 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Intracellular microbial rhodopsin-based optogenetics to control metabolism and cell signaling†
Anastasiia D.
Vlasova
 a,
Siarhei M.
Bukhalovich
a,
Siarhei M.
Bukhalovich
 a,
Diana F.
Bagaeva
a,
Diana F.
Bagaeva
 a,
Aleksandra P.
Polyakova
a,
Aleksandra P.
Polyakova
 a,
Nikolay S.
Ilyinsky
a,
Nikolay S.
Ilyinsky
 a,
Semen V.
Nesterov
a,
Semen V.
Nesterov
 a,
Fedor M.
Tsybrov
a,
Fedor M.
Tsybrov
 a,
Andrey O.
Bogorodskiy
a,
Andrey O.
Bogorodskiy
 a,
Egor V.
Zinovev
a,
Egor V.
Zinovev
 a,
Anatolii E.
Mikhailov
a,
Anatolii E.
Mikhailov
 a,
Alexey V.
Vlasov
a,
Alexey V.
Vlasov
 ab,
Alexander I.
Kuklin
ab,
Alexander I.
Kuklin
 ab,
Valentin I.
Borshchevskiy
ab,
Valentin I.
Borshchevskiy
 ab,
Ernst
Bamberg
ab,
Ernst
Bamberg
 c,
Vladimir N.
Uversky
c,
Vladimir N.
Uversky
 *d and
Valentin I.
Gordeliy
*d and
Valentin I.
Gordeliy
 *e
*e
aResearch Center for Molecular Mechanisms of Aging and Age-Related Diseases, Moscow Institute of Physics and Technology, Dolgoprudny, Russia
bFrank Laboratory of Neutron Physics, Joint Institute for Nuclear Research, Dubna, Russia
cMax Planck Institute of Biophysics, Frankfurt am Main, Germany
dDepartment of Molecular Medicine and Byrd Alzheimer's Research Institute, Morsani College of Medicine, University of South Florida, Tampa, FL, USA. E-mail: vuversky@usf.du
eInstitut de Biologie Structurale Jean-Pierre Ebel, Université Grenoble Alpes-Commissariat à l’Energie Atomique et aux Energies Alternatives-CNRS, 38027 Grenoble, France. E-mail: valentin.gordeliy@ibs.fr
First published on 23rd February 2024
Abstract
Microbial rhodopsin (MRs) ion channels and pumps have become invaluable optogenetic tools for neuroscience as well as biomedical applications. Recently, MR-optogenetics expanded towards subcellular organelles opening principally new opportunities in optogenetic control of intracellular metabolism and signaling via precise manipulations of organelle ion gradients using light. This new optogenetic field expands the opportunities for basic and medical studies of cancer, cardiovascular, and metabolic disorders, providing more detailed and accurate control of cell physiology. This review summarizes recent advances in studies of the cellular metabolic processes and signaling mediated by optogenetic tools targeting mitochondria, endoplasmic reticulum (ER), lysosomes, and synaptic vesicles. Finally, we discuss perspectives of such an optogenetic approach in both fundamental and applied research.
Introduction
Optogenetics is a rapidly developing interdisciplinary field focused on the control of cellular physiological processes using light.1–3 Initially, optogenetics was developed as an instrument to control excitable cells, especially neurons.4,5 Microbial rhodopsin (MR)-based optogenetic tools are expressed in the plasma membrane of excitable cells, where they are capable of inducing firing or silencing of neuronal cells in culture, tissue or brain of living animals.6,7 A vast variety of genes of MRs (more than 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) were found in all domains of life and even in viruses.8,9 This natural variability offers considerable potential for further search and development of new MR optogenetic tools.
000) were found in all domains of life and even in viruses.8,9 This natural variability offers considerable potential for further search and development of new MR optogenetic tools.
MR-based optogenetic tools differ in light sensitivity, ion permeability, pumping selectivity, kinetics, spectral properties, level and selectivity of expression. The toolkit of the optogenetic approach, targeting the plasma membrane, can be expanded by a selective targeting of optogenetic tools to membrane organelles of eukaryotic cells. Targeted expression is a promising emerging approach of precise control of membrane permeability and ion gradients in specific organelles. It provides the means of noninvasive, fast, and reversible control of cell physiology. Although artificial selective targeting of membrane proteins is challenging, successful expression opens new ways to investigate organelle functions and dysfunctions in pathologies, including age-related diseases. The organelle optogenetic approach might be potentially implemented for studies of cancer, cardiovascular, and metabolic disorders.
Recent studies support the amazing potential of MRs in optogenetic applications. Namely, MRs can provide control of a number of cellular processes through organelle optogenetics with low-invasiveness, high time/spatial resolution, and unique selectivity.2 Non-rhodopsin optogenetics is also a rapidly expanding field,10,11 which uses other photosensitive proteins to selectively control intermolecular interactions, providing an ability to control a plethora of processes by light. These processes include transcription,12 receptor positioning,13 activation,14 signal transduction,15 cytoskeletal dynamics and organelle positioning,16 programmed cell death,17 biogenesis18 and composition of membraneless organelles,19 as well as phase-separation propensities of proteins in living cells,20 and many other processes.10 In contrast, MR-optogenetics allows one to directly control the permeability of biological membranes of subcellular compartments and subsequent ion gradients. Potentially, both approaches can be complementary used for expanding control of cellular functions at subcellular scale.
Light-driven channelrhodopsins have revolutionized neuroscience.21 The progress did not stop there; moreover, as the optogenetic toolkit expanded dramatically and became more diversified, it provides important grounds for the rapid growth of optogenetic applications.3 Several recent reviews describe this progress.2,10,22 However, there is still a lack of comprehensive reviews on MR intracellular optogenetics.
This review focuses on optogenetics based on the targeted expression of MRs in intracellular membrane compartments. This area of research dramatically expands possibilities of cellular physiology control. However, targeting of exogenous membrane proteins in cell organelles remains challenging.23 Therefore, this review describes general strategies for protein targeting based on the addition of signal sequences driving proteins to the membranes of a desirable cellular compartment.24 Finally, the article discusses the perspectives of optogenetics in both fundamental and applied research.
Structure and functions of microbial rhodopsins
A detailed description of the structural features and functionality of proteins utilized in intracellular optogenetics is outside the scope of this article, and interested readers are addressed to multiple corresponding reviews discussing structural and functional peculiarities of rhodopsins, also known as retinylidene proteins (e.g., ref. 25–31). Nevertheless, we present here some basic information on the structural organization of this biologically relevant protein superfamily, members of which are found in Eukaryotes, Bacteria, and Archaea.Despite the fact that MRs can function as pumps, channels, and light-sensors,27 they all have a common structural organization typical for all the rhodopsin superfamily members, where the chromophore retinal is covalently bound to an internal pocket formed by the seven membrane-embedded α-helices (7TM) of the opsin apoprotein.30 Light absorption induces isomerization of all-trans-retinal to 13-cis-retinal in microbial rhodopsins (11-cis-retinal to all-trans-retinal in animal rhodopsins) that culminate with protein conformational changes leading to the rhodopsin activation.30 The unique photophysical and photochemical processes required for optimal light to energy or light to signal conversion in rhodopsins are defined by specific chromophore-protein interactions.27 For example, the 7TM protein scaffold of microbial rhodopsins is suitable for light-driven ion pumps, light-gated ion channels, and light sensors which couple to membrane or soluble transducer proteins.29
In the comprehensive review, Ernst et al.27 emphasized: “While all of the known structures of microbial rhodopsins show a common tight α-helical bundle of 7TM helices surrounding the retinal chromophore, there is substantial variation in the arrangement of side chains, the structure of the interfacial regions and loops, as well as in the positions of internal water molecules”.28,29,32 The most conserved common element in the rhodopsin structure is the retinal binding pocket. Importance of local structural organization for the energy transfer between retinal and opsin is illustrated in Fig. 1 representing the overall structure of bacteriorhodopsin (BR) and showing localization of the functionally important conserved aromatic amino acids.
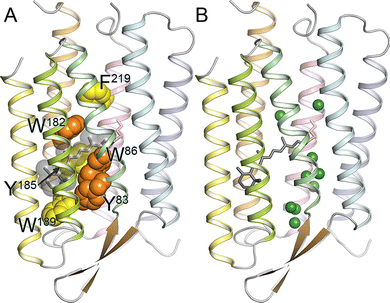 | ||
| Fig. 1 (A) Structure of bacteriorhodopsin (BR), with conserved aromatic residues highlighted (PDB ID: 1QM8). Tyr83, Trp86, and Trp182 are strongly conserved among microbial rhodopsins (orange). Aromatic amino acids are strongly conserved at the position of Tyr185, Trp189, and Phe219 (yellow). In BR, Trp86, Trp182, Tyr185, and Trp189 constitute the chromophore binding pocket for all-trans-retinal (gray). (B) Crystallographically observed internal water molecules of BR (shown as green spheres). Note much higher hydration of the extracellular half (bottom) compared to the cytoplasmic one (top). Adapted from Ernst et al.27 | ||
Here, all-trans-retinal is sandwiched between the strongly conserved Trp86 and Trp182 that form an important part of the chromophore binding site.27 It was suggested that the isomerization pathway from the all-trans to 13-cis-retinal is determined by these bulky groups, whereas the functionally important changes of the helical tilts necessary for BR function are triggered by the interaction of the photoisomerized retinal with Trp182 acting as a mechanical transducer for passing the energy stored in retinal deformation.33 In the BR retinal binding pocket, Tyr185 occupies another important position, participating in hydrogen-bonding stabilization of the protonated retinal Schiff base (RSBH+) counterion.27 As shown in Fig. 1B, all microbial rhodopsins contain protein-bound water molecules near the retinal Schiff base (RSB), contributing to the stabilization of the RSBH+ in the hydrophobic protein interior and thereby playing crucial roles in protein function.27
Abbreviated (un)structural overview of optogenetics
The section below represents an abbreviated glance at the (un)structured biology of MRs discussed in this review. Although the core MR structure is represented by a 7TM helix architecture with three loop regions on both the extracellular and the cytoplasmic sides of the membrane, and although this module is typically used in optogenetics. Fig. 2 shows that many rhodopsins have regions of varying lengths extending beyond their 7TM core architecture (see red segments in Fig. 2a). Curiously, these “extra” pieces represent appendices that are typically expected to be highly disordered (as evaluated by PONDR® VSL234). Fig. 2a also shows that the most disorder in the most extended rhodopsins is concentrated in their C-terminal (cytoplasmic) tails. It is tempting to speculate that these disordered tails might serve as binding platforms for the disorder-based interaction of rhodopsins with specific partners. In line with this hypothesis, each of the four longest rhodopsins discussed in this article contains multiple molecular recognition features, MoRFs, i.e., disordered segments capable of disorder-to-order transitions associated with binding of specific partners, as evaluated using the IUPred2A platform35 (data not shown).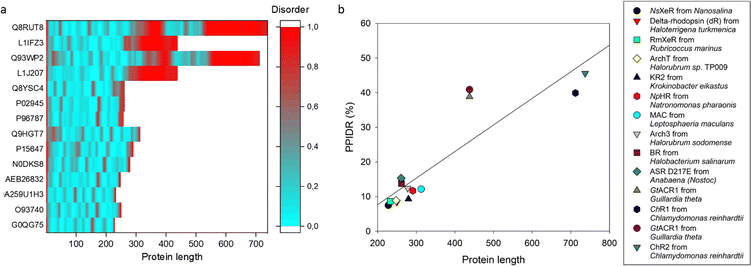 | ||
| Fig. 2 Predicted intrinsic disorder content in the rhodopsins discussed in this article. (a) PONDR® VSL2-based34 evaluation of the intrinsic disorder predispositions. Disorder score of 0.5 is taken as a threshold for intrinsic disorder, where residues/regions with disorder scores exceeding 0.5 are considered disordered (shown in red), whereas residues and regions with disorder scores ranging below 0.5 are considered as ordered (blue). (b) Length dependence of the PONDR® VSL2-predicted levels of intrinsic disorder in the rhodopsins discussed in this article. Disorder levels were evaluated as the percentage of predicted intrinsically disordered residues (PPIDR; i.e., residues with a disorder score of ≥0.5) in a query protein. In both plots, rhodopsins are arranged from the least disordered (at the bottom of the plot) to the most disordered (at the top of the plot) in the following order: NsXeR from Nanosalina (UniProt ID: G0QG75); delta-rhodopsin (dR) from Haloterrigena turkmenica (UniProt ID: O93740); RmXeR from Rubricoccus marinus (UniProt ID: A0A259U1H3); KR2 from Krokinobacter eikastus (NCBI GenBank ID: BAN14808); NpHR from Natronomonas pharaonis (UniProt ID: P15647); MAC from Leptosphaeria maculans (UniProt ID: Q9HGT7); Arch3 from Halorubrum sodomense (UniProt ID: P96787); BR from Halobacterium salinarum (UniProt ID: P02945); ASR D217E from Anabaena (Nostoc) sp. PCC7120 (UniProt ID: Q8YSC4); GtACR1 from Guillardia theta (UniProt ID: L1J207); ChR1 from Chlamydomonas reinhardtii (UniProt ID: Q93WP2); GtACR1 from Guillardia theta (UniProt ID: L1IFZ3); and ChR2 from Chlamydomonas reinhardtii (UniProt ID: Q8RUT8). | ||
Furthermore, Fig. 2b shows the correlation between intrinsic disorder content and protein length and gives a clear indication that most of the “extra” length added to the 7TM core in rhodopsin is materialized in disordered regions. Although the 7TM core is predicted to be mostly ordered, this transmembrane segment includes some flexible regions that correspond to the loops linking the transmembrane helices. Curiously, these loop regions in different rhodopsins show rather different propensities for conformational flexibility (data not shown), suggesting different roles of said conformational flexibility in the functionality of these proteins.
