 Open Access Article
Open Access ArticleUnexpected effect of an axial ligand mutation in the type 1 copper center in small laccase: structure-based analyses and engineering to increase reduction potential and activity†
Jing-Xiang Wang a,
Avery C. Vilbertb,
Lucas H. Williamsc,
Evan N. Mirtsd,
Chang Cuid and
Yi Lu
a,
Avery C. Vilbertb,
Lucas H. Williamsc,
Evan N. Mirtsd,
Chang Cuid and
Yi Lu *abcd
*abcd
aDepartment of Chemistry, The University of Texas at Austin, 105 East 24th Street, Austin, TX 78712, USA. E-mail: yi.lu@utexas.edu
bPacific Northwest National Laboratory, 902 Battelle Boulevard, Richland, WA 99354, USA
cDepartment of Molecular Biosciences, The University of Texas at Austin, 100 East 24th St., Austin, TX 78712, USA
dDepartment of Chemistry, The University of Illinois at Urbana-Champaign, 600 South Matthews Avenue, Urbana, IL 61801, USA
First published on 19th May 2025
Abstract
Type 1 copper (T1Cu) centers are crucial in biological electron transfer (ET) processes, exhibiting a wide range of reduction potentials 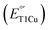 to match their redox partners and optimize ET rates. While tuning
to match their redox partners and optimize ET rates. While tuning  in mononuclear T1Cu proteins like azurin has been successful, it is more difficult for multicopper oxidases. Specifically, while replacing axial methionine to leucine in azurin increased its
in mononuclear T1Cu proteins like azurin has been successful, it is more difficult for multicopper oxidases. Specifically, while replacing axial methionine to leucine in azurin increased its 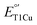 by ∼100 mV, the corresponding M298L mutation in small laccase from Streptomyces coelicolor (SLAC) unexpectedly decreased its
by ∼100 mV, the corresponding M298L mutation in small laccase from Streptomyces coelicolor (SLAC) unexpectedly decreased its  by 12 mV. X-ray crystallography revealed two axial water molecules in M298L-SLAC, leading to the decrease of
by 12 mV. X-ray crystallography revealed two axial water molecules in M298L-SLAC, leading to the decrease of  due to decreased hydrophobicity. Structural alignment and molecular dynamics simulation indicated a key difference in T1Cu axial loop position, leading to the different outcome upon methionine to leucine mutation. Based on structural analyses, we introduced additional F195L and I200F mutations, leading to partial removal of axial waters, a 122-mV increase in
due to decreased hydrophobicity. Structural alignment and molecular dynamics simulation indicated a key difference in T1Cu axial loop position, leading to the different outcome upon methionine to leucine mutation. Based on structural analyses, we introduced additional F195L and I200F mutations, leading to partial removal of axial waters, a 122-mV increase in  , and a 7-fold increase in kcat/KM from M298L-SLAC. These findings highlight the complexity of tuning
, and a 7-fold increase in kcat/KM from M298L-SLAC. These findings highlight the complexity of tuning 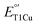 in multicopper oxidases and provide valuable insights into how structure-based protein engineering can contribute to the broader understanding of T1Cu center,
in multicopper oxidases and provide valuable insights into how structure-based protein engineering can contribute to the broader understanding of T1Cu center,  and reactivity tuning for applications, such as in solar energy transfer, fuel cells, and bioremediation.
and reactivity tuning for applications, such as in solar energy transfer, fuel cells, and bioremediation.
Introduction
Type 1 copper (T1Cu) centers play important roles in biological electron transfer (ET) processes from photosynthesis to respiration. They fulfill their roles by exhibiting a wide range of CuII/CuI reduction potentials to match with those of their redox partners in order to tune the ET rates required to perform the functions.1–6 Therefore, understanding structural features responsible for tuning
to match with those of their redox partners in order to tune the ET rates required to perform the functions.1–6 Therefore, understanding structural features responsible for tuning  is important not only to gain deeper insight into T1Cu functions in biology but also to allow for the design of artificial proteins or small molecular redox agents for different applications, from solar energy transfer to fuel cells. Towards this goal, we and other groups have successfully demonstrated the rational tuning of
is important not only to gain deeper insight into T1Cu functions in biology but also to allow for the design of artificial proteins or small molecular redox agents for different applications, from solar energy transfer to fuel cells. Towards this goal, we and other groups have successfully demonstrated the rational tuning of  by >600 mV in azurin (Az), as well as other proteins containing T1Cu sites.7–12 When combined with NiII, a single Az with five mutations can cover the entire physiological range of reduction potentials, from ∼−1 V to ∼1 V.13 In the process, we showed that multiple factors in the primary coordination sphere (PCS) and secondary coordination sphere (SCS) contribute to the tunability of the T1Cu center.8,13–18 Given these successes, it is vital to expand the scope of
by >600 mV in azurin (Az), as well as other proteins containing T1Cu sites.7–12 When combined with NiII, a single Az with five mutations can cover the entire physiological range of reduction potentials, from ∼−1 V to ∼1 V.13 In the process, we showed that multiple factors in the primary coordination sphere (PCS) and secondary coordination sphere (SCS) contribute to the tunability of the T1Cu center.8,13–18 Given these successes, it is vital to expand the scope of  tuning from mononuclear T1Cu proteins that perform interprotein ET functions to other T1Cu-containing proteins and enzymes. Of particular interest are multicopper oxidases, such as laccases, that can carry out a wide variety of reactions from oxygen reduction reaction (ORR) with low overpotential to bioremediation and lignin degradation.19–24
tuning from mononuclear T1Cu proteins that perform interprotein ET functions to other T1Cu-containing proteins and enzymes. Of particular interest are multicopper oxidases, such as laccases, that can carry out a wide variety of reactions from oxygen reduction reaction (ORR) with low overpotential to bioremediation and lignin degradation.19–24
Laccases are members of the multicopper oxidase family containing a T1Cu site for substrate binding and ET and a trinuclear Cu center composed of a type 2 Cu (T2Cu) center and a type 3 Cu (T3Cu) center for the four-electron reduction of O2 to H2O.25–27 Like mononuclear T1Cu proteins, the  of laccases vary widely, ranging from 367 mV in small laccase from Streptomyces coelicolor (SLAC) to ∼800 mV in fungal laccase (all
