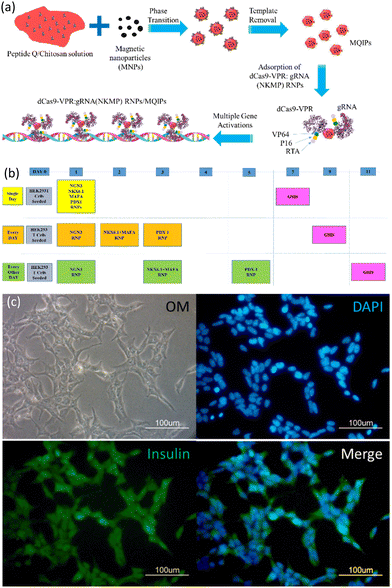
Nanoparticle-mediated CRISPR/dCas9a activation of multiple transcription factors to engineer insulin-producing cells†
Mei-Hwa
Lee
a,
James L.
Thomas
b,
Chien-Yu
Lin
c,
Yi-Chen Ethan
Li
d and
Hung-Yin
Lin
 *c
*c
aDepartment of Materials Science and Engineering, I-Shou University, Kaohsiung 84001, Taiwan
bDepartment of Physics and Astronomy, University of New Mexico, Albuquerque, NM 87131, USA
cDepartment of Chemical and Materials Engineering, National University of Kaohsiung, Kaohsiung 81148, Taiwan. E-mail: linhy@ntu.edu.tw
dDepartment of Chemical Engineering, Feng Chia University, Taichung 40724, Taiwan
First published on 6th February 2023
Abstract
Insulin may help to control blood glucose levels in diabetes; however, the long-term release of insulin is important for therapy. In this work, four guide RNAs (gRNA) for factors that promote specification and maturation of insulin-producing cells were synthesized: pancreatic and duodenal homeobox 1 (PDX1), protoendocrine factor (neurogenin 3, NGN3), NK6 homeobox 1 (NKX6.1), and musculoaponeurotic fibrosarcoma oncogene family A (MAFA). These gRNAs were used to form ribonucleoproteins (RNPs) with tracRNA and dCas9-VPR, and were then immobilized on magnetic peptide-imprinted chitosan nanoparticles, which enhanced transfection. The production and release of insulin from transfected cells were then measured using ELISA and staining with anti-insulin antibodies. The expression of the genes was evaluated using qRT-PCR; this was also used to investigate the cascade of additional transcriptional regulators. The magnitude and duration of insulin production were evaluated for single and repeated transfections (using different transfection schedules) to identify the most promising protocol.
Introduction
The National Diabetes Statistics Report (Centers for Disease Control and Prevention, CDC) estimates that approximately 37.3 million people in the US have diabetes (11.3% of the population), of which 28.7 million people (including 28.5 million adults) are diagnosed and 8.5 million people (23.0% of the adults with diabetes) are undiagnosed. The prevalence of diabetes globally has increased dramatically from 10.3% in 2001–2004 to 13.2% in 2017–2020. About 96 million adults in the US (38.0%) and 26.4 million people aged 65 years or older (48.8%) have prediabetes. In type 1 diabetes (T1D), the body does not produce insulin. In type 2 diabetes the body does not use insulin properly; it is characterized by the dysfunction of pancreatic beta-cells and insulin resistance in peripheral organs. Type 2 represents more than 90% of all cases of diabetes.1The differentiation of embryonic cells into insulin-secreting cells may provide an opportunity for diabetes therapy.2,3 A recent review of glucose-responsive insulin-secreting cells1 summarized the transcription factors that promote the specification and maturation of multipotent progenitor cells into pancreatic β cells.4,5 These factors include pancreatic and duodenal homeobox 1 (PDx1), acting in concert with other transcription factors, such as the protoendocrine factor (neurogenin 3, Ngn3), NK6 homeobox 1 (NKx6.1), and musculoaponeurotic fibrosarcoma oncogene family A (MaFA). Additionally, a cascade of additional transcriptional regulators is required to induce differentiation into a mature β cell.6
