Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Self-assembly of DNA nanogels with endogenous microRNA toehold self-regulating switches for targeted gene regulation therapy†
Jiaqi
Yan‡
abc,
Haixia
Zou‡
a,
Wenhui
Zhou‡
c,
Xiaowan
Yuan
a,
Zhijun
Li
a,
Xiaodong
Ma
c,
Chang
Liu
c,
Yonghui
Wang
c,
Jessica M.
Rosenholm
 c,
Wenguo
Cui
c,
Wenguo
Cui
 b,
Xiangmeng
Qu
b,
Xiangmeng
Qu
 *a and
Hongbo
Zhang
*a and
Hongbo
Zhang
 *bc
*bc
aKey Laboratory of Sensing Technology and Biomedical Instruments of Guangdong Province, School of Biomedical Engineering, Sun Yat-Sen University, 66 Gongchang Road, Shenzhen 518107, China. E-mail: quxm5@mail.sysu.cn
bDepartment of Radiology, Shanghai Institute of Traumatology and Orthopaedics, RuiJin Hospital, Shanghai Jiao Tong University, 149 ChongqingNan Road, Shanghai, 200020, PR China. E-mail: hongbo.zhang@abo.fi
cPharmaceutical Sciences Laboratory and Turku Bioscience Center, Åbo Akademi University, FI-20520 Turku, Finland
First published on 20th June 2022
Abstract
Herein, a smart nanohydrogel with endogenous microRNA-21 toehold is developed to encapsulate gemcitabine-loaded mesoporous silica nanoparticles for targeted pancreatic cancer therapy. This toehold mediated strand displacement method can simultaneously achieve specific drug release and miRNA-21 silencing, resulting in the up-regulation of the expression of tumor suppressor genes PTEN and PDCD4.
Abnormal expression of intracellular genes is the critical cause of tumor heterogeneity, which brings significant challenges to tumor treatment.1 Gene regulation therapy is a promising and essential means of treating genetic or other gene disorder-related diseases. Recent studies exhibit that gene expression can be modulated by many oligonucleotide-based gene regulation tools, including anti-sense oligonucleotides,2 or microRNA.3 Specifically, in recent gene therapy tools, supplementing endogenous down-regulated genes and silencing highly expressed pathogenic genes have become the most popular strategy.4–6 However, the development of reasonable design projects and efficient delivery vectors remain the key technical obstacles in target regulation therapy.7,8
DNA hydrogels are biocompatible, and possess molecular recognition capabilities and nanoscale structure controllability,9–12 which show great potential for target regulation therapy. Further, they are colloidally stable, enable rapid stimuli response, and can passively target the tumors relying on the enhanced permeability and retention effect in tumors.13–15 A microRNA (miRNA) is an endogenous non-coding small RNA with a length of about 22 nucleotides.3,16 It has been found that the expression of multiple miRNAs is related to the development and differentiation of cancer cells.17–20 The construction of many miRNA-based antisense nucleic acid delivery systems is widely studied to achieve gene silencing effects.21 However, in this way, the ability of nucleic acid sequences as vector construction materials is wasted. Therefore, the preparation of miRNA responsive DNA nanohydrogels is a very promising design approach for the targeted regulation therapy of cancer.22 However, DNA exhibits a strong electronegativity, and it is challenging for DNA to be endocytosed by cancer cells.23,24 In addition, the DNA hydrogel bears low hydrophobic drug loading capacity,25–27 which greatly restricts its synergistic application with first-line anticancer drugs.28,29
In this work, we developed a smart nanohydrogel system with effective drug delivery and endogenous microRNA toehold self-regulating switches for on-site targeted regulation therapy in cancer (Fig. 1). The system consists of a protective DNA nanohydrogel-shell (Fig. 1A) with miRNA recognizing ability (Fig. 1B), and a hydrophobic drug reservoir mesoporous silica nanoparticle (MSN),30,31 illustrated in Fig. 1C. By combining MSN and DNA nanohydrogel, a new core–shell structure MSN@DNA will perfectly inherit the advantages of the two materials. The porous structure of MSN can achieve a high hydrophobic drug loading rate, and the DNA nanohydrogel can be used as a gate of the pores of MSN for miRNA responsive decomposition by targeted microRNA toehold self-regulating switches, thereby controlling the release of drugs and in the meantime silencing the targeted gene. In this work, miRNA-21 as a model endogenous target molecule is selected as a pancreatic cancer precise target and chemo/gene combination therapy. Our smart nanohydrogel-based system as a universal platform has great potential to be used for combination therapy of various diseases.