The fact that the cytoplasmic tails of rhodopsins contain high levels of intrinsic disorder is in line with the known peculiarities of disorder distribution within the transmembrane proteins. In fact, it was reported that 50% of transmembrane proteins have at least one intrinsically disordered region (IDR) of 30 amino acids or more, with such disordered domains being preferentially localized to the cytoplasmic side especially of the multipass transmembrane proteins.36 In addition to possessing numerous MoRFs, the IDRs of transmembrane proteins are enriched in sites of various posttranslational modifications (PTMs), including phosphorylation, a crucial signal transduction-related PTM. Therefore, it is very likely that these MoRF- and PTM-enriched IDRs are involved in regulating protein activity, being capable of undergoing disorder-to-order transition at binding to specific binding partners in a PTM-dependent manner.36 This is likely that the presence of MoRFs with PTM sites provides important means for fine tuning of the strength of disorder-based binding, through local structural changes caused by PTMs.36 Furthermore, the presence of disordered regions is typically linked to the capability of proteins to interact with multiple partners, thereby defining their binding promiscuity and multifunctionality, phenomena linked to the protein structure-function continuum model.37
In line with these considerations, published studies reported important roles of the C-terminal tails of rhodopsins in interaction with rhodopsin kinases,38 targeting rhodopsins to specific cellular compartments, such as the outer segment,39 regulation of the photoreceptor degeneration,40 acting as key elements in membrane receptor activation, molecular recognition by signaling molecules, and receptor deactivation,41 playing vital role in maintaining the structure of rhabdomeres, a specialized membrane-rich organelle that serve as a site of phototransduction, in Drosophila,42 and acts as a platform for interaction with various partners, such as arrestin.43
It is clear that a better understanding of the multiple functional and regulatory roles of disordered regions of rhodopsins is necessary for the development of novel applications of these proteins in optogenetics. The lack of knowledge on disorder parts of rhodopsins does not allow us to provide an in-depth discussion of their biological roles and possible meaning for optogenetics. The only what we would like to do here is to pay attention on this fact and stress out that the studies of the disordered parts of rhodopsins are highly important.
Advantages of MR-based optogenetic approaches
Optogenetics offers a rapid and precise manipulation of single cells with light.4,23 This technology was mainly focused on plasma membrane expression of optogenetic tools in excitable cells and provided a methodological revolution in neuroscience.44 The optogenetic approach shed light on the functions of certain neuronal populations in physiologic reactions,44–46 complex behavior47,48 and memory.3,49 The approach is also promising for control of cell physiology with light by selective expression of optogenetic tools in membranes of intracellular organelles.Conventional experimental protocols and therapies for manipulation of organelles physiology are based on biochemical intervention, which have consequent drawbacks. The use of chemicals is usually accompanied by diffusion of the substances in the cells and the body, which greatly affects the selectivity of targeting and the amount of the substance that reaches the targeted place. In addition, biochemical manipulations go along with a poor specificity and a limited exposure time. For example, mitochondrial uncouplers, like carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP) or 2,4-dinitrophenol (DNP), have a toxic effect and an off-target activity, since they reduce the plasma membrane potential and lysosomal acidity simultaneously with mitochondrial inner membrane potential.50,51 This side effect severely limits their use in research and clinics. The same is true for bafilomycin A1,52,53 chloroquine (CQ), or hydroxychloroquine (HCQ) – known inhibitors of autophagy. CQ and HCQ are FDA-approved candidates for clinical use, but they also have undesirable side cytotoxic effects.54
Optogenetics allows selectively influencing cell compartments, certain cells, and even tissues, as well as selectively manipulating their activities with high time and spatial resolutions. Light induced activation of the cell allows for fast response that is reproducible upon repeated stimulation. The frequency of illumination is limited by the photocycle of the protein. If the illumination by short pulses can excite the majority of MRs, then it is possible to decrease the effective power of illumination and reduce a phototoxic damage.55 Naturally, channelrhodopsins are the most convenient tools for rapid changing of plasma membrane potential in excitable cells.56,57 Using MRs with short photocycles allows modulating a membrane potential with frequencies of action potential generation in neurons.58
Moreover, expanding the optogenetic toolkit provides researchers with complementary tools, which can be combined. For example, CrChR2 (cation channel), which is activated by blue light, and NpHR (chloride pump), activated by orange light, were combined to activate and inhibit neural activity.59,60 This pair was also targeted to different subcompartments of retina ganglion cells: N-terminal fusion to ankyrin for soma localization and to postsynaptic density PSD-95-protein for dendrite localization.61 Recently, two other channelrhodopsins, the red-shifted cation-conducting ChrimsonR and the blue-light-absorbing chloride-conducting stGtACR2, were expressed in dopaminergic neurons of the ventral tegmental area in freely-moving mice, allowing bidirectional control of dopamine release, preference-avoidance, and social behavior.62
Optogenetic experiments can be done with wide-field or patterned illumination of tissue. Two-photon excitation has been applied to achieve high spatial resolution and deeper tissue penetration.63 Delivery of light to the tissue is often achieved by optical fiber, but the modern approach is to use implantable LED-devices.62,64,65 An important feature of optogenetics is the control of the frequency and time of light exposure, which could be adjusted for the photocycle of the chosen MR in certain surroundings.66
Thus, organelle optogenetics combines the advantages of the optogenetic approach, including non-invasiveness, selectivity, temporal and spatial resolution, and reversibility of the effect with the specificity of organelle targeting.
Strategies of MR organelle targeting
Targeting of a foreign membrane protein – MR to a certain subcellular compartment is challenging. Despite this, a number of successful examples has been reported (Fig. 3). Plasma membrane targeting of MRs is under investigation since first optogenetic experiments. Fluorescent proteins are often fused to MRs for visualization of subcellular localization of the target. For bacteria and archaea-derived MRs, the addition of N-terminal part from eukaryotic MR at its N-terminus was combined with the addition of a membrane trafficking signal (TS) as well as the ER-export signal at the C-terminus (see detailed description in the section Plasma membrane MRs targeting). Nevertheless, some MRs of archaeal origin could be expressed in the plasma membrane only with TS at the C-terminus. In addition, these constructs comprise a self-cleavage peptide p2A after TS and before a fluorescent protein.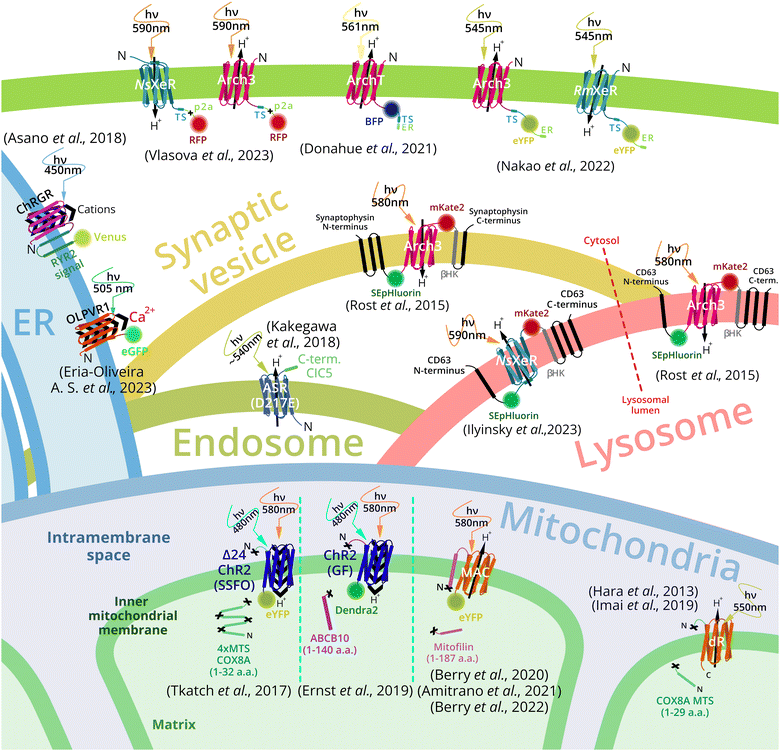 | ||
| Fig. 3 Overview of MR-based organelle optogenetic tools and targeting strategies. Outward proton pumps Arch3 and ArchT, from Halorubrum sodomense and Halorubrum strain TP009, respectively, and inward proton pumps NsXeR from Nanosalina sp. and RmXeR from Rubricoccus marinas were expressed in plasma membrane and used to control cytosol pH.67,68 Outward proton pump Arch3 and inward proton pump NsXeR were exploited for control of lysosomal pH.69 Arch3 was also used for synaptic vesicles acidification.69 Inward proton pump (ASR – anabena sensory rhodopsin mutant D217E) was used for endosome alkalization.70 Channel rhodopsin ChR2 (from Chlamydomonas reinhardtii),71,72 and proton pumps MAC (from Leptosphaeria maculans)73,74 and delta-rhodopsin (from Haloterrigena turkmenica)75,76 were expressed in inner mitochondrial membrane and triggered an optogenetic decrease or increase of the mitochondrial membrane potential respectively. The channelrhodopsin green receiver (ChRGR)77 and type 1 viral channelrhodopsin OLPVR1 (Organic Lake phycodnavirus rhodopsin)78 were expressed in the endoplasmic reticulum to induce Ca2+ release to the cytosol and cell contraction. | ||
The mitochondrial inner membrane (IMM) targeting of MRs was successfully done by N-terminal fusion of MR to mitochondrial localization signal sequence (MLS) or a mitochondrial inner membrane resident protein part, containing MLS. For example, channelrhodopsin 2 (CrChR2 from Chlamydomonas reinhardtii) was targeted to the IMM with four repeats of MLS of the 8th subunit of cytochrome c oxidase (COX8)79 together with an N-terminal truncation of ChR2.30CrChR2 with no deletions was also targeted to the IMM by long signal sequence of the ABC-transporter family protein (ABCB10). For the outward proton pump MAC from Leptosphaeria maculans21 IMM targeting a long N-terminal part, containing MLS, and a transmembrane helix of the IMM-resident protein mitofilin was used.73
Lysosome targeting of the outward proton pump Arch3 was achieved by the chimeric construct called lyso-pHoenix. Arch3 together with fluorescent proteins was inserted between the 1st and the 2nd α-helices of tetraspanin CD63, which was responsible for lysosome localization. The similar construct (pHoenix) comprising a fragment containing Arch3 between the 3rd and the 4th helices of synaptophysin was used for synaptic vesicles targeting.69 For endosome targeting MR ASR (D217E mutant) was fused to the C-terminal cytoplasmic sequence of chloride channel protein 5 (PhotonSABER).77 Also vacuole targeting of proton pumps delta-rhodopsin (dR) from Haloterrigena turkmenica (via fusion to the amino acid transporter Avt6)80 and rhodopsin from Ustilago maydis in Saccharomyces cerevisiae was reported recently.81
For endoplasmic reticulum targeting channelrhodopsin green receiver, ChRGR was fused C-terminally to two transmembrane helices of a mouse ryanodine receptor 2 and the fluorescent protein Venus.77 A viral rhodopsin OLPVR1 was observed in the ER, although no signal sequence was added to the protein.78
This was a brief overview of the signals for organelle targeting of MRs. Next, for each organelle we describe details of targeting and specific physiological effects observed in optogenetic experiments. Targeting strategies and optogenetic effects of MRs are summarized in Table 1 and specific signal sequences, used for targeting, are represented in Table S1 (ESI†).