Recently, genome editing of human pancreatic beta cell models has been employed to understand the molecular mechanisms behind beta cell dysfunction, as recently reviewed.7 Understanding these mechanisms is essential for the development of effective and specific approaches for diabetes care and prevention. The CRISPR/Cas9 system provides a robust and multiplexable genome editing tool, enabling researchers to precisely manipulate specific genomic elements, and facilitating the elucidation of target gene function in disease and general biology.8 A chimeric dCas9 molecule, fused with any functionally active domain, can deliver effector-cargo to specific genome loci, allowing precise manipulations to be conducted with various forms of epigenetic regulation of expression, which is essential for advanced cell reprogramming.9 The modulation of insulin gene expression with CRISPR/Cas9-based transcription factors has been demonstrated with the INS (insulin) gene and the lentivirus.10 The coexpression of the transactivator and combinations of guide RNAs in human cells induces the specific expression of endogenous target genes, demonstrating a simple and versatile means of RNA-guided gene activation.11 The activation of pancreatic β-cell genes by multiplex epigenetic CRISPR-editing using CRISPR/dCas9-VP160, CRISPR/dCas9-TET1 and CRISPR/dCas9-P300 systems and human beta pancreatic genes (PDX1, NEUROG3, PAX4 and INS) has been reported.12 In addition, dCas9 and sgRNA expression plasmids have been used for Lipofectamine™ 2000-mediated HEK293T transfection.12
Results and discussion
Epitope-imprinting technology was developed to generate antibody-like specific recognition sites.13 The use of epitopes as imprinting templates has been examined to circumvent difficulties with whole-protein imprinting, reducing costs and increasing versatility.14 Epitope-imprinted polymers can be used as recognition elements in bioseparation,15 biosensing16–18 and protein delivery.19 Our previous work has demonstrated that imprinted nanoparticles can be used to enhance the delivery of CRISPR/dCas9 ribonucleoproteins, and demonstrated cellular reprogramming with multigene activation.20 In this work, magnetic peptide Q-imprinted chitosan nanoparticles (MQIPs) were used to deliver CRISPR/dCas9 ribonucleoproteins.20 The biocompatibility of such nanoparticles has been demonstrated in our previous work,19 which showed that HEK 293T cells treated with various nanoparticles had viabilities indistinguishable from controls. The template peptide Q is a sequence from dCas9, so that the ribonucleoprotein complexes self-assemble on the MQIPs. The synthesis of the nanoparticles is described in the supporting information and Scheme 1.Fig. S1 (ESI†) presents the properties of the MQIPs with and without RNP complexes bound to them. Fig. S1(a) (ESI†) shows the size distribution of MQIPs with various dCas9-VPR/RNPs bound. The mean size of the MNPs was 88 ± 58 nm (data not shown); that of MQIPs with bound dCas9-VPR was 230 ± 91 nm. When the composite nanoparticles bound dCas9-VPR and INS, NGN3, NKX6.1 + MAFA or PDX1 RNPs, the sizes increased to 241 ± 96, 233 ± 89, 246 ± 99, and 244 ± 98 nm, respectively. Interestingly, the nanoparticles without bound dCas9-VPR have a larger hydrodynamic size (392 ± 46 nm); binding the dCas9-VPR causes the nanoparticle to “condense”. To estimate the MQIP surface area, nitrogen absorption/desorption was measured, for the as-prepared MQIPs (with template still bound), after washing, and after rebinding the peptide Q template (1.0 pg mL−1 for 30 min). The hysteresis area of the MQIPs after template removal is significantly larger than when peptide Q is bound or rebound, which might indicate that the pore volume is increased after template removal. Fig. S1(c) (ESI†) shows the mean specific surface areas of the MQIP, using a BET analysis of the nitrogen adsorption data. Non-imprinted composite nanoparticles (MNIPs) are also shown. MNIPs are smaller than MQIPs (234 ± 29 nm), giving them a larger specific surface area. Notably, while the surface area of MQIPs increases on washing (which removes template peptide) and then decreases again on rebinding peptide Q, the non-imprinted particles show no significant surface area changes on washing and binding peptide Q. Finally, the release of dCas9a from MQIPs is shown in Fig. S1(d) (ESI†), which indicates that the delivery reaches equilibrium after about 12 hr.