of laccases vary widely, ranging from 367 mV in small laccase from Streptomyces coelicolor (SLAC) to ∼800 mV in fungal laccase (all  in this paper are vs. SHE). It has previously been shown that the high catalytic efficiency and low ORR overpotential of fungal laccases are correlated with the high
in this paper are vs. SHE). It has previously been shown that the high catalytic efficiency and low ORR overpotential of fungal laccases are correlated with the high  .28–32 However, fungal laccases display optimal activity under acidic conditions,33 which precludes their effective utilization under physiological conditions for biofuel cells. Additonally, they are also less desirable for other reactions, as many substrates, such as lignin, are more soluble under basic conditions.34 Fungal laccases are also not stable at high temperatures.35 In contrast, SLAC has a much higher thermal stability and optimal activity under neutral or weakly basic conditions.36 However, SLAC has a much lower
.28–32 However, fungal laccases display optimal activity under acidic conditions,33 which precludes their effective utilization under physiological conditions for biofuel cells. Additonally, they are also less desirable for other reactions, as many substrates, such as lignin, are more soluble under basic conditions.34 Fungal laccases are also not stable at high temperatures.35 In contrast, SLAC has a much higher thermal stability and optimal activity under neutral or weakly basic conditions.36 However, SLAC has a much lower  (367 mV).37,38 Therefore, efforts have been devoted to engineer SLAC, as well as a similar small laccase from Streptomyces sviceus (Ssl1), to increase their
(367 mV).37,38 Therefore, efforts have been devoted to engineer SLAC, as well as a similar small laccase from Streptomyces sviceus (Ssl1), to increase their  's, so that they can display high activity while maintaining the high thermal stability and activity at neutral pH.38–41
's, so that they can display high activity while maintaining the high thermal stability and activity at neutral pH.38–41
A major difference between the low 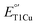 SLAC and the high
SLAC and the high 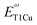 fungal laccase is the presence of a ligand at the axial position of the T1Cu. While SLAC has a Met298 at this position, fungal laccases have hydrophobic residues such as leucine or phenylalanine, which may destabilize CuII over the CuI and thus raise
fungal laccase is the presence of a ligand at the axial position of the T1Cu. While SLAC has a Met298 at this position, fungal laccases have hydrophobic residues such as leucine or phenylalanine, which may destabilize CuII over the CuI and thus raise  .14,15 Consistent with this hypothesis, multiple previous studies have demonstrated that mutation of the axial Met to Leu in different T1Cu proteins all led to an 80–180 mV increase in
.14,15 Consistent with this hypothesis, multiple previous studies have demonstrated that mutation of the axial Met to Leu in different T1Cu proteins all led to an 80–180 mV increase in  because of the increase in hydrophobicity.7,14,42–45 However, in a recent bioRxiv paper, the Liang group reported that when Met298 was mutated to Leu in SLAC, its
because of the increase in hydrophobicity.7,14,42–45 However, in a recent bioRxiv paper, the Liang group reported that when Met298 was mutated to Leu in SLAC, its  was too low to be measured.46 In contrast, a similar M295L mutation in Ssl1 resulted in an 81 mV increase in
was too low to be measured.46 In contrast, a similar M295L mutation in Ssl1 resulted in an 81 mV increase in 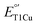 .40 In order to increase
.40 In order to increase  of SLAC, the Met298 was replaced with a noncanonical amino acid, such as cyclopentylalanine or cyclohexylalanine.46 While this work highlights the effectiveness of noncanonical amino acids in tuning
of SLAC, the Met298 was replaced with a noncanonical amino acid, such as cyclopentylalanine or cyclohexylalanine.46 While this work highlights the effectiveness of noncanonical amino acids in tuning  , it does not explain how biology can use natural amino acids to tune
, it does not explain how biology can use natural amino acids to tune  and its functions.
and its functions.
To address the above issues, we herein report that the M298L mutation in SLAC resulted in a 12 mV decrease of  from the native SLAC. To explain this finding, we obtained an X-ray crystal structure of M298L-SLAC, which reveals two water molecules in the axial position of the T1Cu; such water molecules decrease the hydrophobicity of the Cu site and thus lead to a decrease in
from the native SLAC. To explain this finding, we obtained an X-ray crystal structure of M298L-SLAC, which reveals two water molecules in the axial position of the T1Cu; such water molecules decrease the hydrophobicity of the Cu site and thus lead to a decrease in 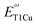 . This change also leads to a decrease in oxidase activity when using 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt (ABTS) as a model substrate. A structural alignment and MD simulations revealed a subtle axial loop difference near the T1Cu site between Az and SLAC, which led to different outcomes upon axial Met298 to Leu mutation, allowing water to bind at the axial position of the T1Cu site. To exclude water and increase
. This change also leads to a decrease in oxidase activity when using 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt (ABTS) as a model substrate. A structural alignment and MD simulations revealed a subtle axial loop difference near the T1Cu site between Az and SLAC, which led to different outcomes upon axial Met298 to Leu mutation, allowing water to bind at the axial position of the T1Cu site. To exclude water and increase  , we introduced F195L and I200F mutations near the axial pocket based on computational modeling. X-ray crystal structure indicates a partial removal of the axial water, which explains a drastic increase in
, we introduced F195L and I200F mutations near the axial pocket based on computational modeling. X-ray crystal structure indicates a partial removal of the axial water, which explains a drastic increase in  of ∼122 mV from M298L-SLAC. The kcat/KM toward ABTS oxidation was also improved by 2.6-fold from wild-type (WT) SLAC, and 7-fold from M298L-SLAC.
of ∼122 mV from M298L-SLAC. The kcat/KM toward ABTS oxidation was also improved by 2.6-fold from wild-type (WT) SLAC, and 7-fold from M298L-SLAC.
