Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Reconfigurable scaffolds for adaptive tissue regeneration
Mingxing
Peng
 ab,
Qilong
Zhao
ab,
Qilong
Zhao
 *a,
Min
Wang
*a,
Min
Wang
 c and
Xuemin
Du
c and
Xuemin
Du
 *a
*a
aInstitute of Biomedical & Health Engineering, Shenzhen Institute of Advanced Technology (SIAT), Chinese Academy of Sciences (CAS), Shenzhen, 518055, China. E-mail: ql.zhao@siat.ac.cn; xm.du@siat.ac.cn
bUniversity of Chinese Academy of Sciences, China
cDepartment of Mechanical Engineering, The University of Hong Kong, Pokfulam Road, Hong Kong
First published on 1st March 2023
Abstract
Tissue engineering and regenerative medicine have offered promising alternatives for clinical treatment of body tissue traumas, losses, dysfunctions, or diseases, where scaffold-based strategies are particularly popular and effective. Over the decades, scaffolds for tissue regeneration have been remarkably evolving. Nevertheless, conventional scaffolds still confront grand challenges in bio-adaptions in terms of both tissue-scaffold and cell-scaffold interplays, for example complying with complicated three-dimensional (3D) shapes of biological tissues and recapitulating the ordered cell regulation effects of native cell microenvironments. Benefiting from the recent advances in “intelligent” biomaterials, reconfigurable scaffolds have been emerging, demonstrating great promise in addressing the bio-adaption challenges through altering their macro-shapes and/or micro-structures. This mini-review article presents a brief overview of the cutting-edge research on reconfigurable scaffolds, summarizing the materials for forming reconfigurable scaffolds and highlighting their applications for adaptive tissue regeneration. Finally, the challenges and prospects of reconfigurable scaffolds are also discussed, shedding light on the bright future of next-generation reconfigurable scaffolds with upgrading adaptability.
1. Introduction
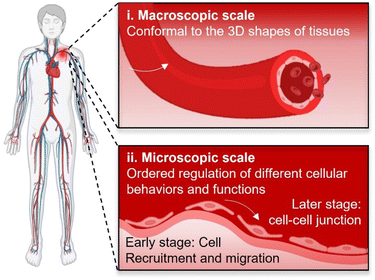
There are urgent and abundant clinical demands for tissue regeneration for reconstituting/reconstructing body tissues with large defects, especially for those lacking sufficient innate renewal potential such as cartilage, nerve, and cardiovascular systems. Since formally introduced by Professor Robert Langer and Professor Joseph Vacanti for the first time in 1993,1 tissue engineering that involves synergistic efforts of biomaterials, cells, and bio-signals has attracted much attention owing to its great potential for assisting tissue regeneration. To date, multiple tissue engineering strategies have been developed and investigated, among which scaffold-based strategies exhibited the highest versatility.2–4 Scaffold-based tissue engineering involves building scaffolds with tailored biophysical and biochemical cues suiting for facilitating cell attachment, proliferation, and differentiation.5 As an important prerequisite for supporting cellular growth, biomaterial scaffolds must be first biocompatible. Over the past two decades, scaffolds have also been continuing to evolve to be biodegradable and bioactive,6 with the aim to avoid secondary surgeries for the removal of implants, stimulating cellular activities and functions, and finally facilitating in situ tissue regeneration by harnessing the innate renewal capability of body tissues.7However, facilitating in situ tissue regeneration using appropriate scaffolds remains greatly challenging, which will require excellent interplays between implanted scaffolds and target tissues/cells. Upon implantation, both tissue–scaffold and cell–scaffold interplays occur at their biointerfaces.8 For attaining desirable tissue–scaffold and cell–scaffold interplays, biointerfaces shall comply with the 3D shapes of target tissues at the macroscopic scale as well as be capable of ordered regulation of different cellular behaviours and functions at the microscopic scale for meeting the requirements of dynamic cell manipulation at diverse tissue regeneration stages (Fig. 1), for example initially promoting the recruitment/migration of endothelial cells and subsequently assisting the formation of cell–cell junctions throughout the vascular remodelling process.9 Therefore, higher requirements of bio-adaption would be laid on scaffolds, including not only conformal interfacing with target tissues, but also ordered regulation of cellular behaviours and functions at their biointerfaces. However, it would be a grand challenge for conventional scaffolds. On one hand, conventional scaffolds with pre-defined planar or curved geometries would be difficult to be perfectly conformal to biological tissues possessing complicated three-dimensional (3D) and even sometimes continuously changing shapes.10 On another hand, it would also be extremely challenging to manipulate cellular functions in programmable manners for adapting to a specific tissue regeneration process by the scaffolds lacking dynamic biointerfaces.11,12
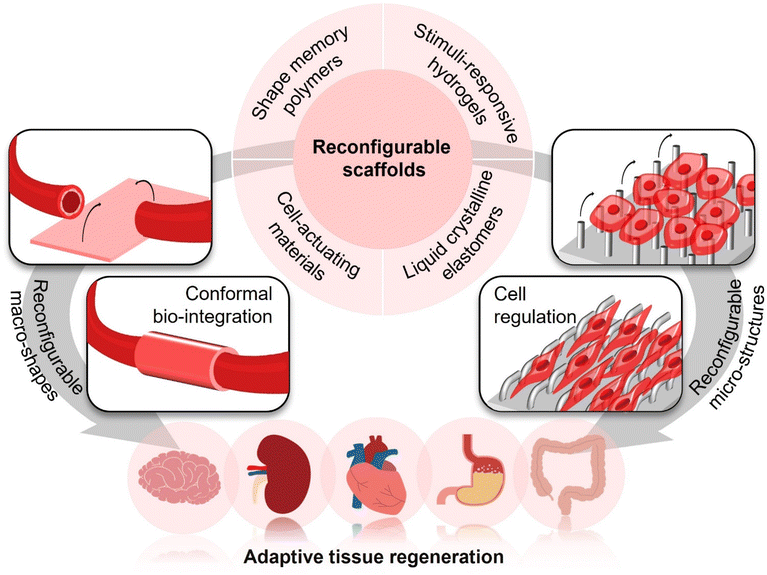
To address the above bio-adaption challenges, reconfigurable scaffolds have recently emerged with the advances in “intelligent” polymers. They have brought the opportunities of enhancing conformal bio-integration with 3D-shaped tissues and manipulating cellular functions on demand by altering their macro-shapes and micro-structures, respectively (Fig. 2). Through programming their macro-shapes by application of a stimulus from either the body itself (e.g., body temperature, body fluids, biomolecules, and cells) or external fields (e.g., light, magnet, and ultrasound), reconfigurable scaffolds could adapt to the 3D shapes of target tissues in an actively compliant manner. Such active shape adaptation would generate less stress concentration in comparison with the conventional passively compliant shaping process,13 which would be particularly important for forming conformal and stable biointerfaces between scaffolds and tissues. Meanwhile, since cells sense the micro-structures at scaffold biointerfaces and react according to the corresponding biomechanical cues,14–16 scaffolds with reconfigurable micro-structures, which could also be modulated by body's environments or external stimuli, could manipulate cellular behaviours and functions in a stage-specific manner by offering dynamic biomechanical induction, subsequently meeting the requirements of ordered cell regulation in specific tissue regeneration processes.
With the significant merits of conformal interfacing and ordered regulation of cellular behaviours and functions, reconfigurable scaffolds have received increasing attention over the past decade, opening a new avenue for adaptive tissue regeneration. Here, we will present a brief overview of the recent advances in reconfigurable scaffolds, introducing popular materials for forming them, highlighting their applications for adaptive tissue regeneration, as well as discussing the current challenges and future direction, with the aim of inspiring a broad range of attention from multidisciplinary fields to develop next-generation scaffolds with upgrading bio-adaption performances for better accommodating complicate and dynamic tissue regeneration processes.