| MR | Targeting strategy | Optogenetic effects on cell culture/organism | Ref. |
|---|---|---|---|
| Plasma membrane | |||
| ArchT from Halorubrum sp. TP009 | ArchT_BFP2_TS_ERex: TS – membrane trafficking signal | Cytosol alkalization by 0.3 pH-units in retinal pigment epithelial cells. Activation of localized plasma membrane ruffling. | 67 |
| BFP2 – blue fluorescent protein | |||
| ERex – ER-export signal | |||
| Arch3 from Halorubrum sodomense | Arch3_TS_eYFP_ERex: TS – membrane trafficking signal | Cytosol alkalization by 1.1 pH-units in HeLa cells. Cell shrinking and apoptosis via a mitochondrial-mediated pathway. Reduction of chemotaxis of C. elegans (Arch3 expression in amphid sensory neurons). | 68 |
| eYFP – enhanced yellow fluorescent protein | |||
| ERex – ER-export signal | |||
| RmXeR from Rubricoccus marinus | RmXeR_TS_eYFP_ERex: TS – membrane trafficking signal | Cytosol acidification in an alkaline medium (pH 9.0). Reduction of cell shrinking and apoptosis. | 68 |
| eYFP – enhanced yellow fluorescent protein | |||
| ERex – ER-export signal | |||
| Arch3 from Halorubrum sodomense | Arch3_TS_p2A_RFP: | Cytosol alkalization by 0.3 pH-units in HeLa cells under physiological conditions (in standard cell culture medium). | 82 |
| TS – membrane trafficking signal | |||
| RFP – red fluorescent protein | |||
| NsXeR from Nanosalina sp. | NsXeR_TS_p2A_RFP: TS – membrane trafficking signal | Cytosol acidification by 0.6 pH-units in HeLa cells under physiological conditions (in standard cell culture medium). | 82 |
| RFP – red fluorescent protein | |||
| Mitochondrial inner membrane | |||
| Mutants of ChR2 from Chlamydomonas reinhardtii (ChR2: I197S, C128A/H134R, C128S/D156A – SSFO, Tr) | mt/2mt/4mt-ChR2(mut)-YFP: mt – cytochrome C oxidase 8 presequence (first 32 a.a.), 2mt, 4mt – tandem repeats of mt, YFP – yellow fluorescent protein | No significant localization in mitochondria. | 71 |
| 4mt-Δ24ChR-YFP: Δ24ChR2 – deletion of the first 24 a.a | Mitochondrial localization in HEK293T, HeLa, GA-PLXR, human cardiomyocytes, pancreatic β-cells | ||
| Mutant of ChR2 from Chlamydomonas reinhardtii ChR2 (C128S/D156A – SSFO) | 4mt-Δ24ChR2(SSFO)-YFP: | Mitochondrial localization in HEK293T, HeLa, GA-PLXR, human cardiomyocytes, pancreatic β-cells. Optogenetic stop of cardiomyocytes beating. Optogenetic decrease of glucose-bursted ATP production. Increased oxygen consumption in HeLa cells | 72 |
| Mutant of ChR2 from Chlamydomonas reinhardtii ChR2(H134R – GF) | mito-ChR2(GF)-eYFP: mito-ABCB10 presequence (first 140 a.a.), eYFP – enhanced yellow fluorescent protein | Mitochondrial localization in H9C2, HeLa, human-induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs). Programmed cell death (apoptosis) of 80% of HeLa cells, triggered by acute illumination. Mitochondrial utilization by mitophagy, the preconditioning effect of low-intensity illumination. | |
| Mutant of ChR2 from Chlamydomonas reinhardtii ChR2(H134R – GF) | mito-GF-eYFP: mito – renal outer medullary (ROMK) presequence | No mitochondrial localization. | 83 |
| eYFP – enhanced yellow fluorescent protein | |||
| BR from Halobacterium salinarum | COXIV-BR: COXIV – cytochrome c-oxidase subunit IV (COX4) presequence from Saccharomyces cerevisiae | Mitochondrial localization in yeast cells (Schizosaccharomyces pombe), operating status and orientation confirmed. | |
| BR – bacterioopsin gene after presequence | |||
| BR from Halobacterium salinarum | ATP2-BR, RIP1-BR: ATP2 – β subunit of the F1F0-ATPase from Schizosaccharomyces pombe | No significant localization in the mitochondria of yeast cells (Schizosaccharomyces pombe). | 75 |
| RIP1 – Fe/S RISC protein signal sequence | |||
| Delta-rhodopsin (dR) from Haloterrigena turkmenica | mito-dR-myc: mito – mitochondrial presequence of human cytochrome c oxidase subunit VIII (first 29a.a.) | Mitochondrial localization in SH-SY5Y and CHO-K1 cells. SH-SY5Y cells: cell survival upon inhibition of the electron transport chain, suppression of MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine) induced cell death. CHO-K1 cells: suppression of cell death induced by rotenone. | |
| myc – human c-Myc proto-oncogene, epitope tag | |||
| Delta-rhodopsin (dR) from Haloterrigena turkmenica | mito-dR: mito – mitochondrial presequence of human cytochrome c oxidase subunit VIII (first 29a.a.) | Mitochondrial localization in Drosophila dCHCHD2−/− cells confirmed, MMP increased, ATP synthesis restored, and ROS level reduced No direct confirmation of orientation and pumping direction was reported. Optogenetic decrease of insoluble α-synuclein. In clusters of dopaminergic neurons – decrease in neuron death, an improvement in locomotor activity. | 84 |
| Mutant of delta-rhodopsin (dR) from Haloterrigena turkmenica (D104N/K225A) | mito-dR: mito – mitochondrial presequence of human cytochrome c oxidase subunit VIII (first 29a.a.) | Drosophila's mitochondrial localization, non-functional dR mutant. | |
| MAC from Leptosphaeria maculans | mitofilin (187 a.a.)-MAC-eGFP | Mitochondrial localization in Caenorhabditis elegans, MMP generation, matrix alkalization, and ATP synthesis in mitochondrial suspension. Increased resistance to mitochondrial toxins. Reversal of the protective effect of hypoxic preconditioning. | 85 |
| mitofilin – inner mitochondrial membrane protein | |||
| eGFP – enhanced green fluorescent protein | |||
| MAC from Leptosphaeria maculans | mitofilin(187 a.a.)-MAC-eGFP | Lifespan extension of C. elegans, reduction of age-associated physiological functional decline. | 86 |
| MAC from Leptosphaeria maculans | mitofilin(187 a.a.)-MAC-eGFP | Mitochondrial localization in mouse and human CD8+ T cells, increase in the mitochondrial membrane potential, increase in the mass of mitochondria, and improvement of migration and effector functions of CD8+ T cells. | 74 |
| Lysosomes and endosomes | |||
| Arch3 from Halorubrum sodomense | CD63-pHluorin-Arch3-mKate-βHK-CD63 (pHoenix) | Lysosomal localization in HEK293, HeLa. Light-driven acidification of lysosomes, alkalized by bafilomycin A1. | 69 |
| CD63 – lysosomal membrane glycoprotein | |||
| pHluorin – pH-sensitive green fluorescent protein | |||
| mKate – far-red fluorescent protein | |||
| βHK – transmembrane helix of the rat gastric H+/K+ ATPase β-subunit | |||
| ASR D217E – anabaena sensory rhodopsin from Anabaena (Nostoc) sp. PCC7120 mutant with an inward proton pump activity | ASR(D217E)-ClC5 (PhotonSABER) | Early and late endosomal localization in hippocampal neurons, Purkinje cells. Light-dependent endosomal alkalization and inhibition of AMPA receptor endocytosis and motor learning in Purkinje cells in vivo. | 70 |
| CIC5 – C-terminal cytoplasmic region of chloride channel protein 5 | |||
| Synaptic vesicles | |||
| Arch3 from Halorubrum sodomense | Synaptophysin-pHluorin-Arch3-mKate-βHK-Synaptophysin (pHoenix) | Localization in synaptic vesicles of mouse hippocampal neurons and in organotypic brain slices. Light-driven acidification of synaptic vesicles, alkalized by bafilomycin A1, increased vesicular transmitter accumulation. | 69 |
| Synaptophysin – vesicular protein | |||
| pHluorin – pH-sensitive green fluorescent protein | |||
| mKate – far-red fluorescent protein | |||
| βHK – transmembrane helix of the rat gastric H+/K+ ATPase β-subunit | |||
| Arch3 from Halorubrum sodomense | Synaptophysin-pHluorin-Arch3-mKate-βHK-Synaptophysin (pHoenix) | Localization of the pHoenix in mouse hippocampal or striatal neuron. They showed that proton efflux from GABAergic SVs is initially faster than glutamatergic SVs in intact synapses. Comparison of the filling rate of empty synaptic vesicles with glutamate and GABA. | 87 |
| Endoplasmic reticulum | |||
| ChRGR channelrhodopsin-green receiver, engineered ChR1 from Chlamydomonas reinhardtii | ChRGR-ER-signal-Venus (ChRGRER–Venus): | An increase of cytosol Ca2+ in the C2C12 cells. Induction of calcium-induced calcium release (CICR) and store-operated Ca2+ entry (SOCE). | 77 |
| ER-signal – two transmembrane helices (Gln4765-Ile4866) of mouse ryanodine receptor 2 | |||
| Venus – an improved variant of enhanced yellow fluorescent protein | |||
| OLPVR1 from Organic Lake phycodnavirus | OLPVR1-GFP | Activation of Ca2+ release from intracellular stores, activation of muscle contraction, tail flicking and swimming in Xenopus laevis tadpoles. | 78 |
Optogenetics of organelles
Plasma membrane: membrane potential and metabolic control
In order to better understand organelle optogenetics, we will first describe targeting and optogenetic effects of MRs expressed in the plasma membrane. Historically, plasma membrane was the first target for optogenetics, and plasma membrane-targeted tools are still the major MR-based optogenetic instruments.88 In fact, plasma membrane-directed optogenetics remains to be the main area for which novel MR-based optogenetic tools are deeply studied and improved.55 There are no reports on engineering MRs with novel properties for the use in intracellular compartments, which have not been used previously as optogenetic tools at the plasma membrane. MR-based optogenetics has demonstrated its significance for neuroscience.89 In such cases, specific promoters are used to selectively express optogenetic tools in certain populations of neurons.79,90 Such approach allows neural circuit identification and is of vital importance for studies of brain functions.91–93 Optogenetics also provided a unique ability to manipulate complex behavior and memory in living animals.94,95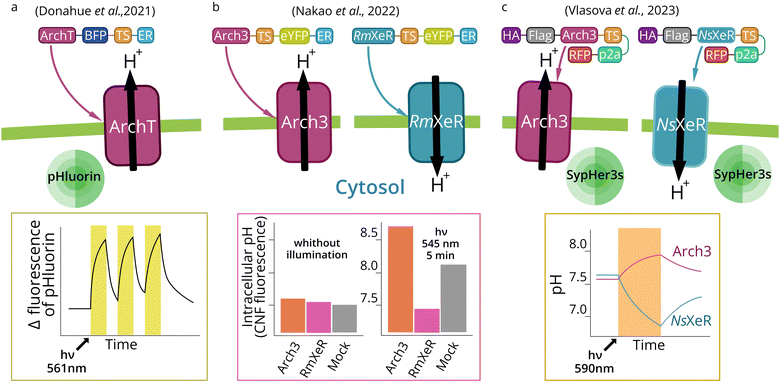 | ||
| Fig. 4 Cytosolic pH manipulation by plasma membrane expression of outward and inward MR proton pumps. Light activation of the outward proton pump ArchT (a) or Arch3 (b) (outward pump from Halorubrum strain TP009 and Halorubrum sodomense, respectively) in the plasma membrane leads to cytosol alkalization in the neutral medium.67,68 Activation of RmXeR (inward pump from Rubricoccus marinas) (b) in the plasma membrane leads to a decrease in cytosol pH in an alkaline (pH 9.0) medium.68 Activation of Arch3 and NsXeR (inward pump from Nanosalina sp.) (c) in the plasma membrane leads to subsequent cytosol alkalization and acidification in a neutral medium.56 | ||
The pH value of the cytoplasm (pH 7.2) and extracellular fluid (pH 7.4) is a fundamental parameter for homeostasis. In cancer cells, the cytosol is constantly alkalized (pH higher than 7.4), which is coupled to acidification of extracellular space (pH lower than 7.1). Higher intracellular pH alters cell phenotype and gene expression profile. For example, cyclin D1 expression is activated by cytosol alkalization.110 An increased intracellular pH may promote cancer cell migration during metastasis,111 likely due to the higher cofilin activity.112 Also, an increased intracellular pH activates WNT-signaling in cells during embryogenesis in vertebrates.113 Expression of inward and outward proton pumps in the plasma membrane may open new opportunities in optogenetic cytosol pH manipulation and control of cell physiology.
Mitochondria: optogenetic control of mitochondrial membrane potential
Mitochondria are double-membraned cellular organelles, mainly serving the energy needs of the cell. They are vital cellular compartments, especially for nerve114 and muscle115 tissues, where energy consumption is high. Certain mitochondrial dysfunctions can lead to deafness, blindness, dementia, diabetes, and cardiomyopathy116 and are involved in cancerogenesis,117 neurodegeneration,118 and aging.119 The state of the mitochondria largely determines the physiology of the cell. The efficiency of mitochondria depends on the mitochondrial membrane potential (MMP) of the IMM.120 This is an important factor that controls the synthesis of ATP, generation of reactive oxygen species (ROS),121 and the functionality of the transport of immature proteins through the IMM.122 Another important and strictly controlled parameter is the concentration of calcium ions in the mitochondria.123 Calcium acts as a signaling molecule and is involved in many important interactions between the mitochondria and the cell, determining the cell's fate.124,125During translation of mitochondrial proteins encoded in the nuclear DNA, N-terminal amino acids often have a signal sequence which directs the protein into the mitochondrion. Most proteins pass through the TOM40 complex of the outer membrane (OM) of mitochondria. Proteins of this complex recognize the signal peptide, recruit it, and send it to the pore of the main subunit of the complex. Transported proteins then may follow several pathways, depending on their signal sequences. If the N-terminal signal is an amphipathic alpha helix, it is recognized by the TIM23 protein complex in the IMM. The charge helps the helix to pass through the membrane, where, on the matrix side, the translocated protein is recruited by an ATP-driven chaperone (mtHsp70), which pulls out the protein from the pore of the TIM23 complex.131 Then, the signal sequence is cleaved by MPP protease in the matrix. If a protein has internal hydrophobic signals, the translocation can be stopped and the protein is laterally released into the IMM.128 If there is no such signal, then the protein will be completely released into the matrix, where it will be picked up by chaperones if required. Then, in the case of a membrane protein, it is targeted into the IMM from the matrix by OXA1, which inserts proteins from both genetic origins (mitochondrial and nuclear).132
Bacteriorhodopsin (BR) was targeted to IMM in yeast cells by using the cytochrome c-oxidase subunit IV (COX4) signal sequence. This presequence is 22 residue-long and has a cleavage site after the 17th amino acid residue (a.a.).134 However, the attempts to do the same with signal sequences of the β subunit of F1/FO-ATPase and Fe/S Rieske protein were unsuccessful.83
CrChR2 mutants were successfully targeted to the inner membrane of mitochondria using a 4-fold tandem repeats of the signal of a mitochondrial inner membrane protein, the 8th subunit of cytochrome c oxidase (COX8, first 32 a.a.)135 with a deletion of the first 24 amino acids of ChR2 (Fig. 5a).71 Also, ChR2 mutants were expressed in the IMM with an unusually long (140 a.a.) signal sequence of the ABC-transporter family protein (ABCB10).72 The first 105 a.a. of this sequence are cleaved in the mitochondria.136 COX8 signal sequences with less than 4 repeats and no protein changes, as well as the signal sequence of ROMK, mitochondrial KATP channel signal sequences, failed to show the required level of localization.72 Also, delta-rhodopsin (dR) and non-functional dR mutant (D104N/K225A) were expressed with COX8 signal sequence (first 29 a.a.).75,76
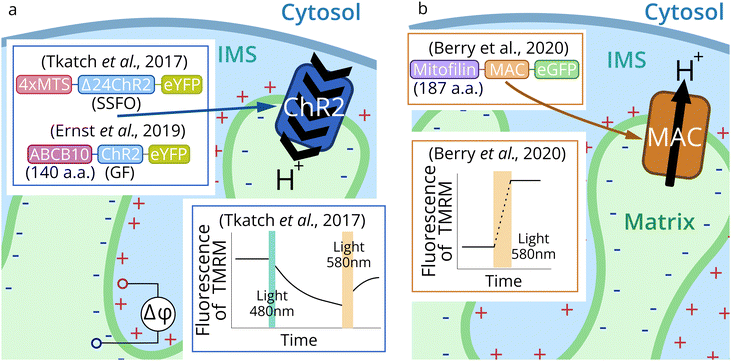 | ||
| Fig. 5 Schematic representation of mitochondria optogenetic approaches. Channelrhodopsin 2 (CrChR2 from Chlamydomonas reinhardtii) was targeted to IMM for optogenetic mitochondrial uncoupling (a).71,72 Proton pump MAC (from Leptosphaeria maculans) (b) was applied for optogenetic increase of MMP.73,74,86 | ||
Proton pump MAC in the latest studies73,74,86 was expressed in mitochondria of C. elegans and mouse T-lymphocytes by N-terminal fusion to the first 187 a.a. of the IMM-resident protein mitofilin (Fig. 5b). This protein has a 34 a.a.-long cleavable signal peptide and the transmembrane part, which appears to remain with the delivered protein.137
First optogenetic tools for mitochondria control were channelrhodopsin 2 mutants (CrChR2: I197S, C128A/H134R, C128S/D156A – SSFO) (Fig. 5a).71CrChR2 and its mutants were targeted to the IMM in HEK293T and HeLa cells, human cardiomyocytes, and melanoma cells (GA-PLXR). The localization and correct orientation in the IMM were confirmed, as well as the functioning of the protein. Activation of the SSFO mutant of CrChR2 was carried out by 495 ± 5 nm blue LED (2 mW mm−2), and the closing of the channel was induced by 595 nm LED (12 mW mm−2). CrChR2-mediated drop in MMP led to reduced mitochondrial calcium influx after histamine stimulation in HeLa cells. It also temporarily stopped the beating of cardiomyocytes. Also, a decrease of ATP production in pancreatic β-cells and increased oxygen consumption in HeLa cells were observed during optogenetic activation of CrChR2 in the IMM.