Results and discussion
Expression and purification of M298L-SLAC resulted in a protein whose electronic absorption spectrum displayed two distinct bands centered at 420 nm and 565 nm, and a shoulder at 330 nm (Fig. 1A). The 330 nm shoulder, which has previously been assigned to be an absorption from the T3Cu center,27 is similar to that of WT-SLAC, suggesting minimal perturbation of the T3Cu center by the M298L mutation. On the other hand, the 565 nm band, which has been assigned to a S(Cys)π to CuII(dx2−y2) charge transfer (CT) transition, is blue-shifted in M298L-SLAC relative to the 590 nm of WT-SLAC, along with a decrease in intensity, while a much stronger absorption appeared at 420 nm than that in WT-SLAC. This 420 nm absorption corresponds to the S(Cys)σ to CuII(dx2−y2) CT transition,5,47,48 with an εσ/π (defined as a ratio of peak intensity of the S(Cys)σ to CuII(dx2−y2) over the S(Cys)π to CuII(dx2−y2) CT transition) of 1.2. The increased intensity for S(Cys)σ to CuII(dx2−y2) CT transition at the expense of S(Cys)π to CuII(dx2−y2) CT transition has been observed before in other perturbed T1Cu sites that have undergone either a tetragonal or tetrahedral distortion from the typical geometry of T1Cu site.49–53 The electron paramagnetic resonance spectroscopy (EPR) spectrum of M298L-SLAC exhibited an increase in g⊥, g‖, and A‖ (Fig. 1B, Table 1 and S1†), further corroborating that this protein contains a distorted T1Cu site. | ||
| Fig. 1 (A) Electronic absorption spectrum and (B) EPR spectrum (black: experimental spectra; red: fitted spectra) of SLAC mutants involved in this study. Data for WT-SLAC is replotted from ref. 38. LF-SLAC stands for F195L/I200F-SLAC. | ||
The electronic absorption and EPR spectral changes observed in M298L-SLAC were not observed in the analogous M121L-Az, which showed an intense S(Cys)π to CuII(dx2−y2) CT transition that is similar to WT-Az and has minimal changes in EPR spectrum from that of WT-Az.42 The electronic absorption spectrum is also distinct from fungal laccase with Leu at the axial position, which displays only a S(Cys)π to CuII(dx2−y2) CT transition at around 610 nm.54
Surprisingly,  of M298L-SLAC was measured to be 354 mV, which is a 12-mV decrease from the 367 mV of WT-SLAC (Fig. 2A and S7†). This is in sharp contrast with other T1Cu proteins, which show an 80–180 mV
of M298L-SLAC was measured to be 354 mV, which is a 12-mV decrease from the 367 mV of WT-SLAC (Fig. 2A and S7†). This is in sharp contrast with other T1Cu proteins, which show an 80–180 mV  increase upon mutation of the axial Met to Leu, attributable to an increase in hydrophobicity. This unexpected decrease in
increase upon mutation of the axial Met to Leu, attributable to an increase in hydrophobicity. This unexpected decrease in  upon introducing a hydrophobic Leu in the axial position of SLAC indicates that there might be other factors playing a role in tuning
upon introducing a hydrophobic Leu in the axial position of SLAC indicates that there might be other factors playing a role in tuning  .
.
 | ||
Fig. 2 (A)  for mutants involved in this paper. All were measured in 50 mM MOPS, 150 mM NaCl, pH 7 buffer. †Data from ref. 38. (B) Relative for mutants involved in this paper. All were measured in 50 mM MOPS, 150 mM NaCl, pH 7 buffer. †Data from ref. 38. (B) Relative  change between each mutant. change between each mutant. | ||
To investigate a structural basis for the perturbed T1Cu and lower  in M298L-SLAC, we obtained a crystal structure of M298L-SLAC at 2.6 Å resolution (Fig. 3A). In addition to coordination by two His and one Cys in a trigonal plane, like in WT-SLAC, the primary coordination sphere of M298L-SLAC has additional electron densities at the axial position, while the Leu residue positioned away from the T1Cu center with a distance of 4.1 Å. The density can be modeled as two water molecules, which are not found in the crystal structure of WT-SLAC. A crystal structure of M298L-SLAC has previously been reported (PDB: 7B4Y),55 but the structure did not include the closest axial water molecule next to the T1Cu, although electron density is found at this position. The presence of the two water molecules near the T1Cu center of the M298L-SLAC tetrahedrally distort the T1Cu site, as can be observed from the distance between T1Cu and the coordination plane of SCys288–NHis231–NHis293. The distance increased from 0.16 Å in WT-SLAC to 0.37 Å in M298L-SLAC. This distortion may be responsible for the change in electronic absorption and EPR spectra from a typical T1Cu center to a perturbed T1Cu center, as similar changes have been observed in T1Cu of Az with a bound exogenous ligand (CN−, N3−, or SCN−).56 These exogenous ligands bind at the axial position of T1Cu center, perturb the geometry and change the orbital interactions between the CuII(dx2−y2) and the S(Cys) from a π interaction to a more predominantly σ interaction. The water molecules at the axial position of the T1Cu also make the T1Cu to experience a more hydrophilic environment and thus can explain the decrease in
in M298L-SLAC, we obtained a crystal structure of M298L-SLAC at 2.6 Å resolution (Fig. 3A). In addition to coordination by two His and one Cys in a trigonal plane, like in WT-SLAC, the primary coordination sphere of M298L-SLAC has additional electron densities at the axial position, while the Leu residue positioned away from the T1Cu center with a distance of 4.1 Å. The density can be modeled as two water molecules, which are not found in the crystal structure of WT-SLAC. A crystal structure of M298L-SLAC has previously been reported (PDB: 7B4Y),55 but the structure did not include the closest axial water molecule next to the T1Cu, although electron density is found at this position. The presence of the two water molecules near the T1Cu center of the M298L-SLAC tetrahedrally distort the T1Cu site, as can be observed from the distance between T1Cu and the coordination plane of SCys288–NHis231–NHis293. The distance increased from 0.16 Å in WT-SLAC to 0.37 Å in M298L-SLAC. This distortion may be responsible for the change in electronic absorption and EPR spectra from a typical T1Cu center to a perturbed T1Cu center, as similar changes have been observed in T1Cu of Az with a bound exogenous ligand (CN−, N3−, or SCN−).56 These exogenous ligands bind at the axial position of T1Cu center, perturb the geometry and change the orbital interactions between the CuII(dx2−y2) and the S(Cys) from a π interaction to a more predominantly σ interaction. The water molecules at the axial position of the T1Cu also make the T1Cu to experience a more hydrophilic environment and thus can explain the decrease in  of M298L-SLAC.
of M298L-SLAC.
