 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Advancements in DNA-PAINT: applications and challenges in biological imaging and nanoscale metrology
Luca
Piantanida
 *a,
Isaac T. S.
Li
*a,
Isaac T. S.
Li
 b and
William L.
Hughes
b and
William L.
Hughes
 a
a
aFaculty of Applied Science, School of Engineering, The University of British Columbia, Kelowna, B.C. V1V 1V7, Canada. E-mail: luca.piantanida@ubc.ca
bDepartment of Chemistry, The University of British Columbia, Kelowna, B.C. V1V 1V7, Canada
First published on 15th May 2025
Abstract
Super-Resolution Microscopy (SRM) has revolutionized bioimaging by breaking the diffraction limit of light, enabling visualization of structures at the nanometer scale. DNA-PAINT (Point Accumulation for Imaging in Nanoscale Topography) is a versatile SRM technique that leverages the programmability of DNA hybridization to achieve high-resolution and multiplexed imaging of molecular targets. This review examines recent advancements in DNA-PAINT, including improvements in imaging resolution, acquisition speed, and imager design, which have enhanced its applications in biological imaging and nanoscale metrology. DNA-PAINT's unique capacities in programming specific interactions have made it indispensable in a range of biological and non-biological settings, from cellular visualization of structure and function to molecular data storage. Here, we highlight recent advancements in DNA-PAINT and its main practical challenges, focusing on how persistent optimization drives innovation. Addressing these challenges continues to drive its expanding role in biological imaging and broader applications across interdisciplinary fields. This review also highlights the interdependence of DNA-PAINT and other techniques that are fundamental to broadening the impact of SRM and shaping the future of biological and biomedical imaging.
Significance
DNA-PAINT advancements have reached a refinement level that has inspired innovative applications across multiple fields. This review focuses on DNA-PAINT's transformative role in SRM, particularly on recent advancements that enhance its precision, versatility, and applicability across multiple disciplines. Through ongoing optimization and integration in diverse fields, DNA-PAINT is advancing cellular and molecular imaging while also expanding the broader potential of SRM to meet future demands. Consequently, this technique is becoming more broadly utilized, and its applications are guiding new advancements in a virtuous cycle that benefits biomedical imaging and nanoscale metrology.1. Introduction
SRM provides the ability to observe molecules below the diffraction limit of light1 and therefore has revolutionized optical microscopy.2 SRM encompasses a variety of imaging modalities that leverage the advantages of different light-path setups to reveal sub-cellular structures in remarkable detail. This has facilitated single-molecule resolution while also maintaining sample preservation, imaging flexibility, and the target specificity characteristics of optical microscopy. SRM techniques utilize diverse imaging mechanisms, from optimizing image acquisition to controlling the fluorescent state of emitters. The imaging techniques that employ widefield, or Total Internal Reflection Fluorescence (TIRF) excitation, and controlled fluorophore blinking to localize individual molecules3–6 are defined as Single-Molecule Localization Microscopy (SMLM).7–9All SMLM techniques require accurate sub-diffraction localization of individual emitters and include methods such as Photoactivation Localization Microscopy (PALM),10,11 Stochastic Optical Reconstruction Microscopy (STORM),12,13 and PAINT.14 The optical image of an ideal point emitter can be represented by its Point Spread Function (PSF). If the PSF of individual emitters does not overlap, their location can be precisely determined with nanometer accuracy. To avoid the overlap, spatially dense emitters are temporally spread out to be sparse within any given time window. This is achieved by allowing individual emitters to “blink” so that only a subset of them fluoresce at any given time, enabling the spatial information to be preserved based on the Nyquist criterion.15,16 By allowing emitters to switch between the OFF (dark) and ON (bright) state stochastically, all binding emitters are observable, given sufficient sampling time.
In PAINT, the emitters do not switch fluorescent states, and the signal ON/OFF cycle is contingent on the alternating of their free diffusion state and their bound state to the target,14 rather than induced or modulated photoswitching17–19 (e.g. PALM and STORM). This allows a sparse subset of the targets to be activated and imaged per frame, enabling high-resolution reconstruction from individual localized events and effectively diluting spatial density over time.
DNA-PAINT is the most widely adopted PAINT method, utilizing the unique programmability of DNA to precisely control imager binding to target molecules.20–23 It typically employs a TIRF optical configuration to excite the molecules within the first hundred nanometers above the surface,24,25 thereby minimizing background fluorescence from the pool of fluorescent imagers (emitters) and improving the signal-to-noise ratio (SNR)26 (Fig. 1a).
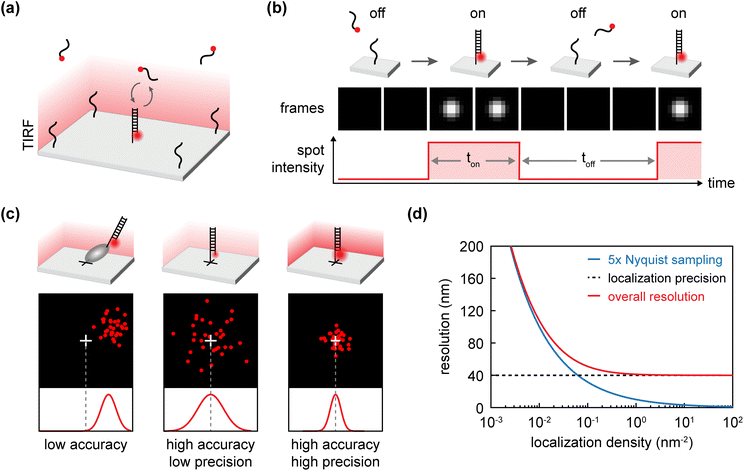 | ||
| Fig. 1 Principles of DNA-PAINT binding cycle, and resolution. (a) Schematic of TIRF-based DNA-PAINT on a DNA origami substrate. (b) Schematic of the on/off cycles of the transient binding of imager to docking strand and their bright (τb) and dark (τd) time. (c) Schematic showing three cases of different localization precision and accuracy. Low accuracy can be caused by offset of imager fluorophore from the actual target location. Precision is related to the photon flux from the imager fluorophore. (d) Relationship between localization density, localization precision and the overall achievable resolution, following the 5× Nyquist criterion.28 | ||
DNA-PAINT creates fluorophore “blinking” through the transient hybridization between complementary single-stranded DNA (ssDNA) oligonucleotides. The “imager” strand is labelled with a fluorophore and diffuses freely in solution, while its complementary “docking” strand is attached to the target. Upon binding, the imager strand generates a localized fluorescent signal. Once unbound, its fast diffusion blurs it into the background. The bright time (τb), or ON time, represents the duration of imager binding, whereas the dark time (τd), or OFF time, indicates the interval between two binding events at the same site (Fig. 1b).
The programmability of the imager-docking DNA hybridization is one main advantage of DNA-PAINT, enabling precise control over target specificity and allowing more advanced multiplexing than other SMLM approaches. Consequently, it has a variety of in vitro and in situ applications, paving the way for performance improvements. These advancements have facilitated the broader integration of DNA-PAINT across many interdisciplinary fields, refining fundamental elements of the technique and driving innovation in bioimaging and nanoscale metrology.
Nanoscale metrology is defined here as the science of measurement at the nanoscale with the goal of characterizing nanomaterials with high precision. This is often challenging when using biomaterials like DNA, as their nanoscale interactions are difficult to control and measure in standard ways. The advancements in DNA-PAINT resolution, speed, and multiplexing have reached a level of precision that has enabled its use in applications well beyond bioimaging. Examples of DNA-PAINT usage in nano-metrology applications alone, while certainly fewer than in bioimaging applications, have increased in recent years.27
This review focuses on the recent advancements and main challenges of DNA-PAINT and how its refinement via application drives innovations in biological and nanoscale metrology.
2. DNA-PAINT advancements
DNA-PAINT has quickly gained interest as an accessible super-resolution imaging technique. However, in its early days, it was limited by slow image acquisitions (up to several hours), challenges in imaging densely labelled targets, and a lack of multi-target detection. Since then, significant efforts have improved its resolution, speed, and multiplexing capability.2.1. Resolution
In DNA-PAINT, spatial resolution is determined by the precise localization of individual emitters. Each fluorescent spot is typically fitted to a 2D Gaussian to determine its position, where the final image is reconstructed from all localization events and the final resolution depends on both localization density and localization precision.29–32 Localization precision is often estimated using the Full Width at Half Maximum (FWHM) of the localization error distribution or the nearest neighbour distribution around a single target.33Challenges arise in regions with high docking strand density, where the distance between docking sites may be smaller than the achievable localization precision.34 To accurately resolve individual docking strands in such environments, three key conditions must be met: (1) high photon count per binding event to minimize localization uncertainty, (2) a high number of binding events per localization site to improve statistical confidence, and (3) shorter blinking cycles to reduce temporal overlap of localizations.35 These conditions enhance localization precision, improve SNR, and reduce imaging artifacts due to falsely localized background noise.35,36
Precision and accuracy are distinct factors in localization. Precision refers to the standard deviation of errors in estimated coordinates, while accuracy measures how close the estimated coordinates are to the true position.3 DNA-PAINT resolution ultimately depends on the interplay between these two factors (Fig. 1c).
The primary constraint on localization precision is photon count per binding event where more photons reduce localization uncertainty and improve precision.35 Factors such as background noise, SNR, camera pixel size, and PSF shape can further influence precision by affecting the error rate of localization algorithms.3
The overall resolution of an SMLM image is determined by both localization density and precision. According to the Nyquist sampling criterion, resolving finer features requires higher localization density (5× Nyquist sampling).28 This concept can be illustrated using the analogy of a tree decorated with lights: if the lights are sparse, the tree's structure remains unresolved; as the density of lights increases, finer details emerge. However, resolution ultimately remains limited by localization precision; features smaller than this limit cannot be resolved, no matter how densely they are labelled (Fig. 1d).
DNA-PAINT resolution can be easily improved by using imaging methodologies from other SRM techniques. One example is its combination with MINimal photon FLUXes (MINFLUX) technology.37 MINFLUX localizes a single fluorophore with nanometer precision by using a doughnut-shaped excitation beam to probe its position. The beam is initially positioned at multiple points around the fluorophore within a defined probing area, and photon flux is recorded at each location. Based on the flux pattern, the beam is iteratively repositioned to refine the fluorophore's location while progressively reducing the probing area. This process minimizes photon flux at the fluorophore's position, ultimately achieving approximately 1 nm resolution.38,39
More recently, the integration of advanced computational processing in DNA-PAINT has allowed researchers to improve resolution by using the sequential imaging of multiple localizations of a single target. By utilizing a DNA barcoding method with orthogonal sequences and performing sequential acquisitions, docking strands separated by just a few angstroms have been resolved40 (see section 3.2). This sets DNA-PAINT's achievable resolution at the highest mark as compared to any other fluorescence microscopy approach. This exceptional resolution opens the possibility for DNA-PAINT applications in research fields where sub-nm resolution is required but was previously unachievable with fluorescence imaging.
