 Open Access Article
Open Access ArticleAn engineered platform to study the influence of extracellular matrix nanotopography on cell ultrastructure†
Shani
Tcherner Elad
a,
Rita
Vilensky
a,
Noa Ben
Asher
 a,
Nataliya
Logvina
a,
Nataliya
Logvina
 a,
Ran
Zalk
a,
Ran
Zalk
 b,
Eyal
Zussman
a and
Leeya
Engel
b,
Eyal
Zussman
a and
Leeya
Engel
 *ac
*ac
aFaculty of Mechanical Engineering, Technion – Israel Institute of Technology, Haifa 3200003, Israel. E-mail: Leeya@technion.ac.il
bIlse Katz Institute for Nanoscale Science and Technology, Ben-Gurion University of the Negev, Beer Sheva, 8410501, Israel
cThe Resnick Sustainability Center for Catalysis, Technion – Israel Institute of Technology, Haifa 3200003, Israel
First published on 27th May 2025
Abstract
Nanoscale fabrication techniques have played an essential role in revealing the impact of extracellular matrix (ECM) nanotopography on cellular behavior. However, the mechanisms by which nanotopographical cues from the ECM influence cellular function remain unclear. To approach these questions, we have engineered a novel class of nanopatterned ECM constructs suitable for cryogenic electron tomography (cryo-ET), the highest resolution modality for imaging frozen hydrated cells in 3D. We electrospun aligned and randomly oriented ECM fibers directly onto transmission electron microscopy (TEM) supports to generate fibrous scaffolds that mimic physiological ECM in healthy (organized ECM) and diseased (disorganized ECM) states. We produced fibers from gelatin without toxic additives and cross-linked them to maintain structural stability in aqueous environments. The electrospun fibers had an average fiber diameter of hundreds of nanometers. We confirmed that the nanopatterned TEM supports can serve as viable cell culture substrates that can influence cell organization and demonstrated their compatibility with plunge freezing and cryo-ET. By enabling nanoscale structural analysis inside cells on substrates with programmable topographies, this platform can be used to study the physical cues necessary for healthy endothelial tissue formation and pathologies that are linked to endothelial dysfunction in diseases such as peripheral arterial disease.
1. Introduction
Engineered ECM microenvironments have dramatically advanced our understanding of how micro- and nano-scale features guide cell organization and modulate cellular function.1–4 Nanotopographical cues influence cell polarity, adhesion,5 migration,6 proliferation,7 and differentiation.8 Disruptions to ECM structure can negatively impact development, tissue repair, and disease progression.9 In endothelial cells, which line all blood and lymphatic vessels, aligned nanofibrillar scaffolds have been shown to induce parallel alignment to mimic the elongated, atherosclerosis-resistant phenotype of cells within straight vessel segments.10,11 This nanofibril alignment reorganizes the cytoskeleton along the nanofibril direction and results in an endothelial phenotype that is resistant to inflammation and flow-induced shear stress.12While micro- and nanostructured cell culture scaffolds inform tissue engineering design choices, there remain gaps in our fundamental understanding of the mechanisms by which nanotopographical cues from the ECM modulate cell behavior and organization.13,14 Cell–ECM adhesion sites interface between the extracellular chemical and physical environment and intracellular cytoskeletal and signaling networks, with mechanotransduction orchestrated primarily by membrane receptors called integrins. Many cell adhesive and mechanotransductive processes are determined at the nanoscale, such as integrin clustering, integrin adhesion complex maturation, and cytoskeletal remodeling. Recent findings have highlighted how strongly nanotopography regulates mechanotransductive processes and how important it is to improve our understanding of these processes at the nanoscale,15,16 for example in cancer progression.17 Combining high-resolution electron microscopy, which surpasses the resolution of even advanced light microscopy techniques, with engineered ECM constructs could contribute to closing these knowledge gaps.