 | ||
| Fig. 3 (A) Crystal structure of M298L-SLAC (PDB: 9NJI) showing the axial water (red spheres), with a 2FO – FC electron density contoured at 1 RMSD; (B) Structure alignment between M298L-SLAC (purple, PDB: 9NJI) and N47S/M121L-azurin (yellow, PDB: 3JT2 (ref. 8)). The red arrow indicates the shift of the loop that holds the axial leucine. The red spheres indicate the axial water in M298L-SLAC. | ||
The presence of the axial water molecule in T1Cu center has been observed previously in a purple cupredoxin from Nitrosopumilus maritimus and the red copper protein nitrosocyanin from Nitrosomonas europaea,57,58 Our crystal structure shows that M298L-SLAC more closely resembles the purple cupredoxin, as the geometry remains a perturbed T1Cu center with a distorted tetrahedral geometry, rather than the tetragonally distorted geometry of nitrosocyanin that is more typical of a T2Cu center. The purple cupredoxin from N. maritimus also exhibits similar electronic absorption and EPR spectral features and to those of M298L-SLAC,57 further corroborating the presence of axial water. Our observation is consistent with a recent report of the presence of axial water molecules in M295A/Y/F/V/I in Ssl1,41 even though the paper did not report M295L mutation. The authors reported an increase in the S(Cys)σ to CuII(dx2−y2) CT transition among all tested mutants, as well as an increase in g‖, indicating a change of energy difference between CuII(dx2−y2) and CuII(dxy), and a change in copper ligand geometry.16
Since the electronic absorption, EPR spectra and 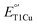 of the T1Cu site of M298L-SLAC are very different from those of the corresponding T1Cu in M121L-Az, we carried out a structural alignment between M298L-SLAC and M121L-Az, revealing a key difference in the loop near the axial Met. The loop in M298L-SLAC is further away from the T1Cu in SLAC than in M121L-Az (Fig. 3B). A similar loop shift is also observed between WT-SLAC and WT-Az (Fig. S1†). We hypothesize that this difference leads to a different preferred conformation of the axial residue in SLAC, allowing water molecules to enter the axial pocket. To test this hypothesis, we conducted molecular dynamics (MD) simulations using the crystal structures of M298L-SLAC and N47S/M121L-Az (PDB: 3JT2 (ref. 8)). We chose N47S/M121L-Az instead of M121L-Az for the simulation because the crystal structure of M121L-Az is not available and Ser47 mimics Thr232 at the corresponding position in SLAC more closely than Asn47 with minimal steric perturbation. Based on our simulations, the axial Leu in M298L-SLAC adopts mainly Conformation I, which matches with the crystal structure M298L-SLAC (Fig. 4A, S2 and S3†). At the same time, the axial Leu could also adopt Conformation II, but at a much lower frequency of around 5%, while its position is also translated outwards due to varying positions of the axial loop in SLAC. In contrast, the axial Leu in N47S/M121L-Az adopts Conformation I much less frequently (about 14%) than Conformation II. Instead, it mainly adopts Conformation II, which aligns well with the geometry in the crystal structure of N47S/M121L-Az (Fig. 4B, S3 and S4†). To ensure that the above finding that M298L-SLAC favors Conformation I was not influenced by the input structure to start the MD simulations, we conducted another simulation using Conformation II as the starting structure and found that the axial Leu in M298L-SLAC is converted back to Conformation I, similar to when the input structure was Conformation I to begin with (Fig. S5†). Together, these results indicate that slight axial loop difference near T1Cu site between Az and SLAC led to different outcome upon axial Met298 to Leu mutation, allowing water to bind at the axial position of the T1Cu site, hence influencing its electronic absorption, EPR and
of the T1Cu site of M298L-SLAC are very different from those of the corresponding T1Cu in M121L-Az, we carried out a structural alignment between M298L-SLAC and M121L-Az, revealing a key difference in the loop near the axial Met. The loop in M298L-SLAC is further away from the T1Cu in SLAC than in M121L-Az (Fig. 3B). A similar loop shift is also observed between WT-SLAC and WT-Az (Fig. S1†). We hypothesize that this difference leads to a different preferred conformation of the axial residue in SLAC, allowing water molecules to enter the axial pocket. To test this hypothesis, we conducted molecular dynamics (MD) simulations using the crystal structures of M298L-SLAC and N47S/M121L-Az (PDB: 3JT2 (ref. 8)). We chose N47S/M121L-Az instead of M121L-Az for the simulation because the crystal structure of M121L-Az is not available and Ser47 mimics Thr232 at the corresponding position in SLAC more closely than Asn47 with minimal steric perturbation. Based on our simulations, the axial Leu in M298L-SLAC adopts mainly Conformation I, which matches with the crystal structure M298L-SLAC (Fig. 4A, S2 and S3†). At the same time, the axial Leu could also adopt Conformation II, but at a much lower frequency of around 5%, while its position is also translated outwards due to varying positions of the axial loop in SLAC. In contrast, the axial Leu in N47S/M121L-Az adopts Conformation I much less frequently (about 14%) than Conformation II. Instead, it mainly adopts Conformation II, which aligns well with the geometry in the crystal structure of N47S/M121L-Az (Fig. 4B, S3 and S4†). To ensure that the above finding that M298L-SLAC favors Conformation I was not influenced by the input structure to start the MD simulations, we conducted another simulation using Conformation II as the starting structure and found that the axial Leu in M298L-SLAC is converted back to Conformation I, similar to when the input structure was Conformation I to begin with (Fig. S5†). Together, these results indicate that slight axial loop difference near T1Cu site between Az and SLAC led to different outcome upon axial Met298 to Leu mutation, allowing water to bind at the axial position of the T1Cu site, hence influencing its electronic absorption, EPR and  .
.
 | ||
| Fig. 4 Two conformations (I and II) that the axial leucine can adopt from MD simulation of. (A) M298L-SLAC; (B) N47S/M121L-Az. | ||
Since the presence of water molecules at the axial position of T1Cu in M298L-SLAC perturbs the T1Cu and lowers  , we further explored additional mutations to occupy the axial pocket and prevent water from entering the axial position. We inspected the crystal structure of M298L-SLAC, found that Phe195 is in proximity to the axial water (Fig. S6†), and proposed that F195L mutation may block water from entering the axial position by occupying the axial pocket with its methyl group protruding into the pocket. A computational model indicated that the F195L mutation to M298L-SLAC still left a small empty space in the pocket, and we added an additional I200F mutation to fill in the space.
, we further explored additional mutations to occupy the axial pocket and prevent water from entering the axial position. We inspected the crystal structure of M298L-SLAC, found that Phe195 is in proximity to the axial water (Fig. S6†), and proposed that F195L mutation may block water from entering the axial position by occupying the axial pocket with its methyl group protruding into the pocket. A computational model indicated that the F195L mutation to M298L-SLAC still left a small empty space in the pocket, and we added an additional I200F mutation to fill in the space.
