Smartly responsive DNA–miRNA hybrids packaged in exosomes for synergistic enhancement of cancer cell apoptosis†
Fan
Zhang
,
Albertina N.
Isak
,
Shiqi
Yang
,
Yuchen
Song
,
Lingjie
Ren
,
Chang
Feng
* and
Guifang
Chen
 *
*
Center for Molecular Recognition and Biosensing, School of Life Sciences, Shanghai University, Shanghai 200444, P. R. China. E-mail: gfchen@shu.edu.cn; cfeng@shu.edu.cn
First published on 30th March 2022
Abstract
Endogenous and exogenous tumor-related microRNAs (miRNAs) are considered promising tumor biomarkers and tumor therapeutic agents. In this work, we propose a miRNA self-responsive drug delivery system (miR-SR DDS), which enables the association between endogenous and exogenous miRNAs, so as to achieve a smart responsive and synergistic drug delivery. The miR-SR DDS consists of DNA–miRNA hybrids of let-7a and the complementary DNA of miR-155, which was packaged in exosomes. In response to the overexpressed miR-155 in breast cancer cells, the hybrids disintegrate and release let-7a and the complementary DNA of miR-155 to inhibit the expression of HMGA1 and relieve the inhibition of SOX1, respectively. Under the dual-targeted gene regulation, results show that the growth, migration and invasion of breast cancer cells can be synergistically inhibited through the Wnt/β-catenin signaling pathway. The concept and successful practice of the miR-SR DDS can be used as a reference for the development of miRNA drugs.
Introduction
MicroRNAs (miRNAs) area class of small non-coding single-stranded RNA molecules that are 18 to 25 nucleotides in length. They play a key role in post-transcriptional regulation of gene expression and participate in a series of life processes, such as cell proliferation, differentiation, angiogenesis, migration and apoptosis.1–3 In recent years, a large number of studies have shown that abnormal miRNA expression levels are related to various types of diseases, especially cancers.4,5 MiRNAs can bind to the 3′-end untranslated region (UTR) of the mRNA of an oncogene or a tumor suppressor gene to inhibit its translation and promote its degradation.6 Delivery of miRNAs or miRNA inhibitors to inhibit the expression of specific oncogenes or activate the expression of tumour suppressor genes by miRNAs has now been shown to complement or enhance radiotherapy and chemotherapy.7–9 However, poor targeting and low gene suppression efficiency that are responsible for the unsatisfactory therapeutic effect are two issues that should be addressed. For the former, targeted nanocarriers and smart stimuli-response release are currently the two most promising options; while for the latter, the combined use of multiple miRNAs or miRNAs with other drugs is an essential approach. Nonetheless, research in related areas is still limited, and simple and well-integrated strategies are still scarce.Different from other exogenous anti-tumor drugs, miRNA is also a naturally occurring endogenous molecule. Its differential expression in tumor cells and normal cells makes it a potential tumor biomarker.10,11 A combination of the expression levels of multiple miRNAs to establish a comprehensive index has been applied in the clinical diagnosis of some cancers.12–14 The differential expression of endogenous miRNA can also be combined with a drug delivery system (DDS) to establish a smart miRNA-responsive DDS. For example, Willner et al. developed a miRNA-responsive DNA-tetrahedral/metal–organic framework conjugate, which could be used as a functional drug carrier and had been proven to have better drug release efficiency in breast cancer cells with high miR-21 expression.15 However, combining endogenous miRNAs and exogenous miRNAs to establish a smart responsive miRNA DDS and so as to address the issues of miRNA delivery and therapeutic effect has not yet been conceived.
In this paper, we propose a miRNA self-responsive drug delivery system (miR-SR DDS) concept, which enables the association between endogenous and exogenous miRNAs, so as to achieve a smart responsive and synergistic gene regulation. In this miR-SR DDS, let-7a, one of the most studied cancer-related miRNAs, is packaged as a miRNA therapeutic agent,16 which has been reported to regulate oncogenes such as Ras,17 c-Myc18 and HMGA1,19,20 and thereby is considered as a tumor suppressor. For example, Powers et al. found that the amplification of oncogene MYCN in neuroblastoma led to high expression of MYCN mRNA, which resulted in the adsorption of let-7 and the up-regulation of downstream molecules.21 The strategy of restoring let-7 may provide a new low-toxicity treatment method for neuroblastoma and other cancers with let-7 deletion. In addition to let-7a, a complementary DNA of miR-155 is also involved in the miR-SR DDS. In contrast to the let-7 family, some miRNAs are overexpressed in tumors. For example, miR-155 has been found to be highly expressed in breast cancer,22 lung cancer,23 gastric cancer24 and pancreatic cancer25 and can promote tumor growth, invasion and metastasis through inhibition of downstream targets such as SHIP1,26 TP53INP127 and SOCS1.28 Therefore, the complementary DNA of miR-155 can be used as a therapeutic agent by inhibiting miR-155. For example, Slack et al. used miR-155 oligonucleotide inhibitor Cobomarsen to slow down the growth rate of tumor cells in vitro and in vivo.29
In the miR-SR DDS, we constructed a responsive DNA–miRNA hybrid structure of let-7a and a complementary DNA of miR-155, which was then packaged in exosomes. It allows the miR-SR DDS to respond to and inhibit miR-155 overexpression in cancer cells, and in parallel smartly release active let-7a for dual-targeted gene regulation. Results show that the up-regulated let-7a will act on its target gene HMGA1 and inhibit the expression of HMGA1, while the down-regulated miR-155 will relieve the inhibition of SOX1. Taken together, both inhibit the growth, migration and invasion of breast cancer cells by synergistically inhibiting the Wnt/β-catenin signaling pathway.
