Layer-by-layer nanoparticle encapsulating all-trans retinoic acid and CpG as a mucosal adjuvant targeting colorectal cancer†
Shiwei
Mi
ab,
Wei
Li
ab,
Yixing
Wen
ab,
Chen
Yang
ab,
Shuai
Liu
a,
Jingjiao
Li
ab,
Xingdi
Cheng
ab,
Yuanyuan
Zhao
ab,
Haonan
Huo
ab,
Haowei
Zu
a and
Xueguang
Lu
 *ab
*ab
aBeijing National Laboratory for Molecular Sciences, CAS Key Laboratory of Colloid, Interface and Chemical Thermodynamics, Institute of Chemistry, Chinese Academy of Sciences, Beijing 100190, China. E-mail: xueguang@iccas.ac.cn
bUniversity of Chinese Academy of Sciences, Beijing 100049, China
First published on 13th March 2024
Abstract
Colorectal cancer (CRC) ranks among the most prevalent cancers globally, demanding innovative therapeutic strategies. Immunotherapy, a promising avenue, employs cancer vaccines to activate the immune system against tumors. However, conventional approaches fall short of eliciting robust responses within the gastrointestinal (GI) tract, where CRC originates. Harnessing the potential of all-trans retinoic acid (ATRA) and cytosine-phosphorothioate-guanine (CpG), we developed layered nanoparticles using a layer-by-layer assembly method to co-deliver these agents. ATRA, crucial for gut immunity, was efficiently encapsulated alongside CpG within these nanoparticles. Administering these ATRA@CpG-NPs, combined with ovalbumin peptide (OVA), effectively inhibited orthotopic CRC growth in mice. Our approach leveraged the inherent benefits of ATRA and CpG, demonstrating superior efficacy in activating dendritic cells, imprinting T cells with gut-homing receptors, and inhibiting tumor growth. This mucosal adjuvant presents a promising strategy for CRC immunotherapy, showcasing the potential for targeting gut-associated immune responses in combating colorectal malignancies.
Introduction
Colorectal cancer (CRC) ranks as the third most prevalent cancer worldwide, accounting for approximately 10% of all cancer cases.1 In recent years, immunotherapy has emerged as a promising approach in both treating cancer and preventing its recurrence.2–5 Cancer vaccines represent an important class of cancer immunotherapies by harnessing the patient's immune system to recognize and combat cancer.6,7 Cancer vaccines typically consist of antigens and adjuvants.8 Antigens are specific molecules or proteins utilized to train the immune system to selectively target cancer cells.9 Adjuvants, on the other hand, function as immune stimulants, enhancing the immune response against tumor antigens.10 The efficacy of cancer vaccines is determined by the selection of antigens and adjuvants.8 To effectively target CRC, vaccines need to provoke immune responses within the intestinal mucosa, the site where tumors originate and progress. However, conventional cancer vaccines, administered through parenteral injection, often fail to trigger robust immune responses in the gastrointestinal (GI) tract due to tissue constraints, resulting in unsatisfactory efficacy in treating CRC.11All-trans retinoic acid (ATRA), a metabolite of vitamin A, plays an important role in intestinal immunity.12–14 During antigen presentation, ATRA activates dendritic cells (DCs) to express CD103, which leads to the induction of gut-homing receptors α4β7 and CCR9 on T cells.15–18 These imprinted T cells have a gut tropism and bolstering intestinal immunity. Therefore, administering vaccines with ATRA is a promising approach to activating anti-tumor immune responses within the gut. However, ATRA is a hydrophobic molecule that requires a delivery vehicle for administration. Similarly, several clinically approved adjuvants for cancer vaccines, like cytosine-phosphorothioate-guanine (CpG), are hydrophilic molecules that also necessitate delivery vehicles to transport them into cells.19–23 To address these challenges, various delivery systems have been developed for the co-delivery of ATRA and CpG. For example, Ma et al. synthesized polymer/lipid nanoparticles (PLNPs) using an oil-in-water emulsion method capable of encapsulating both ATRA and CpG, thereby eliciting robust systemic and intestinal immune responses.19 Sun et al. fabricated mesoporous silica nanoparticles (MSNs) to load antigens and adjuvants first.24 The co-delivery of ATRA and MSNs was achieved by encapsulation within liposomes. Despite the demonstrated efficacy of these co-delivery systems in transporting ATRA and adjuvants, their preparation involves intricate steps and large quantities of carrier materials to deliver the desired doses of ATRA.
The layer-by-layer (LbL) assembly method provides a direct means to incorporate diverse therapeutics into a single nanoparticle. By sequentially layering oppositely charged polyelectrolytes onto a charged core, various therapeutics can be encapsulated within the layers through electrostatic interactions. These LbL nanoparticles have wide-ranging applications in delivering therapeutics for cancer treatment.25–30 The simplicity and adaptability of LbL nanoparticles make them an excellent choice for delivering both ATRA and CpG.
Here, we've developed a straightforward method for preparing LbL nanoparticles capable of co-delivering ATRA and CpG with high loading capacities. The chemical structure of ATRA, comprising a hydrophobic alkyl chain and a hydrophilic carboxylic acid end group, allows it to self-assemble in water, forming negatively charged ATRA nanoparticles (ATRA-NPs). These ATRA nanoparticles served as the core onto which positively charged polyethylenimine (PEI) and negatively charged CpG were sequentially deposited through electrostatic interactions, resulting in CpG- and ATRA-loaded multilayered nanoparticles (ATRA@CpG-NPs). ATRA@CpG-NPs serve as potent mucosal adjuvants to activate DCs and imprint T cells with gut-homing receptors. We demonstrated that administering ATRA@CpG-NPs with a model antigen, ovalbumin peptide (OVA), via intramuscular injections effectively inhibited tumor growth in an orthotopic colorectal tumor model in mice.