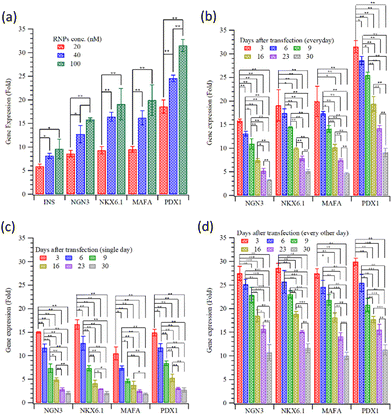
Fig. 1 plots the relative gene expression (compared to non-transfected cells; measured using qRT-PCR) of essential β-cell differentiation genes transfected into HEK293T cells with the combination of the four (NGN3, NKX6.1 + MAFA, and PDX1) transcription factors. Fig. 1(a) shows the dose dependence of gene expression. Cells were treated with MQIPs with 20, 40, or 100 nM of each RNP bound; for four-factor transcription, cells were treated every day for three days. Gene expression increases with dose, but nowhere near in proportion to dose. Different transfection treatment schedules (as shown in Scheme 1) for four-factor transfection are evaluated in Fig. 1(b–d). In Fig. 1(b), cells were treated every day for three days. Expression levels are good, but decay rapidly over time. Scheme 1 also displays immunocytochemistry images of HEK-293T cells that were treated with MQIPs and four (NGN3, NKX6.1 + MAFA, and PDX1) RNPs/dCas9-VPR repeated every day for three days. A comparison of the phase contrast image with the DAPI-staining of the nuclei reveals good cellular viability for the MQIP/RNPs-treated cells. Anti-insulin immunostaining and merged images confirm the expression of insulin in the treated cells. Fig. 1(c) shows that single-day transfection gives significantly lower expression (except perhaps for NGN3), and that expression levels decline rapidly. Fig. 1(d) shows that three transfection treatments, given every other day, gives the best results of the three transfection schedules studied. The initial expression levels are high, 25–30× that of controls, and the expression half-time is about three weeks.
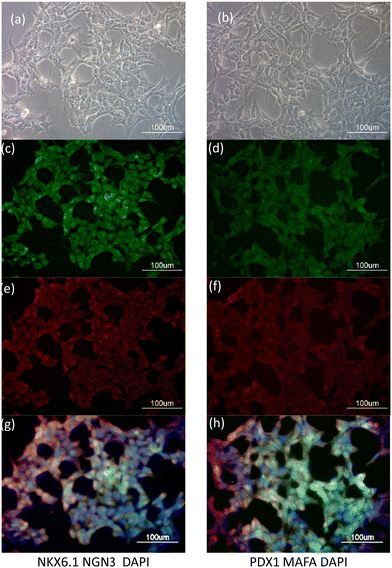
Fig. 2 displays images of DAPI- and anti-MAFA, anti-NGN3, anti-PDX1, and anti-NKX6.1 staining in HEK-293T cells that had been treated with MQIPs/RNPs every other day over five days. Fig. 2(a) and (b) show the phase contrast images of transfected cells. Fig. 2(c–f) show the immunocytochemistry images of DAPI- with anti-MAFA, anti-NGN3, anti-PDX1 or anti-NKX6.1 staining, respectively. Fig. 2(g) and (h) presents merged images of DAPI- with (anti-MAFA and anti-NGN3) or (anti-PDX1 and anti-NKX6.1) staining, respectively. Interestingly, these genes were expressed consistently with the results of qRT-PCR in Fig. 1(d).