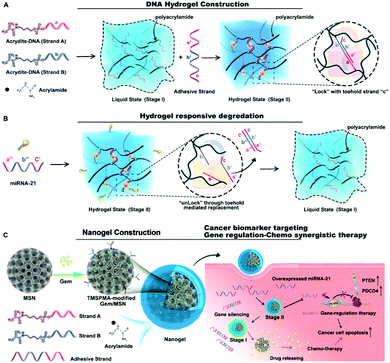 | ||
| Fig. 1 (A) Construction and (B) degradation mechanisms of the designed DNA hydrogel. (C) The combination with MSN for targeted gene regulation therapy. | ||
Two acrydite-modified DNA sequences (strand A and strand B) were grafted onto linear polyacrylic acid (PAA) through free radical reactions to form the hydrogel liquid precursor (stage I), as depicted in Fig. 1A. Then the added adhesive strand linked strands A and B together to promote the formation of hydrogel (stage II). The interior of the hydrogel consisted of a lock structure with toehold strand “c”. When miRNA-21 was used as the key to replace the adhesive strand through toehold-mediated strand displacement, the lock was opened, and the hydrogel returned to the liquid state (Fig. 1B).
Subsequently, gemcitabine (Gem) was loaded into the MSN, and the abovementioned DNA-grafted linear hybrid polymer was anchored to the 3-(trimethoxysilyl)propyl methacrylate (TMSPMA) modified MSN surface based on the free radical reaction mechanism. Then, the miRNA-21 entirely complementary antisense strand anti-miR-21, which partially complements strand A and strand B was added as a “glue” for PAA crosslinking to develop the final nanogel (Gem@MSN@DNA), as shown in Fig. 1C. Anti-miR-21, which served as the adhesive strand, constructed the hydrogel shell in our multifunctional nanosystem. In turn, they were protected by the hydrogel from degradation by the enzyme during blood circulation. Cationic polymer PAA also promoted the endocytosis ability of negatively charged DNA strands. After encountering oncogenic gene miR-21, the DNA adhesive strands inside the nanohydrogel layer were competitively paired by miRNA-21 through the toehold mediated DNA strand displacement mechanism32,33 and specific cancer cell targeting drug release and microRNA-21 regulation therapy were achieved.
When designing the DNA hydrogel layer, the loading content of the miRNA-21 antisense strands is critical for the efficacy of cancer treatment. For pancreatic cancer cell lines, it has been reported that a single concentration (100 nmol L−1) of miRNA-21 antisense oligonucleotides can achieve an obvious gene silencing effect for in vitro cytotoxicity study.34,35 Based on this dosage, 6 tubes that contained the same amount (200 μg) of MSN NPs (Fig. 2A1) were first prepared and numbered from one to six. Different concentrations of strand A and strand B grafted PAA (with concentrations of 10 μM, 50 μM, 100 μM, 150 μM, 200 μM, and 250 μM for each type of strand) were compared. The same Cy3 modified adhesive strand concentration is added to each tube (the final volume and concentration were 50 μL and 200 μM, as shown in Fig. 2A2). After 15 min of incubation at 60 °C, all tubes were centrifuged (Fig. 2A3) and the supernatant was collected into new tubes (Fig. 2A4) for UV-vis absorption detection (Fig. 2B). We found that the Cy3 modified adhesive strand is effectively encapsulated into the DNA nanohydrogel and centrifuged to the bottom of the tube together with MSN NPs. These results indicated that the adhesive strand, as a functional building block for the designed miRNA-21 sensitive hydrogel, can successfully mediate the crosslinking of PAA on the surface of MSN. In the meantime, by increasing the content of A and B strands, 200 μM of adhesion strand has fully assembled inside the hydrogel, verified by the supernatant photos and UV absorbance results. The centrifuged NPs were repeatedly washed and diluted into 1 mL. The final concentration of miRNA-21 antisense strand is 10 μM, which is a hundred times higher than the effective concentration mentioned before, therefore facilitating further investigation with a broad concentration range.