2. Materials for forming reconfigurable scaffolds
Reconfigurable scaffolds, with reconfigurable macro-shapes and/or micro-structures, are usually constructed based on the morphing polymer-based materials capable of dynamically altering their shapes in programmable routes. Considering the applications of tissue engineering and regenerative medicine, the morphing polymer-based materials for forming reconfigurable scaffolds should further be cytocompatible, convenient to be processed/manufactured, and capable of being actuated via a mild approach. For meeting these requirements, shape-memory polymers (SMPs), hydrogels, liquid crystalline elastomers (LCEs), and cell-actuating materials have been widely employed for preparing reconfigurable scaffolds owing to their good cytocompatibility and the capabilities of being conveniently actuated by either body's internal signals or the external stimuli that could safely penetrate through biological tissues. Varying with different morphing mechanisms and characteristics of these materials, the resulting reconfigurable scaffolds will possess different features and will be actuated by different ways, imparting them diverse promise for adaptive tissue regeneration (Table 1).| Classifications | Materials | Scaffold architectures/forms | Manufacturing techniques | Ways to actuate | Response time | Ref. |
|---|---|---|---|---|---|---|
| SMPs | PCL with grafting allyl alcohol | Patterned scaffold | Thermal embossing micro-imprint lithography | Body temperature | ∼40 min | 31 |
| PDLLA-co-TMC | Nanofibrous scaffold | Electrospinning | Body temperature | ∼6 s | 32 and 33 | |
| PDLLA-co-TMC | Porous scaffold | Extrusion-based 3D printing | Body temperature | Not provided | 34 | |
| PLLA/PCL–diacrylate | Porous scaffold | Salt leaching | Body temperature | Not provided | 36 | |
| PU/gelatin | Porous scaffold | Extrusion-based low-temperature 3D printing | Body temperature | Not provided | 37 | |
| Poly(lactic-co-glycolic acid) | Porous scaffold | Hot pressing | Ultrasound | ∼120 s | 39 | |
| PCL/Fe3O4 | Nanofibrous scaffold | Electrospinning | Magnetic field | ∼120 s | 40 | |
| PU/magnesium | Porous scaffold | Extrusion-based low-temperature 3D printing | NIR light | ∼100 s | 41 | |
| CNC–pyridine/PEG–PCL–PU | Film scaffold | Casting | pH | ∼30 min | 42 | |
| PCL/Pellethane® | Nanofibrous scaffold | Electrospinning | Enzyme | ∼7 days | 43 | |
| Keratin protofibril | Hierarchical fibrous scaffold | Self-assembly and extrusion-based 3D printing | Hydration | ∼105 s | 44 | |
| Hydrogels | MA-Alg/MA-HA | Tubular scaffold | Extrusion-based 3D printing and photocrosslinking | Hydration | Not provided | 54 |
| Alginate/polydopamine | Porous scaffold | Extrusion-based 3D printing and ion crosslinking | NIR light | ∼180 s | 55 | |
| PNIPAM/gold nanorods | Patterned scaffold | Microfabrication and photocrosslinking | NIR light | ∼4 s | 59 | |
| Alginate | Patterned scaffold | Casting | Na+/Ca2+ | 7 min | 62 | |
| LCEs | PMHS–DUB–MPBB | Film scaffold | Thermal polymerization | Heating | ∼1 s | 83 |
| MCPAHB–AHOPAHOB–AZ | Film scaffold | Photocrosslinking and thermal polymerization | Green light | ∼150 ms | 84 | |
| Cell-actuating materials | Parylene/NIH-3T3 cells | Film scaffold | Microfabrication | Cell traction force | ∼72 h | 92 |
| NorHA/MSCs | Nanofibrous scaffold | Electrospinning, extrusion-based 3D printing and photocrosslinking | Cell traction force | Not provided | 100 | |
2.1 SMPs
SMPs are one group of polymers with shape-memory effects, possessing the capability of shape recovery from their temporary to permanent shapes upon appropriate stimuli.17 Unlike the elastic recovery of rubber-like materials, SMPs can be locked into specific reshaped temporary geometries, then undergo a controllable morphing process to return their originally permanent shapes. Such shape-morphing capabilities of SMPs result from their unique networks, containing permanent netpoints and stimuli-responsive switchable phases.18 Switchable phases can be formed by either physical interactions or reversible covalent bonding, contributing to locking the SMPs to be temporary shapes after reshaping them into any intended geometries. Once tuning the switchable phases by specific stimulations, it will trigger the shape recovery of SMPs.18Poly(ε-caprolactone) (PCL), poly(urethane) (PU), poly(lactide) (PLA), and their copolymers, which possess excellent biocompatibility and biodegradability, are popular SMPs that have been widely used for various biomedical applications such as controlled drug delivery,19 wound closure,20 intervention stents,21 and neuroelectronics.22,23 Given that these biodegradable SMPs are semi-crystalline with the crystalline regions as the permanent netpoints and switchable phases formed based on physical interactions,24 heating to trigger the melting or glass transition is the common approach for actuating their shape recovery. To achieve reconfigurations within human bodies, it is necessary to tune their melting temperature (Tm) or glass transition temperature (Tg) approximating to the physiological temperature (37 °C). However, the temperatures for actuating the shape recovery of pure PCL (Tm: 59–64 °C),25 PU (Tg: 50–90 °C),26 and PLA (Tg: 60–65 °C)27 are much higher than the physiological temperature (37 °C). To reduce the actuating temperature near the range of physiological temperature, methods for reinforcing the netpoints and/or softening the switchable phase within their networks have been applied, which could be realized by means of grafting cross-linkable end groups,28 copolymerization,29 or blending.30 Notably, these modified thermoresponsive biodegradable SMPs are easy to be processed into diverse scaffold forms by different manufacturing techniques. For instance, branched PCL with grafting allyl alcohol groups enabling covalent crosslinking could be facilely manufactured via thermal embossing micro-imprint lithography to be patterned scaffolds, whose surface microstructures could be altered in response to body temperature.31 A PLA-based copolymer, poly(D,L-lactide-co-trimethylene carbonate) (PDLLA-co-TMC) with tuneable Tg in the range from 20–45 °C, has been used for preparing reconfigurable nanofibrous scaffolds via electrospinning for bone and vascular tissue engineering.32,33 Such copolymers could also be suitable for extrusion-based 3D printing,34 subsequently forming porous scaffolds with initially customized geometries.35 The polymer blends, such as poly(L-lactic acid) (PLLA)/PCL–diacrylate and PU/gelatin, could also be simply processed to be porous scaffolds via salt leaching or extrusion-based low-temperature 3D printing, which showed semi-interpenetrating networks and excellent shape-memory effects that could be actuated by body temperature.36–38
Although shape-memory scaffolds made of biodegradable thermoresponsive SMPs that could be actuated by body temperature have shown numerous promises in tissue engineering, challenges remain such as controlling the scaffold reconfigurations in a programmable spatiotemporal fashion. Once implanted into human bodies, such scaffolds would undergo spontaneous shape recovery. To enhance the controllability of deformation, SMP-based reconfigurable scaffolds that could be actuated by ultrasound,39 magnet,40 and near-infrared (NIR) light41 have been developed through the incorporation of functional nano-/micro-materials. Shape transformations of these scaffolds are also realized on the basis of thermoresponsive SMPs. However, instead of directly heating via body temperature, the energies generated from high-intensity focused ultrasound, the hyperthermia of magnetic nanoparticles under an alternating magnetic field, or photothermal nano-/micro-agents (gold nanorods, carbon nanotubes, black phosphors nanosheets, magnesium nano-/micro-particles, MXenes, polydopamine, etc.) under NIR irradiation are used for actuating the shape recovery of SMPs, subsequently resulting in remotely controlled shape-memory reconfigurable scaffolds.
Apart from biodegradable thermoresponsive SMPs, some new types of SMPs with innovative molecular designs have also been studied for preparing reconfigurable scaffolds with versatile responsive behaviours. For example, through blending pyridine-functionalized cellulose nanocrystals (CNCs) and poly(ethylene glycol–co-caprolactone–co-urethane) (PEG–PCL–PU), the hydrogen bonding between the pyridine moistures and carboxyl groups in PEG–PCL–PU would be stabilized under a high-pH condition yet dissociated at a low pH value. Using such blends, pH-responsive shape-memory scaffolds have been prepared via casting.42 Besides, through forming interpenetrating networks between enzymatically degradable PCL (as the switchable phase) and enzymatically stable Pellethane® (as the permanent netpoint), the resultant polymer hybrids could obtain enzyme-responsive shape-memory capabilities, which could be used for forming scaffolds undergoing slow shape transformations within a long period of 7 days in the presence of a lipase.43 Recently, a natural material, keratin protofibril derived from animal's hair, has also been investigated for forming a shape-memory reconfigurable scaffold consisting of self-assembled long-range ordered keratin molecules.44 The transition of the keratin secondary structure from α-helix to β-sheet upon hydration would actuate the macro-shape transformations of 3D printed keratin-based scaffolds, holding promise in fitting for sophisticated 3D geometries of body tissues.
SMPs have offered important candidates for constructing reconfigurable scaffolds that could be actuated by interior body temperature or an applied external stimulus. The SMP-based reconfigurable scaffolds with excellent shape-memory effects would exhibit high reliability in shape transformations as their permanent shapes could be pre-determined. However, they usually show limited freedom in shape transformations owing to the normal one-way actuation of SMPs. With the advances of two-way and even multi-way SMPs,45–47 it will be expected to develop shape-memory reconfigurable scaffolds with enhanced freedom of shape transformation for a better perspectives of adaptive tissue regeneration.
2.2 Hydrogels
Hydrogels are commonly hydrophilic networks containing significant amounts of water. The water-rich networks of hydrogels make them not only highly friendly to biological systems but also mechanically compatible with biological tissues owing to their tissue-like viscoelasticity.48 These outstanding features have made hydrogels excellent candidates for tissue engineering scaffolds, for example, engineering biomimetic cell microenvironments for 3D cell culture.49 In addition, the water-rich networks also make the hydrogels easy to tune their volumes via controllable swelling/shrinkage,50 therefore offering opportunities for preparing hydrogel-based reconfigurable scaffolds.Taking advantage of the common phenomena that hydrogels undergo volumetric expansion/shrinkage during hydration/dehydration, hydrogel-based reconfigurable structures enabling programmable shape transformations through controlling their swelling behaviours have been widely studied. The programmable shape transformations from the as-fabricated two-dimensional (2D) states to certain target 3D geometries through swelling can be realized by modulating the spatial swelling gradients (either through the thickness or in the plane) by employing different components with varying swelling ratios51 or designing mono-component hydrogels with different crosslinking densities.52 Alternatively, shape transformations of a swollen hydrogel could also be programmed by simply controlling the dimensions (thickness, length, and width) of its original shape.53 Such hydration-actuated shape-morphing strategies are universal for a variety of hydrogels. Under this principle, reconfigurable cell-laden hydrogel scaffolds made of two modified natural polymers, methacrylated alginate (MA-Alg) and methacrylated hyaluronic acid (MA-HA) have been developed via four-dimensional (4D) bioprinting.54 Bioprinted 2D cell-laden films with a crosslinking gradient through the thickness of the film scaffolds formed by top-down photocrosslinking and mild drying could be self-folded into vessel-like 3D tubes upon immersion in water, phosphate buffered saline (PBS), or cell culture medium. In addition to the hydration-induced mechanism, shape transformations of hydrogel-based reconfigurable scaffolds could also be realized by controlling the dehydration for inducing volumetric shrinkage. For example, a bilayer scaffold made of alginate hydrogels and a photothermal agent, polydopamine, has been fabricated by extrusion-based 3D printing and ion crosslinking.55 Upon NIR irradiation, the incorporated polydopamine would convert light energy to heat, subsequently inducing the shape transformations of the scaffolds along with the thermos-induced dehydration of the alginate hydrogels.