Experimental section
Materials
All DNA strands (HPLC purified, sequences are shown in Table S1†) were synthetically purified by Sangon Biotech Co., Ltd (Shanghai). All RNA strands (RNase-free HPLC purified) were obtained from Genscript Biotech Corporation. TRIzol and TransScript® miRNA first-strand cDNA synthesis SuperMix were purchased from Beijing TransGen Biotech Co., Ltd, and Hiseff® qPCR SYBR® Green Master Mix (No ROX) was purchased from Yisheng Biotechnology Co., Ltd (Shanghai). A PI/AnnexinV-FITC cell apoptosis detection kit was provided by BD Biosciences. MCF-7 cells (human breast cancer cells), MDA-MB-231 cells (human breast cancer cells), HEK-293T cells (human embryonic kidney cells) and L02 cells (human normal liver cells) were obtained from the Cell Bank of the Committee on Type Culture Collection of the Chinese Academy of Sciences. Fetal bovine serum (FBS) and cell culture medium were purchased from GIBCO Co., Ltd. The antibodies of SOX1, HMGA1 and GAPDH were purchased from Abcam Trading Co., Ltd (Shanghai). The goat anti-rabbit IgG (HRP) antibody was purchased from Shanghai Epizyme Biotech Co., Ltd. Phosphate-buffered saline (PBS) instant granules were purchased from Vazyme Biotech Co., Ltd. All solutions were prepared using RNase-free distilled water.Construction of tweezer (Tw)/let-7a hybrids
500 nM Tw with different sequence designs (Tw-Gap2, Tw-Gap4, Tw-Gap6, Tw-Tail2, Tw-Tail4, and Tw-Tail6) were mixed with 500 nM let-7a, respectively and incubated at 37 °C for 2 h.Electrophoresis analysis
A 500 nM Tw-Gap4/let-7a hybrid sample was prepared as previously described. The sample was mixed with a 5× loading buffer at a ratio of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to form a 10 μL system, loaded on a 15% polyacrylamide gel, run in 1× Tris borate EDTA (TBE) buffer at 110 V for 60 min, and finally used for gel imaging (Bio-Rad, USA).
1 to form a 10 μL system, loaded on a 15% polyacrylamide gel, run in 1× Tris borate EDTA (TBE) buffer at 110 V for 60 min, and finally used for gel imaging (Bio-Rad, USA).
Fluorescence measurements
The fluorescence emission spectra were obtained using an F-7000 spectrofluorometer (Hitachi High-Technologies, Japan). The main parameters of the instrument are set as follows: the gap width is 5 nm, and the excitation light voltage is 750 V. The modified Cy3 fluorophore chain is excited at 552 nm, and its emission wavelength range is 560–660 nm. The modified FAM fluorophore chain is excited at 492 nm, and its emission wavelength range is 500–600 nm. 300 nM BHQ1-modified Tw-Gap4 was mixed with 300 nM FAM-modified let-7a and incubated at 37 °C for 2 h. Afterwards, a series of concentration gradients (0, 1, 10, 30, 100, 300, 500 and 1000 nM) of miR-155 were added and incubated at 37 °C for 1.5 h, and the fluorescence intensity was measured using a spectrofluorometer. A series of concentration gradients (0, 1, 10, 30, 100, 300, 500 and 1000 nM) of BHQ1-modified Tw-Gap4 were mixed with let-7a (the concentration ratio is 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and incubated at 37 °C for 2 h. Then, 300 nM Cy3-modified miR-155 was added, incubated at 37 °C for 1.5 h, and the fluorescence intensity was measured using a spectrofluorometer.
1) and incubated at 37 °C for 2 h. Then, 300 nM Cy3-modified miR-155 was added, incubated at 37 °C for 1.5 h, and the fluorescence intensity was measured using a spectrofluorometer.
Cell culture
MCF-7 cells and HEK-293T cells were cultured in a high-glucose DMEM medium containing 1% penicillin–streptomycin and 10% FBS at 37 °C in humidified air containing 5% CO2. L02 cells were cultured in RPMI-1640 medium and MDA-MB-231 cells were cultured in Leibovitz's L15 medium under the same culture conditions.Extraction and characterization of exosomes
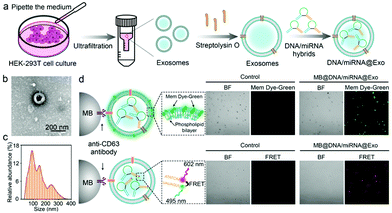
The exosomes were extracted by ultrafiltration centrifugation. The HEK-293T cell culture solution was collected into 50 mL centrifuge tubes and centrifuged at 2000g for 20 min at 4 °C. The supernatant was transferred to a new centrifuge tube and centrifuged at 5000g for 15 min at 4 °C. The obtained supernatant was transferred to a new centrifuge tube and centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 30 min at 4 °C. The supernatant was filtered into an ultrafiltration centrifuge tube with a 0.22 μm filter membrane and centrifuged at 4000g for 10 min at 4 °C. The final supernatant consists of exosomes. The supernatant was pipetted to a new tube and stored at −80 °C. The extracted exosomes were characterized by transmission electron microscopy (TEM) (FEI, USA) and using a nanoparticle tracking analyzer (NTA) (Malvern, UK).
000g for 30 min at 4 °C. The supernatant was filtered into an ultrafiltration centrifuge tube with a 0.22 μm filter membrane and centrifuged at 4000g for 10 min at 4 °C. The final supernatant consists of exosomes. The supernatant was pipetted to a new tube and stored at −80 °C. The extracted exosomes were characterized by transmission electron microscopy (TEM) (FEI, USA) and using a nanoparticle tracking analyzer (NTA) (Malvern, UK).
Exosomes loaded with Tw-Gap4/let-7a hybrids
0.2 μL of 0.5 M Tris(2-carboxyethyl) phosphine (TCEP) was mixed with 10 μL of 8.4 × 104 U mL−1 streptolysin O (SLO) and incubated for 20 min at 37 °C to activate SLO. After SLO is activated, the SLO is diluted into 0.2 U mL−1 for exosome loading. Exosomes were mixed with 0.2 U mL−1 SLO and incubated at 37 °C for 1.5 h.Fluorescence microscopy analysis of Tw-Gap4/let-7a@Exo
The slides and coverslips are cleaned with detergent and 95% alcohol to remove surface stains and then wiped clean with lens cleaning paper. The above exosomes were captured with anti-CD63 antibody-functionalized magnetic beads, and 10 μL of the sample was dropped on a slide. The slide was mounted and observed under a laser scanning confocal microscope (LSCM) (Zeiss, Germany).Fluorescence microscopy analysis of intracellular Tw-Gap4/let-7a hybrids
MDA-MB-231, MCF-7 and L02 cells were pre-cultured on a confocal small dish with a cell concentration of 3 × 104 cells and cultured for 24 h, and then the medium was removed and washed twice with PBS. 200 μl of different samples (including Exo, Tw-Gap4/let-7a, Tw-Gap4@Exo, let-7a@Exo, and Tw-Gap4/let-7a@Exo) were incubated with cells at various times (0, 1, 2, 3, 4, 5 and 6 h) at 37 °C. The cells were fixed with 4% paraformaldehyde (PFA) for 10 min, DAPI stained for 5 min, and then washed with PBS. Confocal fluorescence images of cells at different incubation times were obtained with a confocal microscope.Quantitative real-time polymerase chain reaction (qRT-PCR) analysis of miR-155 in cells
MDA-MB-231, MCF-7, L02 cells (2 × 105 cells per well) were pre-cultured in a 6-well plate and placed in an incubator for 24 h. When the number of cells reaches 1 × 106, the total RNA was extracted from MDA-MB-231, MCF-7 and L02 cells, using TRIzol reagent, and stored at −80 °C for further study. The concentration and purity of total RNA were assessed using aNanodrop (Thermo Scientific, USA). Reverse transcription of miRNA was performed using a reverse transcription kit to obtain a cDNA sample. Afterwards, the miRNA level was quantified by qRT-PCR with an SYBR Green PCR master mix. The internal reference of miRNA is U6. The results were analyzed using the relative quantitative (2−ΔΔCT) method.Cytotoxicity assay
MDA-MB-231, MCF-7, and L02 cells were each placed in a 96-well plate at a rate of about 1 × 104 cells per well and pre-cultured for 24 h under the abovementioned culture conditions. After that, the culture medium was discarded and the cells were incubated with different samples (PBS, Exo, Tw-Gap4/let-7a, let-7a@Exo, Tw-Gap4@Exo, and Tw-Gap4/let-7a@Exo) in an incubator for 4 h. After the fresh medium replaced the old medium for 36 h, the prepared CCK-8 solution was added, and the absorbance at 450 nm was measured after 2 h of incubation in the cell culture environment.Cell apoptosis assay
MDA-MB-231, MCF-7 and L02 cells (2 × 105 cells per well) were pre-cultured in 6-well plates and incubated for 24 h. After removing the medium, cells were incubated with various samples (PBS, Exo, Tw-Gap4/let-7a, let-7a@Exo, Tw-Gap4@Exo, and Tw-Gap4/let-7a@Exo) for 36 h. After trypsinization, the cells were collected into centrifuge tubes, centrifuged twice at 1000 rpm, and placed on ice. The cells were stained with Annexin-V FITC/PI for 15 min, and cell apoptosis was detected by flow cytometry (Beckman Coulter, USA).Wound healing assay
MDA-MB-231, MCF-7, and L02 cells (2 × 105 cells per well) were pre-cultured in a 6-well plate and pre-cultured for 24 h. After removing the medium, the cells were incubated with various samples (PBS, Exo, Tw-Gap4/let-7a, let-7a@Exo, Tw-Gap4@Exo, and Tw-Gap4/let-7a@Exo) for 24 h. Next, a sterile plastic pipette tip was used to scrape the cells from the central shaft, loose cells were washed away with PBS, and the cells were incubated in a serum-free medium at 37 °C. Photographs of the wound healing area were taken using a microscope (Nikon, Japan).Invasion assay
MDA-MB-231, MCF-7, and L02 cells were respectively pre-cultured into a 24-well Transwell chamber with Matrigel (Corning, USA), and the cells were incubated with various samples (PBS, Exo, Tw-Gap4/let-7a, let-7a@Exo, Tw-Gap4@Exo, and Tw-Gap4/let-7a@Exo) in a cell culture environment for 24 h. Then, the cells in the upper chamber were fixed with 4% PFA and washed with PBS. Then the cells on the lower surface of the membrane were stained with 0.1% crystal violet and imaged under a microscope.Western blot analysis
MDA-MB-231, MCF-7, and L02 cells were pre-cultured in a 6-well plate in an incubator for 24 h. The cells were incubated with various samples (PBS, Exo, Tw-Gap4/let-7a, let-7a@Exo, Tw-Gap4@Exo, Tw-Gap4/let-7a@Exo) for 4 h. After replacing the medium, the cells were incubated in the culture environment for 24 h and washed twice with PBS. The cells were lysed on ice using 500 μL of RIPA lysis buffer (containing 1% PMSF, 1% protease inhibitor, and 1% phosphatase inhibitor) to extract total protein. The protein is then quantified and boiled in a metal bath. Proteins were separated by electrophoresis and then transferred to the polyvinylidene fluoride (PVDF) membrane under a constant current of 200 mA. The PVDF membrane was blocked at room temperature for 1 h and then incubated with rabbit anti-SOX1 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), anti-HMGA1 (1
1000), anti-HMGA1 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25
25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), anti-c-Myc (1
000), anti-c-Myc (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) and anti-GAPDH antibodies (1
1000) and anti-GAPDH antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25
25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) respectively at 4 °C overnight. After washing 3 times with PBST, the membrane was incubated with a HRP-conjugated secondary antibody (1
000) respectively at 4 °C overnight. After washing 3 times with PBST, the membrane was incubated with a HRP-conjugated secondary antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000). A chemiluminescence chromogenic solution was added, and the protein band was detected with an imager.
5000). A chemiluminescence chromogenic solution was added, and the protein band was detected with an imager.
Results and discussion
Construction of DNA/miRNA hybrids
As shown in Fig. 1a, the Tw/let-7a hybrid forms a “tweezer” structure and is composed of two domains responsible for binding (loop) and for releasing (stem), respectively. The binding region of the DNA is complementary to miR-155, while partial hybridization occurs between the DNA and let-7a in the releasing region. Upon binding with miR-155, free energy drives the Tw/let-7a hybrids to unfold and release let-7a (Fig. 1b). It should be noted here, a gap or a tail of let-7a is left to unhybridize with the DNA, so that the fully complementary miR-155 can rival with let-7a to bind with the DNA (Fig. 1c).We first optimize the length of the gap or the tail, because it is critical for the formation and response of the hybrid structure. Fig. 1d and Fig. S1† show the electrophoretic results of the products of the Tw/let-7a hybrids with different lengths of the gap or the tail as well as its response to miR-155. Results show that the hybrids can successfully assemble in all the cases of different lengths of the gap and the tail. But in the presence of miR-155, there is a gradient difference in the efficiency of binding to miR-155 and releasing let-7a: Tw-Gap4 > Tw-Tail6 > Tw-Tail4 > Tw-Gap6 > Tw-Gap2. Therefore, the hybrids with a 4 nt gap (Tw-Gap4/let-7a) were adopted as optimized onest for all subsequent experiments.
Next, the concentration dependence and response dynamics of the Tw-Gap4/let-7a hybrids were studied by labeling the let-7a with a fluorophore (FAM) and the Tw-Gap4 with a quencher (BHQ1) to form a beacon. The release of let-7a caused by the binding of miR-155 to the Tw-Gap4 will result in the recovery of the fluorescence signals. Fig. 1e and f show that with the concentration of miR-155, the fluorescence increases, suggesting a typical concentration-dependent releasing process. The curve reaches a plateau when the concentration of miR-155 is equal to or higher than the concentration of the Tw-Gap4/let-7a hybrids, suggesting a “1-to-1” binding and releasing. To confirm that the release of let-7a is originated from the binding of miR-155 to Tw-Gap4, another couple of hybrids was adopted (Cy3-labeled miR-155 and BHQ1-labeled Tw-Gap4). Upon binding to the Tw-Gap4, the fluorescence of miR-155 will be quenched. Fig. 1g and h show that with the concentration of the Tw-Gap4/let-7a hybrids, the fluorescence of miR-155 decreases and finally reaches a valley, suggesting the successful “1-to-1” binding of miR-155 to Tw-Gap4. The dynamics of the Tw-Gap4/let-7a hybrids were then studied. By adopting the same two couples of hybrids (Fig. 1i and j: FAM-labeled let-7a and BHQ1-labeled Tw-Gap4; Fig. 1k and l: Cy3-labeled miR-155 and BHQ1-labeled Tw-Gap4), we show that the fluorescence signals of the two groups increase or decrease sharply in the first dozen of minutes respectively, and then reach a plateau at about 80 min, indicating that the release of let-7a and the binding of miR-155 are synchronized and can be completed within dozens of minutes.
Encapsulation of Tw-Gap4/let-7a in exosome
Exosomes have been widely reported to be a promising vehicle for drug delivery due to their suitable size, good packaging ability and excellent biocompatibility.30,31 Here, we encapsulate the Tw-Gap4/let-7a into exosomes by using an SLO strategy (Fig. 2a).32 The exosomes extracted from the culture medium of HEK-293T cells through ultrafiltration centrifugation were first characterized using TEM and NTA, showing a typical spherical shape with a size ranging from 80 to 150 nm (Fig. 2b and c). After encapsulation of the Tw-Gap4/let-7a and subsequent enrichment using immunomagnetic beads that can bind to the CD63 on the exosomal membrane, the exosomes were also characterized under LSCM. By using a dye (Mem Dye-Green) to stain the exosomal membrane, clear green fluorescence can be observed on the surface of the magnetic beads, suggesting the successful enrichment of the exosomes (Fig. 2d). By using fluorophore-labeled Tw-Gap4/let-7a (ROX-labeled Tw-Gap4 and FAM-labeled let-7a), clear red fluorescence resonance energy transfer (FRET) signals between the two fluorophores can be observed (Fig. 2d). This result indicates that the Tw-Gap4/let-7a hybrids have been successfully encapsulated, and the structure remains intact since the FRET signals only appear when the fluorophore ROX and the fluorophore FAM are close enough.Uptake of Tw-Gap4/let-7a@Exo by the cells
After successfully constructing Tw-Gap4/let-7a@Exo, we studied its uptake by different cells. Human breast cancer cells (MDA-MB-231 and MCF-7) and human normal liver cells (L02) were adopted. FRET signals as well as the individual signals of the fluorophore-labeled Tw-Gap4/let-7a (ROX-labeled Tw-Gap4 and FAM-labeled let-7a) were measured using LSCM to trace the intracellular localization of the Tw-Gap4/let-7a hybrids as shown in Fig. 3a. In the case of cancer cells, in the first 3 to 4 h, the intracellular FRET signal increases, indicating that the intact Tw-Gap4/let-7a hybrids have been taken up gradually by the cells. Following the 3 to 4 h, the FRET signals start to attenuate, and at the same time, the individual signals of the ROX-labeled Tw-Gap4 and FAM-labeled let-7a start to increase (Fig. 3b). The above results indicate that the Tw-Gap4/let-7a hybrids have undergone two processes, namely cell uptake and intracellular disintegration, and as time goes by, the latter gradually dominates. But in the case of normal cells, in the same time window, no attenuation of the FRET signal was observed, suggesting that the Tw-Gap4/let-7a hybrids remain intact in the cells. The differential performance of Tw-Gap4/let-7a hybrids in cancer cells and normal cells is expected to be attributed to the different expression levels of intracellular miR-155. The qRT-PCR results from Fig. S2† confirm this speculation by showing that there is a gradient of the content of miR-155: MFC-7 > MDA-MB-231 ≫ L02, which is well in line with the results of intracellular disintegration performance of the Tw-Gap4/let-7a hybrids. We then upregulated miR-155 in L02 with miR-155 mimic or downregulated miR-155 in MCF-7 and MDA-MB-231 with miR-155 inhibitor to further confirm that the signal differences between different cells are due to the difference in miR-155 expression rather than the difference in cell type or other heterogeneity. Fig. S3† shows the expected results that the intracellular FRET and FAM fluorescence intensities were significantly enhanced in the case of upregulated miR-155 or decreased in the case of downregulated miR-155, suggesting the effective response of the Tw-Gap4/let-7a hybrids to the intracellular miR-155.Taking the above in vitro results together, it should be noted that the high expression of miR-155 in cancer cells promotes the disintegration of the responsible Tw-Gap4/let-7a hybrids and the release of let-7a.
Effect of Tw-Gap4/let-7a@Exo on the cell physiological activity
On the basis of the successful delivery of the Tw-Gap4/let-7a hybrids into cells by exosomes, we then studied the effect of Tw-Gap4/let-7a@Exo on the cell physiological activity, including viability, apoptosis, migration and invasion. The cell viability was measured by using a CCK-8 method. As shown in Fig. 4a, the viability of MCF-7 and MDA-MB-231 was observed to be significantly inhibited (37.6% and 39.7% reduction respectively) when incubated with the Tw-Gap4/let-7a@Exo. In the control groups by using Tw-Gap4@Exo or let-7a@Exo only, the viability was reduced by 25.1% and 31.3% respectively for MCF-7 and 13.1% and 22.2% respectively for MDA-MB-231. Without the exosome vehicle, Tw-Gap4/let-7a cannot act on the viability, and neither can exosomes only. It is also interesting that all the groups have no significant effect on the cell viability of L02. The above results indicate that the Tw-Gap4/let-7a@Exo can selectively inhibit the viability of cell lines with high expression of miR-155, and the effect is better when both Tw-Gap4 and let-7a are presented. Apoptosis analysis shows similar results (Fig. 4b and 5c): the Tw-Gap4/let-7a@Exo selectively induces the apoptosis of MCF-7 and MDA-MB-231, while Tw-Gap4@Exo and let-7a@Exo have an inferior effect, and Tw-Gap4/let-7a or Exo only cannot induce apoptosis. Similar results can also be obtained for cell migration (Fig. 5a and b) and cell invasion (Fig. 5c and d), which is also consistent with literature that the inhibition of miR-155 and the supplement of let-7a are capable of inhibiting the migration and invading breast cancer cells.20,33It was reported that the opposite effects of miR-155 and let-7a on cell migration and invasion are mainly realized by acting on SOX1 mRNA and HMGA1 mRNA,20,33 respectively. Thus, we studied the mechanism of the synergistic action of the Tw-Gap4/let-7a hybrids in breast cancer cells by using western blot to analyse the expression of the target proteins of SOX1 and HMGA1. Results show that the Tw-Gap4/let-7a hybrids encapsulated in exosomes indeed affect the expression of these two proteins by up-regulating SOX1 through the inhibition of miR-155 and down-regulating HMGA1 through the supplement of let-7a (Fig. 5e and f). It is also reported that SOX1 and HMGA1 jointly participate in the Wnt/β-catenin signaling pathway, which is responsible for the growth and invasion of cancer cells.33,34 Our results show that after cancer cells are treated by Tw-Gap4/let-7a@Exo, the key downstream protein c-Myc in the Wnt/β-catenin signaling pathway is down-regulated (Fig. 5e and f). The above results together indicate that the responsive Tw-Gap4/let-7a encapsulated in exosomes can synergistically enhance the therapeutic effect on cancer cells through the Wnt/β-catenin signaling pathway (Fig. 5g).
