Exploration of degrons and their ability to mediate targeted protein degradation
Timothy J.
Harris Jr.
 a and
Darci J.
Trader
a and
Darci J.
Trader
 *ab
*ab
aDepartment of Pharmaceutical Sciences, University of California, Irvine, California 92617, USA. E-mail: dtrader@uci.edu
bDepartment of Chemistry, University of California, Irvine, California 92617, USA
First published on 1st January 2025
Abstract
Degrons are short amino acid sequences that can facilitate the degradation of protein substrates. They can be classified as either ubiquitin-dependent or -independent based on their interactions with the ubiquitin proteasome system (UPS). These amino acid sequences are often found in exposed regions of proteins serving as either a tethering point for an interaction with an E3 ligase or initiating signaling for the direct degradation of the protein. Recent advancements in the protein degradation field have shown the therapeutic potential of both classes of degrons through leveraging their degradative effects to engage specific protein targets. This review explores what targeted protein degradation applications degrons can be used in and how they have inspired new degrader technology to target a wide variety of protein substrates.
Introduction
Protein degradation of ubiquitinated proteins is required for cell proliferation. The amount or rate of the degradation of proteins varies from cell type to cell type, and can vary depending on the life cycle state.1 Proteins that are damaged or currently not required for cell proliferation must be degraded to prevent apoptosis. Proteins can be degraded through two cellular pathways, the ubiquitin-proteasome system (UPS) or through autophagy. The proteasome is made up of two main regions: the 19S regulatory particle (19S RP) and 20S core particle (20S CP).2 The 19S RP is commonly referred to as the cap of the proteasome, and it has many responsibilities including recognizing ubiquitinated-protein substrates, unwinding the protein to reduce its tertiary structure, and shuttling the protein into the catalytic core to be hydrolyzed.3For a protein to be degraded via the UPS pathway, it needs to ubiquitinated. Ubiquitin (Ub) is a small 9 kDa protein that can be covalently appended to a lysine of the protein of interest (POI) to be degraded, Fig. 1.4 The conjugation of Ub to a POI requires the use of several other proteins: E1, E2, and E3. Other reviews describe the importance of the different linkages of Ub to the POI to lead to degradation by the proteasome.5–7 This brief review describes how E3 ligases can recognize a region of a POI to attach a chain of Ub moieties utilizing an intrinsic degron region. The discovery of the different types of degrons and E3 ligases provided the inspiration for the development of bifunctional molecules and molecular glues for targeted protein degradation as new therapeutic agents, Fig. 2.
N-end degron
N-end degrons are a set of N-terminal residues that signal for the degradation of a protein. These N-end degrons follow the “N-end rule” which governs the relative half-life of a respective protein. Originally discovered by the Varshavsky group in 1986, N-end degrons play a vital role in the regulation of protein homeostasis in eukaryotes, yeast, bacteria, and plants. The scope of this review will focus more specifically on their role in eukaryotes.8–10In eukaryotes, N-end degrons play a major part in the ubiquitin proteasome system (UPS).11–16 In many contexts, these sites specifically interact with an E3 ubiquitin ligase such as UBR1 and UBR2 known as N-recognins. Many N-recognins exist as adaptor proteins that bind to E3 ligase complexes and mediate the recruitment of a specific N-terminal degron.17–20 Once bound to the N-degron, the conjugated E2 ligase within the E3 complex polyubiquitinates the target protein at an exposed lysine residue. The ubiquitinated protein is then processed and degraded via the UPS.
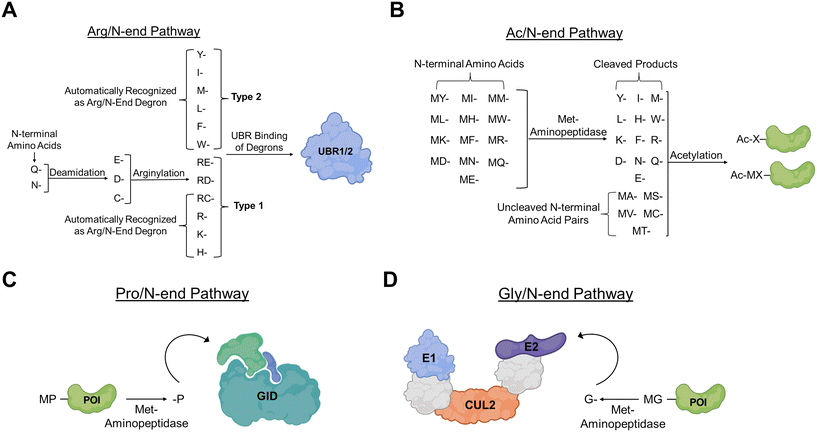
Arg/N-end degrons
There are two main N-end degrons pathways that have been discovered in eukaryotes with the Arg/N-end rule pathway being the most prominent. The Arg/N-end rule pathway utilizes the post translational modification (PTM) arginylation to append the amino acid arginine to a target protein to later be identified and degraded via an N-recognin (Fig. 3A). Two types of N-terminal amino acids fall within the Arg/N-end rule pathway, those that are directly recognized and bound by N-recognins known as type 1s, and those that are either endogenously basic or that must undergo a post translation modification to be converted into an N-terminal arginine.21 The bulky nonpolar and basic amino acids Lys, His, Arg, Tyr, Leu, Phe, Trp, and Ile all serve as primary destabilizing Arg/N-end degrons and are directly recruited by N-recognins without PTMs. On the other hand, the amino acids Asp, Glu, Asn, Gln, and Cys can only be bound by N-recognins and function as primary destabilizing degrons after undergoing arginine transformation from arginine-transferases.Arg/N-end degrons have been found to be pivotal in the regulation of protein homeostasis with many cases of these exposed amino acid residues coinciding with oxidative damage or misfolding of the target protein.22,23 Proteins that have undergone N-terminal arginylation have also been shown to serve as substrates for degradation via macroautophagy.24 Instead of ubiquitin recruitment and subsequent proteolysis by the UPS, the protein substrate is targeted by the autophagic adaptor protein p62. Upon binding to the ubiquitin chain, p62 sequesters the target protein trafficking it to a forming phagosome for lysosomal degradation.
Though Arg/N-end degrons are widely recognized for their role in protein degradation, it has been shown that these degrons and their binding partners (UBR1 and UBR2) also regulate other biological processes.25,26 Some notable examples include regulation of transcription and translation in human cells, where UBR1 and UBR2 double knock-out cells showed increased levels of glucocorticoid receptors, suggesting that these receptors are physiological substrates of the Arg/N-end degron pathway. Continuation of the knock-out study showed lower levels of the intracellular proteins neurofilament L (NF-L) and neurofilament-M (NF-M) despite up-regulation of their mRNAs, suggesting that Arg/N-end can also modulate the translation of specific mRNAs.12
Ac/N-end degrons
The other main branch of N-terminal degrons is known as the Ac/N-end degron pathway (Fig. 3B) and it was discovered in 2010 by the Varshavsky group.27 This pathway relies on acetylation of target proteins to initiate N-recognin recruitment and protein degradation.15,28,29 The acetylation of N-terminal protein residues has been shown to proceed through a different mechanism than the one more commonly observed in the acetylation of internal amino acid residues.30 In many cases, N-terminal methionine residues signal for the acetylated degron through Met-aminopeptidase-mediated cleavage.28 Through this mechanism, the remaining amino acids are then “screened” for capability of acetylation based on the size of their adjacent amino acids. Typically, the N-terminal acetylation occurs in Ala, Val, Ser, Thr, Cys, and Met (in the case of bulky amino acids), though in rare cases the acetylation of N-terminal Gly and Pro has been observed. Additionally, it has been reported that N-terminal acetylation is irreversible and that no N-terminal deacetylases have been identified to this point.31 This irreversibility showcases the finality of this degron pathway and could be directly implicated in why Ac/N-end degrons are relevant to the proteolysis of approximately 80% of the proteome.11,32 Adding this N-terminal acetyl group to a target protein increases its hydrophobicity and introduces a new potential protein binding surface that can be used as a “docking” position to mediate protein complex formation between the multiprotein E2–E3 ligases and the target protein. It has been shown that antagonizing this acetyl-amide binding location can impede the conjugation of the ubiquitin like protein NEDD8 and cause subsequent protein accumulation.11,33 Additionally, N-terminal acetylation not only promotes the degradation of proteins via the UPS, but degrons also have been shown to regulate autophagy. N-terminal acetylation by B-type N-terminal amino transferase (NatB) promotes the formation of actin filaments. These filaments facilitate the trafficking of target proteins to phospholipid scramblase containing vesicles for autophagosome formation, and subsequent acetylation of Vps1 promotes autophagosome-vacuole fusion.34Alternatively, a small subset of Ac/N-end degrons have been implicated in nonproteolytic activities, namely their emerging role in histone modification and protein trafficking. N-alpha-acetyltransferase 40 (NAA40) has been shown to acetylate a terminal serine in histone H4 which affects gene expression with misregulation of this event being implicated in cancer progression.35–38 N-terminal acetylation of the neuronal protein α-synuclein stabilizes its terminal α-helix and increases its affinity for synaptic vesicles regulating neurotransmitter release.39–42
Pro/N-end degrons
The Pro/N-end degron pathway is a less prevalent pathway of N-end degrons that recognizes a Pro residue at the N-terminus or at position 2 of the target protein (Fig. 3C). This pathway, like the Ac/N-end pathway, utilizes Met-aminopeptidase to remove Met groups and an N-terminal Pro residue. The main N-recognin for this pathway is the glucose-induced degradation (GID) E3 ubiquitin-protein ligase complex that mediates the ubiquitination and subsequent proteasomal degradation of the protein substrate. Glucose-induced degradation protein 4 (Gid4) serves as the substrate recognition subunit of the GID and directly binds the N-terminal proline residue. It has also been shown that GID4 can recognize hydrophobic amino acids such as N-terminal Gly and Ile though it has the highest affinity for the peptide sequence Pro-Gly-Leu-Trp.43–45 Solved crystal structures of human Gid4 support its affinity for N-terminal Pro residues.46,47 An orthosteric site glutamic acid residue (Glu237) engages in hydrogen bonding with the secondary amine of Pro.Gly/N-end degrons