Though controlling hydration/dehydration has offered convenient and universal strategies for programming hydrogel shapes between their dry and swollen states, such methods still confront challenges in dynamically modulating the scaffold shapes in biological environments containing cell culture medium or body fluids. To realize controllable shape transformations of hydrogels among their different equilibrium swelling states, stimuli-responsive hydrogels that can respond to diverse environmental and/or external stimuli have received numerous attention in developing hydrogel-based reconfigurable scaffolds.56 One representative example is poly(N-isopropylacrylamide) (PNIPAM), which undergoes a reversible sol–gel phase transition above its lower critical solution temperature (∼32 °C). The sol–gel phase transition can alter the hydrophilic/hydrophobic states of polymer backbones, therefore resulting in thermoresponsive volumetric changes of the PNIPAM hydrogels among different equilibrium swelling states.57 Through incorporating photothermal agents such as gold nanorods, the shape transformations of PNIPAM-based hydrogels can further be remotely controlled by light.58 On the basis of the responsive capabilities, PNIPAM-based hydrogels have been used for preparing scaffolds with light-controlled changing topographies for the dynamic manipulation of cell focal adhesion.59 Besides, some other polymers such as poly(acrylic acid) (PAA),60 chitosan,53 poly[2-(dimethylamino) ethyl methacrylate] (PDMAEMA),61 and their derivates could also be used for preparing hydrogels, enabling controllable shape transformations among their different equilibrium swelling states in response to different pH values. These polymers would undergo protonation/deprotonation with changing pH values, therefore affecting the net charges of polymer backbones and subsequently their electrostatic repulsion/attraction to induce different swelling states of the hydrogels. However, a few studies reported reconfigurable scaffolds made of pH-responsive hydrogels because of normally a narrow range of pH values for cell culture medium and the physiological environments within the human body. Recently, a biopolymer, alginate, has also been found to show stimuli-responsive behaviours in response to the concentrations of two ions (Na+/Ca2+) abundant in physiological environments, which thereby leads to the inside-out 3D reversible shape transformations of alginate hydrogels.62 It can be envisioned that the development of hydrogel-based reconfigurable scaffolds that can be actuated under mild conditions compatible with cells and biological tissues would pave new avenues for diverse tissue engineering applications such as 4D bioprinting and engineering biomimetic environments for cell manipulation.63,64
2.3 LCEs
Liquid crystal (LC) is an inter-mediate phase, referring to a condensed fluid state with a crystal-like long-range order.65 The mobility of LC molecules brings LC materials with tuneable multiple mesophases, which can be simply realized by controlling the molecular orientations between ordered and disordered states. LCEs, with LC moieties being either a part of the polymer backbone or pendant groups to the main chain, can obtain combinational properties of tuneable orientation orders of LC moieties and rubber-like elasticity of elastomers. Based on that, LCEs have shown attractive stimuli-responsive behaviours, i.e., reversibly changing their bulk shapes and surface microstructures in response to various external stimuli.66,67LCEs, with stimuli-responsive shape-morphing properties, have demonstrated great promise for the applications of actuation,68–70 soft robotics,71–73 and controllable drug delivery.74–76 In the field of tissue engineering and regenerative medicine, LCEs have also received intensive attention first because of their biomimetic architectures.77 By engineering the orientations of LC moieties to resemble the anisotropic organizations between cells and extracellular matrix (ECM) fibrils in some biological tissues such as muscle, it could achieve directional alignments of cells on LCE-based scaffolds,78–80 Diverse cellular functions (e.g., cellular function maturation81 and stem cell differentiation82) for facilitating tissue regeneration could be subsequently modulated. Second, LCEs would also pave new avenues for preparing reconfigurable scaffolds to modulate cell behaviours and functions towards intended directions through engineering dynamic cell microenvironments. For example, a LCE-based scaffold has been prepared based on the combinations of an elastomer, poly(methyl hydrosiloxane) (PMHS), a crosslinker, 4-di(10-undecenyloxy)benzene (DUB), and a reactive LC mesogen, 4-methoxyphenyl 4-(3-butenyloxy)benzoate (MPBB).83 Through transient heating via an integrated resistive heater, the PMHS–DUB–MPBB film scaffold would undergo fully reversible, fast-response (about 1 s) and large-amplitude (beyond 30% in strain) shape changes along with a nematic-to-isotropic transition of the LC moieties, which could subsequently direct uniaxial contraction and elongation of neonatal rat ventricular myocytes via cyclic strain. By introducing photo responsive segments, e.g., an organic dye azobenzene (AZ), within the LC networks formed by the polymerization of 4-[(4-methoxyphenoxy)carbonyl] phenyl 4-[[6-(acryloyloxy)hexyl]oxy]benzoate (MCPAHB, mesogen) and 4-[[6-(acryloyloxy)hexyl]oxy]phenyl 4-[[6-(acryloyloxy)hexyl] oxy]benzoate (AHOPAHOB, crosslinker), the LCE-based scaffold (MCPAHB–AHOPAHOB–AZ) could even realize light-controlled shape transformations upon the irradiation of a green laser (514–525 nm).84 Such shape-morphing features of the LCE-based reconfigurable scaffold could assist cardiac contraction via active tension in a non-contact fashion. Despite the great promise of engineering dynamic cell microenvironments, conventional LCE-based reconfigurable scaffolds still suffer from complicated synthesis procedures and low bioactivity. Recently, a new type of natural LCE has been emerging. Cytoskeletal fibrous components, e.g., actin filaments (F-actin), which exhibit sufficient mechanical stability to sustain cellular shape and simultaneously sufficient fluidity for cellular remodelling, can show essential properties of an active nematic LC,85 holding promise for preparing next-generation living reconfigurable scaffolds enabling enhanced integrations with cells and biological tissues.
2.4 Cell-actuating materials
“Intelligent” polymers enabling volumetric changes upon the application of an external stimulus have provided excellent candidates in forming reconfigurable scaffolds. However, they still confront challenges in a relatively low energy conversion efficiency and lack the capability of close-hooped sensing/actuation in dynamic physiological environments. With the evolution for billions of years, biological systems, e.g., cells, have demonstrated various high-efficiency sensing and actuating capabilities, which have offered alternative building blocks for constructing reconfigurable scaffolds with enhanced bio-adaptive features.Cells can generate traction force in the scale of dozens of nano-Newtons for each during their geometric changes, which has proven sufficient to bend elastic micro-pillars.86 Though the traction force of one single cell is difficult to induce reconfigurations of macro-shapes for bulk materials, their collective behaviours will generate a large amplitude of mechanical force up to dozens of micro-Newtons enough to reshape flexible materials or power actuators.87,88 By employing cells derived from biological tissues enabling cyclic contraction/relaxation such as myocardium and skeletal muscles, numerous biohybrid robots that could be actuated by cell traction force have been constructed through the combinations of these living cells and synthetic flexible substrates.89–91 Accordingly, it was also reasonable to employ cell traction force to actuate the reconfigurations of tissue-engineering scaffolds. For example, through culturing NIH-3T3 cells onto a flexible parylene sheet with predesigned hinges, the traction force of the adherent cells would trigger the reconfigurations of the cell-laden sheet from a 2D planar shape to various designated 3D configurations by just adjusting the geometries of the sheet and/or patterns of the hinges.92 Such reconfigurations could be implemented along with cellular remodelling in physiological environments in a non-supervised fashion. Nevertheless, reshaping a flexible scaffold via cell traction force by simply seeding cells exhibits low controllability. Neither the degree nor the orientation of deformation could be precisely programmed. To address this challenge, advanced methodologies such as computer-aided reverse engineering design and optogenetic tools have been introduced,93,94 significantly enhancing the controllability of actuation by programming the cell traction force in controllable spatiotemporal manners.
Notably, cells as one type of living “component” further endow the cell-actuating materials with self-adaptable potentials, corresponding to the ability of feedback control over cellular behaviours and functions during the reconfiguration process.95 It has been well known that the cellular shape affects a variety of cellular behaviours and functions such as adhesion, migration, proliferation, differentiation, and apoptosis via mechanotransduction.96,97 When reshaping a cell-laden scaffold, the mechanical forces generated accompanying with the scaffold deformation process would impose on cellular shapes and subsequently induce feedback modulation effects on the synthetic scaffold in return along with cellular remodelling.98 Such close-loop control of cellular behaviours and functions resemble the cell–ECM interactions during tissue morphogenesis and regeneration, which would mediate the reorganization of cellular cytoskeletons and the following a cascade of downstream gene transcriptions via biomechanical signalling pathways.99 Self-adaptable cell-actuating materials have been developed accordingly through incorporating mesenchymal stem cells (MSCs) within a nanofibrous norbornene-modified hyaluronic acid (NorHA) hydrogel scaffold.100 Due to cell contraction, the attached MSCs within the scaffold could cause the re-assembly/re-arrangements of hydrogel nanofibers and subsequent changes of scaffold macro-shapes. In return, the changes of scaffold macro-shapes would cause re-programming of stem cell fate. In addition to the biomechano-mediating modulation, cell behaviours and functions could also be programmed by other biophysical cues, e.g., electrical stimulations. Recently, flexible piezoelectric materials that can generate electrical signals under cell traction force have attracted broad attention in the field of tissue engineering and regenerative medicine, offering new opportunities for forming cell-actuating materials capable of feedback cell modulation via the electrical stimulation actuated by cell traction force.101–103 Through the combinations of appropriate cells and the “intelligent” polymers introduced above, it will be likely to prepare reconfigurable scaffolds promising for specific tissue engineering applications of interest by provoking self-adaptable interactivity between materials and cells. Moreover, through integrating rationally designed cell-actuating materials and recently emerging bioengineering technologies, such as synthetic biology,104 it will be possible to further develop “living” tissue engineering scaffolds with programmable cellular behaviours and functions in the future that can better accommodate for complicated and dynamic physiological environments.
2.4 Design rationale for reconfigurable scaffolds using different materials
The functions of scaffolds are highly dependent on the properties of constituting materials. When seeking to construct reconfigurable scaffolds suitable for specific tissue engineering applications, it would be greatly important to choose and use materials with appropriate responsive properties. For the scaffolds requiring reliable one-way reconfiguration, it would be favourable to simply trigger their spontaneous shape transformations via internal signals within human bodies, such as body temperature, bio-fluids, or enzymes. Such scaffolds were designed to be deformed into target macro-shapes/micro-structures in pre-determined routes once implanted into bodies, without the needs of any additional facilities. Thermoresponsive SMPs are representative examples that have been commonly used in forming reconfigurable scaffolds whose shapes could undergo spontaneous transformations upon being placed in physiological environments at body temperature.31–37 Some SMPs and hydrogels whose shapes could be deformed upon hydration have also provided candidates for fabricating one-way reconfigurable scaffolds to be triggered by bio-fluids abundant within human bodies.44,54 Besides, enzyme-responsive SMPs would be promising for forming reconfigurable scaffolds enabling spontaneous shape transformations in specific pathophysiological environments within a relatively long time period (∼7 days).43 In addition, cell-actuated materials could also be used for preparing reconfigurable scaffolds with the potential of spontaneous shape transformations within bodies actuated by cell traction force,92 whereas their reconfigurations would be less controllable and reliable than those formed by using SMPs and hydrogels.As for constructing reconfigurable scaffolds requiring alteration in their macro-shapes/micro-structures even after implantation, it should be necessary to introduce materials whose shapes could be controlled upon applications of specific external fields. The reconfigurations of such scaffolds would be expected to be actuated or modulated by specific external stimuli in homeostatic physiological environments with the assistances of appropriate stimuli-responsive materials. To obtain high-efficiency actuation within bodies, the materials used for forming the scaffolds should be able to respond to the external stimuli that can effectively and safely penetrate biological tissues, including NIR light, magnetic field, and ultrasound. SMPs, hydrogels, and LCEs all could obtain field-controlled responsiveness through the incorporation of field-responsive agents, which could convert the energies from the external fields to some specific stimuli, usually in the form of heat, capable of actuating the shape transformations of composite materials. Some hydrogel- and LCE-based composite materials could even realize reversible and/or dynamic reconfigurations controlled by specific external stimuli,51,59,84 offering the possibilities of constructing reconfigurable scaffolds capable of adjusting their macro-shapes/micro-structures on demand for meeting the requirements of dynamic bio-adaptions in changing physiological environments.