Conclusions
In conclusion, we developed DNA–miRNA hybrids, which can respond to miR-155 and smartly release let-7a. After being encapsulated in exosomes, the DNA–miRNA hybrids can be delivered into cancer cells to respond to and inhibit the expression of endogenous miR-155 and supply let-7a in parallel. By up-regulating SOX1, down-regulating HMGA1 and jointly inhibiting the Wnt/β-catenin signaling pathway, the responsive DNA–miRNA hybrids can suppress cell viability, induce cell apoptosis and inhibit cell migration and invasion. Owing to the responsive capability, the DNA–miRNA hybrids only work in cancer cells but have little effect on normal cells, making this system have good biocompatibility. By further developing DNA–miRNA hybrids, engineered exosomes, and combination drugs that simultaneously inhibit multiple genes, this system holds promise for developing miRNA drugs and treating solid tumors.Author contributions
F. Zhang – conceptualization, writing original draft, validation, visualization, and investigation. A. Isak – investigation and validation. S. Yang – investigation. Y. Song – investigation. L. Ren – investigation. C. Feng – project administration and review & editing. G. Chen – funding acquisition and review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant no. 81873539 and 82070518) and the Shanghai Pujiang Program (2019PJD014).Notes and references
- M. Y. Qu, X. J. Zou, F. Fang, S. Y. Wang, L. Xu, Q. Zeng, Z. Fan, L. Chen, W. Yue, X. Y. Xie and X. T. Pei, Nat. Commun., 2020, 11, 4964 CrossRef CAS PubMed.
- Y. Wu, M. H. Yuan, H. T. Wu, W. J. Chen, M. L. Zhang, Q. Q. Ye, J. Liu and G. J. Zhang, Cell Death Dis., 2020, 11, 912 CrossRef CAS PubMed.
- D. Parashar, A. Geethadevi, M. R. Aure, J. Mishra, J. George, C. L. Chen, M. K. Mishra, A. Tahiri, W. Zhao, B. Nair, Y. Lu, L. S. Mangala, C. Rodriguez-Aguayo, G. Lopez-Berestein, A. K. S. Camara, M. Y. Liang, J. S. Rader, R. Ramchandran, M. You, A. K. Sood, V. N. Kristensen, G. B. Mills, S. Pradeep and P. Chaluvally-Raghavan, Cell Rep., 2019, 29, 4389–4406 CrossRef CAS PubMed.
- Z. Q. Wang, M. M. Fu and Y. J. Li, Can. J. Physiol. Pharmacol., 2020, 98, 314–323 CrossRef CAS PubMed.
- M. Maryam, M. Naemi and S. S. Hasani, J. Genet., 2021, 100, 15 CrossRef PubMed.
- D. P. Bartel, Cell, 2009, 136, 215–233 CrossRef CAS PubMed.
- S. Bajan and G. Hutvagner, Cells, 2020, 9, 137 CrossRef CAS PubMed.
- H. F. Nie, X. D. Xie, D. D. Zhang, Y. Zhou, B. F. Li, F. Q. Li, F. Y. Li, Y. L. Cheng, H. Mei, H. Meng and L. Jia, Nanoscale, 2020, 12, 877–887 RSC.
- Y. Xie, Y. Hang, Y. Wang, R. Sleightholm, D. P. Prajapati, J. Bader, A. Yu, W. M. Tang, L. Jaramillo, J. Li, R. K. Singh and D. Oupicky, ACS Nano, 2020, 14, 255–271 CrossRef CAS PubMed.
- J. Hayes, P. P. Peruzzi and S. Lawler, Trends Mol. Med., 2014, 20, 460–469 CrossRef CAS PubMed.
- Z. Y. Lu, Q. T. He, J. F. Liang, W. G. Li, Q. Su, Z. J. Chen, Q. Wan, X. F. Zhou, L. Cao, J. J. Sun, Y. Wu, L. Liu, X. M. Wu, J. S. Hou, K. Q. Lian and A. X. Wang, Mol. Ther. – Nucleic Acids, 2019, 16, 471–480 CrossRef CAS PubMed.
- K. Qu, X. Zhang, T. Lin, T. Liu, Z. X. Wang, S. S. Liu, L. Zhou, J. C. Wei, H. L. Chang, K. Li, Z. Wang, C. Liu and Z. Wu, Sci. Rep., 2017, 7, 1692 CrossRef PubMed.
- C. Y. Li, Y. H. Yin, X. Liu, X. J. Xi, W. X. Xue and Y. Q. Qu, Oncotarget, 2017, 8, 24564–24578 CrossRef PubMed.
- S. Anfossi, A. Babayan, K. Pantel and G. A. Calin, Nat. Rev. Clin. Oncol., 2018, 15, 541–563 CrossRef PubMed.
- P. Zhang, Y. Ouyang, Y. S. Sohn, R. Nechushtai, E. Pikarsky, C. H. Fan and I. Willner, ACS Nano, 2021, 15, 6645–6657 CrossRef CAS PubMed.
- W. P. Tsang and T. T. Kwok, Apoptosis, 2008, 13, 1215–1222 CrossRef CAS PubMed.