Another mutant CrChR2(H134R-GF) was used to trigger autophagy and apoptosis by light.72 Depending on the degree of IMM optogenetic decoupling, the effect can be exactly the opposite. Acute illumination (475 nm LED, 5 mW mm−2, 24 h) triggered the programmed cell death of 80% of HeLa cells, which was predominantly realized by the apoptotic pathway. However, low intensity illumination (0.2 mW mm−2, 2 h) induced mitophagy and mediated a preconditioning effect: cell viability after acute illumination was two-fold higher in a preconditioned group compared to control cells.
Another proton pump – dR was expressed in mitochondria of CHO-K1 and SH-SY5Y cells. Mitochondrial localization was confirmed and optogenetic reduction of rotenone and 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-induced cell death was demonstrated.75 Nevertheless, the direction of proton pumping by dR was not shown, and the direct confirmation of MMP increase in cell culture was not made. Later, dR was expressed in Drosophila mitochondria.76 The CHCHD2 (dCHCHD2) knockout flies (a model of Parkinson's disease) with the mitochondrial dR were subjected to prolonged illumination. The MMP and ATP synthesis were restored. Maintaining the functional potential bypassing the electron transport chain reduces the formation of ROS both directly and by normalizing calcium concentration. As a result, there was an increase of mitochondrial calcium accumulation in dopaminergic neurons of dCHCHD2−/− flies and a decrease in the number of insoluble inclusions of overexpressed α-synuclein in neurons. In addition, the expression of dR in dopaminergic neurons showed an optogenetic decrease in neuronal death, as well as an improvement in locomotor activity in flies expressing dR in comparison with the non-functional mutant.
Taken together, two types of mitochondrial optogenetic tools were proposed: CrChR2 to optogenetically reduce MMP and proton pumps MAC and dR to increase MMP. A variety of physiological outcomes can be controlled by light using these tools from ATP synthesis and mitophagy up to the whole-organism lifespan.
Lysosomes and endosomes: optogenetic control of pH and proteolysis
Lysosomes are small single-membrane organelles containing a number of enzymes that can break down macromolecules.140–143 For the proper physiological function of lysosomes, it is essential to accumulate protons within the lumen in order to maintain an optimal acidic pH (4.5–5.0) for enzyme function. This is achieved by a proton pump vesicular proton ATPase (V-ATPase) under the assistance of other transporters and channels that regulate ion concentrations in lysosomes.144 Dysregulation of acidification leads to the changes in intraluminal pH, which can result in lysosomal storage diseases and is associated with Alzheimer's disease, Parkinson's disease, and some others.145,146 Recently, the participation of lysosomes in many intracellular signaling and metabolic pathways was demonstrated.147 Involvement of altered lysosome function in the pathogenesis of metabolic disorders148 and cancer149,150 was also found.Membrane dynamics, which affects the degradation and recycling of proteins within cells, plays a critical role in maintaining homeostasis.160 Endosomes are a dynamic system that undergoes morphological and biochemical changes associated with the transport of vesicles.161 Among endosomes, two pools are distinguished: early and late endosomes. Early endosomes are small vesicles that emerge from clathrin-coated pits derived from the plasma membrane.162,163 Early endosomes have a slightly acidic intraluminal pH of about 6.2–6.3.164 One of the best-studied proteins associated with early endosomes is Rab5. Early endosomes must then turn into late endosomes, which is accompanied by a change in Rab5 to Rab7.165 The transition to late endosomes is also accompanied by acidification of the internal lumen of endosomes up to 5.0–5.5.164 Some late endosomes may have a complex internal vesicular structure and are also referred to as multivesicular bodies (MVB).166 Finally, late endosomes fuse with lysosomes to degrade intravesicular cargo.167 Defects in the endocytic pathway are primary reason of storage diseases168,169 as well as are associated with many pathological conditions such as cancer and neurodegenerative diseases.170,171 Regulation of endocytic and secretory pathways is important, for example, for neurons to ensure normal synapse function.172
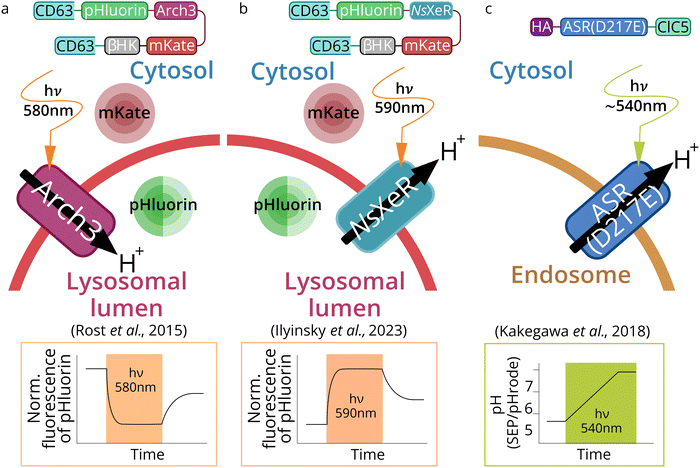 | ||
| Fig. 6 Schematic representation of lysosome- and endosome-based optogenetic approaches. Three types of optogenetic tools were targeted to lytic compartments: outward proton pump Arch3 (from Halorubrum sodomense) for lysosomes acidification (a),69 inward proton pump NsXeR (from Nanosalina sp.) for lysosomes alkalization (b)70 and inward proton pump ASR (Anabena Sensory Rhodopsin mutant D217E) (PhotonSABER) for endosomes alkalization (c).173 | ||
In another study, Kakegawa et al.70 targeted Anabena Sensory Rhodopsin mutant (ASR D217E) which acts as an inward proton pump to endosomes, the construct was called PhotonSABER (Fig. 6c). To target ASR to early endosomes, it was fused to the C-terminal cytoplasmic sequence of chloride channel protein 5 (ClC5). ClC5 has been shown to localize predominantly in early endosomes.174 However, immunocytochemical staining showed that this tool is expressed in early and late endosomes simultaneously.
Alkalization of lysosomes was performed by inward proton pump NsXeR (Fig. 6b). The effectiveness of this Lyso-NsXeR optogenetic approach was demonstrated through its ability to inhibit the activity of lysosome proteolytic enzymes.173
For PhotonSABER, it was demonstrated that light-dependent endosomal alkalization inhibits endocytosis of the AMPA-type glutamate receptor; the latter, in turn, blocks long-term depression in parallel fibers of Purkinje cells synapses (PF-LTD) (Fig. 6c).70 This tool was used to study learning and memory. Unfortunately, PhotonSABER is not selective for endosome subcompartments targeting and therefore requires further development and optimization.
Optogenetic acidification or alkalization of lysosomes and endosomes is a promising way for metabolic control in living cells. Due to the fact that lysosomes and endosomes are relatively small compartments, with a large surface/volume ratio, dramatic changes in luminal pH could be achieved by activation of rhodopsin-proton pumps (more than 1 pH unit). Therefore, the concept of using optogenetic tools to control pH for various types of endosomes and lysosomes is an important and relevant task.
Synaptic vesicles: optogenetic control of neurotransmission
Optogenetic control of neurotransmission was recently demonstrated.69 In neurons, neurotransmitter molecules are stored in and released to the synaptic cleft by synaptic vesicles (SVs). Neurotransmitters interact with a certain receptor on the postsynaptic membrane, leading to excitatory or inhibitory postsynaptic potentials, allowing the transmission of signals between neurons. SVs are small membranous vesicles filled with neurotransmitters.179 The loading of neurotransmitters into SVs is critically dependent on the activity of the V-ATPase, which provides an acidic environment by pumping protons into the lumen of the vesicles, allowing the transporters to load them with neurotransmitters.180,181Targeting MRs to SVs was done by inserting Arch3 proton pump between the 3rd and the 4th helix of tetraspanin–synaptophysin.191 The chimeric construct was similar to lyso-pHoenix except substitution of CD63 to synaptophysin.69
Endoplasmic reticulum: optogenetic control of calcium signaling
Endoplasmic reticulum (ER) is a membrane system that plays a vital role in protein synthesis, sorting, and folding. Translation of proteins of the secretory pathway begins when mRNA connects to the ribosome. Emerging proteins are transported to the ER through the interaction of the N-terminal signal sequence and the signal recognition particle (SRP). The SRP complex contacts the SRP receptor, the ER resident membrane protein. Later translation continues in ER and the polypeptide can co-translationally enter the ER lumen through the translocon, a channel containing Sec proteins.192 Besides co-translational translocation described above, there is also a post-translational translocation when proteins are transported after the completion of their synthesis.193 ER is also responsible for the synthesis and transport of lipids, and the accumulation of Ca2+ and its regulated release into the cytosol.194 Intracellular Ca2+ is a major secondary messenger in the cell, and optogenetic control of ER Ca2+-release is a new promising approach that allows controlling calcium signaling cascades and processes which depend on calcium concentration (myogenesis, muscle contraction, neurons excitation and morphology).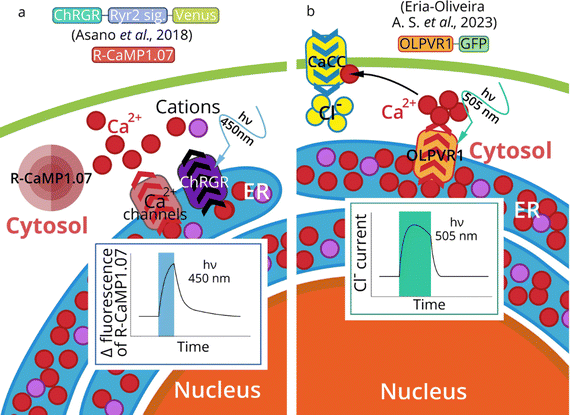 | ||
| Fig. 7 Optogenetics of ER. Activation of ChRGR in the ER of mouse skeletal myoblast cells triggers ER-calcium release (measured by R-CaMP1.07) (a).195 ChRGR – chimeric protein CrChR1 (channelrhodopsin-1 from Chlamydomonas reinhardtii) with the sixth helix domain from CrChR2 (channelrhodopsin-2 from Chlamydomonas reinhardtii).138 Activation of ER-expressed OLPVR1 (Organic Lake phycodnavirus rhodopsin 1) leads to a rise of intracellular Ca2+ by a release from IP3-dependent Ca2+ stores. Subsequent activation of Ca2+-dependent chloride channels (CaCC) in plasma membrane leads to in Cl− influx in Xenopus oocytes (b). | ||
The general strategy of MRs targeting to subcellular organelles is the addition of the proper signal sequences. However, sometimes no signal sequence is needed to achieve ER targeting. Thus, A.-S. Eria-Oliveira et al.78 demonstrated that type 1 viral channelrhodopsin OLPVR1 (Organic Lake phycodnavirus rhodopsin, construct OLPVR1-GFP) localizes intracellularly in the ER in HEK293T cells (Fig. 7b).
In summary, it is possible to manipulate the Ca2+ release from intracellular stores such as ER/SR using optical stimulation. The organelle optogenetic approach would make it possible to understand the importance of intracellular calcium dynamics with unprecedented spatiotemporal precision in various cells. Optical regulation of internal calcium signaling cascades, initiated by a calcium-induced calcium release or the store-operated Ca2+ entry, is a promising idea for the nearest future.
Challenges of MR-based optogenetic approaches
In order to plan and optimize an optogenetic experiment, one must be aware of possible problems and “side effects”. One of the major challenges in organelle optogenetics is the organelle targeting efficiency of chosen optogenetic tools. MRs directed to any cell compartment require an efficient signal sequence or fusion to an organelle resident protein or its part. Though recently native organelle-targeting motifs were predicted in a variety of rhodopsins.198 There is still no universal strategy for organelle targeting, though a large number of signal sequences have been described, such as those targeting the plasma membrane or the mitochondrial matrix. However, problems could occur due to the complexity of the protein targeting process. In accordance with current knowledge, the use of a particular approach is determined not only by the signal sequence, but also by the protein itself, its length, charge, and hydrophobicity of its regions, etc.128 For example, the N-terminal signal sequence from COX8 has been widely used to target proteins to the mitochondrial matrix.199 Interestingly, the N-terminal addition of one, two, or three repeats of the COX8 signal sequence was not sufficient to target ChR2 to IMM.71,72 It has been confirmed that the use of four repeats of the COX8 signal sequence and the deletion of the first 24 a.a. of rhodopsin is the best option to achieve IMM targeting of ChR2.71,72 In case of successful targeting, the functionality of a MR in subcellular compartments also should be carefully examined. Works describing rhodopsins usually consider their activity in model systems. This gives an idea of the characteristics of the protein. However, it is widely known that the lipid environment200 and ion concentrations201 significantly influence rhodopsins functioning. One should consider the photophysical properties of the rhodopsin, its compatibility with the cellular and organelle environment, and the specific requirements of the experimental design (e.g., desired speed of response, wavelength of activation, etc.). Therefore, one cannot 100% guarantee the desired localization and unambiguously describe the features of protein functioning in a specific organelle. Another potential problem is that an overexpression of proteins in membranes of intracellular organelles which have complex three-dimensional organization (for example, IMM) could potentially lead to structural disruption and dysfunction of organelles.Another limitation is related to the choice of transgene delivery vehicle. The most commonly used are adeno-associated viral (AAV) vectors.5 This type of vectors has a wide range of serotypes and unique tropism; however, their small DNA packaging size is a barrier for some tasks because the AAV genome does not exceed 4,800 bases.202 In addition, AAV could induce an immune response and formation of anti-AAV antibodies, and some serotypes appear to be inefficient for transduction, likely due to preexisting immunity.203 Another problem slowing down the potential clinical application of optogenetics is the immunogenicity of the optogenetic tools themselves, because these proteins are derived from non-human (bacterial, archaeal, and eukaryotic) origin, and humanization of these tools is also necessary.204
Most of the optogenetic results obtained with animals were carried out using classical models – C. elegans, zebrafish, or rodents.205–207 The ease of use of nematodes and zebrafish is due to their rapid development cycle, as well as the almost transparent properties of their body, which allows light to be easily delivered into their tissues. As for rodents, the stimulation of their brain neurons is easily achievable due to a smaller amount of brain tissue however, transferring technologies to the human brain requires significant optimization.208
As mentioned above, the parameters of light activation play an important role in optogenetics. The association between light parameters (such as wavelength, intensity, and duration of illumination) and rhodopsin activity is quite well-established.31 Rhodopsins undergo a photocycle upon photon absorption, which is a critical aspect of their functionality. The specifics of this photocycle, including its duration and the resulting biological effect, vary among different rhodopsin types. The precision of the control is influenced by the specific characteristics of the rhodopsin, the light source used, and the biological context. The response time can vary, but typically, rhodopsin activation occurs rapidly upon light exposure. Thus, to determine the duration of illumination for rhodopsin activation and maintenance of its activity one should consider the photocycle duration of the specific rhodopsin, the desired biological outcome, and the cellular context.
