Quantum effects in CH activation with [Cu2O2]2+ complexes†
Selin
Bac
 a and
Shaama
Mallikarjun Sharada
a and
Shaama
Mallikarjun Sharada
 *ab
*ab
aMork Family Department of Chemical Engineering and Materials Science, University of Southern California, Los Angeles, CA 90089, USA. E-mail: ssharada@usc.edu
bDepartment of Chemistry, University of Southern California, Los Angeles, CA 90089, USA
First published on 4th November 2024
Abstract
We investigate the mechanism of primary alkane CH bond activation with dioxo-dicopper ([Cu2O2]2+) complexes, which serve as model catalysts for enzymes capable of activating CH bonds under mild conditions. As large H/D kinetic isotope effects (KIEs) are observed in enzymes and their synthetic mimics, we employ density functional theory along with variational transition-state theory with multidimensional tunneling to estimate reaction rate coefficients. By systematically varying ligand electrophilicity and substrate chain length, we examine trends in rate coefficients and kinetic isotope effects for the two proposed CH activation pathways – one-step oxo-insertion and two-step radical recombination. Although larger tunneling transmission coefficients are obtained for the radical pathway, the oxo-insertion mechanism yields higher rate coefficients on account of lower activation barriers. The question of the preferred CH activation mechanism, however, remains open: excellent agreement is observed between the predicted and known experimental KIE results for the radical pathway, while calculated Hammett slopes for the oxo-insertion pathway closely mirror experiments.
1 Introduction
Efforts to streamline the conversion of natural gas into methanol as a cleaner alternative to traditional syngas-based methods have long been pursued, with the vision of establishing a methanol economy that could revolutionize energy and chemical industries.1 A promising avenue is the development of catalysts capable of selectively activating strong C–H bonds, mimicking the functionality of enzymes found in nature, specifically that of the enzyme particulate methane monooxygenase (pMMO).2–5 While significant progress has been made in elucidating the reactivity of oxygen-activated metal complexes, specifically iron and copper complexes, challenges persist in interpreting the preferred mechanisms of CH activation.6–10Two mechanisms have been proposed for CH activation reactions catalyzed by dioxo-dicopper ([Cu2O2]2+) complexes: (i) one-step oxo-insertion where the activation of a C–H bond occurs in a single concerted step alongside the insertion of an oxygen atom into the metal–carbon bond, and (ii) two-step radical recombination pathway, in which the first step involves homolytic cleavage of C–H bond (hydrogen atom transfer, HAT), leading to the formation of a methyl radical and subsequent recombination with a hydroxyl group.11–15 Computational studies, primarily based on density functional theory (DFT), have contributed valuable insights into these mechanisms but often face challenges in capturing spin-dependence and treating multireference character of these systems.16–18
Experimental studies probing the effects of substrate and catalyst variations on reaction kinetics can help validate computational predictions of the preferred reaction pathways. For instance, Hammett studies have shown enhanced reaction rates following substitutions of aromatic substrates with electron-donating groups.11,19,20 However, the observed experimental barriers in these studies often fall within a narrow range (<10 kJ mol−1), making it challenging for density functional approximations to resolve energy differences accurately. To overcome this limitation, we previously proposed a strategy that yields a wider range of barriers via multiple ligand substitutions in the catalyst instead.14 This method enables a semi-quantitative comparison between the proposed pathways and experimental Hammett plots,11,19–21 aiding in elucidating the true CH activation mechanism.
The kinetic isotope effect (KIE) serves as another means to probe mechanisms and contrast experiments with theory.22 CH functionalization reactions are characterized by large kinetic isotope effects originating in large part from hydrogen tunneling.22–30 The accurate quantification of tunneling effects is crucial for discerning the preferred mechanism, as tunneling can substantially contribute to catalytic activity. Traditional one-dimensional approximations are cost-effective31 but often are inaccurate due to their inability to capture the shape of the effective potential for tunneling and the phenomenon of “corner-cutting,” where the tunneling path deviates from the minimum energy path (MEP).32 One needs to employ approaches such as variational transition state theory with multidimensional tunneling (VTST/MT) to quantify these effects.33–35
Our objective is to contrast rate coefficients obtained using VTST/MT with conventional transition state theory (without tunneling) to examine the importance of multidimensional tunneling in both proposed pathways for CH activation with imidazole-substituted [Cu2O2]2+ complexes, and probe the sensitivity of reaction kinetics to systematic variations in the catalyst and substrate. Although the radical pathway yields significantly larger tunneling transmission coefficients than the oxo-insertion mechanism, the latter yields larger rate coefficients on account of lower activation barriers. Comparing experimental Hammett slopes with computations indicates preference for the oxo-insertion type mechanism with a partially cationic substrate in the transition state. On the other hand the magnitudes of kinetic isotope effects predicted for the radical pathway are in better agreement with experiments. Owing to these conflicting outcomes, the question of which of the two mechanisms is preferred remains unanswered. That being said, this work shows that multidimensional tunneling corrections are necessary to capture kinetics at low reaction temperatures.
