Open Access Article
Open Access ArticlePlasmonic nanoparticle sensors: current progress, challenges, and future prospects
Krishna
Kant
ab,
Reshma
Beeram
c,
Yi
Cao
d,
Paulo S. S.
dos Santos
e,
Lara
González-Cabaleiro
a,
Daniel
García-Lojo
 a,
Heng
Guo
f,
Younju
Joung
g,
Siddhant
Kothadiya
hi,
Marta
Lafuente
a,
Heng
Guo
f,
Younju
Joung
g,
Siddhant
Kothadiya
hi,
Marta
Lafuente
 jk,
Yong Xiang
Leong
l,
Yiyi
Liu
m,
Yuxiong
Liu
m,
Sree Satya Bharati
Moram
c,
Sanje
Mahasivam
jk,
Yong Xiang
Leong
l,
Yiyi
Liu
m,
Yuxiong
Liu
m,
Sree Satya Bharati
Moram
c,
Sanje
Mahasivam
 n,
Sonia
Maniappan
o,
Daniel
Quesada-González
n,
Sonia
Maniappan
o,
Daniel
Quesada-González
 p,
Divakar
Raj
q,
Pabudi
Weerathunge
n,
Xinyue
Xia
r,
Qian
Yu
g,
Sara
Abalde-Cela
s,
Ramon A.
Alvarez-Puebla
p,
Divakar
Raj
q,
Pabudi
Weerathunge
n,
Xinyue
Xia
r,
Qian
Yu
g,
Sara
Abalde-Cela
s,
Ramon A.
Alvarez-Puebla
 tu,
Rizia
Bardhan
tu,
Rizia
Bardhan
 hi,
Vipul
Bansal
hi,
Vipul
Bansal
 n,
Jaebum
Choo
n,
Jaebum
Choo
 g,
Luis C. C.
Coelho
ev,
José M. M. M.
de Almeida
g,
Luis C. C.
Coelho
ev,
José M. M. M.
de Almeida
 ew,
Sergio
Gómez-Graña
a,
Marek
Grzelczak
x,
Pablo
Herves
a,
Jatish
Kumar
ew,
Sergio
Gómez-Graña
a,
Marek
Grzelczak
x,
Pablo
Herves
a,
Jatish
Kumar
 o,
Theobald
Lohmueller
o,
Theobald
Lohmueller
 y,
Arben
Merkoçi
y,
Arben
Merkoçi
 pz,
José Luis
Montaño-Priede
x,
Xing Yi
Ling
pz,
José Luis
Montaño-Priede
x,
Xing Yi
Ling
 l,
Reyes
Mallada
jkaa,
Jorge
Pérez-Juste
l,
Reyes
Mallada
jkaa,
Jorge
Pérez-Juste
 a,
María P.
Pina
a,
María P.
Pina
 jkaa,
Srikanth
Singamaneni
jkaa,
Srikanth
Singamaneni
 m,
Venugopal Rao
Soma
cab,
Mengtao
Sun
m,
Venugopal Rao
Soma
cab,
Mengtao
Sun
 d,
Limei
Tian
d,
Limei
Tian
 f,
Jianfang
Wang
f,
Jianfang
Wang
 r,
Lakshminarayana
Polavarapu
r,
Lakshminarayana
Polavarapu
 *a and
Isabel Pastoriza
Santos
*a and
Isabel Pastoriza
Santos
 *a
*a
aCINBIO, Department of Physical Chemistry, Universidade de Vigo, 36310 Vigo, Spain. E-mail: pastoriza@uvigo.gal; lakshmi@uvigo.gal
bDepartment of Biotechnology, School of Engineering and Applied Sciences, Bennett University, Greater Noida, UP, India
cAdvanced Centre of Research in High Energy Materials (ACRHEM), DRDO Industry Academia – Centre of Excellence (DIA-COE), University of Hyderabad, Hyderabad 500046, Telangana, India
dSchool of Mathematics and Physics, University of Science and Technology Beijing, Beijing 100083, P. R. China
eINESC TEC—Institute for Systems and Computer Engineering, Technology and Science, Rua Dr Alberto Frias, 4200-465 Porto, Portugal
fDepartment of Biomedical Engineering, and Center for Remote Health Technologies and Systems, Texas A&M University, College Station, TX 77843, USA
gDepartment of Chemistry, Chung-Ang University, Seoul 06974, South Korea
hDepartment of Chemical and Biological Engineering, Iowa State University, Ames, IA 50011, USA
iNanovaccine Institute, Iowa State University, Ames, IA 50012, USA
jDepartment of Chemical & Environmental Engineering, Campus Rio Ebro, C/Maria de Luna s/n, 50018 Zaragoza, Spain
kInstituto de Nanociencia y Materiales de Aragón (INMA), CSIC-Universidad de Zaragoza, 50009 Zaragoza, Spain
lDivision of Chemistry and Biological Chemistry, School of Chemistry, Chemical Engineering and Biotechnology, Nanyang Technological University, Singapore 637371, Singapore
mDepartment of Mechanical Engineering and Materials Science, Washington University in St. Louis, St. Louis, MO 63130, USA
nSir Ian Potter NanoBioSensing Facility, NanoBiotechnology Research Laboratory, School of Science, RMIT University, Melbourne, VIC 3000, Australia
oDepartment of Chemistry, Indian Institute of Science Education and Research (IISER) Tirupati, Tirupati 517 507, India
pCatalan Institute of Nanoscience and Nanotechnology (ICN2), CSIC and BIST, Campus UAB, Bellaterra, 08193, Barcelona, Spain
qDepartment of Allied Sciences, School of Health Sciences and Technology, UPES, Dehradun, 248007, India
rDepartment of Physics, The Chinese University of Hong Kong, Shatin, Hong Kong SAR 999077, China
sInternational Iberian Nanotechnology Laboratory (INL), 4715-330 Braga, Portugal
tDepartment of Physical and Inorganic Chemistry, Universitat Rovira i Virgili, Tarragona, Spain
uICREA—Institució Catalana de Recerca i Estudis Avançats, 08010, Barcelona, Spain
vFCUP, University of Porto, Rua do Campo Alegre, 4169-007 Porto, Portugal
wDepartment of Physics, University of Trás-os-Montes e Alto Douro, 5001-801 Vila Real, Portugal
xCentro de Física de Materiales (CSIC-UPV/EHU) and Donostia International Physics Center (DIPC), Paseo Manuel de Lardizabal 5, 20018 Donostia San-Sebastián, Spain
yChair for Photonics and Optoelectronics, Nano-Institute Munich, Department of Physics, Ludwig-Maximilians-Universität (LMU), Königinstraße 10, 80539 Munich, Germany
zCatalan Institution for Research and Advanced Studies (ICREA), Passeig de Lluís Companys, 23, Barcelona 08010, Spain
aaNetworking Research Center on Bioengineering, Biomaterials and Nanomedicine, CIBER-BBN, 28029 Madrid, Spain
abSchool of Physics, University of Hyderabad, Hyderabad 500046, Telangana, India
First published on 20th August 2024
Abstract
Plasmonic nanoparticles (NPs) have played a significant role in the evolution of modern nanoscience and nanotechnology in terms of colloidal synthesis, general understanding of nanocrystal growth mechanisms, and their impact in a wide range of applications. They exhibit strong visible colors due to localized surface plasmon resonance (LSPR) that depends on their size, shape, composition, and the surrounding dielectric environment. Under resonant excitation, the LSPR of plasmonic NPs leads to a strong field enhancement near their surfaces and thus enhances various light–matter interactions. These unique optical properties of plasmonic NPs have been used to design chemical and biological sensors. Over the last few decades, colloidal plasmonic NPs have been greatly exploited in sensing applications through LSPR shifts (colorimetry), surface-enhanced Raman scattering, surface-enhanced fluorescence, and chiroptical activity. Although colloidal plasmonic NPs have emerged at the forefront of nanobiosensors, there are still several important challenges to be addressed for the realization of plasmonic NP-based sensor kits for routine use in daily life. In this comprehensive review, researchers of different disciplines (colloidal and analytical chemistry, biology, physics, and medicine) have joined together to summarize the past, present, and future of plasmonic NP-based sensors in terms of different sensing platforms, understanding of the sensing mechanisms, different chemical and biological analytes, and the expected future technologies. This review is expected to guide the researchers currently working in this field and inspire future generations of scientists to join this compelling research field and its branches.
1. Introduction
The development of sensors for ultrasensitive detection of biologically active molecules and chemical substances (organic and inorganic) is crucial for early diagnosis, probing biological processes, and environmental safety.1–16 Early diagnosis is critical for the prevention of spreading the disease to other parts of the body or other people.17–21 For instance, we witnessed the fast contagion of COVID-19 and its impact on the health system across the globe.22 The early detection of the virus has played a significant role in the prevention of its spreading.23,24 Similarly, the early detection of diseases like cancer not only prevents mortality but also reduces the treatment cost.21 In connection with this, various molecular diagnostics techniques have been developed based on the detection and quantification of nucleic acids (DNA and RNA), proteins, peptides, or antibodies using polymerase chain reaction (PCR),25 enzyme-linked immunosorbent assays (ELISA),26 immunofluorescence,27etc. On the other hand, various analytical chemistry techniques such as gas/liquid chromatography, mass spectrometry, nuclear magnetic resonance spectroscopy, and atomic/emission absorption spectroscopy have been developed for the detection of chemical contaminants in water or air.28 Although modern analytical and molecular diagnostics techniques are precise and reliable, they are expensive and time-consuming due to complex instrumentation. Nevertheless, in the last three decades, various types of nanoparticles (NPs) have been extensively exploited in analytical chemistry, molecular diagnostics. This has led to the development of new research fields so-called nanosensors and nanobiosensors, where the sensing platforms are constructed using nanomaterials.29–33 Generally, nanoparticles act as optical or electrochemical sensors, offering efficiency, ease of use, and cost-effectiveness.Among all, plasmonic NPs are one of the most studied materials over the last three decades in the field of nanoscience and nanotechnology and have emerged at the forefront of chemical and biosensors with a detection capability of fast, efficient, point-of-care, and cost-effective.34–36 In particular, gold (Au) and silver (Ag) NPs have received significant attention due to their tunable optical properties in the visible to near-infrared (NIR) range.10,37–40 Over the years, plasmonic NPs have been greatly exploited in a wide range of applications including photonics, light harvesting, chemical and biological sensing, imaging, and therapy.34,35,41 They exhibit unique optical properties due to strong localized surface plasmon resonance (LSPR) that arises at the surface of NPs. LSPR refers to the collective oscillations of conduction band electrons at the interface of plasmonic NPs and their surrounding medium (typically dielectric) upon their interaction with electromagnetic radiation.38,40 The electron cloud oscillations confine on plasmonic NPs according to their dimensions and thus the wavelength of the LSPR band in the extinction spectra of the NPs strongly depends on their size, shape, composition, interparticle distance, and refractive index of the surrounding medium.37,40 Because of intense efforts from researchers across the globe, currently, we are in a position to precisely control the size, shape and composition of plasmonic NPs through colloid chemistry.42–45 A few milestones of the colloidal synthesis of plasmonic NPs include the Turkevich synthesis method,46,47 seed-mediated synthesis of Au & Ag nanorods (NRs),48,49 polyol synthesis of Ag nanocubes (NCs),50 Ag nanoplates (NPTs),51 Au nanoshells,52 Au nanostars (NSTs),53,54etc. have opened doors for precise shape control of plasmonic NPs. The developments in the colloidal synthesis of plasmonic NPs have enabled the tunability of their LSPR in the visible to NIR. Recently, there has been growing interest in obtaining plasmonic NPs with chiroptical response by shaping or assembling them into chiral morphology (twisted or helical).55,56 Such chiral NPs exhibit circular dichroism signals at their LSPR position, which are tunable from visible to NIR.
The LSPR strongly enhances the light–matter interactions on the surface of NPs by focusing the incident light at a nanometer scale through the incident enhancement of electromagnetic field by several orders of magnitude. These properties have made the plasmonic NPs highly attractive for ultrasensitive optical sensing of various analytes ranging from inorganic ions and small organic molecules to biomacromolecules by refractive index sensitivity, colorimetry (based on an analyte-induced aggregation of NPs), and LSPR-enhanced techniques such as surface enhanced Raman scattering (SERS) and surface enhanced fluorescence (SEF).10,37–40,57,58 The first breakthrough study of DNA–Au NP interactions was published in 1996 by Mirkin et al.59 and Alivisatos et al.,60 reporting the reversible aggregation of oligonucleotides-capped Au NPs by the addition of complementary DNA. These reports have laid the foundation for not only DNA sensing but also sensing of other biomolecules using plasmonic NPs. The sensing is usually based on the change in LSPR of plasmonic NPs by selective aggregation of NPs or change in surrounding dielectric constant upon binding to a target molecule.61 As the plasmonic NPs exhibit intense colors in the visible region due to high extinction coefficients, the change in color (usually red to blue or violet for Au NPs) upon analyte-induced aggregation makes them suitable for naked-eye detection.61
On the other hand, SERS has evolved as one of the most sensitive analytical techniques for ultrasensitive detection (even quantification to some extent) of various analytes.13,62–64 Previous studies have demonstrated the capability of detecting signals from single molecules by SERS.65–68 Moreover, it is a non-invasive technique, and it doesn’t require specific binding of analytes to plasmonic NPs. It is based on enhanced Raman scattering of molecules that are placed near or on the surface of NPs mainly due to the electric field enhancement caused by LSPR. This technique has been extensively investigated in the last two decades for sensing a wide range of analytes using plasmonic NPs of different shapes and their assemblies.13 Besides, SEF has also gained significant attention for enhancing signal-to-noise ratio and improving the sensitivity of fluorescence-based bioassays, where the fluorescence enhancement strongly depends on the size of plasmonic NPs.57,69 Due to their interesting optical properties, plasmonic NPs have emerged at the forefront of materials for chemical and bio-sensing.35 Over the yars, numerous review articles have already been published on various aspects of plasmonic NP sensors, especially focusing on SERS, LSPR shifts, and colorimetric-based sensing.6,9–11,13,35,36,64,66,70–75 However, as the field is well-established, there is a need to discuss the current progress in terms of fundamental understanding, technological advances, and challenges remaining to be addressed, along with prospects in different aspects of plasmonic sensors. Therefore, researchers of different expertise in plasmonic sensors have joined to provide a state-of-the-art overview of various subtopics of plasmonic sensors. As outlined in Scheme 1, this review covers the research progress on different aspects of plasmon NP-based sensors: (1) shape-controlled synthesis of plasmonic NPS with tunable optical properties, (2) different methods of sensing using the plasmonic NPs, (3) SERS (fundamentals, different types of substrates, and sensing different analytes and sensing reaction intermediates), (4) LSPR sensing (fundamentals, methods and different analytes), (5) colorimetric sensing, and (6) plasmonic chiroptical sensors. Finally, a brief outlook is provided on the challenges that need to be addressed soon to realize the real-world applications of plasmonic NP-based sensors.
