Nucleic acid amplification-integrated single-molecule fluorescence imaging for in vitro and in vivo biosensing
Fei
Ma†
ab,
Chen-Chen
Li†
ac and
Chun-Yang
Zhang
 *a
*a
aCollaborative Innovation Center of Functionalized Probes for Chemical Imaging in Universities of Shandong, Key Laboratory of Molecular and Nano Probes, Ministry of Education, Shandong Provincial Key Laboratory of Clean Production of Fine Chemicals, College of Chemistry, Chemical Engineering and Materials Science, Shandong Normal University, Jinan 250014, China. E-mail: cyzhang@sdnu.edu.cn
bSchool of Chemistry and Chemical Engineering, Southeast University, Nanjing 211189, China. E-mail: fei@seu.edu.cn
cKey Laboratory of Optic-electric Sensing and Analytical Chemistry for Life Science, MOE, Shandong Key Laboratory of Biochemical Analysis, College of Chemistry and Molecular Engineering, Qingdao University of Science and Technology, Qingdao 266042, China
First published on 22nd October 2021
Abstract
Single-molecule fluorescence imaging is among the most advanced analytical technologies and has been widely adopted for biosensing due to its distinct advantages of simplicity, rapidity, high sensitivity, low sample consumption, and visualization capability. Recently, a variety of nucleic acid amplification approaches have been developed to provide a straightforward and highly efficient way for amplifying low abundance target signals. The integration of single-molecule fluorescence imaging with nucleic acid amplification has greatly facilitated the construction of various fluorescent biosensors for in vitro and in vivo detection of DNAs, RNAs, enzymes, and live cells with high sensitivity and good selectivity. Herein, we review the advances in the development of fluorescent biosensors by integrating single-molecule fluorescence imaging with nucleic acid amplification based on enzyme (e.g., DNA polymerase, RNA polymerase, exonuclease, and endonuclease)-assisted and enzyme-free (e.g., catalytic hairpin assembly, entropy-driven DNA amplification, ligation chain reaction, and hybridization chain reaction) strategies, and summarize the principles, features, and in vitro and in vivo applications of the emerging biosensors. Moreover, we discuss the remaining challenges and future directions in this area. This review may inspire the development of new signal-amplified single-molecule biosensors and promote their practical applications in fundamental and clinical research.
1 Introduction
Accurate and sensitive detection of biomolecules of interest is essential for both fundamental biological research and clinical applications. To achieve this goal, a series of bioanalytical methods have been developed including electrophoretic mobility shift assay,1 enzyme-linked immunosorbent assay,2 immunohistochemistry,3 microarrays,4 and northern5 and western blotting.6 Despite their wide applications, these traditional methods are often limited by time-consuming and laborious protocols, large sample consumption, semi-quantification, and insufficient sensitivity. The development of new biosensing technologies with improved sensitivity and easy operation is still urgently required. Currently, single-molecule fluorescence imaging represents one of the most advanced analytical tools.7–9 After direct or indirect labelling of molecules of interest with suitable fluorophores, the target signal can be easily quantified by measuring the fluorescence signal emitted from a single fluorophore with fluorescence microscopy such as total internal reflection fluorescence (TIRF)10 and confocal microscopy.11 Superior to the traditional bulk assays which can only measure the ensemble average, single-molecule imaging can measure the information and behaviour of individual molecules, providing an ideal biosensing platform with the characteristics of simplicity, rapidity, high sensitivity, low sample consumption, and visualization ability.12–14An emerging trend in single-molecule fluorescent biosensor development is the introduction of nucleic acid amplification to further improve the assay performance. Besides the function of genetic information storage, nucleic acids (including both DNAs and RNAs) are attractive materials for bioengineering due to the unique selective and predictable DNA Watson–Crick base pairing.15 In comparison with other biomolecules, the nucleic acid signal can be easily amplified through different mechanisms such as polymerization, cleavage, ligation, and strand displacement, and a series of nucleic acid amplification technologies have been well-established and adopted for amplified bioanalysis.16–18 The integration of single-molecule fluorescence imaging with nucleic acid amplification has greatly facilitated the construction of various fluorescent biosensors for in vitro and in vivo detection of DNAs, RNAs, enzymes, and live cells with high sensitivity and good selectivity. In this review, we review the advances in the development of fluorescent biosensors with the integration of single-molecule fluorescence imaging with nucleic acid amplification and summarize the principles, features and in vitro and in vivo applications of the emerging biosensors. Moreover, we discuss the remaining challenges and future directions in this area.
2 Nucleic acid amplification-assisted single-molecule biosensors for in vitro biosensing
Nucleic acid amplification catalyzed by polymerases (including DNA polymerases and RNA polymerases) and nucleases (including exonucleases and endonucleases), and enzyme-free strategies have been integrated with single-molecule fluorescence imaging for in vitro biosensing.2.1 DNA polymerases
DNA polymerases mediate nucleic acid amplification by catalyzing the incorporation of dNTPs into the 3′ end of a DNA primer to produce a longer DNA,19–21 and a series of DNA polymerase-based amplification methods such as rolling circle amplification,22 strand displacement amplification,23 and loop-mediated isothermal amplification24 have been rationally designed. Among them, DNA polymerase-catalyzed rolling circle amplification (RCA) is the most popular isothermal nucleic acid amplification strategy due to its simplicity and high amplification efficiency. RCA uses a circular template to produce a long repeating ssDNA through the polymerization of a short ssDNA primer by a DNA polymerase with strand displacement activity.25–27 RCA has been integrated with single-molecule detection for quantification of DNA and enzyme activities. As shown in Fig. 1A, Schopf and Chen developed a RCA-assisted single-molecule biosensor for DNA assays.28 The capture probe is conjugated with Sepharose beads, which can capture the circular primer in the presence of the target DNA through the formation of a capture probe/target DNA/circular primer sandwich hybrid. The addition of the phi29 DNA polymerase initiates RCA with the target DNA as the primer, and the RCA products coil up to form a cluster. Subsequently, Cy3-labeled reporter probes hybridize with the cluster, enabling the achievement of a high signal/background ratio larger than 10. The target DNA can be quantitatively detected by single-molecule counting of the fluorescent spots on the surface of Sepharose beads. This biosensor eliminates the complicated thermal cycling involved in the traditional polymerase chain reaction, enabling isothermal and sensitive DNA detection with a low detection limit of 1 amol.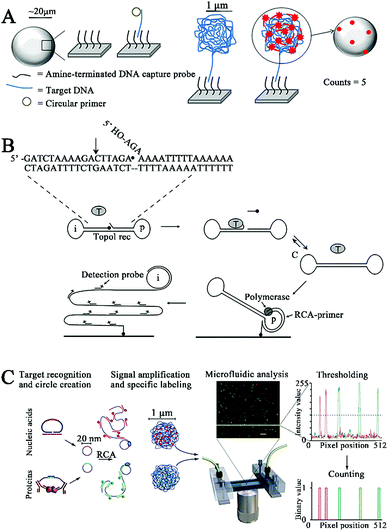 | ||
| Fig. 1 (A) RCA-Integrated single-molecule biosensor for DNA detection. Reprinted from ref. 28 with permission from Elsevier. (B) RCA-Integrated single-molecule biosensor for hTopoI activity detection. Reprinted from ref. 29 with permission from American Chemical Society. (C) RCA-Integrated single-molecule biosensor for bacterial pathogen DNA detection. Reprinted from ref. 30 with permission from Nature Publishing Group. | ||
Stougaard et al. employed RCA-assisted single-molecule detection to measure topoisomerase I (hTopoI).29 hTopoI participates in the regulation of the DNA topology structure to facilitate the DNA replication and transcription process through a cooperative cleavage–ligation reaction.31 hTopoI has emerged as a valuable biomarker for disease diagnosis and prognosis,32,33 and the inhibition of hTopoI activity is an effective way for treatment of diverse diseases including cancer.34–36 As shown in Fig. 1B, the target hTopoI cleaves the 3 base overhang at the 3′ end of the dumbbell substrate, facilitating the annealing of the 3 base overhang at the 5′ end of the dumbbell substrate to the stem region. Subsequently, hTopoI ligates the 3′- and 5′-end to form a closed DNA circle. The DNA circle can serve as a circular template and bind to a DNA primer immobilized on the glass surface to activate the Phi29 DNA polymerase-catalyzed RCA reaction, producing a long ssDNA product which can hybridize with FITC-labeled detection probes and be quantified by single-molecule fluorescence imaging. This biosensor exhibits high sensitivity with a low detection limit of 0.1 fmol, which is 100-fold higher than that obtained by the SYBR Gold-based ensemble measurement. This biosensor can be applied for accurate detection of hTopoI in crude yeast cell extracts.
Despite their good assay performance, the above biosensors require complicated protocols for the immobilization of probes onto the solid surface (e.g., Sepharose beads28 and glass slides29) and the separation of unbound probes. To overcome these limitations, Jarvius et al. constructed a homogeneous RCA-assisted single-molecule biosensor for the detection of bacterial pathogen DNA.30 As shown in Fig. 1C, the target DNA can bind with the padlock probe to induce the ligation of the padlock probe, forming a circular DNA. The resultant circular DNA can serve as a template to activate the RCA reaction, generating a long ssDNA product. The RCA product spontaneously collapses into a random coil, and the subsequent binding of multiple Alexa Fluor488-labeled signal probes generates an amplified fluorescence signal, which can be quantified by single-molecule counting using a confocal microscope coupled with a microfluidic platform. This biosensor can function in a homogeneous manner without the need for separation of unbound fluorophores, and it can sensitively detect the bacterial pathogen Vibrio cholerae with a detection limit of 8 bacterial genomes. Moreover, this biosensor can simultaneously detect multiple bacterial pathogens (i.e., V. fischeri and V. cholerae) using the corresponding padlock probes and signal probes.