2 Models and methods
2.1 Models
Along similar lines to previous studies by our group, the bis-(μ-oxo) isomeric form of the dicopper-dioxo active site is employed.14,36,37 We point the reader to our earlier work justifying this choice over the (multireference) peroxo active site as well as the choice of the singlet spin-potential energy surface.14 The Cu(III) centers are each bound to 2 N-donor imidazole ligands as shown in Fig. 1. We expand on prior work examining the impact of ligand electrophilicity on CH4 activation barriers to include their role in governing multidimensional tunneling and kinetic isotope effects. This is carried out by substituting the hydrogen atom at the carbon atom positioned between the two nitrogen atoms in imidazole with OCH3, CH3, CF3, or NO2. The Hammett parameters for para-substitution to benzoic acid, σp, are reported to be −0.268, −0.17, 0, 0.54, 0.78 for OCH3, CH3, H, CF3, and NO2, respectively.38,39 A negative (positive) Hammett parameter indicates electron-donating (withdrawing) character of the substituent. To examine the impact of alkane chain length on the barrier to primary CH activation and tunneling transmission coefficients, we examine reactants ranging from CH4 to C5H12.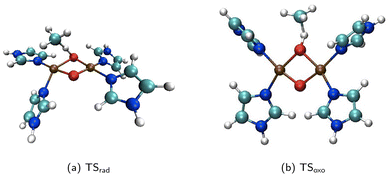 | ||
| Fig. 1 Transition structures for the first step of the (a) two-step radical recombination (TSrad) and (b) one-step oxo-insertion mechanisms (TSoxo). The active site is coordinated to imidazole (labeled ‘H’ in the ‘catalyst’ column of Table 1), and the substrate is CH4. Color scheme: cyan, carbon; dark blue, nitrogen; red, oxygen; brown, copper; and white, hydrogen. The visualizations are created using the visual molecular dynamics (VMD) software.40 | ||
2.2 Minimum energy paths
All density functional theory simulations are carried out using the ab initio quantum chemistry software, Q-Chem.41 Gas phase calculations are carried out using the ωB97X-D/def2-SVP42,43 level of theory and the def2-ECP effective core potential for Cu that includes scalar relativistic corrections.44 Natural bond orbital (NBO) analysis45,46 and energy decomposition analysis (EDA)47,48 are used to characterize initial and transition structures. The latter are determined using the freezing string method49,50 and Hessian-free optimization51 and verified using vibrational analysis. We calculate minimum energy paths (MEPs) in mass-weighted coordinates, referred to as intrinsic reaction coordinates (IRCs), employing the gradient-based predictor-corrector algorithm implemented in Q-Chem.52–54 IRCs are calculated to ensure that the TS indeed connects the intended reactant and product and to construct a reaction path for VTST/MT rate coefficient calculations. The maximum IRC step size is deliberately chosen to ensure a smooth path, set at 0.1 or 0.125 atomic units (a.u.). The convergence threshold for IRC (labeled “RPATH_TOL_DISPLACEMENT” in Q-Chem) is set between 0.001–0.005 a.u. The threshold is chosen in such a way that the vibrational analyses of the IRC end points yield either all real frequencies or only a small number of imaginary frequencies, all smaller than 100i cm−1.We perform vibrational analysis for all geometries constituting the IRC to compute free energies and couplings necessary for multidimensional (small-curvature) tunneling corrections, described below. As has been shown in prior work by our group,14,36 wavefunction stability analysis reveals that stationary points (except the initial state) typically suffer from spin instability. While a spin-correction scheme such as that proposed by Yamaguchi and coworkers55 is necessary to correct the instability and obtain spin-pure solutions, we limit ourselves to using unstable solutions. This is because spin-corrected energies are noisy and can lead to artificial peaks on the MEP (Fig. S1 of ESI†). Given the large number of systems examined in this study and the absence of a systematic protocol for the elimination of points yielding anomalous energies, we elect to use spin-unstable energies, while emphasizing that the reported rate coefficients do not represent true values. Trends in barriers remain largely unaffected by this choice (Table S1 of the ESI†).
2.3 Kinetic isotope effects
The kinetic isotope effect (KIE) is a unitless ratio that quantifies the change in reaction rate coefficient when one or more atoms in the reactants are replaced by its isotope. For reactions involving the transfer of H, the KIE is typically defined as the ratio of the rate coefficient for the reaction with the lighter isotope (protium) to the rate constant for the reaction with the heavier isotope (deuterium), expressed as: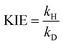 | (1) |
In experiments designed to elucidate mechanisms, the isotopic substitution of protium with deuterium is only carried out for the specific hydrogen atom(s) in the substrate participating in the reaction. Since Q-Chem only allows all hydrogen atoms to be substituted with deuterium in IRC calculations, our current approach involves perdeuteration. While this may limit the direct comparison of predicted KIEs with experimental results, we expect that trends arising from variations in ligands and chain lengths obtained using complete isotopic substitution will be similar to those observed when only the substrate hydrogen atoms are substituted. To precisely capture tunneling corrections, we perform IRC calculations and subsequent vibrational analyses for both the all-protium and all-deuterium systems.
2.4 Reaction rate coefficients
It is not guaranteed that the potential energies of the end-points of the protiated and deuterated IRCs are identical. It is also not necessary that the energies of the IRC end points exactly match those of the reactant and product minima employed to initiate the TS search. We therefore describe the free energy barrier, ΔG‡, as the difference in Gibbs free energies between the highest point on the free energy profile and the initial state free energy.To capture trends in kinetics across catalyst and substrate variations, we choose the initial (or reactant) state free energy as the sum of free energies of isolated catalyst and substrate. This is because energy decomposition analysis (EDA) reveals that the interaction energies between catalyst and substrate fragments in the reactant state described by a pre-association complex span a wide range (−12.9 to −48.0 kJ mol−1) when the alkane chain length is varied. Directly comparing rate coefficients by assigning the reactant as a pre-association complex will therefore not capture the overall kinetics of CH activation. For this reason, we employ what is akin to an apparent free energy barrier that is often reported for heterogeneous catalytic systems.
Rate coefficients obtained from conventional transition state theory, referred to simply as TST,56 are contrasted with variational transition state theory with multidimensional tunneling (VTST/MT).32 The key distinction between the two approaches, in this work, lies in the treatment of tunneling. Conventional TST does not account for quantum mechanical tunneling, and therefore, κ is unity. Within the VTST/MT framework, we calculate two types of tunneling transmission coefficients: (i) zero-curvature tunneling (ZCT, κZCT), which accounts for multidimensional tunneling but ignores the curvature of the reaction path,57–59 and (ii) small-curvature tunneling (SCT, κSCT), which incorporates the reaction path curvature, accounting for corner-cutting effects.60 The procedure is identical to that reported in the Pilgrim software.61 The approach employs splines to interpolate the effective potential energy along the minimum-energy path. Gaussian quadrature is utilized for integration. Our Python implementation is tested by showing that the transmission coefficients for test systems employed in Pilgrim are within 5% of the Pilgrim values.