N-terminal Gly residues have also been shown to be degrons capable of recruiting the Cullin-RING E3 ligases (CRLs) for target protein ubiquitination (Fig. 3D). Mammals express seven canonical Cullin proteins (Cul1, Cul2, Cul3, Cul4A-B, Cul5 and Cul7) that form multi-subunit CRLs.48 Cullin-RING E3 ligases are composed of three mains subunits: the Cullin subunit itself, the E2 binding region, and the substrate binding region. The Cullin subunit serves as the main scaffold for the entire protein complex and is composed of three Cullin repeats. This 3-mer protein subunit then attaches itself to the E2 binding region via a globular carboxy-terminal domain (CTD) that has a highly conserved stretch of approximately 200 amino acids.49–51 This CTD interface allows the RING protein to bind and through subsequent activation of ubiquitin through the E1, E2, and E3 cascade, places a degradation tag on the target protein. Recognition of the N-terminal Gly degron is carried out by the Cul2 adaptor proteins ZYG11B and ZER1.52 It was found that both these adaptor proteins have overlaps in protein substrates with ZYG11B being widely able to recognize shorter N-terminal Gly degrons while ZER1 can target more specific degron substrates upstream of the Gly with preferential binding towards more bulky amino acids. The Gly/N-end degron role has seen implications in a variety of cellular process regulation contexts, particularly, the use of N-terminal Gly residues to regulate N-myristylation has been studied in eukaryotic cells. N-Myristylation refers to a PTM that attaches the 14-carbon fatty acid myristate to the N-terminal glycine of a protein. This attachment aids in plasma targeting, subcellular trafficking, and localization of the target protein, ultimately influencing the proteins functionality in cells.53–55 Gly/N-end degrons recruitment can serve as “quality control” for this process signaling for the degradation of proteins that failed to complete the PTM still bearing the N-terminal Gly residue.52C-end degron
Though N-end degrons were originally discovered in 1986 and it took approximately 30 years for scientists to identify C-end degrons. Found originally in 2018 by the Elledge and Yen labs,56 C-end degrons differ from N-end degrons in their mechanism of action and localization, but ultimately are analogous in their ability to signal for degradation of specific protein substrates (Fig. 4). As their name suggests, C-end degrons are carboxylic acid terminus amino acids that can be present in full length proteins, protein fragments, and prematurely terminated proteins. The responses to C-end degrons stem from the protein's inability to protect their C-terminus fast enough through folding or binding, showcasing the protein loss of function and subsequent need for proteolysis.Gly/C-end degrons
Similar to N-terminal Gly, C-terminal Gly residues can also serve as degradation signals for proteins. Identical to N-terminal Gly binding, the Cul2 E3 complex remains the key N-recognin in the ubiquitination of C-terminal Gly residues. It has been found that this Gly/C-end degron exhibits the highest substrate preference to disubstituted C-terminal Gly residues (Gly-Gly) with the N-recognin KLHDC2 serving as a protein substrate receptor in the Cul2 E3 complex.57,58 KLHDC2 is a part of a related family of the substrate adaptor proteins known as the Kelch family, where each N-recognin has specific selectivity towards C-terminal Gly [associated 2-mer peptide fragments (KLHDC3: RG and KG and KLHDC10: AG, WG, and PG)].56Gly/C-end degrons also play alternative biological roles outside of solely signaling for protein degradation. Like Gly/N-end degrons, C-terminal glycine residues are implicated in the regulation of myristylation, where proteins that fail to undergo the C-terminal PTM are targeted for termination.59 Gly/C-end degrons also have been shown to have roles in viral protein evasion of proteolytic cleavage. Coronavirus non-structural proteins (NSPs) are synthesized as polypeptide chains that are proteolytically cleaved into individual NSP that aid in viral cell replication by viral proteases. Cleavage of NSP1-NSP3 by the viral protease PLpro releases a C-terminal diGly residue that could be potentially targeted by the KLHDC2 pathway.60 It was found that the stability of GFP affixed with the final six residues of the SARS-CoV NSP1 protein has much greater stability compared to those ending only with a diGly residue. This suggests that viral NSP proteins have adapted resistance to proteolytic attack via host cells and the evasion of this Gly/C-end degron pathway is pivotal in viral protein proliferation and infection of host cells.60
Gln/C-end degrons
The study of C-end degrons is currently in its early stages with new pathways being identified rapidly, one of which being the Gln/C-end degron pathway. The C-terminal Gln residue in this pathway is targeted by the protein TRIM7 that belongs to the tripartite-motif- containing (TRIM) protein family of RING-type E3 ligases. Crystal structures of TRIM7 further elucidate its selectivity in binding to Gln residues.61,62 The basic amines in the C-terminal Gln fit easily into the positively charged binding pocket of TRIM7. Additionally, the carbonyl group of Gln forms several hydrogen bonding interactions with TRIM7 making it highly specific for the Gln residue not allowing other substitutions at the C-terminus.The Gln/C-end degron pathway has also been found to be implicated in the immune response. The expression of TRIM proteins is upregulated in response to interferons, and they play various roles in the innate-immunity related pathway such as regulating cytokine secretion, preventing autophagic degradation of cGAS and STING, and facilitating the autophagy meditated degradation of pathogenic proteins.63–68
EE/C-end degron
Cul4, a member of the Cullin family of E3 ligases is a known N-recognin of the EE/ C-end pathway. Specifically, the adaptor protein DCAF12 recognizes the EE degron and targets its respective protein for degradation.69–71 It has been found that DCAF12 can broadly recognize degron substrates bearing a Glu residue at position two with the C-terminus residue being ambiguous, although a pair of C-terminal glutamic acid residues are preferentially targeted.11R-3/C-end degrons
Cul4 is also implicated in the R-3/C-end degron pathway, where degrons with Arg at position three relative to the C-terminus (RXX-COOH) are recognized by the adaptor protein TRPC4AP. Several important protein substrates have been identified for the Cul4-TRPC4AP complex, most notably the transcription regulator protein N-Myc was found to be targeted by the N-recognin via its R-3 C-terminal degron motif.56,72Arg/C-end degrons
Like the N-terminal Arg pathway discussed above, a similar Arg/C-end degron pathway also exists for C-terminal Arg residues. All three mammalian FEM1 proteins, FEM1A, FEM1B and FEM1C, have demonstrated the ability to serve as CRL2 adapters recognizing the C-terminal Arg degron. FEM1A and FEM1C have been shown to bind to peptides that end in the more specific sequence RXXR or RXR, where X is any amino acid.73 Additionally, scientists have found that FEM1B binds to C-terminal Arg residues with a different specificity than FEM1A and FEM1B, having a roughly 5-fold increase in selectivity towards an amino acid sequence derived from the C-terminus of the protein cyclin-dependent kinase 5 activator 1 (CDK5R1).74Small molecule binders of degron motifs
The discovery of degrons marked a pivotal moment in understanding biological pathways that regulate protein degradation. Over the years, this research has laid the groundwork for a variety of degron binding small molecules, primarily the molecule thalidomide and its derivatives. Originally discovered in the early 1950s, the molecule thalidomide was marketed for its antiemetic properties but was later discontinued for its association to birth defects and neuropathies. It was later determined that thalidomide exhibited anti-inflammatory properties being used for the treatment of acute erythema nodosum leprosum, an inflammatory symptom associated with leprosy.75 Because of its effects, thalidomide was later discovered to be efficacious in the treatment of cancer76,77 (multiple myeloma) and this spearheaded scientists to begin structure–activity relationship studies to find new thalidomide analogues. After further mechanism of action studies,78–81 thalidomide derivatives known as immunomodulatory drugs (IMiDs) have been shown to engage the protein target cereblon (CRBN).82–87 CRBN is a Cullin RING E3 ligase that plays an important role within the UPS.75,88–90 IMiDs have been used to leverage this protein's abilities, with the molecules serving as recruiters for the E3 ligase in bifunctional molecule applications.91 Co-crystal structures of CRBN in the IMiDs thalidomide, lenalidomide and pomalidomide show that the molecules bind a rather shallow hydrophobic pocket at the surface of the protein (Fig. 5).92 All three IMiD molecules show very little variation in their binding orientation, with each forming key interactions with His380 and Trp382 via their glutarimide ring. Co-crystal structures of the IMiDs also reveal that the additional chemical moieties of the molecules do not interact with the binding pocket of the protein and instead remain solvent-exposed. Many studies suggest that these solvent-exposed regions can serve as sites for binding other protein substrates, allowing thalidomide and its derivatives to function as molecular glues, simultaneously binding CRBN and other protein substrates. The Ward and Tonnelle groups have demonstrated that treatment with IMiDs leads to the degradation of the Ikaros family of proteins (Ikaros, Aiolos, Helios, Eos, and Pegasus).93,94Ikaros proteins are zinc finger (ZF) transcription factors that participate in a complex network of interactions with gene regulators to control gene expression and proliferation via chromatin remodeling. These proteins primarily regulate hematopoiesis and are important in the development of the adaptive immune system. Members of the Ikaros protein class are characterized by their “zinc finger” motifs that consist of a zinc atom that is in specific coordination with four other amino acids, typically histidine and cysteine. This protein family has two sets of highly conserved C2H2-type zinc finger motifs.95,96 The first set is located at the N-terminus of the protein and mediates binding to specific DNA sequences and the second set is found at the C-terminus of the protein, enables dimerization with other Ikaros proteins and serves as a binding location to mediate interactions with other transcription regulator proteins. Studies have indicated that the second C2H2 ZF domains of both Ikaros (IKZF1) and Aiolos (IKZF3) are drug-inducible degrons. Many other IMiD protein targets that don't share similar primary sequences have been shown to be degraded via this molecular glue mechanism and emerging crystal structures illustrate that many ZF-containing proteins with diverse amino acid sequences bind the same IMiD interface.97–99
Sivers et al. later in 2018 found that ZF degrons bind a complimentary groove on the CRBN surface that fits the overall shape of the C2H2 ZF domain, bringing different amino acids from different ZF degrons in contact with the same solvent exposed region of the respective IMiD and the CRBN protein surface, showing that this degron recruitment isn't based on traditional small-molecule–protein interactions and more heavily relies on the “shape” and pseudo secondary structure of the ZF-containing region.