Materials and methods
All-trans retinoic acid was purchased from Bide Pharmatech Ltd (Shanghai, China). Hyperbranched PEI with a molecular weight of 25 kDa and all other materials were purchased from Sigma-Aldrich unless specifically noted. CpG oligonucleotide (5′-TCCATGACGTTCCTGACGTT-3′), fluorescein-labeled CpG, tumor necrosis factor-α (TNF-α) forward primer (5′-GGTGCCTATGTCTCAGCCTCTT-3′), reverse primer (5′-GCCATAGAACTGATGAGAGGGAG-3′). Interleukin-6 (IL-6) forward primer (5′-TACCACTTCACAAGTCGGAGGC-3′), reverse primer (5′-CTGCAAGTGCATCATCGTTGTTC-3′), IL-12p40 forward primer (5′-TTGAACTGGCGTTGGAAGCACG-3′), reverse primer (5′-CCACCTGTGAGTTCTTCAAAGGC-3′), GAPDH forward primer (5′-TGCACCACCAACTGTTTAGC-3′), reverse primer (5′-GGCATGGACTGTGGTCATGAG-3′) were purchased from Sangon Biotech (Shanghai, China). OVA257-264 (SIINFEKL) peptide was purchased from Top-peptide Biotechnology Co. Ltd (Shanghai, China). The anti-mouse CD16/32 antibody, FITC-anti-mouse CD80 antibody, PE-anti-mouse CD11c antibody, APC-anti-mouse CD86 antibody, FITC-anti-mouse CD3 antibody, PerCP/Cy5.5-anti-mouse CD8 antibody, PE-anti-mouse CCR9 antibody, APC-anti-mouse α4β7 antibody were purchased from Biolegend.Preparation of ATRA@CpG-NPs
ATRA was weighed and dissolved in dimethyl sulfoxide (DMSO). The ATRA solution in DMSO was then added dropwise into Nanopure water with viscous stirring. The volume ratio of water to DMSO is 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. After addition, the mixture was dialyzed against Nanopure water using a dialysis membrane (MWCO = 12–14 kDa, Biorigin, Beijing, China) over 4 hours to obtain ATRA-NPs. To fabricate ATRA@PEI-NPs, 1 mg mL−1 PEI in water was added to ATRA-NPs in a centrifuge tube. The mixture was shaken at room temperature for 30 min before being centrifuged at 12
1. After addition, the mixture was dialyzed against Nanopure water using a dialysis membrane (MWCO = 12–14 kDa, Biorigin, Beijing, China) over 4 hours to obtain ATRA-NPs. To fabricate ATRA@PEI-NPs, 1 mg mL−1 PEI in water was added to ATRA-NPs in a centrifuge tube. The mixture was shaken at room temperature for 30 min before being centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 15 min. The supernatant containing free PEI was discarded. The resulting ATRA@PEI-NPs were washed three times with water to remove unabsorbed PEI. To prepare ATRA@CpG-NPs, CpG in water was added into ATRA@PEI-NPs. The reaction mixture was incubated at room temperature for 30 min and purified by centrifugation. The hydrodynamic diameter, polydispersity index, and zeta potential of nanoparticles were measured using a Zetasizer Nano ZSP instrument (Malvern Instruments) at 25 °C in water. The TEM images were obtained on a JEM-100GX2 microscope at an accelerated voltage of 100 kV.
000g for 15 min. The supernatant containing free PEI was discarded. The resulting ATRA@PEI-NPs were washed three times with water to remove unabsorbed PEI. To prepare ATRA@CpG-NPs, CpG in water was added into ATRA@PEI-NPs. The reaction mixture was incubated at room temperature for 30 min and purified by centrifugation. The hydrodynamic diameter, polydispersity index, and zeta potential of nanoparticles were measured using a Zetasizer Nano ZSP instrument (Malvern Instruments) at 25 °C in water. The TEM images were obtained on a JEM-100GX2 microscope at an accelerated voltage of 100 kV.
Quantification of ATRA by HPLC
Standard ATRA solutions were prepared at concentrations of 1.5625, 3.215, 6.25, 12.5, 25, 50, and 100 μg mL−1 in acetonitrile. ATRA solutions were then subjected to high-performance liquid chromatography (HPLC) analysis using a Waters Breeze HPLC system equipped with a SunFire® C18 column (5 μm, 4.6 × 150 mm) and a photodiode array detector. The mobile phases were acetonitrile containing 0.1% of trifluoroacetic acid (TFA) and triethylamine acetate buffer (0.1 M) containing 0.1% of TFA. A standard curve between the concentration and area under the curve of ATRA was established. To quantify ATRA in ATRA@CpG-NPs, ATRA@CpG-NPs were dissolved in acetonitrile and analyzed by HPLC. The concentration of ATRA was then calculated according to the standard curve.Cellular uptake in vitro
DC2.4 cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% fetal bovine serum (FBS), 100 U mL−1 of penicillin, and 100 μg mL−1 of streptomycin at 37 °C in 5% CO2. To study the cellular uptake, DC2.4 cells were seeded in a 48-well plate with 5 × 104 cells per well or in a confocal dish with 2 × 105 cells per well. Cells were incubated for 24 h before adding PBS, free fluorescein-labeled CpG, ATRA@CpG-NPs in a complete medium. 0.15 μg of CpG was added to each well if applicable. The cells were incubated at 37 °C with 5% CO2 for 2 h. Then, the cells were collected, washed with PBS, and analyzed by a CytoFLEX flow cytometer (Beckman Coulter). For confocal laser scanning microscopy (CLSM) analysis, cells in the confocal dish were washed with PBS three times before being stained by Hoechst 33342 for 10 min. The cells were then washed with PBS three times before fixation with 4% paraformaldehyde for 20 min at room temperature. The prepared samples were imaged using an FV3000 confocal laser scanning microscope (Olympus) at excitation wavelengths of 408 nm (Hoechst 33342) and 488 nm (fluorescein).Animals
All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Peking University and approved by the Animal Ethics Committee of Peking University Institutional Animal Care and Use (LA2021284). Female C57BL/6 mice were purchased from SPF Biotechnology (Beijing, China).Activation of BMDCs
Bone marrow-derived dendritic cells were isolated from C57BL/6 mice (female, six-week-old). Briefly, bone marrow was firstly separated from the femur and tibia bones, the single-cell suspensions were then obtained by syringe and cultured in RPMI 1640 medium supplemented with 10% FBS, 1% of penicillin/streptomycin, and 20 ng mL−1 granulocyte-macrophage colony-stimulating factor (GM-CSF). The medium was half replaced every 2 days. On day 6, unadhered immature dendritic cells were collected and plated at a density of 5 × 105 cells per well in a 48-well plate. Bone-marrow-derived dendritic cells (BMDCs) were incubated for 24 h at 37 °C before being treated with PBS, free CpG with OVA, ATRA@CpG-NPs, or ATRA@CpG-NPs with OVA. The doses of ATRA, CpG, and OVA are 1.95 μg, 0.5 μg, and 10 μg respectively per well if applicable. After incubation for 24 h, BMDCs were collected, washed with FACS buffer (2% FBS in PBS), blocked with anti-CD16/32, and stained with PE-anti-CD11c antibody, FITC-anti-CD80 antibody, and APC-anti-CD86 antibody for 30 min at 4 °C. The cells were then washed with PBS two times and analyzed using a CytoFLEX flow cytometer (Beckman Coulter). To analyze the inflammatory cytokines of BMDCs, the total RNA of treated BMDCs was extracted using Trizol Total RNA Extraction Reagent (Beyotime) according to the manufacturer's instructions. The cDNA is then synthesized using HiScript III 1st Strand cDNA Synthesis Kit (+gDNA wiper) (Vazyme). Real-time reverse transcription-polymerase chain reaction (RT-qPCR) was performed using Taq Pro Universal SYBR·qPCR master mix (Vazyme) on an Archimed X4 instrument (RocGene).Activation of DCs in vivo
C57BL/6 mice were randomly divided into four groups (n = 4). Mice were intramuscularly injected with PBS, free CpG with OVA, ATRA@CpG-NPs, or ATRA@CpG-NPs with OVA. Each mouse received 20 μg of OVA, 15 μg of CpG, and 70 μg of ATRA per dose. After 24 h, ipsilateral inguinal lymph nodes were collected and digested to single-cell suspensions for flow cytometry. After incubation with anti-CD16/32 antibody, the cell suspensions were stained with PE anti-CD11c antibody, FITC anti-CD80 antibody, and APC anti-CD86 antibody for 30 min at 4 °C. The cells were then washed with PBS two times and analyzed using a CytoFLEX flow cytometer (Beckman Coulter).Quantification of CCR9 and α4β7 on T cells
Splenocytes were isolated from female C57BL/6 mice and seeded into 96-well plates at a density of 1 × 105 cells per well. Splenocytes were cultured in RPMI 1640 medium supplemented with 10% of FBS, 1% of penicillin/streptomycin, 1% of non-essential amino acids, 1% of sodium pyruvate, 0.1% of 2-mercaptoethanol, and 100 U mL−1 of Interleukin-2 (Gibco). Cells were then incubated with a mouse T cell activation/expansion Kit (Miltenyi). ATRA@CpG-NPs with OVA, ATRA@CpG-NPs, and ATRA-NPs in water, or free ATRA in DMSO were added to each well at the same concentrations of ATRA (80 nM). After 5 days of incubation, cells were collected and stained with mouse FITC-anti-CD3, PerCP/Cy5.5-anti-CD8, PE-anti-CCR9, and APC-anti-α4β7antibodies for 30 min at 4 °C. T cells were then washed with PBS for two times and analyzed using a CytoFLEX flow cytometer (Beckman Coulter).OT-1 cells isolation and co-culture with ATRA@CpG-NPs
The interferons-γ (IFN-γ) and IL-2 in OT-1 CD8+ T cells were evaluated by enzyme-linked immunosorbent assay (ELISA). The spleen of OT-1 mice was isolated and digested to single-cell suspensions at 1 × 108 cells per mL in PBS containing 2% FBS and 1 mM EDTA. CD8+ T cells were extracted using EasySep™ Mouse CD8+ T Cell Isolation Kit. OT-1 CD8+ T cells were collected and plated at a density of 3 × 105 cells per well in a 96-well plate and co-incubated with PBS, ATRA@CpG-NPs, or ATRA@CpG-NPs with OVA at 37 °C. The doses of ATRA, CpG, and OVA are 0.375 μg mL−1, 0.1 μg mL−1, and 2 μg mL−1 (200 μL), respectively. After incubation for 48 h, the supernatants were collected after centrifugation (300g, 5 min) for ELISA detection according to the manufacturer's protocols (Invitrogen). The results were measured using a microplate reader (BioTek Synergy H1) at 450 nm and 570 nm.Therapeutic efficacy in tumor-bearing mice
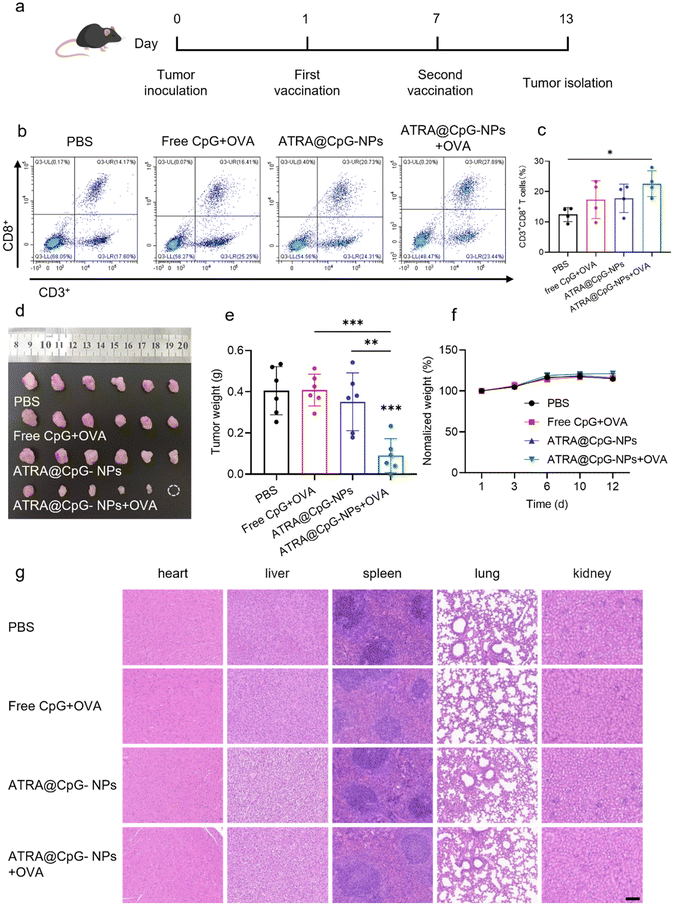
A mouse colon adenocarcinoma cell line MC38 that was stably transfected with OVA expression was purchased from Tongpai Biotechnology (Shanghai). MC38-OVA cells were cultured in Dulbecco's Modified Eagle's medium (DMEM) supplemented with 10% FBS, 100 μg mL−1 of penicillin, and streptomycin. C57BL/6 mice were subcutaneously injected with 1.0 × 106 of MC38-OVA cells in 200 μL of PBS into the right flank. Mice were euthanized when the tumor grew to approximately 500 mm3. The tumors were isolated and divided into 3 to 4 mg pieces. The tumor tissues were then transplanted to the cecum of healthy C57BL/6 mice to establish orthotopic colorectal tumors according to a previously published protocol.37 The mice were intramuscularly vaccinated with PBS, free CpG with OVA, ATRA@CpG-NPs, or ATRA@CpG-NPs with OVA on days 1 and 7 post-tumor inoculations. Each mouse received 20 μg of OVA, 15 μg of CpG, and 70 μg of ATRA per dose. The body weight of mice was measured every three days post-tumor inoculation. Seven days after the last vaccination, the tumor of mice was harvested and digested to single-cell suspensions for flow cytometric analysis. After incubation with anti-CD16/CD32 antibody, the cells from tuomrs were stained with PE anti-CD3 antibody, FITC anti-CD8 antibody, and PerCP/Cy5.5 anti-CD4 antibody to analyze CD8+ T cells. The cells were then washed with PBS two times and analyzed using a CytoFLEX flow cytometer (Beckman Coulter). To evaluate the tumor inhibition efficacy, the tumors on the cecum and the major organs including the heart, liver, spleen, lung, and kidney were isolated at day 13 post-tumor inoculation. The tumors of each group were imaged and weighed. The major organs were fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned into 5 μm slices for hematoxylin and eosin (H&E) staining.In vitro cytotoxicity assay
DC 2.4 cells were incubated with free CpG, free ATRA and ATRA@CpG-NPs with different concentrations of CpG and ATRA for 24 h. The medium containing ATRA@CpG-NPs was then replaced with a culture medium. The cell viability was measured using the cell counting kit-8 (CCK-8) assay according to the manufacturer's instructions (Vazyme). The results were measured using a microplate reader (BioTek Synergy H1) at 450 nm.Statistical analysis
GraphPad Prism was used for statistical analysis. Two-tailed Student's t-test or one-way analysis of variance (ANOVA) with a Tukey's post hoc test was used for comparing two groups or more than two groups, respectively. Data were significantly different if P < 0.05 (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). The specific statistical methods were indicated in the figure legend.Results and discussion
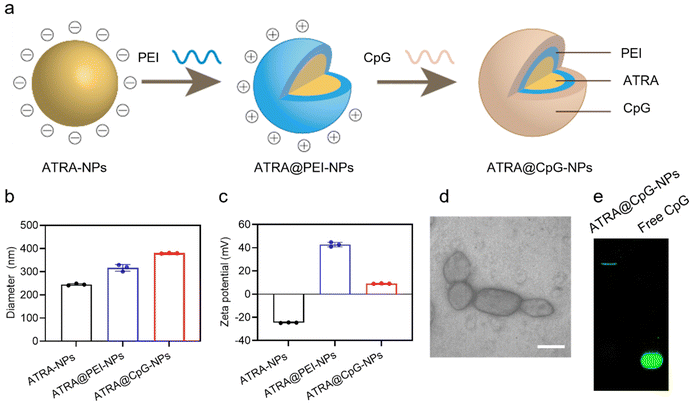
Instead of using carrier materials to deliver ATRA, we prepared ATRA-NPs by themselves through nanoprecipitation (Fig. 1a). ATRA dissolved in DMSO was added to water dropwise and stirred viscously to allow the formation of ATRA-NPs. Previous research has highlighted the impact of solute concentration in organic solvents on the resultant nanoparticle sizes in water. Therefore, we evaluated three ATRA concentrations in DMSO (3, 6, and 10 mg mL−1). Both 3 mg mL−1 and 6 mg mL−1 concentrations of ATRA in DMSO successfully produced ATRA-NPs through nanoprecipitation. However, the higher concentration of 10 mg mL−1 ATRA led to the formation of large aggregates in water. Analysis using dynamic light scattering (DLS) revealed that ATRA-NPs generated from 3 mg mL−1 and 6 mg mL−1 concentrations in DMSO exhibited hydrodynamic diameters of approximately 226 nm and 245 nm (Fig. 1b and S1†), respectively, with narrow polydispersity indexes (<0.25). We selected ATRA-NPs prepared using 6 mg mL−1 ATRA for subsequent experiments due to its higher ATRA concentration in water. Transmission electron microscopy confirmed that these ATRA-NPs manifested as nanocrystals with a cross-sectional diameter of approximately 169 ± 28 nm (Fig. S2†), consistent with the DLS findings. The surface potential of ATRA-NPs is approximately −24.6 mV (Fig. 1c). The negative surface charge likely originated from the carboxylic acid groups of ATRA.After synthesizing ATRA-NPs with negative surface charge, we proceeded to coat them with PEI, which is a commonly used cationic polymer for nucleic acid delivery. An excess amount of PEI is added to ensure successful coating. The stoichiometry between nitrogen in PEI to carboxylic acid group in ATRA is 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. After incubation and purification through centrifugation, the resulting ATRA@PEI-NPs exhibit a hydrodynamic diameter of approximately 316 nm and a surface potential of 42.7 mV (Fig. 1b and c). The increased size and sharp change of zeta potential suggest successful coating of PEI onto ATRA-NPs. We then coated fluorescein-labeled CpG strands onto ATRA@PEI-NPs. We added different amounts of CpG to ATRA@PEI-NPs. The stoichiometries of nitrogen in PEI to phosphate groups in CpG (N/P ratio) were adjusted to 111
1. After incubation and purification through centrifugation, the resulting ATRA@PEI-NPs exhibit a hydrodynamic diameter of approximately 316 nm and a surface potential of 42.7 mV (Fig. 1b and c). The increased size and sharp change of zeta potential suggest successful coating of PEI onto ATRA-NPs. We then coated fluorescein-labeled CpG strands onto ATRA@PEI-NPs. We added different amounts of CpG to ATRA@PEI-NPs. The stoichiometries of nitrogen in PEI to phosphate groups in CpG (N/P ratio) were adjusted to 111![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 55
1, 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 33
1, and 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Such calculation is based on the overall amount of PEI used for preparing ATRA@PEI-NPs. The purification step will remove uncoated PEI. Therefore, the actual N/P ratios are lower than the calculated values. As shown in Fig. S3,† the hydrodynamic diameter of ATRA@CpG-NPs increases with increasing amount of CpG. The Zeta potentials of ATRA@CpG-NPs decrease with increasing amounts of CpG. The high CpG loading results in greatly increased hydrodynamic diameters and nearly neutral surface charge. Such large particles with neutral surface charge could compromise cellular uptake ability. Therefore, we selected ATRA@CpG-NPs with an N/P ratio of 55
1. Such calculation is based on the overall amount of PEI used for preparing ATRA@PEI-NPs. The purification step will remove uncoated PEI. Therefore, the actual N/P ratios are lower than the calculated values. As shown in Fig. S3,† the hydrodynamic diameter of ATRA@CpG-NPs increases with increasing amount of CpG. The Zeta potentials of ATRA@CpG-NPs decrease with increasing amounts of CpG. The high CpG loading results in greatly increased hydrodynamic diameters and nearly neutral surface charge. Such large particles with neutral surface charge could compromise cellular uptake ability. Therefore, we selected ATRA@CpG-NPs with an N/P ratio of 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 which exhibited a hydrodynamic diameter of approximately 380 nm and surface potential of 9.1 mV for the following studies (Fig. 1b and c). TEM analysis showed that ATRA@CpG-NPs exhibit nanocrystal structures (Fig. 1d). Agarose gel electrophoresis showed that ATRA@CpG-NPs move significantly slower compared to free CpG, suggesting the successful loading of CpG in ATRA@CpG-NPs (Fig. 1e). The amount of ATRA in ATRA@CpG-NPs was quantified by HPLC, revealing a mass ratio of approximately 3.75
1 which exhibited a hydrodynamic diameter of approximately 380 nm and surface potential of 9.1 mV for the following studies (Fig. 1b and c). TEM analysis showed that ATRA@CpG-NPs exhibit nanocrystal structures (Fig. 1d). Agarose gel electrophoresis showed that ATRA@CpG-NPs move significantly slower compared to free CpG, suggesting the successful loading of CpG in ATRA@CpG-NPs (Fig. 1e). The amount of ATRA in ATRA@CpG-NPs was quantified by HPLC, revealing a mass ratio of approximately 3.75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
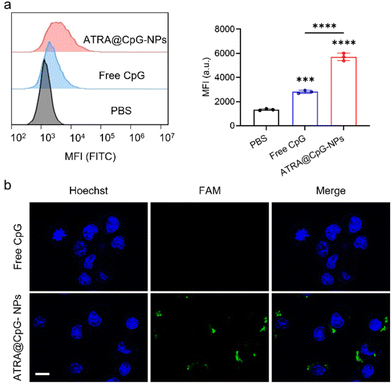
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for ATRA to CpG within the particles (Fig. S4†). We next evaluated the cellular uptake ability of ATRA@CpG-NPs. Free fluorescein-labeled CpG or ATRA@CpG-NPs were incubated with DC2.4 cells. Flow cytometry analysis showed that the median fluorescence intensity (MFI) of cells treated with ATRA@CpG-NPs is significantly higher than that of free CpG-treated cells (Fig. 2a). Confocal laser scanning microscopy images also showed that ATRA@CpG-NPs exhibit much brighter fluorescein signals compared to free CpG (Fig. 2b). These data demonstrated that ATRA@CpG-NPs effectively transport CpG into cells.
1 for ATRA to CpG within the particles (Fig. S4†). We next evaluated the cellular uptake ability of ATRA@CpG-NPs. Free fluorescein-labeled CpG or ATRA@CpG-NPs were incubated with DC2.4 cells. Flow cytometry analysis showed that the median fluorescence intensity (MFI) of cells treated with ATRA@CpG-NPs is significantly higher than that of free CpG-treated cells (Fig. 2a). Confocal laser scanning microscopy images also showed that ATRA@CpG-NPs exhibit much brighter fluorescein signals compared to free CpG (Fig. 2b). These data demonstrated that ATRA@CpG-NPs effectively transport CpG into cells.
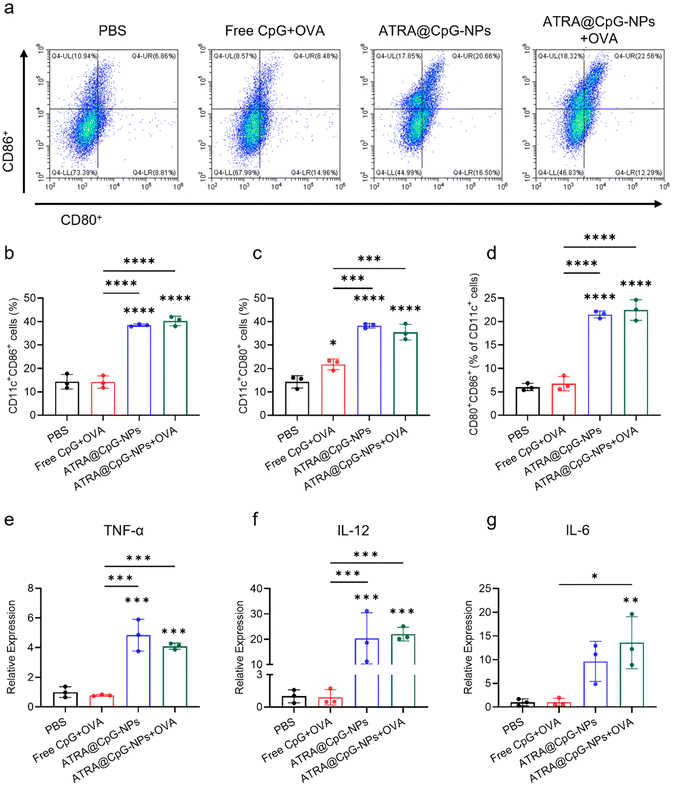
We next evaluated the adjuvant efficacy of ATRA@CpG-NPs in activating the immune systems. We isolated BMDCs from C57BL/6 mice and incubated ATRA@CpG-NPs or ATRA@CpG-NPs with a model antigen ovalbumin peptide. Free CpG with OVA and PBS were used as negative controls. The activation of BMDCs is determined by quantifying the expression of costimulatory markers CD80 and CD86 using flow cytometry. As shown in Fig. 3a–d and S5,† both ATRA@CpG-NPs and ATRA@CpG-NPs + OVA greatly increased the population of CD86+CD11c+, CD80+CD11c+, and CD80+CD86+CD11c+ cells compared to PBS treatment, suggesting effective activation of DCs. In contrast, treatment with free CpG with OVA did not trigger DC activation. ATRA@CpG-NPs with or without OVA showed similar DC activation efficacy. The DC activation in the inguinal lymph nodes of mice following one intramuscular injection showed similar results (Fig. S6†). We hypothesized that DC activation by free OVA is overshadowed by the potent adjuvant.31–33 Previous studies and our results have shown that free OVA antigen without adjuvant has poor DC activation efficacy.34
Activated BMDCs could secrete proinflammatory cytokines that recruit and activate T cells, instigating adaptive immune responses. We further analyzed the mRNA levels of several proinflammatory cytokines including TNF-α, interleukin-12p40 (IL-12p40), and interleukin-6 (IL-6) in BMDCs following different treatments. RT-PCR analysis showed that ATRA@CpG-NPs and ATRA@CpG-NPs + OVA significantly increased TNF-α, IL-12p40, and IL-6 in mRNA levels compared to PBS and free CpG with OVA treatments (Fig. 3e to g). These results suggest that the enhanced cellular uptake of ATRA@CpG-NPs contributes to their superior efficiency in activating DCs compared to free CpG.
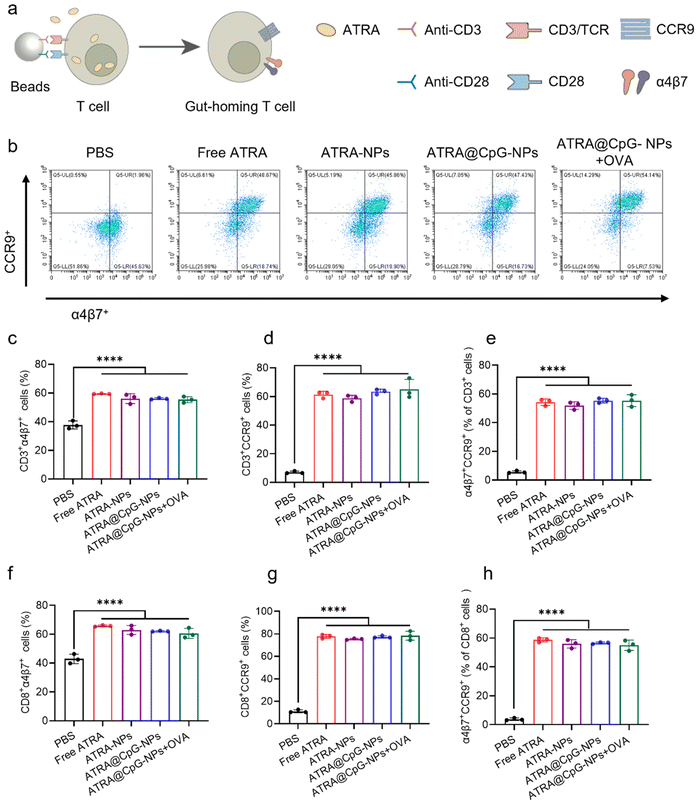
During antigen presentation of DCs to T cells, the presence of ATRA was found to induce the expression of CCR9 and α4β7 on activated T cells, thereby imparting gut tropism to the T cells. To simulate the antigen-presenting process of DCs to T cells, we isolated T cells from the spleen of female C57BL/6 mice and stimulated them using magnetic beads coated with anti-CD3/CD28 antibodies (Fig. 4a). During stimulation, free ATRA, ATRA-NPs, ATRA@CpG-NPs, or ATRA@CpG-NPs with OVA were introduced to T cells. It's worth noting that due to ATRA's poor water solubility, free ATRA was dissolved in DMSO and added to cells, while all other samples were dispersed in water. Utilizing ATRA-NPs improved the water solubility of free ATRA, eliminating the need for the toxic organic solvent. We then examined the expression of CCR9 and α4β7 receptors on activated T cells using flow cytometry. Remarkably, ATRA-NPs, ATRA@CpG-NPs, and ATRA@CpG-NPs with OVA all effectively induced the expression of CCR9 and α4β7 receptors on activated T cells, exhibiting similar efficacy compared to free ATRA dissolved in DMSO (Fig. 4b–e and S7†). The gut-homing of CD8+ T cells plays an important role in regulating colorectal tumor growth since CD8+ T cells are primary tumor-killing cells. We further analyzed CCR9+ and α4β7+ cells among CD3+CD8+ cells. As shown in Fig. 4f–h, ATRA@CpG-NPs greatly increased the population of CD3+CD8+CCR9+, CD3+CD8+α4β7+, and CD3+CD8+CCR9+α4β7+ cells compared to PBS treatment. The results demonstrated that ATRA within ATRA@CpG-NPs maintains its efficacy in inducing gut-homing receptors in T cells.
The ability of ATRA@CpG-NPs to induce gut-homing receptors on CD8+ T cells positions them as a promising mucosal adjuvant for cancer vaccines. To evaluate whether ATRA@CpG-NPs + OVA could activate antigen-specific T cells, we extracted CD8+ T cells from the spleen of OT-1 mice. These CD8+ T cells recognize OVA antigen.35,36 We treated these CD8+ T cells using ATRA@CpG-NPs with or without OVA and detected the content of IL-2 and IFN-γ in the supernatants. Cells treated with ATRA@CpG-NPs + OVA secrete significantly higher amounts of IL-2 and IFN-γ compared to those treated with ATRA@CpG-NPs and PBS treatment, suggesting that ATRA@CpG-NPs + OVA could activate antigen-specific CD8+ T cells (Fig. S8†).
We next tested the combination of OVA antigen and ATRA@CpG-NPs adjuvant as a therapeutic cancer vaccine to treat orthotopic colorectal cancer in mice. We used a mouse colorectal cancer cell line MC38 that was stably transfected to express OVA. To establish orthotopic colorectal tumors, we transplanted a small piece of subcutaneous OVA-MC38 tumor (3–4 mg) onto the cecum of C57BL/6 mice following a previously published method37 (Fig. 5a). Mice were vaccinated with ATRA@CpG-NPs through intramuscular injections on day 1 and 7 post-tumor inoculation. Free CpG with OVA and ATRA@CpG-NPs without OVA were used as controls to study the effect of the nanoparticle and antigen. Each mouse received 20 μg of OVA, 15 μg of CpG, and 70 μg of ATRA per dose. On the 6th day after the second vaccination, we euthanized mice and isolated the orthotopic tumors. As shown in Fig. 5b, c and S9,† ATRA@CpG-NPs with OVA induced a higher population of CD3+CD8+ T cells compared to PBS and free OVA + CpG treatment, suggesting that ATRA@CpG-NPs with OVA increase CD8+ T cell infiltration into orthotopic colorectal tumors. ATRA@CpG-NPs with OVA greatly inhibited orthotopic tumor growth as demonstrated by the reduced tumor size and weight compared to PBS treatment (Fig. 5d and e). In contrast, free CpG with OVA did not show any therapeutic effect, highlighting the crucial role of the nanoparticle in facilitating CpG uptake and ATRA in inducing T-cell homing to the gut. Even though ATRA@CpG-NPs without OVA could induce the expression of gut-homing receptors on T cells (Fig. 4), they are still ineffective in inhibiting tumor growth due to a lack of tumor specificity. Collectively, these data suggest that ATRA@CpG-NPs could serve as a potent mucosal adjuvant to activate the immune system, imprint antigen-specific T cells with gut-homing receptors, and effectively inhibit orthotopic colorectal tumor growth when combined with a tumor antigen.
ATRA is a natural metabolite of vitamin A. ATRA has been clinically approved for the treatment of acne and acute promyelocytic leukemia. The oral dose of ATRA is 10 mg per capsule, which is greatly higher than the potential dose needed for vaccination purposes. We first evaluated the cytotoxicity of ATRA@CpG-NPs in DC 2.4 cells. Our result showed that ATRA@CpG-NPs did not induce additional cytotoxicity compared to free CpG or free ATRA (Fig. S10†). To assess the in vivo toxicity of ATRA@CpG-NPs, we monitored the body weights of mice after vaccination during cancer treatment. Throughout the treatment period, we observed no significant changes in body weight in all groups (Fig. 5f). H&E staining of major organs, including heart, liver, spleen, lungs, and kidneys, showed no significant morphological changes (Fig. 5g), indicating the good safety profile of ATRA@CpG-NPs at therapeutic relevant doses in mice.
Conclusions
In this study, we employed a layer-by-layer assembly method to create nanoparticles capable of co-delivering ATRA and CpG. After optimizing the preparation conditions, ATRA@CpG-NPs were successfully synthesized with high ATRA and CpG loading. These nanoparticles efficiently transported CpG into cells, activated BMDCs, induced proinflammatory cytokine production, and imprinted activated T cells with gut-homing receptors CCR9 and α4β7. As a mucosal adjuvant, the administration of ATRA@CpG-NPs with OVA effectively inhibited orthotopic colorectal tumor growth in mice without inducing noticeable side effects. ATRA@CpG-NPs are also capable of loading more CpG by increasing an additional layer of PEI/CpG or achieving codelivery of CpG and OVA (Fig. S11 and S12†). The simple preparation, high loading capacity, and great efficacy of ATRA@CpG-NPs make them an attractive candidate as a mucosal adjuvant for diverse antigens targeting colorectal cancer and other infectious diseases in the gut.Author contributions
S. M. and X. L. conceived the idea, analyzed the data, and wrote the manuscript. S. M., W. L., Y. W., C. Y., S. L., J. L., X. C., Y. Z., H. H., H. Z. performed experiments. All authors contributed to the discussion and editing of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the National Natural Science Foundation of China (No. 22175188), and start-up funding from the Institute of Chemistry, Chinese Academy of Sciences.References
- R. L. Siegel, K. D. Miller, H. E. Fuchs and A. Jemal, CA Cancer J. Clin., 2022, 72, 7–33 CrossRef PubMed
.
- K. Palucka and J. Banchereau, Nat. Rev. Cancer, 2012, 12, 265–277 CrossRef CAS PubMed
.
- I. Mellman, G. Coukos and G. Dranoff, Nature, 2011, 480, 480–489 CrossRef CAS PubMed
.
- J. Banchereau and A. K. Palucka, Nat. Rev. Immunol., 2005, 5, 296–306 CrossRef CAS PubMed
.
- P. Sharma, S. Hu-Lieskovan, J. A. Wargo and A. Ribas, Cell, 2017, 168, 707–723 CrossRef CAS PubMed
.
- F. S. Hodi, S. J. O'Day, D. F. McDermott, R. W. Weber, J. A. Sosman, J. B. Haanen, R. Gonzalez, C. Robert, D. Schadendorf, J. C. Hassel, W. Akerley, A. J. M. van den Eertwegh, J. Lutzky, P. Lorigan, J. M. Vaubel, G. P. Linette, D. Hogg, C. H. Ottensmeier, C. Lebbé, C. Peschel, I. Quirt, J. I. Clark, J. D. Wolchok, J. S. Weber, J. Tian, M. J. Yellin, G. M. Nichol, A. Hoos and W. J. Urba, N. Engl. J. Med., 2011, 365, 687–696 Search PubMed
.
- L. Milling, Y. Zhang and D. J. Irvine, Adv. Drug Delivery Rev., 2017, 114, 79–101 CrossRef CAS PubMed
.
- M. Saxena, S. H. van der Burg, C. J. M. Melief and N. Bhardwaj, Nat. Rev. Cancer, 2021, 21, 360–378 CrossRef CAS PubMed
.
- T. Fan, M. Zhang, J. Yang, Z. Zhu, W. Cao and C. Dong, Signal Transduction Targeted Ther., 2023, 8, 450 CrossRef CAS PubMed
.
- B. Pulendran, P. S. Arunachalam and D. T. O'Hagan, Nat. Rev. Drug Discovery, 2021, 20, 454–475 CrossRef CAS PubMed
.
- I. Spadoni, G. Fornasa and M. Rescigno, Nat. Rev. Immunol., 2017, 17, 761–773 CrossRef CAS PubMed
.
- P. Czarnewski, S. Das, S. M. Parigi and E. J. Villablanca, Nutrients, 2017, 9, 68 CrossRef PubMed
.
- J. A. Hall, J. R. Grainger, S. P. Spencer and Y. Belkaid, Immunity, 2011, 35, 13–22 CrossRef CAS PubMed
.
- A. Bos, M. van Egmond and R. Mebius, Mucosal Immunol., 2022, 15, 562–572 CrossRef CAS PubMed
.
- M. Iwata, A. Hirakiyama, Y. Eshima, H. Kagechika, C. Kato and S. Y. Song, Immunity, 2004, 1, 527–538 CrossRef PubMed
.
- J. R. Mora, M. Iwata, B. Eksteen, S. Y. Song, T. Junt, B. Senman, K. L. Otipoby, A. Yokota, H. Takeuchi, P. Ricciardi-Castagnoli, K. Rajewsky, D. H. Adams and U. H. Von Andrian, Science, 2006, 314, 1157–1160 CrossRef CAS PubMed
.
- S. I. Hammerschmidt, M. Friedrichsen, J. Boelter, M. Lyszkiewicz, E. Kremmer, O. Pabst and R. Förster, J. Clin. Invest., 2011, 121, 3051–3061 CrossRef CAS PubMed
.
- G. Bakdash, L. T. C. Vogelpoel, T. M. M. Van Capel, M. L. Kapsenberg and E. C. De Jong, Mucosal Immunol., 2015, 8, 265–278 CrossRef CAS PubMed
.
- Y. Du, Y. Xia, Y. Zou, Y. Hu, J. Fu, J. Wu, X. D. Gao and G. Ma, ACS Nano, 2019, 13, 13809–13817 CrossRef CAS PubMed
.
- A. Riccomi, G. Piccaro, D. Christensen, C. Palma, P. Andersen and S. Vendetti, Front. Immunol., 2019, 10, 1–13 CrossRef PubMed
.
- G. Stary, A. Olive, A. F. Radovic-Moreno, D. Gondek, D. P. A. Alvarez, M. Basto, V. D. Perro, A. M. Vrbanac, J. Tager, J. A. Shi, O. C. Yethon, R. Farokhzad, M. N. Langer, U. Starnbach and H. Von Andrian, Science, 2015, 348, aaa8205 CrossRef PubMed
.
- J. E. Dahlman, C. Barnes, O. F. Khan, A. Thiriot, S. Jhunjunwala, T. E. Shaw, Y. Xing, H. B. Sager, G. Sahay, L. Speciner, A. Bader, R. L. Bogorad, H. Yin, T. Racie, Y. Dong, S. Jiang, D. Seedorf, A. Dave, K. S. Sandhu, M. J. Webber, T. Novobrantseva, V. M. Ruda, A. K. R. Lytton-Jean, C. G. Levins, B. Kalish, D. K. Mudge, M. Perez, L. Abezgauz, P. Dutta, L. Smith, K. Charisse, M. W. Kieran, K. Fitzgerald, M. Nahrendorf, D. Danino, R. M. Tuder, U. H. Von Andrian, A. Akinc, D. Panigrahy, A. Schroeder, V. Koteliansky, R. Langer and D. G. Anderson, Nat. Nanotechnol., 2014, 9, 648–655 CrossRef CAS PubMed
.
- S. J. Paston, V. A. Brentville, P. Symonds and L. G. Durrant, Front. Immunol., 2021, 12, 1–21 Search PubMed
.
- X. Zhong, G. Du, X. Wang, Y. Ou, H. Wang, Y. Zhu, X. Hao, Z. Xie, Y. Zhang, T. Gong, Z. Zhang and X. Sun, Small, 2022, 18, 2105530 CrossRef CAS PubMed
.
- Z. Poon, D. Chang, X. Zhao and P. T. Hammond, ACS Nano, 2011, 5, 4284–4292 CrossRef CAS PubMed
.
- Z. Poon, J. B. Lee, S. W. Morton and P. T. Hammond, Nano Lett., 2011, 11, 2096–2103 CrossRef CAS PubMed
.
- S. W. Morton, Z. Poon and P. T. Hammond, Biomaterials, 2013, 34, 5328–5335 CrossRef CAS PubMed
.
- P. T. Hammond, Nanomedicine, 2012, 7, 619–622 CrossRef CAS PubMed
.
- Z. J. Deng, S. W. Morton, E. Ben-Akiva, E. C. Dreaden, K. E. Shopsowitz and P. T. Hammond, ACS Nano, 2013, 7, 9571–9584 CrossRef CAS PubMed
.
- E. C. Dreaden, S. W. Morton, K. E. Shopsowitz, C. Jae-Hyeok, Z. J. Deng, N.-J. Cho and P. T. Hammond, ACS Nano, 2014, 8, 8374–8382 CrossRef CAS PubMed
.
- Y. Xu, S. Ma, J. Zhao, H. Chen, X. Si, Z. Huang, Z. Yu, W. Song, Z. Tang and X. Chen, Biomaterials, 2022, 284, 121489 CrossRef CAS PubMed
.
- Y. Hu, L. Lin, J. Chen, A. Maruyama, H. Tian and X. Chen, Biomaterials, 2022, 252, 120114 CrossRef PubMed
.
- A. M. Weiss, J. Ajit, T. J. Albin, N. Kapoor, S. Maroju, A. Berges, L. Pill, J. Fairman and A. P. Esser-Kahn, Sci. Rep., 2021, 11, 6267 CrossRef CAS PubMed
.
- N. I. Ho, L. G. M. H. In ‘t Veld, T. K. Raaijmakers and G. J. Adema, Front. Immunol., 2018, 9, 2874 CrossRef CAS PubMed
.
- E. C. Lerner, K. I. Woroniecka, V. M. D'Anniballe, D. S. Wilkinson, A. A. Mohan, S. J. Lorrey, J. Waibl-Polania, L. P. Wachsmuth, A. M. Miggelbrink, J. D. Jackson, X. Cui, J. A. Raj, W. H. Tomaszewski, S. L. Cook, J. H. Sampson, A. P. Patel, M. Khasraw, M. D. Gunn and P. E. Fecci, Nat. Cancer, 2023, 4, 1258–1272 CrossRef CAS PubMed
.
- D. E. Jæhger, M. L. Hübbe, M. K. Kræmer, G. Clergeaud, A. V. Olsen, C. Stavnsbjerg, M. N. Wiinholt, A. Kjær, J. R. Henriksen, A. E. Hansen and T. L. Andresen, Sci. Rep., 2021, 11, 19794 CrossRef PubMed
.
- D. Mucida, Y. Park, G. Kim, O. Turovskaya, I. Scott, M. Kronenberg and H. Cheroutre, Science, 2007, 317, 256–260 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4bm00026a |
| This journal is © The Royal Society of Chemistry 2024 |