We report rate coefficients (s−1) using the relation:
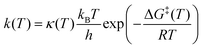 | (2) |
3 Results
Table 1 reports kinetics data obtained for the two proposed mechanisms by varying the ligands coordinated to the [Cu2O2]2+ active site and the chain length of the alkane substrate. To understand the impact of tunneling at both low and room temperatures, transmission coefficients, free energies, and kinetic isotope effects are reported at low (200 K) and ambient temperatures (300 K).| Mechanism | Catalyst | Substrate | ΔE‡ | 200 K | 300 K | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ΔG‡ | κ ZCT | κ SCT | KIESCT | ΔG‡ | κ ZCT | κ SCT | KIESCT | ||||
| Rad | H | C5H12 | 148.1 | 144.1 | 4.8 | 5.4 | 5.5 | 138.8 | 1.4 | 1.4 | 2.5 |
| C4H10 | 148.3 | 143.8 | 1.3 × 102 | 1.4 × 102 | 31.8 | 138.1 | 3.2 | 3.3 | 5.4 | ||
| C3H8 | 151.2 | 146.8 | 5.8 × 102 | 6.2 × 102 | 26.0 | 141.2 | 2.7 | 2.8 | 3.3 | ||
| C2H6 | 154.7 | 150.9 | 3.2 × 102 | 3.5 × 102 | 27.1 | 145.7 | 1.9 | 1.9 | 2.6 | ||
| CH4 | 174.0 | 171.9 | 2.1 × 104 | 2.4 × 104 | 30.4 | 168.4 | 7.4 | 7.8 | 1.4 | ||
| OCH3 | 171.7 | 168.7 | 5.1 × 102 | 5.8 × 102 | 45.7 | 164.6 | 2.7 | 2.8 | 2.4 | ||
| CH3 | 172.4 | 170.8 | 3.1 × 104 | 3.4 × 104 | 58.5 | 167.9 | 6.6 | 6.9 | 2.5 | ||
| CF3 | 163.9 | 161.9 | 9.2 × 102 | 1.2 × 103 | 43.5 | 158.7 | 3.4 | 3.6 | 2.0 | ||
| NO2 | 169.0 | 166.8 | 7.8 × 102 | 9.1 × 102 | 36.7 | 163.5 | 3.1 | 3.2 | 1.7 | ||
| Oxo | H | C5H12 | 44.0 | 37.9 | 3.4 | 4.4 | 8.4 | 30.5 | 1.6 | 1.8 | 3.4 |
| C4H10 | 47.2 | 40.6 | 3.7 | 4.6 | 9.2 | 32.9 | 1.7 | 1.9 | 3.7 | ||
| C3H8 | 52.1 | 46.0 | 4.4 | 5.6 | 8.1 | 38.5 | 1.8 | 2.0 | 3.1 | ||
| C2H6 | 54.2 | 49.0 | 4.0 | 5.1 | 7.3 | 42.2 | 1.7 | 1.9 | 2.5 | ||
| CH4 | 95.6 | 91.3 | 8.2 | 10.7 | 5.2 | 85.6 | 2.2 | 2.4 | 1.7 | ||
| OCH3 | 98.0 | 92.8 | 9.7 | 12.6 | 8.2 | 86.3 | 2.3 | 2.5 | 2.4 | ||
| CH3 | 105.4 | 102.5 | 10.7 | 13.9 | 2.3 | 98.1 | 2.4 | 2.6 | 0.8 | ||
| CF3 | 77.2 | 72.9 | 7.2 | 9.1 | 5.0 | 67.4 | 2.2 | 2.4 | 1.9 | ||
| NO2 | 66.0 | 62.0 | 4.8 | 5.9 | 4.5 | 56.8 | 2.0 | 2.1 | 1.8 | ||
3.1 Radical recombination and oxo-insertion mechanisms
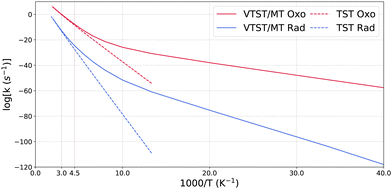
Fig. 1 shows the transition structures corresponding to the oxo-insertion (‘oxo’) mechanism and the first, rate-limiting step of the radical recombination (‘rad’) mechanism for CH4 activation with a complex consisting of four imidazole N-donor ligands (catalyst labeled ‘H’ in Table 1). In the oxo-insertion pathway, methanol is formed in a single step, with the transition state (TSoxo) involving simultaneous Cu–O and C–H cleavage alongside C–O and O–H bond formation. TSoxo exhibits a bent C–H–O configuration (122°) and is associated with an apparent barrier (ΔE‡) of 95.8 kJ mol−1. The radical pathway encompasses a two-step process, with the rate-limiting step involving CH cleavage to form radical species, followed by a recombination step to produce methanol. The first step (TSrad) displays a nearly linear C–H–O configuration (171°) with an associated barrier of 174 kJ mol−1. The imaginary frequencies corresponding to the reaction coordinate in the TSs are 879i and 1981i cm−1 for TSoxo and TSrad, respectively. These values are consistent with our earlier study where we report 1075i and 1858i cm−1 for oxo and rad TSs, respectively.14 Natural bond orbital (NBO) analysis reveals a difference in the total natural charge on CH4 between the transition states and initial states of +0.59 and +0.11 for the oxo and rad mechanisms, respectively, in close agreement with previous work (+0.65 and +0.18).14 On account of its charged character, the oxo TS is therefore expected to be more sensitive to the electrophilicity of ligands coordinated to the metal center.Fig. 2 presents the Arrhenius plot of temperature-dependence of the rate coefficients for the two mechanisms of the imidazole-substituted catalyst with CH4 as the reactant. In the temperature range of 25 K to 500 K, the rate coefficients for oxo are higher than rad. Quantum mechanical tunneling dominates the oxo and rad pathways below 220 K and 330 K, respectively, based on the sharp changes in slopes in Fig. 2. The rad pathway possesses tunneling transmission coefficients that are larger than oxo, with κSCT nearly 3 orders of magnitude larger at 200 K.