Similar to ImiDs, sulfonamides are another class of small molecule binders of E3 ligases that have been found to additionally function as molecular glues for protein substrates. Sulfonamides are highly diverse small molecules that have been used for a wide variety of therapeutics including antimicrobial therapies, carbonic anhydrase inhibitors, anti-thyroid medications and more.100 In the late 1990s, studies indicated that aryl sulfonamides such as indisulam and tasisulam could be used to inhibit the proliferation of cancer cells (Fig. 6).101 Further explorations of the molecular mechanisms of these compounds showed that they targeted a wide scope of oncogenic disease states, showing lower cell viability in colorectal cancer, glioblastoma, non-small cell cancer, acute leukemia and other tumor types after administration. It wasn't until 2017 that scientists discovered that molecules like indisulam were functioning as molecular glues, bridging a target protein and the E3 ligase DCAF15.102 Specifically, it was found that the molecule indisulam inhibits cellular growth by degrading the RNA binding motif protein 39 (RBM39) that serves as an essential splicing factor of RNA regulating protein transcript levels. Bussiere et al. found that indisulam binds a well-defined pocket formed by DCAF15 making key hydrogen binding interactions with both Ala234 and Phe235 of the protein.103 It was also found that the nitrogen within the central sulfonamide of indisulam forms two water-mediated hydrogen bonds with both Thr262 and Asp264 of RMB39, stabilizing the recruitment of the additional protein to the DCAF15. The interaction between the DCAF15 and RBM39 mediated by indisulam binding also extends to other protein substrates, specifically another member of the RNA binding motif protein family RBM23. Both RBM23 and RBM39 share a conserved α-helical sequence (X1XXM4XXG7XXEP11) that is defined as a degron motif allowing the target proteins to be recruited and degraded via ubiquitination by DCAF15.103
A more recent example of small molecule recognition of a degron motif comes from a study conducted by Nurix Therapeutics in 2019.104 This research showcased the ability to target the protein β-catenin for degradation via a molecular glue binding mechanism. β-Catenin is a Wnt signaling effector protein that is often dysregulated and stabilized in cancers. In healthy cells, β-catenin is highly regulated and maintained at very low cytoplasmic levels through a multi-step process including phosphorylation, ubiquitylation, and subsequent degradation by the UPS. The phosphorylation is carried out by the cytoplasmic destruction complex (DC) consisting of suppressor proteins axin and adenomatous polyposis coli (APC), and the serine–threonine kinases glycogen synthase kinase 3 (GSK3) and casein kinase 1 (CK1). The protein β-catenin is initially phosphorylated on Ser45 by CK1, followed by sequential phosphorylation at Thr41, Ser37, and Ser33.105–108 In cancerous cells however, this sequential phosphorylation cascade is not essential for degradation of the protein. The phosphorylated Ser33 and Ser37 of β-catenin have been found to be part of a phosphodegron sequence (DSPGXXSPP) that binds the F box protein β-TrCP that complexes with the Cullin E3 ligase Skp1 (SCF), leading to ubiquitination by the E3 ligase complex and degradation.109 In most colorectal cancers, β-catenin is stabilized due to mutations in Ser37 accounting for roughly 10% of known β-catenin mutations. This mutation impairs β-catenin's ability to bind to β-TrCP.110 Further structural characterization of mutated β-catenin lacking Ser37 phosphorylation reveals that the monophosphorylated degron (Ser33P) binds to β-TrCP in a similar orientation as the wild-type β-catenin.111 These studies also elucidated that the removal of phosphorylated Ser37 disrupted several key electrostatic and hydrogen bonding interactions between both β-catenin and β-TrCP, reducing binding affinity, but also exposing a small hydrophobic binding pocket between the two protein interfaces. After conducting a library screening and hit optimization of over 350![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 compounds, the authors developed compound NX-252114 that increased the binding affinity of the mutant Ser37 β-catenin to β-TrCP 100-fold after administration (Fig. 7).
000 compounds, the authors developed compound NX-252114 that increased the binding affinity of the mutant Ser37 β-catenin to β-TrCP 100-fold after administration (Fig. 7).
Additionally, other small molecules have been identified that increase the binding affinity of phosphodegron motifs to E3 ligases. c-Myc is a protein that is known to function as a transcription regulator in cells, and its misregulation is implicated in a variety of different cancer subtypes.112 c-Myc is widely an elusive drug target because of its unique protein structure, lacking a clear binding pocket for small molecules. Like β-catenin, c-Myc also has two phosphodegron motifs (LLPTPPPLSPP and EETPPPTTPS) that once phosphorylated can be recruited to the F-box and WD repeat domain-containing 7 protein (FBW7) that complexes with the Cullin E3 ligase SCF for subsequent ubiquitination and degradation.113 FBW7 mutations can occur in cancers subtypes that dysregulate binding to Myc causing its stabilization and accumulation in cells.114 A recent study identified a new ligand (KI-FBX-001) that binds at the interface of c-Myc and FBW7 stabilizing the protein partners and aiding in the proteolysis of the c-Myc protein substrate.115 The effects of both NX-252114 and KI-FBX-001 are current working examples of utilizing small molecules to rebuild target protein's encoded degron motifs that were otherwise only natively accessible for targeted protein degradation.
Targeted protein degradation utilizing “degron like” motifs
Over the years, methods for degron-mediated targeted protein degradation have evolved from exploiting endogenous degron motifs within proteins to chemically installing “degron-like” moieties into small molecules that can signal the degradation of target proteins. A noteworthy example comes from the Rao lab, which demonstrated the use of N-terminal amino acids (N-end rule degraders) to induce the degradation of the BCR-ABL protein complex.116 The N-end rule pathway determines protein stability based on the identity of the N-terminal amino acid residue. Specific amino acids, such as basic residues (Arg, Lys, His) and hydrophobic residues (Val, Leu, Ile, Phe, Trp, Tyr), are directly recognized by UBR1/2 E3 ligases, which facilitate substrate ubiquitination and subsequent proteasomal degradation. Leveraging this pathway, the authors designed molecules incorporating a single N-terminal amino acid, a small polyethylene glycol (PEG) linker, and dasatinib, a tyrosine kinase inhibitor, as a protein of interest (POI) recruiter (Fig. 8). Dasatinib targets the Abelson proto-oncogene (ABL), which, in hematological malignancies, forms a fusion with the breakpoint cluster region (BCR) protein, creating the oncogenic BCR–ABL complex.117–120 This fusion protein exhibits heightened stability and kinase activity, driving the uncontrolled proliferation of myeloid cells. Dasatinib locks BCR–ABL into an inactive conformation, halting its activity.121,122 Using the N-terminal amino acids as signaling degrons the authors tested their single amino acid bifunctional molecules and showed that they were able to lower the cellular levels of the BCR–ABL complex in vitro. Further testing showed that the most effective degrader utilized Arg as the N-terminal amino acid, achieving a DC50 of 0.85 nM and an IC50 of 0.36 nM for BCR–ABL degradation. This study is interesting because it is one of the first instances of direct attachment of a recognized degron to a protein of interest (POI) resulting in its degradation. Further expansion on this promising work, such as extending this technology to target other protein substrates, would greatly expand the field of protein degradation and provide scientists with a better understanding of the capacity of chemically installed degrons to signal for target protein degradation.Additional methods of employing “degron like” motifs for targeted protein degradation have been recently reported. HyT-PDs (hydrophobic tag-based protein degradation) represent an alternative approach for targeted protein degradation. HyT-PDs are bifunctional molecules composed of a (POI) binding moiety, a linker, and a highly hydrophobic group known as the hydrophobic tag. This hydrophobic tag acts similarly to a protein's degron by mimicking a damaged or misfolded protein. One of the most prevalent moieties used as hydrophobic tag is adamantane, an organic compound consisting of three fused cyclohexane rings. In 2017, the first instance of a targeted approach for HyT-PDs was published by the Li group.123 In this work, the authors developed a bifunctional degrader consisting of the hydrophobic tag adamantane, a peptide recognition motif for the microtubule associated protein tau, and a poly-arginine cell penetrating peptide tail (Fig. 9). It was found that these series of HyT-PDs were able to effectively lower the concentration of Alzheimer's disease-causing protein tau in both in vitro and in vivo screenings.
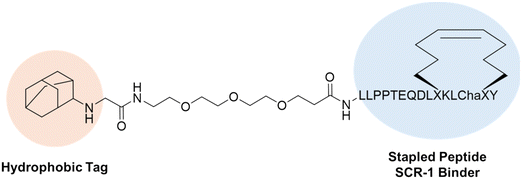
Hydrophobic tags have also been featured in comparative studies to the heavily studied proteolysis targeting chimeras (PROTACs), showing increased efficacy and drug-like properties. Choi et al. synthesized both a PROTAC (ND1-YL2) and HyT-PD (YL2-HyT6) for the target ribonucleic protein SCR-1 (Fig. 10). It was found that the HyT-PD increased the degradation of the POI two-fold, lowering the DC50 from 5 μM as compared to the PROTAC. This observed increase can stem from various factors, but it is hypothesized that the enhanced “drug-likeness” due to improved ADME properties may play a significant role in these increases.124 HyT-PDs typically have lower molecular weights, greater lipophilicity, and longer half-lives compared to PROTACs, contributing to their efficacy.125 For example, the YLS-HyT6 demonstrated better cellular uptake and lower plasma metabolic rates in media compared to the PROTAC BD1-YL2, which may be attributed to these advantageous properties.