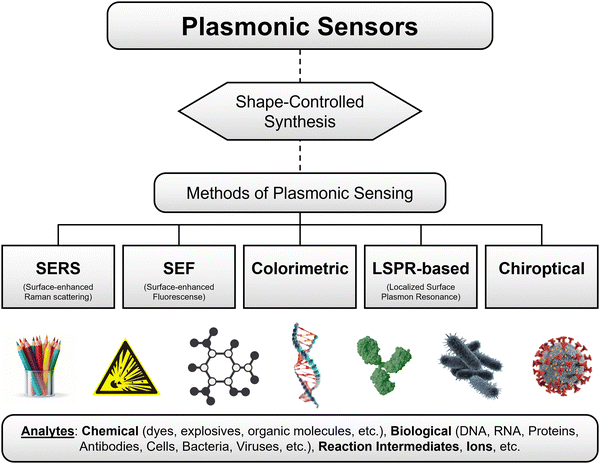 | ||
| Scheme 1 Overview of the contents of the review, which includes research progress of shape-controlled synthesis followed by different methods of sensing for sensing different analytes. | ||
2. Methods of plasmonic sensing
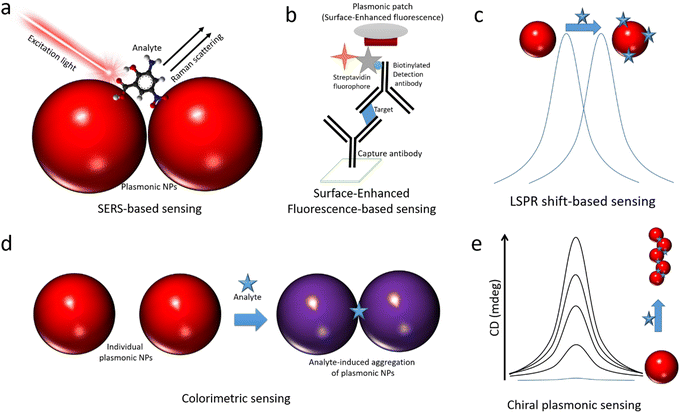
This section provides a brief overview of different techniques used for plasmonic sensing, including SERS, SEFSEF, LSPR, colorimetric, and chiroptical methods. The basic working principles of these methods are schematically illustrated in Fig. 1. For each method, we briefly describe the working principle and various sensing strategies and substances. A detailed overview and state of the art for each method are provided in the following sections.SERS
It is one of the most extensively exploited methods in the field of plasmonic sensors.13,76,77 This is based on the enhancement of inelastic scattering of molecules by several orders of magnitude (106–109 or more) through strong electric field enhancement when they are adsorbed on the surface of plasmonic NPs (Fig. 1a).13 SERS was first observed from pyridine adsorbed on rough silver electrodes by Fleischmann et al. in 1974.78 The enhancement was attributed to a surface area-related phenomenon. Later, this was observed independently by Jeanmaire et al. and Albrecht et al. in 1977.79,80 The enhancement was assigned to the resonant Raman scattering of molecules adsorbed on rough metal surfaces by the interaction with surface plasmons. The detailed history of the discovery of SERS was discussed in previous reviews.13,64,66,81,82 Following these early works, SERS has been greatly studied in terms of fundamental understanding of the enhancement mechanisms, optimization of enhancement factors, exploring various shapes and sizes of plasmonic NPs, development of different substrates, and sensing a wide range of analytes.13,64,82 The SERS enhancement factors are strongly dependent on the chemical composition of plasmonic NPs, the distance between the surface of NPs and the analyte molecule, the excitation wavelength, the position of LSPR, and the resonance of the analyte with the excitation light.83 The sensitivity of this technique can be as high as single molecule detection in some cases.65,66,68,84,85 Over the years, researchers have found that the aggregated NPs and the NPS with sharp tips exhibit strong electric field enhancements and thus better SERS signals at the resonant excitation wavelength. SERS sensing can be performed in different ways, such as in a solution phase, on a solid substrate, in a microfluidic flow channel, and inside a tissue, depending on the type of analyte and the plasmonic substrate.13,14,61,64,86SEF
The concept SEF is similar to that of SERS, however, instead of the Raman signals, the fluorescence of molecules significantly increases upon placing them in proximity to a plasmonic NP surface.57,69,87–95 The enhancement is caused by the interaction of fluorophores with surface plasmons, and thus the fluorophores experience a strong electric field enhancement, leading to enhanced fluorescence intensity. This phenomenon is also called metal-enhanced fluorescence or plasmon-enhanced fluorescence.90,93,94 The enhancement of fluorescence is strongly dependent on the overlap of the optical absorption spectra of fluorophore and extinction spectra of NPs, size of NPs, and the distance between the fluorophore and metal NP surface to overcome the quenching by Főrster resonance energy transfer (FRET).87,94 In addition, plasmonic NPs enhance the intrinsic radiative decay of fluorophores and thus a reduction in lifetime. The mechanisms of surface-enhanced fluorescence have been greatly studied by Lakowicz and co-workers, and they proposed strategies for radiative decay engineering of fluorophores using metal surfaces. SEF has been greatly exploited in the sensing of various analytes.96–99 The working principle of a typical SEF-based sensor is illustrated in Fig. 1c.69,87,95 It is based on the enhancement of the fluorescence intensity of fluorophores that are used as labels in recognizing the binding, thus SEF significantly improves the sensitivity of the biosensors. This has been widely used in fluorescence-linked immunoassays and is compatible with other immunoassays, flow cytometry, etc.69,95,100,101 Moreover, it can shorten overall assay times and lower sample volumes, and it can be combined with lateral flow techniques. In addition, SEF has been explored in single-molecular spectroscopy, bioimaging, DNA hybridization sensing, and beyond.102–105 However, one of the major challenges is to control the interactions between plasmonic NPs and fluorophores to overcome the quenching effect.LSPR-shift
The extinction spectra of plasmonic NPs not only dependent on the morphology but also the dielectric constant of the surrounding medium.39,40 Thus, the change in the surrounding medium leads to a redshift or blue shift of the LSPR peak.39,58,106–108 This has been exploited to detect the molecular interactions near the nanoparticle surface, as illustrated in Fig. 1c.38,39,58,108,109 The NPs either in colloidal form or on a substrate exhibit LSPR shifts upon interacting with target analyte molecules.37,39,108 The interactions can be specific or non-specific depending on the experimental configuration. The extent of LSPR shift depends on the concentration of the molecules that interact with the NP surface.15 Based on the LSPR shifts, a range of analytes have been detected in different sensing configurations. Among all, LSPR-shift-based immunoassays,110 LSPR optical fiber sensors110 and label-free sensing techniques have received significant attention.37,109Colorimetric sensing
Sensing based on analyte-induced color changes of probe materials has been significantly explored in modern science and technology due to its low cost and point-of-care testing ability.111–115 Moreover, it doesn’t require sophisticated instrumentation and skilled manpower. One of the most interesting features that makes colloidal plasmonic NPs highly attractive is their visible colors due to strong surface plasmon resonance. Their extinction coefficients are 1000 times higher than those of conventional organic dye molecular probes used in colorimetric sensing, thus making them highly attractive as colorimetric probes for high-sensitivity detection.116,117 The colorimetric plasmonic sensing is based on the selective aggregation of gold NPs upon binding to an analyte (Fig. 1d), the colloidal solution color changes from red to blue due to surface plasmon coupling between particles in the aggregate.61,118–120 Thus, plasmonic NPs offer naked-eye detection of analytes, and the color changes can be quantified using a mobile device to obtain quantitative information about the analytes.121 The colorimetric plasmonic sensing can be either through specific or non-specific interaction with the analyte.119,122–127 This method has been extensively applied for the detection of metal ions, biomolecules, and proteins.120,123,127Chiral plasmonic sensing
Chirality, a commonly observed feature in biological systems, is one of the most fascinating properties that nature has given us.128 On the one hand, organic chiral molecules are being greatly exploited regarding their synthesis and applications, and inorganic chiral nanomaterials have received significant attention in the last decade.129–131 Chirality in inorganic NPs arises due to their morphology which has no mirror or inversion symmetry.129–134 Thus, they exhibit higher dissymmetry (g) factors (∼0.2) compared to organic systems (∼10−3).131,134 Among all, chiral plasmonic NRs have received significant attention due to their tunable chiroptical signal in the visible range and the g-factors.134 Chirality in plasmonic NPs has been achieved either by shaping them into helical/twisted morphology or through chiral self-assembly using chiral templates.134,135 The strength of chirality (g-factor) strongly depends on the helicity of the individual or assembled NPs.134,136 Thus, chiral plasmonic sensing is generally based on the analyte-induced chirality of plasmonic NPs, as illustrated in Fig. 1e.137,138 The chiroptical activity increases with increasing the analyte concentration, thus enabling its quantitative determination.2.1. Different types of plasmonic metal NPs used in sensing
The shape of plasmonic metal NPs, which are mostly made of Au and Ag, is determined by their crystalline structure, facet, and anisotropy.139 High uniformity in size and shape can ensure the investigation of the physical properties and wide applications of plasmonic NPs. The bottom-up chemical synthesis methods, including chemical reduction, electrochemical deposition, photochemical reaction, and multiphase-based synthesis, have been well-developed for the preparation of high-quality NPsP with superior plasmonic properties. Among the chemical reduction methods, seed-mediated growth is the most common technique for synthesizing high-quality NPs. Two steps are involved in this method: nucleation of nanoscale seeds and atomistic growth of the seeds to give final NPs.140 Once the nucleated clusters lock into well-defined structures, the nanoscale seeds are formed.140 Different nanoscale seeds can be classified according to their crystalline structure, defects, and shape. The typical diameters of nanoscale seeds are in the range of ∼1–15 nm. There are three major categories of nanoscale seeds: single-crystalline, multiply twinned, and plate-like (Fig. 2a, top). Single-crystalline seeds are made of the face-centered cubic lattice (fcc) and are typically by eight {111}and six {100} facets. Due to their small size, their facets are usually difficult to identify and lack defects. Multiply twinned seeds are enclosed typically by {111} facets with multiple twin boundaries in a decahedral shape. When a layer of the fcc lattice is missed or added in stacking order, plate-like seeds are formed. By controlling the concentration of nanoscale seeds, the type and concentration of the capping agent, and the reaction conditions, differently shaped NCs can be synthesized even from the same type of nanoscale seed. The typical shapes of final plasmonic metal NCs obtained from single-crystalline seeds include octahedron, cube, cuboctahedron, and octagonal rod (Fig. 2a, left panel). The shape of final NCs is highly impacted by the defect of nanoscale seeds as well. The defect generated in nanoscale seeds broadens the diversity of final NCs. Decahedron, icosahedron, pentagonal rod, and pentagonal bipyramid can be synthesized from multiply twinned nanoscale seeds (Fig. 2a, middle panel). Plate-like nanoscale seeds enable the growth of two-dimensional-like NCs, such as triangular and hexagonal plates (Fig. 2a, right panel).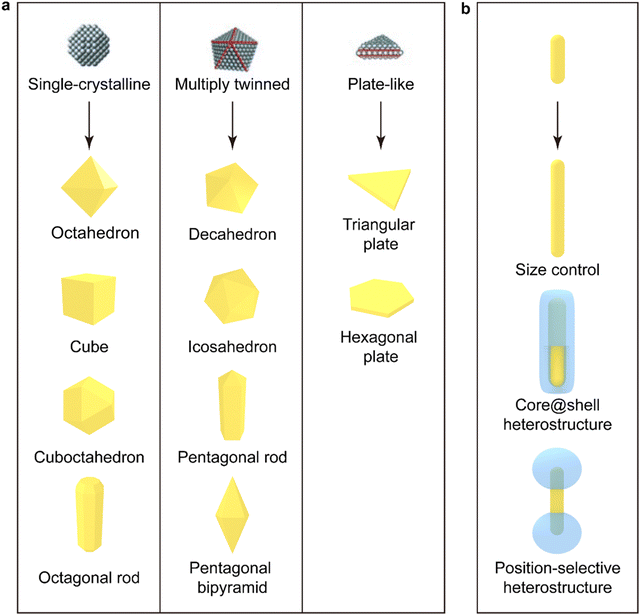 | ||
| Fig. 2 Seed-mediated growth of plasmonic metal NPs. (a) Schematics of three representative types of nanoscale seeds (top) and the faceted NCs (bottom) produced from the seeds by seed-mediated growth. The red lines in the nanoscale seeds indicate twin boundaries and stacking faults.140,141 Reproduced from ref. 140 with permission from Wiley-VCH publisher, copyright 2009. (b) Schematics showing the overgrowth of homo- and hetero-structures with the faceted NPs as seeds. Metal nanorods are used as a representative example for controlling sizes and constructing core@shell and position-selective heterostructures. The illustrated plasmonic NPs are typically made of Au and Ag. | ||
The obtained large metal NCs equipped with unique stacking order and facets can be used as seeds for the further overgrowth of homogeneous and heterogeneous components. Size control is one of the purposes (Fig. 2b, top). In order to maintain high uniformity in size and shape and avoid the production of byproducts, nanoparticles up to hundreds of nanometers in size are synthesized step by step by controlling the concentrations of the seeds and the metal salt precursor.142–144 The obtained metal NCs can also be used to construct heterostructures through seed-mediated growth. Core–shell A@B heterostructures (Fig. 2b, middle) have been designed to satisfy diverse demands, such as plasmon-assisted biomedical applications, solar energy harvesting and photocatalysis. The shell can be made of another plasmonic nanoparticle, semiconductor (metal oxides, metal sulfides, metal selenides), silica, polymer, lanthanide-doped nanomaterial, or metal–organic framework.145 Moreover, the shell component can also be selectively deposited at different positions on the plasmonic metal core where the plasmonic near-field enhancement reaches maximum to optimize the effect of the plasmonic enhancement by the metal core. The position-selective heterostructures can also allow both components to interact with the surrounding environment. As shown in Fig. 2b (bottom), dumbbell-like heterostructures are the most common position-selective heterostructures. However, limited types of position-selective heterostructures have so far been prepared because the conditions to control the deposition location is usually very delicate.145
Shape and size are the most important factors affecting the plasmon properties of metal NPs. The incident light can be confined to different regions on the plasmonic NP surface because of their specific morphology. Specific plasmon modes (LSPR) can be supported on plasmonic metal NPs and interestingly the wavelengths of these plasmon modes can be precisely tuned by varying the morphology and size. The optical properties of these plasmonic NPs determine their applications. Nanospheres (NSs) have attracted the most attention during the past decades among diverse nanoparticles owing to their simplicity in synthesis, spherical symmetry, and readiness in assembly.146 The isotropic spherical geometry of NSs also allows for the easy analytical solving of Maxwell's equations and the calculation of the absorption, scattering, and extinction cross-sections of NSs, which is known as Mie theory.146 Taking Au NSs as an example. They can be synthesized uniformly by seed-mediated growth, as shown in Fig. 3a.147 The dipolar plasmon mode (Fig. 4, solid yellow line) can be tuned from 500 nm to 600 nm by precisely controlling the diameter of Au NSs from 24 nm to 103 nm. With a diameter larger than 130 nm, both quadrupolar and dipolar plasmon modes can be sustained. However, the spectral tunability of the plasmon modes is limited for metal NSs. Nanocubes (NCs), another type of highly symmetric structure, are formed when the round surfaces of an NS are evenly transformed into four flat faces (Fig. 3b). In comparison to NSs, the dipolar plasmon mode of Ag NCs split into two peaks (Fig. 4, solid pink line) owing to their sharp corners.148 In addition, complex plasmon modes can be localized not only to the edges of nanocubes but also to the sharp corners and flat faces when nanocubes are deposited on both dielectric and metallic substrates. The corner plasmon modes of NCs can even split into distal and proximal ones in the presence of a substrate.149
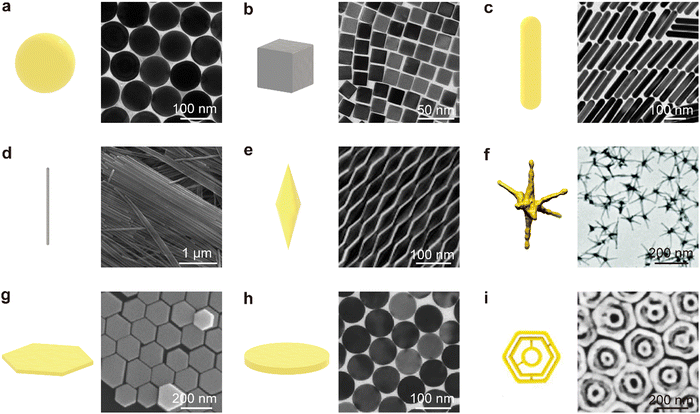 | ||
| Fig. 3 Schematics of representative individual NPs and their corresponding transmission electron microscopy (TEM)/scanning electron microscopy (SEM) images. (a) Au NSs, TEM. Reproduced from ref. 147 with permission from Wiley-VCH publisher, Copyright 2014. (b) Ag@Au NCs, TEM. Reproduced from ref. 150 with permission from ACS publisher, Copyright 2019 (c) Au NRs, TEM Reproduced from ref. 151 with permission ACS publisher, Copyright 2015 (d) Ag NWs, SEM. Reproduced from ref. 152 with permission from ACS publisher, Copyright 2015 (e) Au NBPs, TEM. Reproduced from ref. 153 with permission from Springer Nature publisher, Copyright 2015 (f) Au nanostars, TEM. Reproduced from ref. 154 with permission from RSC publisher, Copyright 2019 (g) hexagonal Au NPLs, SEM. Reproduced from ref. 144 with permission from Wiley-VCH publisher, Copyrights 2016 (h) Circular Au NPLs, TEM. Reproduced from ref. 155 with permission from Wiley-VCH publisher, Copyrights 2018 (i) Intertwined triple Au nanoscale rings, SEM. Reproduced from ref. 156 with permission from ACS publisher, Copyright 2021. | ||
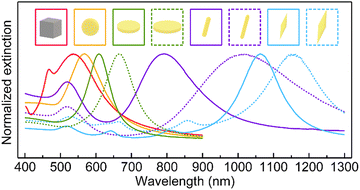 | ||
| Fig. 4 Representative extinction spectra for Ag NCs (73 nm in edge length, solid pink line), Au NSs (88 nm in diameter, solid yellow line), circular Au NPLs (79 nm in diameter, 22 nm in thickness, solid green line; 99 nm in diameter, 22 nm in thickness, dotted green line), Au NRs (14 nm in diameter, 59 nm in length, solid purple line; 10 nm in diameter, 63 nm in length, dotted purple line), Au NBPs (59 nm in waist diameter, 230 nm in length, solid blue line; 70 nm in waist diameter, 276 nm in length, dotted blue line). All samples are dispersed in aqueous solutions for extinction measurements. The data of Au NSs, Au NPLs and Au NBPs are reproduced from ref. 146, 155, 157 with permission from Wiley-VCH publisher, Copyright 2014, 2018 and 2015, respectively. The data of Au NRs are reproduced from ref. 158 with permission from ACS publisher, Copyright 2006. | ||
Anisotropic structures are highly desired because they provide extra size parameters to control the plasmon modes. When NSs are elongated along one direction, NRs are obtained. NRs have longer lengths than diameters, giving rise to an anisotropic shape and a specific length-to-diameter aspect ratio (Fig. 3c). The wavelengths of the plasmon modes are highly sensitive to the aspect ratio of NRs (Fig. 4, solid and dotted purple lines). The longitudinal plasmon wavelength of plasmonic metal NRs can be synthetically varied from the visible to the IR region. Another attractive property of NRs is their different responses to incident light linearly polarized along the transverse and longitudinal directions. The odd and even longitudinal multipolar plasmon modes have been observed on Ag NRs with high aspect ratios. The multipolar plasmon modes can endow NRs with a color routing capability.159 With emitters sandwiched between Ag NRs and a dielectric substrate, the emission of two-dimensional (2D) excitons was found to be routed to the two ends by the multipolar plasmon modes.160 Nanowires (NWs) are obtained when the aspect ratio is further increased (Fig. 3d). The excellent electrical conductivity and transparency of plasmonic metal NWs make them useful as transparent conductive electrodes for flexible optoelectronics.161 At the longitudinal plasmon resonance, the field enhancement is localized at the two ends for one-dimensional nanostructures. The electromagnetic (EM) enhancement around the ends is limited because of the hemispherical shape. By sharpening the ends of NRs, nanobipyramids (NBPs) (Fig. 3e) are formed. Higher-order longitudinal plasmon resonances (Fig. 4, solid and dotted blue lines) can be sustained on NBPs owing to the sharp ends.157 Such sharp ends induce electric field enhancement about three times larger than that of NRs, promoting the generation of hot electrons.162 The local temperature around the tip has been proven to reach dozens of degrees Celsius because of the enhanced near-field.163 To fully use the advantages offered by the sharp ends, nanostars of different types have been developed. Nanostars typically have more than three sharp branches (Fig. 3f). Their plasmon modes are dependent on the number and length of their branches, among other parameters. The drastic electric field enhancement occurs around the sharp ends and tips at the LSPR for NBPs and nanostars, which allows them to be used in SERS, sensing, and photocatalysis.162,164 The sharp ends and tips of NBPs and NSTs can also induce local strain on 2D semiconductors in contact with the NPs, leading to the development of single-photon quantum emitters.165 When NSs are compressed along a certain direction, 2D nanostructures such as hexagonal (Fig. 3g) and circular (Fig. 3h) nanoplates (NPLs) are produced. Hexagonal NPLs can be synthesized by controlling the concentrations of the metal salt precursor and the seeds. Circular NPLs with different diameters can be produced through chemical etching on hexagonal NPLs by adjusting the etching time.144
The thickness is barely affected during the etching process. NPLs, therefore, provide a more flexible method for tuning the plasmon mode by independently varying their thickness and diameter (Fig. 4, solid and dotted green lines). Benefiting from the large atomically flat facets in the lateral direction, the EM field can be confined into a large face-contact area between NPLs and substrates.166 Especially in NPL-on-mirror structures, rich optical properties, including magnetic and anapole plasmon resonances, have been observed within the dielectric layer sandwiched between the NPL and the metal film.166 Fabry–Peŕot nanoresonators constructed out of Au NPLs and an Au film sandwiched with a high-refractive-index dielectric layer possess high quality factors of up to 76. They also benefit from the atomically flat surface of Au NPLs.167 To combine the maximized field enhancement regions of 2D nanostructures and strongly amplified near-field focusing in dimers, trimers, and multimers, complex plasmonic nanostructures have recently been designed and synthesized. For example, Fig. 3i, displays intertwined triple ring structures whose near-field focusing can be tailored through the variation of the intragap distance between the nanoscale rings. Such complex 2D nanostructures have enabled single-particle SERS with large enhancement factors (EF) up to ∼109.156
3. Plasmonic metal nanoparticles-based SERS sensing
3.1. Fundamentals of SERS
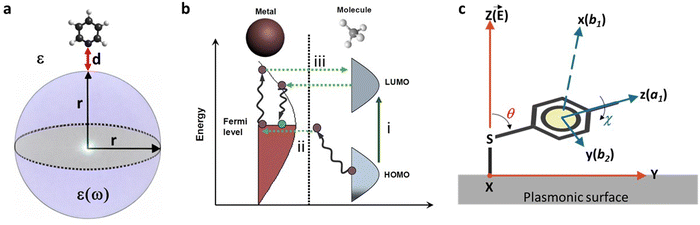
SERS spectroscopy is based on the amplification of Raman signals from molecules sorbed on plasmonic surface, usually gold or silver.168 The discovery of the SERS effect can be attributed to Fleischman, Hendra, and McQuillan in 1974.78 They observed an unusually strong Raman signal from pyridine molecules absorbed on a roughened silver electrode. These authors interpreted the effect as a local increase of the surface concentration of the analyte due to the increase of the surface area of the electrode due to its deterioration because of the consecutive reduction–oxidation cycles. Consequently, the physical effect was incorrectly named SERS spectroscopy. However, subsequent independent reports by Jean Maire and Van Duyne,80 Albrecht and Creighton,79 both published in 1977, together with the seminal work by Moskovits169 demonstrated that this intensity could not be accounted for an increased surface. Conversely, they reported SERS as an eminent electromagnetic (EM) effect derived from the excitation of LSPRs upon their excitation when illuminating nanostructured metals with the appropriate light, as previously theorized by Bohm170 and demonstrated by Otto,86 as a solution of the Maxwell equations. Also, they highlighted that the scattering intensity from the adsorbed molecules was 105–106 times stronger than the conventional Raman signal. Notably, until 1979, all SERS experiments were carried out only on electrodes being Creighton, Blatchford, and Albretch171 the first in report SERS experiments in colloidal Au and Ag nanostructures. In 1980 Otto reported the effect of the charge transfer and the resonance at the surface, the so-called chemical effects on the intensification of the SERS signal.172 In 1983/84 the surface selection rules were developed independently by Creighton173 and Moskovits.174 No matter these advances and as reported by Moskovits in one of the most influential reviews in SERS,63 the research and use of this technique became mostly academic until 1997. In that year, two different papers demonstrated the capability of SERS for the detection of single molecules.68,85 This fact not only fueled the fundamental research in the field but also in closed areas such as nanofabrication optical theory and the development of applications in biology, medicine, environmental science, or catalysis. Thus, today an extraordinary effort led by a myriad of research groups is performed to transform SERS into a real live tool especially in the field of biosciences.The fundamental components in SERS include a molecule, a plasmonic material, and EM radiation. The adsorption of the molecule onto the plasmonic surface is typically classified according to the strength of bonding into either physisorption (weak interaction) or chemisorption (chemical bonding). When this molecule-nanoparticle system is illuminated by EM radiation, the incident photons can induce substrate excitations such as electron–hole pairs, surface plasmons, or surface phonons that contribute to the enhancement. In particular, the nanostructure's absorption of light can generate potent local electric fields at the location of the adsorbed molecular species. This local field significantly influences the optical properties of the adsorbate, thereby causing the SERS enhancement. Moreover, the interaction between the incident radiation and the adsorbed molecule may result in photodissociation, photoreactions, or photo-desorption, each of which leaves unique traces in the resultant SERS spectrum. On the other hand, the interaction of light with the metallic nanostructure depends on the value of the complex dielectric function at the excitation wavelength, which determines the enhancement observed at a given excitation frequency. Particle absorption and scattering are influenced by the shape and size of the metal nanostructure, thus affecting SERS intensities. Furthermore, the excitations in nanostructures are strongly influenced by the medium's dielectric constant. The absorption and scattering of light by metallic nanoparticles (smaller than the wavelength of the incident light) are crucial properties that yield SERS and provide the theoretical basis for the explanation of the EM enhancement.62,83,86,175–180 Although LSPR of nanostructures is necessary for large enhancement factors of SERS, the chemical interactions between analytes and substrates can lead to Raman enhancement via the chemical enhancement mechanism that has often been observed for semiconductor substrates.13,181
For a single isolated plasmonic sphere (Fig. 5a), the extinction and scattering can be fully explained by the Mie theory.182,183 When the light resonates with the LSPR, the metal sphere emits its dipolar field (ESP). The magnitude of that field at a nearby molecule depends on the sphere's radius (r), its distance (d) from the molecule, the dielectric constants of the metal (ε) and the surrounding medium (ε0), as well as the strength of the incident field (E0). Therefore, the molecule experiences an enhanced local field (EM) which includes both electric-field magnitudes: E0 + ESP. The light field enhancement A (ν) is determined by the ratio of the field at the molecule's position to the incoming field.