2.2 Nucleases
Nucleases can cleave the phosphodiester bonds between the nucleotides of nucleic acids. Nucleases include exonucleases that digest DNA from its end and endonucleases that cleave the DNA strand at its middle region.37,38 Exonuclease III (exo III)-, lambda exonuclease (λexo)-, and endonuclease IV (endo IV)-mediated signal amplification has been integrated into a single-molecule biosensor for sensitive detection of enzymes and DNA methylation.Terminal deoxynucleotidyl transferase (TdT) randomly incorporates nucleotides into the DNA strand from the 3′-OH terminus, and it is an important biomarker of leukemia diseases (e.g., acute lymphoblastic leukemia and chronic myelogenous leukemia).39,40 Wang et al. constructed a quantum dot (QD)-based Exo III amplification-integrated single-molecule biosensor for TdT activity detection.41 QDs are novel fluorescent nanomaterials with the characteristics of super brightness, excellent photostability, and a long lifetime.42,43 Moreover, the broad absorption and narrow size-tuneable emission of QDs enable the homogeneous detection of various targets through fluorescence resonance energy transfer (FRET) with QDs as fluorescent donors.44,45 As shown in Fig. 2A, the A-rich probe is labeled with Cy5 and assembled onto the QD surface. Because Exo III is a duplex DNA specific nuclease that catalyzes the cleavage of DNA from the 3′ end,46 no cleavage of the Cy5-labeled probe occurs upon Exo III treatment in the absence of TdT, leading to efficient FRET from QDs to Cy5. In contrast, TdT incorporates dTTPs into the primer to form a T-rich polymerization product. The subsequent binding of the T-rich polymerization product with the A-rich Cy5-labeled probe activates the cyclic cleavage of the Cy5-labeled probe by Exo III, resulting in the release of Cy5 moieties from the QDs and the decrease of FRET, which can be quantified by single-molecule detection. The detection can be completed within 20 min, and the biosensor exhibits a detection limit as low as 1 × 10−6 U μL−1. Moreover, it can measure the endogenous TdT activity in human leukemic Molt-4 cells with a detection limit down to 5 cells.
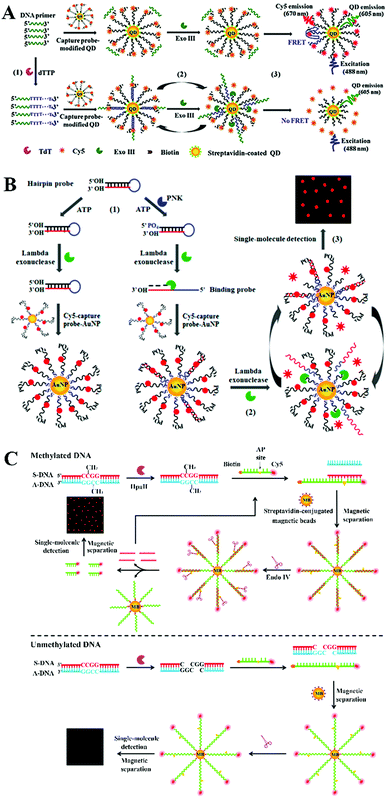 | ||
| Fig. 2 (A) Exo III amplification-integrated single-molecule biosensor for TdT activity detection. Reprinted from ref. 41 with permission from The Royal Society of Chemistry. (B) λexo amplification-integrated single-molecule biosensor for PNK activity detection. Reprinted from ref. 52 with permission from American Chemical Society. (C) Endo IV amplification-integrated single-molecule biosensor for DNA methylation detection. Reprinted from ref. 59 with permission from The Royal Society of Chemistry. | ||
Polynucleotide kinase (PNK) can transfer the phosphate from ATP to the 5′ OH end of the nucleic acid to generate a 5′ P end and plays key roles in DNA recombination and DNA damage repair.47–49 The dysregulated PNK activity has been closely associated with various human diseases including neurodegenerative diseases and spinocerebellar ataxia.50,51 Wang et al. developed a gold nanoparticle (AuNP)-based λexo amplification-integrated single-molecule biosensor for PNK activity detection.52 Gold AuNPs are one of the most popular nanomaterials for biosensing because they can be easily synthesized and exhibit distinct properties including a large surface area, distance-dependent colour changes, and efficient fluorescence quenching capability.53–56 As shown in Fig. 2B, Cy5-labeled capture probes can assemble on the gold nanoparticle (AuNP) surface, resulting in the quenching of Cy5 fluorescence by AuNPs. The target PNK catalyzes the transfer of phosphate from ATP to the 5′ OH end of a hairpin probe to form a 5′ phosphorylated hairpin probe which can be cleaved by λexo (λexo removes nucleotides from the 5′-phosphorylated end of dsDNA57) to generate a single-stranded binding probe. The binding probe can hybridize with capture probes to trigger the lambda exonuclease-catalyzed cyclic digestion of the capture probes, leading to the release of numerous Cy5 moieties from AuNPs. The released Cy5 fluorescent molecules can be quantitatively measured by single-molecule detection. This biosensor exhibits high sensitivity with a detection limit of 9.77 × 10−8 U μL−1, and it can detect the cellular PNK activity in human embryonic kidney HEK293T cells with a detection limit of 9.64 × 10−6 U μL−1.
The above exonuclease-based biosensors have the advantage of simplicity resulting from the sequence-independent activity of exonucleases, but they often suffer from a high background signal due to the nonspecific digestion of the ssDNA probe in the absence of a target. Alternatively, an endonuclease with the capability of recognizing a specific restriction site enables more accurate biosensing with a low background signal.58 Zhang et al. integrated endo IV-assisted signal amplification with single-molecule detection for sensitive detection of DNA methylation.59 Endo IV is an endonuclease that can cleave the DNA phosphodiester backbone at the apurinic/apyrimidinic (AP) site.58 DNA methylation is one of the main epigenetic modification types and it participates in post-transcriptional gene regulation.60–62 The dysregulation of DNA methylation has been closely linked to a variety of human diseases.63–65 As shown in Fig. 2C, DNA methylation protects the dsDNA target from digestion by the endonuclease HpaII, and the remaining sense DNA strand (S-DNA) will hybridize with the biotin- and Cy5-dual labeled signal probe to form the S-DNA/signal probe duplex which can assemble onto the magnetic bead surface through biotin–streptavidin binding. Because the signal probe has an AP site, endonuclease IV can cyclically cleave the signal probe in the duplex and release abundant Cy5 moieties from the magnetic beads. After magnetic separation, the released Cy5 fluorescent molecules can be monitored by single-molecule detection. In contrast, the unmethylated DNA will be completely cleaved by HpaII, and no S-DNA is present. As a result, the signal probe remains as ssDNA and it cannot be digested by dsDNA-specific endonuclease IV for the generation of the Cy5 signal. This biosensor displays a low detection limit of 7.3 × 10−17 M and it can detect a 0.01% methylation level in the mixture. This biosensor can be further applied for the detection of DNA methylation in genomic DNA samples with single-cell sensitivity.
2.3 DNA ligases
DNA ligases can catalyze the ligation of two short DNA strands to form a new longer DNA.66 The DNA ligase-catalyzed ligation amplification reaction (LAR) and ligation chain reaction (LCR) have been integrated into a single-molecule biosensor for the detection of enzymes and DNA methylation.The ligation amplification reaction can achieve efficient signal amplification through thermostable DNA ligase-catalyzed repeat of the ligation events and it has significant advantages such as high fidelity and high ligation efficiency.67,68 Alkaline phosphatase (ALP) displays hydrolytic activity and it can remove the phosphate group from diverse biomolecules.69,70 An abnormal endogenous ALP level can serve as a promising biomarker for the diagnosis of various human diseases (e.g., hypophosphatasia, rickets, and cancer).71–73 Ma et al. developed a ligase amplification reaction-based single-molecule biosensor for ALP activity detection.74 As shown in Fig. 3A, the target ALP can remove the phosphate group from the ssDNA detection probe, and the resultant detection probe with a 3′-hydroxyl terminus can be ligated to the assistant probe by DNA ligase using a DNA template to generate a secondary template. The secondary template will facilitate the ligation between the biotinylated probe and the Cy5-labeled probe to generate a Cy5- and biotin-labeled signal probe. Notably, repeat ligation can be triggered through thermal cycling, resulting in the production of abundant signal probes. The obtained signal probes can assemble onto the QD surface, inducing efficient FRET from the QDs to Cy5 and the emission of the Cy5 signal which can be monitored by single-molecule detection. Superior to other signal amplification technologies with the requirement of multiple enzymes, this biosensor needs only a single DNA ligase to complete signal amplification and exhibits a low detection limit of 5.63 × 10−7 U mL−1. This biosensor can be further applied for the detection of endogenous ALP in human embryonic kidney HEK293T cells and human breast adenocarcinoma MCF-7 cells.