Targeted protein degradation by direct proteasome recruitment
Throughout this review, we have explored various methods of targeted protein degradation that rely on E3 ligase-based mechanisms. However, scientific research has also identified proteolysis mechanisms that function through direct substrate recruitment to the proteasome. One example of this is the hydrophobic tag Boc3Arg, which consists of three tert-butoxycarbonyl groups protecting all side-chain amines in the Arg residue. This highly hydrophobic group was found to avoid ubiquitin dependent degradation of GST-π when tagged to the GST-π inhibitor ethacrynic acid.126,127 It is hypothesized that the Boc3Arg used in the hydrophobic tag might act as a ligand for the 20S CP of the proteasome, facilitating its degradation directly to the catalytic core and avoiding the 19S and the ubiquitin-dependent degradation system. Other proteins have also been successfully targeted for degradation using the hydrophobic tag Boc3Arg such as E. coli dihydrofolate reductase (eDHFR), through the same ubiquitin independent pathway.126More recently, scientists have demonstrated that direct recruitment of target proteins to the full 26S proteasome complex can effectively induce their degradation. In a study conducted by Genentech in 2023, they reported the discovery of a new peptide macrocycle, MC1, which binds directly to the 26S proteasome subunit alpha-2.128 Scientists then conducted cryo-EM with MC1 and found that the molecule binds within 70 Å to the proteasome's unfoldase pore. After binding site conformation, scientists synthesized a bifunctional molecule consisting of the macrocyclic peptide MC1 and the BRD4 inhibitor JQ1 (Fig. 11). It was found that molecule was able to facilitate the degradation of the POI BRD4 in a proximity mediated manner. This was the first instance of direct degradation of a POI by the 26S proteasome via bifunctional molecule recruitment and provided an important foundation for the expansion of this degradation technology.
Ongoing research has expanded upon bifunctional molecules that can recruit a POI directly to the 26S proteasome.129 A new class of bifunctional molecules have been described that bind Rpn-13, a 19S RP subunit, and a POI.130 Like the macrocyclic peptide MC1, TCL-1 binds in close proximity to the proteasome's substrate channel interacting with the ubiquitin receptor Rpn-13 (Fig. 12). Similar findings were also found in a recent study by the Kodadek lab, which employed a similar approach to degrade BRD2/4 and CDK9 using a HaloTag recruitment system. In this work, the ubiquitin receptor Rpn13 was conjugated to a HaloTag, which served as a tether to a chloroalkane moiety on the bifunctional molecule.131 This method demonstrated the degradation of all three target proteins, showcasing the potential applicability of this mechanism. However, further exploration is needed to determine which other 26S proteasome subunits can be effectively targeted. Additionally, it remains unclear which types of proteins are suitable for degradation using this new 26S proteasome-targeting methodology.
 | ||
| Fig. 12 A bifunctional molecule that can increase BRD4 degradation by binding to Rpn-13, a 26S proteasome subunit. | ||
Conclusion
Collectively, these findings illustrate how the UPS exploits the unique properties of protein termini to facilitate selective protein degradation. Despite intense research interest in the N-degron pathways over the past three decades, important new insights continue to be uncovered. We anticipate that the coming years will bring similar advances in our understanding of C-degron pathways, but already some common principles are emerging: for example, recognition of C-terminal degrons is achieved by tandem repeat domains.132 A focus of future work will be to define the cellular contexts in which these pathways operate, and to understand how dysregulation of these pathways may be relevant to specific disease states.Given the research progress in the past two decades and the recent level of interest and investment from both academia and industry, it is clear that targeted protein degradation will become a key therapeutic modality. As noted above, protein degraders that interact with an E3 ligase are already in clinics. New targeted protein degradation methods are also being explored, including those that target the autophagy pathway.133,134 The continuous discoveries regarding the ability of cells to naturally degrade proteins will continue to help guide research in harnessing target protein degradation for a desired therapeutic response.
Data availability
No primary research results, software or code have been included and no new data were generated or analyzed as part of this review.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was supported by a start-up package from the UCI School of Pharmacy and the UCI Chao Family Comprehensive Cancer Center (P30CA062203). It was also supported by a NIH-NIAID grant (1R01AI50847).References
- G. A. Collins and A. L. Goldberg, The Logic of the 26S Proteasome, Cell, 2017, 169(5), 792–806, DOI:10.1016/j.cell.2017.04.023
.
- O. Coux, K. Tanaka and A. L. Goldberg, Structure and Functions of the 20S and 26S Proteasomes, Annu. Rev. Biochem., 1996, 65, 801–847, DOI:10.1146/annurev.bi.65.070196.004101
.
- C. Muli, W. Tian and D. Trader, Small Molecule Inhibitors of the Proteasome's Regulatory Particle, ChemBioChem, 2019, 20(14), 1739–1753, DOI:10.1002/cbic.201900017
.
- D. R. Squair and S. Virdee, A New Dawn beyond Lysine Ubiquitination, Nat. Chem. Biol., 2022, 18(8), 802–811, DOI:10.1038/s41589-022-01088-2
.
- M. E. French, C. F. Koehler and T. Hunter, Emerging Functions of Branched Ubiquitin Chains, Cell Discovery, 2021, 7(1), 1–10, DOI:10.1038/s41421-020-00237-y
.
- R. B. Damgaard, The Ubiquitin System: From Cell Signalling to Disease Biology and New Therapeutic Opportunities, Cell Death Differ., 2021, 28(2), 423–426, DOI:10.1038/s41418-020-00703-w
.
- C. M. Pickart and M. J. Eddins, Ubiquitin: Structures, Functions, Mechanisms, Biochim. Biophys. Acta, Mol. Cell Res., 2004, 1695(1), 55–72, DOI:10.1016/j.bbamcr.2004.09.019
.
- A. Bachmair, D. Finley and A. Varshavsky, In Vivo Half-Life of a Protein Is a Function of Its Amino-Terminal Residue, Science, 1986, 234(4773), 179–186, DOI:10.1126/science.3018930
.
- A. Varshavsky, The N-End Rule: Functions, Mysteries, Uses, Proc. Natl. Acad. Sci. U. S. A., 1996, 93(22), 12142–12149, DOI:10.1073/pnas.93.22.12142
.
- A. Varshavsky, Discovery of Cellular Regulation by Protein Degradation, J. Biol. Chem., 2008, 283(50), 34469–34489, DOI:10.1074/jbc.X800009200
.
- A. Varshavsky, N-Degron and C-Degron Pathways of Protein Degradation, Proc. Natl. Acad. Sci. U. S. A., 2019, 116(2), 358–366, DOI:10.1073/pnas.1816596116
.
- T. T. M. Vu, D. C. Mitchell, S. P. Gygi and A. Varshavsky, The Arg/N-Degron Pathway Targets Transcription Factors and Regulates Specific Genes, Proc. Natl. Acad. Sci. U. S. A., 2020, 117(49), 31094–31104, DOI:10.1073/pnas.2020124117
.
- A. Varshavsky, The N-End Rule Pathway and Regulation by Proteolysis, Protein Sci., 2011, 20(8), 1298–1345, DOI:10.1002/pro.666
.
- A. Shimshon, K. Dahan, M. Israel-Gueta, D. Olmayev-Yaakobov, R. T. Timms, A. Bekturova, Y. Makaros, S. J. Elledge and I. Koren, Dipeptidyl Peptidases and E3 Ligases of N-Degron Pathways Cooperate to Regulate Protein Stability, J. Cell Biol., 2024, 223(8), e202311035, DOI:10.1083/jcb.202311035
.
- K. T. Nguyen, S.-H. Mun, C.-S. Lee and C.-S. Hwang, Control of Protein Degradation by N-Terminal Acetylation and the N-End Rule Pathway, Exp. Mol. Med., 2018, 50(7), 1–8, DOI:10.1038/s12276-018-0097-y
.
- L. T. H. L. Le, S. Park, J. H. Lee, Y. K. Kim and M. J. Lee, N-Recognins UBR1 and UBR2 as Central ER Stress Sensors in Mammals, Mol. Cells, 2024, 47(1), 100001, DOI:10.1016/j.mocell.2023.12.001
.
- D. A. Dougan, D. Micevski and K. N. Truscott, The N-End Rule Pathway: From Recognition by N-Recognins, to Destruction by AAA + Proteases, Biochim. Biophys. Acta, Mol. Cell Res., 2012, 1823(1), 83–91, DOI:10.1016/j.bbamcr.2011.07.002
.
- F. Müller and T. Bange, Identification of N-Degrons and N-Recognins Using Peptide Pull-Downs Combined with Quantitative Mass Spectrometry, Methods Enzymol., 2023, 686, 67–97, DOI:10.1016/bs.mie.2023.02.007
.
- A. Melnykov, S.-J. Chen and A. Varshavsky, Gid10 as an Alternative N-Recognin of the Pro/N-Degron Pathway, Proc. Natl. Acad. Sci. U. S. A., 2019, 116(32), 15914–15923, DOI:10.1073/pnas.1908304116
.
- S. T. Kim, Y. J. Lee, T. Tasaki, J. Hwang, M. J. Kang, E. C. Yi, B. Y. Kim and Y. T. Kwon, The N-Recognin UBR4 of the N-End Rule Pathway Is Required for Neurogenesis and Homeostasis of Cell Surface Proteins, PLoS One, 2018, 13(8), e0202260, DOI:10.1371/journal.pone.0202260
.
- J.-H. Oh, J.-Y. Hyun, S.-J. Chen and A. Varshavsky, Five Enzymes of the Arg/N-Degron Pathway Form a Targeting Complex: The Concept of Superchanneling, Proc. Natl. Acad. Sci. U. S. A., 2020, 117(20), 10778–10788, DOI:10.1073/pnas.2003043117
.
- F. Eisele and D. H. Wolf, Degradation of Misfolded Protein in the Cytoplasm Is Mediated by the Ubiquitin Ligase Ubr1, FEBS Lett., 2008, 582(30), 4143–4146, DOI:10.1016/j.febslet.2008.11.015
.
- J. W. Heck, S. K. Cheung and R. Y. Hampton, Cytoplasmic Protein Quality Control Degradation Mediated by Parallel Actions of the E3 Ubiquitin Ligases Ubr1 and San1, Proc. Natl. Acad. Sci. U. S. A., 2010, 107(3), 1106–1111, DOI:10.1073/pnas.0910591107
.
- Y. D. Yoo, S. R. Mun, C. H. Ji, K. W. Sung, K. Y. Kang, A. J. Heo, S. H. Lee, J. Y. An, J. Hwang, X.-Q. Xie, A. Ciechanover, B. Y. Kim and Y. T. Kwon, N-Terminal Arginylation Generates a Bimodal Degron That Modulates Autophagic Proteolysis, Proc. Natl. Acad. Sci. U. S. A., 2018, 115(12), E2716–E2724, DOI:10.1073/pnas.1719110115
.