In order to achieve the desired effect and reduce photo- and thermal tissue damage, the light delivery strategy, spectrum, output power, and other parameters must be precisely controlled.5 When operating at the level of tissues or the whole organism, the factor of light transmission should be taken into account. The intensity in the visible range drops by about 10 times when passing one millimeter deep into tissues.209 A red-shift of light for optogenetic stimulation allows deeper penetration into tissue layers and causes less photodamage.210 In some cases, it is possible to activate optogenetic tools with two-photon or even three-photon excitation211 using infrared light with a particularly precise way of focusing. In this case, most of the radiation is scattered over a large volume, and the greatest intensity occurs in a small area at a selected point in the tissue. This approach allows one to perform an optogenetic manipulation within a small area, up to a single cell, and has been used, for example, to map neurons in a brain of living mice.212 However, even infrared light does not penetrate well into a human brain, therefore some light delivery devices may assist in overcoming this limitation. Thus, bringing light to deep-lying tissues is a rather difficult task. Such integration into tissues can be accompanied by cell damage, scarring, or infection, as well as technical difficulties during manipulation.213 Nevertheless, a part of these problems can be avoided by creating thinner and more flexible optical fibers intended for implantation in tissues214 or by using miniaturized, biocompatible, implantable LEDs.62,64,65 A good example is an epidural implant to deliver light into the spinal cord.214 This implant was well tolerated by mice and did not affect their behavioral and motor activity.
Taken together, the organelle optogenetic-approach shares challenges with classical optogenetics: light delivery, gene modification of targeted cells, and others, and has some specific problems concerning organelle targeting of optogenetic tools. Despite this, unique opportunities for organelle manipulation with light are already promising for fundamental research and clinics.
Organelle optogenetics for basic research
Subcellular optogenetics is an emerging new field. It is considerably younger than optogenetics of excitable cells. Nevertheless, it is already clear that organelle optogenetics offers unique opportunities for basic research on cell physiology through the control of physicochemical cellular parameters. Fast and reversible manipulation of organelle ion gradients by light provides unique opportunities to study processes which are difficult to model solely by classical chemical approaches.Cytosol pH regulation by plasma membrane-expressed MR proton pumps is an attractive approach to studying the role of intracellular pH in various processes. Local plasma membrane dynamics and cell motility could be studied.67 In addition, the role of pH in cell differentiation,60 cell cycle progression112 and cell death induction68 could be deciphered.
In mitochondria, expression of channelrhodopsins is an attractive tool for influencing OXPHOS and ATP production, modelling mitochondrial preconditioning, and mitochondrial-induced cell death.72 Optogenetic influence on the mitochondrial calcium levels could become a perspective tool to study the role of mitochondria in calcium signaling and calcium storage in neurons and other cells.71 Proton pump expression in IMM provides an opportunity to energize mitochondria in order to study its role in cell differentiation, stress-tolerance,73 cell death,72 and aging.215
Lysosomal proton pumps are an attractive tool to study lysosome function and a number of cellular processes, controlled by lysosomes.147,173 Endosomes alkalization by MR optogenetics interfered with the internalization of the AMPA receptor at active synapses and blocked motor learning.70 MRs, expressed in synaptic vesicles, is a new tool for studying mechanisms of neurotransmission and the control of function of certain synapses.69
The expression of channelrhodopsins in ER allows manipulating the concentration of intracellular calcium, one of the most important secondary messengers in the cell.78 The intracellular Ca2+ release from ER/SR, and the subsequent activation of other internal Ca2+ signaling cascades such as calcium-induced calcium release or the store-operated Ca2+ entry can be studied.77 In addition, it will be possible to study more accurately the impact of optogenetic changes of intracellular calcium levels on the process of myogenesis.109 In addition, ER optogenetics allows studying the importance of calcium stores in dendritic spines in more detail, and it may provide insight into synaptic plasticity,216 the role of dendritic spines during development, in stress, and neurodegenerative diseases.217,218 Thus, further improvements in the field of organelle optogenetics methodology will allow the manipulation of intracellular Ca2+ in neurobiology as well.
Organelle optogenetics provides researchers with unique tools for cell physiology studies under normal and pathologic conditions. Cell lines with organelle-expressed MRs could become a suitable model of organelle-related pathologies for studying their molecular mechanisms and testing potential therapies.
Clinical perspectives of organelle (intracellular) optogenetics
A visual function recovery is now an impressive example of the successful implementation of optogenetic technologies in clinics. It is an important step towards the medical application of optogenetics.219 Attempts to restore photosensitivity in retinal degenerative disease models based on channelrhodopsin ChR2 or halorhodopsin eNpHR have been made since the beginning of optogenetics.220–222 Several clinical trials are currently underway on the use of optogenetic tools in the treatment of retinitis pigmentosa and retinal dystrophy using ChR2 (NCT02556736), Chrimson (NCT03326336), and Chronos (NCT04278131). Most recently, the first case of partial vision recovery in a patient after optogenetic therapy was reported.223Hearing recovery is another promising application of optogenetics which may be considered as a potential alternative to electrical cochlear implants.224,225 Successful restoration of auditory activity has been shown in rodents through the expression of ChR264,226 and Chronos.227 Using animal models, optogenetics was utilized to stimulate regeneration of muscles, recovery of movements, treatment of muscle paralysis,228,229 treatment of heart diseases using light-induced stimulation of the heart muscle,230–233 and even offered new opportunities for the treatment of neurodegenerative diseases.234
Recently, a photosynthetic thylakoid-based system was efficiently used to increase cellular ATP levels and provide the regeneration of mice joint tissues.235 It may be considered as a proof-of-principle study showing that light-induced ATP synthesis in mammals is very promising. Such stimulation of bioenergetics may be also achieved by mitochondria-targeted MRs. Their targeting into energy-deficient tissues is another perspective application.
Organelle-based optogenetics is now at an earlier phase of development than its plasma-membrane-based counterpart. Still, there are some intriguing results that may be developed towards clinical applications in the near future. Using mitochondrial ChR2, researchers were able to manipulate the spontaneous beating of cardiomyocytes and stop it by light, which could be a step towards optogenetic cardiac stimulation approaches.71 Another application of mitochondrial MRs, proton pumps, which in contrast to ChR2 increase MMP, was proposed to activate mitochondria in T-lymphocytes, the cells which are devoted to cancer cell elimination but suffer from a low-oxygen environment inside a tumor.74 This strategy is a potential optogenetic tool for the improvement of the chimeric antigen receptor T-cell therapy.236 MRs, especially proton pumps, expressed in lysosomes, could be tested as a potential optogenetic strategy to compensate for lysosome dysfunction in certain lysosomal storage diseases.69 ER-targeted MRs are perspective tools for optogenetic control of muscle contraction.77 Finally, synaptic vesicles-directed MRs could potentially improve neurotransmission.69
Taking all of this into account, we can conclude that an organelle MR-based optogenetic approach may help to develop novel therapeutic strategies for a number of cardiac, metabolic, and neurologic disorders, as well as cancer.
Thus, organelle optogenetics is a relatively young, but rapidly developing approach offering amazing opportunities to control different cell functions in a precise and non-invasive way. Optogenetic manipulations with MMP, pH of lysosomes, endosomes, synaptic vesicles, cytosol, mitochondrial, and ER calcium concentration were already demonstrated and proved their efficiency. The field has tremendous potential for further development, despite some challenges in protein targeting, light penetration or light delivery. Certainly, an expanding toolkit will considerably enhance the development of intracellular optogenetics.
Author contributions
A. D. V. designed and wrote the manuscript. S. M. B. contributed to the “Mitochondria: optogenetic control of mitochondrial membrane potential” section. D. F. B. and N. S. I. contributed to the “Lysosomes and endosomes: optogenetic control of pH and proteolysis” and “Synaptic vesicles: optogenetic control of neurotransmission” sections. A. P. P., A. V. V. and A. E. M. contributed to the “Endoplasmic reticulum: optogenetic control of calcium signaling” section. V. N. U. and E. V. Z. contributed to the “Structure and functions of microbial rhodopsins” and “Abbreviated (un)structural overview of optogenetics” sections. F. M. T. contributed to the “Advantages of MR-based optogenetic approaches” section. S. M. B. and F. M. T. contributed to the “Challenges of MR-based optogenetic approaches” section. S. M. B., D. F. B. and A. P. P. contributed to the “Organelle optogenetics for basic research” and “Clinical perspectives of organelle (intracellular) optogenetics” sections. S. V. N., A. O. B., and A. V. V., A. I. K., V. N. U., E. B. and V. I. B. edited the text of the manuscript. V. N. U., A. V. V., A. I. K., and V. I. B. managed the project. E. B. provided general support for this work. V. I. G. supervised the project and contributed to all the sections of the manuscript.Conflicts of interest
The authors declare that they have no conflict of interest.Acknowledgements
Preparation of the sections “Plasma membrane: membrane potential and metabolic control”, “Mitochondria: optogenetic control of mitochondrial membrane potential”, “Strategies of MR organelle targeting”, “Advantages of MR-based optogenetic approaches” and “Challenges of MR-based optogenetic approaches” was done with the support from the Russian Science Foundation (RSF) Project 21-64-00018. A. V. V. and E. V. Z. acknowledge the support from the Ministry of Science and Higher Education of the Russian Federation (agreement # 075-03-2024-117, project FSMG-2021-0002, sections “Endoplasmic reticulum: optogenetic control of calcium signaling” and “Structure and functions of microbial rhodopsins”). N. S. I. acknowledges the Ministry of Science and Higher Education of the Russian Federation (agreement # 075-03-2024-117, project FSMG-2024-0012, section “Lysosomes and endosomes: optogenetic control of pH and proteolysis” and “Synaptic vesicles: optogenetic control of neurotransmission”). A. D. V. acknowledges the support from the Ministry of Science and Higher Education of the Russian Federation (Agreement # 075-01593-23-04, Project 720000F.99.1.BN62AA40000, sections “Organelle optogenetics for basic research” and “Clinical perspectives of organelle (intracellular) optogenetics”).References
- K. Deisseroth, Nat. Methods, 2011, 8, 26–29 CrossRef CAS PubMed.
- B. R. Rost, F. Schneider-Warme, D. Schmitz and P. Hegemann, Neuron, 2017, 96, 572–603 CrossRef CAS PubMed.
- A. Alekseev, V. Gordeliy and E. Bamberg, Rhodopsin, 2022, pp. 71–100 Search PubMed.
- E. S. Boyden, F. Zhang, E. Bamberg, G. Nagel and K. Deisseroth, Nat. Neurosci., 2005, 8, 1263–1268 CrossRef CAS PubMed.
- V. Emiliani, E. Entcheva, R. Hedrich, P. Hegemann, K. R. Konrad, C. Lüscher, M. Mahn, Z. H. Pan, R. R. Sims, J. Vierock and O. Yizhar, Nat. Rev. Methods Primers, 2022 DOI:10.1038/s43586-022-00136-4.
- G. Nagel, T. Szellas, W. Huhn, S. Kateriya, N. Adeishvili, P. Berthold, D. Ollig, P. Hegemann and E. Bamberg, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 13940–13945 CrossRef CAS PubMed.
- K. Deisseroth, Nat. Neurosci., 2015, 18, 1213–1225 CrossRef CAS PubMed.
- I. Gushchin and V. Gordeliy, Subcell. Biochem., 2018, 87, 19–56 CAS.
- V. Gordeliy, K. Kovalev, E. Bamberg, F. Rodriguez-Valera, E. Zinovev, D. Zabelskii, A. Alekseev, I. Rosselli, R. Gushchin and I. Okhrimenko, Rhodopsin, 2022, pp. 1–52 Search PubMed.
- P. Tan, L. He, Y. Huang and Y. Zhou, Physiol. Rev., 2022, 102, 1263–1325 CrossRef CAS PubMed.
- A. A. M. Fischer, M. M. Kramer, G. Radziwill and W. Weber, Curr. Opin. Chem. Biol., 2022, 70, 102196 CrossRef CAS PubMed.
- G. Rivera-cancel, L. B. Motta-mena and K. H. Gardner, Biochemistry, 2012, 51, 10024–10034 CrossRef CAS PubMed.
- B. L. Sinnen, A. B. Bowen, J. S. Forte, B. G. Hiester, K. C. Crosby, E. S. Gibson, M. L. Dell’Acqua and M. J. Kennedy, Neuron, 2017, 93, 646–660.e5 CrossRef CAS PubMed.
- L. J. Bugaj, D. P. Spelke, C. K. Mesuda, M. Varedi, R. S. Kane and D. V. Schaffer, Nat. Commun., 2015, 6 DOI:10.1038/ncomms7898.
- P. Van Bergeijk, M. Adrian, C. C. Hoogenraad and L. C. Kapitein, Nature, 2015, 518, 111–114 CrossRef CAS PubMed.
- J. E. Toettcher, O. D. Weiner and W. A. Lim, Cell, 2013, 155, 1422–1434 CrossRef CAS PubMed.
- R. M. Hughes, D. J. Freeman, K. N. Lamb, R. M. Pollet, W. J. Smith and D. S. Lawrence, Angew. Chem., Int. Ed., 2015, 54, 12064–12068 CrossRef CAS PubMed.
- E. Dine and J. E. Toettcher, Biochemistry, 2018, 57, 2432–2436 CrossRef CAS PubMed.
- E. Alghoul, J. Basbous and A. Constantinou, STAR Protoc., 2021, 2, 100677 CrossRef CAS PubMed.
- A. Rademacher, F. Erdel, R. Weinmann and K. Rippe, Methods Mol. Biol., 2023, 2563, 395–411 CrossRef PubMed.
- B. Y. Chow, X. Han, A. S. Dobry, X. Qian, A. S. Chuong, M. Li, M. A. Henninger, G. M. Belfort, Y. Lin, P. E. Monahan and E. S. Boyden, Nature, 2010, 463, 98–102 CrossRef CAS PubMed.
- B. R. Rost, J. Wietek, O. Yizhar and D. Schmitz, Nat. Neurosci., 2022, 25, 984–998 CrossRef CAS PubMed.