Cryo-ET is a rapidly developing cryogenic transmission electron microscopy (TEM) technique uniquely capable of visualizing the interior of intact cells in 3D at the nano- and subnanometer scale without the need for fixation, dehydration, or staining.18–21 This technique is revolutionizing our structural understanding of the cell interior with major implications for mechanobiology, for example in elucidating the architecture of the cytoskeleton22 and cell–ECM contacts.23,24
To culture adherent cells for cryo-ET, cells are typically grown on electron microscopy (EM) grids, small metal mesh disks, 3.05 mm in diameter, that are overlaid with a porous foil of carbon, silicon dioxide, or gold. The foil typically features a periodic array of 1–2 μm wide holes (e.g., Quantifoil), through which the electron beam passes during imaging.25 Adherent mammalian cells cultured on EM grids can span several holes, enabling imaging of multiple intact regions of the cell in situ. EM grids were originally developed for cryo-TEM of purified proteins; thus, their synthetic porous thin film coating offers none of the rich physical cues that are present in vivo. EM grids can be coated with ECM proteins prior to cell seeding to promote cell attachment. It can be challenging to achieve even distributions of cells on EM grids as the cells often attach to the grid bars, making them inaccessible to the electron beam for imaging. In addition, clumping of cells can create thick regions that do not vitrify properly during plunge freezing. We and others have developed methods to micropattern EM grids with two-dimensional islands of ECM proteins to direct cell shape and distribution on EM grids for structural analysis with cryo-ET.26–29 In a previous study we applied EM grid micropatterning to streamline preparation of endothelial cell-cell contacts for cryo-ET.27 ECM micropatterning can recapitulate certain mechanical aspects of the cell microenvironment (e.g., spatial constraints from neighboring cells), but it cannot modulate substrate nanotopography. In addition, because micropatterning relies on photo-patterning, it is limited to a resolution of approximately 1–1.5 μm.
Electrospinning is a versatile method for producing polymer fibers by subjecting droplets of viscoelastic polymer solution to high voltage electrostatic fields.30 It has been applied to generate fibrous scaffolds for tissue engineering.23,31,32 Electrospun fiber morphology depends on the viscoelasticity of the solution, the charge density carried by the jet, and the surface tension of the solution.33
Here we present a new class of nanofibrillar supports customized for imaging frozen hydrated cells with cryo-ET (see Fig. 1). We generate nanofibers using electrospinning and align them directly onto EM grids with a rotating collection disk.34 Our ability to tune the degree of organization of the gelatin nanofibers by varying the rotation speed of the disk faciliates 3D high-resolution imaging of the interior of endothelial cells cultured on thin films with different nanotopographies. This platform can be utilized to study the nanoscale underpinnings of cellular sensitivity to substrate nanotopography as well as the downstream effects of disruptions to ECM organization on cell ultrastructure.
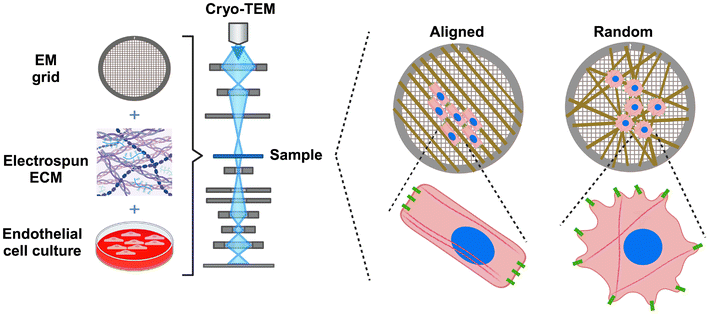 | ||
| Fig. 1 Schematic depicts a platform for high resolution cryo-TEM imaging of cells grown on ECM with different topographical cues. | ||
2. Materials and methods
2.1. Electrospinning gelating on EM grids
We electrospun gelatin fibers on gold grids coated with a porous gold thin film (UltrAuFoil R2/2, Quantifoil). EM grids are available in a range of conductive materials such as copper, but gold is preferred for cell culture due to its low toxicity. Grids with gold foil were used as they mitigate the movement of frozen specimens during imaging relative to non-conductive foils such as carbon.35 We cleaned the grids using atmospheric plasma treatment using a benchtop plasma system (PE-50, Plasma Etch, Inc.) for 30 seconds, assisted by a custom holder (see Fig. 2). To secure the grids during plasma exposure and electrospinning, we applied polydimethylsiloxane (PDMS) in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 ratio on top of a scanning electron microscopy (SEM) stub and cured it at room temperature for 24 hours to create an adhesive surface onto which the grids were mounted.
10 ratio on top of a scanning electron microscopy (SEM) stub and cured it at room temperature for 24 hours to create an adhesive surface onto which the grids were mounted.