Strand A and strand B are partially complementary to the two ends of the adhesive strand and are riveted on the PAA linear polymer (Table S1†). Therefore, the adhesive strand will adhere to the A/B strand and promote the crosslinking of the polymer. We confirmed this anticipation through a polyacrylamide gel experiment. As shown in Fig. 2C, a strong band is observed at around 20 bp when strand A or strand B are added together with the adhesive strand and labelled with GelRed, accompanied by the disappearance of the band of the A/B strand. This phenomenon demonstrated the successful combination between each strand. However, the newly formed double-strand band seems to overlap with the previous adhesive strand, resulting in unobvious results. Since we cannot compare the moving speed of the single-strand and hybrid double-strand inside the polyacrylamide gel, another dye DAPI was chosen to verify the synthesis of new double-strands because DAPI can only bind to A–T base-pair regions compared to GelRed. As shown in Fig. 2C, after three single strands were added together, there was indeed only one new band formed by using DAPI as the labelling dye.
Due to the coincidence of the positions of the newly generated bands and the excess bands after the reaction of the three strands (strand A, B and adhesive strand), we used the NUPACK web application36 to specifically analyze the possible reactions, as elucidated in Fig. 2D. We found that when each strand with 1 mM was added together, in addition to the 0.94 mM product, there was 0.017 mM strand A remaining, which explains that the band at around 14 bp was strand A (Fig. 2C). Meanwhile, 0.017 mM strand A–adhesive strand and 0.017 mM strand B–adhesive strand were also generated (Fig. 2D), where the two strands share the same location as the band of the final product (Fig. 2C).
Then, to verify the “unlock” system of our designed hydrogel through the toehold mediated strand replacement mechanism, we first prepared bulk hydrogels to facilitate observation of their macroscopic changes (Fig. 2E and F). We can clearly see that the DNA hydrogel changed (Fig. 2E) to a transparent milky white liquid (Fig. 2F). This result proved that miRNA-21 could competitively bind to the adhesive chains inside the hydrogel, leading to the disintegration of the hydrogel.
Subsequently, DLS data and TEM results are explored to confirm the successful combination between the DNA hydrogel and pure MSN NPs, and the miRNA-21 sensitivity of the hydrogel shell. We found that when the MSN surface was modified with the DNA nanogel, its particle size increased from 120 nm to about 150 nm, which was reflected in Fig. 3A. Moreover, the particle size of Gem@MSN@DNA NPs did not change after one week of storage in PBS buffer (Fig. S1†). However, when incubated with 5 nM miRNA-21 for 4 h, its particle size returned to 120 nm, which indicates that the nanogel shell has an excellent miRNA-21 responsive degradation capability. Meanwhile, from the zeta potential results, we can see that the TMSPMA modified MSN (Gem@MSN@DNA + miR-21 group) is negatively charged at around −33 mV. After being covered with DNA hydrogel, the zeta potential of the final NPs becomes neutral (Fig. 3B), which can promote the phagocytosis ability of negatively charged NPs by cancer cells.37 Also, the formation of a DNA hydrogel layer is confirmed by TEM (Fig. 3C). We can clearly see a hydrogel layer grown on the surface of MSN NPs. More importantly, after incubation with 5 nM miRNA-21 for 4 hours, the polymer layer on MSN surface disappeared (Fig. 3D), indicating the successful construction of a miRNA-responsive gating switch.
The Gem-loaded MSN NPs were protected by the “gate material” DNA nanogel, to prevent premature drug release before accumulating into the tumor tissue (Fig. 3D). The release profiles of Gem and the Cy3-labeled adhesive strand from DNA hydrogel-covered MSN were tested. Toehold mediated strand displacement between miRNA-21 and the adhesive strand is elucidated in Fig. 3E. When strands A and B were combined with the adhesive strand, their free energy of the secondary structure, calculated using NUPACK software, was −25.20 kcal mol−1, which was higher than the combination between miRNA-21 and adhesive strand (−35.28 kcal mol−1), as shown in Fig. 3E. These simulated results proved that the NPs could realize the responsive disintegration of miRNA-21. Furthermore, along with the degradation of NPs, miRNA-21 will also be consumed and silenced by adhesion strands (antisense sequences), realizing gene/chemo combination therapy.