- M. Garita-Hernandez, L. Guibbal, L. Toualbi, F. Routet, A. Chaffiol, C. Winckler, M. Harinquet, C. Robert, S. Fouquet, S. Bellow, J. A. Sahel, O. Goureau, J. Duebel and D. Dalkara, Front. Neurosci., 2018, 12, 789 CrossRef PubMed.
- V. Gradinaru, F. Zhang, C. Ramakrishnan, J. Mattis, R. Prakash, I. Diester, I. Goshen, K. R. Thompson and K. Deisseroth, Cell, 2010, 141, 154–165 CrossRef CAS PubMed.
- M. Engelhard, Methods Mol. Biol., 2022, 2501, 53–69 CrossRef CAS PubMed.
- E. G. Govorunova, O. A. Sineshchekov, H. Li and J. L. Spudich, Annu. Rev. Biochem., 2017, 86, 845–872 CrossRef CAS PubMed.
- O. P. Ernst, D. T. Lodowski, M. Elstner, P. Hegemann, L. S. Brown and H. Kandori, Chem. Rev., 2014, 114, 126–163 CrossRef CAS PubMed.
- T. Kouyama and M. Murakami, Photochem. Photobiol. Sci., 2010, 9, 1458–1465 CrossRef CAS PubMed.
- J. P. Klare, I. Chizhov and M. Engelhard, Results Probl. Cell Differ., 2008, 45, 73–122 CAS.
- J. L. Spudich, C. S. Yang, K. H. Jung and E. N. Spudich, Annu. Rev. Cell Dev. Biol., 2000, 16, 365–392 CrossRef CAS PubMed.
- V. Gordeliy, K. Kovalev, E. Bamberg, F. Rodriguez-Valera, E. Zinovev, D. Zabelskii, A. Alekseev, R. Rosselli, I. Gushchin and I. Okhrimenko, Methods Mol. Biol., 2022, 2501, 1–52 CrossRef CAS PubMed.
- L. S. Brown, Biochim. Biophys. Acta, 2014, 1837, 553–561 CrossRef CAS PubMed.
- H. Luecke, B. Schobert, J.-P. Cartailler, H.-T. Richter, A. Rosengarth, R. Needleman and J. K. Lanyi, J. Mol. Biol., 2000, 300, 1237–1255 CrossRef CAS PubMed.
- Z. Obradovic, K. Peng, S. Vucetic, P. Radivojac and A. K. Dunker, Proteins: Struct., Funct., Bioinf., 2005, 61, 176–182 CrossRef CAS PubMed.
- B. Mészáros, G. Erdős and Z. Dosztányi, Nucleic Acids Res., 2018, 46, W329–W337 CrossRef PubMed.
- J. Bürgi, B. Xue, V. N. Uversky and F. G. Van Der Goot, PLoS One, 2016 DOI:10.1371/JOURNAL.PONE.0158594.
- V. N. Uversky, Prog. Mol. Biol. Transl. Sci., 2019, 166, 1–17 CrossRef CAS PubMed.
- Q. Chen, M. Plasencia, Z. Li, S. Mukherjee, D. Patra, C. L. Chen, T. Klose, X. Q. Yao, A. A. Kossiakoff, L. Chang, P. C. Andrews and J. J. G. Tesmer, Nature, 2021, 595, 600–605 CrossRef CAS PubMed.
- X. Fang, A. A. Peden, F. J. M. van Eeden and J. J. Malicki, J. Cell Sci., 2021, 134(6) DOI:10.1242/JCS.254995.
- D. Feng, Z. Chen, K. Yang, S. Miao, B. Xu, Y. Kang, H. Xie and C. Zhao, J. Biol. Chem., 2017, 292, 1735–17386 Search PubMed.
- T. Y. Kim, T. Schlieter, S. Haase and U. Alexiev, Eur. J. Cell Biol., 2012, 91, 300–310 CrossRef CAS PubMed.
- S. T. Ahmad, M. Natochin, N. O. Artemyev and J. E. O’Tousa, FASEB J., 2007, 21, 449–455 CrossRef CAS PubMed.
- M. Ascano and P. R. Robinson, Biochemistry, 2006, 45, 2398–2407 CrossRef CAS PubMed.
- A. V. Gourine, V. Kasymov, N. Marina, F. Tang, M. F. Figueiredo, S. Lane, A. G. Teschemacher, K. M. Spyer, K. Deisseroth and S. Kasparov, Science, 2010, 329, 571–575 CrossRef CAS PubMed.
- Y. Oka, M. Ye and C. S. Zuker, Nature, 2015, 520, 349–352 CrossRef CAS PubMed.
- C. Anaclet, L. Ferrari, E. Arrigoni, C. E. Bass, C. B. Saper, J. Lu and P. M. Fuller, Nat. Neurosci., 2014, 17, 1217–1224 CrossRef CAS PubMed.
- P. S. Kunwar, M. Zelikowsky, R. Remedios, H. Cai, M. Yilmaz, M. Meister and D. J. Anderson, eLife, 2015, 2015, 1–30 Search PubMed.
- Z. Wu, A. E. Autry, J. F. Bergan, M. Watabe-uchida and C. G. Dulac, Nature, 2014, 509, 325–330 CrossRef CAS PubMed.
- D. S. Roy, A. Arons, T. I. Mitchell, M. Pignatelli, T. J. Ryan and S. Tonegawa, Nature, 2016, 531, 508–512 CrossRef CAS PubMed.
- B. M. Kenwood, J. L. Weaver, A. Bajwa, I. K. Poon, F. L. Byrne, B. A. Murrow, J. A. Calderone, L. Huang, A. S. Divakaruni, J. L. Tomsig, K. Okabe, R. H. Lo, G. C. Coleman, L. Columbus, Z. Yan, J. J. Saucerman, J. S. Smith, J. W. Holmes, K. R. Lynch, K. S. Ravichandran, S. Uchiyama, W. L. Santos, G. W. Rogers, M. D. Okusa, D. A. Bayliss and K. L. Hoehn, Mol. Metab., 2014, 3, 114–123 CrossRef CAS PubMed.
- Y. N. Antonenko, L. S. Khailova, D. A. Knorre, O. V. Markova, T. I. Rokitskaya, T. M. Ilyasova, I. I. Severina, E. A. Kotova, Y. E. Karavaeva, A. S. Prikhodko, F. F. Severin and V. P. Skulachev, PLoS One, 2013, 8, 1–13 CrossRef PubMed.
- N.-E. L. Saris, M. A. Andersson, R. Mikkola, L. C. Andersson, V. V. Teplova, P. A. Grigoriev and M. S. Salkinoja-Salonen, Toxicol. Ind. Health, 2009, 25, 441–446 CrossRef CAS PubMed.
- K. A. I. T. Sundquist and S. C. Marks, J. Bone Miner. Res., 1994, 9, 1575–1582 CrossRef CAS PubMed.
- C. Doyno, D. M. Sobieraj and W. L. Baker, Clin. Toxicol., 2021, 59, 12–23 CrossRef CAS PubMed.
- D. R. Hochbaum, Y. Zhao, S. L. Farhi, N. Klapoetke, C. A. Werley, V. Kapoor, P. Zou, J. M. Kralj, D. MacLaurin, N. Smedemark-Margulies, J. L. Saulnier, G. L. Boulting, C. Straub, Y. K. Cho, M. Melkonian, G. K. S. Wong, D. J. Harrison, V. N. Murthy, B. L. Sabatini, E. S. Boyden, R. E. Campbell and A. E. Cohen, Nat. Methods, 2014, 11, 825–833 CrossRef CAS PubMed.
- H. Kandori, Front. Mol. Biosci., 2015, 2, 150981 Search PubMed.
- E. G. Govorunova, O. A. Sineshchekov and J. L. Spudich, Front. Cell. Neurosci., 2022, 15, 800313 CrossRef PubMed.
- J. Prigge and M. Wietek, Optogenetics, 2016, pp. 141–165 Search PubMed.
- F. Zhang, L. Wang, M. Brauner, J. F. Liewald, K. Kay, N. Watzke, P. G. Wood, E. Bamberg, G. Nagel, A. Gottschalk and K. Deisseroth, Nature, 2007, 446, 633–641 CrossRef CAS PubMed.
- X. Han and E. S. Boyden, PLoS One, 2007 DOI:10.1371/journal.pone.0000299.
- K. P. Greenberg, A. Pham and F. S. Werblin, Neuron, 2011, 69, 713–720 CrossRef CAS PubMed.
- L. Li, L. Lu, Y. Ren, G. Tang, Y. Zhao, X. Cai, Z. Shi, H. Ding, C. Liu, D. Cheng, Y. Xie, H. Wang, X. Fu, L. Yin, M. Luo and X. Sheng, Nat. Commun., 2022, 13, 1–14 Search PubMed.
- E. Papagiakoumou, Biol. Cell, 2013, 105, 443–464 CrossRef CAS PubMed.
- D. Keppeler, M. Schwaerzle, T. Harczos, L. Jablonski, A. Dieter, B. Wolf, S. Ayub, C. Vogl, C. Wrobel, G. Hoch, K. Abdellatif, M. Jeschke, V. Rankovic, O. Paul, P. Ruther and T. Moser, Sci. Transl. Med., 2020, 12(553) DOI:10.1126/scitranslmed.abb8086.
- L. Yin, M. J. Rasch, Q. He, S. Wu, F. Dou and Y. Shu, Cereb. Cortex, 2015, bhv245 CrossRef PubMed.
- O. A. Shemesh, D. Tanese, V. Zampini, C. Linghu, K. Piatkevich, E. Ronzitti, E. Papagiakoumou, E. S. Boyden and V. Emiliani, Nat. Neurosci., 2017, 20, 1796–1806 CrossRef CAS PubMed.
- C. E. T. Donahue, M. D. Siroky and K. A. White, J. Am. Chem. Soc., 2021, 143, 18877–18887 CrossRef CAS PubMed.
- S. Nakao, K. Kojima and Y. Sudo, J. Am. Chem. Soc., 2022, 144, 3771–3775 CrossRef CAS PubMed.
- B. R. Rost, F. Schneider, M. K. Grauel, C. Wozny, C. G. Bentz, A. Blessing, T. Rosenmund, T. J. Jentsch, D. Schmitz, P. Hegemann and C. Rosenmund, Nat. Neurosci., 2015, 18, 1845–1852 CrossRef CAS PubMed.
- W. Kakegawa, A. Katoh, S. Narumi, E. Miura, J. Motohashi, A. Takahashi and K. Kohda, Neuron, 2018, 99, 985–998.e6 CrossRef CAS PubMed.
- T. Tkatch, E. Greotti, G. Baranauskas, D. Pendin, S. Roy, L. I. Nita, J. Wettmarshausen, M. Prigge, O. Yizhar, O. S. Shirihai, D. Fishman, M. Hershfinkel, I. A. Fleidervish, F. Perocchi, T. Pozzan and I. Sekler, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E5167–E5176 CrossRef CAS PubMed.
- P. Ernst, N. Xu, J. Qu, H. Chen, M. S. Goldberg, V. Darley-Usmar, J. J. Zhang, B. O’Rourke, X. Liu and L. Zhou, Biophys. J., 2019, 117, 631–645 CrossRef CAS PubMed.
- B. J. Berry, A. J. Trewin, A. S. Milliken, A. Baldzizhar, A. M. Amitrano, Y. Lim, M. Kim and A. P. Wojtovich, EMBO Rep., 2020, 21, 1–14 CrossRef PubMed.
- A. M. Amitrano, B. J. Berry, K. Lim, K. Do Kim, R. E. Waugh, A. P. Wojtovich and M. Kim, Front. Immunol., 2021, 12 Search PubMed.
- K. Y. Hara, T. Wada, K. Kino, T. Asahi and N. Sawamura, Sci. Rep., 2013, 3, 1–4 Search PubMed.
- Y. Imai, T. Inoshita, H. Meng, K. Shiba-Fukushima, K. Y. Hara, N. Sawamura and N. Hattori, Commun. Biol., 2019, 2, 1–11 CrossRef PubMed.
- T. Asano, H. Igarashi, T. Ishizuka and H. Yawo, Front. Neurosci., 2018, 12, 1–8 Search PubMed.
- A.-S. Eria-Oliveira, M. Folacci, A. A. Chassot, S. Fedou, N. Theze, D. Zabelskii, A. Alekseev, E. Bamberg, V. Gordeliy and G. Sandoz, Nat. Commun., 2024, 15, 65 CrossRef PubMed.
- C. Smith, T. Frolinger, J. Brathwaite, S. Sims and G. M. Pasinetti, Commun. Biol., 2018, 1, 1–8 CrossRef CAS PubMed.
- K. M. Daicho, Y. Hirono-Hara, H. Kikukawa, K. Tamura and K. Y. Hara, Microb. Cell Fact., 2024, 23, 4 CrossRef CAS PubMed.
- A. Peterson, K. Baskett, W. C. Ratcliff and A. Burnetti, Curr. Biol., 2024, 34(3), 648–654 CrossRef CAS PubMed.
- A. Vlasova, A. Polyakova, A. Gromova, S. Dolotova, S. Bukhalovich, D. Bagaeva, N. Bondarev, F. Tsybrov, K. Kovalev, A. Mikhailov, D. Sidorov, A. Bogorodskiy, N. Ilyinsky, A. Kuklin, A. Vlasov, V. Borshchevskiy and V. Ivanovich, Int. J. Biol. Macromol., 2023, 242, 124949 CrossRef CAS PubMed.
- A. Hoffmann, V. Hildebrandt, J. Heberle and G. Boldt, Proc. Natl. Acad. Sci. U. S. A., 1994, 91, 9367–9371 CrossRef CAS PubMed.
- Y. Imai, T. Inoshita, H. Meng, K. Shiba-Fukushima, K. Y. Hara, N. Sawamura and N. Hattori, Commun. Biol., 2019, 2, 1–11 CrossRef PubMed.
- B. J. Berry, A. J. Trewin, A. M. Amitrano, M. Kim and A. P. Wojtovich, J. Mol. Biol., 2018, 430, 3873–3891 CrossRef CAS PubMed.
- B. J. Berry, A. Vodičková, A. Müller-eigner, C. Meng, C. Ludwig, M. Kaeberlein, S. Peleg and A. P. Wojtovich, Nat. Aging, 2023, 3, 157–161 CrossRef PubMed.
- D. J. Klionsky, et al. , Autophagy, 2012, 8, 445–544 CrossRef CAS PubMed.
- V. Gradinaru, F. Zhang, C. Ramakrishnan, J. Mattis, R. Prakash, I. Diester, I. Goshen, K. R. Thompson and K. Deisseroth, Cell, 2010, 141, 154–165 CrossRef CAS PubMed.