We produced smooth fibers by electrospinning a 28% gelatin solution made of 0.84 g of porcine skin-derived gelatin (Sigma-Aldrich, Type A, Bloom Strength 170–190 g) dissolved in 0.6 mL of acetic acid and 2.4 mL of DI water (8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 v/v).36,37 The solution was stirred for 30–60 minutes on a hot plate maintained at approximately 39 °C. It is essential to keep the solution warm to prevent gelation prior to electrospinning. Because higher concentrations of gelatin reduce the formation of beads in the fibers, we initially used a 30% gelatin solution following literature precedent.36 However, in our electrospinning setup, the 30% solution was excessively viscous and difficult to spin. Reducing the gelatin concentration to 28% yielded a stable electrospinning process with a smooth fiber morphology, free from “beads-on-string” type defects.38
2 v/v).36,37 The solution was stirred for 30–60 minutes on a hot plate maintained at approximately 39 °C. It is essential to keep the solution warm to prevent gelation prior to electrospinning. Because higher concentrations of gelatin reduce the formation of beads in the fibers, we initially used a 30% gelatin solution following literature precedent.36 However, in our electrospinning setup, the 30% solution was excessively viscous and difficult to spin. Reducing the gelatin concentration to 28% yielded a stable electrospinning process with a smooth fiber morphology, free from “beads-on-string” type defects.38
Electrospinning was performed using a rotating collector made of an aluminum disc with a diameter of 20 cm. The sharp edge of the disc serves to focus the electrical field and direct the polymer jet onto the EM grid that is affixed to it (see setup in Fig. 2).34 The tangential velocity of the disc was varied to alter the degree of fiber alignment on the grids. Typically, two stubs holding eight grids were used per experiment. A 1 mL syringe with a 21 g needle (0.337 mm inner diameter) was employed for gelatin extrusion. The needle-to-grid distance was maintained at 7 cm. The potential difference between the syringe tip and the wheel was maintained at 13 kV with a flow rate set to 0.3 mL h−1. For the collection of aligned fibers, the tangential velocity of the disk was 11 m s−1 for a duration of 3.5 minutes; for randomly oriented fibers, it was set to 2.2 m s−1 for 2.5 minutes (see Fig. S1†). Experiments were conducted at room temperature (23 ± 1 °C) and relative humidity of 67 ± 4%. Following electrospinning, a heated metal biopsy punch (approximately 4 mm in diameter) was used to carefully excise the gelatin fibers around each grid, facilitating grid removal without damaging the fibers.
2.2. Cross-linking
To prevent the gelatin fibers from dissolving in an aqueous environment they must be crosslinked (see Fig. S2†). Prior to electrospinning, a 0.05 M solution of N-(3-dimethylaminopropyl)-N-ethylcarbodiimide hydrochloride (EDC) was prepared by dissolving 0.2 g of EDC in 16.7 mL of ethanol and 4.17 mL of deionized (DI) water (8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 v/v). The solution was then stored at 4 °C. After electrospinning, the grids were immersed in the cold EDC 0.05 M solution and left overnight at 4 °C in glass dishes sealed with Parafilm.39 EDC cross-links gelatin by reacting with the carboxyl groups of aspartic and glutamic residues in the gelatin molecule, forming an O-acylisourea intermediate. This intermediate undergoes nucleophilic attack by lysine's amine groups, forming an amide bond between gelatin polymer chains. Ultimately, this cross-linking reaction establishes a gelatin hydrogel network. The EDC cross-linking method offers notable advantages, including high conversion efficiency, mild reaction conditions, and excellent preservation of gelatin's biocompatibility.40 It is critical that the EDC solution remain cold during the cross-linking process.39
2 v/v). The solution was then stored at 4 °C. After electrospinning, the grids were immersed in the cold EDC 0.05 M solution and left overnight at 4 °C in glass dishes sealed with Parafilm.39 EDC cross-links gelatin by reacting with the carboxyl groups of aspartic and glutamic residues in the gelatin molecule, forming an O-acylisourea intermediate. This intermediate undergoes nucleophilic attack by lysine's amine groups, forming an amide bond between gelatin polymer chains. Ultimately, this cross-linking reaction establishes a gelatin hydrogel network. The EDC cross-linking method offers notable advantages, including high conversion efficiency, mild reaction conditions, and excellent preservation of gelatin's biocompatibility.40 It is critical that the EDC solution remain cold during the cross-linking process.39
To remove cross-linking residues, we created an array of three DI water droplets (100–200 μL each) on a Parafilm sheet. Grids were gently passed through the droplets before being transferred to a glass Petri dish filled with DI water and incubated at 37 °C for 24 hours for the final rinse step.39
2.3. Characterization of gelatin fibers
We utilized high-resolution scanning electron microscopy (HRSEM) to analyze gelatin fiber morphology and topographic structure (Zeiss Ultra Plus HRSEM). First we deposited an ultra-thin layer of conductive metal onto the specimens by sputter-coating to prevent charging during imaging. We chose iridium for sputter coating, due to its high secondary electron yield and resistance to oxidation. The sputtered film thickness ranges from 5 to 7 nm, sufficient for excellent conductivity without significantly altering fiber topography.We used Fiji software,41 along with the Directionality plugin, to analyze fiber orientation.42 This plugin finds the preferred fiber orientation by analyzing the input image and computing a directional histogram highlighting structures’ predominant orientation. The peaks indicate the preferred direction. Fiber diameters were measured manually using Fiji software.