- J. Rageul, J. J. Park, U. Jo, A. S. Weinheimer, T. T. M. Vu and H. Kim, Conditional Degradation of SDE2 by the Arg/N-End Rule Pathway Regulates Stress Response at Replication Forks, Nucleic Acids Res., 2019, 47(8), 3996–4010, DOI:10.1093/nar/gkz054
.
- D. Leboeuf, T. Abakumova, T. Prikazchikova, L. Rhym, D. G. Anderson, T. S. Zatsepin and K. I. Piatkov, Downregulation of the Arg/N-Degron Pathway Sensitizes Cancer Cells to Chemotherapy In Vivo, Mol. Ther., 2020, 28(4), 1092–1104, DOI:10.1016/j.ymthe.2020.01.021
.
- C.-S. Hwang, A. Shemorry and A. Varshavsky, N-Terminal Acetylation of Cellular Proteins Creates Specific Degradation Signals, Science, 2010, 327(5968), 973–977, DOI:10.1126/science.1183147
.
- H.-K. Kim, R.-R. Kim, J.-H. Oh, H. Cho, A. Varshavsky and C.-S. Hwang, The N-Terminal Methionine of Cellular Proteins as a Degradation Signal, Cell, 2014, 156(1–2), 158–169, DOI:10.1016/j.cell.2013.11.031
.
- A. Shemorry, C.-S. Hwang and A. Varshavsky, Control of Protein Quality and Stoichiometries by N-Terminal Acetylation and the N-End Rule Pathway, Mol. Cell, 2013, 50(4), 540–551, DOI:10.1016/j.molcel.2013.03.018
.
- R. Ree, S. Varland and T. Arnesen, Spotlight on Protein N-Terminal Acetylation, Exp. Mol. Med., 2018, 50(7), 1–13, DOI:10.1038/s12276-018-0116-z
.
- H. Aksnes, R. Ree and T. Arnesen, Co-Translational, Post-Translational, and Non-Catalytic Roles of N-Terminal Acetyltransferases, Mol. Cell, 2019, 73(6), 1097–1114, DOI:10.1016/j.molcel.2019.02.007
.
- H. Aksnes, A. Drazic, M. Marie and T. Arnesen, First Things First: Vital Protein Marks by N-Terminal Acetyltransferases, Trends Biochem. Sci., 2016, 41(9), 746–760, DOI:10.1016/j.tibs.2016.07.005
.
- S. M. Kienle, T. Schneider, C. Bernecker, J. Bracker, A. Marx, M. Kovermann, M. Scheffner and K. Stuber, Biochemical and Structural Consequences of NEDD8 Acetylation, ChemBioChem, 2024, e202400478, DOI:10.1002/cbic.202400478
.
- D. C. Scott, J. T. Hammill, J. Min, D. Y. Rhee, M. Connelly, V. O. Sviderskiy, D. Bhasin, Y. Chen, S.-S. Ong, S. C. Chai, A. N. Goktug, G. Huang, J. K. Monda, J. Low, H. S. Kim, J. A. Paulo, J. R. Cannon, A. A. Shelat, T. Chen, I. R. Kelsall, A. F. Alpi, V. Pagala, X. Wang, J. Peng, B. Singh, J. W. Harper, B. A. Schulman and R. K. Guy, Blocking an N-Terminal Acetylation–Dependent Protein Interaction Inhibits an E3 Ligase, Nat. Chem. Biol., 2017, 13(8), 850–857, DOI:10.1038/nchembio.2386
.
- K. Hole, P. Van Damme, M. Dalva, H. Aksnes, N. Glomnes, J. E. Varhaug, J. R. Lillehaug, K. Gevaert and T. Arnesen, The Human N-Alpha-Acetyltransferase 40 (hNaa40p/hNatD) Is Conserved from Yeast and N-Terminally Acetylates Histones H2A and H4, PLoS One, 2011, 6(9), e24713, DOI:10.1371/journal.pone.0024713
.
- D. Pavlou and A. Kirmizis, Depletion of Histone N-Terminal-Acetyltransferase Naa40 Induces P53-Independent Apoptosis in Colorectal Cancer Cells via the Mitochondrial Pathway, Apoptosis: Int. J. Program. Cell Death, 2016, 21(3), 298–311, DOI:10.1007/s10495-015-1207-0
.
- C. Demetriadou, D. Pavlou, F. Mpekris, C. Achilleos, T. Stylianopoulos, A. Zaravinos, P. Papageorgis and A. Kirmizis, NAA40 Contributes to Colorectal Cancer Growth by Controlling PRMT5 Expression, Cell Death Dis., 2019, 10(3), 236, DOI:10.1038/s41419-019-1487-3
.
- Y.-H. Ho and R. Huang, Effects of Oncohistone Mutations and PTM Crosstalk on the N-Terminal Acetylation Activities of NatD, ACS Chem. Biol., 2023, 18(4), 693–700, DOI:10.1021/acschembio.1c00840
.
- M. Runfola, A. De Simone, M. Vendruscolo, C. M. Dobson and G. Fusco, The N-Terminal Acetylation of α-Synuclein Changes the Affinity for Lipid Membranes but Not the Structural Properties of the Bound State, Sci. Rep., 2020, 10(1), 204, DOI:10.1038/s41598-019-57023-4
.
- R. Bell, R. J. Thrush, M. Castellana-Cruz, M. Oeller, R. Staats, A. Nene, P. Flagmeier, C. K. Xu, S. Satapathy, C. Galvagnion, M. R. Wilson, C. M. Dobson, J. R. Kumita and M. Vendruscolo, N-Terminal Acetylation of α-Synuclein Slows down Its Aggregation Process and Alters the Morphology of the Resulting Aggregates, Biochemistry, 2022, 61(17), 1743–1756, DOI:10.1021/acs.biochem.2c00104
.
- I. Dikiy and D. Eliezer, N-Terminal Acetylation Stabilizes N-Terminal Helicity in Lipid- and Micelle-Bound α-Synuclein and Increases Its Affinity for Physiological Membranes, J. Biol. Chem., 2014, 289(6), 3652–3665, DOI:10.1074/jbc.M113.512459
.
- J. Burré, The Synaptic Function of α-Synuclein, J. Parkinson’s Dis., 2015, 5(4), 699–713, DOI:10.3233/JPD-150642
.
- S.-J. Chen, X. Wu, B. Wadas, J.-H. Oh and A. Varshavsky, An N-End Rule Pathway That Recognizes Proline and Destroys Gluconeogenic Enzymes, Science, 2017, 355(6323), eaal3655, DOI:10.1126/science.aal3655
.
- C. Dong, S.-J. Chen, A. Melnykov, S. Weirich, K. Sun, A. Jeltsch, A. Varshavsky and J. Min, Recognition of Nonproline N-Terminal Residues by the Pro/N-Degron Pathway, Proc. Natl. Acad. Sci. U. S. A., 2020, 117(25), 14158–14167, DOI:10.1073/pnas.2007085117
.
- J. Chrustowicz, D. Sherpa, J. Teyra, M. S. Loke, G. M. Popowicz, J. Basquin, M. Sattler, J. R. Prabu, S. S. Sidhu and B. A. Schulman, Multifaceted N-Degron Recognition and Ubiquitylation by GID/CTLH E3 Ligases, J. Mol. Biol., 2022, 434(2), 167347, DOI:10.1016/j.jmb.2021.167347
.
- C. Dong, H. Zhang, L. Li, W. Tempel, P. Loppnau and J. Min, Molecular Basis of GID4-Mediated Recognition of Degrons for the Pro/N-End Rule Pathway, Nat. Chem. Biol., 2018, 14(5), 466–473, DOI:10.1038/s41589-018-0036-1
.
- J. S. Shin, S. H. Park, L. Kim, J. Heo and H. K. Song, Crystal Structure of Yeast Gid10 in Complex with Pro/N-Degron, Biochem. Biophys. Res. Commun., 2021, 582, 86–92, DOI:10.1016/j.bbrc.2021.10.007
.
- H. C. Nguyen, W. Wang and Y. Xiong, Cullin-RING E3 Ubiquitin Ligases: Bridges to Destruction, Subcell. Biochem., 2017, 83, 323–347, DOI:10.1007/978-3-319-46503-6_12
.
- D. M. Duda, D. C. Scott, M. F. Calabrese, E. S. Zimmerman, N. Zheng and B. A. Schulman, Structural Regulation of Cullin-RING Ubiquitin Ligase Complexes, Curr. Opin. Struct. Biol., 2011, 21(2), 257–264, DOI:10.1016/j.sbi.2011.01.003
.
- A. Sarikas, T. Hartmann and Z.-Q. Pan, The Cullin Protein Family, Genome Biol., 2011, 12(4), 220, DOI:10.1186/gb-2011-12-4-220
.
- C. P. Lin and E. A. Komives, Diversity of Structure and Function in Cullin E3 Ligases, Curr. Opin. Struct. Biol., 2024, 88, 102879, DOI:10.1016/j.sbi.2024.102879
.
- R. T. Timms, Z. Zhang, D. Y. Rhee, J. W. Harper, I. Koren and S. J. Elledge, A Glycine-Specific N-Degron Pathway Mediates the Quality Control of Protein N-Myristoylation, Science, 2019, 365(6448), eaaw4912, DOI:10.1126/science.aaw4912
.
- H. Harada, K. Moriya, H. Kobuchi, N. Ishihara and T. Utsumi, Protein N-Myristoylation Plays a Critical Role in the Mitochondrial Localization of Human Mitochondrial Complex I Accessory Subunit NDUFB7, Sci. Rep., 2023, 13(1), 22991, DOI:10.1038/s41598-023-50390-z
.
- M. Yuan, Z. Song, M. Ying, H. Zhu, Q. He, B. Yang and J. Cao, N-Myristoylation: From Cell Biology to Translational Medicine, Acta Pharmacol. Sin., 2020, 41(8), 1005–1015, DOI:10.1038/s41401-020-0388-4
.