- J. Deubner, P. Coulon and I. Diester, Curr. Opin. Struct. Biol., 2019, 57, 157–163 CrossRef CAS PubMed.
- L. H. Tai, A. M. Lee, N. Benavidez, A. Bonci and L. Wilbrecht, Nat. Neurosci., 2012, 15, 1281–1289 CrossRef CAS PubMed.
- J. D. Lenz and M. K. Lobo, Behav. Brain Res., 2013, 255, 44–54 CrossRef CAS PubMed.
- R. T. LaLumiere, Front. Behav. Neurosci., 2014, 8, 1–7 Search PubMed.
- D. Riga, M. R. Matos, A. Glas, A. B. Smit, S. Spijker and M. C. Van den Oever, Front. Syst. Neurosci., 2014, 8, 1–19 Search PubMed.
- X. Liu, S. Ramirez, P. T. Pang, C. B. Puryear, A. Govindarajan, K. Deisseroth and S. Tonegawa, Nature, 2012, 484, 381–385 CrossRef CAS PubMed.
- G. Etter, S. van der Veldt, F. Manseau, I. Zarrinkoub, E. Trillaud-Doppia and S. Williams, Nat. Commun., 2019, 10, 1–11 CrossRef CAS PubMed.
- V. Gradinaru, K. R. Thompson and K. Deisseroth, Brain Cell Biol., 2008, 36, 129–139 CrossRef PubMed.
- I. Nilsson, P. Lara, T. Hessa, A. E. Johnson, G. Von Heijne and A. L. Karamyshev, J. Mol. Biol., 2015, 427, 1191–1201 CrossRef CAS PubMed.
- C. Grimm, A. Silapetere, A. Vogt, Y. A. Bernal Sierra and P. Hegemann, Sci. Rep., 2018, 8, 1–12 Search PubMed.
- V. Shevchenko, T. Mager, K. Kovalev, V. Polovinkin, A. Alekseev, J. Juettner, I. Chizhov, C. Bamann, C. Vavourakis, R. Ghai, I. Gushchin, V. Borshchevskiy, A. Rogachev, I. Melnikov, A. Popov, T. Balandin, F. Rodriguez-Valera, D. J. Manstein, G. Bueldt, E. Bamberg and V. Gordeliy, Sci. Adv., 2017, 3, 1–11 Search PubMed.
- K. Inoue, Bull. Chem. Soc. Jpn., 2016, 89, 1416–1424 CrossRef CAS.
- C. Bamann, G. Nagel, E. Bamberg and J. W. Goethe-universita, Curr. Opin. Neurobiol., 2010, 20, 610–616 CrossRef CAS PubMed.
- I. Gushchin, V. Shevchenko, V. Polovinkin, K. Kovalev, A. Alekseev, E. Round, V. Borshchevskiy, T. Balandin, A. Popov, T. Gensch, C. Fahlke, C. Bamann, D. Willbold, G. Büldt, E. Bamberg and V. Gordeliy, Nat. Struct. Mol. Biol., 2015, 22, 390–396 CrossRef CAS PubMed.
- X. Han, B. Y. Chow, H. Zhou, N. C. Klapoetke, A. Chuong, R. Rajimehr, A. Yang, J. W. Baratta, V. Michael, R. Desimone and E. S. Boyden, Front. Syst. Neurosci., 2011, 5, 1–8 Search PubMed.
- D. Zabelskii, N. Dmitrieva, O. Volkov, V. Shevchenko, K. Kovalev, T. Balandin, D. Soloviov, R. Astashkin, E. Zinovev, A. Alekseev, E. Round, V. Polovinkin, I. Chizhov, A. Rogachev, I. Okhrimenko, V. Borshchevskiy, V. Chupin, G. Büldt, N. Yutin, E. Bamberg, E. Koonin and V. Gordeliy, Commun. Biol., 2021, 4(1) DOI:10.1038/s42003-021-02326-4.
- E. G. Govorunova, O. A. Sineshchekov, R. Janz, X. Liu and J. L. Spudich, Science, 2015, 349, 647–650 CrossRef CAS PubMed.
- A. Berndt, S. Y. Lee, C. Ramakrishnan and K. Deisseroth, Science, 2014, 23, 1509–1511 Search PubMed.
- E. G. Govorunova, Y. Gou, O. A. Sineshchekov, H. Li, X. Lu, Y. Wang, L. S. Brown, F. St-pierre, M. Xue and J. L. Spudich, Nat. Neurosci., 2022, 25, 967–974 CrossRef CAS PubMed.
- T. Asano, T. Ishizua and H. Yawo, Biotechnol. Bioeng., 2012, 109, 199–204 CrossRef CAS PubMed.
- T. Asano, T. Ishizuka, K. Morishima and H. Yawo, Sci. Rep., 2015, 5, 8317 CrossRef CAS PubMed.
- L. M. Koch, E. S. Birkeland, S. Battaglioni, X. Helle, M. Meerang, S. Hiltbrunner, A. J. Ibáñez, M. Peter, A. Curioni-Fontecedro, I. Opitz and R. Dechant, Nat. Metab., 2020, 2, 1212–1222 CrossRef CAS PubMed.
- C. Stock and A. Schwab, Pflugers Arch., 2009, 458, 981–992 CrossRef CAS PubMed.
- C. Frantz, G. Barreiro, L. Dominguez, X. Chen, R. Eddy, J. Condeelis, M. J. S. Kelly, M. P. Jacobson and D. L. Barber, J. Cell Biol., 2008, 183, 865–879 CrossRef CAS PubMed.
- M. Oginuma, Y. Harima, O. A. Tarazona, M. Diaz-Cuadros, A. Michaut, T. Ishitani, F. Xiong and O. Pourquié, Nature, 2020, 584, 98–101 CrossRef CAS PubMed.
- M. P. Mattson, M. Gleichmann and A. Cheng, Neuron, 2008, 60, 748–766 CrossRef CAS PubMed.
- H. N. Carter, C. C. W. Chen and D. A. Hood, Physiology, 2015, 30, 208–223 CrossRef CAS PubMed.
- R. K. Naviaux, Eur J. Pediatr., 2000, 159, 219–226 CrossRef PubMed.
- S. Vyas, E. Zaganjor and M. C. Haigis, Cell, 2016, 166, 555–566 CrossRef CAS PubMed.
- M. T. Lin and M. F. Beal, Nature, 2006, 443, 787–795 CrossRef CAS PubMed.
- K. Boengler, M. Kosiol, M. Mayr, R. Schulz and S. Rohrbach, J. Cachexia Sarcopenia Muscle, 2017, 349–369 CrossRef PubMed.
- L. D. Zorova, V. A. Popkov, E. Y. Plotnikov, D. N. Silachev, I. B. Pevzner, S. S. Jankauskas, V. A. Babenko, S. D. Zorov, A. V. Balakireva, M. Juhaszova, S. J. Sollott and D. B. Zorov, Anal. Biochem., 2018, 552, 50–59 CrossRef CAS PubMed.
- J. Suski, M. Lebiedzinska, M. Bonora, P. Pinton, J. Duszynski and M. R. Wieckowski, in Mitochondrial Bioenergetics, ed. C. M. Palmeira and A. J. Moreno, 2018, vol. 1782 Search PubMed.
- K. Demishtein-zohary and A. Azem, Cell Tissue Res., 2017, 33–41 CrossRef CAS PubMed.
- C. Giorgi, S. Marchi and P. Pinton, Nat. Rev. Mol. Cell Biol., 2018, 19(11), 713–730 CrossRef CAS PubMed.
- S. Orrenius, B. Zhivotovsky and P. Nicotera, Nat. Rev. Mol. Cell Biol., 2003, 4, 552–565 CrossRef CAS PubMed.
- H. K. Baumgartner, J. V. Gerasimenko, C. Thorne, P. Ferdek, T. Pozzan, A. V. Tepikin, O. H. Petersen, R. Sutton, A. J. M. Watson and O. V. Gerasimenko, J. Biol. Chem., 2009, 284, 20796–20803 CrossRef CAS PubMed.
- E. Kummer and N. Ban, Nat. Rev. Mol. Cell Biol., 2021, 22, 307–325 CrossRef CAS PubMed.
- T. Kleine, U. G. Maier and D. Leister, Annu. Rev. Plant Biol., 2009, 60, 115–138 CrossRef CAS PubMed.
- N. Wiedemann and N. Pfanner, Annu. Rev. Biochem., 2017, 86, 685–714 CrossRef CAS PubMed.
- M. C. Avendaño-Monsalve, J. C. Ponce-Rojas and S. Funes, Biol. Chem., 2020, 401, 645–661 CrossRef PubMed.
- N. Pfanner, B. Warscheid and N. Wiedemann, Nat. Rev. Mol. Cell Biol., 2019, 20, 267–284 CrossRef CAS PubMed.
- D. Mokranjac and W. Neupert, Biochim. Biophys. Acta, Bioenerg., 2010, 1797, 1045–1054 CrossRef CAS PubMed.
- B. Homberg, P. Rehling and L. D. Cruz-Zaragoza, Trends Cell Biol., 2023, 33, 765–772 CrossRef CAS PubMed.
- B. J. Berry and A. P. Wojtovich, FEBS J., 2020, 287, 4544–4556 CrossRef CAS PubMed.
- E. C. Hurt, B. Pesold-Hurt and G. Schatz, FEBS Lett., 1984, 178, 306–310 CrossRef CAS PubMed.
- A. E. Palmer, M. Giacomello, T. Kortemme, S. A. Hires, V. Lev-Ram, D. Baker and R. Y. Tsien, Chem. Biol., 2006, 13, 521–530 CrossRef CAS PubMed.
- S. A. Graf, S. E. Haigh, E. D. Corson and O. S. Shirihai, J. Biol. Chem., 2004, 279, 42954–42963 CrossRef CAS PubMed.
- C. Gieffers, F. Korioth, P. Heimann, C. Ungermann and J. Frey, Exp. Cell Res., 1997, 232, 395–399 CrossRef CAS PubMed.
- K. Y. Ma, M. R. Fokkens, F. Reggiori, M. Mari and D. S. Verbeek, Transl. Neurodegener, 2021, 10, 1–17 CrossRef PubMed.
- S. Demine, P. Renard and T. Arnould, Cells, 2019, 8(8) DOI:10.3390/cells8080795.
- E. J. Blott and G. M. Griffiths, Nat. Rev. Mol. Cell Biol., 2002, 3, 122–131 CrossRef CAS PubMed.
- J. P. Luzio, P. R. Pryor and N. A. Bright, Nat. Rev. Mol. Cell Biol., 2007, 8, 622–632 CrossRef CAS PubMed.
- P. Saftig and J. Klumperman, Nat. Rev. Mol. Cell Biol., 2009, 10, 623–635 CrossRef CAS PubMed.
- S. R. Bonam, F. Wang and S. Muller, Nat. Rev. Drug Discovery, 2019, 18, 923–948 CrossRef CAS PubMed.
- J. A. Mindell, Annu. Rev. Physiol., 2012, 74, 69–86 CrossRef CAS PubMed.
- Y. B. Hu, E. B. Dammer, R. J. Ren and G. Wang, Transl. Neurodegener, 2015, 4, 1–10 CrossRef PubMed.
- Q. Song, B. Meng, H. Xu and Z. Mao, Transl. Neurodegener., 2020, 9, 1–14 CrossRef PubMed.
- A. Ballabio and J. S. Bonifacino, Nat. Rev. Mol. Cell Biol., 2019, 21, 101–118 CrossRef PubMed.
- J. Gilleron, J. M. Gerdes and A. Zeigerer, Traffic, 2019, 20, 552–570 CrossRef CAS PubMed.
- P. Boya and G. Kroemer, Oncogene, 2008, 27, 6434–6451 CrossRef CAS PubMed.
- H. Appelqvist, P. Wäster, K. Kågedal and K. Öllinger, J. Mol. Cell Biol., 2013, 5, 214–226 CrossRef CAS PubMed.
- T. Braulke and J. S. Bonifacino, Biochim. Biophys. Acta, Mol. Cell Res., 2009, 1793, 605–614 CrossRef CAS PubMed.
- S. R. Carlsson, J. Roth, F. Piller and M. Fukuda, J. Biol. Chem., 1988, 263, 18911–18919 CrossRef CAS PubMed.
- Y. Zhao, J. Ren, S. Padilla-Parra, E. E. Fry and D. I. Stuart, Nat. Commun., 2014, 5(1) DOI:10.1038/ncomms5321.
- M. S. Pols and J. Klumperman, Exp. Cell Res., 2009, 315, 1584–1592 CrossRef CAS PubMed.
- A. Mironov and M. Pavelka, The Golgi Apparatus, 2008, vol. 13 Search PubMed.
- J. S. Bonifacino and L. M. Traub, Annu. Rev. Biochem., 2003, 72, 395–447 CrossRef CAS PubMed.
- J. S. Bonifacino and J. Lippincott-Schwartz, Nat. Rev. Mol. Cell Biol., 2003, 4, 409–414 CrossRef CAS PubMed.
- D. L. Hogue, C. Nash, V. Ling and T. C. Hobman, Biochem. J., 2002, 365, 721–730 CrossRef CAS PubMed.
- S. Storch, S. Pohl and T. Braulke, J. Biol. Chem., 2004, 279, 53625–53634 CrossRef CAS PubMed.
- H. Schulze, T. Kolter and K. Sandhoff, Biochim. Biophys. Acta, Mol. Cell Res., 2009, 1793, 674–683 CrossRef CAS PubMed.
- J. L. Jeger, Mol. Biol. Rep., 2020, 47, 9801–9810 CrossRef CAS PubMed.
- J. Huotari and A. Helenius, EMBO J., 2011, 30, 3481–3500 CrossRef CAS PubMed.
- A. Conte and S. Sigismund, The Ubiquitin Network in the Control of EGFR Endocytosis and Signaling, Elsevier Inc., 2016, ch. 6, vol. 141 Search PubMed.
- F. R. Maxfield and D. J. Yamashiro, Adv. Exp. Med. Biol., 1987, 225, 189–198 CrossRef CAS PubMed.
- J. Rink, E. Ghigo, Y. Kalaidzidis and M. Zerial, Cell, 2005, 122, 735–749 CrossRef CAS PubMed.
- Y. G. Zhao, P. Codogno and H. Zhang, Nat. Rev. Mol. Cell Biol., 2021, 22, 733–750 CrossRef CAS PubMed.
- J. P. Luzio, S. R. Gray and N. A. Bright, Biochem. Soc. Trans., 2010, 38, 1413–1416 CrossRef CAS PubMed.