Encouraged by the above computer modeling, we first obtained F195L/I200F-SLAC (called LF-SLAC hereafter) without the M298L mutation to investigate the effect of the two mutations on WT-SLAC. The electronic absorption spectrum of LF-SLAC showed similar absorption features as WT-SLAC (Fig. 1A), with a strong absorption band at ∼590 nm and a slight increase at ∼400 nm. The EPR spectrum can be simulated by a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of T1Cu and T2Cu, with g‖ for T1Cu slightly decreased from 2.229 in WT-SLAC to 2.217 in LF-SLAC, and A‖ for T1Cu slightly increased from 67.0 × 10−4 cm−1 in WT-SLAC to 70.3 × 10−4 cm−1 in LF-SLAC (Fig. 1B, Table 1 and S1†). These results indicate a minimal effect of the F195L/I200F mutation on the T1Cu site. After combining F195L/I200F mutations with M298L, LF-M298L-SLAC exhibited two strong absorption bands centered at 420 nm and 580 nm, similar to those of M298L-SLAC. However, the εσ/π decreased slightly to from 1.2 in M298L-SLAC to 0.9, suggesting that the F195L/I200F mutation resulted in a less distorted T1 site than that observed in M298L-SLAC (Fig. 1A). Fitting the EPR spectrum of LF-M298L-SLAC using two components (T1Cu and T2Cu) was worse than fitting the spectrum with three components at a ratio of 1(T1Cu): 1(T1Cu): 2(T2Cu) (Table S1 and Fig. S10†). Therefore, the EPR spectral simulation suggests that the LF-M298L-SLAC has two species that contain T1Cu at a ratio of 1
1 mixture of T1Cu and T2Cu, with g‖ for T1Cu slightly decreased from 2.229 in WT-SLAC to 2.217 in LF-SLAC, and A‖ for T1Cu slightly increased from 67.0 × 10−4 cm−1 in WT-SLAC to 70.3 × 10−4 cm−1 in LF-SLAC (Fig. 1B, Table 1 and S1†). These results indicate a minimal effect of the F195L/I200F mutation on the T1Cu site. After combining F195L/I200F mutations with M298L, LF-M298L-SLAC exhibited two strong absorption bands centered at 420 nm and 580 nm, similar to those of M298L-SLAC. However, the εσ/π decreased slightly to from 1.2 in M298L-SLAC to 0.9, suggesting that the F195L/I200F mutation resulted in a less distorted T1 site than that observed in M298L-SLAC (Fig. 1A). Fitting the EPR spectrum of LF-M298L-SLAC using two components (T1Cu and T2Cu) was worse than fitting the spectrum with three components at a ratio of 1(T1Cu): 1(T1Cu): 2(T2Cu) (Table S1 and Fig. S10†). Therefore, the EPR spectral simulation suggests that the LF-M298L-SLAC has two species that contain T1Cu at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Species 1 exhibits a small A‖ at 65.7 × 10−4 cm−1 that is closer to 67.0 × 10−4 cm−1 in WT-SLAC, while Species 2 had a much larger A‖ of 109.1 × 10−4 cm−1 that is closer to type 1.5 Cu (Fig. 1B and Table 1).59
1. Species 1 exhibits a small A‖ at 65.7 × 10−4 cm−1 that is closer to 67.0 × 10−4 cm−1 in WT-SLAC, while Species 2 had a much larger A‖ of 109.1 × 10−4 cm−1 that is closer to type 1.5 Cu (Fig. 1B and Table 1).59
To obtain more insights into the T1Cu site of LF-M298L-SLAC, we obtained its crystal structure at 2.6 Å resolution. The overall scaffold, especially the loops near T1Cu site aligns well with those of M298L-SLAC, indicating that the introduction of F195L/I200F mutations did not perturb the folding of the protein. However, even though the electron density at the axial position of T1Cu can still be fitted by two water molecules as in M298L-SLAC, the electron density is decreased significantly and can now be fitted with an occupancy of ∼0.5. The Leu at position 195 could be fitted by two conformations, with one conformation having the methyl group to occupy the axial pocket and blocks the axial water from entering the pocket (Fig. 5A), while the other conformation having the methyl group rotated away from the axial pocket and allows two water molecules to occupy the position (Fig. 5B). These two conformations at Leu195 also led to two conformations for nearby Met223. These results may explain the EPR results, as Species 1 is a species that contains a T1Cu without axial water, while Species 2 contains axial water, like M298L-SLAC, and the T1Cu of LF-M298L-SLAC is in equilibration between the two species. The two conformations also explain that the mutations resulted in only 50% water occupancy next the T1Cu, as only one of the two conformations is capable of removing water.
 | ||
| Fig. 5 Crystal structure of LF-M298L-SLAC (PDB: 9NJJ) showing the two F195L conformations in subfigure (A) and (B). The axial water is shown in red spheres. | ||
Since the decrease of  in M298L-SLAC is due to the presence of water at the axial position of T1Cu, this partial removal of the axial water should result in an increase of
in M298L-SLAC is due to the presence of water at the axial position of T1Cu, this partial removal of the axial water should result in an increase of  . Indeed, we found LF-M298L-SLAC displayed a drastic increase in
. Indeed, we found LF-M298L-SLAC displayed a drastic increase in 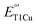 , by 122 mV from 355 to 477 mV (Fig. 2A and S8†). To confirm that the increase in
, by 122 mV from 355 to 477 mV (Fig. 2A and S8†). To confirm that the increase in  is due to the effect of the F195L/I200F mutations on M298L-SLAC, but not on WT-SLAC, the
is due to the effect of the F195L/I200F mutations on M298L-SLAC, but not on WT-SLAC, the  of F195L/I200F-SLAC was also measured to be 392 mV (Fig. 2A and S9†). It was only a 25 mV increase from that of WT-SLAC, significantly less than the 122 mV increase from that of M298L-SLAC. Likewise, M298L decreased
of F195L/I200F-SLAC was also measured to be 392 mV (Fig. 2A and S9†). It was only a 25 mV increase from that of WT-SLAC, significantly less than the 122 mV increase from that of M298L-SLAC. Likewise, M298L decreased  of WT-SLAC by 12 mV, while increased
of WT-SLAC by 12 mV, while increased  of LF-SLAC by 85 mV (Fig. 2B). These results indicate that the large
of LF-SLAC by 85 mV (Fig. 2B). These results indicate that the large  increase in LF-M298L-SLAC is due to the effect of F195L/I200F mutations on M298L-SLAC, likely arising from partial removal of the axial water to increase the hydrophobicity of T1Cu site.
increase in LF-M298L-SLAC is due to the effect of F195L/I200F mutations on M298L-SLAC, likely arising from partial removal of the axial water to increase the hydrophobicity of T1Cu site.