2.2. Speed
The speed of DNA-PAINT imaging is governed by the time required to accumulate sufficient localization events for a given localization density, which in turn affects the overall resolution (Fig. 1d). This speed is inherently limited by the kinetics of imager-docking hybridization. The average duration of the imager's bright and dark states determines the total length of the blinking cycle (OFF/ON/OFF) (Fig. 1b) and sets the optimal imaging frame rate. To maximize signal collection, the frame rate should be slow enough to capture sufficient photons per binding event but fast enough to minimize the likelihood of recording multiple binding events at the same localization site (Fig. 2). In general, longer τb improve localization precision, while shorter τd accelerate image acquisition. Therefore, for high-precision and high-speed imaging, τb should be long enough for sufficient photon collection, while τd should remain as short as possible without creating too many overlap events41 (Fig. 2). | ||
| Fig. 2 Principles of DNA-PAINT speed. Representation of the interdependence between kon and koff to create high event rate while avoiding signal overlap towards faster DNA-PAINT speed. | ||
Tuning blinking kinetics enables further control over imaging speed. The bright time is primarily dictated by the stability of the imager-docking duplex, which can be adjusted by modifying the imager sequence. Longer DNA sequences stabilize hybridization, reducing the dissociation rate (koff = 1/τb). Generally, an 8–10 nucleotide (nt) imager exhibits a τb ranging from 0.3 to 10 seconds, depending on its cytosine–guanine (CG) content.41,42 In contrast, the dark time is dependent on the concentration of the imager (Ci) and on rate (τd = (Cikon)−1). Increasing the imager concentration shortens τd, accelerating the binding cycle and enhancing acquisition speed.42
Buffer composition plays a critical role in modulating binding kinetics and has been extensively optimized since DNA-PAINT's inception.22,43 Researchers have analyzed the influence of buffer salinity on imager-docking kinetics, identifying optimal buffer compositions that enhance the blinking cycle performance and improve the acquisition speed.42 Another effective modification involves using low concentrations of ethylene carbonate, which accelerates duplex de-hybridization by enhancing the solubility of hydrophobic DNA bases.44 While ethylene carbonate reduces τb, it does not significantly alter hybridization efficiency, making it a valuable tool for tuning the koff for faster imaging speed.
Imager design modifications are not solely restricted to sequence length; the association rate of the imager can also be enhanced by employing specific proteins as molecular carriers.45 This protein-assisted delivery can effectively increase the binding rate in particular biological environments. However, it is essential to consider the size of the imager when linked to the protein, as steric hindrance might impede imaging in densely packed docking strand environments.
Furthermore, the imager can be engineered to incorporate Förster Resonance Energy Transfer (FRET) between two fluorophores. This implementation increases acquisition speed due to significant background suppression, as well as the enhanced photobleaching effect on the acceptor dye.46 The background reduction using fluorogenic imagers ultimately contributes towards a higher imaging speed by enabling short bright times without the risk of capturing non-specific adsorption of diffusing imager strands47 (see section 3.3.2). It also further enables deeper imaging and illumination volume.
Lastly, modifications to the docking strand design also contribute to faster imaging. Concatenating repeated binding motifs along the docking sequence increases binding frequency, reducing τd and enabling faster acquisition. This approach allows imaging at lower imager concentrations, further improving the SNR while maintaining efficient binding kinetics.48 While imaging speed has historically been DNA-PAINT's main weakness, these improvements have led to practical imaging times, reinforcing the value of DNA-PAINT in cell imaging and supporting better performance achievable in live-cell imaging.
2.3. Multiplexing
The ability to detect multiple distinct target molecules within the same sample remains a key advantage of DNA-PAINT. To this end, researchers have developed a variety of approaches for multiplexed detection using DNA-PAINT (Fig. 3).Employing spectrally distinct fluorophores linked to specific imager strands with orthogonal sequences is one straightforward way to introduce multiplexing (Fig. 3a). The detection of different targets relies on recording various emission wavelengths from the same sample, however this spectral multiplexing is limited by the overlapping emission spectra of the fluorophores, which restricts the number of distinguishable dyes that can be used within the visible range.49 Implementing multiple colour detection channels can be costly and may necessitate the selection of fluorophores with suboptimal photophysical properties, such as brightness and quantum efficiency. As a result single-colour detection is often preferred.
A second strategy to address the limitation generated by overlapping spectra is to introduce orthogonal imager strands in a sequential mode, allowing for the detection of distinct targets with a single fluorophore (Fig. 3b). In this approach, termed Exchange-PAINT, washing steps are performed each time a new imager strand is introduced to detect a different target on the sample. This method has demonstrated the capability to detect multiple cell organelles in a single overlaid image generated from sequential acquisitions.50–52
The fundamental concept underlying this technique is that imager strands bind to target molecules proportionally to the number of docking strands attached to them, demonstrating how DNA-PAINT can be used quantitatively.53,54 The ability to count molecules through the binding frequency of imager and docking strands forms the basis of analysing kinetic equilibrium.
Researchers have extensively analysed the transient binding characteristics of imager strands to enhance multiplexing by modulating their kinetics (Fig. 3c). One approach to achieve modulation is by modifying the imager-docking strand sequence design. Target molecules can be labelled with docking strands that feature specific repetitions of binding domains complementary to the imager strands. The combination of different domain lengths creates numerous possibilities for optimization. This “barcoding” technique has been widely explored by researchers to improve multiplexing efficiency.51 Depending on the number of repetitions and their lengths, the binding frequency can be engineered to create kinetic barcoding, which permits the detection of up to 124 different frequencies, each assigned a specific colour in the final image.55
Variations of this kinetic barcoding concept have equipped researchers with additional tools to optimize multiplexing, speed, and SNR in DNA-PAINT. For instance, binding domains can have overlapping sequences to fine-tune their kinetics, enhancing both multiplexing and speed.48 Alternatively, sequence mismatches can be introduced between imager and docking strands to enhance their binding kinetics.56
In addition to sequence modifications, the structural design of imager-docking strands can be refined. As an example, coupling imager strands with a quencher–fluorophore pair can enhance fluorescent emission,56 while domain repeats on the docking strands can be utilized to improve binding specificity.57 Both strategies have been shown to reduce the fluorescent background from free imager strands, thereby optimizing the SNR for more effective sampling.
Recently, researchers developed two distinct methods to achieve virtually unlimited multiplexing capabilities for single protein imaging. While using different multiplexing strategies, these methods commonly take advantage of a secondary labelling of the primary docking strand attached to the target. In this case, the imager does not bind directly to the docking strand but instead hybridizes to an additionally supplied DNA strand, enabling an extra molecular level for barcoding.
The first method, called Secondary label-based Unlimited Multiplexed PAINT (SUM-PAINT), uses a DNA strand complementary to the docking strand as secondary label.58 This strand carries a toehold sequence for signal extinction and a sequence complementary to the imager. The possibility of removing this secondary label (by strand displacement and/or blocking) enables the implementation of sequential rounds of barcoding. In each round, six speed-optimized sequences are used as imagers, allowing a six-step Exchange-PAINT readout per barcoding round. Using SUM-PAINT, researchers have been able to image 30 proteins at single-molecule level unveiling distinct heterogeneity in neuronal synapses.58
In the second method, known as Fluorogenic Labelling in conjunction with Adapter-mediated Switching for High-throughput DNA-PAINT (FLASH-PAINT), the secondary label strand is not removed by strand displacement and so does not carry a toehold sequence.59 Instead this so called “transient adapter” (that, similarly to SUM-PAINT, binds the primary docking strand) is removed by the addition of an “eraser” strand between imaging rounds. The eraser strand carries a higher affinity than the docking strand, hence enabling the sequestering of the transient adapter from the target. In this way, another unique adapter sequence can be added for the detection of the next target in the following imaging round. The use of transient adapters and eraser strands allows rapid and efficient switching between secondary label strands using the same imager sequence, avoiding the need for Exchange-PAINT's time-consuming washing steps.59 In general, using DNA secondary label strands is very promising for high-throughput single-molecule multiplexing DNA-PAINT imaging, especially for cell environments, but it does expose the different DNA strands to crosstalk and non-specific binding.
3. Challenges and applications
Thanks to its multiplexing potential and the programmability of its transient binding equilibrium, DNA-PAINT has been successfully applied in both biological and non-biological environments without compromising resolution. Advances in DNA-PAINT have been demonstrated through various approaches aimed at exploring its achievable limits as well as enhancing its practical applications. This interdependence between advancements in the technique and its broadening applications has created an ideal environment for inspiring new applications, made possible by the recent imaging achievements.Reaching this stage of development is rare for an imaging technique and has largely been facilitated by the exceptional programmability of DNA nanotechnology. Many DNA-PAINT studies specifically rely on DNA nanodevices to test and showcase their applicability. In the following sections, we review a selection of key studies that highlight the advancements of DNA-PAINT in both biological and non-biological environments. To better understand the impact of these advancements, this review also illustrates the main imaging challenges relative to both environments (Table 1).
| Imaging challenge | Conditioning factors | Main limitation | Compensation strategy | |
|---|---|---|---|---|
| General challenges | Insufficient number of photons collected per binding event | Short bright time due to a weak imager-docking binding | Poor localization precision and consequent imaging blurry | -Increase binding stability and so, the bright time by using: |
| • Long binding domain | ||||
| • Stronger secondary structure | ||||
| • Stronger chemical composition (LNA, BNA, etc.) | ||||
| Overlapping PSFs | Adjacent sites binding events are recorded simultaneously | Artificial blurry in dense array | -Filtering out not matching PSFs | |
| -Use “multi-emitter” algorithms | ||||
| Imager photobleaching | Photochemical alteration of the dye (accelerated by oxygen in solution) | Decreasing of the number of photons per event over time and so, of the localization precision | -Implement an enzymatic oxygen-scavenging system in the imaging solution | |
| Sample drift | Sample spatial adjustment over time relative to the objective | Ambient thermal fluctuations | -Increase equilibration time | |
| -Addition of fiducial markers | ||||
| -Implement real-time drift adjustment system | ||||
| Poor strand incorporation in DNA nanostructures | Weak incorporation design on specific spots | High number of false negative sites | -Reinforce DNA nanostructure design by optimizing length, position, and composition of the docking strand | |
| Off-target/false localizations | Imagers non-specific adsorption, confined diffusion within illumination volume, or unbound imagers localizations from fast imaging. | High number of false positives sites | -Implement fluorogenic imager designs | |
| -Surface passivation | ||||
| Biological environment challenges | Linkage error | High steric hindrance of the target's label linkage moiety | Biased resolution especially in dense target environments | -Reduce the steric hindrance of the label and optimize its affinity by using: |
| • Nanobodies | ||||
| • SOMAmers | ||||
| • Affimer | ||||
| • Small peptide labels | ||||
| • Peptide-binding proteins | ||||
| Imager diffusion in the dense cell environment | Non-specific binding of imager to cellular DNA/RNA | High number of false positives and slow acquisition time | -Increase the DNA hybridization specificity by using: | |
| • Longer binding sequence | ||||
| • Left-DNA (L-DNA) | ||||
| • Chemical composition | ||||
| Enzymatic degradation of the imager | Low number of binding events per target localization | Poor resolution | -Increase the enzymatic resistance by changing the chemical composition (LNA, BNA) | |
| Laser phototoxicity | Over production of reactive oxygen species | Cell morphology and physiology artificial changes | -Reduce laser intensity | |
| Dynamicity of the live cell environment | Active target drift over time | Poor localization precision and consequent blurry | -Use fixed cells where possible |
3.1. General challenges
Fundamental challenges inherent to the technique are closely tied to imager design and data acquisition conditions. The relative simplicity of DNA origami platforms is often utilized by researchers to address these main constraints before progressing to more complex cellular environments, providing a valuable step for assessing DNA-PAINT performance.The number of photons collected during a single binding event directly affects localization precision and is dependent on the imager's τb. An imager strand with a short τb will not yield enough photons for adequate localization precision, thereby impairing image resolution. Issues like this arise from DNA sequence designs that result in imagers with weak hybridization stability. Typically, short hybridization domain sequences and non-optimized DNA base compositions lead to weak and consequently short binding times. Any strategy that improves the stability of the imager strand's bound state will enhance τb, allowing for the collection of an adequate number of photons. Conversely, excessively long bright times caused by elongated imager strand sequences can also complicate imaging, as they prevent sufficiently rapid binding rates lead to slow acquisition times. Maintaining an appropriate range of association and dissociation rates is vital for the success of DNA-PAINT imaging.22,35 Although these ranges are not narrow, they do somewhat constrain the design possibilities for imager and docking strands.