Upon loading Gem into MSN, a drug loading degree of 22.95% is reached; that is 1 mg MSN can hold about 300 μg of Gem (Table S2†). Subsequently, the miRNA-21 responsive release experiment was performed, and 0 nM, 1 nM and 5 nM were selected based on the previous study.38,39 The Gem inside NPs exhibited an accelerated release profile when the concentration of miRNA-21 in the release environment increased. We found that within 40 hours, with a 5 nM concentration of miRNA-21 present in the release medium, the release content of Gem could reach more than 80%, but in the presence of less than 1 nM miRNA-21, only 30% of Gem was released within the same time frame. Negligible drug release was observed when there was no miRNA-21 in the release media, which demonstrated the NPs’ excellent miRNA-21 recognition ability in controlling the drug release (Fig. 3F). Notably, the results of the release of Cy3-labeled adhesive strand were consistent with the gemcitabine release profile, which showed a miRNA-21 responsive release ability (Fig. 3G). However, Cy3 was released more rapidly and at a higher rate, and Cy3-anti-miRNA-21 was released more than 95% at 4 h with 5 nM miRNA-21, since the outermost nanogel shell can be easily degraded when encountering the miRNA-21.
Next, the endocytosis of NPs in pancreatic cancer cell PANC-1 and pancreatic islet B epithelial cell Min6 are studied. PANC-1 has a high expression of miRNA-21, and it has also been reported that after Gem administration, the miRNA-21 was greatly up-regulated, which could significantly reduce the efficacy of chemotherapy.40 Compared with PANC-1 cells, the Min6 cells had a very low miRNA-21 expression,41 as shown in Fig. S2,† so it is selected as a negative control to evaluate the NPs’ selectivity towards miRNA-21. We first loaded the FITC fluorescent dye inside NPs for tracking the endocytosis of NPs by different cells through confocal microscopy and flow cytometry experiments (Fig. 3H and I).
For the PANC-1 cell line, the results showed that with the extension of the incubation time, the amount of NP uptake by cells gradually increased. Within 10 hours, 100% of the cells had taken up NPs, which indicated that the NPs could be effectively endocytosed. Interestingly, due to high miRNA-21 content in PANC-1 cells, we have observed that the FITC dye inside the NPs was more likely to diffuse throughout the cells, which indirectly indicated that the DNA hydrogel shell was disintegrated and facilitated the FITC release. In the meantime, Min6 cells also displayed efficient endocytosis ability of NPs, 100% of the cells had taken up within 10 h. This may be because we did not modify the cell-targeting ligands on the surface of NPs, different cells possessed relatively the same endocytosis effect on NPs. However, the FITC inside the nanoparticles did not seem to diffuse after 16 h, but rather accumulated at a certain location in the cytoplasm. This phenomenon reflected that with less miRNA-21 content, it was difficult for FITC to diffuse through the intact DNA-hydrogel layer. Furthermore, since healthy tissue around the tumor can also internalize the nanosystem, therefore, within a certain period, the miRNA hydrogel layer would monitor the miRNA-21 content of healthy cells, and treat them in a timely manner when their miRNA-21 content showed abnormal change.
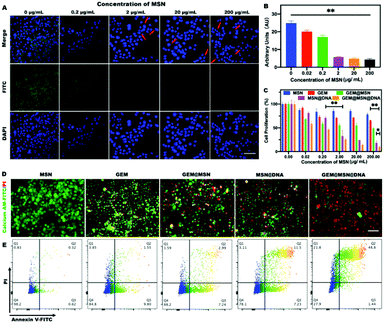
The fluorescence in situ hybridization (FISH) method was subsequently utilized to study the silencing effect of miRNA-21 in PANC-1 cells. The FITC green, fluorescent probe was used to observe the content of miRNA-21. We found that the cytoplasm of PANC-1 cells exhibited bright green fluorescence without the addition of NPs, indicating the presence of a large number of proliferation miRNA-21 genes (Fig. 4A). Upon the addition of NPs, the content of miRNA-21 was significantly inhibited (Fig. 4B), especially when the concentration of NPs exceeded 0.2 μg mL−1 (adhesive strand concentration equal to 10 nM). In addition, when the concentration of NPs reached more than 2 μg mL−1, the cells gradually entered the apoptotic stage, and the red arrow indicates the occurrence of nuclear fragmentation (Fig. 4A). The permeability of the nucleus was also found to increase and green fluorescence was observed inside the nucleus, since during the process of cell apoptosis, the nuclear membrane permeability will be increased.42,43 These results fully proved the miRNA-21 recognition ability of the DNA hydrogel layer of the NPs and the silencing function of the miRNA-21 antisense building block inside the hydrogel.