- N. Hayashi and K. Titani, N-Myristoylated Proteins, Key Components in Intracellular Signal Transduction Systems Enabling Rapid and Flexible Cell Responses, Proc. Jpn. Acad. Ser. B Phys. Biol. Sci., 2010, 86(5), 494–508, DOI:10.2183/pjab.86.494
.
- I. Koren, R. T. Timms, T. Kula, Q. Xu, M. Z. Li and S. J. Elledge, The Eukaryotic Proteome Is Shaped by E3 Ubiquitin Ligases Targeting C-Terminal Degrons, Cell, 2018, 173(7), 1622–1635.e14, DOI:10.1016/j.cell.2018.04.028
.
- D. C. Scott, M. T. King, K. Baek, C. T. Gee, R. Kalathur, J. Li, N. Purser, A. Nourse, S. C. Chai, S. Vaithiyalingam, T. Chen, R. E. Lee, S. J. Elledge, G. Kleiger and B. A. Schulman, E3 Ligase Autoinhibition by C-Degron Mimicry Maintains C-Degron Substrate Fidelity, Mol. Cell, 2023, 83(5), 770–786.e9, DOI:10.1016/j.molcel.2023.01.019
.
- D.-V. Rusnac, H.-C. Lin, D. Canzani, K. Tien, T. R. Hinds, A. F. Tsue, M. F. Bush, H.-C. S. Yen and N. Zheng, Recognition of Diglycine C-End Degron by CRL2KLHDC2 Ubiquitin Ligase, Mol. Cell, 2018, 72(5), 813–822.e4, DOI:10.1016/j.molcel.2018.10.021
.
- D. Sherpa, J. Chrustowicz and B. A. Schulman, How the Ends Signal the End: Regulation by E3 Ubiquitin Ligases Recognizing Protein Termini, Mol. Cell, 2022, 82(8), 1424–1438, DOI:10.1016/j.molcel.2022.02.004
.
- C. Yeh, W. Huang, P. Hsu, K. Yeh, L. Wang, P. W. Hsu, H. Lin, Y. Chen, S. Chen, C. Yeang and H. S. Yen, The C-degron Pathway Eliminates Mislocalized Proteins and Products of Deubiquitinating Enzymes, EMBO J., 2021, 40(7), e105846, DOI:10.15252/embj.2020105846
.
- Y. Ru, X. Yan, B. Zhang, L. Song, Q. Feng, C. Ye, Z. Zhou, Z. Yang, Y. Li, Z. Zhang, Q. Li, W. Mi and C. Dong, C-Terminal Glutamine Acts as a C-Degron Targeted by E3 Ubiquitin Ligase TRIM7, Proc. Natl. Acad. Sci. U. S. A., 2022, 119(30), e2203218119, DOI:10.1073/pnas.2203218119
.
- X. Liang, J. Xiao, X. Li, Y. Liu, Y. Lu, Y. Wen, Z. Li, X. Che, Y. Ma, X. Zhang, Y. Zhang, D. Jian, P. Wang, C. Xuan, G. Yu, L. Li and H. Zhang, A C-Terminal Glutamine Recognition Mechanism Revealed by E3 Ligase TRIM7 Structures, Nat. Chem. Biol., 2022, 18(11), 1214–1223, DOI:10.1038/s41589-022-01128-x
.
- Y. Liu, L. Jiang, X. Sun, Y. Song, Y. Liu and L. Zhang, Interplay between TRIM7 and Antiviral Immunity, Front. Cell. Infect. Microbiol., 2023, 13 DOI:10.3389/fcimb.2023.1256882
.
- W. Fan, K. B. Mar, L. Sari, I. K. Gaszek, Q. Cheng, B. M. Evers, J. M. Shelton, M. Wight-Carter, D. J. Siegwart, M. M. Lin and J. W. Schoggins, TRIM7 Inhibits Enterovirus Replication and Promotes Emergence of a Viral Variant with Increased Pathogenicity, Cell, 2021, 184(13), 3410–3425.e17, DOI:10.1016/j.cell.2021.04.047
.
- M. Lu, A. Ma, J. Liu, W. Zhou, P. Cao, T. Chu and L. Fan, Study on the Expression of TRIM7 in Peripheral Blood Mononuclear Cells of Patients with Sepsis and Its Early Diagnostic Value, BMC Infect. Dis., 2022, 22(1), 865, DOI:10.1186/s12879-022-07874-6
.
- M. Li, J. Yan, H. Zhu, C. Guo, X. Jiang, Y. Gao, X. Liu, P. Jiang and J. Bai, TRIM7 Inhibits Encephalomyocarditis Virus Replication by Activating Interferon-β Signaling Pathway, Vet. Microbiol., 2023, 281, 109729, DOI:10.1016/j.vetmic.2023.109729
.
- C. Yuan, J. Liu, L. Liu, H. Jia, Q. Gao, X. Wang and J. Zhao, TRIM7 Suppresses Cell Invasion and Migration through Inhibiting HIF-1α Accumulation in Clear Cell Renal Cell Carcinoma, Cell Biol. Int., 2022, 46(4), 554–567, DOI:10.1002/cbin.11750
.
- R. Ji, Y. Gu, J. Zhang, C. Gao, W. Gao, X. Zang and Y. Zhao, TRIM7 Promotes Proliferation and Migration of Vascular Smooth Muscle Cells in Atherosclerosis through Activating C-Jun/AP-1, IUBMB Life, 2020, 72(2), 247–258, DOI:10.1002/iub.2181
.
- R. T. Timms, E. L. Mena, Y. Leng, M. Z. Li, I. A. Tchasovnikarova, I. Koren and S. J. Elledge, Defining E3 Ligase–Substrate Relationships through Multiplex CRISPR Screening, Nat. Cell Biol., 2023, 25(10), 1535–1545, DOI:10.1038/s41556-023-01229-2
.
- T. Lidak, N. Baloghova, V. Korinek, R. Sedlacek, J. Balounova, P. Kasparek and L. Cermak, CRL4-DCAF12 Ubiquitin Ligase Controls MOV10 RNA Helicase during Spermatogenesis and T Cell Activation, Int. J. Mol. Sci., 2021, 22(10), 5394, DOI:10.3390/ijms22105394
.
- G. L. Righetto, Y. Yin, D. M. Duda, V. Vu, M. M. Szewczyk, H. Zeng, Y. Li, P. Loppnau, T. Mei, Y.-Y. Li, A. Seitova, A. N. Patrick, J.-F. Brazeau, C. Chaudhry, D. Barsyte-Lovejoy, V. Santhakumar and L. Halabelian, Probing the CRL4DCAF12 Interactions with MAGEA3 and CCT5 Di-Glu C-Terminal Degrons, PNAS Nexus, 2024, 3(4), pgae153, DOI:10.1093/pnasnexus/pgae153
.
- S. H. Choi, J. B. Wright, S. A. Gerber and M. D. Cole, Myc Protein Is Stabilized by Suppression of a Novel E3 Ligase Complex in Cancer Cells, Genes Dev., 2010, 24(12), 1236–1241, DOI:10.1101/gad.1920310
.
- X. Chen, S. Liao, Y. Makaros, Q. Guo, Z. Zhu, R. Krizelman, K. Dahan, X. Tu, X. Yao, I. Koren and C. Xu, Molecular Basis for Arginine C-Terminal Degron Recognition by Cul2FEM1 E3 Ligase, Nat. Chem. Biol., 2021, 17(3), 254–262, DOI:10.1038/s41589-020-00704-3
.
- A. G. Manford, E. L. Mena, K. Y. Shih, C. L. Gee, R. McMinimy, B. Martínez-González, R. Sherriff, B. Lew, M. Zoltek, F. Rodríguez-Pérez, M. Woldesenbet, J. Kuriyan and M. Rape, Structural Basis and Regulation of the Reductive Stress Response, Cell, 2021, 184(21), 5375–5390.e16, DOI:10.1016/j.cell.2021.09.002
.
- Q. Shi and L. Chen, Cereblon: A Protein Crucial to the Multiple Functions of Immunomodulatory Drugs as Well as Cell Metabolism and Disease Generation, J. Immunol. Res., 2017, 2017, 9130608, DOI:10.1155/2017/9130608
.
- D. Weber, K. Rankin, M. Gavino, K. Delasalle and R. Alexanian, Thalidomide Alone or with Dexamethasone for Previously Untreated Multiple Myeloma, J. Clin. Oncol., 2003, 21(1), 16–19, DOI:10.1200/JCO.2003.03.139
.
- A. Palumbo, L. Giaccone, A. Bertola, P. Pregno, S. Bringhen, C. Rus, S. Triolo, E. Gallo, A. Pileri and M. Boccadoro, Low-Dose Thalidomide plus Dexamethasone Is an Effective Salvage Therapy for Advanced Myeloma, Haematologica, 2001, 86(4), 399–403 Search PubMed
.
- T. Paravar and D. J. Lee, Thalidomide: Mechanisms of Action, Int. Rev. Immunol., 2008, 27(3), 111–135, DOI:10.1080/08830180801911339
.
- N. Vargesson, Thalidomide-Induced Teratogenesis: History and Mechanisms, Birth Defects Res., Part C, 2015, 105(2), 140–156, DOI:10.1002/bdrc.21096
.
- T. Ito and H. Handa, Molecular Mechanisms of Thalidomide and Its Derivatives, Proc. Jpn. Acad. Ser. B Phys. Biol. Sci., 2020, 96(6), 189–203, DOI:10.2183/pjab.96.016
.
- K. C. Anderson, Lenalidomide and Thalidomide: Mechanisms of Action--Similarities and Differences, Semin. Hematol., 2005, 42(4 Suppl 4), S3–S8, DOI:10.1053/j.seminhematol.2005.10.001
.
- T. Mori, T. Ito, S. Liu, H. Ando, S. Sakamoto, Y. Yamaguchi, E. Tokunaga, N. Shibata, H. Handa and T. Hakoshima, Structural Basis of Thalidomide Enantiomer Binding to Cereblon, Sci. Rep., 2018, 8(1), 1294, DOI:10.1038/s41598-018-19202-7
.
- J. Yamamoto, T. Ito, Y. Yamaguchi and H. Handa, Discovery of CRBN as a Target of Thalidomide: A Breakthrough for Progress in the Development of Protein Degraders, Chem. Soc. Rev., 2022, 51(15), 6234–6250, 10.1039/D2CS00116K
.