- F. Liu, Y. Tai and J. Q. Ma, Cancer Biol. Ther., 2018, 19, 534–542 CrossRef CAS PubMed.
- C. Y. Luo, J. Y. Zhang, Y. Zhang, X. Zhang, Y. N. Chen and W. M. Fan, Cell Cycle, 2020, 19, 1983–1993 CrossRef CAS PubMed.
- Y. S. Lee and A. Dutta, Genes Dev., 2007, 21, 1025–1030 CrossRef CAS PubMed.
- K. Liu, C. F. Zhang, T. Li, Y. L. Ding, T. Tu, F. F. Zhou, W. K. Qi, H. B. Chen and X. C. Sun, Internet J. Oncol., 2015, 46, 2526–2534 CrossRef CAS PubMed.
- J. T. Powers, K. M. Tsanov, D. S. Pearson, F. Roels, C. S. Spina, R. Ebright, M. Seligson, Y. D. deSoysa, P. Cahan, J. Theissen, H. Tu, A. Han, K. C. Kurek, G. S. LaPier, J. K. Osborne, S. J. Ross, M. Cesana, J. J. Collins, F. Berthold and G. Q. Daley, Nature, 2016, 535, 246–251 CrossRef CAS PubMed.
- S. Jiang, H. W. Zhang, M. H. Lu, X. H. He, Y. Li, H. Gu, M. F. Liu and E. D. Wang, Cancer Res., 2010, 70, 3119–3127 CrossRef CAS PubMed.
- M. M. Yang, H. C. Shen, C. Qiu, Y. Ni, L. G. Wang, W. Dong, Y. D. Liao and J. J. Du, Eur. J. Cancer, 2013, 49, 604–615 CrossRef CAS PubMed.
- L. B. Yin, T. Liu, C. Y. Li, G. Q. Yan, C. Li, J. T. Zhang and L. Wang, Cancer Cell Int., 2020, 20, 303 CrossRef CAS PubMed.
- M. Gironella, M. Seux, M. J. Xie, C. Cano, R. Tomasini, J. Gommeaux, S. Garcia, J. Nowak, M. L. Yeung, K. T. Jeang, A. Chaix, L. Fazli, Y. Motoo, Q. Wang, P. Rocchi, A. Russo, M. Gleave, J. C. Dagorn, J. L. Iovanna, A. Carrier, M. J. Pebusque and N. J. Dusetti, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 16170–16175 CrossRef CAS PubMed.
- S. S. Mortazavi-Jahromi, M. Aslani, S. Omidian, A. Ahmadzadeh, Z. Rezaieyazdi and A. Mirshafiey, Drug Dev. Res., 2020, 81, 295–304 CrossRef CAS PubMed.
- C. M. Zhang, J. Zhao and H. Y. Deng, J. Biomed. Sci., 2013, 20, 79 CrossRef CAS PubMed.
- W. Zhang, C. J. Chen and G. L. Guo, Pharmacol. Sci., 2018, 22, 7323–7332 CAS.
- E. Anastasiadou, A. G. Seto, X. Beatty, M. Hermreck, M. E. Gilles, D. Stroopinsky, L. C. Pinter-Brown, L. Pestano, C. Marchese, D. Avigan, P. Trivedi, D. Escolar, A. L. Jackson and F. J. Slack, Clin. Cancer Res., 2021, 27, 1139–1149 CrossRef CAS PubMed.
- X. Zhang, H. B. Zhang, J. M. Gu, J. Y. Zhang, H. Shi, H. Qian, D. Q. Wang, W. R. Xu, J. M. Pan and H. A. Santos, Adv. Mater., 2021, 33, 2005709 CrossRef CAS PubMed.
- Y. Y. Li, Y. T. Zhang, Z. Li, K. Zhou and N. P. Feng, Sci. Eng., 2019, 5, 4870–4881 CAS.
- J. H. Lee, J. A. Kim, M. H. Kwon, J. Y. Kang and W. J. Rhee, Biomaterials, 2015, 54, 116–125 CrossRef CAS PubMed.
- Y. Han, Med. Sci., 2017, 46, 555 Search PubMed.
- L. Q. Song, D. Liu, J. J. He, X. J. Wang, Z. J. Dai, Y. Zhao, H. F. Kang and B. F. Wang, APMIS, 2016, 124, 547–555 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Electrophoretic patterns of the Tw/let-7a hybrid with different lengths of a tail, relative miR-155 expression of different cells by quantitative real-time PCR (qRT-PCR), fluorescence images of different cells after the treatment of Tw-Gap4/let-7a@Exo, and sequences of oligonucleotides. See DOI: https://doi.org/10.1039/d1nr08539e |
| This journal is © The Royal Society of Chemistry 2022 |