Another fundamental challenge related to imager design is the potential overlap of captured PSFs of binding events. This overlap commonly occurs when two or more PSFs from adjacent localization spots are recorded simultaneously, making them temporally inseparable. This phenomenon is typical in high-density regions of docking strands or when utilizing fast-kinetic imagers for accelerated imaging. During image reconstruction, PSF overlap can lead to inaccurate localization determinations between adjacent molecules and contribute to artificial blurring.34,62 While this effect can be mitigated by filtering out mismatched PSFs and utilizing multi-emitter fitting algorithms that accommodate multiple PSFs, completely avoiding overlap remains challenging and poses a fundamental difficulty in high-density hybridization domains.
Introducing nanoparticles that remain fluorescent throughout the acquisition period, serving as fiducial markers, addresses this problem. These markers allow the drift to be calculated across frames and subsequently subtracted during image reconstruction. Drift correction can also be achieved through cross-correlation of localizations between consecutive image frames, using common image post-processing software.63,64 While sample drift can be effectively corrected computationally, the best results are obtained when drift is also corrected through active, real-time adjustments of the microscope stage position.65,66 This is accomplished by tracking fiducial markers in the camera's field of view with software connected to a closed-loop system, which drives piezo-electric actuators to finely adjust the stage position in the x and y directions.
Another challenge, particularly during long acquisition times, is photobleaching of the fluorophores attached to the imagers. While the TIRF optical setup significantly reduces photobleaching by concentrating illumination on a depth of only a few hundred nanometres, photobleaching of molecules in solution remains a concern.67 As the finite number of imagers in the solution diffuse toward the illuminated surface, the risk of photobleaching increases. When no fresh imagers remain, photobleaching leads to a decline in localization events over time, which limits localization precision in subsequent image frames. High laser power and prolonged acquisition times exacerbate this effect.68 Since oxygen in the solution accelerates photobleaching, incorporating an enzymatic oxygen-scavenging system in the imaging buffer can help mitigate this problem. Several effective oxygen-scavenging mixtures have been identified that enhance the photostability of the dye, allowing for longer acquisition times.69 Although these measures assist in combating photobleaching, they do not eliminate the effect entirely, so minimizing laser power should also be considered during extended acquisitions.70
When using DNA origami to benchmark DNA-PAINT performance, researchers must also carefully consider the rate of strand incorporation into the platform. Docking strands are DNA sequences that are only partially hybridized to the main structure, leaving their exposed binding domains available to the imager. The exposed portions and incomplete hybridization make them susceptible to incorporation failures during the assembly process and may result in structurally weaker spots prone to damage.71,72 The incorporation rate and accessibility of the docking strands are influenced by several factors, including the design of the DNA origami, their position within the grid, and their length.73 A low incorporation rate of docking strands can lead to false negatives, which may bias single and multiplex detection. Increasing the concentration of strands during the assembly process can enhance their incorporation rate.71 Additionally, analysing and addressing potential weak points in the nanostructure design can improve incorporation efficiency and increase accessibility for the imager, thereby resolving issues of ineffective site detection.
3.2. Imaging DNA nanostructures
DNA-PAINT imaging in cellular environments presents challenges due to their dense and dynamic nature, which interfere with the diffusion of imaging strands and their targeting efficiency. Consequently, the limits of the technique are validated on DNA nanostructure devices before being tested in biological conditions.Among the most used platforms are 2D and 3D DNA origami,74 which offer high addressability and programmability, allowing for the precise arrangement of docking strands in nanometer-scale patterns (Fig. 4a). These artificial nanostructures are essential for benchmarking different designs of imagers and testing their peak performance before further evaluation. Recently, utilizing a DNA barcoding method with orthogonal sequences and performing sequential acquisitions, researchers were able to resolve two docking strands that were placed just 1 base pair (bp) apart thanks to the programmability of a DNA origami platform40 (Fig. 4b).
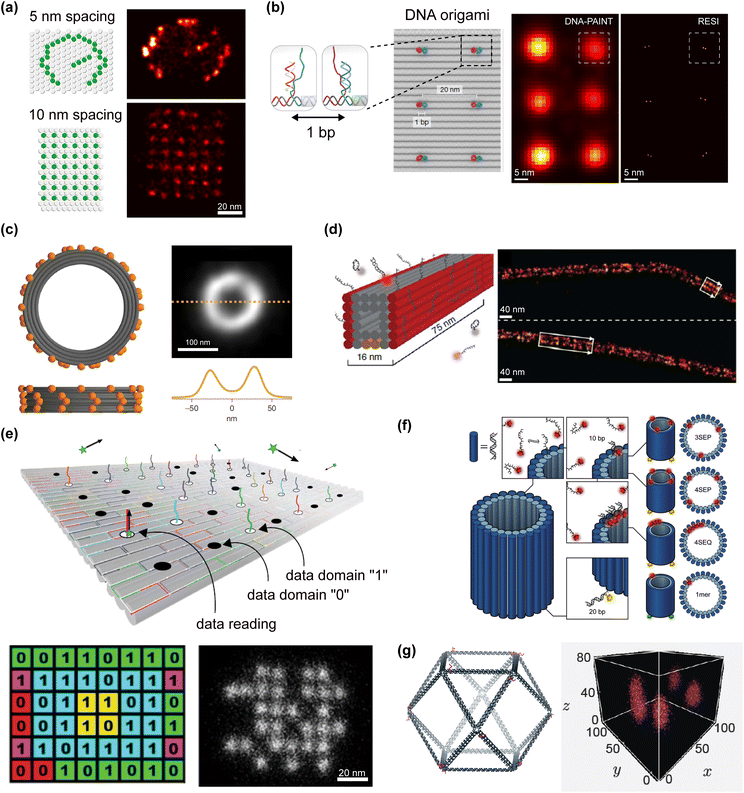 | ||
| Fig. 4 DNA-PAINT advancements and applications of DNA nanostructures imaging. (a) 2D DNA origami structures are exceptional tools to test DNA-PAINT resolution enabling the dockings strands positioning in dense arrays (adapted from Schueder et al.42 with permission from Springer Nature, copyright 2019), (b) with a possible precision of 1 base pair (bp) (adapted from Reinhardt et al.40via Creative Commons CC BY 4.0 licence). (c and d) 3D DNA origami structures can mimic cellular components (nuclear pore complex and microtubule respectively) shape and size to optimize DNA-PAINT performance before imaging in cells (adapted from Chung et al.56 and Jungmann et al.50 with permissions from Springer Nature, copyright 2014, and 2022). (e) DNA-PAINT imaging on DNA origami platforms enables innovative application in DNA data storage (adapted from Dickinson et al.60via Creative Commons CC BY 4.0 licence). (f) The use of DNA origami nanotubes can help to analyse the stochiometric quantification of tightly positioned targets (adapted from Baker et al.54 with permission from the Royal Society of Chemistry). (g) DNA-PAINT imaging on 3D DNA wireframe structures additionally promotes application in DNA origami data cryptography (adapted from Wisna et al.61via Creative Commons CC-BY-NC-ND 4.0 licence). | ||
Rectangular DNA origami platforms are deposited onto a surface using protein surface passivation (e.g. biotin/streptavidin) or cationic buffer solution to ensure adhesion. This fixed position enables effective imaging while the sample is flushed with solutions to introduce imager strands or wash away excess molecules. This approach has been instrumental in benchmarking key DNA-PAINT optimization achievements, including imaging in dense array patterns and enhancing resolution through imager strand sequence design optimization.35,42 DNA origami 2D platforms have also been valuable for benchmarking multiplexing capabilities and quantifying the transient binding dynamics between docking and imager strands.48,53,57 New docking strands can be modified in length, position, and sequence, allowing for easy substitution into the DNA origami platform without the need to redesign the structure. This flexibility is particularly advantageous for testing various sequences and patterns to determine optimal configurations for specific biomolecular targets.
Additionally, the versatility of the DNA origami technique in 3D scaffolding has been successfully utilized to analyse DNA-PAINT's ability to capture signals along the z-axis, enabling 3D imaging. This is accomplished by using a cylindrical lens that introduces astigmatism into the emission beam path of the optical setup.50 Several types of 3D DNA structures can be generated to mimic the shapes of biological molecules of interest, facilitating the testing of 3D DNA-PAINT advancements prior to application in biological settings. For example, cylindrical structures can be designed to replicate the diameter of Nuclear Pore Complexes (NPCs) (Fig. 4c) or to precisely quantify imagers cross-reactivity (Fig. 4f). Moreover, parallelepiped structures (Fig. 4d) can mimic the thickness and length of microtubules while precisely positioning docking strands at specific distances from the surface.50,54,56,75 3D structures can be also used to tune the position and density of docking strands along the z-axis. This is crucial for optimizing properties such as binding frequency and maximum achievable resolution. Although various 3D structures can be employed to evaluate the performance of 3D DNA-PAINT,75,76 the use of 3D DNA origami offers additional customization and functionalization potential.