- M. D. Hartmann, I. Boichenko, M. Coles, F. Zanini, A. N. Lupas and B. Hernandez Alvarez, Thalidomide Mimics Uridine Binding to an Aromatic Cage in Cereblon, J. Struct. Biol., 2014, 188(3), 225–232, DOI:10.1016/j.jsb.2014.10.010
.
- S. A. Holstein and P. L. McCarthy, Immunomodulatory Drugs in Multiple Myeloma: Mechanisms of Action and Clinical Experience, Drugs, 2017, 77(5), 505–520, DOI:10.1007/s40265-017-0689-1
.
- H. Quach, D. Ritchie, A. Stewart, P. Neeson, S. Harrison, M. Smyth and H. Prince, Mechanism of Action of Immunomodulatory Drugs (IMiDS) in Multiple Myeloma, Leukemia, 2010, 24(1), 22–32, DOI:10.1038/leu.2009.236
.
- J. B. Bartlett, K. Dredge and A. G. Dalgleish, The Evolution of Thalidomide and Its IMiD Derivatives as Anticancer Agents, Nat. Rev. Cancer, 2004, 4(4), 314–322, DOI:10.1038/nrc1323
.
- C. Shen, A. Nayak, L. R. Neitzel, A. A. Adams, M. Silver-Isenstadt, L. M. Sawyer, H. Benchabane, H. Wang, N. Bunnag, B. Li, D. T. Wynn, F. Yang, M. Garcia-Contreras, C. H. Williams, S. Dakshanamurthy, C. C. Hong, N. G. Ayad, A. J. Capobianco, Y. Ahmed, E. Lee and D. J. Robbins, The E3 Ubiquitin Ligase Component, Cereblon, Is an Evolutionarily Conserved Regulator of Wnt Signaling, Nat. Commun., 2021, 12(1), 5263, DOI:10.1038/s41467-021-25634-z
.
- L. E. Franssen, I. S. Nijhof, S. Couto, M.-D. Levin, G. M. J. Bos, A. Broijl, S. K. Klein, Y. Ren, M. Wang, H. R. Koene, A. C. Bloem, A. Beeker, L. M. Faber, E. van der Spek, R. Raymakers, R. J. Leguit, P. Sonneveld, S. Zweegman, H. Lokhorst, T. Mutis, A. Thakurta, X. Qian and N. W. C. J. van de Donk, Cereblon Loss and Up-Regulation of c-Myc Are Associated with Lenalidomide Resistance in Multiple Myeloma Patients, Haematologica, 2018, 103(8), e368–e371, DOI:10.3324/haematol.2017.186601
.
- P. P. Chamberlain, L. A. D'Agostino, J. M. Ellis, J. D. Hansen, M. E. Matyskiela, J. J. McDonald, J. R. Riggs and L. G. Hamann, Evolution of Cereblon-Mediated Protein Degradation as a Therapeutic Modality, ACS Med. Chem. Lett., 2019, 10(12), 1592–1602, DOI:10.1021/acsmedchemlett.9b00425
.
- T. Ito, Y. Yamaguchi and H. Handa, Exploiting Ubiquitin Ligase Cereblon as a Target for Small-Molecule Compounds in Medicine and Chemical Biology, Cell Chem. Biol., 2021, 28(7), 987–999, DOI:10.1016/j.chembiol.2021.04.012
.
- E. S. Fischer, K. Böhm, J. R. Lydeard, H. Yang, M. B. Stadler, S. Cavadini, J. Nagel, F. Serluca, V. Acker, G. M. Lingaraju, R. B. Tichkule, M. Schebesta, W. C. Forrester, M. Schirle, U. Hassiepen, J. Ottl, M. Hild, R. E. J. Beckwith, J. W. Harper, J. L. Jenkins and N. H. Thomä, Structure of the DDB1–CRBN E3 Ubiquitin Ligase in Complex with Thalidomide, Nature, 2014, 512(7512), 49–53, DOI:10.1038/nature13527
.
- M. Dijon, F. Bardin, A. Murati, M. Batoz, C. Chabannon and C. Tonnelle, The Role of Ikaros in Human Erythroid Differentiation, Blood, 2008, 111(3), 1138–1146, DOI:10.1182/blood-2007-07-098202
.
- L. B. John and A. C. Ward, The Ikaros Gene Family: Transcriptional Regulators of Hematopoiesis and Immunity, Mol. Immunol., 2011, 48(9), 1272–1278, DOI:10.1016/j.molimm.2011.03.006
.
- Q. L. Sievers, G. Petzold, R. D. Bunker, A. Renneville, M. Słabicki, B. J. Liddicoat, W. Abdulrahman, T. Mikkelsen, B. L. Ebert and N. H. Thomä, Defining the Human C2H2 Zinc-Finger Degrome Targeted by Thalidomide Analogs through CRBN, Science, 2018, 362(6414), eaat0572, DOI:10.1126/science.aat0572
.
- Y. Fan and D. Lu, The Ikaros Family of Zinc-Finger Proteins, Acta Pharm.
Sin. B, 2016, 6(6), 513–521, DOI:10.1016/j.apsb.2016.06.002
.
- J. Krönke, E. C. Fink, P. W. Hollenbach, K. J. MacBeth, S. N. Hurst, N. D. Udeshi, P. P. Chamberlain, D. R. Mani, H. W. Man, A. K. Gandhi, T. Svinkina, R. K. Schneider, M. McConkey, M. Järås, E. Griffiths, M. Wetzler, L. Bullinger, B. E. Cathers, S. A. Carr, R. Chopra and B. L. Ebert, Lenalidomide Induces Ubiquitination and Degradation of CK1α in Del(5q) MDS, Nature, 2015, 523(7559), 183–188, DOI:10.1038/nature14610
.
- J. Krönke, N. D. Udeshi, A. Narla, P. Grauman, S. N. Hurst, M. McConkey, T. Svinkina, D. Heckl, E. Comer, X. Li, C. Ciarlo, E. Hartman, N. Munshi, M. Schenone, S. L. Schreiber, S. A. Carr and B. L. Ebert, Lenalidomide Causes Selective Degradation of IKZF1 and IKZF3 in Multiple Myeloma Cells, Science, 2014, 343(6168), 301–305, DOI:10.1126/science.1244851
.
- J. An, C. M. Ponthier, R. Sack, J. Seebacher, M. B. Stadler, K. A. Donovan and E. S. Fischer, pSILAC Mass Spectrometry Reveals ZFP91 as IMiD-Dependent Substrate of the CRL4CRBN Ubiquitin Ligase, Nat. Commun., 2017, 8(1), 15398, DOI:10.1038/ncomms15398
.
- T. H. Maren, Relations Between Structure and Biological Activity of Sulfonamides, Annu. Rev. Pharmacol. Toxicol., 1976, 16, 309–327, DOI:10.1146/annurev.pa.16.040176.001521
.
- T. Owa, H. Yoshino, T. Okauchi, K. Yoshimatsu, Y. Ozawa, N. H. Sugi, T. Nagasu, N. Koyanagi and K. Kitoh, Discovery of Novel Antitumor Sulfonamides Targeting G1 Phase of the Cell Cycle, J. Med. Chem., 1999, 42(19), 3789–3799, DOI:10.1021/jm9902638
.
- T. Han, M. Goralski, N. Gaskill, E. Capota, J. Kim, T. C. Ting, Y. Xie, N. S. Williams and D. Nijhawan, Anticancer Sulfonamides Target Splicing by Inducing RBM39 Degradation via Recruitment to DCAF15, Science, 2017, 356(6336), eaal3755, DOI:10.1126/science.aal3755
.
- D. E. Bussiere, L. Xie, H. Srinivas, W. Shu, A. Burke, C. Be, J. Zhao, A. Godbole, D. King, R. G. Karki, V. Hornak, F. Xu, J. Cobb, N. Carte, A. O. Frank, A. Frommlet, P. Graff, M. Knapp, A. Fazal, B. Okram, S. Jiang, P.-Y. Michellys, R. Beckwith, H. Voshol, C. Wiesmann, J. M. Solomon and J. Paulk, Structural Basis of Indisulam-Mediated RBM39 Recruitment to DCAF15 E3 Ligase Complex, Nat. Chem. Biol., 2020, 16(1), 15–23, DOI:10.1038/s41589-019-0411-6
.
- K. R. Simonetta, J. Taygerly, K. Boyle, S. E. Basham, C. Padovani, Y. Lou, T. J. Cummins, S. L. Yung, S. K. von Soly, F. Kayser, J. Kuriyan, M. Rape, M. Cardozo, M. A. Gallop, N. F. Bence, P. A. Barsanti and A. Saha, Prospective Discovery of Small Molecule Enhancers of an E3 Ligase-Substrate Interaction, Nat. Commun., 2019, 10(1), 1402, DOI:10.1038/s41467-019-09358-9
.
- C. Liu, Y. Li, M. Semenov, C. Han, G.-H. Baeg, Y. Tan, Z. Zhang, X. Lin and X. He, Control of β-Catenin Phosphorylation/Degradation by a Dual-Kinase Mechanism, Cell, 2002, 108(6), 837–847, DOI:10.1016/S0092-8674(02)00685-2
.
- V. S. W. Li, S. S. Ng, P. J. Boersema, T. Y. Low, W. R. Karthaus, J. P. Gerlach, S. Mohammed, A. J. R. Heck, M. M. Maurice, T. Mahmoudi and H. Clevers, Wnt Signaling through Inhibition of β-Catenin Degradation in an Intact Axin1 Complex, Cell, 2012, 149(6), 1245–1256, DOI:10.1016/j.cell.2012.05.002
.
- N.-C. Ha, T. Tonozuka, J. L. Stamos, H.-J. Choi and W. I. Weis, Mechanism of Phosphorylation-Dependent Binding of APC to β-Catenin and Its Role in β-Catenin Degradation, Mol. Cell, 2004, 15(4), 511–521, DOI:10.1016/j.molcel.2004.08.010
.