With our deeper understanding of the effects of M298L, F195L, I200F mutations on the electronic absorption, EPR, and 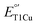 along with structural insight, we further examined the oxidase activities of M298L-SLAC, LF-SLAC and LF-M298L-SLAC using ABTS as a substrate and obtained their Michaelis–Menten parameters (Fig. 6A and S11–S13†). While
along with structural insight, we further examined the oxidase activities of M298L-SLAC, LF-SLAC and LF-M298L-SLAC using ABTS as a substrate and obtained their Michaelis–Menten parameters (Fig. 6A and S11–S13†). While  of M298L-SLAC is 12 mV lower than that of WT-SLAC, its kcat decreased by 9 folds, and kcat/KM decreased by 3 folds over that of WT-SLAC (Fig. 6B and C). This decrease in ABTS oxidation activity is consistent with a previous report, although at different conditions.55 Interestingly, introducing F195L/I200F mutations, while having 2-fold decrease of kcat over WT-SLAC and similar kcat/KM with WT-SLAC, increased kcat of M298L-SLAC by 12 folds and kcat/KM of M298L-SLAC by 7 folds (Fig. 6B and C), indicating a successful increase of the oxidase activity for SLAC by rational designing the T1Cu secondary coordination sphere to remove water in the axial position.
of M298L-SLAC is 12 mV lower than that of WT-SLAC, its kcat decreased by 9 folds, and kcat/KM decreased by 3 folds over that of WT-SLAC (Fig. 6B and C). This decrease in ABTS oxidation activity is consistent with a previous report, although at different conditions.55 Interestingly, introducing F195L/I200F mutations, while having 2-fold decrease of kcat over WT-SLAC and similar kcat/KM with WT-SLAC, increased kcat of M298L-SLAC by 12 folds and kcat/KM of M298L-SLAC by 7 folds (Fig. 6B and C), indicating a successful increase of the oxidase activity for SLAC by rational designing the T1Cu secondary coordination sphere to remove water in the axial position.
 | ||
| Fig. 6 (A) Michaelis–Menten parameters for mutants involved in this paper using ABTS as substrate. †Data from ref. 38. (Michaelis–Menten parameters were measured in 50 mM MOPS, 150 mM NaCl, pH 7 at 310 K.) Histogram demonstrating (B) kcat, and (C) kcat/KM for different mutants. | ||
Conclusions
In this work, we investigated how the mutation of axial Met to Leu tunes the of SLAC. Even though the axial Met to Leu mutation in azurin and many other T1Cu-containing proteins resulted in an increase of
of SLAC. Even though the axial Met to Leu mutation in azurin and many other T1Cu-containing proteins resulted in an increase of  , the same mutation in M298L-SLAC decreased the
, the same mutation in M298L-SLAC decreased the  due to the presence of water molecules at the axial position of the T1Cu. The crystal structure of M298L-SLAC shows a rotation of axial Leu, which creates a space for two water molecules in the T1Cu axial pocket. By rationally introducing F195L/I200F mutation, we were able to partially remove the axial water, as supported by spectroscopic and crystallographic studies. The resulting LF-M298L-SLAC variant displays a drastic increase in
due to the presence of water molecules at the axial position of the T1Cu. The crystal structure of M298L-SLAC shows a rotation of axial Leu, which creates a space for two water molecules in the T1Cu axial pocket. By rationally introducing F195L/I200F mutation, we were able to partially remove the axial water, as supported by spectroscopic and crystallographic studies. The resulting LF-M298L-SLAC variant displays a drastic increase in  by 122 mV relative to WT-SLAC and catalytic activity towards an oxidase model complex ABTS with 12-fold greater kcat and 7-fold greater kcat/KM than M298L-SLAC, demonstrating the power of rational design in tuning enzyme activity. Together, this work highlighted that the same mutation in different proteins of the same class may lead to completely different outcomes. To achieve the goals of increasing
by 122 mV relative to WT-SLAC and catalytic activity towards an oxidase model complex ABTS with 12-fold greater kcat and 7-fold greater kcat/KM than M298L-SLAC, demonstrating the power of rational design in tuning enzyme activity. Together, this work highlighted that the same mutation in different proteins of the same class may lead to completely different outcomes. To achieve the goals of increasing  and enzymatic activity, careful spectroscopic and structural studies, along with computational modeling are required. Moreover, this work also provided insights into the relationship between protein structure and T1Cu tunability, and guiding future protein designs for tuning the enzymatic activity.
and enzymatic activity, careful spectroscopic and structural studies, along with computational modeling are required. Moreover, this work also provided insights into the relationship between protein structure and T1Cu tunability, and guiding future protein designs for tuning the enzymatic activity.
Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
J.-X. W.: conceptualization, formal analysis, investigation, writing – original draft preparation. A. C. V.: formal analysis, investigation, writing – original draft preparation. L. H. W.: investigation. E. N. M.: conceptualization, investigation. Y. L.: conceptualization, project administration, supervision, writing – review & editing, and funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is supported by the National Science Foundation (CHE-2420683 for Y.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) L.). It was also part of the Chemical Transformation Initiative at Pacific Northwest National Laboratory (PNNL), conducted under the Laboratory Directed Research and Development Program at PNNL, a multiprogram national laboratory operated by Battelle for the U.S. Department of Energy. We also thank the Robert A. Welch Foundation (Grant F-0020) for its support of the Lu group research program at the University of Texas at Austin. Crystallography data were acquired at the beamline 8.2.1 at the Advanced Light Source (ALS) (proposal ALS-12445) of the Lawrence Berkeley National Laboratory, and the beamline 23-ID-B at the Advanced Photon Source (APS) of the Argonne National Laboratory. We would like to thank Dr Yan Jessie Zhang for offering us the use of their APS beamtime and advice on X-ray crystallography, Lisa Phan for her guidance on MD simulation, and Dr Casey Van Stappen and Dr Yunling Deng for their help in revising the manuscript.
L.). It was also part of the Chemical Transformation Initiative at Pacific Northwest National Laboratory (PNNL), conducted under the Laboratory Directed Research and Development Program at PNNL, a multiprogram national laboratory operated by Battelle for the U.S. Department of Energy. We also thank the Robert A. Welch Foundation (Grant F-0020) for its support of the Lu group research program at the University of Texas at Austin. Crystallography data were acquired at the beamline 8.2.1 at the Advanced Light Source (ALS) (proposal ALS-12445) of the Lawrence Berkeley National Laboratory, and the beamline 23-ID-B at the Advanced Photon Source (APS) of the Argonne National Laboratory. We would like to thank Dr Yan Jessie Zhang for offering us the use of their APS beamtime and advice on X-ray crystallography, Lisa Phan for her guidance on MD simulation, and Dr Casey Van Stappen and Dr Yunling Deng for their help in revising the manuscript.
References
- H. B. Gray, B. G. Malmström and R. J. P. Williams, J. Biol. Inorg. Chem., 2000, 5, 551–559 CrossRef CAS PubMed.