- G. Parenti, D. L. Medina and A. Ballabio, EMBO Mol. Med., 2021, 13(2), e12836 CrossRef CAS PubMed.
- F. M. Platt, A. D’Azzo, B. L. Davidson, E. F. Neufeld and C. J. Tifft, Nat. Rev. Dis. Primers, 2018, 4(1), 27 CrossRef PubMed.
- D. S. Rao, S. V. Bradley, P. D. Kumar, T. S. Hyun, D. Saint-Dic, K. Oravecz-Wilson, C. G. Kleer and T. S. Ross, Cancer Cell, 2003, 3, 471–482 CrossRef CAS PubMed.
- R. A. Nixon, Neurobiol. Aging, 2005, 26, 373–382 CrossRef CAS PubMed.
- D. Wang, C. C. Chan, S. Cherry and P. R. Hiesinger, Cell. Mol. Life Sci., 2013, 70, 2919–2934 CrossRef CAS PubMed.
- N. S. Ilyinsky, S. M. Bukhalovich, D. F. Bagaeva, S. V. Nesterov, A. A. Alekseev, F. M. Tsybrov, A. O. Bogorodskiy, O. V. Moiseeva, A. D. Vlasova, K. V. Kovalev, A. E. Mikhailov, A. V. Rogachev, E. Bamberg, V. Ivanovich and V. I. Borshchevskiy, bioRxiv, 2023, preprint, DOI:10.1101/2023.08.02.551716.
- A. Vandewalle, F. Cluzeaud, K. C. Peng, M. Bens, A. Lüchow, W. Günther and T. J. Jentsch, Am. J. Physiol.: Cell Physiol., 2001, 280(2) DOI:10.1152/AJPCELL.2001.280.2.C373.
- H. Xu and D. Ren, Annu. Rev. Physiol., 2015, 77, 57–80 CrossRef CAS PubMed.
- P. Li, M. Gu and H. Xu, Trends Biochem. Sci., 2019, 44, 110–124 CrossRef CAS PubMed.
- P. C. Trivedi, J. J. Bartlett and T. Pulinilkunnil, Cells, 2020, 9, 1–35 CrossRef PubMed.
- C. Meyer-Schwesinger, Cell Tissue Res., 2021, 385, 371–392 CrossRef CAS PubMed.
- T. C. Südhof, Cell, 2006, 127, 671–673 CrossRef PubMed.
- K. W. Beyenbach and H. Wieczorek, J. Exp. Biol., 2006, 209, 577–589 CrossRef CAS PubMed.
- M. Forgac, Nat. Rev. Mol. Cell Biol., 2007, 8, 917–929 CrossRef CAS PubMed.
- J. Burré and W. Volknandt, J. Neurochem., 2007, 101, 1448–1462 CrossRef PubMed.
- S. Wittig, M. Ganzella, M. Barth, S. Kostmann, D. Riedel, Á. Pérez-Lara, R. Jahn and C. Schmidt, Nat. Commun., 2021, 12, 858 CrossRef CAS PubMed.
- Y. Xu, X. Song, D. Wang, Y. Wang, P. Li and J. Li, Mol. Brain, 2021, 14, 37 CrossRef PubMed.
- Z. Taoufiq, M. Ninov, A. Villar-Briones, H.-Y. Wang, T. Sasaki, M. C. Roy, F. Beauchain, Y. Mori, T. Yoshida, S. Takamori, R. Jahn and T. Takahashi, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 33586–33596 CrossRef CAS PubMed.
- S.-H. Song and G. J. Augustine, Mol. Cells, 2015, 38, 936–940 CrossRef CAS PubMed.
- S. L. Jackman, J. Turecek, J. E. Belinsky and W. G. Regehr, Nature, 2016, 529, 88–91 CrossRef CAS PubMed.
- H. Liu, H. Bai, E. Hui, L. Yang, C. S. Evans, Z. Wang, S. E. Kwon and E. R. Chapman, eLife, 2014, 3 DOI:10.7554/eLife.01524.
- A. D. Blagoveshchenskaya, E. W. Hewitt and D. F. Cutler, Di-Leucine Signals Mediate Targeting of Tyrosinase and Synaptotagmin to Synaptic-like Microvesicles within PC12 Cells, 1999, vol. 10 Search PubMed.
- V. F. Prado and M. A. M. Prado, Signals Involved in Targeting Membrane Proteins to Synaptic Vesicles, 2002, vol. 22 Search PubMed.
- D. J. Adams, C. P. Arthur and M. H. B. Stowell, Sci. Rep., 2015, 5, 1–9 Search PubMed.
- D. S. Schwarz and M. D. Blower, Cell. Mol. Life Sci., 2016, 73, 79–94 CrossRef CAS PubMed.
- T. A. Rapoport, Nature, 2007, 450, 663–669 CrossRef CAS PubMed.
- D. Burdakov, O. H. Petersen and A. Verkhratsky, Cell Calcium, 2005, 38, 303–310 CrossRef CAS PubMed.
- J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov, O. Ronneberger, K. Tunyasuvunakool, R. Bates, A. Žídek, A. Potapenko, A. Bridgland, C. Meyer, S. A. A. Kohl, A. J. Ballard, A. Cowie, B. Romera-Paredes, S. Nikolov, R. Jain, J. Adler, T. Back, S. Petersen, D. Reiman, E. Clancy, M. Zielinski, M. Steinegger, M. Pacholska, T. Berghammer, S. Bodenstein, D. Silver, O. Vinyals, A. W. Senior, K. Kavukcuoglu, P. Kohli and D. Hassabis, Nature, 2021, 596, 583–589 CrossRef CAS PubMed.
- R. Coronado, J. Morrissette, M. Sukhareva and D. M. Vaughan, Am. J. Physiol.: Cell Physiol., 1994, 266, C1485–C1504 CrossRef CAS PubMed.
- L. Wen, H. Wang, S. Tanimoto, R. Egawa, Y. Matsuzaka, H. Mushiake, T. Ishizuka and H. Yawo, PLoS One, 2010, 5(9) DOI:10.1371/journal.pone.0012893.
- K. Sushmita, S. Sharma, M. S. Kaushik and S. Kateriya, Biophys. Physicobiol., 2023, 20, e201008 CrossRef CAS PubMed.
- N. Wiedemann, A. E. Frazier and N. Pfanner, J. Biol. Chem., 2004, 279, 14473–14476 CrossRef CAS PubMed.
- O. Soubias, W. E. Teague, K. G. Hines and K. Gawrisch, Biophys. J., 2015, 108, 1125–1132 CrossRef CAS PubMed.
- I. S. Okhrimenko, K. Kovalev, L. E. Petrovskaya, N. S. Ilyinsky, A. A. Alekseev, E. Marin, T. I. Rokitskaya, Y. N. Antonenko, S. A. Siletsky, P. A. Popov, Y. A. Zagryadskaya, D. V. Soloviov, I. V. Chizhov, D. V. Zabelskii, Y. L. Ryzhykau, A. V. Vlasov, A. I. Kuklin, A. O. Bogorodskiy, A. E. Mikhailov, D. V. Sidorov, S. Bukhalovich, F. Tsybrov, S. Bukhdruker, A. D. Vlasova, V. I. Borshchevskiy, D. A. Dolgikh, M. P. Kirpichnikov, E. Bamberg and V. I. Gordeliy, Commun. Chem., 2023, 6, 1–16 CrossRef PubMed.
- S. R. De Silva and A. T. Moore, J. Physiol., 2022, 21, 4623–4632 CrossRef PubMed.
- S. D. Mendoza, Y. El-Shamayleh and G. D. Horwitz, J. Neurophysiol., 2017, 117, 2004–2013 CrossRef PubMed.
- J. Wu, D. L. Prole, Y. Shen, Z. Lin, A. Gnanasekaran, Y. Liu, L. Chen, H. Zhou, S. R. W. Chen, Y. M. Usachev, C. W. Taylor and R. E. Campbell, Biochem. J., 2014, 464, 13–22 CrossRef CAS PubMed.
- A. Prabhakar, D. Vujovic, L. Cui, W. Olson and W. L. Id, PLoS One, 2019, 2, 1–11 Search PubMed.
- Y. Kimura, C. Satou, S. Fujioka, W. Shoji, K. Umeda, T. Ishizuka and H. Yawo, Curr. Biol., 2013, 23, 843–849 CrossRef CAS PubMed.
- J. F. Liewald, M. Brauner, G. J. Stephens, M. Bouhours, C. Schultheis, M. Zhen and A. Gottschalk, Nat. Methods, 2008, 5, 895–902 CrossRef CAS PubMed.
- Y. Shen, R. E. Campbell, D. C. Cote and M.-E. Paquet, Front. Neural Circuits, 2020, 14, 1–11 CrossRef PubMed.
- W. Yang, L. Carrillo-Reid, Y. Bando, D. S. Peterka and R. Yuste, eLife, 2018, 7, 1–21 CAS.
- K. Lehtinen, M. S. Nokia and H. Takala, Front. Cell. Neurosci., 2022, 15, 1–20 Search PubMed.
- W. Zong, R. Wu, M. Li, Y. Hu, Y. Li, J. Li, H. Rong, H. Wu, Y. Xu, Y. Lu, H. Jia, M. Fan, Z. Zhou, Y. Zhang, A. Wang, L. Chen and H. Cheng, Nat. Methods, 2017, 14, 713–719 CrossRef CAS PubMed.
- K. T. Newmaster, F. A. Kronman, Y. T. Wu and Y. Kim, Front. Neuroanat., 2022, 15, 1–19 Search PubMed.
- R. P. Bonin, F. Wang and M. Desrochers, Mol. Pain, 2016, 12, 1–11 CrossRef CAS PubMed.
- M. R. B. Kim Tae-il, J. G. McCall, Y. Hwan Jung, X. Huang, E. R. Siuda, Y. Li, J. Song, Y. Min Song, H. An Pao, R.-H. Kim, C. Lu, S. Dan Lee, I.-S. Song, G. Shin, R. Al-Hasani, S. Kim, M. Peun Tan and Y. Huang, Science, 2013, 340, 211–216 CrossRef PubMed.
- B. J. Berry, A. Vodičková, A. Müller-eigner, C. Meng, C. Ludwig, M. Kaeberlein, S. Peleg and A. P. Wojtovich, Nat. Aging, 2023, 3, 157–161 CrossRef PubMed.
- T. Collin, A. Marty and I. Llano, Curr. Opin. Neurobiol., 2005, 15, 275–281 CrossRef CAS PubMed.
- M. Segal and E. Korkotian, Neuroscientist, 2016, 22, 477–485 CrossRef CAS PubMed.
- M. Segal and E. Korkotian, Front. Neuroanat., 2014, 8, 1–7 Search PubMed.
- A. Bansal, S. Shikha and Y. Zhang, Nat. Biomed. Eng., 2023, 7(4) DOI:10.1038/s41551-021-00829-3.
- V. Busskamp, J. Duebel, D. Balya, M. Fradot, T. J. Viney, S. Siegert, A. C. Groner, E. Cabuy, V. Forster, M. Seeliger, M. Biel, P. Humphries, M. Paques, S. Mohand-Said, D. Trono, K. Deisseroth, J. A. Sahel, S. Picaud and B. Roska, Science, 2010, 329, 413–417 CrossRef CAS PubMed.
- P. S. Lagali, D. Balya, G. B. Awatramani, T. A. Münch, D. S. Kim, V. Busskamp, C. L. Cepko and B. Roska, Nat. Neurosci., 2008, 11, 667–675 CrossRef CAS PubMed.
- A. Bi, J. Cui, Y.-P. Ma, E. Olshevskaya, M. Pu, A. M. Dizhoor and Z.-H. Pan, Neuron, 2006, 50, 23–33 CrossRef CAS PubMed.
- J. A. Sahel, E. Boulanger-Scemama, C. Pagot, A. Arleo, F. Galluppi, J. N. Martel, S. D. Esposti, A. Delaux, J. B. de Saint Aubert, C. de Montleau, E. Gutman, I. Audo, J. Duebel, S. Picaud, D. Dalkara, L. Blouin, M. Taiel and B. Roska, Nat. Med., 2021, 27, 1223–1229 CrossRef CAS PubMed.
- J. A. Brant, D. O. Adewole, F. Vitale and D. K. Cullen, Curr. Opin. Biotechnol, 2021, 72, 131–138 CrossRef CAS PubMed.
- A. Dieter, D. Keppeler and T. Moser, EMBO Mol. Med., 2020, 12, 1–16 Search PubMed.
- V. H. Hernandez, A. Gehrt, K. Reuter, Z. Jing, M. Jeschke, A. M. Schulz, G. Hoch, M. Bartels, G. Vogt, C. W. Garnham, H. Yawo, Y. Fukazawa, G. J. Augustine and E. Bamberg, J. Clin. Invest., 2014, 124, 1114–1129 CrossRef PubMed.
- A. Mittring, T. Moser and A. Huet, Brain Stimul, 2023, 16, 466–483 CrossRef PubMed.
- T. Bruegmann, T. Van Bremen, C. C. Vogt, T. Send, B. K. Fleischmann and P. Sasse, Nat. Commun., 2015, 6, 1–8 Search PubMed.
- P. Magown, B. Shettar, Y. Zhang and V. F. Rafuse, Nat. Commun., 2015, 6(1) DOI:10.1038/ncomms9506.
- U. Nussinovitch and L. Gepstein, Nat. Biotechnol., 2015, 33, 750–754 CrossRef CAS PubMed.
- A. Arrenberg, D. Stainier, H. Baier and J. Huisken, Science, 2010, 330, 971–974 CrossRef CAS PubMed.
- T. Bruegmann, D. Malan, M. Hesse, T. Beiert, C. J. Fuegemann, B. K. Fleischmann and P. Sasse, Nat. Methods, 2010, 7, 897–900 CrossRef CAS PubMed.
- P. Sasse, M. Funken, T. Beiert and T. Bruegmann, Front. Physiol., 2019, 10 DOI:10.3389/fphys.2019.00675.
- P. Mirzayi, P. Shobeiri, A. Kalantari, G. Perry and N. Rezaei, Mol. Brain, 2022, 15, 1–14 CrossRef PubMed.
- P. Chen, X. Liu, C. Gu, P. Zhong, N. Song, M. Li, Z. Dai, X. Fang, Z. Liu, J. Zhang, R. Tang, S. Fan and X. Lin, Nature, 2022, 612, 546–554 CrossRef CAS PubMed.
- A. M. Amitrano and M. Kim, Immune Netw., 2023, 23, e9 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3cs00699a |
| This journal is © The Royal Society of Chemistry 2024 |