3. Adaptive tissue regeneration assisted by scaffolds with reconfigurable macro-shapes
Emerging “intelligent” polymers and “living” cell-actuated materials have laid the foundations for developing reconfigurable scaffolds whose macro-shapes and/or micro-structures could be modulated on demand, subsequently bringing exciting promise in the field of tissue engineering and regenerative medicine from different aspects. First, the scaffold with reconfigurable macro-shapes has offered an innovative alternative for intervention therapies via minimally invasive injection.105 Specifically, using an elastic and biodegradable SMP poly[octamethylene maleate (anhydride) citrate] (POMAC), a cardiomyocyte-laden microfabricated scaffold has been prepared, capable of altering their macro-shapes via shape recovery triggered by body temperature. Thanks to their reconfigurable macro-shapes, the cardiomyocyte-laden scaffold could be first reconfigured into a small-diameter tube suitable for injection into targeted body sites using a syringe in a minimally invasive manner via a small orifice with a diameter down to 1 mm. Upon minimally invasive injection to the epicardium, the shape of the scaffold would recover to its permanent shape (a planar patch with a size of 1 cm × 1 cm) for treating myocardial infarction (Fig. 3a). Owing to the mild conditions to trigger shape recovery, the viability and functions of the incorporated cardiomyocytes were well preserved during the reconfiguration process. Benefitting from the well-preserved cell viability and functions, as well as a less risky intervention approach of minimally invasive injection, this shape-memory cardiomyocyte-laden scaffold with reconfigurable macro-shapes has significantly improved cardiac functions in both small-animal (rat) and large-animal (porcine) myocardial infarction models.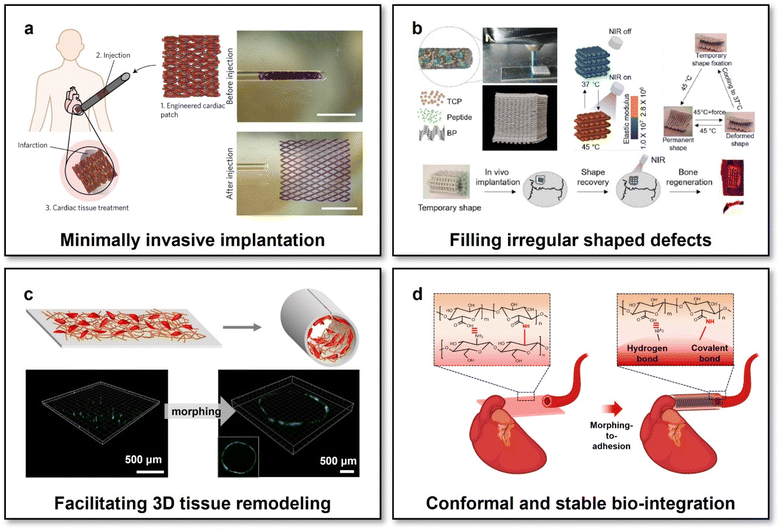 | ||
| Fig. 3 Scaffolds with reconfigurable macro-shapes for adaptive tissue regeneration. (a) Shape-memory scaffolds with incorporated cardiomyocytes for cardiac tissue treatment via minimally invasive implantation. This figure has been reproduced from ref. 105 with permission from Spring Nature; copyright: 2017. (b) NIR-controlled reconfigurable bone tissue engineering scaffolds for filling irregular shaped cranial defects. This figure has been reproduced from ref. 106 with permission from IOP Publishing; copyright: 2020. (c) Shape-memory bilayer scaffolds facilitating facile 3D remodelling of endothelium tissue. This figure has been reproduced from ref. 107 with permission from the WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim; copyright: 2018. (d) Reconfigurable scaffolds based on multifunctional hydrogels achieving conformal and stable interfacing with high-curvature biological tissues via a morphing-to-adhesion way. This figure has been reproduced from ref. 112 with permission from the American Chemical Society; copyright: 2022. | ||
Second, the reconfigurable macro-shapes of scaffolds have brought them the conveniences of fitting irregular shaped defects of biological tissues, which should be particularly difficult, if possible, for conventional scaffolds with pre-defined geometries. For treating irregular shaped cranial bone defects, a nanocomposite scaffold with NIR-controlled reconfigurable capabilities has been developed through the integrations of shape-memory PDLLA-co-TMC, photothermal black phosphorus nanosheets, osteoconductive β-tricalcium phosphate, and osteoinductive peptides.106 The scaffold could be initially compressed into a small-size temporary shape for minimally invasive injection. Upon implanted into bone defect sites and the accompanying NIR irradiation for heating up the scaffold to reach the Tg of PDLLA-co-TMC (45 °C), the scaffold would be softened and concurrently self-expanded its shape to fill an irregular shaped bone defect via shape recovery (Fig. 3b). The low elastic modulus of the scaffold during the shape filling process made it feasible to perfectly match the zigzag borders of irregular shaped cranial bone defects in a rat model yet cause negligible additional stress. The NIR-irradiated reconfiguration process has also been proven safe for preserving the bioactivities of incorporated osteoconductive and osteoinductive agents. Moreover, NIR irradiation could trigger the pulsed release of osteoinductive agents, imparting the reconfigurable scaffolds with coupling multi-functions for adaptive bone integration.
Third, scaffolds with reconfigurable macro-shapes have offered the possibility of adapting to 3D shapes of biological tissues in an actively compliant fashion, which would significantly facilitate 3D tissue-scaffold interplays at the macroscopic biointerfaces. To enhance the interplays between endothelial cells and tissue engineering vascular grafts for adaptive vascular regeneration, Zhao et al. developed a reconfigurable scaffold consisting of a shape-memory actuating layer and an electrospun nanofibrous functional layer made of PDLLA-co-TMC and PCL/gelatin methacrylate (GelMA) blends, respectively.107 The electrospun PCL/GelMA nanofibrous functional layer possessed desirable mechanical properties and bioactivity for supporting the focal adhesion and intercellular junction of endothelial cells.108 The scaffold, possessing reconfigurable capabilities and desirable microenvironments for endothelial cells, could therefore achieve high-efficiency incorporations of endothelial cells at its initially planar shapes and facile 3D endothelium remodelling after the body-temperature-triggered shape recovery of the scaffold to its permanent 3D tubular shapes (Fig. 3c). Through adjusting the permanent tubular shapes of the 3D reconfigurable scaffolds via different template-assisted fabrication, it could make the scaffolds compliable with the surfaces of blood vessels of different diameters, ranging from 3 to 7 mm for meeting the respective therapeutic requirements of small-diameter and large-diameter blood vessels in the proof-of-concept studies. In addition to conformal interfacing with 3D-shaped biological tissues, the scaffolds with programmable reconfigurable macro-shapes would further be available for fitting the changing shapes of growing tissues. By using a viscoplastic material, referring to a type of material showing strain-rate-dependent mechanical properties (the liquid-like flowable state at low strain rates and the stiff state at high strain rates), to form the flexible substrate of bioelectronics, the macro-shapes of bioelectronics could be reconfigured and maintained conformal to gradually growing sciatic nerves without any mechanical constraints to the tissues.109 Thanks to the viscoplasticity of the flexible substrate, nearly zero stress was generated for the bio-integrated bioelectronics during the low-rate strain of the gradual tissue growing process. This study also offered the inspiration of developing scaffolds with self-adaptable reconfigurable macro-shapes for adapting to growing 3D-shaped biological tissues.
Besides conformal interfacing, one additional factor affecting tissue-scaffold interplays should be their interfacial stability. In the field of bioelectronics, it has been demonstrated that mechanically stable biointerfaces formed assisted by a biocompatible bio-adhesive functional layer were effective to significantly enhance the reliability of implanted bioelectronics.110,111 Recently, we have developed a multifunctional tissue engineering scaffold with coupled reconfigurable and bio-adhesive capabilities.112 The scaffold was formed based on two types of naturally derived polysaccharide hydrogels, i.e., chitosan and alginate. The two polysaccharide hydrogels at the two scaffold layers exhibited distinct different swelling behaviours upon immersion in water or bio-fluids, therefore endowing the scaffolds with swelling-triggered reconfigurable capabilities. Through functionalizing alginate with N-hydroxysuccinimide ester (NHS), the activated carboxylic acid groups of NHS-functionalized alginate could form instant binding with the terminal primary amine groups, in either chitosan hydrogels or biological tissues, via hydrogen and covalent bonds. Accordingly, the morphing-to-adhesion scaffold could not only be self-sealed after the programmable reconfigurations from an initially 2D planar sheet to 3D tubes with tuneable diameters but also formed conformal and mechanically stable interfaces with diverse tubular tissues in a wide range of surface curvatures (2.8 × 102–1.3 × 103 m−1) (Fig. 3d). Such a scaffold with superior adaptability over the scaffolds with just reconfigurable functions in both interfacial conformality and stability paves new avenues for facilitating tissue–scaffold interplays.
Despite the various promises of the scaffolds with reconfigurable macro-shapes in the field of tissue engineering and regenerative medicine, it is somewhat interesting to note that most of the established scaffolds with reconfigurable macro-shapes could only undergo a one-way shape transformation. It does not mean that just one-way reconfigurations of macro-shapes are “ideal” enough for adaptive tissue regeneration. To investigate cellular responses under reversible mechanical stimulations, a NIR-controlled PNIPAM-based hydrogel has been developed,113 realizing rapidly reversible reconfigurations of its macro-shapes for programming stem cells in mechanically and geometrically dynamic microenvironments. It has been revealed that the reversible changes of scaffold macro-shapes could determine stem cell fates in different ways compared with their non-reversible counterparts. In the future, it can be expected to develop reconfigurable scaffolds capable of reversibly and dynamically adjusting their macro-shapes to direct the cellular behaviours and functions in a designed spatiotemporal manner for enhanced adaptive tissue regeneration.
4. Adaptive tissue regeneration assisted by scaffolds with reconfigurable micro-structures
Engineering scaffolds with defined micro-structures has emerged as a popular method for directing the cellular morphology and subsequently a cascade of gene expressions via activating specific mechanotransduction signalling pathways by topographical cues.114 Topography-mediated effects have been proven vital to affect cell traction force orientation, and then determining a series of cellular functions essential for tissue regeneration, for example, cellular focal adhesion,115 cell maturation,116 stem cell differentiation,117 and macrophage polarization.118 However, tissue regeneration usually requires dynamically programming cellular functions at different stages, which would be challengeable for scaffolds with pre-defined static micro-structures. It should be therefore of great value to develop scaffolds with reconfigurable micro-structures and to explore whether cellular functions could be programmed on demand by dynamic topographical cues when altering the micro-structures of scaffolds.Using a stimuli-responsive material with dynamic host–guest interactions, a scaffold with reconfigurable micro-structures has been fabricated by 3D laser lithography.119 Upon application or removal of soluble competitive guests, the micro-structures of the scaffold could be reversibly changed in physiological environments, exerting programmable mechanical stretching on individual cells to provoke the reorganizations of their actin cytoskeleton (Fig. 4a). In addition to the morphology of a single cell, the cellular focal adhesion, highly critical for cell mechanobiology, could also be modulated in vitro through controlling the reconfigurations of scaffold micro-structures.59 A nanofabricated scaffold for the dynamic manipulation of cellular focal adhesion was formed based on a thermoresponsive PNIPAM hydrogel and photothermal gold nanorods. Upon application of NIR irradiation, initially upstanding sub-micron posts would be actuated and bent due to the shrinkage of the hydrogel, thereby manipulating the focal adhesion of attached cells in a sub-cellular high resolution (Fig. 4b).
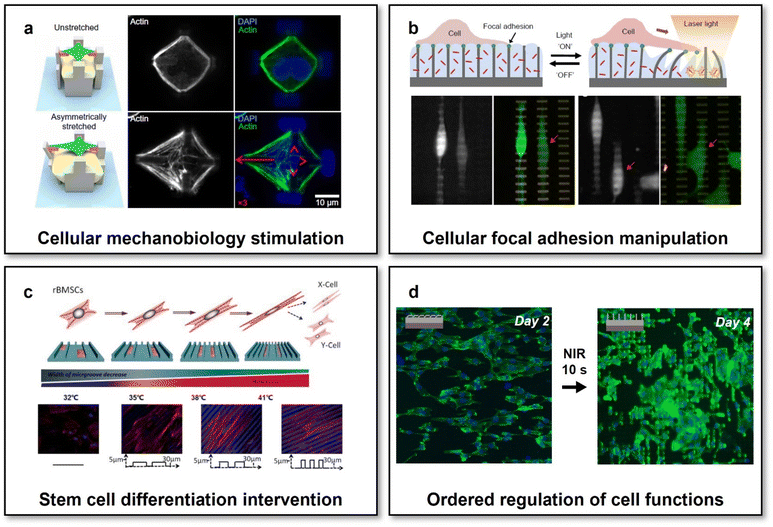 | ||
| Fig. 4 Scaffolds with reconfigurable micro-structures for adaptive tissue regeneration. (a) Reversible mechanical stimulation of one single cell by the reconfigurable micro-structures of a 3D scaffold. This figure has been reproduced from ref. 119 with permission from American Association for the Advancement of Science; copyright: 2020. (b) NIR-controlled reconfigurable microstructures for manipulating cellular focal adhesion in vitro. This figure has been reproduced from ref. 59 with permission from Spring Nature; copyright: 2017. (c) Shape-memory scaffolds for programming the differentiation of stem cells. This figure has been reproduced from ref. 31 with permission from the WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim; copyright: 2014. (d) Reconfigurable scaffolds with NIR-controlled microstructures for the state-specific manipulation of vascular endothelial cells. This figure has been reproduced from ref. 130 with permission from Oxford Academic; copyright: 2020. | ||
Though the feasibility of altering the morphology and focal adhesion of individual cell through programming the reconfigurations of scaffold micro-structures has been widely investigated and the potential outcomes of evoking the cascade of gene expression via mechanotransduction could be predicted, it is still difficult to assume the similar effects on the collective behaviours and functions of the amounts of cells involving complicated cell–cell interactions.120,121 By using SMPs and micro-fabrication/micro-embossing methods, a variety of scaffolds with temperature-actuated reconfigurable microstructures, enabling reconfigurations either from micro-/nano-topographies to flat or in a reverse way, have been developed and employed to investigate the mechanotransduction of dynamic topographical cues on collective cellular behaviours and functions.28,122–125 For different cell models such as NIH-3T3 fibroblast cell lines,28,122 primary mouse embryonic fibroblasts,122 and human-derived stem cells,123,124 the reconfigurations of scaffold micro-structures could always cause the switches of cellular shapes and reorientations of cytoskeletons for a large number of cells. Therefore, it was reasonable to hypothesize that collective cellular functions could also be programmed by the reconfigurable microstructures of the scaffolds, which has then been confirmed by controllable collective migration,126 contraction,127 and differentiation31 on shape-memory scaffolds with reconfigurable micro-structures. Specifically, using shape-memory PU, electrospun scaffolds whose nanofiber arrangements could be programmed in unaligned-to-aligned or aligned-to-unaligned manners. The rearrangements of nanofiber orientations via the shape recovery of PU actuated by altering environmental temperatures (from 30 to 37 °C) would cause changes in the cellular morphology and thereby the differential polarized motility of cells.126 On a nanofabricated shape-memory PCL scaffold with dynamic anisotropic nanogrooves, primary cardiac muscle cell functions such as cytoskeletal developments, focal adhesion alignment, and collective contraction orientation in cell monolayers could also be regulated through controlling the reconfigurations of its micro-structures (altering the orientation of nanogrooves by 90°) via shape recovery by tuning the environmental temperature from 32 to 37 °C.127 As for rat bone marrow-derived mesenchymal stem cells (rBMSCs), the reconfigurations of micro-structures realized by a PCL-based SMP scaffold have been proven visible to promote the myogenic differentiation of rBMSCs (Fig. 4c).31 When increasing the environmental temperature from 32 to 41 °C, the engineered surface micro-structures of the scaffold could be tuned gradually via shape recovery from temporary smooth states to permanent microgroove arrays. Such reconfigurations of micro-structures would exert dynamic mechanical forces on the attached rBMSCs and thereby mimicking the functions of the native ECM in the muscular developmental and regenerative processes.128 Through gene expression and western blot analyses, it was revealed that such biomechanically dynamic microenvironments could significantly enhance the directional differentiation of rBMSCs to a myogenic phenotype even without the additions of any induction factors.
Despite demonstrated the effectiveness of the above-introduced SMP-based scaffolds for manipulating collective cellular functions through controlling the reconfigurations of their micro-structures, there has been two factors remaining that might limit their practical translational applications, i.e., (1) the approach to actuate the reconfigurations (usually actuated by altering the overall environmental temperature) and (2) the match between dynamically controlled cellular functions and specific requirements of ordered cell regulation in specific tissue regeneration processes. Though cell viability has been reported to be negligibly affected by the changes in the environmental temperature during the reconfigurations of micro-structures for the above-mentioned SMP-based scaffolds, it would be still preferred to develop a reconfigurable scaffold whose micro-structures could be programmed in a remote-controlled fashion, particularly for in vivo applications. Through embedding magnetic nanoparticles within a microfabricated elastic scaffold, the micro-structures of the scaffold could be maintained in stable physiological environments and actively actuated by a gradient magnetic field in a remote-controlled manner. It has been proven effective to prevent microorganism attachment and even remove the established biofilm by using such a scaffold, enabling magnet-actuated high-frequency and reversible reconfigurations of its microstructures.129 Our research group has recently developed a scaffold with reconfigurable microstructures that could be remotely actuated by NIR light at the physiological temperature.130 The scaffold was designed to consist of two layers: one microfabricated actuating upper layer made of a poly(L-lactide-co-D,L-lactide) (PLLADLLA) SMP and one photothermal bottom layer containing gold nanorods. The surface micro-structures of the scaffold could be shaped into temporary anisotropic topography with oriented bent micropillar arrays, which could be well preserved when immersed in a 37 °C cell culture medium due to the high Tg of PLLADLLA (∼53 °C). Upon exposure to biosafe NIR irradiation (with a power density of 5 W cm−2) for just 10 s, the heat generated in the bottom photothermal layer would trigger the shape recovery of PLLADLLA and then the reconfigurations of the micro-structures at the upper layer of the scaffold to be permanent upstanding micropillar arrays. Owing to the relatively short time of NIR irradiation and thermal diffusion gradient arising from the bilayer scaffold design, the NIR-triggered actuation process was found to show negligible influences on the viability and functions of cells grown on the upper actuating layer. Moreover, we observed the NIR-controlled changes in the surface micro-structures of the scaffold could effectively induce the transitions of cell morphology and cytoskeleton (F-actin) development from initially anisotropic to later isotropic states (Fig. 4d). Under the mechanotransduction effects resulting from the shifted cellular morphology and F-actin arrangements, the scaffold with NIR-programmable reconfigurable micro-structures achieved enhanced migration of endothelial cells at the initially anisotropic state and improved focal adhesion and intercellular junction of endothelial cells in the monolayer at the later isotropic state. Such ordered regulation of endothelial cells would meet the dynamic requirements of endothelium remodelling, i.e., first recruiting endothelial cells from surroundings and then directing the formation of a stable confluent endothelial cell monolayer, enabling such reconfigurable scaffolds with specific implications to adaptive vascular regeneration. This study would also inspire to develop reconfigurable scaffolds capable of manipulating the behaviours and functions of other cell types on demand for different tissue engineering and regenerative medicine applications.
5. Summary and outlook
Over the past decade, reconfigurable scaffolds, showing obvious superiorities in adaptability over conventional static non-responsive scaffolds by offering an additional temporal dimension for tuning their properties and features, have been rapidly emerging and promoted remarkable advances in tissue engineering and regenerative medicine. With the assistance of “intelligent” polymers, it has been possible to program the macro-shapes of scaffolds compliable with the 3D shapes of biological tissues for optimizing tissue-scaffold interplays at bio-interfaces. Meanwhile, the dynamic manipulation of different cellular functions has also been realized via mechanotransduction for meeting the requirements of ordered cell regulation within specific tissue developmental and regenerative processes through programming the micro-structures of scaffolds to attain dynamic cell-scaffold interplays. Moreover, some efforts have been recently made to construct multi-scale responsive scaffolds enabling combinational reconfigurations in both macro-shapes and micro-structures. Not only oriented cellular alignments could further be stimulated by the combinational guidance of reconfigurable macro-shapes and micro-structures,131 but also conformal integration with 3D biological tissues and directional programming of cellular fates could be simultaneously achieved by the multi-scale responsive scaffolds.132 We are witnessing emerging reconfigurable scaffolds with upgrading adaptability “re-shaping” the field of tissue engineering and regenerative medicine. Additionally, reconfigurable scaffolds further hold promise to facilitate some emerging biotechnologies in tissue engineering and regenerative medicine, such as organoid engineering.133 As far as functional organoids were conventionally built on the Matrigel possessing inherent drawbacks in batch-to-batch variability and immunogenicity, reconfigurable scaffolds might provide promising alternatives for programming stem cells to functional organoids via offering adaptable in vitro microenvironments with well-defined compositions and functions for reliably assisting morphogenesis.134,135Despite the great promise in adaptive tissue regeneration, reconfigurable scaffolds, in the endeavour of perfect bio-adaptions to complicated and dynamic pathophysiological environments, still confront challenges, such as spatiotemporally controllable modulations of cell behaviours and functions as compared with the native ECM. In the spatial scale, it remains challenging to control the reconfigurations of scaffolds with a high resolution down to the nanoscale, which would be important for precise manipulation of cellular behaviours and functions via specific biomolecule-mediated approaches, for example tuning integrin binding to modulate the mechanobiology of cells.136 In the temporal scale, on one hand, for either macro-shapes or micro-structures of the scaffolds, realizing transient and cyclic (>1 Hz) reconfigurations with excellent mechanical robustness which resembles some natural microenvironments such as a beating heart is still difficult. On another hand, it is also extremely hard to realize ultra-slow reconfigurations accommodating for the developmental or growing processes of biological tissues within an actuating period from several days to even several months. It will be expected that innovative multidisciplinary efforts, particularly innovations in materials sciences,137,138 nanoscience,139 and engineering technologies,140 should be made for addressing these challenges of reconfigurable scaffolds. In addition, through coupling multiple biophysical cues and/or introducing sensing capabilities to enhance the environmental interactivity of scaffolds,16,141–144 it can even be envisioned that reconfigurable scaffolds will further obtain superior adaptability, even the “intelligence” of closed-loop control, for matching the extreme physiological complexities among different body tissues.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge the financial support provided by the National Key R&D Program of China (2017YFA0701303), the National Natural Science Foundation of China (52173148, 52022102, 51903245, and 52003287), the National Natural Science Foundation of China/RGC Joint Research Scheme (52261160380), the Youth Innovation Promotion Association of CAS (2022368, 2019353), the Guangdong Regional Joint Fund-Key Project (2021B1515120076), the Natural Science Foundation of Guangdong Province (2114050001067), and the Shenzhen Science and Technology Innovation Committee (JCYJ20220818101800001).References
- R. Langer and J. P. Vacanti, Science, 1993, 260, 920–926 CrossRef CAS PubMed.
- E. S. Place, J. H. George, C. K. Williams and M. M. Stevens, Chem. Soc. Rev., 2009, 38, 1139–1151 RSC.
- M. P. Lutolf and J. A. Hubbell, Nat. Biotechnol., 2005, 23, 47–55 CrossRef CAS PubMed.
- S. J. Hollister, Nat. Mater., 2005, 4, 518–524 CrossRef CAS PubMed.
- J. Gao, X. Yu, X. Wang, Y. He and J. Ding, Engineering, 2022, 13, 31–45 CrossRef CAS.
- L. L. Hench and J. M. Polak, Science, 2002, 295, 1014–1017 CrossRef CAS PubMed.
- A. K. Gaharwar, I. Singh and A. Khademhosseini, Nat. Rev. Mater., 2020, 5, 686–705 CrossRef CAS.
- Q. Zhao and X. Du, Smart Mater. Med., 2022, 3, 37–40 CrossRef.
- S. Hauser, F. Jung and J. Pietzsch, Trends Biotechnol., 2017, 35, 265–277 CrossRef CAS PubMed.
- Q. Zhao, C. Li, H. C. Shum and X. Du, Lab Chip, 2020, 20, 4321–4341 RSC.
- S. E. Iismaa, X. Kaidonis, A. M. Nicks, N. Bogush, K. Kikuchi, N. Naqvi, R. P. Harvey, A. Husain and R. M. Graham, npj Regener. Med., 2018, 3, 6 CrossRef PubMed.
- E. S. Place, N. D. Evans and M. M. Stevens, Nat. Mater., 2009, 8, 457–470 CrossRef CAS PubMed.
- A. Nojoomi, J. Jeon and K. Yum, Nat. Commun., 2021, 12, 603 CrossRef CAS PubMed.
- P. Cai, B. Hu, W. R. Leow, X. Wang, X. J. Loh, Y. L. Wu and X. Chen, Adv. Mater., 2018, 30, 1800572 CrossRef PubMed.
- G. Brusatin, T. Panciera, A. Gandin, A. Citron and S. Piccolo, Nat. Mater., 2018, 17, 1063–1075 CrossRef CAS PubMed.
- X. Wan, Z. Liu and L. Li, Adv. Funct. Mater., 2021, 31, 2010626 CrossRef CAS.
- M. D. Hager, S. Bode, C. Weber and U. S. Schubert, Prog. Polym. Sci., 2015, 49–50, 3–33 CrossRef CAS.
- H. Ramaraju, R. E. Akman, D. L. Safranski and S. J. Hollister, Adv. Funct. Mater., 2020, 30, 2002014 CrossRef CAS.
- A. Melocchi, M. Uboldi, M. Cerea, A. Foppoli, A. Maroni, S. Moutaharrik, L. Palugan, L. Zema and A. Gazzaniga, Adv. Drug Delivery Rev., 2021, 173, 216–237 CrossRef CAS PubMed.
- A. Lendlein and R. Langer, Science, 2002, 296, 1673–1676 CrossRef PubMed.
- Y. Deng, B. Yang, F. Zhang, Y. Liu, J. Sun, S. Zhang, Y. Zhao, H. Yuan and J. Leng, Biomaterials, 2022, 291, 121886 CrossRef CAS PubMed.
- Y. C. Zhang, N. Zheng, Y. Cao, F. L. Wang, P. Wang, Y. J. Ma, B. W. Lu, G. H. Hou, Z. Z. Fang, Z. W. Liang, M. K. Yue, Y. Li, Y. Chen, J. Fu, J. Wu, T. Xie and X. Feng, Sci. Adv., 2019, 5, eaaw1066 CrossRef CAS PubMed.
- J. Wang, Q. Zhao, Y. Wang, Q. Zeng, T. Wu and X. Du, Adv. Mater. Technol., 2019, 4, 1900566 CrossRef CAS.
- G. I. Peterson, A. V. Dobrynin and M. L. Becker, Adv. Healthcare Mater., 2017, 6, 1700694 CrossRef PubMed.
- M. A. Woodruff and D. W. Hutmacher, Prog. Polym. Sci., 2010, 35, 1217–1256 CrossRef CAS.
- Z. G. Wei, R. Sandstrom and S. Miyazaki, J. Mater. Sci., 1998, 33, 3743–3762 CrossRef CAS.
- L. S. Nair and C. T. Laurencin, Prog. Polym. Sci., 2007, 32, 762–798 CrossRef CAS.
- M. Ebara, K. Uto, N. Idota, J. M. Hoffman and T. Aoyagi, Adv. Mater., 2012, 24, 273–278 CrossRef CAS PubMed.
- M. Saatchi, M. Behl, U. Nochel and A. Lendlein, Macromol. Rapid Commun., 2015, 36, 880–884 CrossRef CAS PubMed.
- J. Wang, Q. Zhao, H. Cui, Y. Wang, H. Chen and X. Du, J. Mater. Chem. A, 2018, 6, 24748–24755 RSC.
- T. Gong, K. Zhao, G. Yang, J. Li, H. Chen, Y. Chen and S. Zhou, Adv. Healthcare Mater., 2014, 3, 1608–1619 CrossRef CAS PubMed.
- M. Bao, X. X. Lou, Q. H. Zhou, W. Dong, H. H. Yuan and Y. Z. Zhang, ACS Appl. Mater. Interfaces, 2014, 6, 2611–2621 CrossRef CAS PubMed.
- L. Han, Y. Wang, L. Wu, Z. Wu, Y. He, H. Mao and Z. Gu, ACS Biomater. Sci. Eng., 2023, 9, 520–530 CrossRef CAS PubMed.
- T. Kivijarvi, G. Oyvind, M. A. Yassin, S. Jain, S. Yamada, A. Morales-Lopez, K. Mustafa and A. Finne-Wistrand, Mater. Today Bio, 2022, 17, 100483 CrossRef PubMed.
- X. Chen, S. Y. Han, W. H. Wu, Z. H. Wu, Y. Yuan, J. Wu and C. Liu, Small, 2022, 18, 2106824 CrossRef CAS PubMed.
- M. R. Pfau, K. G. McKinzey, A. A. Roth, L. M. Graul, D. J. Maitland and M. A. Grunlan, J. Mater. Chem. B, 2021, 9, 3826–3837 RSC.
- L. N. Woodard, K. T. Kmetz, A. A. Roth, V. M. Page and M. A. Grunlan, Biomacromolecules, 2017, 18, 4075–4083 CrossRef CAS PubMed.
- Y. J. Wang, U. S. Jeng and S. H. Hsu, ACS Biomater. Sci. Eng., 2018, 4, 1397–1406 CrossRef CAS PubMed.
- M. Bao, Q. H. Zhou, W. Dong, X. X. Lou and Y. Z. Zhang, Biomacromolecules, 2013, 14, 1971–1979 CrossRef CAS PubMed.
- T. Gong, W. B. Li, H. M. Chen, L. Wang, S. J. Shao and S. B. Zhou, Acta Biomater., 2012, 8, 1248–1259 CrossRef CAS PubMed.
- Y. C. Zhang, C. R. Li, W. Zhang, J. J. Deng, Y. Y. Nie, X. F. Du, L. Qin and Y. X. Lai, Bioact. Mater., 2022, 16, 218–231 CrossRef CAS PubMed.
- Y. Li, H. M. Chen, D. Liu, W. X. Wang, Y. Liu and S. B. Zhou, ACS Appl. Mater. Interfaces, 2015, 7, 12988–12999 CrossRef CAS PubMed.
- S. L. Buffington, J. E. Paul, M. M. Ali, M. M. Macios, P. T. Mather and J. H. Henderson, Acta Biomater., 2019, 84, 88–97 CrossRef CAS PubMed.
- L. Cera, G. M. Gonzalez, Q. H. Liu, S. Choi, C. O. Chantre, J. Lee, R. Gabardi, M. C. Choi, K. Shin and K. K. Parker, Nat. Mater., 2021, 20, 242–249 CrossRef CAS PubMed.
- T. Xie, Nature, 2010, 464, 267–270 CrossRef CAS PubMed.
- J. J. Li, W. R. Rodgers and T. Xie, Polymer, 2011, 52, 5320–5325 CrossRef CAS.
- T. Chung, A. Rorno-Uribe and P. T. Mather, Macromolecules, 2008, 41, 184–192 CrossRef CAS.
- S. Correa, A. K. Grosskopf, H. Lopez Hernandez, D. Chan, A. C. Yu, L. M. Stapleton and E. A. Appel, Chem. Rev., 2021, 121, 11385–11457 CrossRef CAS PubMed.
- J. Z. Lou and D. J. Mooney, Nat. Rev. Chem., 2022, 6, 726–744 CrossRef CAS.
- S. J. Jeon, A. W. Hauser and R. C. Hayward, Acc. Chem. Res., 2017, 50, 161–169 CrossRef CAS PubMed.
- Z. J. Wang, C. N. Zhu, W. Hong, Z. L. Wu and Q. Zheng, Sci. Adv., 2017, 3, e1700348 CrossRef PubMed.
- A. S. Gladman, E. A. Matsumoto, R. G. Nuzzo, L. Mahadevan and J. A. Lewis, Nat. Mater., 2016, 15, 413–418 CrossRef PubMed.
- H. Hu, C. Huang, M. Galluzzi, Q. Ye, R. Xiao, X. Yu and X. Du, Research, 2021, 2021, 9786128 CrossRef PubMed.
- A. Kirillova, R. Maxson, G. Stoychev, C. T. Gomillion and L. Ionov, Adv. Mater., 2017, 29, 1703443 CrossRef PubMed.
- Y. Luo, X. Lin, B. Chen and X. Wei, Biofabrication, 2019, 11, 045019 CrossRef CAS PubMed.
- M. A. Mohamed, A. Fallahi, A. M. A. El-Sokkary, S. Salehi, M. A. Akl, A. Jafari, A. Tamayol, H. Fenniri, A. Khademhosseini, S. T. Andreadis and C. Cheng, Prog. Polym. Sci., 2019, 98, 101147 CrossRef CAS PubMed.
- M. A. Haq, Y. L. Su and D. J. Wang, Mater. Sci. Eng., C, 2017, 70, 842–855 CrossRef PubMed.
- X. Du, H. Cui, B. Sun, J. Wang, Q. Zhao, K. Xia, T. Wu and M. S. Humayun, Adv. Mater. Technol., 2017, 2, 1700120 CrossRef.
- A. Sutton, T. Shirman, J. V. Timonen, G. T. England, P. Kim, M. Kolle, T. Ferrante, L. D. Zarzar, E. Strong and J. Aizenberg, Nat. Commun., 2017, 8, 14700 CrossRef PubMed.
- O. E. Philippova, D. Hourdet, R. Audebert and A. R. Khokhlov, Macromolecules, 1997, 30, 8278–8285 CrossRef CAS.
- A. Zengin, G. Karakose and T. Caykara, Eur. Polym. J., 2013, 49, 3350–3358 CrossRef CAS.
- X. Du, H. Cui, Q. Zhao, J. Wang, H. Chen and Y. Wang, Research, 2019, 2019, 6398296 CAS.
- R. Y. Tam, L. J. Smith and M. S. Shoichet, Acc. Chem. Res., 2017, 50, 703–713 CrossRef CAS PubMed.
- S. Miao, N. Castro, M. Nowicki, L. Xia, H. Cui, X. Zhou, W. Zhu, S. J. Lee, K. Sarkar, G. Vozzi, Y. Tabata, J. Fisher and L. G. Zhang, Mater. Today, 2017, 20, 577–591 CrossRef CAS PubMed.
- C. Tschierske, Chem. Soc. Rev., 2007, 36, 1930–1970 RSC.
- T. J. White and D. J. Broer, Nat. Mater., 2015, 14, 1087–1098 CrossRef CAS PubMed.
- G. Babakhanova, T. Turiv, Y. B. Guo, M. Hendrikx, Q. H. Wei, A. P. H. J. Schenning, D. J. Broer and O. D. Lavrentovich, Nat. Commun., 2018, 9, 456 CrossRef PubMed.
- A. M. Rather, Y. Xu, Y. Chang, R. L. Dupont, A. Borbora, U. I. Kara, J. C. Fang, R. Mamtani, M. Zhang, Y. X. Yao, S. Adera, X. P. Bao, U. Manna and X. G. Wang, Adv. Mater., 2022, 34, 2110085 CrossRef CAS PubMed.
- J. A. Lv, Y. Liu, J. Wei, E. Chen, L. Qin and Y. Yu, Nature, 2016, 537, 179–184 CrossRef CAS PubMed.
- T. Guin, M. J. Settle, B. A. Kowalski, A. D. Auguste, R. V. Beblo, G. W. Reich and T. J. White, Nat. Commun., 2018, 9, 2531 CrossRef PubMed.
- H. R. Liu, H. M. Tian, X. M. Li, X. L. Chen, K. Zhang, H. Y. Shi, C. H. Wang and J. Y. Shao, Sci. Adv., 2022, 8, eabn5722 CrossRef CAS PubMed.
- J. H. Lee, J. Bae, J. H. Hwang, M. Y. Choi, Y. S. Kim, S. Park, J. H. Na, D. G. Kim and S. K. Ahn, Adv. Funct. Mater., 2022, 32, 2110360 CrossRef CAS.
- M. Camacho-Lopez, H. Finkelmann, P. Palffy-Muhoray and M. Shelley, Nat. Mater., 2004, 3, 307–310 CrossRef CAS PubMed.
- W. K. Fong, T. Hanley and B. J. Boyd, J. Controlled Release, 2009, 135, 218–226 CrossRef CAS PubMed.
- R. Huang, R. Lan, C. Shen, Z. Zhang, Z. Wang, J. Bao, Z. Wang, L. Zhang, W. Hu, Z. Yu, S. Zhu, L. Wang and H. Yang, ACS Appl. Mater. Interfaces, 2021, 13, 59221–59230 CrossRef CAS PubMed.
- A. Zabara and R. Mezzenga, J. Controlled Release, 2014, 188, 31–43 CrossRef CAS PubMed.
- B. Gurboga, E. B. Tuncgovde and E. Kemiklioglu, Biotechnol. Bioeng., 2022, 119, 1047–1052 CrossRef CAS PubMed.
- D. Martella, L. Pattelli, C. Matassini, F. Ridi, M. Bonini, P. Paoli, P. Baglioni, D. S. Wiersma and C. Parmeggiani, Adv. Healthcare Mater., 2019, 8, 1970009 CrossRef.
- A. Nasajpour, A. Mostafavi, A. Chlanda, C. Rinoldi, S. Sharifi, M. S. Ji, M. Ye, S. J. Jonas, W. Swieszkowski, P. S. Weiss, A. Khademhosseini and A. Tamayol, ACS Mater. Lett., 2020, 2, 1067–1073 CrossRef CAS.
- J. Jiang, N. P. Dhakal, Y. Guo, C. Andre, L. Thompson, O. Skalli and C. Peng, Adv. Healthcare Mater., 2020, 9, 2000487 CrossRef CAS PubMed.
- D. Martella, P. Paoli, J. M. Pioner, L. Sacconi, R. Coppini, L. Santini, M. Lulli, E. Cerbai, D. S. Wiersma, C. Poggesi, C. Ferrantini and C. Parmeggiani, Small, 2017, 13, 1702677 CrossRef PubMed.
- E. Sleep, B. D. Cosgrove, M. T. McClendon, A. T. Preslar, C. H. Chen, M. H. Sangji, C. M. R. Perez, R. D. Haynes, T. J. Meade, H. M. Blau and S. I. Stupp, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E7919–E7928 CrossRef CAS PubMed.
- A. Agrawal, O. Adetiba, H. Kim, H. Chen, J. G. Jacot and R. Verduzco, J. Mater. Res., 2015, 30, 453–462 CrossRef CAS.
- C. Ferrantini, J. M. Pioner, D. Martella, R. Coppini, N. Piroddi, P. Paoli, M. Calamai, F. S. Pavone, D. S. Wiersma, C. Tesi, E. Cerbai, C. Poggesi, L. Sacconi and C. Parmeggiani, Circ. Res., 2019, 124, e44–e54 CrossRef CAS PubMed.
- T. B. Saw, W. Xi, B. Ladoux and C. T. Lim, Adv. Mater., 2018, 30, 1802579 CrossRef PubMed.
- J. Fu, Y. K. Wang, M. T. Yang, R. A. Desai, X. Yu, Z. Liu and C. S. Chen, Nat. Methods, 2010, 7, 733–736 CrossRef CAS PubMed.
- J. Yoon, T. W. Eyster, A. C. Misra and J. Lahann, Adv. Mater., 2015, 27, 4509–4515 CrossRef CAS PubMed.
- A. W. Feinberg, A. Feigel, S. S. Shevkoplyas, S. Sheehy, G. M. Whitesides and K. K. Parker, Science, 2007, 317, 1366–1370 CrossRef CAS PubMed.
- L. Sun, Y. Yu, Z. Chen, F. Bian, F. Ye, L. Sun and Y. Zhao, Chem. Soc. Rev., 2020, 49, 4043–4069 RSC.
- C. Appiah, C. Arndt, K. Siemsen, A. Heitmann, A. Staubitz and C. Selhuber-Unkel, Adv. Mater., 2019, 31, 1807747 CrossRef PubMed.
- L. Ricotti, B. Trimmer, A. W. Feinberg, R. Raman, K. K. Parker, R. Bashir, M. Sitti, S. Martel, P. Dario and A. Menciassi, Sci. Robot., 2017, 2, eaaq0495 CrossRef PubMed.
- K. Kuribayashi-Shigetomi, H. Onoe and S. Takeuchi, PLoS One, 2012, 7, e51085 CrossRef CAS PubMed.
- S. J. Park, M. Gazzola, K. S. Park, S. Park, V. Di Santo, E. L. Blevins, J. U. Lind, P. H. Campbell, S. Dauth, A. K. Capulli, F. S. Pasqualini, S. Ahn, A. Cho, H. Yuan, B. M. Maoz, R. Vijaykumar, J. W. Choi, K. Deisseroth, G. V. Lauder, L. Mahadevan and K. K. Parker, Science, 2016, 353, 158–162 CrossRef CAS PubMed.
- J. C. Nawroth, H. Lee, A. W. Feinberg, C. M. Ripplinger, M. L. McCain, A. Grosberg, J. O. Dabiri and K. K. Parker, Nat. Biotechnol., 2012, 30, 792–797 CrossRef CAS PubMed.
- T.-C. Tang, B. An, Y. Huang, S. Vasikaran, Y. Wang, X. Jiang, T. K. Lu and C. Zhong, Nat. Rev. Mater., 2020, 6, 332–350 CrossRef.
- C. S. Chen, M. Mrksich, S. Huang, G. M. Whitesides and D. E. Ingber, Science, 1997, 276, 1425–1428 CrossRef CAS PubMed.
- T. C. von Erlach, S. Bertazzo, M. A. Wozniak, C. M. Horejs, S. A. Maynard, S. Attwood, B. K. Robinson, H. Autefage, C. Kallepitis, A. Del Rio Hernandez, C. S. Chen, S. Goldoni and M. M. Stevens, Nat. Mater., 2018, 17, 237–242 CrossRef CAS PubMed.
- L. Q. Li, J. Eyckmans and C. S. Chen, Nat. Mater., 2017, 16, 1164–1168 CrossRef CAS PubMed.
- M. S. Sakar, J. Eyckmans, R. Pieters, D. Eberli, B. J. Nelson and C. S. Chen, Nat. Commun., 2016, 7, 11036 CrossRef CAS PubMed.
- M. D. Davidson, M. E. Prendergast, E. Ban, K. L. Xu, G. Mickel, P. Mensah, A. Dhand, P. A. Janmey, V. B. Shenoy and J. A. Burdick, Sci. Adv., 2021, 7, eabi8157 CrossRef CAS PubMed.
- Z. Liu, M. Cai, X. Zhang, X. Yu, S. Wang, X. Wan, Z. Wang and L. Li, Adv. Mater., 2021, 33, 2106317 CrossRef CAS PubMed.
- X. Zhang, X. Cui, D. Wang, S. Wang, Z. Liu, G. Zhao, Y. Zhang, Z. Li, Z. L. Wang and L. Li, Adv. Funct. Mater., 2019, 29, 1900372 CrossRef.
- X. Wan, Y. Zhao, Z. Li and L. Li, Exploration, 2022, 2, 20210029 CrossRef.
- J. Larouche and C. A. Aguilar, Trends Biotechnol., 2019, 37, 604–617 CrossRef CAS PubMed.
- M. Montgomery, S. Ahadian, L. Davenport Huyer, M. Lo Rito, R. A. Civitarese, R. D. Vanderlaan, J. Wu, L. A. Reis, A. Momen, S. Akbari, A. Pahnke, R. K. Li, C. A. Caldarone and M. Radisic, Nat. Mater., 2017, 16, 1038–1046 CrossRef CAS PubMed.
- C. Wang, H. Yue, J. Liu, Q. Zhao, Z. He, K. Li, B. Lu, W. Huang, Y. Wei, Y. Tang and M. Wang, Biofabrication, 2020, 12, 045025 CrossRef CAS PubMed.
- Q. Zhao, J. Wang, H. Cui, H. Chen, Y. Wang and X. Du, Adv. Funct. Mater., 2018, 28, 1801027 CrossRef.
- Q. Zhao, H. Cui, J. Wang, H. Chen, Y. Wang, L. Zhang, X. Du and M. Wang, ACS Appl. Mater. Interfaces, 2018, 10, 23583–23594 CrossRef CAS PubMed.
- Y. Liu, J. Li, S. Song, J. Kang, Y. Tsao, S. Chen, V. Mottini, K. McConnell, W. Xu, Y. Q. Zheng, J. B. Tok, P. M. George and Z. Bao, Nat. Biotechnol., 2020, 38, 1031–1036 CrossRef CAS PubMed.
- C. Wang, X. Chen, L. Wang, M. Makihata, H.-C. Liu, T. Zhou and X. Zhao, Science, 2022, 377, 517–523 CrossRef CAS PubMed.
- J. Deng, H. Yuk, J. Wu, C. E. Varela, X. Chen, E. T. Roche, C. F. Guo and X. Zhao, Nat. Mater., 2020, 20, 229–236 CrossRef PubMed.
- S. Wang, Q. Zhao, J. Li and X. Du, ACS Appl. Mater. Interfaces, 2022, 14, 42420–42429 CrossRef CAS PubMed.
- Y. Chandorkar, A. Castro Nava, S. Schweizerhof, M. Van Dongen, T. Haraszti, J. Kohler, H. Zhang, R. Windoffer, A. Mourran, M. Moller and L. De Laporte, Nat. Commun., 2019, 10, 4027 CrossRef PubMed.
- M. F. A. Cutiongco, B. S. Jensen, P. M. Reynolds and N. Gadegaard, Nat. Commun., 2020, 11, 1384 CrossRef CAS PubMed.
- A. K. Yip, A. T. Nguyen, M. Rizwan, S. T. Wong, K. H. Chiam and E. K. F. Yim, Biomaterials, 2018, 181, 103–112 CrossRef CAS PubMed.
- Z. Wang, W. J. Lee, B. T. H. Koh, M. Hong, W. Wang, P. N. Lim, J. Feng, L. S. Park, M. Kim and E. S. Thian, Sci. Adv., 2018, 4, eaat4537 CrossRef CAS PubMed.
- S. Zhang, B. Ma, F. Liu, J. Duan, S. Wang, J. Qiu, D. Li, Y. Sang, C. Liu, D. Liu and H. Liu, Nano Lett., 2018, 18, 2243–2253 CrossRef CAS PubMed.
- Y. Z. Zhu, H. Liang, X. M. Liu, J. Wu, C. Yang, T. M. Wong, K. Y. H. Kwan, K. M. C. Cheung, S. L. Wu and K. W. K. Yeung, Sci. Adv., 2021, 7, eabf6654 CrossRef CAS PubMed.
- M. Hippler, K. Weissenbruch, K. Richler, E. D. Lemma, M. Nakahata, B. Richter, C. Barner-Kowollik, Y. Takashima, A. Harada, E. Blasco, M. Wegener, M. Tanaka and M. Bastmeyer, Sci. Adv., 2020, 6, eabc2648 CrossRef CAS PubMed.
- R. Mayor and S. Etienne-Manneville, Nat. Rev. Mol. Cell Biol., 2016, 17, 97–109 CrossRef CAS PubMed.
- C. L. Marchant, A. N. Malmi-Kakkada, J. A. Espina and E. H. Barriga, Nat. Mater., 2022, 21, 1314–1323 CrossRef CAS PubMed.
- M. Ebara, M. Akimoto, K. Uto, K. Shiba, G. Yoshikawa and T. Aoyagi, Polymer, 2014, 55, 5961–5968 CrossRef CAS.
- L. F. Tseng, P. T. Mather and J. H. Henderson, Acta Biomater., 2013, 9, 8790–8801 CrossRef CAS PubMed.
- D. M. Le, K. Kulangara, A. F. Adler, K. W. Leong and V. S. Ashby, Adv. Mater., 2011, 23, 3278–3283 CrossRef CAS PubMed.
- K. A. Davis, K. A. Burke, P. T. Mather and J. H. Henderson, Biomaterials, 2011, 32, 2285–2293 CrossRef CAS PubMed.
- J. Wang, A. Quach, M. E. Brasch, C. E. Turner and J. H. Henderson, Biomaterials, 2017, 140, 150–161 CrossRef CAS PubMed.
- P. Y. Mengsteab, K. Uto, A. S. T. Smith, S. Frankel, E. Fisher, Z. Nawas, J. Macadangdang, M. Ebara and D. H. Kim, Biomaterials, 2016, 86, 1–10 CrossRef CAS PubMed.
- R. O. Hynes, Science, 2009, 326, 1216–1219 CrossRef CAS PubMed.
- H. Gu, S. W. Lee, J. Carnicelli, T. Zhang and D. Ren, Nat. Commun., 2020, 11, 2211 CrossRef CAS PubMed.
- Q. Zhao, J. Wang, Y. Wang, H. Cui and X. Du, Natl. Sci. Rev., 2020, 7, 629–643 CrossRef CAS PubMed.
- K. Uto, T. Aoyagi, C. A. DeForest, A. S. Hoffman and M. Ebara, Adv. Healthcare Mater., 2017, 6, 1601439 CrossRef PubMed.
- D. You, G. Chen, C. Liu, X. Ye, S. Wang, M. Dong, M. Sun, J. He, X. Yu, G. Ye, Q. Li, J. Wu, J. Wu, Q. Zhao, T. Xie, M. Yu and H. Wang, Adv. Funct. Mater., 2021, 31, 2103920 CrossRef CAS.
- M. Hofer and M. P. Lutolf, Nat. Rev. Mater., 2021, 6, 402–420 CrossRef CAS PubMed.
- A. Chrisnandy, D. Blondel, S. Rezakhani, N. Broguiere and M. P. Lutolf, Nat. Mater., 2022, 21, 479–487 CrossRef CAS PubMed.
- E. Karzbrun, A. H. Khankhel, H. C. Megale, S. M. K. Glasauer, Y. Wyle, G. Britton, A. Warmflash, K. S. Kosik, E. D. Siggia, B. I. Shraiman and S. J. Streichan, Nature, 2021, 599, 268–272 CrossRef CAS PubMed.
- S. H. D. Wong, B. Yin, B. Yang, S. Lin, R. Li, Q. Feng, H. Yang, L. Zhang, Z. Yang, G. Li, C. H. J. Choi and L. Bian, Adv. Funct. Mater., 2019, 29, 1806822 CrossRef.
- F. Wang, M. Liu, C. Liu, C. Huang, L. Zhang, A. Cui, Z. Hu and X. Du, Natl. Sci. Rev., 2023, 10, nwac164 CrossRef PubMed.
- F. Wang, M. Liu, C. Liu, Q. Zhao, T. Wang, Z. Wang and X. Du, Sci. Adv., 2022, 8, eabp9369 CrossRef CAS PubMed.
- S. Z. Sharabani, N. Edelstein-Pardo, M. Molco, N. B. Schwartz, M. Morami, A. Sivan, Y. G. Rom, R. Evental, E. Flaxer and A. Sitt, Adv. Funct. Mater., 2022, 32, 2111471 CrossRef.
- L. R. Zhang, B. R. Liu, C. W. Wang, C. Xin, R. Li, D. W. Wang, L. Q. Xu, S. Y. Fan, J. Zhang, C. C. Zhang, Y. L. Hu, J. W. Li, D. Wu, L. Zhang and J. R. Chu, Nano Lett., 2022, 22, 5277–5286 CrossRef CAS PubMed.
- Y. Wang, H. Cui, Q. Zhao and X. Du, Matter, 2019, 1, 626–638 CrossRef.
- X. Du, H. Cui, T. Xu, C. Huang, Y. Wang, Q. Zhao, Y. Xu and X. Wu, Adv. Funct. Mater., 2020, 30, 1909202 CrossRef CAS.
- H. Cui, Q. Zhao, L. Zhang and X. Du, Adv. Intell. Syst., 2020, 2, 2000138 CrossRef.
- H. Chen, L. Wang, Y. Lu and X. Du, Microsyst. Nanoeng., 2020, 6, 58 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |