. Funct. Mater., 2009, 19, 842–852 CrossRef CAS.
- T. Xia, M. Kovochich, J. Brant, M. Hotze, J. Sempf, T. Oberley, C. Sioutas, J. I. Yeh, M. R. Wiesner and A. E. Nel, Comparison of the Abilities of Ambient and Manufactured Nanoparticles To Induce Cellular Toxicity According to an Oxidative Stress Paradigm, Nano Lett., 2006, 6, 1794–1807 CrossRef CAS PubMed.
- T. C. Long, N. Saleh, R. D. Tilton, G. V. Lowry and B. Veronesi, Titanium Dioxide (P25) Produces Reactive Oxygen Species in Immortalized Brain Microglia (BV2): Implications for Nanoparticle Neurotoxicity, Environ. Sci. Technol., 2006, 40, 4346–4352 CrossRef CAS PubMed.
- A. L. Neal, What can be inferred from bacterium–nanoparticle interactions about the potential consequences of environmental exposure to nanoparticles?, Ecotoxicology, 2008, 17, 362 CrossRef CAS PubMed.
- F. Thevenod, Catch me if you can! Novel aspects of cadmium transport in mammalian cells, BioMetals, 2010, 23, 857–875 CrossRef CAS PubMed.
- S. Sugiharto, T. M. Lewis, A. J. Moorhouse, P. R. Schofield and P. H. Barry, Anion-cation permeability correlates with hydrated counterion size in glycine receptor channels, Biophys. J., 2008, 95, 4698–4715 CrossRef CAS PubMed.
- R. Epsztein, E. Shaulsky, M. Qin and M. Elimelech, Activation behavior for ion permeation in ion-exchange membranes: Role of ion dehydration in selective transport, J. Membr. Sci., 2019, 580, 316–326 CrossRef CAS.
- S. Adapa and A. Malani, Role of hydration energy and co-ions association on monovalent and divalent cations adsorption at mica-aqueous interface, Sci. Rep., 2018, 8, 12198 CrossRef PubMed.
- Z. Qu and H. C. Hartzell, Anion permeation in Ca(2+)-activated Cl(−) channels, J. Gen. Physiol., 2000, 116, 825–844 CrossRef CAS PubMed.
- Y. Mu, F. Wu, Q. Zhao, R. Ji, Y. Qie, Y. Zhou, Y. Hu, C. Pang, D. Hristozov, J. P. Giesy and B. Xing, Predicting toxic potencies of metal oxide nanoparticles by means of nano-QSARs, Nanotoxicology, 2016, 10, 1207–1214 CrossRef CAS PubMed.
- G. E. Brown, V. E. Henrich, W. H. Casey, D. L. Clark, C. Eggleston, A. Felmy, D. W. Goodman, M. Grätzel, G. Maciel, M. I. McCarthy, K. H. Nealson, D. A. Sverjensky, M. F. Toney and J. M. Zachara, Metal Oxide Surfaces and Their Interactions with Aqueous Solutions and Microbial Organisms, Chem. Rev., 1999, 99, 77–174 CrossRef CAS PubMed.
- M. Balali-Mood, K. Naseri, Z. Tahergorabi, M. R. Khazdair and M. Sadeghi, Toxic Mechanisms of Five Heavy Metals: Mercury, Lead, Chromium, Cadmium, and Arsenic, Front. Pharmacol., 2021, 12, 643972 CrossRef CAS PubMed.
- A. Manuja, B. Kumar, R. Kumar, D. Chhabra, M. Ghosh, M. Manuja, B. Brar, Y. Pal, B. N. Tripathi and M. Prasad, Metal/metal oxide nanoparticles: Toxicity concerns associated with their physical state and remediation for biomedical applications, Toxicol. Rep., 2021, 8, 1970–1978 CrossRef CAS PubMed.
- V. Forest, J.-F. Hochepied, L. Leclerc, A. Trouvé, K. Abdelkebir, G. Sarry, V. Augusto and J. Pourchez, Towards an alternative to nano-QSAR for nanoparticle toxicity ranking in case of small datasets, J. Nanopart. Res., 2019, 21, 95 CrossRef.
- M. Jaishankar, T. Tseten, N. Anbalagan, B. B. Mathew and K. N. Beeregowda, Toxicity, mechanism and health effects of some heavy metals, Interdiscip. Toxicol., 2014, 7, 60–72 CrossRef PubMed.
- S. J. Stohs and D. Bagchi, Oxidative mechanisms in the toxicity of metal ions, Free Radical Biol. Med., 1995, 18, 321–336 CrossRef CAS PubMed.
- A. Abdal Dayem, M. K. Hossain, S. B. Lee, K. Kim, S. K. Saha, G.-M. Yang, H. Y. Choi and S.-G. Cho, The Role of Reactive Oxygen Species (ROS) in the Biological Activities of Metallic Nanoparticles, Int. J. Mol. Sci., 2017, 18, 120 CrossRef PubMed.
- R. R. Crichton, in Metal Chelation in Medicine, The Royal Society of Chemistry, 2017, pp. 1–23, 10.1039/9781782623892-00001.
- N. Mei, Y. Zhang, Y. Chen, X. Guo, W. Ding, S. F. Ali, A. S. Biris, P. Rice, M. M. Moore and T. Chen, Silver nanoparticle-induced mutations and oxidative stress in mouse lymphoma cells, Environ. Mol. Mutagen., 2012, 53, 409–419 CrossRef CAS PubMed.
- F. Chen, L. Wu, X. Xiao, L. Rong, M. Li and X. Zou, Mixture toxicity of zinc oxide nanoparticle and chemicals with different mode of action upon Vibrio fischeri, Environ. Sci. Eur., 2020, 32, 41 CrossRef CAS.
- D. Wang, S. Wang, L. Bai, M. S. Nasir, S. Li and W. Yan, Mathematical Modeling Approaches for Assessing the Joint Toxicity of Chemical Mixtures Based on Luminescent Bacteria: A Systematic Review, Front. Microbiol., 2020, 11, 1651 CrossRef PubMed.
- M. Na, S. H. Nam, K. Moon and J. Kim, Development of a nano-QSAR model for predicting the toxicity of nano-metal oxide mixtures to Aliivibrio fischeri, Environ. Sci.: Nano, 2023, 10, 325–337 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4en01183j |
| This journal is © The Royal Society of Chemistry 2025 |