To further explore the cytotoxicity of Gem-loaded and DNA hydrogel-wrapped Gem@MSN@DNA NPs, different drugs and NP groups were studied with the PANC-1 cells and Min-6 cells, as shown in Fig. 4C and Fig. S3.† For PANC-1 cells, pure Gem showed no obvious difference when Gem concentration exceeded 0.2 μM (calculated based on the loading degree). After Gem was loaded into MSN, the efficacy of Gem increased but still changed modestly when the MSN concentration was higher than 0.2 μg mL−1. On the other hand, MSN@DNA showed an excellent therapeutic effect, especially when the MSN concentration exceeded 2 μg mL−1 (miRNA-21 antisense strand equal to 100 nM). Finally, the Gem@MSN@DNA group showed a powerful cancer cell suppressing effect. The Gem@MSN@DNA group killed more than 50% of the cells only at the MSN concentration of 0.2 μg mL−1 (Gem and miRNA-21antisense DNA concentration equal to 0.2 μM and 10 nM) and killed more than 80% of the cells at the highest concentration (Gem and miRNA-21 antisense DNA concentration equal to 200 μM and 10 mM). These results confirmed that the nanosystem could achieve excellent synergistic effects for both chemo and gene therapy. Then, for Min6 cells, the Gem@MSN group showed serious toxicity when the MSN concentration becomes 2 μg mL−1 (Gem concentration equal to 2 μM). However, when the hydrogel is wrapped on the surface of the nanoparticle, we can see that the Gem@MSN@DNA group possessed significantly higher safety than the pure Gem and Gem@MSN groups. These results indicated that the DNA hydrogel-encapsulated nanoformulations are highly selective for miRNA-21. Only under a high concentration of miRNA-21 can the NPs release gemcitabine and miRNA-21 antisense sequence together.
We then conducted the live/dead cell assay for the PANC-1 cell line with different groups, while green represented live cells and red represented apoptotic cells (Fig. 4D). The results exhibited that the GEM/MSN/DNA group possessed enhanced cytotoxicity, due to the synergistic effect of chemotherapy and gene therapy. At the same time, the experimental results of apoptosis (Fig. 4E) were consistent with those of the cytotoxicity assay, and over 70% of the cells in the Gem@MSN@DNA group went into the late apoptotic stage after 24 h of treatment. Gemcitabine can stimulate cells to further produce excessive miRNA-21,40 which is one of the reasons for chemo-resistance. Therefore, the nano-system we designed has great synergistic therapeutic significance.
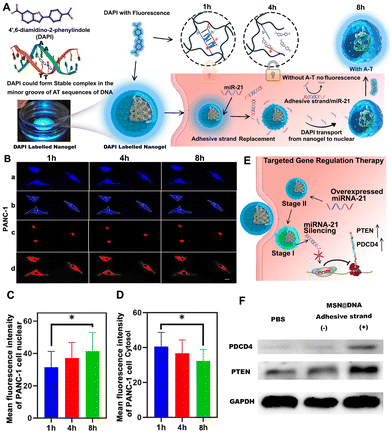
Later, the intracellular disintegration process of the hydrogel layer is verified through confocal microscopy. DAPI is used to mark the AT base pairs of the double-stranded building block inside the NPs (Fig. 5A). Through this method, the unwinding process of double-stranded DNA from the hydrogel shell of NPs can be observed. Because DAPI is toxic to cells, we only observed the fluorescence changes in the cytoplasm and nucleus within 8 h. After DAPI-labelled NPs are internalized into PANC-1 cells, DAPI will be released due to the unwinding between the adhesive strand, strand A and strand B. The released DAPI will increase the fluorescence in the nucleus, as elucidated in Fig. 5A. We confirmed this phenomenon in PANC-1 cells, as shown in Fig. 5B. Software Image J was used to quantify the fluorescence image (Fig. 5Ba), and the nucleus and cytoplasm were manually distinguished (Fig. 5Bb), and fluorescence intensity analysis was performed separately (Fig. 5Bc and d). Mean fluorescence intensity of PANC-1 cell nucleus showed that the DAPI blue fluorescence in the nucleus gradually increased during incubation within 6 h (Fig. 5C). Furthermore, the blue fluorescence in the cytoplasm gradually decreased (Fig. 5D), indicating that DAPI is released due to the degradation of the hydrogel shell from NPs and entered the nucleus. However, this phenomenon was not observed in Min6 cells (Fig. S4A–C†), indicating that the Gem@MSN@DNA nano-platform had great miRNA-21 targeting specificity.
The silencing of miRNA-21 can cause changes in the expression of various genes in pancreatic cancer cells (Fig. 5E). It is reported that miR-21 targets and inhibits PDCD4 and PTEN tumor suppressor genes.34,35 By inhibiting miRNA-21, we discovered that the tumor suppressor genes PTEN and PDCD4 in the PANC-1 cells significantly increased through western blot assay (Fig. 5F). It is worth mentioning that in the process of constructing a DNA hydrogel, we introduced strand A and strand B, and these two strands have the same sequence of partial miRNA-21. Hence, they may have the same function as miRNA-21. Therefore, MSN@DNA NPs with or without adhesive strands were prepared in comparative experiments, to ensure that strand A and strand B will not have the same function as miRNA-21 to cause cell proliferation (Fig. 5F). Of note, strands A and B can also be anchored to the surface groups of NPs without the adhesion strand by the aforementioned free radical reaction. The results found that the MSN@DNA (without the adhesive strand) group is the same as the PBS group and had no effect on PDCD4 and PTEN expression. However, for the adhesive strand group, PTEN and PDCD4 are significantly up-regulated, demonstrating the successful inhibition of the miRNA-21 gene (Fig. S5†).
Conclusions
In conclusion, a simple yet multifunctional targeting strategy is successfully developed by encapsulating the Gem-loaded MSN with an miRNA-21 responsive DNA hydrogel through a simple in situ growth technology for pancreatic cancer-targeted chemo/gene synergistic treatment. The antisense strand of miRNA-21 is used as a recognition building block to participate in the cross-linking of DNA hydrogels. The targeting strategy applied by the Gem@MSN@DNA nanosystem is completely different from the receptor-mediated and normal tumor microenvironment-mediated targeting strategies (such as pH or redox). Its targeting is based on the unique oncogene gene of pancreatic cancer cells, and thus will not be heavily interfered with by protein adsorption during blood circulation. Only after the nanocarrier encounters miRNA-21, which is highly expressed by the pancreatic cancer cells, can the hydrogel layer achieve responsive disintegration through the toehold mediated strand displacement mechanism. Subsequently, with the degradation of the DNA protective layer, the Gem inside MSN is gradually released, which greatly enhanced the bioavailability of Gem. Moreover, the degradation of the DNA hydrogel resulted in a significant consumption and silencing of miRNA-21, leading to an enhancement of the expression of tumor suppressor genes PTEN and PDCD4, which promoted cell apoptosis and realized gene/chemotherapy synergistic therapy. Finally, it is theoretically possible to synthesize DNA hydrogel structures responsive to any miRNA by designing different DNA sequences. Thereby different miRNA targeted therapeutic systems can be designed.Author contributions
Jiaqi Yan, Haixia Zou and Wenhui Zhou conceived the study. Xiangmeng Qu and Hongbo Zhang supervised the project. Wenguo Cui designed the experiments. Jiaqi Yan, Xiaodong Ma, Chang Liu, and Yonghui Wang performed the experiments. Xiaowan Yuan and Zhijun Li, analyzed the experimental results. Jiaqi Yan wrote the manuscript. Jessica M. Rosenholm edited the manuscript.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was funded by National Natural Science Foundation of China (81871472 and 21705048), the Natural Science Foundation of Guangdong Province (2021A1515012333), Research Fellow (Grant No. 328933), the Solution for Health Profile (336355), and the InFLAMES Flagship (337531) grants from the Academy of Finland, and the Finland China Food and Health International Pilot Project funded by the Finnish Ministry of Education and Culture. The Key Laboratory of Sensing Technology and Biomedical Instruments of Guangdong Province (2020B1212060077). Ruijin Hospital Guangci Introducing Talent Project. Electron microscopy samples were processed and analyzed in the Electron Microscopy Laboratory, University of Turku; confocal/flow cytometry were performed at the Cell Imaging Core, Turku Bioscience Centre. Biocenter Finland is acknowledged for providing support and infrastructure.References
- J. Li, C. Zheng, S. Cansiz, C. Wu, J. Xu, C. Cui, Y. Liu, W. Hou, Y. Wang, L. Zhang, I. t. Teng, H.-H. Yang and W. Tan, J. Am. Chem. Soc., 2015, 137, 1412–1415 CrossRef CAS PubMed.
- H. Qiao, L. Zhang, D. Fang, Z. Zhu, W. He, L. Hu, L. Di, Z. Guo and X. Wang, Chem. Sci., 2021, 12, 4547–4556 RSC.
- L. Wu, W. Zhou, L. Lin, A. Chen, J. Feng, X. Qu, H. Zhang and J. Yue, Bioact. Mater., 2021, 7, 292–323 CrossRef PubMed.
- S. Banerjee, H. Cui, N. Xie, Z. Tan, S. Yang, M. Icyuz, V. J. Thannickal, E. Abraham and G. Liu, J. Biol. Chem., 2013, 288, 35428–35436 CrossRef CAS PubMed.
- V. Baumann and J. Winkler, Future Med. Chem., 2014, 6, 1967–1984 CrossRef CAS PubMed.
- C. F. Bennett, A. R. Krainer and D. W. Cleveland, Annu. Rev. Neurosci., 2019, 42, 385–406 CrossRef CAS PubMed.
- J. Shen, L. Zhang, J. Yuan, Y. Zhu, H. Cheng, Y. Zeng, J. Wang, X. You, C. Yang, X. Qu and H. Chen, Anal. Chem., 2021, 93, 15033–15041 CrossRef CAS PubMed.
- Y. Zhang, J. Tu, D. Wang, H. Zhu, S. K. Maity, X. Qu, B. Bogaert, H. Pei and H. Zhang, Adv. Mater., 2018, 30, e1703658 CrossRef PubMed.
- Y. S. Zhang and A. Khademhosseini, Science, 2017, 356(6337), eaaf3627 CrossRef PubMed.
- R. Zhong, Q. Tang, S. Wang, H. Zhang, F. Zhang, M. Xiao, T. Man, X. Qu, L. Li, W. Zhang and H. Pei, Adv. Mater., 2018, 30, e1706887 CrossRef PubMed.
- N. Li, X. Y. Wang, M. H. Xiang, J. W. Liu, R. Q. Yu and J. H. Jiang, Anal. Chem., 2019, 91, 2610–2614 CrossRef CAS PubMed.
- Z. Li, L. Jiang, Y. Jiang, X. Yuan, M. Aizitiaili, J. Yue and X. Qu, Adv. Intell. Syst., 2020, 2, 2000086 CrossRef.
- T. Yata, Y. Takahashi, M. Tan, H. Nakatsuji, S. Ohtsuki, T. Murakami, H. Imahori, Y. Umeki, T. Shiomi, Y. Takakura and M. Nishikawa, Biomaterials, 2017, 146, 136–145 CrossRef CAS PubMed.
- J. Song, K. Im, S. Hwang, J. Hur, J. Nam, G. O. Ahn, S. Hwang, S. Kim and N. Park, Nanoscale, 2015, 7, 9433–9437 RSC.
- J. Zhao, P. Zhang, Z. He, Q. H. Min, E. S. Abdel-Halim and J. J. Zhu, Chem. Commun., 2016, 52, 5722–5725 RSC.
- S. Griffiths-Jones, R. J. Grocock, S. van Dongen, A. Bateman and A. J. Enright, Nucleic Acids Res., 2006, 34, D140–D144 CrossRef CAS PubMed.
- Y. H. Feng and C. J. Tsao, Biomed. Rep., 2016, 5, 395–402 CrossRef CAS PubMed.
- F. Sicard, M. Gayral, H. Lulka, L. Buscail and P. Cordelier, Mol. Ther., 2013, 21, 986–994 CrossRef CAS PubMed.
- S. R. Pfeffer, C. H. Yang and L. M. Pfeffer, Drug Dev. Res., 2015, 76, 270–277 CrossRef CAS PubMed.
- M. Aizitiaili, Y. Jiang, L. Jiang, X. Yuan, K. Jin, H. Chen, L. Zhang and X. Qu, Nano Lett., 2021, 21, 2141–2148 CrossRef CAS PubMed.
- L. Chen, G. Li, X. Wang, J. Li and Y. Zhang, ACS Nano, 2021, 15, 11929–11939 CrossRef CAS PubMed.
- Y. Chen, X. Gong, Y. Gao, Y. Shang, J. Shang, S. Yu, R. Li, S. He, X. Liu and F. Wang, Chem. Sci., 2021, 12, 15710–15718 RSC.
- T. Kim, K. Nam, Y. M. Kim, K. Yang and Y. H. Roh, ACS Nano, 2021, 15, 1942–1951 CrossRef CAS PubMed.
- S. Dobres, G. Mula, J. Sauer and D. Zhu, Eng. Regen., 2022, 3, 13–23 Search PubMed.
- J. Gačanin, C. V. Synatschke and T. Weil, Adv. Funct. Mater., 2020, 30, 1906253 CrossRef.
- S. Liu, W. Su, Y. Li, L. Zhang and X. Ding, Biosens. Bioelectron., 2018, 103, 1–5 CrossRef CAS PubMed.
- K. Shi, B. Dou, C. Yang, Y. Chai, R. Yuan and Y. Xiang, Anal. Chem., 2015, 87, 8578–8583 CrossRef CAS PubMed.
- G. Brachi, F. Bussolino, G. Ciardelli and C. Mattu, Front. Bioeng. Biotechnol., 2019, 7, 307 CrossRef PubMed.
- X. Fu, X. Zhang, D. Huang, L. Mao, Y. Qiu and Y. Zhao, Chem. Eng. J., 2022, 431, 133362 CrossRef CAS.
- C. Chen, W. Yao, W. Sun, T. Guo, H. Lv, X. Wang, H. Ying, Y. Wang and P. Wang, Int. J. Biol. Macromol., 2019, 122, 1090–1099 CrossRef CAS PubMed.
- Z. Lu, J. Wang, L. Qu, G. Kan, T. Zhang, J. Shen, Y. Li, J. Yang, Y. Niu, Z. Xiao, Y. Li and X. Zhang, Bioact. Mater., 2020, 5, 1127–1137 CrossRef PubMed.
- Q. Hu, H. Li, L. Wang, H. Gu and C. Fan, Chem. Rev., 2019, 119, 6459–6506 CrossRef CAS PubMed.
- S. Jiang, Z. Ge, S. Mou, H. Yan and C. Fan, Chem, 2021, 7, 1156–1179 CAS.
- T. A. Mace, A. L. Collins, S. E. Wojcik, C. M. Croce, G. B. Lesinski and M. Bloomston, J. Surg. Res., 2013, 184, 855–860 CrossRef CAS PubMed.
- Y. Li, Y. Chen, J. Li, Z. Zhang, C. Huang, G. Lian, K. Yang, S. Chen, Y. Lin, L. Wang, K. Huang and L. Zeng, Cancer Sci., 2017, 108, 1493–1503 CrossRef CAS PubMed.
- B. R. Wolfe, N. J. Porubsky, J. N. Zadeh, R. M. Dirks and N. A. Pierce, J. Am. Chem. Soc., 2017, 139, 3134–3144 CrossRef CAS PubMed.
- J. Yan, X. Xu, J. Zhou, C. Liu, L. Zhang, D. Wang, F. Yang and H. Zhang, ACS Appl. Bio Mater., 2020, 3, 1216–1225 CrossRef CAS PubMed.
- W. Mao, C. Hu, H. Zheng, J. Xie, X. Shi, Y. Du and F. Wang, Mol. Ther.–Nucleic Acids, 2020, 22, 27–37 CrossRef CAS PubMed.
- J. Liu, W. Liu, K. Zhang, J. Shi and Z. Zhang, Adv. Healthcare Mater., 2020, 9, e1901316 CrossRef PubMed.
- E. Giovannetti, N. Funel, G. J. Peters, M. Del Chiaro, L. A. Erozenci, E. Vasile, L. G. Leon, L. E. Pollina, A. Groen, A. Falcone, R. Danesi, D. Campani, H. M. Verheul and U. Boggi, Cancer Res., 2010, 70, 4528–4538 CrossRef CAS PubMed.
- S. R. Filios and A. Shalev, Diabetes, 2015, 64, 3631–3644 CrossRef CAS PubMed.
- C. Strasser, P. Grote, K. Schauble, M. Ganz and E. Ferrando-May, Nucleus, 2012, 3, 540–551 CrossRef PubMed.
- L. Lindenboim, H. Zohar, H. J. Worman and R. Stein, Cell Death Discovery, 2020, 6, 29 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2bm00640e |
| ‡ Equal concentration. |
| This journal is © The Royal Society of Chemistry 2022 |