Swelling of cross-linked, electrospun nanofibrous mats was measured by immersing them in deionized water at room temperature for 30 minutes and weighing them before and after. The swelling ratio was calculated using the equation: Swelling ratio (%) = ((Ws − Wd)/Wd) × 100, where Wd and Ws are the weights of the mats in the dry and swollen states, respectively.36 We also imaged cross-linked gelatin fibers electrospun onto glass slides in a dry state, and after swelling in water at room temperature for 30 minutes (Fig. S2†).
2.4. Cell culture
Human umbilical vein endothelial cells (HUVECs, Sciencell 8000-SCL) were cultured in EGM-2 MV Microvascular Endothelial Cell Growth Medium, supplemented with penicillin, streptomycin, and various growth factors. Cells were grown on dishes precoated with 0.2% gelatin and were maintained at 37 °C and 5% CO2. Experiments were carried out using cells from passages 7 to 12.27Before cell seeding, we prepared glass-bottom dishes with a custom silicone sticker, creating wells that are 4 mm in diameter to comfortably accommodate a 3 mm EM grid. We placed the grids fiber-coated side up in 20–30 μL droplets of cell culture medium in the wells. We then added 5 μL from a cell suspension of approximately 1 million cells per mL, with the aim of depositing approximately two cells per grid-square. After the cells began to adhere to the fibers (up to several hours from the time of cell seeding), 2 mL of warm media was added to each dish and they were left in the incubator overnight at 37 °C and 5% CO2. In our hands, seeding the cells into a small droplet of media contained in a well above the EM grid promoted a more even distribution of cells on the grid compared to filling the entire dish containing an EM grid with cells suspended in medium. The silicone wells also served to prevent the four EM grids per dish from sliding over each other during transportation to the microscope and plunge-freezer.
2.5. Immunofluorescence
To fix the cells, we prepared a 4% paraformaldehyde (PFA) solution in PBS from a 16% PFA stock. After aspirating the culture media, the sample was rinsed with PBS to remove residual media. Following washing, we incubated the sample in 4% PFA at room temperature for 15 minutes, then rinsed it three times with PBS. The samples were then stored at 4 °C, in PBS, until immunostaining.For immunostaining, cells were permeabilized using an Antibody Dilution Buffer (ADB) containing 1% bovine serum albumin (BSA) and 0.1% Triton X. Following a 1-hour blocking step with ADB, anti-paxillin staining (Abcam, ab32084, used at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200) was applied for three hours. After rinsing, a secondary staining solution containing anti-rabbit Alexa Fluor 488 conjugate antibody (Cell signaling 4412, used at 1
200) was applied for three hours. After rinsing, a secondary staining solution containing anti-rabbit Alexa Fluor 488 conjugate antibody (Cell signaling 4412, used at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500), ActinRed 555 (Thermo-Fisher Scientific R37112, 2 drops were used in 500 μL of ADB), and Hoechst 33342 (1
500), ActinRed 555 (Thermo-Fisher Scientific R37112, 2 drops were used in 500 μL of ADB), and Hoechst 33342 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) was applied for overnight incubation at 4 °C, followed by further rinses with PBS. For Fig. 4, cells were imaged on an Olympus IX83 inverted microsope, using a 40× objective. Fig. S2–S10† were imaged on a Nikon ECLIPSE Ti2-E spinning disk microscope using 20× air and 60× oil immersion objectives.
1000) was applied for overnight incubation at 4 °C, followed by further rinses with PBS. For Fig. 4, cells were imaged on an Olympus IX83 inverted microsope, using a 40× objective. Fig. S2–S10† were imaged on a Nikon ECLIPSE Ti2-E spinning disk microscope using 20× air and 60× oil immersion objectives.