- A. Donaire, B. Jiménez, C. O. Fernández, R. Pierattelli, T. Niizeki, J.-M. Moratal, J. F. Hall, T. Kohzuma, S. S. Hasnain and A. J. Vila, J. Am. Chem. Soc., 2002, 124, 13698–13708 CrossRef CAS PubMed.
- E. I. Solomon, R. K. Szilagyi, S. DeBeer George and L. Basumallick, Chem. Rev., 2004, 104, 419–458 CrossRef CAS PubMed.
- J. J. Warren, K. M. Lancaster, J. H. Richards and H. B. Gray, J. Inorg. Biochem., 2012, 115, 119–126 CrossRef CAS PubMed.
- E. I. Solomon, D. E. Heppner, E. M. Johnston, J. W. Ginsbach, J. Cirera, M. Qayyum, M. T. Kieber-Emmons, C. H. Kjaergaard, R. G. Hadt and L. Tian, Chem. Rev., 2014, 114, 3659–3853 CrossRef CAS PubMed.
- J. Liu, S. Chakraborty, P. Hosseinzadeh, Y. Yu, S. Tian, I. Petrik, A. Bhagi and Y. Lu, Chem. Rev., 2014, 114, 4366–4469 CrossRef CAS PubMed.
- A. J. Di Bilio, T. K. Chang, B. G. Malmström, H. B. Gray, B. Göran Karlsson, M. Nordling, T. Pascher and L. G. Lundberg, Inorganica Chim. Acta, 1992, 198–200, 145–148 CrossRef CAS.
- N. M. Marshall, D. K. Garner, T. D. Wilson, Y.-G. Gao, H. Robinson, M. J. Nilges and Y. Lu, Nature, 2009, 462, 113–116 CrossRef CAS PubMed.
- K. M. Lancaster, K. Yokoyama, J. H. Richards, J. R. Winkler and H. B. Gray, Inorg. Chem., 2009, 48, 1278–1280 CrossRef CAS PubMed.
- S. M. Berry, M. H. Baker and N. J. Reardon, J. Inorg. Biochem., 2010, 104, 1071–1078 CrossRef CAS PubMed.
- J. Szuster, A. J. Leguto, U. A. Zitare, J. P. Rebechi, A. J. Vila and D. H. Murgida, Bioelectrochemistry, 2022, 146, 108095 CrossRef CAS PubMed.
- A. Singha, A. Sekretareva, L. Tao, H. Lim, Y. Ha, A. Braun, S. M. Jones, B. Hedman, K. O. Hodgson, R. D. Britt, D. J. Kosman and E. I. Solomon, J. Am. Chem. Soc., 2023, 145, 13284–13301 CrossRef CAS PubMed.
- P. Hosseinzadeh, N. M. Marshall, K. N. Chacón, Y. Yu, M. J. Nilges, S. Y. New, S. A. Tashkov, N. J. Blackburn and Y. Lu, Proc. Natl. Acad. Sci. U.S.A., 2016, 113, 262–267 CrossRef CAS PubMed.
- S. M. Berry, M. Ralle, D. W. Low, N. J. Blackburn and Y. Lu, J. Am. Chem. Soc., 2003, 125, 8760–8768 CrossRef CAS PubMed.
- D. K. Garner, M. D. Vaughan, H. J. Hwang, M. G. Savelieff, S. M. Berry, J. F. Honek and Y. Lu, J. Am. Chem. Soc., 2006, 128, 15608–15617 CrossRef CAS PubMed.
- R. G. Hadt, N. Sun, N. M. Marshall, K. O. Hodgson, B. Hedman, Y. Lu and E. I. Solomon, J. Am. Chem. Soc., 2012, 134, 16701–16716 CrossRef CAS PubMed.
- Y. Yu, N. M. Marshall, D. K. Garner, M. J. Nilges and Y. Lu, J. Inorg. Biochem., 2022, 234, 111863 CrossRef CAS PubMed.
- C. Van Stappen, Y. Deng, Y. Liu, H. Heidari, J.-X. Wang, Y. Zhou, A. P. Ledray and Y. Lu, Chem. Rev., 2022, 122, 11974–12045 CrossRef CAS PubMed.
- C.-W. Lee, H. B. Gray, F. C. Anson and B. G. Malmström, J. Electroanal. Chem. Interfacial Electrochem., 1984, 172, 289–300 CrossRef CAS.
- D. M. Mate and M. Alcalde, Microb. Biotechnol., 2017, 10, 1457–1467 CrossRef CAS PubMed.
- D. Singh and N. Gupta, Biologia, 2020, 75, 1183–1193 CrossRef CAS.
- J. A. Cracknell, K. A. Vincent and F. A. Armstrong, Chem. Rev., 2008, 108, 2439–2461 CrossRef CAS PubMed.
- C. J. Rodgers, C. F. Blanford, S. R. Giddens, P. Skamnioti, F. A. Armstrong and S. J. Gurr, Trends Biotechnol., 2010, 28, 63–72 CrossRef CAS PubMed.
- A. A. Gewirth and M. S. Thorum, Inorg. Chem., 2010, 49, 3557–3566 CrossRef CAS PubMed.
- E. I. Solomon, U. M. Sundaram and T. E. Machonkin, Chem. Rev., 1996, 96, 2563–2605 CrossRef CAS PubMed.
- K. Nakamura and N. Go, Cell. Mol. Life Sci., 2005, 62, 2050–2066 CrossRef CAS PubMed.
- S. M. Jones and E. I. Solomon, Cell. Mol. Life Sci., 2015, 72, 869–883 CrossRef CAS PubMed.
- F. Xu, W. Shin, S. H. Brown, J. A. Wahleithner, U. M. Sundaram and E. I. Solomon, Biochim. Biophys. Acta, Protein Struct. Mol. Enzymol., 1996, 1292, 303–311 CrossRef PubMed.
- I. Mateljak, E. Monza, M. F. Lucas, V. Guallar, O. Aleksejeva, R. Ludwig, D. Leech, S. Shleev and M. Alcalde, ACS Catal., 2019, 9, 4561–4572 CrossRef CAS.
- H. Schweiger, E. Vayner and A. B. Anderson, Electrochem. Solid-State Lett., 2005, 8, A585–A587 CrossRef CAS.
- S. Shleev, A. Jarosz-Wilkolazka, A. Khalunina, O. Morozova, A. Yaropolov, T. Ruzgas and L. Gorton, Bioelectrochemistry, 2005, 67, 115–124 CrossRef CAS PubMed.