3.2 Chain length effects
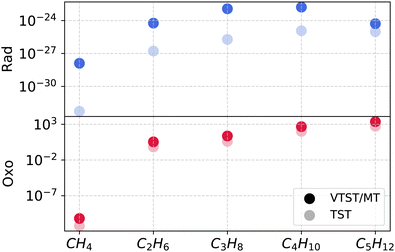
Fig. 3 depicts the alkane chain length dependence of the VTST/MT and TST rate coefficients at 200 K for the two mechanisms. In the case of the oxo-insertion mechanism, barriers drop steeply from CH4 to C2H6, followed by a gradual decline. ZCT and SCT coefficients vary less strongly with chain length, with C2–C5 values about half of those obtained for CH4 activation. As a result, the TST and VTST/MT rate coefficients are close to each other for C2–C5 and higher than the value obtained for CH4.TST rate coefficients are smaller than VTST/MT in the rad mechanism because multidimensional tunneling plays a more important role when compared to the oxo mechanism. There is a sharp decline in the barrier from CH4 to C2H6 followed by a more gradual decrease from C2H6 to C5H12, reflected in the TST rate coefficients in Fig. 3. The trend in VTST/MT rate coefficients, however, is not monotonic because the increase associated with a drop in barrier is partially offset by the general decrease in tunneling transmission coefficients from C1 to C5.
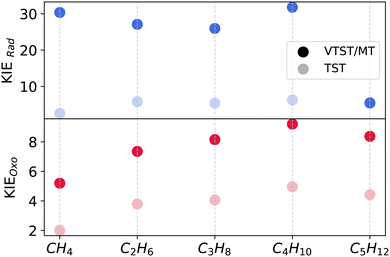
We explore the trends in kinetic isotope effects for both mechanisms in Fig. 4 across varying substrate chain lengths at 200 K. Owing to larger contributions from multidimensional tunneling, the KIE values for the rad mechanism are consistently larger than those for the oxo mechanism. The TST-based KIEs are much smaller than the VTST/MT KIEs. While there are no significant differences in KIEs observed across C1–C4 substrates, the KIE drops to 5.1 in C5 due to smaller tunneling contributions. For the oxo mechanism, KIE values increase from C1 to C4, reaching their highest value for C4, and then show a slight decrease for C5. Unlike the rad mechanism, there is no sharp change in KIE between C4 and C5.
3.3 Ligand effects
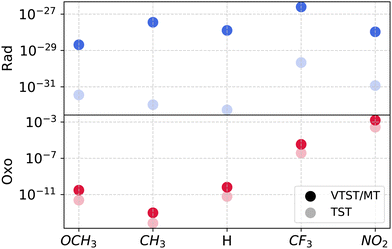
We report the dependence of rate coefficients on ligand electrophilicity in Fig. 5. Oxo barriers exhibit pronounced ligand dependence, evidenced by a large ZPE-corrected apparent activation barrier range (39.3 kJ mol−1). With the exception of OCH3, increasing electron-withdrawing character, quantified by the Hammett parameter for para- substitution σp, leads to lower barriers and therefore higher rate coefficients. We attribute the decrease in the barrier to the stabilization of the transition state by the electron-withdrawing ligand via charge transfer interactions.14 The tunneling transmission coefficients across these ligand-substituted systems all lie within an order of magnitude of each other and are smaller than 15 at 200 K. As a result, there are very small differences between TST and VTST/MT rate coefficients with varying ligand electrophilicity. We note that (with the exception of OCH3), there is a monotonic decreases in both κZCT and κSCT with increasing substituent electron-withdrawing character, i.e., CH3 > H > CF3 > NO2.As reported in an earlier study, rad is less sensitive to the ability of a ligand to push/pull electron density, and therefore, the barriers exhibit a narrower range (10.0 kJ mol−1). The TST rate coefficients, therefore, vary by less than 2 orders of magnitude across the ligand-substituted systems. VTST/MT rate coefficients are higher than those obtained from TST, owing to large contributions from κZCT/SCT.
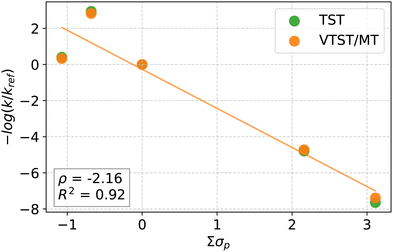
Fig. 6 depicts the Hammett plots for the oxo-insertion pathway using the σp Hammett parameter, with the σ values on the x-axis scaled by a factor of four to reflect the number of ligand substitutions.14 Due to smaller tunneling coefficients observed for the oxo-insertion mechanism, the linear fit to TST and VTST/MT data are similar. In contrast, shown in Fig. S6 of the ESI,† the radical pathway shows no dependence on ligands, resulting in poor linear fit for VTST/MT (R2 < 0.03) and only a modest improvement for TST (R2 = 0.5, ρ = −0.56).
Despite differences in the choice of ligands and substrate, the slope of the Hammett plot for oxo-insertion, ρ = −2.16, is consistent with an electrophilic attack on the substrate reported in previous experimental studies (examining activation of weaker CH bonds) as well as prior work in our group.11,14,19,20 If we employ the meta-substituted σm Hammett parameter, the slope becomes −3.12 with an R2 value of 0.96 (Fig. S5, ESI†). With σm as the electrophilicity descriptor, OCH3 is electron-withdrawing (σm,OCH3 = 0.12) and no longer an outlier, and the fit is improved.
The impact of ligand electrophilicity on KIEs for oxo and rad mechanisms are shown in Fig. 7. Due to larger tunneling contributions, we observe larger VTST/MT KIEs for the rad pathway compared to oxo across all ligands. There are no discernible trends in oxo KIEs with variation in ligand electrophilicity.