2.6. Vitrification, cryo-ET, and tomogram reconstruction
We vitrified the grids in a Leica EM GP2 plunge freezer, using a blotting time of 13 seconds. We dipped each grid in a droplet of PBS prior to loading it into the tool. Following freezing, we clipped the grids into autogrids and loaded them into a Thermo-Fisher Scientific Glacios cryogenic transmission electron microscope (cryo-TEM) with an X-FEG electron source operated at 200 kV. Tilt series were acquired in counting mode at a calibrated pixel size of 3.063 Å (39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000× nominal magnification) by a Falcon 4i direct detector coupled to a Selectris X energy filter set to ±5 eV around zero-loss peak. Fiducials-free, dose-symmetric tilt series of 61 exposures from −60° to +60° at 3° intervals and a total dose of ∼120 ē Å−2 were collected with Tomo (Thermo-Fisher Scientific) at a defocus range between −2 μm and −9 μm. We reconstructed the tomograms using patch tracking in IMOD software package, version 4.9.1.43 We segmented the tomogram volume and prepared the images in Fig. 5C using Amira (FEI/Thermo Fisher Scientific) software.
000× nominal magnification) by a Falcon 4i direct detector coupled to a Selectris X energy filter set to ±5 eV around zero-loss peak. Fiducials-free, dose-symmetric tilt series of 61 exposures from −60° to +60° at 3° intervals and a total dose of ∼120 ē Å−2 were collected with Tomo (Thermo-Fisher Scientific) at a defocus range between −2 μm and −9 μm. We reconstructed the tomograms using patch tracking in IMOD software package, version 4.9.1.43 We segmented the tomogram volume and prepared the images in Fig. 5C using Amira (FEI/Thermo Fisher Scientific) software.
3. Results and discussion
We successfully engineered nanopatterned ECM constructs that are compatible with cryo-ET by electrospinning 28% gelatin on gold EM grids (Fig. 3 and S1 and ESI Movies 1–5†). Plasma treating the EM grids prior to electrospinning was critical for maintaining adhesion between the gelatin fibers and the gold EM grid surface. Cross-linking of the gelatin fibers was critical to preventing their dissolution in (aqueous) cell culture medium (Fig. S2 and S3†). We measured an average swelling ratio of 227% for spin-coated cross-linked gelatin fiber mats in deionized water.We observed a higher degree of fiber orientation when the collection wheel was rotating at a faster rate (aligned condition) versus the slower rate of rotation (random condition) (Fig. 3A and B, respectively and Fig. S1†). We measured an average fiber diameter of 417.7 ± 9.0 nm (SEM) for the aligned condition and 479.6 ± 10.3 nm (SEM) for the random condition (Fig. 3C and ESI Table 1†). Furthermore, there was no difference in mean, max, or min pore area between aligned and random fibers, but porosity was higher in random fibers (0.43) than in aligned fibers (0.34). In vivo, ECM is composed of various proteins that can bundle into larger structures ranging from 200 nm to several microns in diameter;44,45 thus, the gelatin fibers we produced are within the physiological range. The slight disparity in diameter between the conditions may be due to the fibers in the aligned condition undergoing more elongation due to tensile stress as the rotating disk collected the fibers at a higher tangential velocity for that condition. EM grids with electrospun fibers had a shelf-life of at least four months when stored in a desiccator (Fig. 3F).
The electrospun EM grids serve as viable cell culture platforms. We observed multiple instances of cells dividing on EM grids coated with electrospun gelatin fibers after 24 hours in culture (Fig. S4–S10†). Our immunofluorescence results show that the shape and orientation of endothelial cells grown on the electrospun TEM grids are influenced by the fiber direction (Fig. 4), consistent with previous reports of cells cultured on engineered aligned nanofibrillar ECM constructs11,12 or nanogrooves.46 Specifically, cells on the aligned fibers display an elongated morphology and align with the fiber direction. In contrast, on the randomly oriented fibers, the cells adopt a rounder, star-shaped morphology and F-actin-rich protrusions appear to follow the random orientation of the gelatin nanofibers. We did not perform proliferation or cell migration studies in this work, but Bettinger et al. previously showed that endothelial cells responded to 600 nm sized features in ridge-groove gratings by aligning and elongating along the grating axis, reducing proliferation, and increasing migration.46
We demonstrate that our engineered ECM constructs are compatible with vitrification by plunge freezing and cryo-ET (see Fig. 5 and ESI Movies 1–6†). Despite the added thickness of the gelatin fibers, the grids were readily clipped into standard Autogrids for automatic handling within the cryo-TEM where they were imaged. Our reconstructed tomograms show nanoscale cellular features such as mitochondria, actin filaments, microtubules, granules, ribosomes, and vesicles in 3D, some of which are segmented in Fig. 5 (see ESI Movies 1–6† for full tomograms). The parallel orientation of the microtubules in Fig. 5G–I may have been guided by the aligned gelatin fibers on which the cells were cultured.47
 | ||
| Fig. 5 Cryo-ET of endothelial cells cultured on electrospun gelatin fibers. (A and B) Cryo-TEM images of gelatin fibers, with a close up in (B) showing an F-actin-rich cellular region proximal to the gelatin (dark gray). (C) Tomographic slice showing vesicles and a microtubule (see ESI Movie 1† for full tomogram). (D–F) Tomographic slices overlaid with segmentation of select features in (F) showing a double-membranes organelle (membranes in blue and violet), microtubules (green), and vesicles (yellow) (see ESI Movie 2† for full tomogram). (G–I) Tomographic slices overlaid with segmentation of select features in (F) showing a double-membranes organelle (membranes in blue and violet) and microtubules (green) (see ESI Movie 3† for full tomogram). (J–L) Tomographic slices overlaid with segmentation of select features in (L) showing depicting mitochondria (blue), granules or ribosomes (violet), and vesicles (yellow) (see ESI Movie 4† for full tomogram). Scale bars, 100 nm. | ||
In this study, we were limited to imaging thin (several 100s of nanometers thick) regions of the cells suspended between the gelatin fibers, especially given that we were using a 200 kV (vs. 300 kV) cryo-TEM for imaging.48,49 Cryo-focused ion beam milling (cryo-FIB) can be used in future studies to generate thin lamellae from thicker regions of the frozen cells to facilitate their imaging with cryo-ET.50,51 Because the gelatin fibers themselves are hundreds of nanometers thick, cryo-FIB will be particularly important for targeting and studying the nanoscale organization of focal adhesions, located at the cell–ECM fiber interface with cryo-ET. Given their role in regulating migration, morphogenesis, and sensitivity to mechanical forces, visualizing focal adhesions on engineered nanotopographies could reveal key mechanisms of topography-driven cell behavior, such as spatial biasing.4 As it preserves the cell membrane and associated structures, our platform can be particularly suitable for studying recently discovered curved adhesions, cell–ECM adhesions that selectively form on curved membranes (e.g., at cell–ECM fiber interfaces) that are molecularly and functionally distinct from focal adhesions and clathrin-containing adhesions and are mediated by integrin v5.15
Previous methods for producing aligned collagen fibrillar films, such as extrusion, are not readily adapted to cryo-TEM because resulting ECM mats are on the order of hundreds of micrometers thick. Electrospinning allows us to deposit a layer of nanofibers on the EM grids that is sufficient to orient the cells, but thin enough for vitrification by plunge freezing (<10 μm).
This new class of cryo-ET cell culture supports can be used to investigate the structural organization of the cell–ECM adhesions that underlie the mechanosensitivity of endothelial cells to ECM topography.
Studying nanoscale structures at the cell–ECM interface may help resolve long-standing questions in tissue engineering, such as why specific fibril dimensions better promote cell survival in engineered tissues. We anticipate that the fibers will provide a valuable means for locating and identifying the cell–ECM adhesions. This can ultimately improving our understanding of the mechanical cues required for the formation of healthy tissue and of the pathologies that result from defects in endothelial tissue organization associated with peripheral arterial disease.
Finally, by mimicking the physical cues present in physiological ECM, these EM grids create a more in vivo-like environment that can improve the translational relevance of cryo-ET studies of mammalial cell ultrastructure and support the growth of cell types that are challenging to culture (e.g., tissue-derived primary cells or stem cells) on EM grids.
4. Conclusions
We have developed a new class of supports for cryo-ET that are coated with thin films of aligned ECM fibers to provide cells with programmed nanotopographical cues that mimic the in vivo ECM topography. We controlled the degree of alignment between the fibers in the ECM thin films by varying the electrospinning parameters, specifically the speed of the rotating collector wheel. This platform can be used in future cryo-ET studies to probe how physical cues in the cell microenvironment influence junctional and cytoskeletal architecture.Author contributions
Shani Tcherner Elad: conceptualization, methodology, software, formal analysis, investigation, data curation, writing – original draft, visualization, project administration. Rita Vilensky: methodology, investigation. Noa Ben Asher: methodology, investigation, writing – review & editing. Nataliya Logvina: software, formal analysis, data curation, writing – review & editing, visualization. Ran Zalk: software, investigation. Eyal Zussman: resources, writing – review & editing, supervision. Leeya Engel: conceptualization, methodology, investigation, writing – original draft, supervision, project administration, funding acquisition.Data availability
The data supporting this article have been included as part of the ESI.† Tomograms and raw electron microscopy data have been uploaded to the EMDB database (accession code: EMD-53927).Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Israel Science Foundation (ISF) Personal Research Grant No. 1925/23. We thank Dr Ngan Huang, Dr Doron Elad, and Dr Alex Dunn for helpful advice and Dr Olga Kleinerman for HRSEM imaging, which was performed at the Technion Center for Electron Microscopy of Soft Matter. The authors are grateful for the generous support from the Guzik Foundation to BGU's Cryo-electron microscopy unit, where cryo-ET was performed. L.E. is a Diane and Guilford Glazer Foundation Faculty Fellow. Fig. 1 and 2C were created with Biorender.com.References
- D.-H. Kim, P. P. Provenzano, C. L. Smith and A. Levchenko, J. Cell Biol., 2012, 197, 351–360 CrossRef PubMed.
- C. S. Ranucci and P. V. Moghe, J. Biomed. Mater. Res., 2001, 54, 149–161 CrossRef.
- M. J. Dalby, N. Gadegaard, R. Tare, A. Andar, M. O. Riehle, P. Herzyk, C. D. Wilkinson and R. O. Oreffo, Nat. Mater., 2007, 6, 997–1003 CrossRef PubMed.
- C. J. Bettinger, R. Langer and J. T. Borenstein, Angew. Chem., Int. Ed., 2009, 48, 5406–5415 CrossRef PubMed.
- A. Curtis, B. Casey, J. Gallagher, D. Pasqui, M. Wood and C. Wilkinson, Biophys. Chem., 2001, 94, 275–283 CrossRef PubMed.
- K. Diehl, J. Foley, P. Nealey and C. Murphy, J. Biomed. Mater. Res., Part A, 2005, 75, 603–611 CrossRef PubMed.
- M. Dalby, M. Riehle, H. Johnstone, S. Affrossman and A. Curtis, Tissue Eng., 2002, 8, 1099–1108 CrossRef PubMed.
- M. J. Dalby, N. Gadegaard and R. O. Oreffo, Nat. Mater., 2014, 13, 558–569 CrossRef PubMed.
- A. Naba, Nat. Rev. Mol. Cell Biol., 2024, 25, 865–885 CrossRef PubMed.
- E. S. Lai, N. F. Huang, J. P. Cooke and G. G. Fuller, Regener. Med., 2012, 7, 649–661 CrossRef PubMed.
- N. F. Huang, E. S. Lai, A. J. Ribeiro, S. Pan, B. L. Pruitt, G. G. Fuller and J. P. Cooke, Biomaterials, 2013, 34, 2928–2937 CrossRef PubMed.
- K. H. Nakayama, V. N. Surya, M. Gole, T. W. Walker, W. Yang, E. S. Lai, M. A. Ostrowski, G. G. Fuller, A. R. Dunn and N. F. Huang, Nano Lett., 2016, 16, 410–419 CrossRef PubMed.
- J. Luo, M. Walker, Y. Xiao, H. Donnelly, M. J. Dalby and M. Salmeron-Sanchez, Bioact. Mater., 2022, 15, 145–159 CAS.
- K. M. Yamada, A. D. Doyle and J. Lu, Trends Cell Biol., 2022, 32, 883–895 CrossRef CAS PubMed.
- W. Zhang, C.-H. Lu, M. L. Nakamoto, C.-T. Tsai, A. R. Roy, C. E. Lee, Y. Yang, Z. Jahed, X. Li and B. Cui, Nat. Cell Biol., 2023, 25, 1453–1464 CrossRef CAS.
- M. Chighizola, T. Dini, S. Marcotti, M. D'Urso, C. Piazzoni, F. Borghi, A. Previdi, L. Ceriani, C. Folliero and B. Stramer, et al. , J. Nanobiotechnol., 2022, 20, 418 CrossRef CAS PubMed.
- J. C. Ashworth and T. R. Cox, Nat. Rev. Cancer, 2024, 24, 461–479 CrossRef CAS.
- O. Medalia, I. Weber, A. S. Frangakis, D. Nicastro, G. Gerisch and W. Baumeister, Science, 2002, 298, 1209–1213 CrossRef CAS PubMed.
- S. Asano, B. D. Engel and W. Baumeister, J. Mol. Biol., 2016, 428, 332–343 CrossRef CAS PubMed.
- L. Gan and G. J. Jensen, Q. Rev. Biophys., 2012, 45, 27–56 CrossRef CAS PubMed.
- L. N. Young and E. Villa, Annu. Rev. Biophys., 2023, 52, 573–595 CrossRef.
- S. Chakraborty, M. Jasnin and W. Baumeister, Protein Sci., 2020, 29(6), 1302–1320 CrossRef.
- A. Martins, R. Reis and N. Neves, Int. Mater. Rev., 2008, 53, 257–274 CrossRef.
- I. Patla, T. Volberg, N. Elad, V. Hirschfeld-Warneken, C. Grashoff, R. Fässler, J. P. Spatz, B. Geiger and O. Medalia, Nat. Cell Biol., 2010, 12, 909–915 CrossRef PubMed.
- E. Ermantraut, K. Wohlfart and W. Tichelaar, Ultramicroscopy, 1998, 74, 75–81 CrossRef.
- L. Engel, G. Gaietta, L. P. Dow, M. F. Swift, G. Pardon, N. Volkmann, W. I. Weis, D. Hanein and B. L. Pruitt, J. Micromech. Microeng., 2019, 29, 115018 CrossRef.
- L. Engel, C. G. Vasquez, E. A. Montabana, B. M. Sow, M. P. Walkiewicz, W. I. Weis and A. R. Dunn, J. Struct. Biol., 2021, 213, 107791 CrossRef PubMed.
- L. Engel, R. Held, C. G. Vasquez, W. I. Weis and A. R. Dunn, protocols.io, 2022, DOI:10.17504/protocols.io.bz22p8ge.
- M. Toro-Nahuelpan, I. Zagoriy, F. Senger, L. Blanchoin, M. Théry and J. Mahamid, Nat. Methods, 2020, 17, 50–54 CrossRef PubMed.
- S. Theron, E. Zussman and A. Yarin, Polymer, 2004, 45, 2017–2030 CrossRef.
- N. Nseir, O. Regev, T. Kaully, J. Blumenthal, S. Levenberg and E. Zussman, Tissue Eng., Part C, 2013, 19, 257–264 CrossRef.
- Y. Zhang, C. T. Lim, S. Ramakrishna and Z.-M. Huang, J. Mater. Sci.: Mater. Med., 2005, 16, 933–946 CrossRef.
- H. Fong, I. Chun and D. H. Reneker, Polymer, 1999, 40, 4585–4592 CrossRef.
- A. Theron, E. Zussman and A. L. Yarin, Nanotechnology, 2001, 12, 384 CrossRef.
- C. J. Russo and L. A. Passmore, Science, 2014, 346, 1377–1380 CrossRef.
- K. Jalaja and N. R. James, Int. J. Biol. Macromol., 2015, 73, 270–278 CrossRef PubMed.
- P. Sajkiewicz and D. Kołbuk, J. Biomater. Sci., Polym. Ed., 2014, 25, 2009–2022 CrossRef.
- P. P. Bhat, S. Appathurai, M. T. Harris, M. Pasquali, G. H. McKinley and O. A. Basaran, Nat. Phys., 2010, 6, 625–631 Search PubMed.
- J. Li, A. He, J. Zheng and C. C. Han, Biomacromolecules, 2006, 7, 2243–2247 CrossRef.
- C. E. Campiglio, N. Contessi Negrini, S. Farè and L. Draghi, Materials, 2019, 12, 2476 CrossRef PubMed.
- J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld and B. Schmid, et al. , Nat. Methods, 2012, 9, 676–682 CrossRef.
- Z.-Q. Liu, Appl. Opt., 1991, 30, 1369–1373 CrossRef.
- J. R. Kremer, D. N. Mastronarde and J. R. McIntosh, J. Struct. Biol., 1996, 116, 71–76 CrossRef PubMed.
- A. Padhi and A. S. Nain, Ann. Biomed. Eng., 2020, 48, 1071–1089 CrossRef PubMed.
- C. G. Jones, T. Huang, J. H. Chung and C. Chen, ACS Biomater. Sci. Eng., 2021, 7, 1600–1607 CrossRef PubMed.
- C. J. Bettinger, Z. Zhang, S. Gerecht, J. T. Borenstein and R. Langer, Adv. Mater., 2008, 20, 99–103 CrossRef PubMed.
- C. Oakley and D. Brunette, J. Cell Sci., 1993, 106, 343–354 CrossRef.
- M. W. Martynowycz, M. T. Clabbers, J. Unge, J. Hattne and T. Gonen, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2108884118 CrossRef.
- P. Szwedziak , bioRxiv, 2024, 2024–2012.
- C. Berger, N. Premaraj, R. B. Ravelli, K. Knoops, C. López-Iglesias and P. J. Peters, Nat. Methods, 2023, 20, 499–511 CrossRef PubMed.
- A. Rigort, F. J. Bäuerlein, E. Villa, M. Eibauer, T. Laugks, W. Baumeister and J. M. Plitzko, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 4449–4454 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nr05508j |
| This journal is © The Royal Society of Chemistry 2025 |