- Y. Kamitaka, S. Tsujimura, K. Kataoka, T. Sakurai, T. Ikeda and K. Kano, J. Electroanal. Chem., 2007, 601, 119–124 CrossRef CAS.
- L. I. Ramírez-Cavazos, C. Junghanns, N. Ornelas-Soto, D. L. Cárdenas-Chávez, C. Hernández-Luna, P. Demarche, E. Enaud, R. García-Morales, S. N. Agathos and R. Parra, J. Mol. Catal. B: Enzym., 2014, 108, 32–42 CrossRef.
- L. Delugeau, S. Grelier and F. Peruch, ChemSusChem, 2025, e202402377 CrossRef CAS PubMed.
- K. Hildén, T. K. Hakala and T. Lundell, Biotechnol. Lett., 2009, 31, 1117–1128 CrossRef PubMed.
- M. Sherif, D. Waung, B. Korbeci, V. Mavisakalyan, R. Flick, G. Brown, M. Abou-Zaid, A. F. Yakunin and E. R. Master, Microb. Biotechnol., 2013, 6, 588–597 CrossRef PubMed.
- M. C. Machczynski, E. Vijgenboom, B. Samyn and G. W. Canters, Protein Sci., 2004, 13, 2388–2397 CrossRef CAS PubMed.
- J. Wang, A. C. Vilbert, C. Cui, E. N. Mirts, L. H. Williams, W. Kim, J. Y. Zhang and Y. Lu, Angew. Chem., Int. Ed., 2023, 62, e202314019 CrossRef CAS PubMed.
- M. Gunne, A. Höppner, P.-L. Hagedoorn and V. B. Urlacher, FEBS J., 2014, 281, 4307–4318 CrossRef CAS PubMed.
- A. C. Olbrich, J. N. Schild and V. B. Urlacher, J. Inorg. Biochem., 2019, 201, 110843 CrossRef CAS PubMed.
- A. C. Olbrich, S. Mielenbrink, V. P. Willers, K. Koschorreck, J. A. Birrell, I. Span and V. B. Urlacher, Chem. - Eur. J., 2024, e202403005 Search PubMed.
- B. G. Karlsson, R. Aasa, B. G. Malmström and L. G. Lundberg, FEBS Lett., 1989, 253, 99–102 CrossRef CAS.
- L. D. Kanbi, S. Antonyuk, M. A. Hough, J. F. Hall, F. E. Dodd and S. S. Hasnain, J. Mol. Biol., 2002, 320, 263–275 CrossRef CAS PubMed.
- S. DeBeer George, L. Basumallick, R. K. Szilagyi, D. W. Randall, M. G. Hill, A. M. Nersissian, J. S. Valentine, B. Hedman, K. O. Hodgson and E. I. Solomon, J. Am. Chem. Soc., 2003, 125, 11314–11328 CrossRef CAS PubMed.
- M. Choi, N. Sukumar, A. Liu and V. L. Davidson, Biochemistry, 2009, 48, 9174–9184 CrossRef CAS PubMed.
- S. Fischer, A. Natter Perdiguero, K. Lau and A. Deliz Liang, bioRxiv, 2025, DOI:10.1101/2025.02.09.636911.
- L. B. LaCroix, D. W. Randall, A. M. Nersissian, C. W. G. Hoitink, G. W. Canters, J. S. Valentine and E. I. Solomon, J. Am. Chem. Soc., 1998, 120, 9621–9631 CrossRef CAS.
- E. I. Solomon, Inorg. Chem., 2006, 45, 8012–8025 CrossRef CAS PubMed.
- Y. Lu, L. B. LaCroix, M. D. Lowery, E. I. Solomon, C. J. Bender, J. Peisach, J. A. Roe, E. B. Gralla and J. S. Valentine, J. Am. Chem. Soc., 1993, 115, 5907–5918 CrossRef CAS.
- A. Messerschmidt, L. Prade, S. J. Kroes, J. Sanders-Loehr, R. Huber and G. W. Canters, Proc. Natl. Acad. Sci. U.S.A., 1998, 95, 3443–3448 CrossRef CAS PubMed.
- Comprehensive Coordination Chemistry II: From Biology to Nanotechnology, ed. J. A. McCleverty and T. J. Meyer, Elsevier Pergamon, Amsterdam; Boston, 2nd edn, 2004, vol. 8 Search PubMed.
- S. Ghosh, X. Xie, A. Dey, Y. Sun, C. P. Scholes and E. I. Solomon, Proc. Natl. Acad. Sci. U.S.A., 2009, 106, 4969–4974 CrossRef CAS PubMed.
- K. M. Clark, Y. Yu, N. M. Marshall, N. A. Sieracki, M. J. Nilges, N. J. Blackburn, W. A. van der Donk and Y. Lu, J. Am. Chem. Soc., 2010, 132, 10093–10101 CrossRef CAS PubMed.
- R. P. Bonomo, A. M. Boudet, R. Cozzolino, E. Rizzarelli, A. M. Santoro, R. Sterjiades and R. Zappal, J. Inorg. Biochem., 1998, 71, 205–211 CrossRef CAS PubMed.
- K. Zovo, H. Pupart, A. Van Wieren, R. E. Gillilan, Q. Huang, S. Majumdar and T. Lukk, ACS Omega, 2022, 7, 6184–6194 CrossRef CAS PubMed.
- N. Bonander, B. G. Karlsson and T. Vänngård, Biochemistry, 1996, 35, 2429–2436 CrossRef CAS PubMed.
- P. Hosseinzadeh, S. Tian, N. M. Marshall, J. Hemp, T. Mullen, M. J. Nilges, Y.-G. Gao, H. Robinson, D. A. Stahl, R. B. Gennis and Y. Lu, J. Am. Chem. Soc., 2016, 138, 6324–6327 CrossRef CAS PubMed.
- R. L. Lieberman, D. M. Arciero, A. B. Hooper and A. C. Rosenzweig, Biochemistry, 2001, 40, 5674–5681 CrossRef CAS PubMed.
- S. J. Kroes, C. W. G. Hoitink, C. R. Andrew, J. Ai, J. Sanders-Loehr, A. Messerschmidt, W. R. Hagen and G. W. Canters, Eur. J. Biochem., 1996, 240, 342–351 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures, molecular dynamics simulation details, electron paramagnetic resonance simulation parameters, Nernst plot, Michaelis–Menten curve; the crystal structures of M298L-SLAC and LF-M298L-SLAC have been deposited in the PDB with accession codes 9NJI and 9NJJ. See DOI: https://doi.org/10.1039/d5sc02177d |
| This journal is © The Royal Society of Chemistry 2025 |