The use of DNA origami structures to analyse and assess DNA-PAINT imaging performance has proven to be highly effective and relatively straightforward to implement. As demand has increased, the DNA nanostructures themselves have undergone refinements in design, shape, and docking array incorporation. This continuous optimization has elevated DNA origami as a key player in the field, as demonstrated by the meticulous attention given to perfecting their components.41,71
The exploration of DNA origami structures in relation to DNA-PAINT has provided the research community with a powerful, ready-to-use tool for nanoscale imaging. This development has also spurred innovative applications beyond traditional biological settings.77,78 For instance, 2D DNA origami platforms have successfully facilitated the prototyping of molecular data storage technologies, nanoscale metrology, and encryption for molecular cryptography36,60,61 (Fig. 4e). These DNA origami rectangles serve as efficient tools to translate ON and OFF signal localization sites into “1” and “0” digital bits, offering robust combinatorial platforms for encoding algorithms.36,60 Moreover, the use of 3D DNA wireframe structures can facilitate the signal readout in DNA data cryptography applications (Fig. 4g). The advances achieved by optimizing DNA-PAINT performance for biological environments have, at the same time, promoted innovation in nanoscale metrology applications.
3.3. Biological environment challenges
The application of DNA-PAINT in biological settings encounters significant challenges due to its complex and dense nature. Imaging molecules deep within thick samples or tissues is particularly difficult using a TIRF optical setup, which effectively precludes the application of DNA-PAINT to more complex biological systems. Consequently, DNA-PAINT's focus has largely remained at the cellular level. One of the primary challenges in cell imaging is the specific labeling of target molecules, which is complicated by high molecular density and typically lower achievable resolution compared to imaging DNA nanostructures.To address the limitations imposed by steric hindrance, alternative strategies have been developed for labelling target molecules in cellular environments. For example, nanobodies, which are derived by cleaving the constant regions from antibody molecules, are substantially smaller than full antibodies, thereby minimizing steric hindrance. Their use reduces the linkage error and allows for approximately 8–10 nm localization precision. However, their affinity for endogenous proteins is often limited, typically necessitating that target molecules be genetically modified to include epitopes recognizable by the nanobodies.82,83
Another strategy to maintain a small label size while avoiding the need for genetic modification involves using modified aptamers and small peptides. Slow-Off-rate Modified Aptamers (SOMAmers) are designed to mimic antibody epitopes and can be extended with a docking strand, demonstrating DNA-PAINT imaging resolution less than 8 nm.84 Affimers are small peptides that have been screened for high target affinity, stability, and small size.85 Their coupling with a docking strand enables imaging of actin fibres with an 18 nm resolution.86 Although these modified aptamers and small peptide labels simplify chemical construction, their implementation across a broad range of targets may require extensive screening processes to identify suitable imagers for specific target molecules.
Existing peptide-binding proteins can assist in overcoming this screening challenge, though they may also necessitate genetically modified tags to ensure adequate specificity.80,87 Ultimately, the selection of the most appropriate labelling strategy for DNA-PAINT imaging in cellular environments depends significantly on the researchers’ expertise and the specific objectives of the study.
Moreover, in live cell environments the laser phototoxicity damage phenomenon can alter cell morphology, inducing artificial physiology changes that can lead to misinterpretation of the imaging results. The main mechanism of phototoxicity is the generation of Reactive Oxygen Species (ROS) of which overproduction can affect cell compartments and molecular targets. The amount of generated ROS species depends on exposure duration and laser intensity. For this reason, in applications where reducing phototoxicity is paramount, fluorogenic imager designs can be used as a solution as they allow the use of minimal laser power.47,56,92 In live cells, the constant variation of their content represents an imaging challenge as well. The dynamic nature of cellular trafficking and the rearrangement of cytoskeletal proteins pose significant challenges, particularly when long acquisition times are required.93,94
As a result of these challenges, DNA-PAINT imaging in biological environments tends to yield lower resolution compared to imaging in non-biological settings. While the use of nanobodies and SOMAmers has achieved FWHM values of approximately 8 nm in fixed cell,84 the majority of cell imaging studies report values ranging from 10 to 30 nm. Although these values are promising for DNA-PAINT in cellular contexts, they are easily surpassed by the achievable sub-5 nm resolution when imaging on DNA origami platforms.
3.4. Imaging biological structures
Numerous advancements in DNA-PAINT have enhanced its performance in terms of speed, multiplexing, and resolution. However, achieving these performances in biological samples is challenging due to complex and dynamic environments. Consequently new findings have primarily been tested and applied in fixed cells, where conditions are more controllable. The cell cytoskeleton, microtubules, mitochondria, and NPCs are among the most imaged cellular structures.87 Fixing cells allows these organelles to remain stable throughout the required acquisition time, ensuring optimal performance while the cell membrane's permeability facilitates the arrival of imagers to their targets.88 Recent advancements in DNA-PAINT have also reduced acquisition times, making them compatible with live-cell imaging.98–100As mentioned, one practical method for targeting subcellular components is to link the DNA imager strand to primary-secondary antibody pairs, which provide tagging specificity and variability. This established method is applicable to a variety of targets, including membrane receptor single synapses95,101 and intracellular organelles50,52,80 (Fig. 5a). However, the linkage error (see section 3.3.1) and consequent high steric hindrance from antibodies can limit the achievable labeling density in dense cellular environments.79 To overcome this limitation, single domain antibodies (or nanobodies) have been successfully employed to label targets in various subcellular organelles at approximately 20 nm resolution83,102 (Fig. 5b). Furthermore, the cyclic exchange of imager strands and washing solutions, facilitated by Exchange-PAINT, allows for the creation of informative overlaid images with nanoscale resolution of multiplexed targets.50,52,103
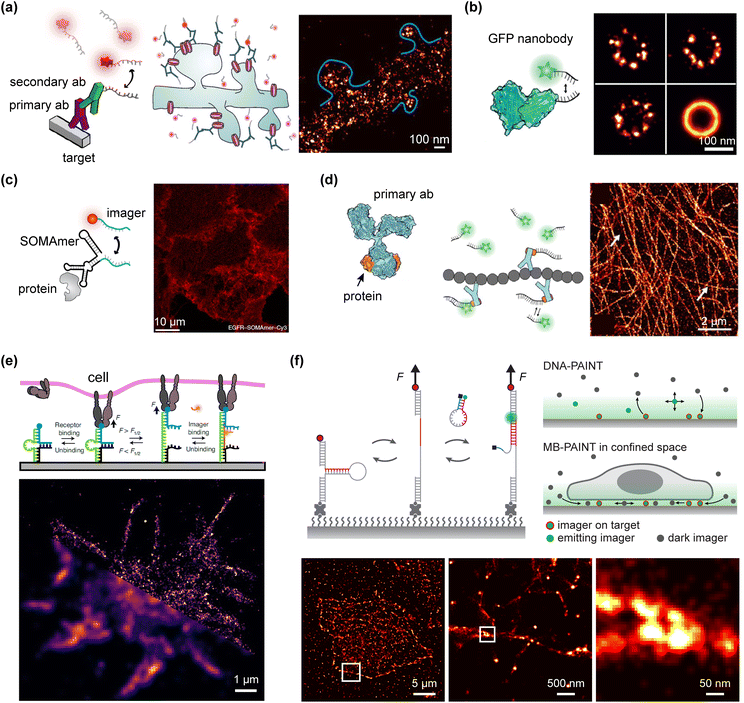 | ||
| Fig. 5 DNA-PAINT advancements in biological imaging applications. (a) Common primary-secondary antibodies DNA-PAINT labelling are used to image membrane receptors in synapses (adapted from Schnitzbauer et al.43 with permission from Springer Nature, copyright 2017; and from Böger et al.95via Creative Commons CC BY 4.0 licence). (b) Single-domain antibodies or nanobodies can provide 20 nm resolution in imaging nuclear pore complexes (adapted from Schlichthaerle et al.96via Creative Commons CC BY 4.0 licence). (c) Modified aptamers SOMAmers reduce the linkage error improving cell imaging resolution (adapted from Strauss et al.84 with permission from Springer Nature, copyright 2018). (d) Bacterial-derived small proteins (orange) that bind primary antibodies provide high affinity and low linkage error (adapted from Schlichthaerle et al.97 with permission from John Wiley & Sons, copyright 2019). (e) Imager strands with modified secondary structure can be used for DNA-PAINT live-cell dynamic tension imaging (adapted from Brockman et al.98 with permission from Springer Nature, copyright 2020). (f) Molecular beacons DNA-PAINT probes can detect forces exerted by extracellular matrix proteins during cell movement as well (adapted from Kim et al.47 with permission from John Wiley & Sons, copyright 2023). | ||
The trend toward smaller and simpler imagers without compromising labeling specificity has propelled DNA-PAINT cell imaging approaches that offer high labeling capacity and remarkable resolution. Notable examples include the conjugation of imager strands to natural small proteins82,86 (Affimers), and modified aptamers (SOMAmers)84 (Fig. 5c). The small size of these labels (7–30 kDa), combined with their high target affinity, has enabled DNA-PAINT cell imaging with sub-10 nm resolution and quantitative multiplexing.86,104,105 Additionally, bacterial-derived small proteins have been utilized as binders for primary antibodies97 (Fig. 5d). Depending on their bacterial origin, these proteins exhibit high affinity for various immunoglobulin proteins across different mammalian species, providing versatile binding tools for a broad range of primary antibodies. Their smaller size compared to secondary antibodies also reduces linkage distance and minimizes steric hindrance, providing tools for resolution improvement.
Moreover, the successful application of protein-based specificity in DNA-PAINT cell imaging has inspired strategies to enhance imaging speed. Proteins conjugated to imager strands can generate faster binding kinetics, thereby shortening acquisition times.45 This principle holds true for small peptides linked to antibodies that label microtubule proteins;106 the rapid interaction of these short peptides can mimic the transient binding dynamics of DNA-PAINT, resulting in faster kinetics in cell imaging conditions.106 Additionally, non-DNA-based transient linking mechanisms can be employed in PAINT imaging to analyse specific cellular uptake kinetics.107 For instance, by utilizing intrinsically weak and reversible carbohydrate–lectin interactions, known as Glyco-PAINT, researchers can analyse the kinetic and diffusion parameters of multiple ligands. This enables direct imaging of single molecules on the cell membrane or in living cells,99 demonstrating the versatility of target labeling in cellular environments, broadening the technique's utilization capabilities in bioimaging.
The significance of adapting target labeling techniques is further highlighted by other PAINT-based imaging analyses. Specifically designed oligonucleotides and fluorescent molecules have enabled the visualization of chromosomal domains and nucleoid-associated proteins in bacteria108 as well as in situ tracking of human genome regions.109 Additionally, adaptation of the imager strand design has facilitated the use of DNA-PAINT imaging to analyse the tension exerted by cells on extracellular matrix proteins during movement on surfaces98 (Fig. 5e). Modifications to the DNA secondary structure can create molecular beacons that are quenched in solution, emitting fluorescence only upon unfolding and binding to their targets. This capability efficiently suppresses background noise when imaging narrow adhesion spaces of cells, where both bound and unbound imager strands are closely confined to the surface. Consequently, DNA-PAINT can be employed to reveal the tension forces exerted by living cells47 (Fig. 5f). These examples serve to highlight DNA-PAINT's transformative capacity upon adoption in alternative fields.
4. Integration with other techniques
DNA-PAINT is characterized by fluorophore transient binding onto target structures, but imaging of such dynamic binding is not limited to TIRF. By combining DNA-PAINT with other imaging modalities, its imaging effectiveness is enhanced, integrating it with different methodologies augments the obtained information reciprocally. For instance, single-molecule FRET can be combined with DNA-PAINT to enable multiplexed imaging with specificity. This combination offers an alternative approach for multiplexed DNA-PAINT imaging while reducing background noise caused by non-specific binding of imagers.115 Furthermore, single-molecule FRET correlated with DNA-PAINT imaging can be applied to several established DNA-PAINT protocols.Another example of how DNA-PAINT's integration with other techniques mutually enhances their performance is its combination with MINFLUX (see section 2.1) to achieve higher resolution. By leveraging MINFLUX's fast acquisition and sub-nanometer precision with DNA-PAINT's high-density imaging and multiplexing capability,37 this combination has become one of the most powerful approaches in SRM. The recent advancement of incorporating Graphene Energy Transfer (GET) to achieve sub-nanometer 3D localization precision further showcases this.116 GET enables axial resolution below 0.3 nm by exploiting fluorescence lifetime quenching near graphene, while MINFLUX provides lateral resolution at the nanometer scale. This approach has enabled direct visualization of 3 nm-spaced docking strands on DNA origami and demonstrated rapid 3D DNA-PAINT imaging with Local PAINT (L-PAINT), a method that increases local imager concentration to boost imaging speed and reduce background noise. Together, these advances further solidify DNA-PAINT's role in pushing the boundaries of molecular-resolution imaging.116
Advancements in the application of artificial neural networks to DNA-PAINT image reconstruction is yet another example of how combining techniques from different fields can amplify DNA-PAINT's capabilities. The power of neural networks lies in their ability to extract complex features from a finite training dataset,117 accelerating image acquisition by reconstructing super-resolution images from a limited number of localizations.118,119 A well-trained neural network can predict the positions of imager strands in highly dense DNA-PAINT images, enabling acquisition times as short as a few minutes, even for large sample areas.120 Given DNA-PAINT's multiplexing capabilities, this combination with neural networks could pave the way for large-scale, high-throughput microscopy.
4.1. Correlative microscopy
Correlating the same sample area with two or more microscopy techniques can reveal additional insights. By combining different microscopy approaches, the limitations of each method can be overcome, allowing researchers to benefit from the strengths of both.121,122 Despite its promise, correlative microscopy has seen limited application in conjunction with DNA-PAINT imaging, primarily due to the incompatibility of different imaging environments and the complex alignment required to precisely correlate features between two fields of view.High-resolution correlative microscopy combining Atomic Force Microscopy (AFM) and DNA-PAINT has been used to analyse the addressability of docking strands on a DNA origami platform. AFM provides high-resolution characterization of structural defects resulting from poor strand incorporation, while DNA-PAINT confirms the functional activity of the docking strands72,123 (Fig. 6a). This correlative approach allows for the identification of defects at the single-molecule level, which could previously only be analysed statistically across groups of DNA structures. The combination of nanoscale topography from AFM with the super-resolution optical localization of dye-labelled strands offers promising prospects for nanoscale metrology applications.
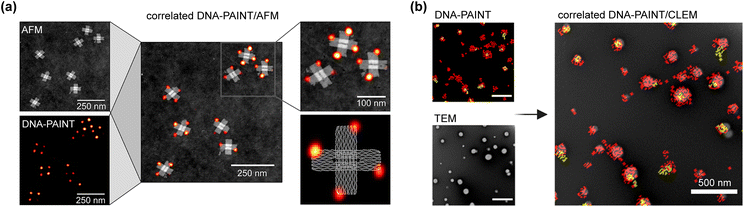 | ||
| Fig. 6 Main advancements of DNA-PAINT imaging using correlated microscopy. (a) Precise correlation between DNA-PAINT and AFM imaging allows to distinguish defect types in docking sites (adapted with permission from Green et al.72 Copyright 2021, American Chemical Society). (b) Correlative DNA-PAINT and TEM imaging can help to characterize the size and ligand number on nanoparticles (adapted from Andrian et al.110via Creative Commons CC-BY-NC-ND 4.0 licence). | ||
Electron Microscopy (EM) provides higher resolution than fluorescence microscopy but lacks molecular specificity. Combining EM with optical/fluorescence microscopy—known as Correlated Light and Electron Microscopy (CLEM)—is another example of how correlation can enhance characterization.124 Fluorescence microscopy's molecular specificity, paired with the high-resolution, widefield capabilities of EM, enables ultrastructural analysis of cellular and sub-cellular contexts.124 Several studies have extended this combination to SMLM, including iPALM and STORM, to characterize plasma membrane proteins and subcellular components at the nanoscale.124,125 More recently, DNA-PAINT has been applied to CLEM, where it has been correlated with Transmission Electron Microscopy (TEM) in a super-resCLEM method. This approach quantifies the number of surface ligands on nanoparticles while achieving nanometre precision in size and morphology110 (Fig. 6b). By combining the precision of both techniques, researchers have shown how size heterogeneity affects ligand distribution, and how significant nanoparticle populations would have gone undetected using only a single technique.110 Correlative methods like these are poised to enable precise, multiparametric characterization of nanomaterials, which is not possible with individual techniques alone, thus further broadening DNA-PAINT utilization.
4.2. Advanced optics
Applying DNA-PAINT to whole cells and thick samples presents several challenges, particularly due to sample thickness, density, and poor permeability to imagers. Advanced optical setups have been explored to overcome these limitations, with a focus on improving SNR, reducing background noise, and achieving super-resolution imaging in micrometer-thick samples.126 3D imaging in whole cells can be achieved by acquiring different sections along the z-axis, facilitated by illumination astigmatism introduced by a cylindrical lens.50 This optical sectioning has been further enhanced by combining DNA-PAINT with Spinning Disk Confocal (SDC) Microscopy.111 The use of a spinning disk that introduces a parallel array of pinholes provides an alternative to classical confocal microscopy, offering sectioning capabilities while retaining the advantage of camera-based (widefield) detection (Fig. 7a). By accumulating ∼500 nm-thick imaging sections through this setup, researchers have achieved whole-cell imaging with multiplexed, quantitative super-resolution.111 They demonstrated that DNA-PAINT imaging can be performed away from the coverslip, achieving ∼20 nm lateral and ∼80 nm axial resolution, thus extending the technique to a wider range of biological samples. | ||
| Fig. 7 Main advancements of DNA-PAINT imaging using advanced optics. (a) Confocal imaging combined with a spinning disk optical sectioning can be enhanced towards whole-cell imaging (adapted from Schueder et al.111via Creative Commons CC-BY 4.0 licence). (b) The alternative use of Lattice Light-Sheet microscopy has enabled the 3D DNA-PAINT whole-cell imaging without the need for optical sectioning (adapted from Ghosh et al.112 Reprinted with permission from AAAS). (c) Fluorescent Lifetime microscopy allows effective DNA-PAINT imaging in dense cellular environments detecting multi-target in 3D (adapted from Oleksiievets et al.113via Creative Commons CC BY 4.0 licence). (d) Innovative illumination setups like prism-based TIRF enable the expansion of the FOV allowing DNA-PAINT imaging at the multicellular scale (adapted from Rames et al.114via Creative Commons CC-BY-NC-ND 4.0 licence). | ||
Further advancements in 3D DNA-PAINT imaging have been made through the development of fluorogenic imagers. These imagers, akin to molecular beacons, remain quenched when not hybridized to docking strands, significantly reducing fluorescent background in bulk samples. This innovation has enabled 3D DNA-PAINT imaging of whole cells without the need for optical sectioning.56 DNA-PAINT's localization precision depends on the SNR and, consequently, on the number of photons collected from each imager. In a 4Pi optical setup, fluorescence is collected by two opposing objectives and combined interferometrically, doubling photon-collection efficiency.127 However, classical DNA-PAINT is incompatible with 4Pi imaging setups due to its unachievable optical sectioning. The introduction of fluorogenic imagers intrinsically provides a background reduction that is high enough to replace the background suppression provided by optical sectioning through TIRF illumination and thereby has allowed the combination of these imagers with 4Pi optical microscopy for faster 3D whole-cell DNA-PAINT imaging.56
Advanced optical illumination setups, such as Lattice Light Sheet (LLS) microscopy, offer additional solutions for imaging thick samples.128 In LLS microscopy, a thin sheet of light is used to illuminate the sample perpendicular to the optical axis, generating a sectioning tool for whole cells28 and tissues.129 This approach minimizes exposure to off-focus light, reducing background fluorescence and enhancing contrast. LLS microscopy has been successfully applied to SMLM approaches, including PAINT, achieving multiplexed 3D super-resolution imaging of intracellular membranes.28,130 Recently, LLS was combined with DNA-PAINT to investigate the organization of endogenous CD20 receptors in live B cells, achieving fast, whole-cell imaging112 (Fig. 7b). In addition to LLS, other innovative illumination techniques, such as line-scanning TIRF and prism-based TIRF, have been adapted for SMLM and DNA-PAINT to expand the FOV without sacrificing lateral resolution.114,131 These approaches expand DNA-PAINT imaging to larger sample areas, enabling super-resolution and high-throughput spatial mapping across molecular and multicellular scales (Fig. 7d).
Moreover, advanced hardware for signal acquisition has introduced new imaging dimensions to conventional DNA-PAINT. Recent improvements in photon multiplication technology have led to the development of innovative camera detectors,132 enabling the recording of fluorescence lifetimes (on the nanosecond scale) for individual fluorophores in dye mixtures.133 In Fluorescence Lifetime (FL) DNA-PAINT, targets can be distinguished by the different lifetimes of fluorophores conjugated to imager strands, allowing for effective imaging in dense cellular environments113 (Fig. 7c). This approach enables multi-target detection with fast acquisition times and has significant potential for high-throughput, highly multiplexed bioimaging.
5. Outlook
The significant advancements in DNA-PAINT, as reported in the literature, have elevated this imaging technique to a remarkable level of optimization. Due to these great strides, it is difficult to foresee a substantial margin for progress in DNA-PAINT alone, particularly in terms of imaging speed, resolution, and multiplexing. However, recent synergies between DNA-PAINT and various approaches are driving promising developments in alternative application areas.The exceptional performance of DNA-PAINT is increasingly enabling its use in broader nanoscale metrology applications. A striking example is the use of DNA-PAINT in the engineering of DNA-based materials for innovative molecular data storage. In this method, digital data is encoded onto 2D DNA origami platforms by inserting or omitting docking strands in a pre-arranged dense pattern. DNA-PAINT is then used to “read” the binary data by detecting signals from the specific patterns.60 This integration of DNA-PAINT as a reading technology has created a novel methodology for interpreting binary data, providing an alternative to traditional DNA sequencing technologies.134
The precise addressability of individual docking sites in 3D space has also facilitated the development of DNA-PAINT for DNA origami cryptography. In this approach, binary data is encoded through a 3D pattern of docking strand positions within a wireframe DNA origami structure. DNA-PAINT's 3D accuracy is then employed to assess these docking sites at different z-axis levels. The encoded message can only be decoded by applying specific pattern-matching rules.61 This 3D DNA-PAINT-based method offers a faster and more versatile alternative to AFM-based DNA origami cryptography.135
Thanks to these collective advancements, DNA-PAINT now meets the high standards of accuracy and precision required for nanoscale metrology, expanding its utility to applications beyond biological imaging. As this potential continues to grow, DNA-PAINT is expected to see increased use in fields such as material science, physics, and biophysics. Nevertheless, its primary focus remains on achieving true molecular resolution in ultrastructural cellular contexts, particularly in the imaging of thick samples and tissues. The integration of DNA-PAINT with other techniques and advanced optical setups will play a crucial role in this direction. Recent innovations in optical sectioning, lattice light-sheet microscopy, and fluorescence lifetime microscopy have already demonstrated the feasibility of expanding DNA-PAINT's capabilities to larger FOVs and thicker samples. Imaging large FOVs highlights the importance of having homogenous illumination across all the imaging area. Flat-top TIRF illumination improves the quantification accuracy over large FOV, enabling both homogenous spatial resolution and precise binding kinetics.136 Advance setups like this may become a standard feature for DNA-PAINT cellular imaging. These advancements further solidify DNA-PAINT's influential role in future biological and biomedical imaging applications.
In addition, recent studies have shown a more robust characterization of nanostructures when DNA-PAINT is combined with other microscopy techniques rather than alone.72,110 The augmentation of information demonstrated by these associations of different approaches anticipates the potential of DNA-PAINT correlative microscopy in cellular imaging.
6. Conclusions
DNA-PAINT has emerged as a powerful tool in SRM due to its high precision, programmability, and versatility. By leveraging the principles of DNA hybridization, this technique offers unprecedented control over the localization of fluorescent molecules, allowing for highly detailed imaging at the nanoscale. As advancements in imager design, binding kinetics, and image acquisition continue to refine DNA-PAINT, its applications have expanded beyond traditional biological systems to include broader fields such as nanoscale metrology. Recently, DNA-PAINT has proven useful for data storage and encryption at the molecular level, showing potential in applications orthogonal to biological imaging. These innovations not only enhance the resolution and accuracy of molecular imaging but also pave the way for new, interdisciplinary applications of SRM. As research progresses, DNA-PAINT is poised to remain a key player in the field, driving further innovation in molecular imaging and nanoscale metrology.Author contributions
L. P. conceptualized the idea, designed and wrote the manuscript. L. P., I. T. S. L. and W. L. H. revised and finalized the manuscript.Data availability
No primary research results, software or code have been included and no new data was generated or analysed as part of this review.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We sincerely thank J. Dardis for copy-editing the manuscript. L. P. and W. L. H. acknowledge partial funding support from the National Science Foundation (Award# 2227626) and the Canada Research Chairs Program (Award ID: CRC-2022-001710). I. T. S. L. acknowledges funding support from the Natural Sciences and Engineering Research Council (RGPIN-2024-04352) and the Canada Research Chairs Program (Award ID: CRC-2020-00143).References
- E. Abbe, Arch. Mikrosk. Anat., 1873, 9, 413–418 CrossRef.
- L. Schermelleh, A. Ferrand, T. Huser, C. Eggeling, M. Sauer, O. Biehlmaier and G. P. C. Drummen, Nat. Cell Biol., 2019, 21, 72–84 CrossRef CAS PubMed.
- M. Lelek, M. T. Gyparaki, G. Beliu, F. Schueder, J. Griffié, S. Manley, R. Jungmann, M. Sauer, M. Lakadamyali and C. Zimmer, Nat. Rev. Methods Primers, 2021, 1, 1–27 CrossRef.
- L. von Diezmann, Y. Shechtman and W. E. Moerner, Chem. Rev., 2017, 117, 7244–7275 CrossRef CAS PubMed.
- R. E. Thompson, D. R. Larson and W. W. Webb, Biophys. J., 2002, 82, 2775–2783 CrossRef CAS PubMed.
- K. I. Mortensen, L. S. Churchman, J. A. Spudich and H. Flyvbjerg, Nat. Methods, 2010, 7, 377–381 CrossRef CAS PubMed.
- M. Bates, T. R. Blosser and X. Zhuang, Phys. Rev. Lett., 2005, 94, 108101 CrossRef PubMed.
- M. Bates, B. Huang, G. T. Dempsey and X. Zhuang, Science, 2007, 317, 1749–1753 CrossRef CAS PubMed.
- J. Fölling, M. Bossi, H. Bock, R. Medda, C. A. Wurm, B. Hein, S. Jakobs, C. Eggeling and S. W. Hell, Nat. Methods, 2008, 5, 943–945 CrossRef PubMed.
- S. T. Hess, T. P. K. Girirajan and M. D. Mason, Biophys. J., 2006, 91, 4258–4272 CrossRef CAS PubMed.
- E. Betzig, G. H. Patterson, R. Sougrat, O. W. Lindwasser, S. Olenych, J. S. Bonifacino, M. W. Davidson, J. Lippincott-Schwartz and H. F. Hess, Science, 2006, 313, 1642–1645 CrossRef CAS PubMed.
- M. J. Rust, M. Bates and X. Zhuang, Nat. Methods, 2006, 3, 793–796 CrossRef CAS PubMed.
- M. Heilemann, S. Van De Linde, M. Schüttpelz, R. Kasper, B. Seefeldt, A. Mukherjee, P. Tinnefeld and M. Sauer, Angew. Chem., Int. Ed., 2008, 47, 6172–6176 CrossRef CAS PubMed.
- A. Sharonov and R. M. Hochstrasser, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 18911–18916 CrossRef CAS PubMed.
- F. Strecker, Die elektrische Selbsterregung: mit einer Theorie der aktiven Netzwerke, S.Hirzel-Verlag, Stuttgart, 1947 Search PubMed.
- H. Nyquist, Bell Syst. Tech. J., 1932, 11, 126–147 CrossRef.
- P. Annibale, S. Vanni, M. Scarselli, U. Rothlisberger and A. Radenovic, PLoS One, 2011, 6, e22678 CrossRef CAS PubMed.
- S. H. Lee, J. Y. Shin, A. Lee and C. Bustamante, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 17436–17441 CrossRef CAS PubMed.
- J. Demmerle, E. Wegel, L. Schermelleh and I. M. Dobbie, Methods, 2015, 88, 3–10 CrossRef CAS PubMed.
- J. R. Pyle and J. Chen, Beilstein J. Nanotechnol., 2017, 8, 2296–2306 CrossRef CAS PubMed.
- D. J. Nieves, K. Gaus and M. A. B. Baker, Genes, 2018, 9, 621 CrossRef PubMed.
- R. Jungmann, C. Steinhauer, M. Scheible, A. Kuzyk, P. Tinnefeld and F. C. Simmel, Nano Lett., 2010, 10, 4756–4761 CrossRef CAS PubMed.
- J. Chen, A. Bremauntz, L. Kisley, B. Shuang and C. F. Landes, ACS Appl. Mater. Interfaces, 2013, 5, 9338–9343 CrossRef CAS PubMed.
- E. J. Ambrose, Nature, 1956, 178, 1194–1194 CrossRef CAS PubMed.
- A. L. Mattheyses and D. Axelrod, J. Biomed. Opt., 2006, 11, 014006 CrossRef PubMed.
- W. P. Ambrose, P. M. Goodwin and J. P. Nolan, Cytometry, 1999, 36, 224–231 CrossRef CAS PubMed.
- E. Graugnard, W. L. Hughes, R. Jungmann, M. A. Kostiainen and V. Linko, MRS Bull., 2017, 42, 951–958 CrossRef CAS PubMed.
- W. R. Legant, L. Shao, J. B. Grimm, T. A. Brown, D. E. Milkie, B. B. Avants, L. D. Lavis and E. Betzig, Nat. Methods, 2016, 13, 359–365 CrossRef PubMed.
- M. Sauer and M. Heilemann, Chem. Rev., 2017, 117, 7478–7509 CrossRef CAS PubMed.
- S. Van De Linde, A. Löschberger, T. Klein, M. Heidbreder, S. Wolter, M. Heilemann and M. Sauer, Nat. Protoc., 2011, 6, 991–1009 CrossRef CAS PubMed.
- H. Deschout, F. C. Zanacchi, M. Mlodzianoski, A. Diaspro, J. Bewersdorf, S. T. Hess and K. Braeckmans, Nat. Methods, 2014, 11, 253–266 CrossRef CAS PubMed.
- D. Baddeley and J. Bewersdorf, Ann. Rev. Biochem., 2018, 87, 965–989 CrossRef CAS PubMed.
- R. W. Cole, T. Jinadasa and C. M. Brown, Nat. Protoc., 2011, 6, 1929–1941 CrossRef CAS PubMed.
- J. Min, C. Vonesch, H. Kirshner, L. Carlini, N. Olivier, S. Holden, S. Manley, J. C. Ye and M. Unser, Sci. Rep., 2014, 4, 1–9 Search PubMed.
- M. Dai, R. Jungmann and P. Yin, Nat. Nanotechnol., 2016, 11, 798–807 CrossRef CAS PubMed.
- L. Piantanida, G. D. Dickinson, J. M. Majikes, W. Clay, J. A. Liddle, T. Andersen, E. J. Hayden, W. Kuang and W. L. Hughes, ACS Nano, 2024, 18, 22369–22377 CrossRef CAS PubMed.
- L. M. Ostersehlt, D. C. Jans, A. Wittek, J. Keller-Findeisen, K. Inamdar, S. J. Sahl, S. W. Hell and S. Jakobs, Nat. Methods, 2022, 19, 1072–1075 CrossRef CAS PubMed.
- F. Balzarotti, Y. Eilers, K. C. Gwosch, A. H. Gynnå, V. Westphal, F. D. Stefani, J. Elf and S. W. Hell, Science, 2017, 355, 606–612 CrossRef CAS PubMed.
- R. Schmidt, T. Weihs, C. A. Wurm, I. Jansen, J. Rehman, S. J. Sahl and S. W. Hell, Nat. Commun., 2021, 12, 1–12 CrossRef PubMed.
- S. C. M. Reinhardt, L. A. Masullo, I. Baudrexel, P. R. Steen, R. Kowalewski, A. S. Eklund, S. Strauss, E. M. Unterauer, T. Schlichthaerle, M. T. Strauss, C. Klein and R. Jungmann, Nature, 2023, 617, 711–716 CrossRef CAS PubMed.
- M. Schickinger, M. Zacharias and H. Dietz, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E7512–E7521 CrossRef CAS PubMed.
- F. Schueder, J. Stein, F. Stehr, A. Auer, B. Sperl, M. T. Strauss, P. Schwille and R. Jungmann, Nat. Methods, 2019, 16, 1101–1104 CrossRef CAS PubMed.
- J. Schnitzbauer, M. T. Strauss, T. Schlichthaerle, F. Schueder and R. Jungmann, Nat. Protoc., 2017, 12, 1198–1228 CrossRef CAS PubMed.
- F. Civitci, J. Shangguan, T. Zheng, K. Tao, M. Rames, J. Kenison, Y. Zhang, L. Wu, C. Phelps, S. Esener and X. Nan, Nat. Commun., 2020, 11, 1–8 CrossRef PubMed.
- M. Filius, T. J. Cui, A. N. Ananth, M. W. Docter, J. W. Hegge, J. Van Der Oost and C. Joo, Nano Lett., 2020, 20, 2264–2270 CrossRef CAS PubMed.
- A. Auer, M. T. Strauss, T. Schlichthaerle and R. Jungmann, Nano Lett., 2017, 17, 6428–6434 CrossRef CAS PubMed.
- S. H. Kim and I. T. S. Li, Angew. Chem., Int. Ed., 2023, 62, e202217028 CrossRef CAS PubMed.
- S. Strauss and R. Jungmann, Nat. Methods, 2020, 17, 789–791 CrossRef CAS PubMed.
- E. Lubeck and L. Cai, Nat. Methods, 2012, 9, 743–748 CrossRef CAS PubMed.
- R. Jungmann, M. S. Avendaño, J. B. Woehrstein, M. Dai, W. M. Shih and P. Yin, Nat. Methods, 2014, 11, 313–318 CrossRef CAS PubMed.
- S. S. Agasti, Y. Wang, F. Schueder, A. Sukumar, R. Jungmann and P. Yin, Chem. Sci., 2017, 8, 3080–3091 RSC.
- F. Schueder, M. T. Strauss, D. Hoerl, J. Schnitzbauer, T. Schlichthaerle, S. Strauss, P. Yin, H. Harz, H. Leonhardt and R. Jungmann, Angew. Chem., Int. Ed., 2017, 56, 4052–4055 CrossRef CAS PubMed.
- R. Jungmann, M. S. Avendaño, M. Dai, J. B. Woehrstein, S. S. Agasti, Z. Feiger, A. Rodal and P. Yin, Nat. Methods, 2016, 13, 439–442 CrossRef CAS PubMed.
- M. A. B. Baker, D. J. Nieves, G. Hilzenrat, J. F. Berengut, K. Gaus and L. K. Lee, Nanoscale, 2019, 11, 12460–12464 RSC.
- O. K. Wade, J. B. Woehrstein, P. C. Nickels, S. Strauss, F. Stehr, J. Stein, F. Schueder, M. T. Strauss, M. Ganji, J. Schnitzbauer, H. Grabmayr, P. Yin, P. Schwille and R. Jungmann, Nano Lett., 2019, 19, 2641–2646 CrossRef CAS PubMed.
- K. K. H. Chung, Z. Zhang, P. Kidd, Y. Zhang, N. D. Williams, B. Rollins, Y. Yang, C. Lin, D. Baddeley and J. Bewersdorf, Nat. Methods, 2022, 19, 554–559 CrossRef CAS PubMed.
- A. H. Clowsley, W. T. Kaufhold, T. Lutz, A. Meletiou, L. Di Michele and C. Soeller, Nat. Commun., 2021, 12, 1–10 CrossRef PubMed.
- E. M. Unterauer, S. Shetab Boushehri, K. Jevdokimenko, L. A. Masullo, M. Ganji, S. Sograte-Idrissi, R. Kowalewski, S. Strauss, S. C. M. Reinhardt, A. Perovic, C. Marr, F. Opazo, E. F. Fornasiero and R. Jungmann, Cell, 2024, 187, 1785–1800 CrossRef CAS PubMed.
- F. Schueder, F. Rivera-Molina, M. Su, Z. Marin, P. Kidd, J. E. Rothman, D. Toomre and J. Bewersdorf, Cell, 2024, 187, 1769–1784 CrossRef CAS PubMed.
- G. D. Dickinson, G. M. Mortuza, W. Clay, L. Piantanida, C. M. Green, C. Watson, E. J. Hayden, T. Andersen, W. Kuang, E. Graugnard, R. Zadegan and W. L. Hughes, Nat. Commun., 2021, 12, 2371 CrossRef CAS PubMed.
- G. B. M. Wisna, D. Sukhareva, J. Zhao, D. Satyabola, M. Matthies, S. Roy, C. Wang, P. Šulc, H. Yan and R. F. Hariadi, bioRxiv, preprint, 2023, DOI:10.1101/2023.08.29.555281.
- A. Small and S. Stahlheber, Nat. Methods, 2014, 11, 267–279 CrossRef CAS PubMed.
- J. Cnossen, T. J. Cui, C. Joo and C. Smith, Opt. Express, 2021, 29, 27961–27974 CrossRef CAS PubMed.
- Y. Le Wu, P. Hoess, A. Tschanz, U. Matti, M. Mund and J. Ries, Nat. Methods, 2022, 20, 139–148 Search PubMed.
- S. Coelho, J. Baek, M. S. Graus, J. M. Halstead, P. R. Nicovich, K. Feher, H. Gandhi, J. J. Gooding and K. Gaus, Sci. Adv., 2020, 6, eaay8271 CrossRef CAS PubMed.
- A. Balinovic, D. Albrecht and U. Endesfelder, J. Phys. D: Appl. Phys., 2019, 52, 204002 CrossRef CAS.
- P. Blumhardt, J. Stein, J. Mücksch, F. Stehr, J. Bauer, R. Jungmann and P. Schwille, Molecules, 2018, 23, 3165 CrossRef PubMed.
- K. L. Tosheva, Y. Yuan, P. Matos Pereira, S. N. Culley and R. Henriques, J. Phys. D: Appl. Phys., 2020, 53, 163001 CrossRef CAS PubMed.
- J. Widengren, A. Chmyrov, C. Eggeling, P. Å. Löfdahl and C. A. M. Seidel, J. Phys. Chem. A, 2006, 111, 429–440 CrossRef PubMed.
- C. Kuang, D. Luo, X. Liu and G. Wang, Measurement, 2013, 46, 1393–1398 CrossRef.
- M. T. Strauss, F. Schueder, D. Haas, P. C. Nickels and R. Jungmann, Nat. Commun., 2018, 9, 1–7 CrossRef CAS PubMed.
- C. M. Green, W. L. Hughes, E. Graugnard and W. Kuang, ACS Nano, 2021, 15, 56 Search PubMed.
- A. S. Eklund, A. Comberlato, I. A. Parish, R. Jungmann and M. M. C. Bastings, ACS Nano, 2021, 15, 17668–17677 CrossRef CAS PubMed.
- P. W. K. Rothemund, Nature, 2006, 440, 297–302 CrossRef CAS PubMed.
- R. Lin, A. H. Clowsley, T. Lutz, D. Baddeley and C. Soeller, Methods, 2020, 174, 56–71 CrossRef CAS PubMed.
- R. Iinuma, Y. Ke, R. Jungmann, T. Schlichthaerle, J. B. Woehrstein and P. Yin, Science, 2014, 344, 65–69 CrossRef CAS PubMed.
- S. Kempter, A. Khmelinskaia, M. T. Strauss, P. Schwille, R. Jungmann, T. Liedl and W. Bae, ACS Nano, 2019, 13, 996–1002 CAS.
- F. N. Gür, S. Kempter, F. Schueder, C. Sikeler, M. J. Urban, R. Jungmann, P. C. Nickels and T. Liedl, Adv. Mater., 2021, 33, 2101986 CrossRef PubMed.
- S. M. Früh, U. Matti, P. R. Spycher, M. Rubini, S. Lickert, T. Schlichthaerle, R. Jungmann, V. Vogel, J. Ries and I. Schoen, ACS Nano, 2021, 15, 12161–12170 CrossRef PubMed.
- A. Banerjee, M. Anand and M. Ganji, Nanoscale, 2023, 15, 6563–6580 RSC.
- M. Ganji, T. Schlichthaerle, A. S. Eklund, S. Strauss, R. Jungmann, M. Ganji, T. Schlichthaerle, A. S. Eklund, S. Strauss and R. Jungmann, ChemPhysChem, 2021, 22, 911–914 CrossRef CAS PubMed.
- G. Carrington, D. Tomlinson and M. Peckham, Mol. Biol. Cell, 2019, 30, 2737 CrossRef CAS PubMed.
- F. Schueder, P. Mangeol, E. HoYee Chan, R. Rees, J. Schünemann, R. Jungmann, D. Görlich and F. Schnorrer, eLife, 2023, 12, 79344 CrossRef PubMed.
- S. Strauss, P. C. Nickels, M. T. Strauss, V. Jimenez Sabinina, J. Ellenberg, J. D. Carter, S. Gupta, N. Janjic and R. Jungmann, Nat. Methods, 2018, 15, 685–688 CrossRef CAS PubMed.
- C. Tiede, R. Bedford, S. J. Heseltine, G. Smith, I. Wijetunga, R. Ross, D. Alqallaf, A. P. E. Roberts, A. Balls, A. Curd, R. E. Hughes, H. Martin, S. R. Needham, L. C. Zanetti-Domingues, Y. Sadigh, T. P. Peacock, A. A. Tang, N. Gibson, H. Kyle, G. W. Platt, N. Ingram, T. Taylor, L. P. Coletta, I. Manfield, M. Knowles, S. Bell, F. Esteves, A. Maqbool, R. K. Prasad, M. Drinkhill, R. S. Bon, V. Patel, S. A. Goodchild, M. Martin-Fernandez, R. J. Owens, J. E. Nettleship, M. E. Webb, M. Harrison, J. D. Lippiat, S. Ponnambalam, M. Peckham, A. Smith, P. K. Ferrigno, M. Johnson, M. J. McPherson and D. C. Tomlinson, eLife, 2017, 6, e24903 CrossRef PubMed.
- T. Schlichthaerle, A. S. Eklund, F. Schueder, M. T. Strauss, C. Tiede, A. Curd, J. Ries, M. Peckham, D. C. Tomlinson and R. Jungmann, Angew. Chem., Int. Ed., 2018, 57, 11060–11063 CrossRef CAS PubMed.
- S. J. Sahl, S. W. Hell and S. Jakobs, Nat. Rev. Mol. Cell Biol., 2017, 18, 685–701 CrossRef CAS PubMed.
- R. van Wee, M. Filius and C. Joo, Trends Biochem. Sci., 2021, 46, 918–930 CrossRef CAS PubMed.
- H. J. Geertsema, G. Aimola, V. Fabricius, J. P. Fuerste, B. B. Kaufer and H. Ewers, Nat. Biotechnol., 2021, 39, 551–554 CrossRef CAS PubMed.
- T. Imanishi, S. Obika and L. Pu, Chem. Commun., 2002, 2, 1653–1659 RSC.
- Y. V. Pabon-Martinez, Y. Xu, A. Villa, K. E. Lundin, S. Geny, C. H. Nguyen, E. B. Pedersen, P. T. Jørgensen, J. Wengel, L. Nilsson, C. I. E. Smith and R. Zain, Sci. Rep., 2017, 7, 1–16 CrossRef CAS PubMed.
- W. Kasprzycka, W. Szumigraj, P. Wachulak and E. A. Trafny, BioEssays, 2024, 46, 2300122 CrossRef CAS PubMed.
- C. Niederauer, C. Nguyen, M. Wang-Henderson, J. Stein, S. Strauss, A. Cumberworth, F. Stehr, R. Jungmann, P. Schwille and K. A. Ganzinger, Nat. Commun., 2023, 14, 1–8 Search PubMed.
- F. Stehr, J. Stein, J. Bauer, C. Niederauer, R. Jungmann, K. Ganzinger and P. Schwille, Nat. Commun., 2021, 12, 1–8 CrossRef PubMed.
- C. Böger, A.-S. Hafner, T. Schlichthärle, M. T. Strauss, S. Malkusch, U. Endesfelder, R. Jungmann, E. M. Schuman and M. Heilemann, Neurophotonics, 2019, 6, 035008 Search PubMed.
- T. Schlichthaerle, M. T. Strauss, F. Schueder, A. Auer, B. Nijmeijer, M. Kueblbeck, V. Jimenez Sabinina, J. V. Thevathasan, J. Ries, J. Ellenberg and R. Jungmann, Angew. Chem., Int. Ed., 2019, 58, 13004–13008 CrossRef CAS PubMed.
- T. Schlichthaerle, M. Ganji, A. Auer, O. Kimbu Wade and R. Jungmann, ChemBioChem, 2019, 20, 1032–1038 CrossRef CAS PubMed.
- J. M. Brockman, H. Su, A. T. Blanchard, Y. Duan, T. Meyer, M. E. Quach, R. Glazier, A. Bazrafshan, R. L. Bender, A. V. Kellner, H. Ogasawara, R. Ma, F. Schueder, B. G. Petrich, R. Jungmann, R. Li, A. L. Mattheyses, Y. Ke and K. Salaita, Nat. Methods, 2020, 17, 1018–1024 CrossRef CAS PubMed.
- C. Oi, Z. Gidden, L. Holyoake, O. Kantelberg, S. Mochrie, M. H. Horrocks and L. Regan, Commun. Biol., 2020, 3, 1–10 CrossRef PubMed.
- S. A. Jones, S. H. Shim, J. He and X. Zhuang, Nat. Methods, 2011, 8, 499–505 CrossRef CAS PubMed.
- K. K. Narayanasamy, A. Stojic, Y. Li, S. Sass, M. R. Hesse, N. S. Deussner-Helfmann, M. S. Dietz, T. Kuner, M. Klevanski and M. Heilemann, Front. Synaptic Neurosci., 2021, 13, 671288 CrossRef CAS PubMed.
- S. Sograte-Idrissi, N. Oleksiievets, S. Isbaner, M. Eggert-Martinez, J. Enderlein, R. Tsukanov and F. Opazo, Cells, 2019, 8, 48 CrossRef CAS PubMed.
- E. M. Unterauer and R. Jungmann, Front. Synaptic Neurosci., 2022, 13, 798267 CrossRef PubMed.
- J. Hellmeier, S. Strauss, S. Xu, L. A. Masullo, E. M. Unterauer, R. Kowalewski and R. Jungmann, Nat. Methods, 2024, 21, 1702–1707 CrossRef CAS PubMed.
- T. Nojima, H. Konno, N. Kodera, K. Seio, H. Taguchi and M. Yoshida, PLoS One, 2012, 7, e52534 CrossRef CAS PubMed.
- A. S. Eklund, M. Ganji, G. Gavins, O. Seitz and R. Jungmann, Nano Lett., 2020, 20, 6732–6737 CrossRef CAS PubMed.
- R. Riera, T. P. Hogervorst, W. Doelman, Y. Ni, S. Pujals, E. Bolli, J. D. C. Codée, S. I. van Kasteren and L. Albertazzi, Nat. Chem. Biol., 2021, 17, 1281–1288 CrossRef CAS PubMed.
- G. Nir, I. Farabella, C. Pérez Estrada, C. G. Ebeling, B. J. Beliveau, H. M. Sasaki, S. H. Lee, S. C. Nguyen, R. B. McCole, S. Chattoraj, J. Erceg, J. AlHaj Abed, N. M. C. Martins, H. Q. Nguyen, M. A. Hannan, S. Russell, N. C. Durand, S. S. P. Rao, J. Y. Kishi, P. Soler-Vila, M. Di Pierro, J. N. Onuchic, S. P. Callahan, J. M. Schreiner, J. A. Stuckey, P. Yin, E. L. Aiden, M. A. Marti-Renom and C. T. Wu, PLoS Genet., 2018, 14, e1007872 CrossRef PubMed.
- C. K. Spahn, M. Glaesmann, J. B. Grimm, A. X. Ayala, L. D. Lavis and M. Heilemann, Sci. Rep., 2018, 8, 1–12 CAS.
- T. Andrian, P. Delcanale, S. Pujals and L. Albertazzi, Nano Lett., 2021, 21, 5360–5368 CrossRef CAS PubMed.
- F. Schueder, J. Lara-Gutiérrez, B. J. Beliveau, S. K. Saka, H. M. Sasaki, J. B. Woehrstein, M. T. Strauss, H. Grabmayr, P. Yin and R. Jungmann, Nat. Commun., 2017, 8, 1–9 CrossRef CAS PubMed.
- A. Ghosh, M. Meub, D. A. Helmerich, P. Eiring, K. M. Kortüm, S. Doose and M. Sauer, Science, 2025, 387, eadq4510 CrossRef CAS PubMed.
- N. Oleksiievets, Y. Sargsyan, J. C. Thiele, N. Mougios, S. Sograte-Idrissi, O. Nevskyi, I. Gregor, F. Opazo, S. Thoms, J. Enderlein and R. Tsukanov, Commun. Biol., 2022, 5, 1–8 CrossRef PubMed.
- M. J. Rames, J. P. Kenison, D. Heineck, F. Civitci, M. Szczepaniak, T. Zheng, J. Shangguan, Y. Zhang, K. Tao, S. Esener and X. Nan, Chem. Biomed. Imaging, 2023, 1, 817–830 CrossRef CAS PubMed.
- N. S. Deußner-Helfmann, A. Auer, M. T. Strauss, S. Malkusch, M. S. Dietz, H. D. Barth, R. Jungmann and M. Heilemann, Nano Lett., 2018, 18, 4626–4630 Search PubMed.
- J. Zähringer, F. Cole, J. Bohlen, F. Steiner, I. Kamińska and P. Tinnefeld, Light: Sci. Appl., 2023, 12, 1–8 CrossRef PubMed.
- Y. Lecun, Y. Bengio and G. Hinton, Nature, 2015, 521, 436–444 CrossRef CAS PubMed.
- W. Ouyang, A. Aristov, M. Lelek, X. Hao and C. Zimmer, Nat. Biotechnol., 2018, 36, 460–468 CrossRef CAS PubMed.
- M. Zhu, L. Zhang, L. Jin, J. Chen, Y. Zhang and Y. Xu, Front. Chem., 2022, 10, 864701 Search PubMed.
- K. K. Narayanasamy, J. V. Rahm, S. Tourani and M. Heilemann, Nat. Commun., 2022, 13, 1–11 Search PubMed.
- A. I. Gómez-Varela, D. R. Stamov, A. Miranda, R. Alves, C. Barata-Antunes, D. Dambournet, D. G. Drubin, S. Paiva and P. A. A. De Beule, Sci. Rep., 2020, 10, 1–10 CrossRef PubMed.
- A. Miranda, A. I. Gómez-Varela, A. Stylianou, L. M. Hirvonen, H. Sánchez and P. A. A. De Beule, Nanoscale, 2021, 13, 2082–2099 Search PubMed.
- C. M. Green, K. Schutt, N. Morris, R. M. Zadegan, W. L. Hughes, W. Kuang and E. Graugnard, Nanoscale, 2017, 9, 10205–10211 RSC.
- P. De Boer, J. P. Hoogenboom and B. N. G. Giepmans, Nat. Methods, 2015, 12, 503–513 CrossRef CAS PubMed.
- K. A. Sochacki, G. Shtengel, S. B. Van Engelenburg, H. F. Hess and J. W. Taraska, Nat. Methods, 2014, 11, 305–308 CrossRef CAS PubMed.
- M. Tokunaga, N. Imamoto and K. Sakata-Sogawa, Nat. Methods, 2008, 5, 159–161 CrossRef CAS PubMed.
- J. Wang, E. S. Allgeyer, G. Sirinakis, Y. Zhang, K. Hu, M. D. Lessard, Y. Li, R. Diekmann, M. A. Phillips, I. M. Dobbie, J. Ries, M. J. Booth and J. Bewersdorf, Nat. Protoc., 2020, 16, 677–727 CrossRef PubMed.
- C. H. Lu, W. C. Tang, Y. T. Liu, S. W. Chang, F. C. M. Wu, C. Y. Chen, Y. C. Tsai, S. M. Yang, C. W. Kuo, Y. Okada, Y. K. Hwu, P. Chen and B. C. Chen, Commun. Biol., 2019, 2, 1–10 CrossRef PubMed.
- F. Cella Zanacchi, Z. Lavagnino, M. Perrone Donnorso, A. Del Bue, L. Furia, M. Faretta and A. Diaspro, Nat. Methods, 2011, 8, 1047–1049 CrossRef PubMed.
- F. Wäldchen, J. Schlegel, R. Götz, M. Luciano, M. Schnermann, S. Doose and M. Sauer, Nat. Commun., 2020, 11, 1–6 Search PubMed.
- A. Mau, K. Friedl, C. Leterrier, N. Bourg and S. Lévêque-Fort, Nat. Commun., 2021, 12, 1–11 CrossRef PubMed.
- R. Hartig, Y. Prokazov, E. Turbin and W. Zuschratter, Methods Mol. Biol., 2014, 1076, 457–480 CrossRef CAS PubMed.
- N. Oleksiievets, C. Mathew, J. C. Thiele, J. I. Gallea, O. Nevskyi, I. Gregor, A. Weber, R. Tsukanov and J. Enderlein, Nano Lett., 2022, 22, 6454–6461 CrossRef CAS PubMed.
- L. Ceze, J. Nivala and K. Strauss, Nat. Rev. Genet., 2019, 20, 456–466 CrossRef CAS PubMed.
- Y. Zhang, F. Wang, J. Chao, M. Xie, H. Liu, M. Pan, E. Kopperger, X. Liu, Q. Li, J. Shi, L. Wang, J. Hu, L. Wang, F. C. Simmel and C. Fan, Nat. Commun., 2019, 10, 1–8 CrossRef PubMed.
- F. Stehr, J. Stein, F. Schueder, P. Schwille and R. Jungmann, Nat. Commun., 2019, 10, 1–8 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |