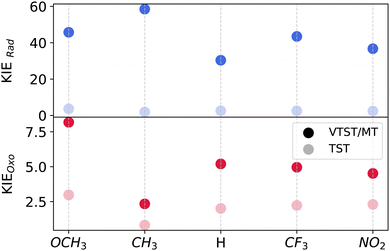 | ||
| Fig. 7 A comparison of VTST/MT (with κSCT) and conventional TST-based kinetic isotope effects calculated at 200 K for oxo and rad mechanisms across different ligand substitutions. | ||
4 Discussion
Our examination of C–H activation by [Cu2O2]2+ complexes reveals that the singlet oxo-insertion pathway is energetically more favorable than the singlet radical pathway despite considerably larger tunneling coefficients and KIEs observed with the latter. We contrast these findings with experimental literature examining the activity of dioxo-dicopper complexes towards activating various CH bonds. A study of intramolecular hydroxylation of the benzyl CH bond with a [Cu(II)2–O2]2+ center reported a substantial KIE of 35.4 at 193 K.63 Similarly, a KIE of 3164 was observed for the oxidation of 10-methyl-9,10-dihydroacridine (ArcH2) at 148 K. A computational study by Kim and co-workers examining the rad mechanism reports a VTST/MT-based KIE of 28 (277 K) for CH4 hydroxylation with the [Cu2O2]2+ active site in which each Cu(III) is bound to three N-donor amine groups. All these values closely resemble our VTST/MT-based KIE of 30.6 at 200 K for the rad mechanism, with the oxo yielding KIE = 5.2.Although KIEs indicate a possible rad pathway, barriers and Hammett plots conclude that the oxo pathway may be preferred. Applying spin- and zero-point corrections, the ΔE‡ value for CH4 activation with imidazole-bound [Cu2O2]2+viaoxo is 25.2 kJ mol−1, significantly lower than the rad barrier of 160.0 kJ mol−1. While experimental Hammett curves are typically generated by varying the electrophilicities of substituents to an aromatic reactant, the resulting ranges in activation barriers are typically too narrow to be meaningfully interpreted using computations. Since each Cu(III) is coordinated to two N-donor ligands in this work, we can quadruple substituent effects by replacing an H- in the imidazole N-donor with an electron-donating or withdrawing group. The resulting range of zero point-corrected barriers for oxo is 39.4 kJ mol−1. A reasonable linear fit and a finite, negative slope are observed only for the oxo-insertion pathway and not the radical pathway (Section S3 of the ESI†). Although the magnitudes of the slopes are also in agreement with experiment (ρ from −2.2 at 148 K to −1.48 at 193 K), this may be fortuitous because experimental studies are carried out for substrates containing weaker CH bonds than CH4 and the Hammett plots are constructed using σp+ values rather than σp.20,21,63
Owing to these contrasting findings and differences in choices of substrates/catalysts in experiments and theory, it is difficult to draw conclusions regarding the preferred mechanism of CH activation. One possibility that we have not yet explored is the fact that the rad barrier in the triplet state is lower in energy than the singlet rad and singlet oxo, reported in an earlier study by our group.14 In other words, a two-state-reactivity (TSR) scenario, in which ligand-dependent crossover occurs from the singlet to the triplet state, remains to be explored.65–67 For instance, a study by Shaik and coworkers, of the rad mechanism-based CH activation with nonheme Fe(IV)-oxo bound to tetramethylcyclam ligands, showed that the unexpected faster kinetics with strong electron-donating ligands originates in a large probability of spin-inversion between the triplet and quintet states as well as enhanced tunneling.68 Such a study for the dicopper system is hampered by the fact that spin-pure singlet states (unlike triplets or quintets) are difficult to determine on account of spin contamination in DFT, making it difficult to accurately identify crossing points with higher spin-states.
To explain the differences in mechanistic conclusions arising from comparing barriers, KIEs, and Hammett slopes, another testable hypothesis is that the distinction between the two mechanisms becomes blurry when the size of the substrate grows. Although rigorous testing of this hypothesis is a topic for future work, with an increase in alkane chain length in this study, we observe small changes in the ‘radical character’ of the TS. The total charge on the alkane fragment in the rad TS, calculated using NBO, increases by 55% from 0.11 (CH4) to 0.17 (C5H12), indicating an increase in cationic character with increasing chain length. In contrast, the alkane fragment charge in the oxo TS increases only by 15% from 0.59 (CH4) to 0.68 (C5H12). The imaginary frequency associated with the rad reaction coordinate decreases from 1981 cm−1 (CH4) to 1733 cm−1 (C5H12), which in part explains the drop in tunneling transmission coefficients and KIEs with increasing chain length at 200 K (Table 1). To resolve the question of preferred mechanism therefore, we need a more extensive analysis of substrate sensitivity than what is reported in this work.
Our examination of ligand and chain length dependence is inspired by studies of CH activation with nonheme Fe(IV)-oxo complexes by Shaik and coworkers.23,68 In addition to TSR, they show that electron-donating ligands enhance tunneling transmission coefficients. If we assume that the substituents behave as if they are meta- substituted, the κ values for oxo decrease monotonically from CH3 to NO2. However, all transmission coefficients are within an order of magnitude of each other, and therefore the substituent effect on tunneling with oxo is not significant enough to impact rate coefficients. While a general decrease is also noted for the rad mechanism, with the CH3-bound catalyst yielding κSCT that is two orders of magnitude larger than the NO2-bound one, the decrease is not monotonic. The decrease in oxo and rad barriers with increasing chain length also leads to a decrease in tunneling transmission coefficients. Contrary to findings by Shaik and coworkers suggesting a volcano-type relationship between KIE and CH bond dissociation energy (BDE),23 we do not find a clear correlation for either reaction pathway when the alkane chain length is varied. We note however that the range of homolytic BDEs is narrow in our work (∼18.8 kJ mol−1) compared to theirs (∼83.7 kJ mol−1).23
Tunneling emerges as the primary factor driving the substantial kinetic isotope effects observed in C–H activation catalyzed by [Cu2O2]2+ complexes. The magnitudes of KIEs and their trends with chain length and ligand substitution exhibit at least a two-fold increase when zero- or small-curvature tunneling is incorporated (Fig. S2 of ESI†). In addition, overlapping KIEs between SCT and ZCT-based rate coefficient calculations indicate that corner-cutting does not significantly impact most systems. Therefore, even though prior computational studies of CH activation with [Cu2O2]2+ complexes estimate κSCT's,22,27 the zero-curvature approximation is expected to suffice. Mandal and Shaik23 also demonstrated, for CH activation with nonheme Fe(IV)-oxo complexes, that a simple Eckart tunneling model yields KIE values in good agreement with experiments and multidimensional models. The parity plot in Fig. S3 of the ESI† also illustrates this, with the exception of systems in which κSCT values exceed 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000.