- S. Amit, A. Hatzubai, Y. Birman, J. S. Andersen, E. Ben-Shushan, M. Mann, Y. Ben-Neriah and I. Alkalay, Axin-Mediated CKI Phosphorylation of β-Catenin at Ser 45: A Molecular Switch for the Wnt Pathway, Genes Dev., 2002, 16(9), 1066–1076, DOI:10.1101/gad.230302
.
- R. J. Deshaies and C. A. P. Joazeiro, RING Domain E3 Ubiquitin Ligases, Annu. Rev. Biochem., 2009, 78, 399–434, DOI:10.1146/annurev.biochem.78.101807.093809
.
- B. Rubinfeld, P. Robbins, M. El-Gamil, I. Albert, E. Porfiri and P. Polakis, Stabilization of β-Catenin by Genetic Defects in Melanoma Cell Lines, Science, 1997, 275(5307), 1790–1792, DOI:10.1126/science.275.5307.1790
.
- G. Wu, G. Xu, B. A. Schulman, P. D. Jeffrey, J. W. Harper and N. P. Pavletich, Structure of a β-TrCP1-Skp1-β-Catenin Complex: Destruction Motif Binding and Lysine Specificity of the SCFβ-TrCP1 Ubiquitin Ligase, Mol. Cell, 2003, 11(6), 1445–1456, DOI:10.1016/S1097-2765(03)00234-X
.
- R. Dhanasekaran, A. Deutzmann, W. D. Mahauad-Fernandez, A. S. Hansen, A. M. Gouw and D. W. Felsher, The MYC Oncogene — the Grand Orchestrator of Cancer Growth and Immune Evasion, Nat. Rev. Clin. Oncol., 2022, 19(1), 23–36, DOI:10.1038/s41571-021-00549-2
.
- M. Welcker, B. Wang, D.-V. Rusnac, Y. Hussaini, J. Swanger, N. Zheng and B. E. Clurman, Two Diphosphorylated Degrons Control C-Myc Degradation by the Fbw7 Tumor Suppressor, Sci. Adv., 2022, 8(4), eabl7872, DOI:10.1126/sciadv.abl7872
.
- K. Shimizu, N. T. Nihira, H. Inuzuka and W. Wei, Physiological Functions of FBW7 in Cancer and Metabolism, Cell. Signalling, 2018, 46, 15–22, DOI:10.1016/j.cellsig.2018.02.009
.
- S. Guo, M. Stilgenbauer, B. Wu, Y. Mankan and A. N. Koehler, Abstract 6587: Discovery of Molecular Glue of FBXW7:MYC Interaction Using Small Molecule Microarray, Cancer Res., 2024, 84(6_Supplement), 6587, DOI:10.1158/1538-7445.AM2024-6587
.
- J. Zhang, C. Ma, Y. Yu, C. Liu, L. Fang and H. Rao, Single Amino Acid–Based PROTACs Trigger Degradation of the Oncogenic Kinase BCR–ABL in Chronic Myeloid Leukemia (CML), J. Biol. Chem., 2023, 299(8), 104994, DOI:10.1016/j.jbc.2023.104994
.
- D. Cilloni and G. Saglio, Molecular Pathways: BCR-ABL, Clin. Cancer Res., 2012, 18(4), 930–937, DOI:10.1158/1078-0432.CCR-10-1613
.
- Z. Yan, K. Shanmugasundaram, D. Ma, J. Luo, S. Luo and H. Rao, The N-Terminal Domain of the Non-Receptor Tyrosine Kinase ABL Confers Protein Instability and Suppresses Tumorigenesis, J. Biol. Chem., 2020, 295(27), 9069–9075, DOI:10.1074/jbc.RA120.012821
.
- D. M. Ross and T. P. Hughes, Treatment-Free Remission in Patients with Chronic Myeloid Leukaemia, Nat. Rev. Clin. Oncol., 2020, 17(8), 493–503, DOI:10.1038/s41571-020-0367-1
.
- G. Q. Daley, R. A. Van Etten and D. Baltimore, Induction of Chronic Myelogenous Leukemia in Mice by the P210bcr/Abl Gene of the Philadelphia Chromosome, Science, 1990, 247(4944), 824–830, DOI:10.1126/science.2406902
.
- G. P. Amarante-Mendes, A. Rana, T. S. Datoguia, N. Hamerschlak and G. Brumatti, BCR-ABL1 Tyrosine Kinase Complex Signaling Transduction: Challenges to Overcome Resistance in Chronic Myeloid Leukemia, Pharmaceutics, 2022, 14(1), 215, DOI:10.3390/pharmaceutics14010215
.
- A. A. Wylie, J. Schoepfer, W. Jahnke, S. W. Cowan-Jacob, A. Loo, P. Furet, A. L. Marzinzik, X. Pelle, J. Donovan, W. Zhu, S. Buonamici, A. Q. Hassan, F. Lombardo, V. Iyer, M. Palmer, G. Berellini, S. Dodd, S. Thohan, H. Bitter, S. Branford, D. M. Ross, T. P. Hughes, L. Petruzzelli, K. G. Vanasse, M. Warmuth, F. Hofmann, N. J. Keen and W. R. Sellers, The Allosteric Inhibitor ABL001 Enables Dual Targeting of BCR-ABL1, Nature, 2017, 543(7647), 733–737, DOI:10.1038/nature21702
.
- N. Gao, T.-T. Chu, Q.-Q. Li, Y.-J. Lim, T. Qiu, M.-R. Ma, Z.-W. Hu, X.-F. Yang, Y.-X. Chen, Y.-F. Zhao and Y.-M. Li, Hydrophobic Tagging-Mediated Degradation of Alzheimer's Disease Related Tau, RSC Adv., 2017, 7(64), 40362–40366, 10.1039/C7RA05347A
.
- S. R. Choi, H. M. Wang, M. H. Shin and H.-S. Lim, Hydrophobic Tagging-Mediated Degradation of Transcription Coactivator SRC-1, Int. J. Mol. Sci., 2021, 22(12), 6407, DOI:10.3390/ijms22126407
.
- K. Hirai, H. Yamashita, S. Tomoshige, Y. Mishima, T. Niwa, K. Ohgane, M. Ishii, K. Kanamitsu, Y. Ikemi, S. Nakagawa, H. Taguchi, S. Sato, Y. Hashimoto and M. Ishikawa, Conversion of a PROTAC Mutant Huntingtin Degrader into Small-Molecule Hydrophobic Tags Focusing on Drug-like Properties, ACS Med. Chem. Lett., 2022, 13(3), 396–402, DOI:10.1021/acsmedchemlett.1c00500
.
- T. Shoda, N. Ohoka, G. Tsuji, T. Fujisato, H. Inoue, Y. Demizu, M. Naito and M. Kurihara, Targeted Protein Degradation by Chimeric Compounds Using Hydrophobic E3 Ligands and Adamantane Moiety, Pharmaceuticals, 2020, 13(3), 34, DOI:10.3390/ph13030034
.
- Y. Shi, M. J. C. Long, M. M. Rosenberg, S. Li, A. Kobjack, P. Lessans, R. T. Coffey and L. Hedstrom, Boc3Arg-Linked Ligands Induce Degradation by Localizing Target Proteins to the 20S Proteasome, ACS Chem. Biol., 2016, 11(12), 3328–3337, DOI:10.1021/acschembio.6b00656
.
- C. Bashore, S. Prakash, M. C. Johnson, R. J. Conrad, I. A. Kekessie, S. J. Scales, N. Ishisoko, T. Kleinheinz, P. S. Liu, N. Popovych, A. T. Wecksler, L. Zhou, C. Tam, I. Zilberleyb, R. Srinivasan, R. A. Blake, A. Song, S. T. Staben, Y. Zhang, D. Arnott, W. J. Fairbrother, S. A. Foster, I. E. Wertz, C. Ciferri and E. C. Dueber, Targeted Degradation via Direct 26S Proteasome Recruitment, Nat. Chem. Biol., 2023, 19(1), 55–63, DOI:10.1038/s41589-022-01218-w
.
- C. A. Loy, C. S. Muli, E. M. H. Ali, D. Xie, M. H. Ahmed, C. Beth Post and D. J. Trader, Discovery of a Non-Covalent Ligand for Rpn-13, a Therapeutic Target for Hematological Cancers, Bioorg. Med. Chem. Lett., 2023, 95, 129485, DOI:10.1016/j.bmcl.2023.129485
.
- E. M. H. Ali, C. A. Loy and D. J. Trader, ByeTAC: Bypassing an E3 Ligase for Targeted Protein Degradation, bioRxiv, January 21, 2024, preprint, p. 2024.01.20.576376, DOI:10.1101/2024.01.20.576376.
- M. Balzarini, J. Tong, W. Gui, I. M. Jayalath, B.-B. Schell and T. Kodadek, Recruitment to the Proteasome Is Necessary but Not Sufficient for Chemically Induced, Ubiquitin-Independent Degradation of Native Proteins, ACS Chem. Biol., 2024, 19(11), 2323–2335, DOI:10.1021/acschembio.4c00422
.
- R. T. Timms and I. Koren, Tying up Loose Ends: The N-Degron and C-Degron Pathways of Protein Degradation, Biochem. Soc. Trans., 2020, 48(4), 1557–1567, DOI:10.1042/BST20191094
.
- C. H. Ji, H. Y. Kim, M. J. Lee, A. J. Heo, D. Y. Park, S. Lim, S. Shin, S. Ganipisetti, W. S. Yang, C. A. Jung, K. Y. Kim, E. H. Jeong, S. H. Park, S. Bin Kim, S. J. Lee, J. E. Na, J. I. Kang, H. M. Chi, H. T. Kim, Y. K. Kim, B. Y. Kim and Y. T. Kwon, The AUTOTAC Chemical Biology Platform for Targeted Protein Degradation via the Autophagy-Lysosome System, Nat. Commun., 2022, 13(1), 904, DOI:10.1038/s41467-022-28520-4
.
- X. Li, Q. Liu, X. Xie, C. Peng, Q. Pang, B. Liu and B. Han, Application of Novel Degraders Employing Autophagy for Expediting Medicinal Research, J. Med. Chem., 2023, 66(3), 1700–1711, DOI:10.1021/acs.jmedchem.2c01712
.
| This journal is © The Royal Society of Chemistry 2025 |