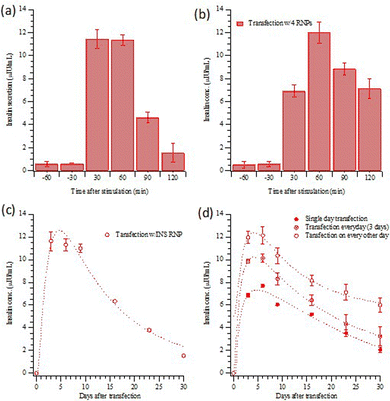
Fig. 3(a) and (b) present the insulin release (as measured by ELISA kits) from transfected HEK293T cells with INS or the four gene combination, stimulated with a high-glucose medium. The results indicate that a very small amount of insulin was released before stimulation. As shown in Fig. 3(a), the insulin from the HEK 293T cells with direct INS gene activation was rapid; after 30 minutes, insulin secretion had already peaked. Although the response with the four-gene activation was slower (Fig. 3(b)), the overall concentration of insulin released was slightly higher than that following only INS gene activation. Insulin release after high-glucose stimulation was examined for 30 days following INS transfection, Fig. 3(c). The stimulated insulin release decayed to half of its initial level in about two weeks. Fig. 3(d) shows the insulin release with the four differentiation transcription factors transfection, with the three different transfection schedules (Scheme 1). The maximum concentrations of insulin released are about 7.71 ± 0.15, 10.13 ± 0.38 and 12.12 ± 0.76 μIU mL−1 for transfections on (1) the first day only, (2) on three consecutive days and (3) every other day for five days. Semi-quantitative measurements of fluorescence are compared in Fig. S2 (ESI†). The alternate day schedule gave the strongest and longest-lasting insulin response, with a half time of four weeks.
Fig. 4 depicts the relative gene expressions from four-factor transfected HEK-293T cells, using the alternate day transfection schedule. The gene expression pattern of HEK293T cells with four-factor transfection is similar to that with single (INS) gene activation, as shown in Fig. S3(a) (ESI†). However, high-glucose stimulation not only increased the expression of c-AMP/PKA but also reduced the expression of PI3K (cf.Fig. 4 and Fig. S3(a), ESI†). The cAMP/PKA signalling pathway regulates glucose homeostasis affecting multiple processes, including insulin and glucagon secretion, glucose uptake, glycogen synthesis and breakdown, gluconeogenesis, and the neural control of glucose homeostasis.21 PKA can directly promote insulin exocytosis by phosphorylating secretory granule-associated proteins and thereby increasing Ca2+ influx.22 The activation of the PI3K/AKT pathway promotes the secretion of insulin from pancreatic β cells.23 The overexpression and constitutive activation of AKT in pancreatic β cells increase the mass, proliferation and size of the β cells, and these processes are mediated by signaling intermediates that are downstream of AKT, such as FoxO and mTOR.24
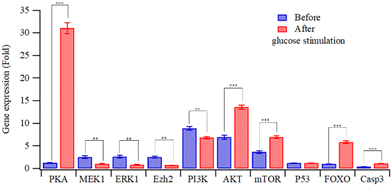 | ||
| Fig. 4 Relative gene expression in HEK293T cells transfected with MQIPs/four RNPs before and after glucose stimulation (**: p < 0.005; ***: p < 0.0005). | ||
Conclusions
Differentiation of the insulin-producing cells is of interest, especially for diabetes therapy. In this study, the precise delivery of multigene activation for inducing the maturation of insulin-producing cells was achieved, using a molecularly imprinted composite nanoparticle for CRISPR delivery. The efficacy of different transfection schedules was examined, with transfection on alternate days over a five-day period producing the strongest and longest-lasting response, when cells were subsequently challenged with high glucose concentrations. Importantly, the cascade of gene activation induced by the four-fold transfection not only increases the amount of insulin released but also extends the duration of release, compared to that achievable with INS transfection alone.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to thank the Ministry of Science and Technology of ROC under Contract no. MOST 109-2221-E-035-007-MY3, MOST 109-2314-B-390-001-MY3 and MOST 110-2221-E-390-003-MY3.References
- S. Gheibi, T. Singh, J. P. M. da Cunha, M. Fex and H. Mulder, Cells, 2020, 9, 2465 CrossRef CAS PubMed.
- M. Cito, S. Pellegrini, L. Piemonti and V. Sordi, Endocr. Connect., 2018, 7, R114–R125 CAS.
- M. Nemati, G. Ranjbar Omrani, B. Ebrahimi and A. Alizadeh, Stem Cells Int., 2021, 2021, 6652915 CrossRef PubMed.
- H. Vethe, L. Ghila, M. Berle, L. Hoareau, Ø. A. Haaland, H. Scholz, J. A. Paulo, S. Chera and H. Ræder, Front. Endocrinol., 2019, 10, 293 CrossRef.