000.
5 Conclusions
By examining the singlet potential energy surface of CH activation with [Cu2O2]2+ complexes, this study aims to uncover the dependence of the rate coefficients, tunneling transmission coefficients, and kinetic isotope effects of two proposed mechanisms – one-step oxo-insertion and two-step radical recombination – on catalyst electrophilicity and substrate CH bond strength. To this end, we employ DFT simulations and contrast rate coefficients and KIEs obtained using conventional TST without tunneling and VTST with multidimensional tunneling. We find that the use of multidimensional tunneling approximations is necessary to capture isotope effects in these systems, although the role of corner-cutting appears to be small. The barriers for the oxo-insertion pathway are always lower than those for radical recombination. However, while calculated Hammett slopes are in agreement with experiment for the oxo pathway, the KIEs for the rad pathway align better with experimentally observed isotope effects. Future work includes exploration of two-state reactivity and expansion in substrate scope to address these conflicting mechanistic conclusions.Author contributions
SB carried out the computational work described in this study. SMS led the design of simulations and both authors contributed to manuscript writing.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Award No. DE-SC0021417 and the USC Viterbi School of Engineering's Ershaghi Center for Energy Transition (E-CET). The authors acknowledge computational resources and support from USC's Center for Advanced Research Computing (CARC) and the National Energy Research Scientific Computing Center (NERSC). The authors thank Kaustubh Rane (USC) for images of the transition structures.Notes and references
- G. A. Olah, A. Goeppert and G. S. Prakash, Beyond oil and gas: The methanol economy, John Wiley & Sons, 2011 Search PubMed.
- C. E. Elwell, N. L. Gagnon, B. D. Neisen, D. Dhar, A. D. Spaeth, G. M. Yee and W. B. Tolman, Chem. Rev., 2017, 117, 2059–2107 CrossRef CAS PubMed.
- E. A. Lewis and W. B. Tolman, Chem. Rev., 2004, 104, 1047–1076 CrossRef CAS PubMed.
- L. M. Mirica, X. Ottenwaelder and T. D. P. Stack, Chem. Rev., 2004, 104, 1013–1046 CrossRef CAS PubMed.
- N. Kitajima and Y. Moro-oka, Chem. Rev., 1994, 94, 737–757 CrossRef CAS.
- P. E. Siegbahn and R. H. Crabtree, J. Am. Chem. Soc., 1997, 119, 3103–3113 CrossRef CAS.
- C. Krebs, D. Galonic Fujimori, C. T. Walsh and J. M. Bollinger Jr, Acc. Chem. Res., 2007, 40, 484–492 CrossRef CAS PubMed.
- W. Nam, Acc. Chem. Res., 2015, 48, 2415–2423 CrossRef CAS PubMed.
- C. J. Cramer, M. Włoch, P. Piecuch, C. Puzzarini and L. Gagliardi, J. Phys. Chem. A, 2006, 110, 1991–2004 CrossRef CAS PubMed.
- C. J. Cramer, A. Kinal, M. Włoch, P. Piecuch and L. Gagliardi, J. Phys. Chem. A, 2006, 110, 11557–11568 CrossRef CAS PubMed.
- S. Mahapatra, J. A. Halfen and W. B. Tolman, J. Am. Chem. Soc., 1996, 118, 11575–11586 CrossRef CAS.
- V. C.-C. Wang, S. Maji, P. P.-Y. Chen, H. K. Lee, S. S.-F. Yu and S. I. Chan, Chem. Rev., 2017, 117, 8574–8621 CrossRef CAS PubMed.
- V. W. Bowry and K. Ingold, J. Am. Chem. Soc., 1991, 113, 5699–5707 CrossRef CAS.
- Z. Lan and S. M. Sharada, Phys. Chem. Chem. Phys., 2018, 20, 25602–25614 RSC.
- Y. Shiota, G. Juhász and K. Yoshizawa, Inorg. Chem., 2013, 52, 7907–7917 CrossRef CAS PubMed.
- K. Yoshizawa and Y. Shiota, J. Am. Chem. Soc., 2006, 128, 9873–9881 CrossRef CAS PubMed.
- Y. Shiota and K. Yoshizawa, Inorg. Chem., 2009, 48, 838–845 CrossRef CAS PubMed.
- S. Ye and F. Neese, Inorg. Chem., 2010, 49, 772–774 CrossRef CAS PubMed.
- L. M. Mirica, M. Vance, D. J. Rudd, B. Hedman, K. O. Hodgson, E. I. Solomon and T. D. P. Stack, Science, 2005, 308, 1890–1892 CrossRef CAS PubMed.
- C. Citek, C. T. Lyons, E. C. Wasinger and T. D. P. Stack, Nat. Chem., 2012, 4, 317–322 CrossRef CAS PubMed.
- K. D. Karlin, M. S. Nasir, B. I. Cohen, R. W. Cruse, S. Kaderli and A. D. Zuberbuehler, J. Am. Chem. Soc., 1994, 116, 1324–1336 CrossRef CAS.
- Y. Kim, B. K. Mai and S. Park, JBIC, J. Biol. Inorg. Chem., 2017, 22, 321–338 CrossRef CAS PubMed.
- D. Mandal and S. Shaik, J. Am. Chem. Soc., 2016, 138, 2094–2097 CrossRef CAS PubMed.
- J. Kaizer, E. J. Klinker, N. Y. Oh, J.-U. Rohde, W. J. Song, A. Stubna, J. Kim, E. Münck, W. Nam and L. Que, J. Am. Chem. Soc., 2004, 126, 472–473 CrossRef CAS PubMed.
- M. P. Meyer and J. P. Klinman, Chem. Phys., 2005, 319, 283–296 CrossRef CAS PubMed.
- E. Derat, D. Kumar, H. Hirao and S. Shaik, J. Am. Chem. Soc., 2006, 128, 473–484 CrossRef CAS PubMed.
- K. Park, Y. Pak and Y. Kim, J. Am. Chem. Soc., 2012, 134, 3524–3531 CrossRef CAS PubMed.
- C. Citek, S. Herres-Pawlis and T. D. P. Stack, Acc. Chem. Res., 2015, 48, 2424–2433 CrossRef CAS PubMed.
- J. P. Klinman, Biochim. Biophys. Acta, Bioenerg., 2006, 1757, 981–987 CrossRef CAS PubMed.
- J. P. Layfield and S. Hammes-Schiffer, Chem. Rev., 2014, 114, 3466–3494 CrossRef CAS PubMed.
- R. T. Skodje and D. G. Truhlar, J. Phys. Chem., 1981, 85, 624–628 CrossRef CAS.
- A. Fernandez-Ramos, B. A. Ellingson, B. C. Garrett and D. G. Truhlar, Rev. Comput. Chem., 2007, 23, 125 CAS.
- D. G. Truhlar and B. C. Garrett, Annu. Rev. Phys. Chem., 1984, 35, 159–189 CrossRef CAS.
- D. G. Truhlar and B. C. Garrett, Acc. Chem. Res., 1980, 13, 440–448 CrossRef CAS.
- J. L. Bao and D. G. Truhlar, Chem. Soc. Rev., 2017, 46, 7548–7596 RSC.
- Z. Lan and S. M. Sharada, Phys. Chem. Chem. Phys., 2020, 22, 7155–7159 RSC.
- Z. Lan and S. M. Sharada, Phys. Chem. Chem. Phys., 2021, 23, 15543–15556 RSC.
- L. P. Hammett, J. Am. Chem. Soc., 1937, 59, 96–103 CrossRef CAS.
- C. Hansch, A. Leo and R. Taft, Chem. Rev., 1991, 91, 165–195 CrossRef CAS.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics, 1996, 14, 33–38 CrossRef CAS PubMed.
- E. Epifanovsky, A. T. B. Gilbert, X. Feng, J. Lee, Y. Mao, N. Mardirossian, P. Pokhilko, A. F. White, M. P. Coons, A. L. Dempwolff, Z. Gan, D. Hait, P. R. Horn, L. D. Jacobson, I. Kaliman, J. Kussmann, A. W. Lange, K. U. Lao, D. S. Levine, J. Liu, S. C. McKenzie, A. F. Morrison, K. D. Nanda, F. Plasser, D. R. Rehn, M. L. Vidal, Z.-Q. You, Y. Zhu, B. Alam, B. J. Albrecht, A. Aldossary, E. Alguire, J. H. Andersen, V. Athavale, D. Barton, K. Begam, A. Behn, N. Bellonzi, Y. A. Bernard, E. J. Berquist, H. G. A. Burton, A. Carreras, K. Carter-Fenk, R. Chakraborty, A. D. Chien, K. D. Closser, V. Cofer-Shabica, S. Dasgupta, M. de Wergifosse, J. Deng, M. Diedenhofen, H. Do, S. Ehlert, P.-T. Fang, S. Fatehi, Q. Feng, T. Friedhoff, J. Gayvert, Q. Ge, G. Gidofalvi, M. Goldey, J. Gomes, C. E. Gonzalez-Espinoza, S. Gulania, A. O. Gunina, M. W. D. Hanson-Heine, P. H. P. Harbach, A. Hauser, M. F. Herbst, M. Hernandez Vera, M. Hodecker, Z. C. Holden, S. Houck, X. Huang, K. Hui, B. C. Huynh, M. Ivanov, A. Jasz, H. Ji, H. Jiang, B. Kaduk, S. Kahler, K. Khistyaev, J. Kim, G. Kis, P. Klunzinger, Z. Koczor-Benda, J. H. Koh, D. Kosenkov, L. Koulias, T. Kowalczyk, C. M. Krauter, K. Kue, A. Kunitsa, T. Kus, I. Ladjanszki, A. Landau, K. V. Lawler, D. Lefrancois, S. Lehtola, R. R. Li, Y.-P. Li, J. Liang, M. Liebenthal, H.-H. Lin, Y.-S. Lin, F. Liu, K.-Y. Liu, M. Loipersberger, A. Luenser, A. Manjanath, P. Manohar, E. Mansoor, S. F. Manzer, S.-P. Mao, A. V. Marenich, T. Markovich, S. Mason, S. A. Maurer, P. F. McLaughlin, M. F. S. J. Menger, J.-M. Mewes, S. A. Mewes, P. Morgante, J. W. Mullinax, K. J. Oosterbaan, G. Paran, A. C. Paul, S. K. Paul, F. Pavosevic, Z. Pei, S. Prager, E. I. Proynov, A. Rak, E. Ramos-Cordoba, B. Rana, A. E. Rask, A. Rettig, R. M. Richard, F. Rob, E. Rossomme, T. Scheele, M. Scheurer, M. Schneider, N. Sergueev, S. Mallikarjun Sharada, W. Skomorowski, D. W. Small, C. J. Stein, Y.-C. Su, E. J. Sundstrom, Z. Tao, J. Thirman, G. J. Tornai, T. Tsuchimochi, N. M. Tubman, S. P. Veccham, O. Vydrov, J. Wenzel, J. Witte, A. Yamada, K. Yao, S. Yeganeh, S. R. Yost, A. Zech, I. Y. Zhang, X. Zhang, Y. Zhang, D. Zuev, A. Aspuru-Guzik, A. T. Bell, N. A. Besley, K. B. Bravaya, B. R. Brooks, D. Casanova, J.-D. Chai, S. Coriani, C. J. Cramer, G. Cserey, A. E. DePrince, R. A. DiStasio, A. Dreuw, B. D. Dunietz, T. R. Furlani, W. A. Goddard, S. Hammes-Schiffer, T. Head-Gordon, W. J. Hehre, C.-P. Hsu, T.-C. Jagau, Y. Jung, A. Klamt, J. Kong, D. S. Lambrecht, W. Liang, N. J. Mayhall, C. W. McCurdy, J. B. Neaton, C. Ochsenfeld, J. A. Parkhill, R. Peverati, V. A. Rassolov, Y. Shao, L. V. Slipchenko, T. Stauch, R. P. Steele, J. E. Subotnik, A. J. W. Thom, A. Tkatchenko, D. G. Truhlar, T. Van Voorhis, T. A. Wesolowski, K. B. Whaley, H. L. Woodcock, P. M. Zimmerman, S. Faraji, P. M. W. Gill, M. Head-Gordon, J. M. Herbert and A. I. Krylov, J. Chem. Phys., 2021, 155, 084801 CrossRef CAS PubMed.
- J.-D. Chai and M. Head-Gordon, Phys. Chem. Chem. Phys., 2008, 10, 6615–6620 RSC.
- J.-D. Chai and M. Head-Gordon, J. Chem. Phys., 2008, 128, 084106 CrossRef PubMed.
- F. Weigend and R. Ahlrichs, Phys. Chem. Chem. Phys., 2005, 7, 3297–3305 RSC.
- J. P. Foster and F. Weinhold, J. Am. Chem. Soc., 1980, 102, 7211–7218 CrossRef CAS.
- G. Frenking, K. Wichmann, N. Fröhlich, C. Loschen, M. Lein, J. Frunzke and V. M. Rayón, Coord. Chem. Rev., 2003, 238, 55–82 CrossRef.
- P. R. Horn, Y. Mao and M. Head-Gordon, Phys. Chem. Chem. Phys., 2016, 18, 23067–23079 RSC.
- Y. Mao, D. S. Levine, M. Loipersberger, P. R. Horn and M. Head-Gordon, Phys. Chem. Chem. Phys., 2020, 22, 12867–12885 RSC.
- A. Behn, P. M. Zimmerman, A. T. Bell and M. Head-Gordon, J. Chem. Phys., 2011, 135, 224108 CrossRef PubMed.
- S. M. Sharada, P. M. Zimmerman, A. T. Bell and M. Head-Gordon, J. Chem. Theory Comput., 2012, 8, 5166–5174 CrossRef PubMed.
- S. M. Sharada, A. T. Bell and M. Head-Gordon, J. Chem. Phys., 2014, 140, 164115 CrossRef PubMed.
- K. Fukui, J. Phys. Chem., 1970, 74, 4161–4163 CrossRef CAS.
- K. Ishida, K. Morokuma and A. Komornicki, J. Chem. Phys., 1977, 66, 2153–2156 CrossRef CAS.
- M. W. Schmidt, M. S. Gordon and M. Dupuis, J. Am. Chem. Soc., 1985, 107, 2585–2589 CrossRef CAS.
- K. Yamaguchi, F. Jensen, A. Dorigo and K. Houk, Chem. Phys. Lett., 1988, 149, 537–542 CrossRef CAS.
- D. G. Truhlar, B. C. Garrett and S. J. Klippenstein, J. Phys. Chem., 1996, 100, 12771–12800 CrossRef CAS.
- A. Kuppermann and D. G. Truhlar, J. Am. Chem. Soc., 1971, 93, 1840–1851 CrossRef CAS.
- B. Garrett, D. Truhlar, R. Grev and A. Magnuson, J. Phys. Chem., 1983, 87, 4554 Search PubMed.
- B. C. Garrett, D. G. Truhlar, R. S. Grev and A. W. Magnuson, J. Phys. Chem., 1980, 84, 1730–1748 CrossRef CAS.
- Y. P. Liu, G. C. Lynch, T. N. Truong, D. H. Lu, D. G. Truhlar and B. C. Garrett, J. Am. Chem. Soc., 1993, 115, 2408–2415 CrossRef CAS.
- D. Ferro-Costas, D. G. Truhlar and A. Fernández-Ramos, Comput. Phys. Commun., 2020, 256, 107457 CrossRef CAS.
- S. Bac, S. J. Quiton, K. J. Kron, J. Chae, U. Mitra and S. M. Sharada, J. Chem. Phys., 2022, 156, 184119 CrossRef CAS PubMed.
- S. Itoh, M. Taki, H. Nakao, P. L. Holland, W. B. Tolman, L. Que, Jr and S. Fukuzumi, Angew. Chem., Int. Ed., 2000, 39, 398–400 CrossRef CAS PubMed.
- C. Citek, B.-L. Lin, T. E. Phelps, E. C. Wasinger and T. D. P. Stack, J. Am. Chem. Soc., 2014, 136, 14405–14408 CrossRef CAS PubMed.
- D. Schröder, S. Shaik and H. Schwarz, Acc. Chem. Res., 2000, 33, 139–145 CrossRef PubMed.
- S. Shaik, S. P. De Visser, F. Ogliaro, H. Schwarz and D. Schröder, Curr. Opin. Chem. Biol., 2002, 6, 556–567 CrossRef CAS PubMed.
- B. Yang, L. Gagliardi and D. G. Truhlar, Phys. Chem. Chem. Phys., 2018, 20, 4129–4136 RSC.
- D. Mandal, R. Ramanan, D. Usharani, D. Janardanan, B. Wang and S. Shaik, J. Am. Chem. Soc., 2015, 137, 722–733 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cp02929a |
| This journal is © the Owner Societies 2024 |