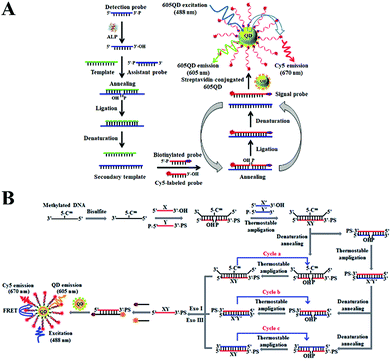 | ||
| Fig. 3 (A) LAR-Integrated single-molecule biosensor of ALP activity detection. Reprinted from ref. 74 with permission from The Royal Society of Chemistry. (B) LCR-Integrated single-molecule biosensor for DNA methylation detection. Reprinted from ref. 75 with permission from The Royal Society of Chemistry. | ||
The above biosensor can only achieve linear amplification by using one set of ligation probes (i.e., a biotinylated probe and a Cy5-labeled probe). To improve the assay performance, Wang et al. developed an exponential LCR-integrated single-molecule biosensor for the detection of DNA methylation by using two sets of ligation probes.75 As shown in Fig. 3B, the methylated DNA remains unchanged after bisulfite treatment, and the remaining DNA can function as a template to promote the DNA ligase-catalyzed ligation of probe X and probe Y. The resultant XY ligation product can also serve as a new template for ligation of probe X′ and probe Y′ to produce the X′Y′ ligation product. In turn, the X′Y′ ligation product further acts as a template to ligate X and Y probes. As a result, the LCR is activated to produce large amounts of XY strands. Subsequently, XY can hybridize with Cy5-labeled reporter probes and biotin-labeled capture probes to form the Cy5–biotin dual-labeled DNA structure which can assemble onto the 605QD to induce efficient FRET from the 605QD to Cy5. The FRET signal can be quantitatively monitored by single-molecule detection. In contrast, when unmethylated DNA is present, cytosine is converted to uracil upon bisulfite treatment, and no ligation reaction can be triggered due to the base mismatch. Consequently, no FRET signal can be detected. This biosensor enables exponential amplification of DNA methylation, and the detection limit can reach a value as low as 1 aM. Moreover, this biosensor can distinguish different DNA methylation levels in non-small-cell lung cancer H157 cells and small-cell lung cancer H209 cells with single-cell sensitivity.
2.4 Combination of multiple enzymes
Besides the use of single enzymes mentioned above, different kinds of enzymes can be combined together to achieve significant signal amplification. The exponential amplification reaction (EXPAR) is an isothermal nucleic acid amplification technology that employs a DNA polymerase and a nicking endonuclease to achieve exponential synthesis of ssDNA with a 106–109 fold amplification efficiency within minutes.48,76,77 Zhang et al. developed an EXPAR-integrated single-molecule biosensor for microRNA detection.78 MicroRNAs are short non-coding RNAs and they play versatile roles in cellular functions, especially in gene regulation, while the dysregulation of microRNA expression may cause various human diseases including cancer, and accurate detection of microRNAs is essential for clinical disease diagnosis, prognosis and drug discovery.79,80 As shown in Fig. 4A, the target microRNA can bind to the X′–X′ template to activate Vent (exo-) DNA polymerase- and Nt.BstNBI nicking enzyme-catalyzed EXPAR to produce a large amount of single-stranded X0. The resultant X0 can bind to an X′–Y′ template to produce a large amount of single-stranded Y through the strand displacement reaction. The obtained Y can hybridize with the Cy5-modified reporter probe and biotin-modified capture probe to produce a biotin- and Cy5-dual labeled hybrid which can assemble onto the 605QD surface to induce efficient FRET from the 605QD to Cy5. The FRET signal can be simply quantified by microfluidic flow-based single-molecule detection. This biosensor exhibits a near-zero background signal and the detection limit can reach 1 aM for the let-7a assay.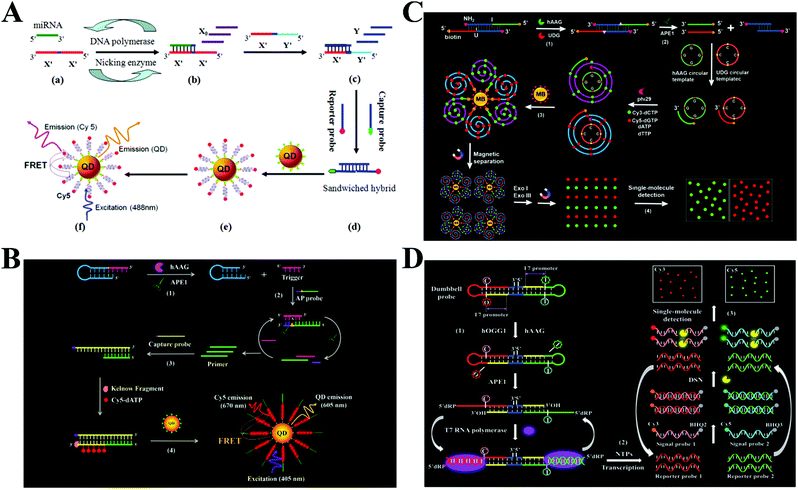 | ||
| Fig. 4 (A) DNA polymerase- and nicking endonuclease-mediated EXPAR-integrated single-molecule biosensor for microRNA detection. Reprinted from ref. 78 with permission from American Chemical Society. (B) APE1- and DNA polymerase-mediated polymerization amplification-integrated single-molecule biosensor for hAAG activity detection. Reprinted from ref. 81 with permission from The Royal Society of Chemistry. (C) RCA- and exonuclease-mediated cyclic amplification-integrated single-molecule biosensor for simultaneous detection of hAAG and UDG activities. Reprinted from ref. 82 with permission from The Royal Society of Chemistry. (D) RNA polymerase-mediated transcription amplification- and DSN-mediated cyclic amplification-integrated single-molecule biosensor for simultaneous detection of hOGG1 and hAAG activities. Reprinted from ref. 83 with permission from The Royal Society of Chemistry. | ||
DNA glycosylases can excise the damaged DNA nucleotides by cleaving the base–sugar (N-glycosylic) bond to maintain the integrity of genomic DNA and regulate the gene expression.84 The dysregulation of DNA glycosylase activities will disturb the gene regulation system and has been closely associated with various human diseases including neurodegeneration, leukaemia, chronic inflammation, and diverse types of cancer. DNA glycosylases have become important biomarkers for clinical diagnosis and monitoring of disease treatment.85,86 Zhang et al. integrated endo IV- and DNA polymerase-mediated signal amplification with single-molecule detection for the measurement of human alkyladenine DNA glycosylase (hAAG) activity.81 As shown in Fig. 4B, the target hAAG can remove the hypoxanthine base of the hairpin probe to produce an AP site, and then the AP site is cleaved by APE1 to release a single-stranded trigger. The trigger strand can subsequently bind with an AP probe containing an AP site to trigger the APE1-mediated cyclic cleavage of AP probes to obtain a single-stranded primer. The primer can hybridize with the biotin-labeled capture probe to initiate Klenow fragment DNA polymerase-mediated incorporation of multiple Cy5–dATPs into the dsDNA products. The assembly of Cy5- and biotin-dual labeled dsDNAs onto the streptavidin-coated QDs induces efficient FRET from the QDs to Cy5. The Cy5 signal can be sensitively quantified by single-molecule detection. This biosensor exhibits high sensitivity with a detection limit down to 4.42 × 10−12 U mL−1. Moreover, it can accurately quantify hAAG activity in different cell lines at the single-cell level.
Simultaneous detection of multiple targets can greatly facilitate high-throughput detection and reduce the assay time, but the above-mentioned biosensors are only suitable for single target detection. Zhang and colleagues integrated RCA- and exonuclease-mediated cyclic amplification with single-molecule detection for simultaneous measurement of multiple DNA glycosylases.82 As shown in Fig. 4C, two DNA glycosylases including hAAG and uracil–DNA glycosylase (UDG) were selected as the model targets. The hAAG and UDG can remove hypoxanthine and uracil bases in the dsDNA probe and release two ssDNA primers with the assistance of apurinic/apyrimidinic endonuclease 1. The two primers can bind with two corresponding circular templates to initiate the RCA reaction catalyzed by the phi29 DNA polymerase, resulting in the incorporation of abundant Cy3–dCTPs and Cy5–dGTPs into the RCA products. After the magnetic separation of fluorescently labelled RCA products from unbound Cy3–dCTPs and Cy5–dGTPs, the Cy3 and Cy5 moieties in RCA products are released upon the exonuclease I and exonuclease treatment. The released Cy3 and Cy5 fluorescent molecules indicate the presence of hAAG and UDG, respectively, and they can be quantified by single-molecule detection. This biosensor shows high sensitivity with a detection limit of 6.10 × 10−9 U mL−1 for hAAG and 1.54 × 10−9 U mL−1 for UDG. Moreover, this biosensor can simultaneously detect hAAG and UDG in cancer cells with single-cell sensitivity, and it can accurately quantify cellular enzyme levels in human lung adenocarcinoma A549 cells, human cervical carcinoma HeLa cells, human colon cancer SW480 cells, and human hepatocyte HL-7702 cells.