- R. Tran, C. Moraes and C. A. Hoesli, Sci. Rep., 2020, 10, 1190 CrossRef CAS PubMed.
- H. M. Tse, V. Kozlovskaya, E. Kharlampieva and C. S. Hunter, Mol. Endocrinol., 2015, 29, 1388–1399 CrossRef CAS PubMed.
- D. Balboa, R. B. Prasad, L. Groop and T. Otonkoski, Diabetologia, 2019, 62, 1329–1336 CrossRef CAS PubMed.
- F. Zhang, Y. Wen and X. Guo, Hum. Mol. Genet., 2014, 23, R40–R46 CrossRef CAS PubMed.
- K. M. Shakirova, V. Y. Ovchinnikova and E. B. Dashinimaev, Front. Bioeng. Biotechnol., 2020, 8, 882 CrossRef PubMed.
- B. Alzhanuly, Z. Y. Mukhatayev, D. M. Botbayev, Y. Ashirbekov, N. D. Katkenov, N. T. Dzhaynakbaev and K. O. Sharipov, Open Access Maced. J. Med. Sci., 2021, 9, 876–881 CrossRef.
- P. Perez-Pinera, D. D. Kocak, C. M. Vockley, A. F. Adler, A. M. Kabadi, L. R. Polstein, P. I. Thakore, K. A. Glass, D. G. Ousterout and K. W. Leong, Nat. Methods, 2013, 10, 973–976 CrossRef CAS PubMed.
- G. C. Alejandra, C. Lucia, H. S. Ho, G. Luis, R. P. Juan and P.-B. Federico, bioRxiv, 2020, preprint DOI:10.1101/2020.07.24.214544.
- K. Yang, S. Li, L. Liu, Y. Chen, W. Zhou, J. Pei, Z. Liang, L. Zhang and Y. Zhang, Adv. Mater., 2019, 31, 1902048 CrossRef CAS PubMed.
- S. P. B. Teixeira, R. L. Reis, N. A. Peppas, M. E. Gomes and R. M. A. Domingues, Sci. Adv., 2021, 7, eabi9884 CrossRef CAS PubMed.
- M.-H. Lee, J. L. Thomas, C.-L. Liao, S. Jurcevic, T. Crnogorac-Jurcevic and H.-Y. Lin, Sep. Purif. Technol., 2018, 192, 213–219 CrossRef CAS.
- M.-H. Lee, J. L. Thomas, C.-L. Liao, S. Jurcevic, T. Crnogorac-Jurcevic and H.-Y. Lin, Microchim. Acta, 2017, 1–8, DOI:10.1007/s00604-017-2169-4.
- M.-H. Lee, J. L. Thomas, Z.-L. Su, W.-K. Yeh, A. S. Monzel, S. Bolognin, J. C. Schwamborn, C.-H. Yang and H.-Y. Lin, Biosens. Bioelectron., 2021, 175, 112852 CrossRef CAS.
- M.-H. Lee, K.-H. Liu, J. L. Thomas, C.-Y. Chen, C.-Y. Chen, C.-H. Yang and H.-Y. Lin, Biosens. Bioelectron., 2022, 200, 113930 CrossRef CAS.
- M.-H. Lee, C.-C. Lin, J. L. Thomas, C.-K. Chan and H.-Y. Lin, Nanotechnology, 2021, 32, 18LT02 CrossRef CAS.
- M.-H. Lee, C.-C. Lin, J. L. Thomas, J.-A. Li and H.-Y. Lin, Mater. Today Bio, 2021, 9, 100091 CrossRef CAS PubMed.
- H. Yang and L. Yang, J. Mol. Endocrinol., 2016, 57, R93–R108 CAS.
- C. Ämmälä, F. M. Ashcroft and P. Rorsman, Nature, 1993, 363, 356–358 CrossRef.
- S. Georgia and A. Bhushan, J. Clin. Invest., 2004, 114, 963–968 CrossRef CAS PubMed.
- E. Bernal-Mizrachi, W. Wen, S. Stahlhut, C. M. Welling and M. A. Permutt, J. Clin. Invest., 2001, 108, 1631–1638 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2tb02431d |
| This journal is © The Royal Society of Chemistry 2023 |