RNA polymerases catalyze the transcription amplification reaction through the polymerization of rNTPs in a 5′ to 3′ direction with specific promoter-containing DNA as the template, generating large amounts of ssRNAs.76,87,88 Duplex specific nuclease (DSN) is a nonspecific endonuclease enzyme and it can cleave the DNA in the DNA–RNA duplex.80,89,90 Wang et al. integrated RNA polymerase-mediated transcription amplification and DSN-mediated cyclic amplification with single-molecule detection for simultaneous detection of multiple DNA glycosylases including human 8-oxoguanine (hOGG1) and hAAG.83 As shown in Fig. 4D, hOGG1 and hAAG catalyze the removal of 8-oxoG and deoxyinosine bases from a dumbbell probe, respectively, consequently unfolding the two loop regions of the probe and exposing two T7 promoters. The exposed T7 promoters can activate the transcription reactions to generate two ssRNAs including reporter 1 and reporter 2. The hybridization of two reporters with two signal probes, which are labelled with fluorophores (Cy3 and Cy5) and quenchers (BHQ2 and BHQ3), respectively, initiates DSN-catalyzed cyclic digestion of signal probes, resulting in the recovery of Cy3 and Cy5 fluorescence signals which can be quantified by single-molecule detection, with Cy3 and Cy5 indicating hOGG1 and hAAG, respectively. The integration of single-molecule detection and transcription amplification endows this biosensor with good specificity and high sensitivity. The detection limit can reach 3.52 × 10−8 U μL−1 for hOGG1 and 3.55 × 10−7 U μL−1 for hAAG. Moreover, this biosensor can measure hAAG and hOOG1 in human serum and human lung adenocarcinoma A549 cells with single-cell sensitivity.
2.5 Enzyme-free strategies
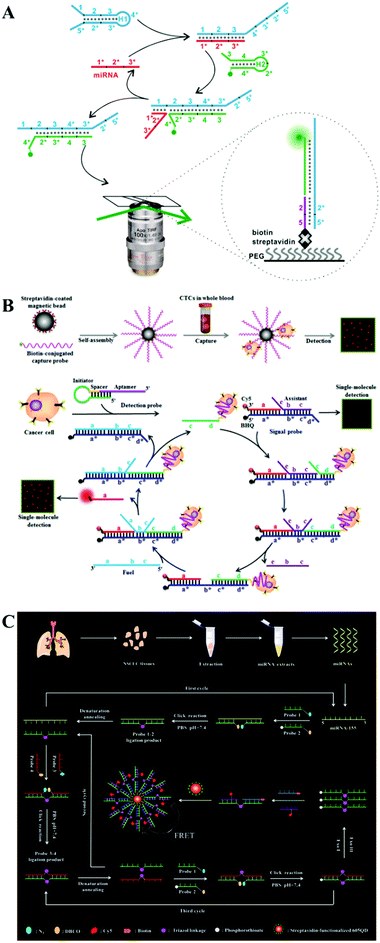
Despite the high efficiency for nucleic acid amplification, protein enzymes are relatively expensive and unstable with a tendency to be interfered by experimental conditions such as temperature, salt concentration, and pH value, which may impair the assay accuracy and reproducibility. Alternatively, enzyme-free strategies allow efficient signal amplification via the programmed DNA self-assembly process91 such as DNA walker92 and DNA circuits.93 A variety of enzyme-free nucleic acid amplification strategies such as catalytic hairpin assembly (CHA), entropy-driven DNA amplification, and click chemistry-based ligation chain reaction (LCR) have been integrated into single-molecule biosensors for sensitive detection of RNA and cancer cells.The CHA catalyzes the self-assembly of two separate hairpin DNA probes to form one DNA duplex through toehold-mediated strand displacement, which has significant advantages such as low background, high catalytic efficiency, and a simple reaction scheme.94,95 Hu et al. developed a CHA-integrated single-molecule biosensor for microRNA detection.96 As shown in Fig. 5A, the target microRNA can hybridize with domain 1 of hairpin probe H1 to trigger the toehold-mediated strand displacement reaction, and the opened H1 in turn binds to Cy3-labeled hairpin probe H2 to trigger the formation of the H1/H2 duplex via the toehold-mediated strand displacement reaction and release the microRNA from H1. The free microRNA can bind to other H1 to activate the cyclic CHA reaction. As a result, numerous H1/H2 duplexes are produced, which contain exposed domain 2*5* that can bind to the biotin-modified capture probe to form a biotin–Cy3 dual-labeled complex. The complex can be captured on a streptavidin-coated glass coverslip, and the immobilized Cy5 signal can be detected by TIRF imaging-based single-molecule detection after washing away unbound probes. This biosensor exhibits a low detection limit of 20 aM and can discriminate different microRNA (including miR-141, miR-21, and miR-16) levels between healthy persons and cancer (prostate cancer and breast cancer) patients.
 | ||
| Fig. 5 (A) CHA-Integrated single-molecule biosensor for microRNA detection. Reprinted from ref. 96 with permission from The Royal Society of Chemistry. (B) Entropy-driven DNA amplification-integrated single-molecule biosensor for cancer cell detection. Reprinted from ref. 99 with permission from American Chemical Society. (C) Click chemistry-based LCR-integrated single-molecule biosensor for microRNA detection. Reprinted from ref. 100 with permission from The Royal Society of Chemistry. | ||
In comparison with CHA that is driven by the negative free energy change of base pair formation, the entropy-driven amplification reaction is an enzyme-free cascade with DNA branch migration reaction being driven by the configurational entropic gain of the liberated DNA strands, and it is much stabler, simpler, and faster than the CHA reaction due to the elimination of unusual secondary structures (e.g., pseudoknots and kissing loops).97,98 Ma et al. constructed an entropy-driven DNA amplification-integrated single-molecule biosensor for cancer cell detection.99 As shown in Fig. 5B, a biotinylated A549 cell aptamer is assembled onto streptavidin-coated magnetic beads, facilitating efficient capture of A549 cells from serum. The binding of the detection probe with the captured A549 cells results in the exposure of an initiator sequence which activates the entropy-driven amplification reaction to release large amounts of Cy5-modified reporter strands from the quenched signal probes. The released Cy5 fluorescent molecules can be visualized and detected by single-molecule imaging. This biosensor can accurately detect rare cancer cells in an enzyme- and antibody-free manner, and it can sensitively detect even 3 cancer cells in complicated serum samples.
Despite their excellent performance, CHA and entropy-driven amplification exhibit a relatively low sensitivity due to their linear amplification nature. To achieve exponential signal amplification, Wang et al. developed a click chemistry-based LCR-integrated single-molecule biosensor for sensitive detection of microRNAs.100 As shown in Fig. 5C, four DNA probes (probes 1, 2, 3, and 4) are designed as the ligation elements. Among them, probe 1 and probe 3 are modified with azide (N3), while probe 2 and probe 4 are modified with dibenzocyclooctyne (DBCO). The hybridization of a microRNA with probe 1 and probe 2 induces the formation of the ligation product of probe 1–2 through click chemistry. Probe 1–2 subsequently templates probe 3 and probe 4 to generate the ligation product of probe 3–4. Both probe 1–2 and probe 3–4 can act as new templates to trigger cyclic ligation of the four DNA probes. As a result of these click chemistry-mediated LCR amplifications, numerous probes 1–2 are generated. The hybridization of probe 1–2 with the biotinylated capture probe and Cy5-labeled reporter probe forms the sandwiched product which can assemble on the streptavidin-coated QDs through biotin–streptavidin binding and induces efficient FRET from the QDs to Cy5. The Cy5 signal can be sensitively quantified by single-molecule imaging. This biosensor employs copper-free click chemistry to eliminate the unstable DNA ligase for LCR initiation, and it can discriminate the single-base mismatched microRNA with fM sensitivity. Moreover, this biosensor can accurately detect the microRNA expression level in tissue samples from both healthy persons and lung cancer patients.
Generally, both enzyme-based and enzyme-free strategies have advantages and disadvantages. Enzyme-based amplification methods are more popular than enzyme-free strategies due to their high signal amplification efficiency and well-studied mechanism, but the enzymes are usually costly, unstable, and easily interfered by environmental conditions. Enzyme-free strategies are attracting increasing attention in biosensing due to their simple reaction scheme, reagentless nature, and high robustness in complicated environments. However, the relatively low amplification efficiency and the requirement of delicate probe design limit their wide applications.
3 Nucleic acid amplification-integrated single-molecule detection for in vivo biosensing
Besides their use for in vitro assays, single-molecule biosensors have also been employed for in vivo biosensing applications. These biosensors enable real-time tracking of the dynamic changes of targets of interest in live cells, facilitating the study of their roles in physiological and pathological processes.3.1 Enzyme-free biosensors
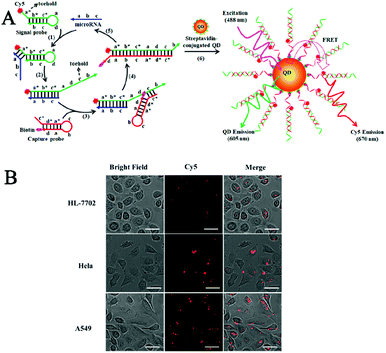
Enzyme-free strategies include CHA and hybridization chain reaction (HCR), and they have been employed to construct in vivo biosensors for microRNA and lncRNA detection.Ma et al. reported the CHA-catalyzed self-assembly of a single QD biosensor for sensitive detection of microRNAs in live cells.101 As shown in Fig. 6A, the binding of the target microRNA with toehold a* of the signal probe exposes its toehold c through the strand displacement reaction, and toehold c can subsequently bind with the capture probe to produce a signal probe/capture probe duplex, resulting in the release of the microRNA from the microRNA/capture probe duplex. The released microRNA can hybridize with a new capture probe to trigger new rounds of strand displacement cascade, generating numerous Cy5- and biotin-labeled signal probe/capture probe duplexes which can assemble onto the streptavidin-coated QDs to induce efficient FRET from the QDs to Cy5. The Cy5 signal can be simply quantified by single-molecule detection. This biosensor does not require any enzymes to achieve efficient signal amplification, and it can sensitively detect miR-21 with fM sensitivity and single-base discrimination capability. Moreover, this biosensor can be used to detect circulating microRNAs in clinical blood samples from lung cancer patients and for real-time imaging of intracellular microRNA expression in live cells including human normal liver HL-7702 cells, human lung adenocarcinoma A549 cells, and human cervical cancer HeLa cells (Fig. 6B).
 | ||
| Fig. 6 (A) CHA-Catalyzed self-assembly of a single QD biosensor for microRNA detection. (B) Imaging of microRNAs in live cells. Reprinted from ref. 101 with permission from American Chemical Society. | ||
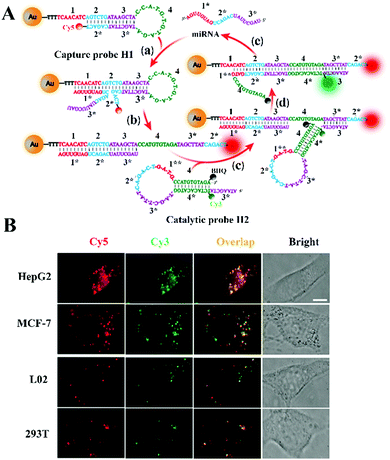
Li et al. constructed a CHA-catalyzed self-assembled AuNP biosensor for single-molecule detection of microRNAs in live cells.102 As shown in Fig. 7A, the assembly of the Cy5-labeled H1 strand onto the AuNP results in the quenching of Cy5 by AuNPs. The target microRNA can hybridize with toehold 1 of H1 to expose toehold 3* of H1 via the strand displacement reaction. The exposed toehold 3* can subsequently bind with the Cy3-labeled H2 strand to initiate the strand displacement reaction, leading to the formation of the H1/H2 duplex and the release of the microRNA. The released microRNA can activate new toehold-mediated strand displacement cascades to produce numerous H1/H2 duplexes, consequently inducing the separation of Cy3 and Cy5 moieties from the AuNPs and the recovery of both Cy3 and Cy5 signals. The colocalized Cy5 and Cy3 signals can be monitored by single-molecule detection. This biosensor employs dual-colour (Cy3 and Cy5) colocalization to greatly reduce the false-positive signal, and it can accurately detect miR-21 with fM sensitivity. Moreover, this biosensor can be applied for single-molecule imaging of different miR-21 expressions in cancer cell lines (e.g., human breast cancer MCF-7 cells and human hepatocellular carcinoma HepG2 cells) and normal cell lines (e.g., human liver L02 cells and human embryonic kidney 293T cells).
 | ||
| Fig. 7 (A) CHA-Catalyzed self-assembly of the AuNP nanosensor for single-molecule detection of microRNAs. (B) Imaging of microRNAs in live cells. Reprinted from ref. 102 with permission from American Chemical Society. | ||
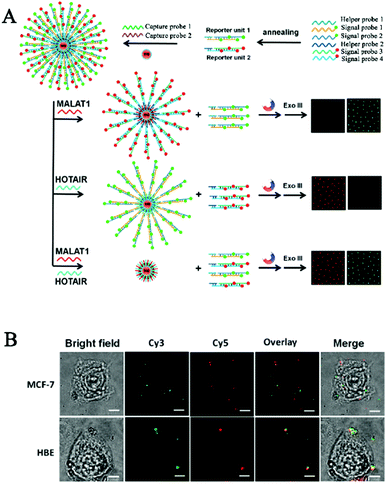
One limitation of the above CHA-based biosensors is the requirement of careful hairpin-structured probe design to avoid nonspecific hybridization. Zhang et al. developed a simpler HCR-based single-molecule biosensor for real-time imaging of lncRNAs in live cells with the involvement of only linear DNA probes.103 Long (>200 nt) noncoding RNAs (lncRNAs) are endogenous RNAs without the protein-coding capability,104,105 and they play pivotal roles in a number of cellular functions such as transcription control, chromatin modelling, and epigenetic regulation.105–107 Abnormal lncRNA levels have been closely associated with diverse diseases and have been regarded as valuable hallmarks of cancer.108–110 As shown in Fig. 8A, the reporter unit consists of long fluorophore (Cy3 or Cy5)-labeled DNA chains constructed by the HCR reaction, and they can assemble onto the streptavidin-modified magnetic beads after hybridization with the corresponding biotin-labeled capture probes, with capture probe 1 for sensing the mammalian metastasis-related lung adenocarcinoma transcript 1 (MALAT1) lncRNA and capture probe 2 for sensing the HOX gene antisense intergenic RNA (HOTAIR). MALAT1 and HOTAIR can specifically bind capture probe 1 and capture probe 2, respectively, inducing the release of reporter unit 1 and reporter unit 2 from the magnetic beads through the strand displacement reaction. After the digestion by exonuclease III, the free Cy3 and Cy5 florescent molecules can be accurately quantified by single-molecule detection, with Cy3 and Cy5 indicating the presence of MALAT1 and HOTAIR, respectively. This biosensor enables the simultaneous detection of two different kinds of lncRNAs with aM sensitivity at constant reaction temperature, and it can be applied for real-time imaging of multiple lncRNAs in human breast cancer MCF-7 cells and normal human breast epithelial HBE cells at a single-particle level (Fig. 8B).
 | ||
| Fig. 8 (A) HCR amplification-integrated single-molecule biosensor for in vitro detection (A) and in vivo imaging (B) of lncRNAs in live cells. Reprinted from ref. 103 with permission from American Chemical Society. | ||
3.2 Enzyme-assisted biosensors
The above in vivo biosensors can transduce the target signal into a freely dispersed fluorescence signal, but they are not suitable for in situ biosensing. Alternatively, DNA polymerase-catalyzed RCA can generate a long DNA from the original site of the target molecule, facilitating in situ biosensing.25–27 RCA-integrated single-molecule biosensors have been applied for the detection of cell membrane proteins, mRNAs, and microRNAs.Cell membrane proteins are involved in many important cellular functions such as cellular signal transduction, uptake of outside materials, and discharge of inside materials, and their abnormal expression and distribution are closely associated with disease progression.112,113 Fan et al. constructed a RCA-integrated single-molecule biosensor for in situ detection of cell membrane proteins in live cells.111 As shown in Fig. 9A, a short DNA strand consisting of an aptamer and a RCA primer sequence was designed as the recognition probe. The recognition probe can bind with the target membrane protein to initiate RCA, producing a long ssDNA strand. The hybridization of RCA products with Cy5-labeled signal probes induces the generation of a high fluorescence signal which can be simply quantified by single-molecule detection. This biosensor can measure two model membrane proteins including the epithelial cell adhesion/activating molecule (EpCAM) and protein tyrosine kinase 7 (PTK7) with good specificity and single-molecule/protein sensitivity, and it can discriminate the expression levels of different membrane proteins in different cancer cell lines including human cervical cancer HeLa cells and human hepatocellular carcinoma HepG2 cells (Fig. 9B). Moreover, this biosensor can be further applied to analyse the downregulation of EpCAM during the epithelial–mesenchymal transition process.
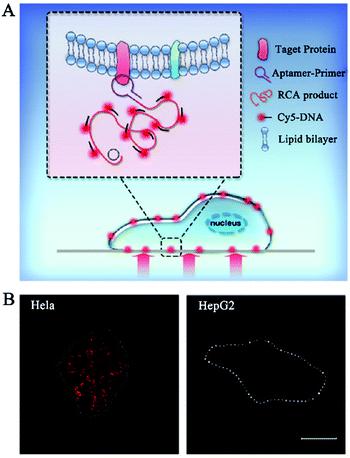 | ||
| Fig. 9 (A) RCA-Integrated single-molecule biosensor for detection of cell membrane proteins. (B) Real-time imaging of cell membrane proteins in live cells. Reprinted from ref. 111 with permission from The Royal Society of Chemistry. | ||
Deng et al. developed a RCA-integrated single-molecule biosensor for in situ detection and imaging of mRNAs.114 As shown in Fig. 10A, the target mRNA can serve as a ligation template to induce the ligation of the padlock probe by SplintR ligase. The formation of a circular template can trigger an efficient phi29 DNA polymerase-catalyzed RCA reaction. Upon hybridization with the fluorophore-labeled detection probe, the RCA products are visible as distinct fluorescent spots which can be quantified by single-molecule counting. This biosensor can discriminate different mRNA targets with single-nucleotide variations, and it can be applied for spatial mapping and correlation analysis of mRNA ACTB expression in single cells (Fig. 10B).
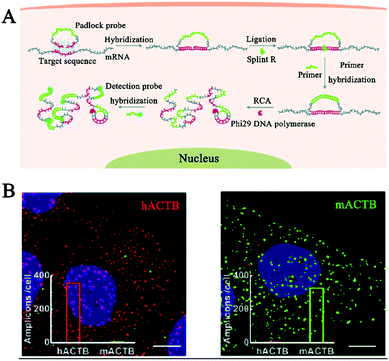 | ||
| Fig. 10 (A) RCA-Integrated single-molecule biosensor for in situ detection of mRNAs. (B) Imaging of mRNAs in single cells. Reprinted from ref. 114 with permission from The Royal Society of Chemistry. | ||
The above method requires SplintR ligase to recognize mRNAs, which inevitably complicates the assay system. Deng et al. developed a toehold-mediated RCA for in situ single-molecule imaging of microRNAs without the need for additional ligases.115 As shown in Fig. 11A, the target microRNA binds to the toehold domain of the dumbbell probe, inducing its switch from the “closed” form to the “activated circular” form through toehold-mediated branch migration. The circular probe subsequently acts as an efficient template to trigger RCA, producing a long ssDNA product. The RCA product can hybridize with the FAM-labeled probe to generate a fluorescent spot which can be well distinguished from the background. In contrast, the mismatched microRNA is unable to open the dumbbell probe, and thus no RCA occurs to produce a detectable signal. This biosensor is very sensitive with a detection limit of 0.72 fM. It can discriminate one-base mismatch and enable single-molecule imaging of the let-7a microRNA in single A549 cells (Fig. 11B).
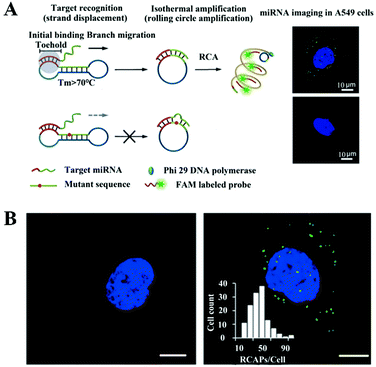 | ||
| Fig. 11 (A) Toehold-mediated RCA-integrated single-molecule biosensor for in situ detection of microRNA. (B) Imaging of microRNAs in A549 cells. Reprinted from ref. 115 with permission from Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. | ||
4 Conclusions and outlook
Benefiting from the advances in fluorescence microscopy and spectroscopy, single-molecule fluorescence detection provides a powerful platform for simple and sensitive quantification of biomolecules of interest. Despite its high sensitivity for single-molecule detection, it is difficult to detect low abundance targets with a concentration lower than the pM levels,116 which severely limits its practical applications in accurate detection of extremely rare targets (e.g., the biomarkers existing in clinical samples). To overcome this issue, a series of nucleic acid amplification technologies have been introduced and integrated with single-molecule detection. Through the utilization of different enzyme (e.g., polymerase,28–30 nuclease,41,52,59 and ligase74,75)-catalyzed amplification and enzyme-free amplification strategies (i.e., CHA and HCR), the target signal can be easily transferred and efficiently amplified, which greatly facilitates the construction of novel single-molecule biosensors for sensitive detection of a variety of disease-related biomarkers including DNAs, DNA methylation, microRNAs, lncRNAs, enzymes, and live cells (Table 1), holding great potential in disease diagnosis and fundamental biomedical research.| Amplification strategy | Target | Detection range | Detection limit | Real sample | Ref. |
|---|---|---|---|---|---|
| DNA polymerase-catalyzed RCA | DNA | 1 amol–1 fmol | 1 × 10−18 mol | — | 28 |
| DNA polymerase-catalyzed RCA | TopoI | 0.1 fmol–3 fmol | 1 × 10−16 mol | Yeast cells | 29 |
| DNA polymerase-catalyzed RCA | Genomes | 10–105 genomes | 8 genomes | Bacterial pathogen | 30 |
| Exo III-catalyzed cyclic cleavage | TdT | 1 × 10−6–1 × 10−3 U mL−1 | 1 × 10−9 U L−1 | HeLa and Molt-4 cells | 41 |
| λ exo-catalyzed cyclic cleavage | PNK | 1 × 10−7–1 × 10−3 U μL−1 | 9.77 × 10−2 U L−1 | HEK293T cells | 52 |
| Endo VI-catalyzed cyclic cleavage | DNA methylation | 0–100 nM | 7.3 × 10−18 M | HepG2, MDA-MB231, MCF-7, HeLa and A549 cells | 59 |
| Ligase amplification reaction | ALP | 1 × 10−6–0.1 U mL−1 | 5.63 × 10−4 U L−1 | HEK and MCF-7 cells | 74 |
| Ligation chain reaction | DNA methylation | 1.0 × 10−18–1.0 × 10−11 M | 1.0 × 10−18 M | H157 and H209 cells | 75 |
| DNA polymerase and nicking endonuclease-catalyzed EXPAR | MicroRNA | 0.1 aM–10 fM | 1.0 × 10−19 M | — | 78 |
| APE 1-catalyzed cyclic cleavage and DNA polymerase-catalyzed polymerization | hAAG | 1 × 10−9–1 × 10−3 U μL−1 | 4.42 × 10−6 U L−1 | A549, HeLa, HL-7702, MRC-5, and HEK-293 cells | 81 |
| DNA polymerase-catalyzed RCA, and Exo I- and Exo III-catalyzed digestion | hAAG, UDG | 1 × 10−10–1 × 10−3 U μL−1 for both targets | 8.69 × 10−5 U L−1 for hAAG, 5.20 × 10−5 U L−1 for UDG | A549 and HeLa cells | 82 |
| RNA polymerase-catalyzed transcription amplification and DSN-catalyzed cyclic cleavage | hOGG1, hAAG | 5 × 10−7–1 × 0.4 U μL−1 for hOGG1, 5 × 10−7–1 × 0.1 U μL−1 for hAAG | 3.52 × 10−2 U L−1 for hOOG1, 3.55 × 10−1 U L−1 for hAAG | HeLa and A549 cells | 83 |
| Catalytic hairpin assembly | MicroRNA | 0.1–10 fM | 1.7 × 10−17 M | Serum from healthy persons and prostate cancer patients | 96 |
| Click chemistry-based LCR | MicroRNA | 1 × 10−16–1 × 10−9 M | 3.87 × 10−17 M | HeLa and MCF-7 cells, tissue from healthy persons and lung cancer patients | 100 |
| Entropy-driven DNA nanomachine | Cancer cells | 0–2000 cells | 1 cell | Cells spiked in whole blood | 99 |
| CHA | MicroRNA | 10 fM–1 pM | 1.0 × 10−15 M | HepG2, MCF-7, L02, and 293T cells | 102 |
| CHA | MicroRNA | 0–1 × 10−9 M | 1.0 × 10−15 M | A549, HeLa, and HL-7702 cells, serum from healthy persons and lung cancer patients | 101 |
| Hybridization chain reaction | lncRNA MALAT1 and HOTAIR | 0.1 aM–1 pM for both targets | 1.0 × 10−19 M for MALAT1, 3.1 × 10−20 M for HOTAIR | MCF7 and HBE cells | 103 |
| DNA polymerase-catalyzed RCA | Membrane proteins | — | — | EpCAM and MCF-7 cells | 111 |
| SplintR ligase- and DNA polymerase-catalyzed RCA | mRNA | — | — | MCF-7 cells | 114 |
| DNA polymerase-catalyzed RCA | MicroRNA | 1 fM–10 nM | 7.2 × 10−16 M | A549 cells | 115 |
Even though great progress has been made in this area, there are several limitations that remain to be solved. First, a lot of other nucleic acid amplification technologies have been established besides those mentioned above, such as loop mediated isothermal amplification,24 helicase-dependent amplification,117 recombinase polymerase amplification,118 and DNAzyme-mediated amplification.119 The adaption of these amplification strategies may further extend the usability of single-molecule biosensors and improve the detection sensitivity. Second, all these biosensors require organic dyes for fluorescence readout, but organic dyes usually suffer from poor stability/solubility, low brightness, and a small Stokes shift, which may impair the assay performance. Although QDs have been used as FRET donors in some cases,41,74,75,78,101 organic dyes still need to be used as FRET acceptors, and thus the limitations of organic dyes cannot be excluded. The development of new fluorescent nanomaterials with the potential to replace organic dyes and be integrated in single-molecule biosensors is highly desired. Third, besides the biomarkers mentioned in this review, a variety of biomolecules (e.g., telomerase, DNA/RNA methyltransferase, and RNA epigenetic modifications) have been discovered as promising biomarkers. The development of nucleic acid amplification-integrated single-molecule biosensors for accurate detection of these biomarkers will greatly facilitate the comprehensive understanding of various diseases and early clinical diagnosis. Fourth, most of the reported biosensors require several separate steps for signal amplification, single-molecule imaging and statistical data analysis, which is relatively complicated and time-consuming. The development of an integrated system with the capability of automatically processing the entire procedures is of great value. With the input of these efforts, the nucleic acid amplification-integrated single-molecule biosensor will find more and more practical applications in clinical disease prediction, diagnosis, and prognosis.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant no. 21735003 and 21705096) and the Award for Team Leader Program of Taishan Scholars of Shandong Province, China.Notes and references
- L. M. Hellman and M. G. Fried, Nat. Protoc., 2007, 2, 1849–1861 CrossRef CAS PubMed
.
- R. M. Lequin, Clin. Chem., 2005, 51, 2415–2418 CrossRef CAS PubMed
.
- H. Yaziji and T. Barry, Adv. Anat. Pathol., 2006, 13, 238–246 CrossRef PubMed
.
- A. Brazma, P. Hingamp, J. Quackenbush, G. Sherlock, P. Spellman, C. Stoeckert, J. Aach, W. Ansorge, C. A. Ball and H. C. Causton, Nat. Genet., 2001, 29, 365–371 CrossRef CAS PubMed
.
- G. S. Pall and A. J. Hamilton, Nat. Protoc., 2008, 3, 1077–1084 CrossRef CAS PubMed
.
- B. T. Kurien and R. H. Scofield, Methods, 2006, 38, 283–293 CrossRef CAS PubMed
.
- P. M. Goodwin, W. P. Ambrose and R. A. Keller, Acc. Chem. Res., 1996, 29, 607–613 CrossRef CAS
.
- Z. Farka, M. J. Mickert, M. Pastucha, Z. Mikušová, P. Skládal and H. H. Gorris, Angew. Chem., Int. Ed., 2020, 59, 10746–10773 CrossRef CAS PubMed
.
- S. Ray, J. R. Widom and N. G. Walter, Chem. Rev., 2018, 118, 4120–4155 CrossRef CAS PubMed
.
- T. P. Burghardt, K. Ajtai and J. Borejdo, Biochemistry, 2006, 45, 4058–4068 CrossRef CAS PubMed
.
- V. Vukojević, M. Heidkamp, Y. Ming, B. Johansson, L. Terenius and R. Rigler, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 18176–18181 CrossRef PubMed
.
- N. Akkilic, S. Geschwindner and F. Höök, Biosens. Bioelectron., 2020, 151, 111944 CrossRef CAS PubMed
.
- F. Ma, Y. Li, B. Tang and C.-Y. Zhang, Acc. Chem. Res., 2016, 49, 1722–1730 CrossRef CAS PubMed
.
- A. B. Taylor and P. Zijlstra, ACS Sens., 2017, 2, 1103–1122 CrossRef CAS PubMed
.
- Y.-J. Chen, B. Groves, R. A. Muscat and G. Seelig, Nat. Nanotechnol., 2015, 10, 748–760 CrossRef CAS PubMed
.
- Y. Zhao, F. Chen, Q. Li, L. Wang and C. Fan, Chem. Rev., 2015, 115, 12491–12545 CrossRef CAS PubMed
.
- P. Gill and A. Ghaemi, Nucleosides, Nucleotides Nucleic Acids, 2008, 27, 224–243 CrossRef CAS PubMed
.
- B. Schweitzer and S. Kingsmore, Curr. Opin. Biotechnol., 2001, 12, 21–27 CrossRef CAS PubMed
.
- K. A. Eckert and T. A. Kunkel, Genome Res., 1991, 1, 17–24 CrossRef CAS PubMed
.
- G. T. Walker, M. C. Little, J. G. Nadeau and D. D. Shank, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 392–396 CrossRef CAS PubMed
.
- K. B. Mullis and H. A. Erlich, Science, 1988, 239, 487–491 CrossRef PubMed
.
- P. M. Lizardi, X. Huang, Z. Zhu, P. Bray-Ward, D. C. Thomas and D. C. Ward, Nat. Genet., 1998, 19, 225–232 CrossRef CAS PubMed
.
- G. T. Walker, M. S. Fraiser, J. L. Schram, M. C. Little, J. G. Nadeau and D. P. Malinowski, Nucleic Acids Res., 1992, 20, 1691–1696 CrossRef CAS PubMed
.
- T. Notomi, H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino and T. Hase, Nucleic Acids Res., 2000, 28, e63 CrossRef CAS PubMed
.
- M. M. Ali, F. Li, Z. Zhang, K. Zhang, D.-K. Kang, J. A. Ankrum, X. C. Le and W. Zhao, Chem. Soc. Rev., 2014, 43, 3324–3341 RSC
.
- S. Jiang, M. Liu, W. Tantai, Q. Xu, X. Zou, F. Ma and C.-Y. Zhang, Chem. Commun., 2021, 57, 2041–2044 RSC
.
- W. Zhao, M. M. Ali, M. A. Brook and Y. Li, Angew. Chem., Int. Ed., 2008, 47, 6330–6337 CrossRef CAS PubMed
.
- E. Schopf and Y. Chen, Anal. Biochem., 2010, 397, 115–117 CrossRef CAS PubMed
.
- M. Stougaard, J. S. Lohmann, A. Mancino, S. Celik, F. F. Andersen, J. Koch and B. R. Knudsen, ACS Nano, 2009, 3, 223–233 CrossRef CAS PubMed
.
- J. Jarvius, J. Melin, J. Göransson, J. Stenberg, S. Fredriksson, C. Gonzalez-Rey, S. Bertilsson and M. Nilsson, Nat. Methods, 2006, 3, 725–727 CrossRef CAS PubMed
.
- L. Stewart, M. R. Redinbo, X. Qiu, W. G. Hol and J. J. Champoux, Science, 1998, 279, 1534–1541 CrossRef CAS PubMed
.
- J. A. Palshof, E. V. S. Høgdall, T. S. Poulsen, D. Linnemann, B. V. Jensen, P. Pfeiffer, L. S. Tarpgaard, N. Brünner, J. Stenvang and M. Yilmaz, BMC Cancer, 2017, 17, 1–10 CrossRef PubMed
.
- M. S. Hede, S. Fjelstrup, F. Lötsch, R. M. Zoleko, A. Klicpera, M. Groger, J. Mischlinger, L. Endame, L. Veletzky and R. Neher, Sci. Rep., 2018, 8, 1–12 CrossRef CAS PubMed
.
- P. D'arpa and L. F. Liu, Biochim. Biophys. Acta, Rev. Cancer, 1989, 989, 163–177 CrossRef
.
- B. K. Sinha, Drugs, 1995, 49, 11–19 CrossRef CAS PubMed
.
- A. Thomas and Y. Pommier, Clin. Cancer Res., 2019, 25, 6581–6589 CrossRef CAS PubMed
.
- W. Yang, Q. Rev. Biophys., 2011, 44, 1–93 CrossRef CAS PubMed
.
- C. M. Dupureur, Curr. Opin. Chem. Biol., 2008, 12, 250–255 CrossRef CAS PubMed
.
- A. Venditti, G. Del Poeta, F. Buccisano, A. Tamburini, M. Cox-Froncillo, G. Aronica, A. Bruno, B. Del Moro, A. Epiceno and A. Battaglia, Leukemia, 1998, 12, 1056–1063 CrossRef CAS PubMed
.
- H. Drexler, C. Sperling and W. Ludwig, Leukemia, 1993, 7, 1142–1150 CAS
.
- L.-J. Wang, M.-L. Luo, Q. Zhang, B. Tang and C.-Y. Zhang, Chem. Commun., 2017, 53, 11016–11019 RSC
.
- F. Ma, C.-C. Li and C.-Y. Zhang, J. Mater. Chem. B, 2018, 6, 6173–6190 RSC
.
- W. C. Chan and S. Nie, Science, 1998, 281, 2016–2018 CrossRef CAS PubMed
.
- I. L. Medintz, A. R. Clapp, H. Mattoussi, E. R. Goldman, B. Fisher and J. M. Mauro, Nat. Mater., 2003, 2, 630–638 CrossRef CAS PubMed
.
- C.-Y. Zhang, H.-C. Yeh, M. T. Kuroki and T.-H. Wang, Nat. Mater., 2005, 4, 826–831 CrossRef CAS PubMed
.
- X. Zuo, F. Xia, Y. Xiao and K. W. Plaxco, J. Am. Chem. Soc., 2010, 132, 1816–1818 CrossRef CAS PubMed
.
- N. K. Bernstein, R. S. Williams, M. L. Rakovszky, D. Cui, R. Green, F. Karimi-Busheri, R. S. Mani, S. Galicia, C. A. Koch and C. E. Cass, Mol. Cell, 2005, 17, 657–670 CrossRef CAS PubMed
.
- M. Liu, F. Ma, Q. Zhang and C.-Y. Zhang, Chem. Commun., 2018, 54, 1583–1586 RSC
.
- C. J. Whitehouse, R. M. Taylor, A. Thistlethwaite, H. Zhang, F. Karimi-Busheri, D. D. Lasko, M. Weinfeld and K. W. Caldecott, Cell, 2001, 104, 107–117 CrossRef CAS PubMed
.
- A. Chatterjee, S. Saha, A. Chakraborty, A. Silva-Fernandes, S. M. Mandal, A. Neves-Carvalho, Y. Liu, R. K. Pandita, M. L. Hegde and P. M. Hegde, PLoS Genet., 2015, 11, e1004749 CrossRef PubMed
.
- L. C. Dumitrache and P. J. McKinnon, Mech. Ageing Dev., 2017, 161, 121–129 CrossRef CAS PubMed
.
- L.-j. Wang, Q. Zhang, B. Tang and C.-Y. Zhang, Anal. Chem., 2017, 89, 7255–7261 CrossRef CAS PubMed
.
- M. Liu, J. G. Qiu, F. Ma and C. Y. Zhang, WIRES Nanomed. Nanobi., 2021, e1716 Search PubMed
.
- R. A. Sperling, P. R. Gil, F. Zhang, M. Zanella and W. J. Parak, Chem. Soc. Rev., 2008, 37, 1896–1908 RSC
.
- K. Saha, S. S. Agasti, C. Kim, X. Li and V. M. Rotello, Chem. Rev., 2012, 112, 2739–2779 CrossRef CAS PubMed
.
- R. Wilson, Chem. Soc. Rev., 2008, 37, 2028–2045 RSC
.
- K. Subramanian, W. Rutvisuttinunt, W. Scott and R. S. Myers, Nucleic Acids Res., 2003, 31, 1585–1596 CrossRef CAS PubMed
.
- E. D. Garcin, D. J. Hosfield, S. A. Desai, B. J. Haas, M. Björas, R. P. Cunningham and J. A. Tainer, Nat. Struct. Mol. Biol., 2008, 15, 515–522 CrossRef CAS PubMed
.
- Y. Zhang, J. Hu, X. Zou, F. Ma, J.-G. Qiu and C.-Y. Zhang, Chem. Commun., 2021, 57, 2073–2076 RSC
.
- L. D. Moore, T. Le and G. Fan, Neuropsychopharmacology, 2013, 38, 23–38 CrossRef CAS PubMed
.
- A. Razin and A. D. Riggs, Science, 1980, 210, 604–610 CrossRef CAS PubMed
.
- Q. Zhang, Y. Wu, Q. Xu, F. Ma and C.-Y. Zhang, Biosens. Bioelectron., 2020, 112712 Search PubMed
.
- P. M. Das and R. Singal, J. Clin. Oncol., 2004, 22, 4632–4642 CrossRef CAS PubMed
.
- M. Kulis and M. Esteller, Adv. Genet., 2010, 70, 27–56 Search PubMed
.
- K. D. Robertson and A. P. Wolffe, Nat. Rev. Gene., 2000, 1, 11–19 CrossRef CAS PubMed
.
- I. R. Lehnman, Science, 1974, 186, 790–797 CrossRef PubMed
.
- D. Y. Wu and R. B. Wallace, Genomics, 1989, 4, 560–569 CrossRef CAS PubMed
.
- W. Cao, Trends Biotechnol., 2004, 22, 38–44 CrossRef CAS PubMed
.
- U. Sharma, D. Pal and R. Prasad, Indian
J. Clin. Biochem., 2014, 29, 269–278 CrossRef CAS PubMed
.
- Y. Han, J. Chen, Z. Li, H. Chen and H. Qiu, Biosens. Bioelectron., 2020, 148, 111811 CrossRef CAS PubMed
.
- J. L. Millán and M. P. Whyte, Calcif. Tissue Int., 2016, 98, 398–416 CrossRef PubMed
.
- S. Vimalraj, Gene, 2020, 754, 144855 CrossRef CAS PubMed
.
- S. Turan, B. Topcu, I. Gökçe, T. Güran, Z. Atay, A. Omar, T. Akçay and A. Bereket, J. Clin. Res. Pediatr. E., 2011, 3, 7 Search PubMed
.
- F. Ma, M. Liu and C.-Y. Zhang, Chem. Commun., 2019, 55, 8963–8966 RSC
.
- Z.-y. Wang, L.-j. Wang, Q. Zhang, B. Tang and C.-y. Zhang, Chem. Sci., 2018, 9, 1330–1338 RSC
.
- F. Ma, Y. Yang and C.-Y. Zhang, Anal. Chem., 2014, 86, 6006–6011 CrossRef CAS PubMed
.
- J. Van Ness, L. K. Van Ness and D. J. Galas, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 4504–4509 CrossRef CAS PubMed
.
- Y. Zhang and C.-y. Zhang, Anal. Chem., 2012, 84, 224–231 CrossRef CAS PubMed
.
- F. Ma, S. Jiang and C.-Y. Zhang, Biosens. Bioelectron., 2020, 157, 112177 CrossRef CAS PubMed
.
- F. Ma, W.-j. Liu, Q. Zhang and C.-Y. Zhang, Chem. Commun., 2017, 53, 10596–10599 RSC
.
- C.-C. Li, W.-X. Liu, J. Hu and C.-Y. Zhang, Chem. Sci., 2019, 10, 8675–8684 RSC
.
- C.-C. Li, H.-Y. Chen, J. Hu and C.-Y. Zhang, Chem. Sci., 2020, 11, 5724–5734 RSC
.
- L.-J. Wang, L. Liang, B.-J. Liu, B. Jiang and C.-Y. Zhang, Chem. Sci., 2021, 12, 5544–5554 RSC
.
- A. Banerjee, W. L. Santos and G. L. Verdine, Science, 2006, 311, 1153–1157 CrossRef CAS PubMed
.
- T. M. Hitchcock, L. Dong, E. E. Connor, L. B. Meira, L. D. Samson, M. D. Wyatt and W. Cao, J. Biol. Chem., 2004, 279, 38177–38183 CrossRef CAS PubMed
.
- L.-J. Wang, M.-L. Luo, X.-Y. Yang, X.-F. Li, Y. Wu and C.-Y. Zhang, Theranostics, 2019, 9, 4450 CrossRef CAS PubMed
.
- V. Zawadzki and H. J. Gross, Nucleic Acids Res., 1991, 19, 1948 CrossRef CAS PubMed
.
- F. Ma, W.-j. Liu, L. Liang, B. Tang and C.-Y. Zhang, Chem. Commun., 2018, 54, 2413–2416 RSC
.
- B.-C. Yin, Y.-Q. Liu and B.-C. Ye, J. Am. Chem. Soc., 2012, 134, 5064–5067 CrossRef CAS PubMed
.
- X. Qiu, H. Zhang, H. Yu, T. Jiang and Y. Luo, Trends Biotechnol., 2015, 33, 180–188 CrossRef CAS PubMed
.
- P. Yin, H. M. Choi, C. R. Calvert and N. A. Pierce, Nature, 2008, 451, 318–322 CrossRef CAS PubMed
.
- B. Li, A. D. Ellington and X. Chen, Nucleic Acids Res., 2011, 39, e110 CrossRef CAS PubMed
.
- C. Jung, P. Allen and A. Ellington, Nat. Nanotechnol., 2016, 11, 157–163 CrossRef CAS PubMed
.
- J. Liu, Y. Zhang, H. Xie, L. Zhao, L. Zheng and H. Ye, Small, 2019, 15, 1902989 CrossRef CAS PubMed
.
- Y. Jiang, B. Li, J. N. Milligan, S. Bhadra and A. D. Ellington, J. Am. Chem. Soc., 2013, 135, 7430–7433 CrossRef CAS PubMed
.
- X. Hu, J. Fan, B. Duan, H. Zhang, Y. He, P. Duan and X. Li, Anal. Chim. Acta, 2018, 1042, 109–115 CrossRef CAS PubMed
.
- D. Y. Zhang, A. J. Turberfield, B. Yurke and E. Winfree, Science, 2007, 318, 1121–1125 CrossRef CAS PubMed
.
- C. P. Liang, P. Q. Ma, H. Liu, X. Guo, B. C. Yin and B. C. Ye, Angew. Chem., Int. Ed., 2017, 56, 9077–9081 CrossRef CAS PubMed
.
- F. Ma, S.-h. Wei and C.-y. Zhang, Anal. Chem., 2019, 91, 7505–7509 CrossRef CAS PubMed
.
- Z.-Y. Wang, D.-L. Li, X. Tian and C.-Y. Zhang, Chem. Sci., 2021, 12, 10426–10435 RSC
.
- F. Ma, Q. Zhang and C.-Y. Zhang, Nano Lett., 2019, 19, 6370–6376 CrossRef CAS PubMed
.
- B. Li, Y. Liu, Y. Liu, T. Tian, B. Yang, X. Huang, J. Liu and B. Liu, ACS Nano, 2020, 14, 8116–8125 CrossRef CAS PubMed
.
- Y. Zhang, C. Wang, X. Zou, X. Tian, J. Hu and C.-Y. Zhang, Nano Lett., 2021, 21, 4193–4201 CrossRef CAS PubMed
.
- J. J. Quinn and H. Y. Chang, Nat. Rev. Gene., 2016, 17, 47–62 CrossRef CAS PubMed
.
- C. Ernst and C. C. Morton, Front. Cell. Neurosci., 2013, 7, 168 Search PubMed
.
- S. Geisler and J. Coller, Nat. Rev. Mol. Cell Biol., 2013, 14, 699–712 CrossRef CAS PubMed
.
- C. Wahlestedt, Nat. Rev. Drug Discovery, 2013, 12, 433–446 CrossRef CAS PubMed
.
- T. Gutschner and S. Diederichs, RNA Biol., 2012, 9, 703–719 CrossRef CAS PubMed
.
- E. A. Gibb, C. J. Brown and W. L. Lam, Mol. Cancer, 2011, 10, 1–17 CrossRef PubMed
.
- R. A. Gupta, N. Shah, K. C. Wang, J. Kim, H. M. Horlings, D. J. Wong, M.-C. Tsai, T. Hung, P. Argani and J. L. Rinn, Nature, 2010, 464, 1071–1076 CrossRef CAS PubMed
.
- Y. Fan, L. Li, M. Lu, H. Si and B. Tang, Chem. Commun., 2019, 55, 4043–4046 RSC
.
- H.-X. Zhou and X. Pang, Chem. Rev., 2018, 118, 1691–1741 CrossRef CAS PubMed
.
- M. B. Stone, S. A. Shelby and S. L. Veatch, Chem. Rev., 2017, 117, 7457–7477 CrossRef CAS PubMed
.
- R. Deng, K. Zhang, Y. Sun, X. Ren and J. Li, Chem. Sci., 2017, 8, 3668–3675 RSC
.
- R. Deng, L. Tang, Q. Tian, Y. Wang, L. Lin and J. Li, Angew. Chem., Int. Ed., 2014, 126, 2421–2425 CrossRef
.
- P. Holzmeister, G. P. Acuna, D. Grohmann and P. Tinnefeld, Chem. Soc. Rev., 2014, 43, 1014–1028 RSC
.
- F. Ma, M. Liu, B. Tang and C.-Y. Zhang, Anal. Chem., 2017, 89, 6182–6187 CrossRef CAS PubMed
.
- R. K. Daher, G. Stewart, M. Boissinot and M. G. Bergeron, Clin. Chem., 2016, 62, 947–958 CrossRef CAS PubMed
.
- F. Wang, J. Elbaz, C. Teller and I. Willner, Angew. Chem., Int. Ed., 2011, 123, 309–313 CrossRef
.
Footnote |
| † These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2021 |