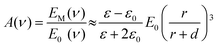 | (1) |
Here, A(ν) is particularly strong when the real part of ε(ν) is equal to −2ε0. Additionally, for a significant EM enhancement, the imaginary part of the dielectric constant needs to be small. These conditions describe the resonant excitation of localized surface plasmons for a metal sphere.184 Similarly, the scattered field will be enhanced if it is in resonance with the particle LSPR. Therefore, considering the enhancing effects for both the laser and the Stokes field, the EM SERS enhancement factor, GSERS (νS), can be expressed as:
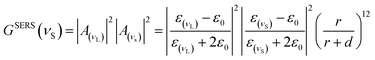 | (2) |
This equation indicates that the enhancement scales with the fourth power (|E|4) of the local field of the metallic nanostructure, and that it is particularly strong when excitation and scattered fields are in resonance with LSPRS. On the other hand, the EM predicts that SERS does not require direct contact between the molecule and the metal, but it is necessary a certain sensing volume. This is because, as shown in eqn (1) and (2), the EM enhancement factor strongly decreases with the distance from the metal surface due to the decay of the dipolar field [1/d]3 to the fourth power, resulting in [1/d]12.
Maximum values for EM enhancement in spherical isolated nanoparticles are on the order of 106. Theory predicts stronger EM enhancement for regions with sharp features and large curvatures, which may exist on silver and gold nanostructures. For instance, the EM SERS enhancement factor can be increased up to 1011 when the particle presents sharp tips or edges. These features localize the electric field within confined regions, enabling extremely high enhancement factors.185,186 Additionally, closely spaced interacting NPs can provide further field enhancement. EM enhancement factors up to 1011 have been estimated for dimers of Ag NPs.187 These substantial enhancements are typically attributed to highly concentrated EM fields associated with strong LSPRs at interstitial sites (often referred to as EM or SERS hotspots) in nanostructures consisting of two or more coupled nanostructured surfaces with closely spaced features. The size of these hotspots is as small as a few nanometers, and the EM field concentration on them strongly depends on the geometry of the nanostructured site where the probed molecules reside, the wavelength, and the polarization of the incident light.185 When optical excitation is localized in such small hot spots, extraordinarily large EM SERS enhancements (proportional to field enhancement to the fourth power) up to 1012 have been theoretically predicted.62
While the EM mechanism is the primary contributor to the SERS signal, early observations noted a dependence of the scattering signal on the electrode potential,188 suggesting an electronic coupling between the molecule and the metal. In addition, the presence of metal in the system can alter the polarizability of adsorbed molecules, thereby potentially increasing the Raman scattering efficiency. On the other hand, experimental observations, such as the effect's dependence on the chemical nature of the molecule and strong molecular selectivity, provide clear indications for the existence of an additional chemical SERS enhancement. Moreover, the best EM SERS enhancement factors (1012) leave a gap of about two or three orders of magnitude to the highest experimentally observed non-resonant SERS enhancement factors on the order of 1015,189 which suggests the existence of additional enhancement mechanism(s) accounting for the missing factors.
Different mechanisms have been proposed to account for the chemical SERS effect, sometimes also referred to as the first-layer effect, as it necessitates direct contact between the molecule and the metal. The electronic coupling between the molecule and metal and the formation of a surface complex can lead to charge transfer (CT) from the metal to the molecule, or vice versa, and within the adsorbed molecule itself, resulting in an increased Raman signal. Fig. 5b displays a typical energy level diagram for a molecule-metal system, where the energies of the highest occupied molecular orbital (HOMO) and the lowest unoccupied molecular orbital (LUMO) are roughly symmetric relative to the Fermi level of the metal, alongside possible CT processes involving molecular states (path (i)) and molecular and metallic states (paths (ii) and (iii)). Generally, the chemical SERS enhancement factor is believed to contribute enhancement factors on the order of 10–103, coexisting with EM enhancement.
The surface selection rules, formulated by Moskovits in the 80's,174 explore the alteration of the relative intensity of bands within a given SERS spectrum as a result of the orientation of the molecule being studied in relation to the plasmonic surface. For example, when considering mercaptobenzene (Fig. 5c),190 and a C2v symmetry for the mercaptobenzene group, its vibrational modes can be classified into in-plane (ip) a1 and b2 modes and out-of-plane (oop) a2 and b1 modes. On the other hand, assuming that the surface electric field, E, effectively has only a normal component (Z direction in Fig. 5c),191 the intensity of a vibrational mode is proportional to the square of the scalar product of the electric field and the dipole moment derivative of the mode, d![[small mu, Greek, vector]](https://www.rsc.org/images/entities/i_char_e0e9.gif) /dQ.192,193
/dQ.192,193
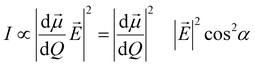 | (3) |
![[E with combining right harpoon above (vector)]](https://www.rsc.org/images/entities/i_char_0045_20d1.gif) and d
and d![[small mu, Greek, vector]](https://www.rsc.org/images/entities/i_char_e0e9.gif) /dQ. Defining θ as the tilt angle of the z axis of the mercaptobenzene unit with the surface normal (Z), and c as the twist angle of the molecular plane around the z axis (which is 0° when y is parallel to the surface). Then, by considering that the ip a1 and b2 modes have dipole moment derivatives along z and y axes, respectively, and the oop b1 modes have dipole moment derivatives perpendicular to the phenyl ring (along x axis), the molecular-fixed axis system xyz can be correlated with the experimental axis system XYZ by the two Eulerian angles, θ and χ. The intensities of a1, b1 and b2 can be then represented as follows from the above equation:38
/dQ. Defining θ as the tilt angle of the z axis of the mercaptobenzene unit with the surface normal (Z), and c as the twist angle of the molecular plane around the z axis (which is 0° when y is parallel to the surface). Then, by considering that the ip a1 and b2 modes have dipole moment derivatives along z and y axes, respectively, and the oop b1 modes have dipole moment derivatives perpendicular to the phenyl ring (along x axis), the molecular-fixed axis system xyz can be correlated with the experimental axis system XYZ by the two Eulerian angles, θ and χ. The intensities of a1, b1 and b2 can be then represented as follows from the above equation:38I(a1) ∝ cos2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ θ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) I0(α1) I0(α1) | (4) |
I(b1) ∝ sin2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ θ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) con2 con2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) χ χ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) I0(b1) I0(b1) | (5) |
I(b2) ∝ sin2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ θ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) sin2 sin2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) χ χ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) I0(b1) I0(b1) | (6) |
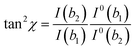 | (7) |
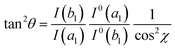 | (8) |
Thus, by assigning the a1, b1 and b2 vibrational modes in the SERS spectra, usually by computational density functional theory (DFT) methods, before and after the coupling, it is possible to know the tilt and twist angles. The deformation of these angles is, however, restricted by the fact that the chemoreceptor is chemically bound to the surface and requires very large analytes to be effective.
3.2. SERS sensing platforms
SERS-based detection of chemical and biological analytes at the trace or single molecule level is extremely desirable in a variety of scientific and technological domains, including analytical chemistry,194 materials science,195 forensics,196 life science,197 food industry,198 explosive detection199 and biomedical diagnostics.200 The advancement of nanofabrication techniques, the ability to obtain desired plasmonic substrates, the tunability of LSPR of plasmonic substrates according to the requirements, and the sensitivity of SERS spectroscopy brought down the detection limits to a single molecule level. In particular, the design and fabrication of SERS substrates using desired plasmonic substrates is a key factor in achieving high-sensitivity detection.201 A wide range of SERS substrates have been reported in the literature with the specific goal of controlling plasmonic hotspots and thus achieving high sensitivity.202 SERS substrates can be broadly classified into two types depending on how the plasmonic NPs are being used in sensing analytes: (1) plasmonic NPs deposited on solid substrates in a controlled manner (2) colloidal solutions of plasmonic NPs with or without Raman reporters decorated on their surface.203,204 To fabricate efficient SERS solid substrates various nanofabrication techniques such as focused ion beam technique,205 soft lithography,206 electron beam lithography,207 stamping,208 nanosphere lithography,209 optical lithography,210 molecular assembly-based lithography, and colloidal self-assembly have been employed.211 Despite tremendous progress in the field, developing reliable and reproducible SERS solid substrates has been challenging. Extensive research has been conducted to optimize SERS substrates by close packing of NPs, using different shapes that exhibit strong field enhancements, positioning the analyte in the hotspot, and integrating SERS spectroscopy with other analytical systems (e.g., microfluidic, optofluidic, and paper-based) to achieve ultrahigh detection sensitivity.212–214 On the other hand, sensing in a liquid medium can performed using SERS tags or pure plasmonic NPs. In particular, SERS tags have been extensively used in the ultrasensitive quantitative detection of a wide range of analytes. SERS tags are commonly prepared by first functionalizing the surface of Au or Ag NPs of different shapes with molecules (Raman reporters) that exhibit strong intrinsic Raman scattering, followed by coating a biocompatible shell, usually a polymer or SiO2. The shell can then be functionalized with a biorecognition system such as an antibody for specific binding to a target analyte. The SERS tags can also be used in solid substrate-based sensing platforms. Besides, the colloidal solutions of bare plasmonic NCs, with or without a biorecognition system have been used for specifically or nonspecifically binding to analytes to induce SERS signals. In the colloidal solution, the SERS detection is also influenced by the Brownian motion of analytes or metal NPs, and it is often used to attain quantitative analysis, but it has a low LOD.215 In some cases, SERS substrates can be one or a few particles that enable the detection of single molecules through strong EM enhancements. On the other hand, they can also be rigid or flexible solid substrates (glass, silicon, paper, plastic, etc.) in which plasmonic NPs are chemically or physically immobilized. The substrates can also be integrated with other technologies such as microfluidics, electrocatalysis, and beyond. In the following section, the detection of various analytes using different types of SERS substrates has been discussed.3.3. Single-molecule SERS (SM-SERS) approaches and requirements
Numerous comprehensive reviews have extensively covered substrates and methodologies aimed at acquiring Raman spectra from individual molecules. Here, our focus shifts towards discussing and highlighting some of the latest advancements and promising strategies in fabricating dependable probes for single-molecule (SM)-SERS in a reproducible manner. Several critical conditions must be met for successful SM-SERS, including the design of a nanostructure capable of sufficiently enhancing the EM field.216 In SERS, both incident and scattered light undergo enhancement. The signal amplification thus scales approximately with the fourth power of the electric field intensity (∼|E|4).217,218 For single-molecule measurements, an EM-field enhancement of at least on the order 107–1010 is required.219The most common approach to obtaining sufficient enhancement factors takes advantage of the electromagnetic “hotspot” formed in the gap between two adjacent Au or Ag NPs. Plasmonic coupling at the particle junction results in a highly localized, strong electromagnetic field if the dimer is excited with light at a wavelength matching the coupled plasmon resonance.220 However, the SERS enhancement obtained from plasmonic dimers is inversely proportional to the interparticle distance. For NSs, this typically requires distances on the order of 1–2 nm. Top-down nanolithography methods such as e-beam lithography or focussed ion beam milling are capable of fabricating nanostructures with such high precision. Nevertheless, benefiting from the EM enhancement requires precise localization of a single analyte in the hotspot region, a task that can be challenging and reliant on a suitable localization strategy.
Bottom-up approaches represent an alternative method, where the analyte is directly assembled together with the NP dimer. For instance, Lim et al. reported the formation of Au NS dimers with a DNA tether, that simultaneously enabled the localization of a single Raman-active dye at the particle junction.221 Growing a Ag shell on the Au particle surface further reduced the interparticle spacing and increased enhancement. However, nanospheres formed by reduction chemistry or metal overgrowth are typically not perfectly spherical and may exhibit facets or edges. This variability can result in variations in the hotspot size, thus hindering quantitative measurements. A strategy for synthesizing nearly perfect spheres through chemical etching was reported by the Schlücker group.222 Structural uniform dimers of ideal spheres were formed by the substrate-supported assembly and exhibited excellent and reproducible plasmonic properties for reliable measurements.223 Nanodimer formation using alkanethiol224 or DNA-linker225 has proven useful for assembling spherical particles. A more versatile approach to dimer formation, with a defined gap size and hotspot targeting, is offered by DNA origami technology. DNA origami is a nanofabrication method that allows the design of three-dimensional structures with nanoscale precision.226 This technique has been employed to assemble complex and multifunctional plasmonic nanostructures, enabling the study of enhanced light–matter interactions in various scientific applications, including SM-SERS.227
The first reports on DNA-origami assembled plasmonic dimers for SERS were published almost ten years ago. Interestingly, all reports employed different structural designs, illustrating the great design flexibility of DNA nanotechnology. For example, Prinz et al. designed gold dimers on triangular DNA origami scaffolds.228 Thacker et al. developed a design to obtain strong plasmonic coupling between two 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nm gold nanoparticles reproducibly held with gaps of 3.3 ± 1
nm gold nanoparticles reproducibly held with gaps of 3.3 ± 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nm on a porous DNA sheet.229 Kühler et al. utilized gold nanodimers linked by a three-layered DNA origami spanning the plasmonic “hotspot”.230 Pilo-Pais et al. employed a DNA origami template where Au NPs were selectively placed on the corners of rectangular origami and subsequently enlarged via solution-based metal deposition.231
nm on a porous DNA sheet.229 Kühler et al. utilized gold nanodimers linked by a three-layered DNA origami spanning the plasmonic “hotspot”.230 Pilo-Pais et al. employed a DNA origami template where Au NPs were selectively placed on the corners of rectangular origami and subsequently enlarged via solution-based metal deposition.231
In recent years, further origami designs, such as nanoforks232 and funnel-spacers,233 have been devised, expanding the methodology towards successful SM measurements. In the first demonstration of SM-SERS with DNA-Origami assembled Au dimer nanoantennas, Simoncelli et al., employed optothermal-induced shrinking of a DNA template to reduce the gap sizes between two 40 nm Au NSs.233 By shrinking the plasmonic hotspot, SERS spectra from single molecules positioned in the NP gap were obtained. However, plasmonic heating is generally not wanted during the measurement. Along with hot carrier generation, high temperatures could lead to carbonization of the analyte.234,235 Carbonization is characterized by the emergence of a broad carbon D-band at ∼1350 cm−1 and a stronger G-band at ∼1580 cm−1 that dominates the time-averaged spectrum. Strategies to avoid carbonization have been demonstrated, with gold being less prone to induce carbon formation compared to silver,236 and carbonization tends to be further suppressed when SERS measurements are conducted in water.237 As briefly mentioned earlier, the strength of DNA origami lies in its flexibility to realize alignments of particles with different shapes and materials. This flexibility allows for the creation of large hotspots capable of accommodating sizable molecules and proteins while still providing sufficient field enhancement. Nanostars and bowtie antennas formed by Au nanotriangles have emerged as alternatives to nanosphere dimers to achieve strong field enhancement with wider gaps. Au NSTs feature sharp tips where the electromagnetic field is concentrated at resonant excitations.238 Recently, Kanehira et al. reported that dimers formed with “nanoflower” particles outperform spheres on “nanofork”-DNA origami nanoantennas.239 Single protein SERS was demonstrated by Tanwar et al., where thrombin was bound to a DNA template and then sandwiched between two bimetallic NSTs (Fig. 6a).240 However, aligning NSTs precisely tip-to-tip can be challenging, even with DNA origami. Furthermore, the sharp tips of nanostars exhibit limited stability under resonant excitation and are prone to melting during measurement. Other methodologies have therefore been explored and reported. Heck et al. demonstrated SM-SERS on biotin/streptavidin with self-assembled “nanolenses” made of silver particles (Fig. 6b).241 Tapoi et al. demonstrated SM-SERS of cytochrome c with DNA origami nanofork antennas (Fig. 6c).232 Zhang et al. demonstrated non-resonant SERS of Cy5 in 5 nm gaps between gold triangles on DNA origami.67
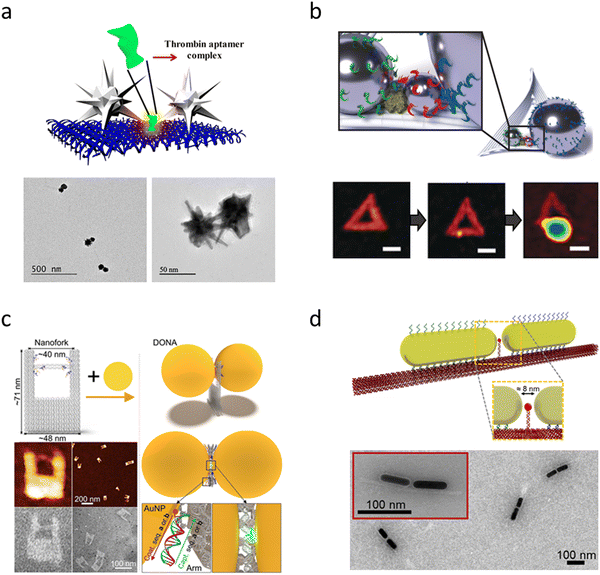 | ||
| Fig. 6 Examples of plasmonic DNA origami nanoantennas for single protein measurements. (a) Schematic of gold/silver nanostar dimers formed around thrombin on a rectangular DNA origami sheet. TEM images of the nanostar dimer structure (Reproduced from ref. 240 with permission from ACS publisher, Copyright 2021). (b) A single streptavidin molecule is positioned in the gap between silver nanoparticles of different sizes, forming a so-called “nanolens“. Reproduced from ref. 241 with permission from Wiley-VCH publisher, Copyright 2018. The sequence of AFM images shows the DNA origami template, the template with streptavidin attached, and the finally formed plasmonic structure. (c) DNA origami nanofork antenna structure. An analyte molecule is tethered to a DNA bridge spanning the particle gap reproduced from ref. 232 with permission from ACS publisher, Copyright 2021 (d) gold nanorods assembled by a DNA origami beam. A capturing strand is located between the nanorod tips to bind single proteins from the solution. Reproduced from ref. 242 with permission from Nature Publisher, Copyright 2023. | ||
A general limitation of most of these DNA origami approaches is a single dye or biomolecule must first be attached to the DNA origami. Subsequently, the plasmonic NPs are assembled around the analyte in a second step prior to measurements. This strategy may not be viable for typical sensing applications, where proteins or biomolecules must be identified from a liquid sample. Ideally, a SERS probe would enable the capture of analytes from the solution. Recently, a DNA origami scaffold with control over the tip-to-tip alignment of Au NRs with an average gap size of 8 nm has been reported (Fig. 6d).243 These gaps were accessible for proteins captured using specific anchoring sites located in the hotspot region. The capability to target specific biologically relevant proteins may facilitate future in situ or in vivo measurements, for example, Sharma et al.244 have reported the SERS detection of single epidermal growth factor (EGF) receptors using DNA origami-assembled gold nanorod dimer nanoantennas. To stabilize the structure during measurements, the origami template could additionally be protected via site-specific silanization without hindering any analyte binding.245 In combination with recent implementations of machine learning for SERS data analysis,246 the design for DNA origami assembled nanostructures for SM-SERS will pave the way for their wider use in biodiagnostics and personalized medicine.
As discussed above placing the analyte in the hotspot is critical for SM-SERS. Another approach that has been exploited for SM-SERS is tip-enhanced Raman spectroscopy (TERS, see Section 3.6 for more details), in which analyte molecules are placed in between the plasmonic tip and substrates.247–249 The apex of the tip localizes the EM field in a confined space through the lightning rod effect.250,251 Thus, the strong coupling between the tip and substrate results in strong EM field enhancement so the SERS EF. The spectral overlap of the SPR of the hotspot and the electronic transition of the analyte is an important factor for SM-SERS.250 This technique has been used in the chemical mapping of single molecules with atomic precision,252in situ probing of catalytic reactions at the SM level, molecular conformations in molecular electronics where SM junctions are used,253 and detection of biomolecules (viruses, DNA, and RNA)248,251,254 with SM precision. Despite significant progress in recent years, the practical applications of SM sensing are yet to be realized. It is still challenging to prove whether the SERS signals come from SM. Moreover, the degradation of analytes under laser beam effects the reproducibility of SM SERS spectra.
3.4. Target analytes
In direct SERS, target analytes are molecules/ions that exhibit Raman active vibrational transitions, undergoing changes in polarizability during these vibrations. Moreover, it is more suitable for molecules with unsaturated bonds and aromatic rings (high polarizability) like polyaromatic hydrocarbons (PAHs), dyes, etc. Nevertheless, other analytes show low Raman cross-section (Raman efficiency) like some gas species, proteins, etc so an indirect SERS methodology is applied for its detection. This type of detection implies using plasmonic substrates/NP functionalized with a Raman active molecule (typically with a large Raman scattering cross section and called SERS tag). Thus, in indirect SERS the interaction of the SERS tag with the target analyte produces a change in the Raman intensity or in the Raman shift. In the section, we summarize the detection of some of the most relevant target analytes, covering explosives and chemical warfare agents, ionic and gaseous species or biomolecules.Tremendous work has been performed to focus on challenges concerning trace explosives detection, such as innovations in flexible substrates that facilitate swabbing or wiping from surfaces, vapor detection, and making cost-effective, durable plasmonic structures. The molecular formula and structure of various explosives and chemical warfare simulants are provided in Fig. 7. Using tagged Au NRs, trace quantification of HMX and RDX has been reported recently based on the nitrite derived during hydrolysis. The study by Guven et al. has also been extended to real-world soil samples for simultaneous detection of HMX and RDX and achieving a recovery of greater than 90%.255 With a novel 2D transition metal nitride substrate as plasmonic material, explosive molecules like PETN, RDX, and HMX were detected on three base substrates: silicon, glass, and paper. Soundiraraju et al. have observed that the paper substrate has outperformed the other two substrates and a commercial substrate in enhancement.256 Similarly, 2D hybrid graphene oxide (GO) and Ag NPs substrates fabricated by laser ablation were used to detect RDX and HMX using SERS with a sensitivity in the order of picomolar concentration.257 Singh and co-workers were ableable to detect DNT and picric acid (PA), and dipicolinic acid (DPA) with nanomolar sensitivity using ultra-thin GO doped with nitrogen and nitrogen sulfur and decorated with Ag NPs.258 Further, Naqvi et al. have achieved laser-ablated AgNP/GO substrates with a dual benefit of chemical and electromagnetic enhancements.259 Ag-NPs embedded in GO have also been used for TNT sensing.260 Liyanage et al. have used flexible gold NTs-based plasmonic sensor for the trace detection of RDX, TNT, and PETN, with a remarkable platforms for reproducibility of 4%, LOD of 56 parts-per-quadrillions and enhancements of EF ∼106. The selectivity of the performance has also been analyzed by detecting explosives directly from fingerprints transferred to the sensor using a benchtop Raman system.261 Silver NTs have also been shown to be advantageous in the trace detection PA with μM sensitivity by Wang et al.262 Different papers (sandpaper, filter paper, and printing paper) loaded with picosecond ablated AgAu alloy NPs were used to detect RDX with enhancement factors 105. Byram and co-workers have demonstrated sample collection through swabbing promising real-world applications in surface examinations.263 Fan and co-workers have utilized plasmonic sticky and flexible SERS substrates to detect HNX, TATB, LLM-105, RDX, HMX, FOX-7, and TNT molecules. HNX traces on bags and fingerprints were also analyzed using the same substrate.264 Gao et al. investigated a novel wrinkled, flexible Au-coated polyester (PET) film prepared by lithography and plasma etching and used it to detect TNT.265 A flexible polydimethylsiloxane (PDMS)-based SERS substrate with Ag NPs has demonstrated TNT detection by wiping a cloth bag by Gao et al.266 Moram et al. have used NaCl as an aggregation agent for AgAu alloy NPs and encompassed them in simple filter papers. The detection of NTO and DNT was achieved by systematically optimizing the soaking time of the filter paper and studying the effects of the concentration of the NaCl.267 Explosive vapor detection using SERS is still a challenge because of the poor adsorption of molecules on the SERS substrate. Overcoming the challenge, gold NCs were used by Ben-Jaber et al. for the trace vapor detection of DNT and RDX, achieving an enhancement factor of 1010, the highest reported for the vapors so far.268 Vapor detection of TNT, RDX, and PETN has been reported with ng sensitivity using a handheld Raman spectrometer.269 A multiscale SERS substrate composed of photonic crystals and Au–silica NPs with core–shell morphology that can detect trace-level vapors rapidly (3 minutes) of DNT with 1 ppm sensitivity using a portable Raman spectrometer.270 Sputtered gold and silver alloy on roughened glass, along with principal component analysis (PCA), has been used for the selective detection of three explosives, TNT, RDX, and PETN.271 Anisotropic and cost-effective Ag dendrites prepared by electrochemical etching were used to detect RDX and AN with an enhancement of 104 and RSD of 9%.272 The study has been further extended using Au-decorated Ag dendrites, which have enhanced 106 times and lower LOD of ∼nM.273 Femtosecond laser ablation patterned micro square arrays on the Si surface. It was applied for trace detection of PA, RDX, and DNT using AgAu alloy NPs as plasmonic material. Their study found that the nanostructure at a particular fluence has yielded higher enhancement in the order of 106 for PA and 104 for RDX. The substrate also demonstrated efficiency in multiplexing along with PCA.274 Picomolar detection of TNT, RDX, and HMX was reported using Au nanogap substrates.275 Trimetallic AgAuCu nanostructures obtained via femtosecond Bessel beam ablation was tested for the trace detection of PETN and TNT in the nanomolar regime. The nanostructures forming a ladder-like periodic structure showed advantagesadvantages over thethe conventional Gaussian beam-structured surfaces.276 Podagatlapalli et al. have employed Bessel beam ablated Ag nanostructures obtained in water for the sensing of hexanitrohexaazaisowurtzitane (CL-20) with an EF of 106.277 Laser-ablated Si nanostructures, which yielded periodic structures on the surface, effectively detected 100 nM of 5-amino-3-nitro-1H-1,2,4-triazole (ANTA) after coating with gold.278 Simultaneous detection of explosives mixtures [PA+ 3-nitro-1,2,4- triazole-5-one (NTO) and PA + DNT] has also been reported using picosecond laser structured SERS substrates by Byram and co-workers. Unique Ag nanoribbons were observed during picosecond laser ablation of Ag under cylindrical focusing at a particular fluence, which was further used for explosive sensing with nanomolar sensitivity.279 Ag NSs fabricated for different angles of incidence during laser ablation have demonstrated femtomolar detection of 1-nitropyrazole (NPZ).280 Superhydrophobic substrates with Si pillars decorated with ZnO–Ag NPTs were used for the detection of NTO (∼10−12 M), TNT (∼10−12 M), and FOX-7 (10−10 M), extending their studies to natural samples as well.281 Ag-decorated ZnO structures were also used to detect ANTA, FOX-7, and CL-20.282 Syed et al. have prepared reusable and cost-effective Cu NSs fabricated by femtosecond and picosecond ablation and have demonstrated trace detection efficiency for CL-20, FOX-7, and ANTA with an EF of 105.283 Subsequently, hybrid nanoparticles, formed by combining laser-induced nanoparticle-embedded periodic surface structures (LINEPSS) on Si via femtosecond (fs) laser ablation followed by 20 nm Ag coating, further improved the EF to ∼107 in the detection of ANTA molecule, further improved the EF to ∼107 in the detection of ANTA molecule.284 Using a combination of laser-ablated structures and chemically synthesized gold NSTs, SERS-based detection of ammonium nitrate (AN) has been reported by Rathod et al. with μM sensitivity.285 The mixture of pesticide molecules (thiram and tetrabenazine (TBZ)) was detected using fs laser patterned Si with Au NSTs.286 Bharti et al. have synthesized alloy NPs by a two-step method of laser ablation followed by irradiation, and they were used for the successful detection of PA and AN with EFs ranging from 104 to 107.287 Core–shell Au@Pd NPs synthesized using the same method were used to detect AN and PA.288 Xiao et al. have used 1-D SPP-supported Ag nanostructures.289 In contrast, hybrid, recyclable SERS substrates based on TiO2 NRs decorated with Au NRs were used by Samransuksamer et al. for the detection of TNT.290 A new method to enhance the sensitivity of detection using a contactless approach with vigorous dielectric microspheres (DMs) embedded within a PDMS film has been proposed. The method was evaluated using 4-aminothiophenol (4-ATP) as the Raman probe. It significantly improved the detection of dipicolinic acid (DPA), an anthrax biomarker, with a 5.7-fold sensitivity improvement on a solid substrate.291 A multi-color fluorescent nanoprobe based on Tb3+ ion with Au nanoclusters was synthesized to detect DPA with a sensitivity of 3.4 nM and high specificity, making it effective, convenient, simple to operate, and possessing broad application prospects.292 Low-cost, scalable polyvinylidene fluoride (PVDF)-based Ag NP decorated SERS substrate has been used to detect DPA with a sensitivity of 1 ppm.293 Femtosecond laser ablated Au NSs decorated with Au NPs were used to detect DPA, DNT, and PA with LODs 0.83 pg L−1, 3.6 pg L−1, and 2.3 pg L−1.294 Bai et al. reported that papain-capped Au NPs tend to undergo well-regulated aggregation upon adding Hg2+ ions and DPA, resulting in ultrasensitive detection of DPA without sample pre-treatment and a low LOD of 67.25 pM.295 A microporous Si substrate coated with an optimized layer of Au was developed by Singh and co-workers for trace detection of PA with LOD in the nM regime, and the enhancement was attributed to the presence of dense hotspot network resulting in coupling between both localized and extended plasmons.296 Kong et al. have demonstrated a highly sensitive photonic crystal-based substrate with diatom frustule and Ag NPs for detecting TNT with 10−10 M sensitivity. Nanolitre of the analyte molecules was carefully delivered to the hotspot area using inkjet printing, resulting in superior sensitivity.297 SERS coupled with digital microfluidics (SERS-DMF) has been proposed for highly sensitive and automated detection of explosives with better reproducibility and efficiency with proof of the concept for TNT and NTO with LODs of 10−7 and 10−8 M, respectively, by Liu et al.298 Using a simple, inexpensive hydrophobic condensation strategy, biomimetic super-hydrophobic Ag micro/nano-pillar array surfaces were prepared as SERS substrates for ultrasensitive sensing of PA and NTO with picomolar sensitivity.299 Chemically produced Ag NWs were used to detect perchlorates, chlorates, nitrates, picric acid, and 2,4-dinitrophenol.300 Monoethanolamine (MEA) based Au NPs were employed to selectively and sensitively detect TNT in real-world samples such as envelopes, luggage, lake water, and clothing, with a detection limit of 21.47 pM by Lin et al.301 AuNR@Ag NCs loaded into bacterial cellulose aerogels were used by Wu et al. for sensitive detection of TNT with enhanced selectivity achieved by using 4-ATP resulting in EF of 108.302 Exotic waxberry-like Au NPs decorated on PDA were fabricated using seed-mediated growth and further used for the detection of DNT by Chen et al.303 Based on Janowsky complex formation of explosives and the SERS of the derivatives, TNT, HNS, and tetryl (2,4,6-trinitrophenyl-n methylnitramine) were detected with a sensitivity of 6.81 ng mL−1, 135.1 ng mL−1, 17.2 ng mL−1, respectively, by Milligan et al.304 To overcome the signal fluctuations in SERS, a deep learning model called neural network-aided SERS has been proposed to bridge the gap between lab and field applications.305 The model employed a signal-to-noise ratio approach to label the spectra, which were further classified by the model, and accurate predictions were made in out-of-sample testing. With the goal of quantification of trace explosives, a low-cost hydrophobic filter paper has been proposed, coated with silicone oil and using laser-ablated Au NPs. The authors have used PCA and support vector regression to quantify traces of PA with an accuracy of 94% within 10 s.306 Working under resonance Raman scattering conditionsconditions of an explosive molecule, PA, trace detection (100 nM) was achieved using AgAu nanostructures.307 Deep learning method based onon the FAB-ResNet and (NLSTM) networks in the successful detection of explosive residues.308 Detection of bio-hazardous materials has also been reported using SERS-based sensing. With the help of Au-coated Si, nanopillars functionalized with a chemical nerve agent antidote, 4-pyridine amide oxime, selectively and sensitively detect nerve agents Tabun, Cyclosarin, and VX in aqueous solutions. The SERS signals of the analytes distinctly discriminate between specific and non-specific binding down to sub-ppm levels, with binding specific SERS response decreasing in the order of Tabun > VX > Cyclosarin.309 Self-assembled Au NPs coated with a citrate layer that acted as a trap for probe molecules were used by Lafuente et al. for the detection of dimethyl methyl phosphonate (DMMP) in the gas phase, achieving a sensitivity of 130 parts-per-billion.310 The core–shell structure Au@ZrO2 has also demonstrated excellent applicability for detecting DMMP with an RSD of 6.8% and a durability of 30 days.311 Using high droplet adhesion that led to a high density of hotspots, femtomolar detection of two nerve agents, VX and Tabun, has been reported by Hakonen et al. using flexible Au decorated Si nanopillars and a handheld Raman system.312 Gas phase detection of DMMP was reported by Lafuente and co-workers using Ag nanoplates on different substrates like Si, stainless steel mesh, and graphite foils with sensitivity down to 2.5 ppmV using a portable Raman spectrometer.313 Huang et al. have demonstrated flexible, uniform cotton swabs coated with Ag NPs that were tagged to detect DMMP specifically have shown good reproducibility of 5.6%, implying practical usage.314 Electrochemically roughened silver foil has been used to detect two nerve agents, DMMP and DIMP in the vapor phase.315 Wang et al. have shown that by concentrating the molecules in the vital hotspot areas on the SERS substrates, a thin water film-confined SERS strategy was proposed, allowing for the successful detection of DMMP molecules weakly interacting with SERS-active substrates.316 Wang and co-workers have achieved sensitive and flexible AuNPs@polyimide SERS heating chips and successfully detected nM TNT.317 There are a few earlier review articles on the technique and materials used for SERS.318–326
 | ||
| Fig. 7 Molecular formula and structure of various explosive molecules and chemical warfare simulants covered in this review. | ||
Trace-level liquid/solid phase detection is essential and highly beneficial for chemical and explosive sensing applications. However, detecting vapor-phase chemicals and explosive compounds is a better solution to overcome the drawbacks of existing physical probing approaches in the present and foreseeable future. It is imperative to create essential technology that facilitates vapor-phase detection of explosive molecules economically with better sensitivity and stability. Explosive vapor sensing has been demonstrated by a few researchers in recent years.327 The small quantity of DNT that emanates from TNT dominates the vapor headspace and aids as a natural tracer for TNT vapor detection. Therefore, detecting volatile impurities of TNT, such as 2,4-DNT, 2,6-DNT, and 1,3-DNB, is desirable. SERS-active substrates' response time and sensitivity are essential for superior DNT vapor detection. TNT vapor detection is limited due to low vapor pressure. Adhikari et al. have achieved rapid and sensitive vapor-phase detection of TNT, DNT, and PETN using nanogap SERS-substrate produced by Au NPs decoration on SiO2 nanopillars and Au layers. The detection capability of TNT, PETN, and DNT for 10−9 atm., 10−11 atm., and 10−7 atm. vapor pressure was estimated at a lower laser power of 0.2 mW, and room temperature.328 Shabtai and co-workers demonstrated the utility of Au NPs modified quartz fibers for gas phase detection of VX and HD. They reported the least detection for VX, and HD at ∼8 ppbv, and ∼0.73 ppbv, respectively.329
Detection of airborne chemicals and explosive molecules using SERS at a standoff distance (unlike the Raman technique) is challenging owing to the confinement of near-field enhancement. Consequently, very few reports on the SERS studies using standoff configurations exist in the literature. Phan-Quang and co-workers have recently demonstrated the ‘first’ in-air SERS detection in a standoff configuration (at a distance of 200 cm).330 They have utilized aerosolized plasmonic colloidosomes as airborne plasmonic hotspots. They prepared a 3D plasmonic cloud (containing ∼109 Ag NCs cm−3) to detect airborne species (methylbenzenethiol (MBT), rhodamine 6G (Rh6G)), and methylene blue (MB). Zhang et al. have demonstrated the possibility of remote chemical detection (4-ATP) using fiber-based SERS substrates. Such studies can be extended to explosives and other hazardous materials.331 Another exciting work demonstrates the possibility of detecting buried explosives. In this case, they could detect buried explosives (2,4-dinitrotoluene) in simulated field conditions.332 They claim that the explosives used in landmines are often composed of TNT. Further, its manufacturing impurities include (a) 2,4-DNT, (b) 2,6-DNT, and (c) 1,3-dinitrobenzene (1,3-DNB). A thorough optimization of efficient SERS substrates and appropriate progress in signal collection and detection optics will make it possible to detect buried chemicals (e.g., in sand). Machine learning/deep learning techniques augment the SERS data (wherever there are limitations).305 A summary of the significant works on SERS-based sensing of explosives reported in recent years is presented in Table 1. In Fig. 8 shows a schematic summary of the SERS-based explosives sensing techniques, materials, and requirements for commercialization, including the future challenges and scope. There is also keen interest in developing novel SERS materials/substrates for detecting explosives.333–338 For example, Kalasung et al. have developed effective Au/ZnO-based sensors to detect PETN.339 Their sensors exhibited an optimal performance at 104 g mL−1 while the LOD was 10−7 g mL−1 with an EF of 3.01 × 106. As discussed elsewhere, flexible SERS substrates are essential for various applications wherein the analyte available is too small and cannot be transported to the lab.338 These substrates facilitate swabbing and collecting the trace analyte molecules from any uneven surface, and further, with the help of a portable Raman spectrometer, can be used to identify the molecule. With advancements in the preparation of novel, efficient, versatile SERS substrates and in the development of miniature spectrometers, explosive sensing is expected to become cost-effective and practical in the near future.
| S. No. | Target molecules | SERS substrates | Preparation technique (s) | LOD | Ref. | Comments |
|---|---|---|---|---|---|---|
| LOD: limit of detection. LCD: lowest concentration detected. EF: enhancement factor. NP: nanoparticle. NPT: nanoplate. NC: nanocube. NR: nanorod. NS: nanosphere. NST: nanostar. ND: nanodendrite. LINEPSS: laser-induced NP-embedded periodic surface structures. PCA: principal component analysis. PVDF: Polyvinylidenefluoride. p-ABT: p-amino benzenethiol. ATS: N1-(3-trimethoxysilylpropyl) diethylenetriamine. MEA: Monoethanolamine. | ||||||
| 1 | RDX (1,3,5-trinitroperhydro-1,3,5-triazine) | Au NRs with Ag NPs | Chemical reduction | 0.39 mg L−1 | 255 | RDX + HMX sensing in clay soil via UV-Visible and SERS |
| Triangular Au NPTs | Self-assembly of Au NPTs onto functionalized glass coverslip, followed by transferring it onto to a flexible adhesive substrate | 50 ppq | 261 | Detection of explosive residues from fingerprints left on solid surfaces | ||
| 2D-titanium nitride (Ti2NTx) (MXene) | Chemically synthesized and deposited on paper, silicon or glass | 1 μM | 256 | The performance of different substrates was compared. | ||
| Silver NCs | Polyol method | 1 nM | 268 | Enhancement factor (EF) ∼9.2 × 1010 | ||
| AuAg film on glass | Sputtering | 40 pg | 271 | Distinction of explosive using PCA | ||
| Ag Nano dendrites (NDs) | Electroless etching process | 5 μM | 272 | EF104 | ||
| Ag NPs decorated-rGO substrate | rGO-modified Hummers’ method; Ag NPs-in situ reduction | 10−12 M | 257 | The size of Ag NPs was around 20 nm and EF was ∼1.6 × 109 | ||
| Si micro squared arrays with AgAu alloy NPs | fs laser ablation | 1 μM | 274 | EF ∼104 | ||
| Au nanogap | e-beam evaporation | 10 pM | 275 | Picomolar detection of TNT, RDX, and HMX | ||
| Ag–Au alloy NPs | ps laser ablation | 100 nM | 263 | Filter Paper loaded with NPs | ||
| Au-coated GaAs nano-ripple | fs laser ablation in liquid | 10 μM | 340 | 70 nm ripples | ||
| 2 | HMX (Cyclotetramethylene-tetranitramine) | TriangularAu NPTs | Self-assembly of Au NPTs onto functionalized glass coverslip, followed by transferring it onto to a flexible adhesive substrate | 0.61 mg L−1 | 261 | Direct detection on a benchtop Raman system |
| 2D-titanium nitride (MXene) | Chemically synthesized and deposited on paper, silicon or glass | 1 μM | 256 | The performance of different substrates was compared. | ||
| Au NPs | Iterative hydroxylamine seeding method | 8.1 × 10−6 M | 269 | ng sensitivity using a portable Raman spectrometer | ||
| Ag NPs decorated-rGO substrate | rGO-modified Hummers' method | 10−12 M | 257 | 20 nm Ag NPs, EF ∼ 1.6 × 109 | ||
| Ag NPs: In situ chemical reduction | ||||||
| Ag nanotubular arrays | Axicon-generated femtosecond | 5 μM | 341 | TNT (50 nM), tetryl (50 nM), RDX (500 nM), superior RSD values, and enhancement factors of ∼106 | ||
| Bessel beam ablation in air | ||||||
| 3 | PETN (Pentaerythritol tetranitrate) | 2D-titanium nitride (MXene) | Chemically synthesized and deposited on paper, silicon or glass | 1 μM | 256 | The performance of different substrates was compared. |
| Au nanogap | e-beam evaporation | 10 pM | 275 | Picomolar detection of TNT, RDX, HMX | ||
| Au NPs on quanrtz or glass fibers or polyurethane sponges | Iterative hydroxylamine seeding method | 1.5 × 10−6 M | 269 | Detection of vapors using portable Raman spectrometer | ||
| AgAuCu | fs laser ablation | 500 μM | 276 | Laser-induced induced AgAuCu periodic surface structures | ||
| 4 | Fox-7 (1,1-diamino-2,2-dinitroethylene) | ZnO//Ag mesoporous nanosheetn on Si micropillars | Atomic layer deposition, chemical bath deposition, ion-sputtering | 10−9 M | 281 | Mesoporous |
| Nanosheets on a Si micropillar array | ||||||
| 5 | CL-20 (Hexanitrohexaazaisowurtzitane) | Ag/ZnO nanostructures | Thermal oxidation | 10 μM | 282 | EF ∼ 104 |
| Ag NPs | Laser ablation in liquid (LAL) | 5 μM | 277 | ∼2 ps and ∼40 fs, 800 nm; Axicon (base angle = 25°). LAL in distilled water with Bessel beam focusing | ||
| Cu NPs | Laser ablation in liquid (LAL) | 5 μM | 283 | ∼2 ps and ∼40 fs, 800 nm; axicon (base angle = 25°); pulse energy 25–400 μJ; LAL in acetone with Gaussian beam focusing | ||
| 6 | Ammonium nitrate (AN) | Ag NDs | Electroless etching process | 1 μM | 272 | EF∼ 104 |
| Ag NPs with Au NSTs | Ag NPs-ps laser ablation | 5 μM | 285 | Bessel beam-axicon lens (base angle of 10°) | ||
| Au NSTs-chemical reduction | ||||||
| Ag–Cu NPs | fs laser ablation | 5 μM | 287 | EFs ranging from 104 to 107 | ||
| Au decorated Ag NDs | Electroless etching process | 100 nM | 273 | Wafer-scale synthesis. Low-cost and high durability | ||
| Shell-wrapped core@shell Au@PdNPs | Pulsed laser ablation | 0.5 μM | 288 | Analysis of EFEF for different morphologies | ||
| 7 | TNT (2,4,6-Trinitrotoluene) | Ag sinusoidal nanograting | SiO2 nanogratingn by two-beam interference (266 nm) followed by Ag evaporation | 10−5 M | 289 | 1-D SPP-supported Ag nanostructures |
| TiO2 NRs with Au NPs | TiO2 NRs: electron-beam evaporation with glancing-angle deposition | 10−7 M | 290 | Hybrid and recyclable substrates | ||
| Au NPs: deposition–precipitation | ||||||
| Triangular Au NPTs | Self-assembled Au TNPTs on amine-functionalized glass, transferred to a flexible adhesive substrate by the stamping technique | 900 ppq | 261 | Direct detection on a benchtop device | ||
| Photonic crystal biosilica with Ag NPs | In situ growth | 10−10 M | 297 | Analyte delivered to the hotspot area using inkjet printing | ||
| Au nanogaps | Electron beam evaporation | 1 pM | 275 | pM detection of TNT, RDX, and HMX | ||
| Flexible PET nanocones with Au coating (30 nm) | Colloidal lithography and oxygen plasma etching | 10−13 M | 265 | Trace detection (10−10 M) on cloth bag | ||
| Nanowrinkles@ micro (zigzag, coral, rectangular) patterned PDMS – Au film/Ag NPs | UV photolithography, oxygen plasma and stretching treatment – Au deposition by electron beam | 10−13 M | 266 | Trace detection (10−9 M) on cloth surface | ||
| Au NPs | Chemical reduction | 1.1 × 10−7 M | 269 | Au NPs on quartz fibers or polyurethane sponges to detect vapors using a handheld device | ||
| Pyramid AgFe NPs embedded rGO | rGO: Hummers' method | μM | 260 | TNT in an envelope, luggage, lake water, and clothing | ||
| AgFe: chemical method | ||||||
| MEA—modified Au NPs | Chemical methods | 21.47 pM | 301 | TNT in an envelope, luggage, lake water, and clothing | ||
| AuNR@Ag NCs on cellulose aerogels | Chemical methods | 8 × 10−12 g L−1 | 302 | p-ABT modified substrates to complex TNT via Meisenheimer complex formation | ||
| Ag nanoribbons | PS laser ablation | 25 nM | 342 | Cylindrical focusing | ||
| AuNPs@ polyimide heating chips | Liquid–liquid self-assembly; sandwich structure | 10−9 M | 317 | In situ collection and detection of TNT in complex environments soil, cloth, fruit | ||
| Digital microfluidic device with Ag NPs | Chip fabrication technique | 100 nM | 298 | Tested for real-world samples and proved good accuracy | ||
| 8 | DNT (2,4-Dinitrotoluene) | Polydopamine-Au nanowaxberry | Seed-mediated synthesis | 1 μM | 303 | DNT spiked in environmental water and methanol |
| AgAu NPs on filter paper | fs laser ablation | 1 μM | 267 | Optimization of the soaking time and studied the effects of NaCl concentration | ||
| Ag NCs | Polyol method | 10−15 M | 268 | Detected DNT vapor in a sealed container | ||
| Diatom photonic crystals and core–shell Au@SiO2 NPs | Chemical methods | 10 ppb | 270 | DNT vapor using stagnant vapor and vapor flow chambers | ||
| Ag NPs/rGO hybrids | rGO: Hummers' method | 10−12 M | 259 | Dual benefit of chemical and EM enhancement | ||
| Ag NPs: Chemical method | ||||||
| Au NSs (linear and crossed pattern) | fs laser ablation in air | 3.6 pg L−1 | 294 | Detection of DPA, DNT and PA with LODs 0.83 pg L−1, 3.6 pg L−1 and 2.3 pg L−1 | ||
| 9 | Tetryl (2,4,6-trinitrophenyl-n methylnitramine) | Ag NPs | Chemical reduction | 17.2 ng mL−1 | 304 | Detection via formation Janowsky complex with 3-mercapto-2-butanone |
| Laser-induced AgAuCu periodic nanostructures | fs laser ablation | 200 nM | 276 | Ladder-like NPs | ||
| Au NPs@Ag NDs | Electroless deposition | 100 nM | 305 | |||
| Au-coated GaAs periodic surface structures | GaAs: fs laser ablation in air | 10−5 M | 343 | EFEF ∼104 | ||
| Au deposition and annealing | ||||||
| 10 | Dipicolinic acid (Pyridine-2,6-dicarboxylic acid) | Au NSs (linear and crossed pattern) | fs laser ablation in air | 0.83 pg L−1 | 294 | Detection of DPA, DNT and PA with LODs 0.83 pg L−1, 3.6 pg L−1 and 2.3 pg L−1 |
| Au NP | Chemical reduction | 0.01 ppb | 295 | 30–100 nm NPs. Controllable aggregation upon addition of Hg2+ ions and DPA | ||
| Au nanoclusters | Chemical reduction | 3.4 nM | 292 | Hybridization of Tb3+ ion | ||
| Ag NPs doped PVDF | Suction filtration | 1 ppm | 293 | PVDF pretreated with various solvents and NaCl | ||
| Ag NPs with nitrogen and nitrogen–Sulphur with GO | GO: Hummers' method | 0.01 ppm | 258 | Nanomolar sensitivity | ||
| Ag NPs: chemical methodmethod | ||||||
| Dielectric microspheres in PDMS film with Ag NPs | Modified photolithography | 0.1 μM | 291 | Proposed a new method called DSERS-based on dielectric materials | ||
| 11 | ANTA (5-amino, 3-nitro, -1H-1,2,4-nitrozole) | Au-coated laser-induced Si periodic surface structures | ps laser ablation | 100 nM | 278 | Focused airy pattern of the p-polarized laser |
| Ag plating on Si LINEPSS structures | fs laser ablation in acetone | 1 μM | 284 | SERS efficiency was improved by Ag/Au NP deposition on LINEPSS | ||
| 12 | NTO (3-nitro-1,2,4-triazol-5-one) | Mesoporous ZnOAg nanosheets on Si micropillar array | Atomic layer deposition, chemical bath deposition, ion-sputtering | 6 × 10−12 M | 281 | SERS efficiency was improved by Ag/Au NPNP deposition on LINEPSS |
| AgAu NPs on filter paper | fs laser ablation | 10 μM | 267 | Detected TNT, NTO, FoX-7 (10−8 M) in real-world samples, like river and tap water | ||
| Digital microfluidic device with Ag NPs | Chip fabrication technique | 10 nM | 298 | Optimization ofof the soaking time and studied the effects of NaCl concentration | ||
| Superhydrophobic Ag micro/nano-pillar array | Electrochemical deposition on structured Si | 10−13 M | 299 | Tested on real samples and proved good accuracy | ||
| 13 | 1-Nitropyrazole (NPZ) | Metal NSs | Laser ablation | fM | 280 | Different angles of incidence during laser ablation have been studied |
| 14 | Urea nitrate (UN) | Au NPs | Chemical reduction | 9.2 × 10−4 M | 269 | Au NPs on quartz fibers or polyurethane sponges were used to detect vapors using a handheld spectrometer |
| 15 | Dimethyl methyl phosphonate (DMMP) | Cotton swab with Ag NPs | ATS—incorporated Ag NPsNPs later. Ag NPs: in situ chemical reduction (NaBH4) | 1 g L−1 | 314 | Flexible and reproducible |
| Citrate-stabilized Au NPs | Chemical reduction and self-assembly | 130 ppb | 310 | Gas phase detection | ||
| Core–shell Au@ZrO2 NPs | Chemical reduction | 100 mg kg−1 | 311 | Also detected methyl parathion and triazophos at 0.01 mg kg−1. On orange peel, LOD of 0.5 and 0.1 mg kg−1, respectively | ||
| Au micro/NSs bowl-like array | Electrodeposition | 10−3 mol L−1 | 316 | Better signal at slower evaporation and stronger excitation power | ||
| Ag NPTs | Chemical reduction | 2.5 ppm V (14 mg m−3) | 313 | Gas phase measurements using a portable Raman spectrometer | ||
| 16 | DIMP (Diisopropyl methylphosphonate) | Silver oxide | Electrochemical | 0.3 torr | 315 | Detection of DMMP vapor at 1.6 torr |
| 17 | VX (O-ethyl S-diisopropylaminomethyl methylphosphonothiolate), TABUN | Au-coated Si nanopillars | Lithography and reactive ion etching | ∼13 and ∼670 fmol | 312 | Flexible Au-decorated Si nanopillars and a handheld Raman system |
| Ag/Au-coated Si nanopillars | mask-less lithography process and electron beam evaporation | 0.4 μM for VX | 309 | Binding-specific SERS response decreases in the order: Tabun >VX >Cyclosarin | ||
3.4.2.1. Metallic cations. Metallic cations, as atomic species, cannot be detected directly by SERS. However, different indirect SERS strategies have been developed for the selective and sensitive detection of metallic cationic species. Different strategies have been reported for cations such as Cd(II),344,345 As(III),346,347 Hg(II),345,348–350 Pb(II),350–352 Cu(II),352 and Zn(II),352 among others. One of the most common strategies involves inducing the aggregation of a colloidal dispersion of plasmonic nanoparticles through the presence of the target metal cation. This process begins with the functionalization of plasmonic nanoparticles, wherein they are initially coated with a Raman-active molecule displaying a specific affinity for the target cation via covalent or not covalent interactions. In the presence of the cation, the nanoparticles aggregate, forming hotspots that significantly enhance the SERS signal of the attached Raman-active molecule on the plasmonic surface (Fig. 9a).347,353 This SERS signal enhancement is directly proportional to the concentration of the target cation, allowing for quantification. However, a key challenge lies in achieving high selectivity, necessitating the careful selection of an appropriate chelation system to avoid undesired interactions. Each system is tailored to a specific metal cation; for instance, Duan et al.344 designed a self-aggregating gold nanoparticle system for the detection of Cd(II), a metal associated with conditions like cancer and cardiovascular diseases. The chelation of Cd(II) is initiated by atom transfer radical polymerization from the nanoparticle surface (SI-ATRP), resulting in immediate nanoparticle aggregation that produces an increase in the SERS signal of the nanoparticle probe. Importantly, this process is selective, as it does not occur in the presence of other cations.
 | ||
| Fig. 9 Ionic SERS detection. (A) Schematic diagram of the detection of As(III) ion by the aggregation of nanoparticles in function of As(III) concentration. Reproduced from ref. 347 with permission from Elsevier Publication copyright 2020. (B) Schematic representation of the Hg(II) ion sensing mechanism based on the replacement of Rhodamine B molecules through the reduction of Hg(II) ions on the surface of gold nanoparticles. Reproduced from ref. 354 with permission from Springer publication, copyright 2009 (C) representative SERS spectra of 4-MBA at different pHs and experimental calibration curve or ratio R = (A695 − A848)/(A695 + A848) as a function of pH. Reproduced from ref. 355 with permission from ACS publication, copyright 2021. | ||
Not all strategies for detecting metallic cation detection via SERS involve the aggregation of colloidal metal nanoparticles; some focus on solid plasmonic substrates functionalized with a DNA probe, which undergoes conformation changes in the presence of the target cation leading to a “turn-on/turn-off” SERS response. This approach has been extensively used for quantification of Hg(II). For instance, Chung et al.356 employed a doubly labeled DNA, incorporating a thiol for binding to Au surface and a Raman-active molecule to provide the SERS signal at opposite ends. In the absence of Hg(II), no significant SERS signals are observed, as the Raman-active molecule remains distant from the Au surface. However, the presence of Hg(II) induced a conformational change of the DNA, causing the approximation of the Raman reporter to the surface, leading to the amplification of the SERS signals. Another strategic approach capitalizes on the strong affinity of certain cations for the plasmonic metal surface, as observed in the case of Hg(II) ions and gold. This high affinity prompts the displacement of a pre-assembled Raman-active molecule from the gold surface leading to a reduction in the SERS signal.354 Moreover, higher concentrations of Hg(II) result in a more substantial replacement of the Raman-active molecule, leading to a further decrease in the SERS signal (Fig. 9b). It is important to note that this method is specifically applicable for identifying cations (or anions) with a pronounced affinity for the plasmonic surface.
Regardless of the approach taken, the development of a strategy for simultaneous detection and quantification of various cations is a challenge. This challenge becomes particularly pronounced when the accurate quantification of each cation is required. To progress in this field, further exploration is essential, potentially involving the development of intricate plasmonic nanostructures. These nanostructures can integrate various chelation agents customized for each specific cation, resulting in the generation of distinct Raman responses as a function of the bound cation.
3.4.2.2. Anions. Similar to cations, atomic anions (such as halogens, S2−, N3−, etc.) cannot be directly detected by Raman. However, unlike cations, the direct or indirect detection of anions is deemed less significant, resulting in less exploration of their SERS detection. A notable exception lies with oxyanions (SO42−, ClO4−, etc.) and other anions such as cyanide (CN−) or thiocyanate (SCN−) which possess moderate Raman cross-sections enabling their direct detection by SERS. Nevertheless, their limited affinity for plasmonic surfaces presents a challenge for direct detection. To overcome this hurdle, a common strategy involves functionalizing the plasmonic substrate to render it positively charged. This modification induces an electrostatic attraction, drawing the anions into close proximity with the SERS substrate, thus facilitating their detection. Du et al.357 employed this strategy to successfully detect anions in water. They modified Ag-based SERS substrates with amino and amide groups to create a positively charged surface, enhancing the substrate's affinity for anions such as SO42−, CN−, SCN−, and ClO4−. As a result of this functionalization, the detection limits reached the ppb regime, showcasing the effectiveness of this approach in improving sensitivity for anion detection. It should be noted that while there are numerous approaches to detecting metallic cations, the methods for characterizing anions are comparatively fewer. This discrepancy likely arises from the lower perceived significance and demand for their detection and quantification.
3.4.2.3. H3O+/HO− ions. The quantification of the H3O+/HO− ions holds significant importance in biological systems due to the implications of pH. Understanding pH is paramount as its value profoundly influences the functionality of cells and organisms across myriad facets. In numerous applications, the necessity for straightforward, intuitive, sensitive, and stable pH detection, encompassing both acidic and basic characteristics inside and outside the cell, is indisputable. Moreover, SERS pH sensors offer the advantage of in situ real-time pH monitoring in a non-invasive and non-destructive manner. Halas et al. were the first to report the effect of the pH medium on the Raman signal of the 4-mercaptobenzoic acid (4-MBA) due to the partial ionization/deionization of the carboxylic group,358 converting this Raman active molecule in a pH indicator that was used for the development of different pH sensors. However, while 4-MBA can accurately predict pH values within the range of 5.0 to 8.0 (Fig. 9C),355 its limited coverage is insufficient for various applications. Consequently, alternative molecules with broader pH ranges have been explored. These include 3,5-dimercaptobenzoic acid, ranging from pH 4.0 to pH 8.0;359 4-mercaptopyridine, from pH 3.0 to pH 8.0;360 4-aminothiophenol, with a range of 3.0 to 9.0;361 Alizarin Yellow R, covering pH 10.04 to 14.04;362 among others. However, to date, no SERS pH sensor has been able to cover the entire pH range simultaneously, thus, further investigation is necessary to develop such a sensor.
The literature offers numerous examples of both extracellular and intracellular pH SERS sensors. For instance, De Marchi et al.355 introduced a noteworthy extracellular pH SERS sensor consisting of a mesoporous plasmonic substrate (Au@Ag@mSiO2 nanorattles functionalized with 4-MBA). This substrate allows for the diffusion of metabolites secreted by bacterial colonies, thereby inducing pH variations, which in turn enables the correlation between metabolite production and the corresponding phenotype discrimination. On a different note, Ren et al.363 developed an intracellular pH SERS sensor based on Au NPs functionalized with 4-mercaptopyridine (4-MPy) as the probe molecule and Bovine serum albumin (BSA) to provide biocompatibility and stability. These NPs exhibited exceptional sensitivity from pH 4 to 9, biocompatibility, and stability. This innovative approach facilitates long-term dynamic monitoring of cellular processes and offers the potential for discriminating between normal and cancerous cells based on their pH distribution.
Detection, identification, and monitoring of ppb to few ppm of toxic and harmful gases in the environment is essential to address societal challenges; but is nonetheless challenging. This means, considering the low density of gases and the SERS surface selectivity, very few molecules per unit volume are available for detection. There are scarce examples of label-free SERS detection of gases, even at ppb level, by direct interaction with the bare metallic surface.373–379 From a methodological point of view, the conventional approach uses rigid or flexible SERS substrates and gas sampling is performed directly from the environment. The experimental LOD and response time are mainly determined by the “sticking” probability of the target to the SERS substrate, highlighting the importance of proper surface functionalization and fluid-dynamics. The other important facing challenge is the small Raman cross section of the gaseous molecules of interest,380i.e. poor Raman scattering efficiency, up to 7 orders of magnitude lower than that of standard SERS probes. Thus, although in SERS the rational design of the metallic nanostructure is relevant, in the case of SERS-based gas sensing, the development of strategies for gas molecules confinement near the plasmonic surface is of paramount importance.381,382
Different approaches are being applied to bring the detection limits down to relevant levels for practical applications, including: analyte condensation by cooling the plasmonic material,383 electrodynamic precipitation of targeted charged molecules,384 the use of 3D matrices for constructing plasmonic nanostructures with efficient access to target analytes,378,379,385–388 hybridization of noble metal-based SERS active substrates with semiconductors369,389,390 for the synergetic coupling of EM and charge-transfer (CT) mechanisms, and surface functionalization. By far, functionalization of the metallic surface (see Fig. 10) is the strategy most widely attempted for enhancing the gas adsorption efficiency by means of: (i) capturing probes capable of non-covalent interactions with large Raman cross-section values for being used as reporters,310,391–393 (ii) ultra-thin layers of sensing materials capable for specific adsorption or eventually “in situ” reaction with the target molecules provoking a solid transformation,394,395 (iii) 2D nanomaterials,389,390 (iv) thin sheets of porous metal oxides396–398 and (v) the extensively studied and reviewed metal–organic frameworks MOFs(mainly ZIF-8).364,366,399–405 Special attention deserves the scarcely explored field of metal-oxide-wrapped plasmonic nanostructures. A SERS substrate based on CuO-coated (6 nm thick) Au-wrapped Si nanocone array was validated for the detection of 2-chloroethyl ethyl sulfide (CEES) in 10 min and the lowest detectable concentration was 10 ppbV.395 The adsorption of CEES molecules on the CuO coating is attributed to the surface-hydroxyl-induced specific binding. Above all, metal-oxide-wrapped metal architecture offers unique opportunities for the development of dual-modal chemoresistive-SERS platforms.397
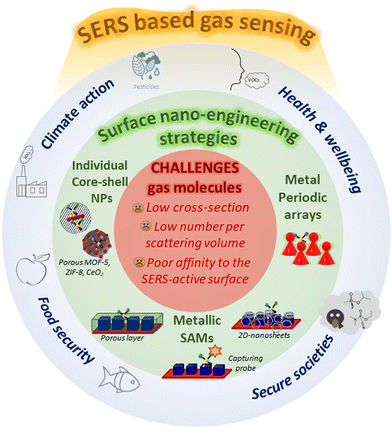 | ||
| Fig. 10 Schematic illustration of SERS applications for gas sensing with focus on challenges and some of the surface nano-engineering strategies discussed in this section. | ||
In addition to the preconcentration based SERS signal amplification, MOFs may also impair selectivity by molecular sieving effect, improve the stability of the metal and prevent the EM field decay along the radial direction.402 Different strategies are described in the literature to unite MOFs with metal nanoparticles for gas detection.406 The most extended is the encapsulation of individual metallic nanostructures into porous frameworks. As an example, a SERS platform based on a close-packed thin film of Au@Ag NRs individually encapsulated within a ZIF-8 framework has been demonstrated for the detection of chemical weapons in gas phase366 (see Fig. 11a). Interestingly, the characteristic Raman peak of ZIF-8 at 685 cm−1, was used as an internal standard for quantitative detection. The performance with a portable Raman instrumentation was validated for 2.5 ppm of DMMP in ambient air, and 76 ppb of CEES; with response times of 21 s and 54 s, respectively. The SERS substrate showed good stability and reusability by degassing at 200 °C. H2S detection by Au NBPs encapsulated in ZIF-8 occurred thanks to indirect detection.401 Before introducing the gas, the peak corresponding to Au–Br (273 cm−1) (cetyltrimethyl ammonium bromide is used as a soft template in the synthesis), is observed and after H2S exposure this peak decreases and at the same time Au–S peak (175 cm−1) increases. The dynamic range for detection was 0.13 ppbV to 1300 ppmV. As proof-of-concept demonstration, this SERS substrate was applied to monitor the release of H2S from spoiled fish samples. Core–shell Ag NC@ZIF-8 nanostructures with cysteamine anchored on the silver surface have been used as SERS probes to build up the Array-SERS chip407 for benzaldehydes detection (recognized as lung cancer biomarkers). The PDMS chip has a prism array patterned in the microfluidic channel to increase the sticking probability of the gaseous molecules and sensing interfaces and lower the LOD to 1 ppb. Through the specific Schiff reaction, a LOD of 10 ppb for aldehydes was achieved on gold super-particle functionalized with 4-ATP and coated with a thick shell of ZIF-8.405 Here, MOF shells play an extra role: precluding big gaseous molecules present in complex VOC systems. In an attempt to accumulate more molecules in the proximity of the plasmonic surface for lowering LOD, hollow ZIF-8 wrapped in gold has been recently investigated.408
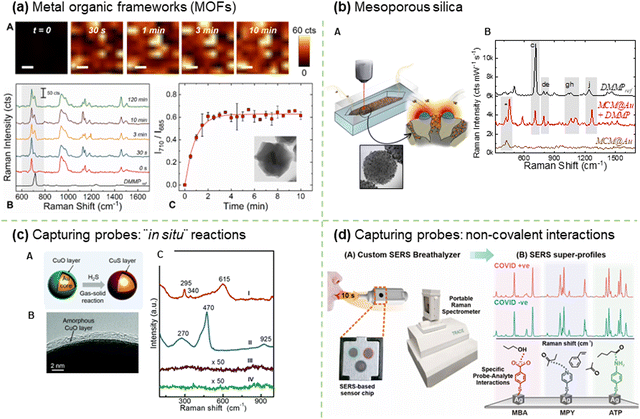 | ||
| Fig. 11 Strategies for enhancing the gas adsorption efficiency. (a) Metal organic frameworks Reproduced from ref. 366 with permission from ACS publisher, copyright 2021 (A) time-resolved SERS detection of gaseous 2500 ppbV DMMP in N2 using Au@Ag@ZIF-8 thin film as sensing platform: SERS intensity mappings; (B) average SERS spectra obtained from SERS mappings; and, (C) SERS intensity ratio I710/I685 between the intensity at 710 cm−1 (DMMP) and the intensity at 685 cm−1 (ZIF-8) as a function of exposure time. Insertion Au@Ag@ZIF-8 nanoparticle. (b) Mesoporous silica Reproduced from ref. 398 with permission from RSC publisher, copyright 2023 (A) plasmonic mesoporous nanoparticles (MCM@Au) allocated in the microfluidic SERS chip illustrating the preconcentration effect; (B) average SERS spectra of DMMP (2.5 ppmV) recorded on the MCM@Au compared to the Raman fingerprint of pure DMMP and blank MCM@Au. (c) Capturing probes: “in situ” reaction Reproduced from ref. 394 with permission RSC publisher, copyright 2020 (A) illustration of the interaction between the Au@CuO nanoparticle and H2S; (B) HRTEM image of Au@CuO; and, (C) Raman spectra of the Au@CuO nanoparticles before (I) and after (II) exposure to H2S gas (100 ppb) for 10 min, and those corresponding to the Au NPs (III) and CuO NPs (IV) substrates after exposure. (d) Capturing probes: non-covalent interactions, reproduced from ref. 393 with permission from ACS publisher, copyright 2022 (A) Overview of the SERS-based strategy to identify COVID-positive individuals using their breath volatile organic compounds (BVOCs); (B) Representative SERS spectra of each molecular receptor (MBA, MPY, ATP) in the presence of COVID-positive and COVID-negative breath samples. | ||
Although less investigated, the opposite configuration, coating of nano-porous particles with metallic nanoparticles also leads to plasmonic architectures with remarkable performance. As an illustrative example, the 2.5 ppm LOD of naked MIL-100 (Fe) crystals for toluene (attributed to π–π interactions and CT mechanism) decreased to 0.48 ppb level upon decoration with Au NPs due to hotspots formation.370 Recently, the detection of DMMP down to the 100 ppb level has been demonstrated with MCM48-Au nanostructures,398 comprising a spherical MCM48 mesoporous silica particle (1237 m2 g−1) as core and a matrix of individual Au NPs randomly distributed on its outer surface (see Fig. 11b) The robust, reliable, and reusable SERS platform resulting from the integration of MCM48@Au nanostructured material in a microfluidic Si chip exhibits promising capabilities for on-field application.
The concept of harmful small gaseous molecules detection by ultrathin layer solid transformation enabled SERS was proposed by Bao et al.394 (see Fig. 11c). They found the existence of an optimal thickness of the CuO wrapping layer and Au@CuO nanoparticles film in terms of LOD and response time. The thickness of the layer has been demonstrated as a crucial parameter in SERS detection for other core–shell structures.395–397 When exposed to H2S, the CuO layer transforms to CuS with a characteristic Raman peak at 470 cm−1. A dynamic range of 0.1 ppb to 1 ppm for H2S with RSD 8.5% with concentration-dependence pre-processing time (i.e. 25 s for 100 ppb of H2S) is found. This versatile concept was extended to other gases, SO2, CS2, CH3SH and HCl by the addition of ultrathin Cu (OH)2 and ZnO layers and the integration of the SERS substrate on a micro-electro-mechanical heating chip to induce those solid transformations that exhibit higher activation energy. The study of alternative substrates based on a thin CuO layer coated onto gold nanoshell films on a polystyrene sphere monolayer demonstrates the universality of the concept. The proposed strategy based on stable inorganic layers with higher Raman cross-section values is well-adapted to the typical excitation power of portable Raman instruments.
Ling and co-workers have been extensively working on array-based SERS platforms based on Ag nanocubes (AgNCs) with different surface functionalities to combine both chemical capture and physical molecular enrichment strategies. Firstly, MOF thin layers on top of multilayer ensemble of AgNCs364 were studied due to beneficial EM field enhancement by short-range static dipolar coupling between metallic nanoparticles. Thanks to the preconcentration-based SERS signal amplification, an optimal ZIF-8 thickness of 146 nm was determined, and the resulting platform was successfully tested with polyaromatic hydrocarbons (PAHs) and VOCs. Recently, the same group reported a 91.7% accuracy for multiplex quantification of SO2 and NO2 within the typical composition of exhaust gases. In this case, AgNCs were functionalized with 4-mercaptopyridine and 4-aminophenyl disulfide392 capturing probes. By introducing additional thiolated receptors, the AgNC SERS platform (32 AgNCs μm−2) was successfully applied for flavor quantification in a machine-learning-driven “SERS taster”, achieving up to 100% accuracy in an artificial wine matrix, containing 5 different flavors.409 There are many diseases (diabetes, halitosis, cancer, pulmonary infections including COVID-19) that could be diagnosed by breath analysis, which is simply a non-invasive way of sampling. The same group393 has developed a multi-probe AgNCs array capable of recording distinct SERS fingerprints after adsorption of exhaled VOCs (see Fig. 11d). Such platform has enabled to identify COVID-19 infected individuals in less than 5 min, achieving >95% sensitivity and specificity across 501 participants regardless of their displayed symptoms.
The simultaneous detection and quantification of 9 VOCs on a single microfluidic platform has been reported by Wang and co-workers.390 This chip, based on her pioneering work,407 is composed of a top PDMS layer with a microcolumn premixer and a bottom PDMS layer with 3 SERS detection areas. Each SERS detection unit is a honeycomb-type 3D hierarchical structure coated with core–shell–shell Au@Ag@Au bimetallic NCs, but is distinctively functionalized (nanosheets of Ti3C2Tx, 2,4-dinitrophenylhydrazine, functionalized Ti3C2Tx layer, labelling with Rhodamine 6G). The so-called “universal gas sensor” was evaluated in two application scenarios, i.e. indoor pollution monitoring and exhaled breath analyses, showing a ppb level detection performance with a dynamic range of three orders of magnitude. In addition, it is quite noteworthy the robustness of the sensing platform, showing a relative standard deviation RSD of 8% in scanned areas of 100 × 100 μm2 besides an RSD of 9% among different chips.
SERS technology for gas detection is a relatively young field, but the last 5 years have seen substantial advances in surface nanoengineering to address the detection performance requirements for closing the gap with real-life applications. Universal concepts and array platforms combined with customized microfluidics chambers, machine learning and affordable handheld Raman equipment, rank SERS gas sensing as a real tool for a broad spectrum of applications with promising market shares. Nevertheless, there still technical aspects to solve, including the gas sampling procedures for on-site sensing,410 the improvement of SERS instrumentation by means of easy plug-in for focusing on SERS substrates avoiding photothermal effects, the reusability of SERS substrates (assisted by UV light,377 Raman laser line411 or annealing366,398,412), an important issue for those that involve a certain complexity and significant cost. Future translations into practical workflows would clearly benefit from (i) the unification of optical and fluidic paths to build optofluidic SERS chips for highly effective detection, and (ii) the integration of less selective multiple transduction signals, thus serving to raise the first alarm before waking up the SERS platform.
3.4.4.1. Nucleic acid (DNA and RNA) biomarker sensing. SERS has emerged as a powerful analytical technique for the detection of nucleic acids (poly-and oligonucleotides based on DNA and RNA) that can act as important biomarkers in diverse scenarios ranging from clinical diagnosis, bioterrorism, forensic cases, environmental analyses, and food safety.75,413–418 One of the most established analytical tools for the detection of these markers is PCR and related molecular assays in variously modified forms, such as those involving the next generation sequencing.75,413,414,419–421 However, the limitations in these methods, namely the prerequisite of specialized settings both in terms of infrastructure and expertise, turnaround time, cross-contamination among samples, and the need for amplification before detection, challenge their ability to be deployed for user-friendly and point-of-care (POC) applications. Thus, faster, and simpler alternative techniques that match the high sensitivity of molecular assays can be of great value for the on-demand sensing of nucleic acid biomarkers. SERS has shown promise in filling this gap by offering ultrahigh sensitivity with the potential to detect even single molecules through the careful design of SERS-active plasmonic nanostructures. Besides, several advantages associated with Raman-based sensing provide a strong justification and rationale for the recent upsurge in seen SERS-based sensors, especially in the context of DNA/RNA detection422,423 These include the ability to use the Raman reporters (i.e., a dye molecule with a specific Raman signal with narrow bands), the multiplexing ability (i.e., detecting multiple targets simultaneously under a single laser excitation), the relative insensitivity of the Raman signal to environmental conditions (e.g., variation in ambient humidity, oxygen levels, and temperatures), low sample volume requirements, and the ability to monitor Raman signal directly from the whole cells without requiring significant sample processing. Thus, this section aims to capture the recent progress made in the relevant field to date while discussing the challenges encountered during operation with the intent to highlight the prospects of plasmonic NPs for the detection of nucleic acids. The progress in this area is subdivided into two main strategies, including the direct (label-free) and indirect (labeled) detection approaches.
3.4.4.2. Direct detection approaches. In the direct approach, the generated Raman signal corresponds to the intrinsic fingerprints of nucleic acids that are brought in proximity to the EM fields enhanced by the plasmonic nanostructures, typically referred to as hotspots. In general, nucleic acids show four distinct Raman spectral regions corresponding to the (i) ring stretching bands at 500–820 cm−1 from purines (adenine and guanine) and pyrimidines (cytosine, thymine and uracil); (ii) deoxyribose-linked phosphodiester vibrations at 820–1150 cm−1 where symmetric stretching of phosphodioxy (νPO2−) gives rise to distinctive band at ca. 1089 cm−1; (iii) in-plane ring variation of nucleobases at 1150–1620 cm−1; and (iv) superimposition of carbonyl stretching modes at 1620–1720 cm−1.75 These intrinsic modes of nucleic acids are measured by obtaining the average SERS signal over several spots deposited on metallic surfaces (bulk SERS) or those arising from EM-field-activated few scattered locations (few/single-molecule SERS).
The direct SERS-based sensors are straightforward and convenient because of no requirement of fabricating and modifying the system with Raman reporters. However, the output signal highly depends on the effective adsorption and trapping of nucleic acids on hotspots in plasmonic NPs or SERS-active substrates. Therefore, reliable strategies that can enhance these binding events become an extremely important prerequisite for fabricating ultrasensitive and reproducible SERS sensors for the detection of nucleic acids.424 The use of external linkers such as thiols in DNA that can spontaneously bond to plasmonic metal NPs and surfaces is one of the most broadly employed methods to enhance this binding.425,426 Since the quality and reproducibility of the SERS signal can be influenced by the conformation (e.g., secondary structures) within nucleic acid molecules and their packing density on the metal surface, a pre-treatment step of, thermal annealing is typically included. This allows nucleic acids to attain a linear confirmation prior to the adsorption on the NPs surface, thus facilitating binding events.426 Although the reproducibility in the SERS output signal is observed to significantly improve with the use of thiolated DNA, the inherent drawbacks such as dependability of the process on DNA modification (thiolation) increases the cost and complexity of operation. Further, the possibility of non-specific binding of nucleobases on metal surfaces that lead to flattened configuration of nucleic acids on the NP surface rather than their placement in a standing-up configuration (despite the thermal annealing step) challenges optimum hybridization in the presence of the nucleic acid target.427
As an alternative strategy, non-specific physical adsorption of non-thiolated DNA on the metal NP surface via electrostatic interaction has been attempted.426,427 This was initially reported with negatively charged Ag colloids and mixing of these NPs with DNA caused displacement of surface ions in NPs to facilitate DNA binding.427 This was followed by the addition of magnesium sulfate (MgSO4) to promote NP aggregation. The relatively low affinity of sulfate ions towards the Ag surface discouraged the displacement of DNA from Ag NPs even at high MgSO4 concentrations. This allowed to produce a DNA sensor with a single base mismatch in a 24-mer short nucleotide sequence with a sensitivity of 10−6 M and a high signal-to-noise (S/N) ratio.427 Subsequently, the same group attained an improved sensitivity (10−9 M) and reproducibility to detect single-stranded (ssDNA) and double-stranded (dsDNA) using negatively charged Ag and Au colloids.428 Further, since the calibration of sensor baseline is a hallmark for reliable target analyte quantification, efforts have been made to employ the signal from the phosphate backbone of DNA as an internal standard to calibrate absolute signal using iodide-modified Ag NPs and MgSO4 in which iodide ions were proposed to function as a cleaning agent.424
Another important aspect of biomolecular detection is the ability to detect secondary structures, as these structures may play important functional roles in various biological processes. An aggregation-based method was described involving citrate-capped Ag NPs and aluminium ions (an aggregation agent) to distinguish the tetramolecular i-motifs (four-stranded DNA structure produced by cytosine-rich sequences) and G-quadruplexes (guanine-rich oligonucleotides) with high sensitivity.429,430 The same group could also discriminate among the four nucleotide bases in DNA (A, T, C, and G) and specific structures of DNA (dsDNA, triple-stranded DNA, and G-quadruplex) with high sensitivity and reproducibility using core–shell Au@Ag particles as the enhancer of Raman signals and Ca2+ ions as the aggregation agent.431 Similarly, titanium ions (as an aggregation agent) and dichloromethane (as an interfacial agent) have been employed for the fabrication of a label-free SERS sensor for the detection of micro-RNAs (miRNA)432 and DNA.433
It is reflected from the above examples that the use of different aggregation agents has remained a common underlying strategy when non-thiolated nucleic acids are employed to capture the target nucleic acid in direct SERS-based sensing strategies. Despite the progress made in aggregation-based direct SERS sensors, the addition of external aggregating agents leads to inconsistent aggregation since the trapping of the target nucleic acid in these aggregates relies on numerous interconnected experimental variables, including the concentration and the type of salt, mode and time of physical mixing, and the sequential order in which the reagents are added during the assay. To address such challenges, a microfluidic chip which provides control over the reagent movements and aggregation in the NPs was proposed.434 The method could detect unlabeled polyadenosine and polycytosine RNA oligonucleotides selectively using nanolitre volume of the samples.
The aggregation agents-free and label-free sensors for DNA and miRNA have also been reported.435–437 One such method involved the use of positively charged spermine-coated Ag NPs that acted as an “electrostatic glue” to adhere DNA on the NP surface. The adsorbed DNA promoted the clustering in NPs, thus trapping DNA on hotspots to deliver vibrant SERS signals. The distinctive difference in the signal with and without the target DNA served as the basis for the sensor which could detect DNA with nanograms per millilitre sensitivity.435 The same group employed microfluidics to enhance sensitivity to picograms level436 and achieved quantification of the base composition in ssDNA and dsDNA.437 The key highlights of these studies are that these did not require pre-amplification and pre-treatment steps, while achieving good sensitivity with a minute amount of the sample.
Even though the above methods could distinguish either one or more nucleotides in the DNA sequences, the presence of the same four nucleotide bases on the target DNA still pose challenges with the identification of specific SERS signals from the target after its hybridization with the capture probe. To overcome this challenge, the adenine base in the capture probe DNA was substituted with 2-aminopurine and the adenine signal which was presented solely in the target DNA could be detected using SERS.438 However, post-modification DNA showed tilted orientation and thus hindered the interaction of all adenine bases with the substrate, limiting the efficacy in SERS detection to less than 5 nm from the surface. To address this issue, peptide nucleic acid (PNA), an artificially generated DNA analogue that contains a pseudo-peptide polymer instead of the deoxyribose phosphate backbone in DNA was used as the capture probe on a glass slide.439 The PNA molecules are uncharged in nature and consist of a flexible polyamide backbone and thus provide a faster hybridization rate and resistance to biological degradation. The hybridization of PNA with the target DNA results in a net negative charge which could facilitate the binding of silver ions to produce Ag NPs in situ through a chemical reaction between AgNO3 and hydroquinone. This Ag enhancement effect was employed to produce a powerful SERS signal.439
Further, the use of amplification techniques which can amplify signal by altering the length of the nucleic acid fragments was also exploited to enhance the signals in direct SERS-based DNA and RNA sensing.440 A recent study evinced the use of iodide-modified Ag NPs and magnetic beads with duplex-specific nuclease (DSN)-based signal amplification for the detection of miRNAs.440 In this method, magnetic NPs with capture probes first hybridized with the target miRNA, followed by DSN-mediated cleavage in the formed DNA–RNA duplex that resulted in the release of miRNA to progress rehybridization with another probe. This step, subsequently followed by cyclic amplification released a higher number of nucleotide fragments from capture DNA to supernatant. The direct relationship between the total phosphate backbone (abundant nucleotide) in the supernatant with miRNA concentration was successfully utilized for the detection of miRNA-21 with a LOD of 42 aM. Furthermore, tip-enhanced Raman scattering (TERS) has also evolved as a promising tool for direct SERS-based DNA and RNA detection with superior performances and achieving single-molecule detection.441
3.4.4.3. Indirect detection approaches. Although the simplicity of experimental procedure for the direct SERS sensing is appealing, indistinguishable signals in the case of complex sample matrices, structural complexity of nucleic acid fragments, reliance on the surface chemistry of plasmonic nanostructures that modulate their affinity to nucleic acids, and the relatively low signal from the target molecules still pose challenges in the real-time applicability of the direct SERS methods.413 To overcome such challenges, an indirect SERS-based strategy has been approached. The method typically involves the use of a Raman reporter as a label and measuring the signal corresponding to the distance between the Raman reporter and SERS-active surface as an indirect estimate of the targeted nucleic acid. The Raman reporter has been integrated in a variety of interesting approaches with a common criterion that the signal intensity of the Raman reporter should offer a direct relationship with the concentration of DNA/RNA of interest.
Sandwich-type construction is one of the broadly used approaches for indirect SERS sensors that uses capture DNA and reporter DNA probe with a Raman reporter. Both DNAs are partially complementary to different regions of the target DNA, which allows the creation of a capture-target-reporter sandwich-type structure post-hybridization. The use of this approach for the determination of six DNA targets, including the hepatitis A virus, Vall7 polyprotein gene, hepatitis B virus surface antigen gene, human immunodeficiency virus, Ebola virus, variola virus, and Bacillus anthracis protective antigen gene has been reported.442 The study employed a chip in a microarray format on which the capture strand that was partially complementary to the target DNA was immobilized. The sensor also utilized Au NP nanoprobes modified with Raman reporter and oligonucleotide sequences that could specifically bind to the target DNA of interest. When the sample was exposed to the chip, the target DNA in the sample could hybridize with the capture probes on the chip, which was followed by the hybridization of Au NP nanoprobes using the overhanging region in the capture-target duplex. Washing steps post-hybridization eliminated the unbound nanoprobes and red colour (visible to the naked eye) was detected at high target concentration (pico to nanomolar). The use of Ag enhancement in which Ag NPs grew as a signal-enhancing layer on the surface of nanoprobes further improved the sensitivity (high attomolar and mid picomolar). Subsequently, the same group highlighted the application of a similar approach for multiple DNA detection in which chips were replaced with glass beads to provide a random-array approach for efficient hybridization kinetics, facile operation, and relatively lower production cost.443 Besides Ag enhancement, the exploitation of numerous nanostructures with different EM enhancement factors has been reported for DNA and RNA sensing using this sandwich-type construction. This includes the use of silicon nanowires decorated with in situ grown Ag NP,12in situ Ag NP on silicon wafer,444 Au particle-on-wire,445 probe-tethered Ag NP,446 Ag NP deposited silica-coated poly(styrene-co-acrylic acid) core,447 and Au NP decorated graphene.448
In addition to the microarray format, modification of plasmonic NPs with the capture probes that are complementary to the target at various locations has also been reported for sandwich-type indirect SERS-based sensing of nucleic acids.449,450 In such cases, in the presence of the target nucleic acid, capture probes hybridize with the target nucleic acid and create links between plasmonic NP. This gives rise to controlled plasmonic coupling in NPs (with and without nucleic acid target), and subsequent determination of the attached Raman reporter at different locations provides an indirect SERS response Further, the use of magnetic NPs in these systems has also seen to provide additional benefits to facilitate cleaning/separation (washing step) of the capture-target-reporter hybrids from the reaction mixture and concentrating the samples to achieve a strong SERS signal.451–454
Fabrication of a SERS-based lateral flow device (LFD) using this sandwich-type method has also been established for detecting DNA related to human immunodeficiency virus type 1 (HIV-1).455 This development could overcome the relatively lower sensitivity in colorimetric LFDs and provided one step closer to practical real-time sensing of DNA using the SERS-based method. The device showed excellent sensitivity of 0.24 pg mL−1, which was claimed to be at least 1000 times greater in comparison to the known colorimetric and fluorometric methods for HIV-1 detection. The same group evinced the multiplexing ability in LFDs by detecting two DNAs related to Kaposi's sarcoma and bacillary angiomatosis with superior sensitivities.456 The intrinsic benefits from multiplexing, high sample throughput, low consumption of sample, and reduced cost per assay make the proposed SERS-based LFD a promising platform for the detection of diseases in its early stage. Different from a sandwich-type assay, the inherent differences in the affinity of ssDNA and dsDNA to metal NPs have also been exploited for indirect SERS-based DNA and RNA detection.457 The underlying principle is similar to that exploited under direct SERS sensing424 with an additional component of a Raman reporter attached to the probe DNA or the nanoparticle. Using similar concept, a PNA-based SERS sensor for DNA detection was also reported.458,459
As a further advancement, the integration of enzyme-free and enzyme-assisted nucleic acid amplification techniques with SERS has been attempted to improve the sensitivity for DNA and RNA detection.459 In this case, sensitivity improvement is achieved via either signal (increased SERS signal by target recycling) or target (increased low amounts of target DNA to a detectable level) amplification. However, the intrinsic limitations of target amplification techniques in terms of potential contamination and non-specific amplification that require strict laboratory control and skilled labor, provide a strong rationale for the preferable use of signal amplification methods over target amplification. The widely employed nucleic acid amplification techniques include both enzyme-assisted and enzyme-free methods. The enzyme-assisted amplification methods include duplex-specific nuclease (DSN) in which a specific enzyme can degrade the dsDNA or DNA–RNA duplex over the ssDNA and double stranded-RNA; and rolling circle amplification (RCA) which is an isothermal polymerase chain reaction producing single-stranded repetitive DNA/RNA. Enzyme-free methods, on the other hand, include hybridization chain reaction (HCR) which is an isothermal and linear amplification method that works based on hybridization between DNA hairpins and interior stands; and catalytic hairpin assembly (CHA) which is a programmable DNA circuit utilizing partially complementary two DNA hairpins and one ssDNA.
The application of DSN-assisted amplification was reported using different SERS-active substrates and Raman reporters. Some representative examples include, Au NPs encapsulated within an AgAu shell,460,461 Au NPs coated with Ag and magnetic NPs,461 Ag-coated Fe3O4 NPs,462 Ag-coated Fe3O4 and Ag-coated Au NPs,463 stimuli-responsive DNA microcapsule with Raman dye,464 and Raman reporter trapped in 3D hydrogels. Similarly, several reports have highlighted the application of RCA with SERS,465 including Raman probes functionalized Au NPs,466 probes functionalized Au NSs on glass slide and Au NRs,467 and magnetic SERS substrate with Ag NP decorated core–shell Co@C.468 Although the use of enzyme-assisted amplification methods has been able to achieve considerably greater sensitivity and wide linear range for detecting nucleic acids, the considerations of assay time, cost, thermostability of the employed enzyme and requirement of resource-rich settings may limit the applicability of these methods for real-time DNA and RNA sensing.
To circumvent those challenges, enzyme-free amplification techniques have been introduced and CHA is one of the commonly reported approaches with a range of Raman reporter-modified SERS-active substrates.468 These include, Au nanodumbells as core and Au NPs as satellite,469 AuAg alloy NPs,470 Ag-coated Au NPs,471 hollow AgAu nanospheres,472 Au nanocages,473 Ag NP-decorated silicon wafer474 and Au NPs.475 Another enzyme-free signal amplification technique that is broadly employed is HCR, which has, for instance, used Raman probes functionalized bifunctional bio-barcode Au NPs,476 DNA probes modified magnetic beads and Au NPs with Raman reporter,476 and DNA-modified Ag NPs.477 In contrast to signal amplification methods discussed above, target amplification techniques involving PCR have also been described for ultrasensitive DNA detection with an LOD of 11.8 aM.478 Moreover, recent progress in the field has highlighted the prospects of coupling amplification-free methods such as clustered regularly interspaced short palindromic repeats (CRISPR) with SERS for the development of ultrasensitive DNA and RNA sensors.479–481
Although the use of SERS as a promising tool for the detection of DNA and RNA biomarkers has been able to accomplish superior sensitivity, some of the inherent challenges with SERS have hampered its applicability for real-time analysis. Reproducibility of the SERS signal perhaps remains the biggest hurdle since the amplitude of the Raman signal can vary substantially with the uneven and randomly distributed hotspots created by the plasmonic nanomaterials. These well-defined hotspots are otherwise critical to attaining a gigantic enhancement factor from a combination of EM and CT enhancement mechanisms. Thus, insufficient attention to standardizing protocols (e.g., synthesis of NPs, the configuration of assembly on surfaces, use of excess chemicals, uncontrolled aggregation, and limited shelf-life of plasmonic crystals and substrates) can lead to vast variations and uncertainties in the generated Raman signals. To overcome these limitations, it is paramount to pay considerable attention to these variables and thus develop methodologies to fabricate robust, stable, and cost-effective Raman substrates within a high precision, reproducibility, is scalability framework. For example, a recent study designed Ag/black phosphorus nanocomposite that utilizes photoreduction to generate hotspots without the need of chemical reagents. This approach could evade background signal interference from the biological fingerprint and accurately detect single molecules with as low as 10−20 M sensitivity.482 In addition, adopting large-scale standardized manufacturing processes, like those in the semiconductor industry, is crucial for producing stable and active SERS substrates with extremely high reproducibility. However, challenges lie ahead in combining colloidal synthesis approaches that are most suitable for producing SERS-active plasmonic NPs with microfabrication processes to produce reproducible surfaces and substrates. More recently, certain antistrophic colloidal NPs (e.g., nanostars and nanorods),483–485 nanoparticle dimers,486 and self-assembled NPs487,488 have seen progress towards uniform and even distribution of larger number of hotspots, which is crucial for greater reproducibility. On the other hand, use of a standard reference in which the signal remained constant relative to the employed signal probe has also been described recently to achieve enviable reproducibility in SERS-based miRNA sensing.489,490 Another recent study reported the influence of the nanocrystal morphology for generating stable, reliable, and reproducible SERS signals using hierarchic Au NCs assembly assay.491 The outcomes of this study evince the better suitability of precisely shaped NCs with the uniform EM field over the spherical NPs (curved interfaces) for stable and reproducible SERS signals. Similarly, the fabrication of SERS-active interior nanogap particles was also reported as a promising method to achieve stable, reliable, and reproducible SERS signal with ultrahigh sensitivity owing to uniformity and narrow distribution in enhancement factors in the as-synthesized nanostructures.65,492–494 These studies suggest that alternative approaches for obtaining favorably stable substrates remain essential for reproducible SERS sensing.
Another challenge with the SERS-based biomolecular sensing is the background noise that stems during measurement of biological samples. The biological matrices, such as tissues, fluids, or cellular components may lead to scattering/fluorescence, while the amorphous carbon resulting from the burning of biomolecules upon laser excitation can generate a strong background signal and thus overshadow, distort, or mask the desired Raman signatures of the biomolecules of interest. To address this undesirable feature, the SERS community is also actively exploring strategies to enhance the signal-to-noise ratio, improve spectral resolution, and employ advanced data analysis algorithms to extract accurate and reliable information. In this context, combining SERS with artificial intelligence and deep machine learning approaches opens a new avenue for the analysis of SERS spectra. Various mathematical and statistical methods, including principal component analysis, discriminant analysis, cluster analysis, and sophisticated approaches such as decision trees, random forests, artificial neural networks (ANN), recurrent neural network (RNN), Bayesian learning, and support vector machines, can effectively extract the intricate characteristics of complex Raman spectra of biomolecules to deliver more accurate classification. These advanced analytical technologies have the potential to propel the practical applications of SERS to a new level. For example, compared to traditional linear spectral analysis methods, ANN can detect nonlinear dependencies, making them well-suited for complex biological samples lacking linear patterns. The use of such ANN models to train SERS spectra of DNA with an accuracy of over 85% in identifying DNA damage was demonstrated.495 Similarly, the use of recurrent neural network (RNN) models to identify RNA was also described using DNA probes which can capture RNA from severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) using Ag NRs.495 The RNN model demonstrated impressive prediction accuracies of 97.2% and 100% for positive and negative samples, respectively.496
Last but not least, the initial cost associated with the instrument and the requirement of skilled personnel to handle and service the instrument limits the use of this technique in resource-poor settings, irrespective of the progress witnessed in the field to date. To address this challenge several studies have evinced the fabrication of SERS-based point-of-care (POC) devices, a technology still in its infancy stage that requires a considerable amount of new research and development efforts.455,456,471,473 Lastly, as the use of SERS is advancing towards in vivo applications,497,498 the toxicity and stability of SERS probes need thorough evaluation and optimization for reliable in situ detection. In summary, addressing the current challenges with SERS-based detection of nucleic acids in terms of reproducibility, non-uniformity of hotspots, background interference, cost of operation, and potential toxicity and stability of probes are crucial for advancing the practical applications of this unique and promising diagnostics platform. Continued research and innovation in not only fabricating new SERS-active materials and substrates with consistent and reproducible SERS signals but also the development of novel strategies to detect DNA and RNA markers, perhaps by combining with innovations in other fields, could enable the widespread adoption of SERS as a mainstream spectroscopic technology in diverse fields, ranging from biomedical diagnostics to environmental monitoring and beyond (Fig. 12).
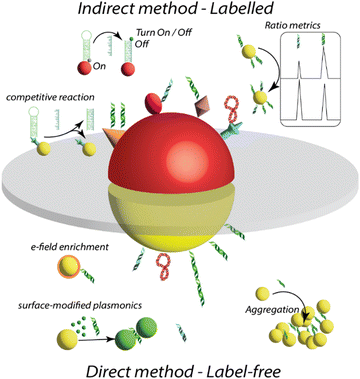 | ||
| Fig. 12 A schematic representation of different SERS-based strategies deployed for detection of nucleic acid biomarkers, as classified under direct and indirect methods. | ||
3.4.4.4. Protein sensing.
3.4.4.4.1. In vivo protein detection and imaging. The past two decades have witnessed a huge surge in the utility of SERS for clinically relevant molecular imaging in vivo and detection of protein biomarkers ex vivo. The ability of SERS to enable rapid, non-invasive real-time tracking in vivo is attributable to its high sensitivity, exceptional spatiotemporal resolution, and multiplexing owing to narrow spectral linewidths of Raman reporters.499 These attributes of SERS are collectively controlled by the optical properties of the plasmonic NPs used,200,500 the molecular characteristics of the Raman reporters used, the targeting ligands on the NP surface such as antibodies, peptides, and aptamers allowing specificity for a target protein,501,502 and protective ligands such as PEG for stability.503,504 Whereas early studies on SERS in vivo primarily focused on tracking one or two protein targets (Fig. 13),505,506 recent work has leveraged multispectral palette with NPs to allow from 5-plex SERS imaging507 to a library of 14 nanoprobes,508 to 26-plex passively accumulated probes in vivo.509 These examples demonstrate that with further signal optimization, SERS has the potential to propel clinically significant applications in vivo. However, a major hurdle in SERS that often limits its imaging potential is the tissue autofluorescence background and the overlap of Raman peaks of reporters in the fingerprint region with the native peaks of biological metabolites.510 The tissue autofluorescence emerges from endogenous fluorophores such as collagen, elastin, nicotinamide adenine dinucleotide (NAD), and flavin adenine dinucleotide (FAD) among others.511 Although using infrared excitations can reduce autofluorescence and self-absorption, it can significantly reduce Raman signal intensity due to the fourth power dependence of the Raman cross-section with excitation frequency. Moreover, the sensitivity of typical NIR detectors is also limited. Besides, the interference of background signals from other issue parts makes it challenging to quantify the real signals. Besides, the direct detection of proteins (without chromophores or functional groups that bind to metal surfaces) that have low Raman cross section and low affinity toward metal surfaces remains a major hurdle. To surmount these challenges, multiple approaches have been implemented including the synthesis of bioorthogonal reporters with peaks in the biological Raman silent region (1740–2800 cm−1) which have minimal overlap with biological metabolites.512 Such Raman reporters have shown excellent targeted multiplexed detection allowing background-free high contrast in a c. Elegans in vivo model.513
In addition to synthesis approaches, improvement in SERS signal in vivo has also resulted from advanced Raman setups that reduce biological noise during data collection. Spatially offset Raman scattering (SORS) is such an approach where the signal from the scattered photons is collected by offsetting the point of the light source from the point of the photon collection.514 This lateral shift allows the collection of signals from the scattered photons originating from deeper in the tissues despite interfering substrates such as bones. Leveraging this approach, Raman spectra have been obtained at high resolution through 7 mm of thick skull tissue that enabled higher signal-to-noise than conventional Raman spectroscopy techniques.515 Another advanced Raman setup used shifted-excitation Raman difference spectroscopy (SERDS) for in vivo imaging where the laser source allows two slightly shifted emission lines similar to the bandwidth of the Raman peaks being studied.516 The spectral data resulting from these emission lines differ in wavenumbers but with negligible change in signal from the background. Therefore, with SERDS the Raman data has minimal interference from the tissue autofluorescence background, and this setup has enabled researchers to pursue highly precise intraoperative imaging with a handheld Raman device.
Whereas SERS in vivo has largely focused on the use of nanoparticles conjugated with Raman reporters, tracking the Raman signal of intrinsic metabolites has also enabled innovations in intradermal glucose detection in situ with microneedle array-based sensors.517 In this unique study, diabetes was induced with streptozotocin in mice fed with a high-fat diet, and the microneedles coated with Ag nanoparticles were gently inserted on mouse skin to measure glucose levels in the subcutaneous interstitial fluid. SERS data was collected through each needle tip of the microneedle arrays with a handheld scanner, and the glucose measured was on par with levels measured with conventional glucometers. Unlike traditional methods to monitor glucose where diabetic patients use fingerstick devices, which is painful and inconvenient, this study demonstrates a clinically significant application of SERS in a point-of-care setting where minimally invasive microneedle arrays allow a painless procedure for glucose tracking with no bleeding. The clinical relevance of SERS has also been enhanced by combining it with clinical imaging modalities to leverage the strengths of SERS while overcoming its limitations in whole-body and depth-resolved imaging.518 The ability to integrate SERS with other imaging techniques is driven by the innovative design of nanoparticles labeled with Raman reporters and other molecules and ligands. Such multifunctional NPs should also maintain their overall biocompatibility, stability, and functionality of each component on the NPs.519,520 Multimodal imaging has the potential to not only enable highly precise biomarker tracking in vivo, but also allow margin assessment before, during, and after surgical procedures.
In this effort, magnetic resonance imaging (MRI) and SERS have been combined with Prussian blue-coated Au NPs where Prussian blue played a dual role-the iron ions allowed MRI contrast and cyanide bridges allowed bioorthogonal SERS signal in the biological Raman silent region.517 By targeting the CD44 receptors with hyaluronic acid ligands, these nanoparticles showed homing in the tumor periphery. In a more ambitious approach, multifunctional NPs were synthesized by Shi et al. that integrated SERS, MRI, and computed tomography (CT) imaging and dual therapeutics including drug delivery and photothermal therapy.521 The authors designed gold core-silica shell gap-enhanced NPs that they decorated with gadolinium to allow MRI contrast and Raman reporters and achieved receptor targeting via folate molecules. They also loaded ibrutinib, which is a small-molecule oral drug used for treating lymphomas. These “all-in-one” NPs enabled preoperative tumor imaging with MR/CT and intraoperative accuracy with SERS while also allowing highly effective multivalent treatment. In addition to MRI, positron emission tomography (PET) imaging is also a mainstay in the clinic facilitating deep tissue whole-body imaging. Single-channel PET with a single radiotracer has been effective when combined with SERS active NPs allowing preoperative planning, intraoperative resection of the lymphatic tissue, and postoperative margin confirmation.520 However, multiplexing cannot be achieved with PET; therefore, integration of SERS in a single functional NP is beneficial for tracking multiple proteins in vivo. In a work by Bardhan and co-workers,522 the authors showed that PET-CT-SERS active multimodal NSTs enable simultaneous detection of multiple immunomarkers in vivo combining the merits of all imaging modalities. In their approach, two sets of Au NSTs were synthesized, each conjugated to a different Raman reporter with minimal spectral overlap to target CD8+ (cluster of differentiation 8) T cells and PD-L1 (programmed cell death ligand 1) expressing cancer cells in vivo. Nanostars were also conjugated with anti CD8 or anti PD-L1 antibodies and 64Cu radiotracer was chelated via DOTA (dodecane tetraacetic acid). The NSTs tracked the two immunomarkers in vivo as well as provided early response to antiPD-L1 + antiCD137 checkpoint blockade immunotherapies in both treatment-responsive and treatment-resistant melanoma tumors. There are now several clinical trials ongoing where multiple imaging modalities have been integrated for diagnostics (e.g., trial # NCT02790853). This suggests that SERS combined with a clinical technique enabled by biocompatible NPs may transition this technology “from bench to bedside” for clinical diagnosis (Fig. 13).523
3.4.4.4.2. Ex vivo protein detection. In addition to in vivo imaging, SERS has also enabled highly precise detection of proteins ex vivo with femtomolar detection limits (Fig. 13).524 SERS-based ex vivo assays are quantitative, allow multiplexing of >30 biomolecules, and are comparatively low cost than fluorescence-based assays.525 In such ex vivo assays, the type of the metal NPs used (Ag, Au, and Al) and the type of the targeting moiety conjugated on the NPs (antibodies, peptides, DNA etc.) govern the sensitivity, specificity, stability, and overall shelf-life of the nanoprobes used for detection.195,526 Whereas sandwich immunoassays have been the most prevalent in ex vivo SERS sensors,527 recent innovations in lateral flow assays (LFAs),528 porous architectures,529 and microfluidics530 demonstrate the promise of these technologies for ultimate use as POC devices for at-home use. Here we will discuss a few examples of ex vivo SERS assays and conclude this section with commercialization of SERS substrates.
In a unique approach by Chen et al., nanoporous anodic aluminum oxide (AAO) was developed as a vertical flow sensing unit, and bimetallic core–shell Au@Ag nanotags encoded with Raman reporters as the labeling unit.531 Through this approach the authors detected multiple inflammatory biomarkers including, procalcitonin, interleukin-6, serum amyloid A, and C reactive protein in patient samples by functionalizing the bimetallic NPs with four distinct Raman reporters and antibodies specific to these proteins. The high surface area to volume ratio and the nanoscale confinement within the AAO pores aided with the highly effective SERS nanoprobes allowed femtomolar LOD of 7.53, 4.72, 48.3, and 53.4 fg mL−1 for the four biomarkers, respectively. In another multiplexing study, Bardhan and co-workers reported a novel sensor portable, reusable, accurate diagnostics with nanostar antennas (PRADA) for detecting biomarkers of cardiac disorders including cardiac troponin 1 (cTn1) and neuropeptide Y (NPY) in cardiac patient serum samples.532 PRADA is a rendition of a sandwich immunoassay with Au NSTs functionalized with targeting peptides and Raman tags as the detection unit, and magnetic microbeads conjugated with polyclonal antibodies as the capture units. Through this unique architecture, PRADA achieved a LOD of 5.5 pg ml−1 of cTnI and 120 pg ml−1 for NPY demonstrating that small peptides can achieve detection sensitivities on par with antibodies while enabling high stability of the nanoprobes. PRADA was reusable where the magnetic microbead bottom probes could be removed with a magnet allowing regeneration of the sensor chip for ∼14 cycles making PRADA amenable to affordable sensing at remote sites. In addition to antibodies and peptides, aptamers conjugated NPs are also excellent in selectively detecting proteins allowing to expand SERS assays to neurological diseases. In this effort, a self-assembled conjugate of Raman tag-encoded polyadenine (poly-A) block oligonucleotides was anchored on Au NPs and coated with targeting aptamers.533 When the protein of interest is bound to the aptamers, it induces aggregation of Au NPs increasing the plasmonic coupling effect and leading to hotspots. These functionalized Au NPs enabled multiplexed detection of Aβ1-42 and Tau proteins, which are biomarkers of Alzheimer's disease enabling LOD of 0.00042 pM for Aβ1-42 and 0.037 nM for Tau proteins, respectively. In addition to the traditional SERS sensor architectures, microfluidic devices have expanded the capabilities of SERS ex vivo assays achieving rapid and ultrasensitive detection attributable to the spatial confinement of nanoprobes and better mixing of nanoprobes with the analytes of interest. For example, Wang et al. designed an extracellular vesicle phenotype analyzer chip (EPAC),534 “nano” mixing enhanced microchip that allowed the authors to track low-abundance tumor-specific extracellular vesicles (EVs) in biological fluids. Multiplexing was achieved by labeling the EVs Au NPs and Raman reporters to detect receptor tyrosine-protein kinase, melanoma cell adhesion molecule, low-affinity nerve growth factor receptor, and melanoma chondroitin sulfate proteoglycan with high sensitivity and specificity. In another study, Tian and co-workers developed a futuristic design (Fig. 13) of a flexible and wearable paper-based microfluidics for quantitative detection of uric acid (UA) in sweat at physiological concentrations.535 The authors used chromatography paper with uniformly distributed Au nanorods to fabricate the plasmonic paper-based microfluidic biochip. The nanorods contributed to high SERS signal and ultrasensitive detection of UA in sweat. The authors also showed that microfluidic biochips are highly flexible and can be stretched and twisted withstanding stress and strain. They also demonstrated that the flexible biochips can be conformally laminated on a human subject and with the utility of a portable Raman spectrometer, rapid and accurate data collection is possible.
These examples show that SERS has progressed rapidly since its advent and has emerged as a mature technology with many commercial sources of SERS substrates that are now available.201,536,537 A recent study compared the efficacy of 7 different commercial SERS substrates in the detection of cocaine in oral biofluids from healthy donors.536 The substrates were purchased from Diagnostic a SERS, JASMAT Optics Corp, Silmeco Aps, FLEW Solutions, Quark Photonics, and Metrohm Australia. The authors found that only one of these substrates excelled in reaching a 1 ng mL−1 LOD for cocaine in buffered solutions, and 10 ng mL−1 in human samples. The authors attributed the superior performance of these substrates to their design where silicon pillars were used as the substrate backing and Ag as the active metal. The performance of the substrate was controlled as the pillars leaned towards each other giving rise to clusters as the solvent evaporated. The nanogap (25–40 nm) created between the tips of the pillars within the clusters enabled intense SERS hotspot and high Raman signal. In a similar study, three different commercial SERS substrates including RAM-SERS-SP from Ocean Optics, QSERS from Nanova Inc., and Hamamatsu from Hamamatsu Photonics were compared with 4-mercaptobenzoic acid (MBA) as a model probe molecule.537 The authors used two different excitation wavelengths (633 and 785 nm) and calculated the LOD and limit of quantification (LOQ) between 1–10 μM with a range of use of only one or two orders of magnitude. The best performing substrate for MBA was Hamamatsu at the 633 nm excitation wavelength. These comparative studies are highly informative in (i) demonstrating the potential and limitations of current commercial SERS substrates, (ii) motivating researchers who have designed custom SERS platforms with exceptional sensitivities to move their technology “from bench to market”, and (iii) inspiring new technological innovations that go beyond the well-established protein targets to sensing unconventional biomolecules in rare diseases that have not been previously probed.
3.4.4.4.3. Cell detection. In recent years SERS strategies to aid in cell detection, analysis, phenotyping or functionality studies have been increasingly used. SERS has been extensively used for the detection of single or multiple cells in cell culture media, plasma, serum, tissues, or even body fluids.538 Lately, particular attention has been given to the detection of circulating tumour cells (CTCs) in different cancer types with SERS.538–541 Ding and co-workers have also reported on a dual strategy combining SERS and fluorescence for the detection of circulating tumor cells (CTCs).542 In this work they used Au nanoflowers onto indium tin oxide (ITO) surfaces, which were functionalized with aptamers able to capture CTCs. Simultaneously, Au NSTs were used as probes having an anti-EpCAM to improve the selectivity of the strategy. After the retention of the CTCs, then by using a complementary aptamer sequence to that of the retention, the originally captured CTCs were released into the medium to perform the dual SERS-fluorescence detection. Remarkably, they could detect down to 5 and 10 cells mL−1 with SERS and fluorescence respectively, ensuring linearity up to 200 cells mL−1. Another example using SERS tags by Oliveira et al. showed that by combining SERS tags paired with membrane protein recognition antibodies and microfluidic platforms, it is possible to perform a multiplex analysis of the surface protein expression at the single cell level.543 For this, they used Au NSTs codified with different Raman reporters, which were subsequently coated with a silica layer and later functionalized with the target antibodies EpCAM, Vimentin, and CD45. Interestingly, by having the individual cells contained within droplets, it is possible to map the surface of those cells in terms of the intensity of the SERS tags, which directly report the presence or absence of the specific proteins on top of the SERS tags. Unlike fluorescence in which a binary reply on whether a protein is present or not, by using these SERS tags it is possible to analyze the distribution of the protein expression along the cell membrane (Fig. 14).
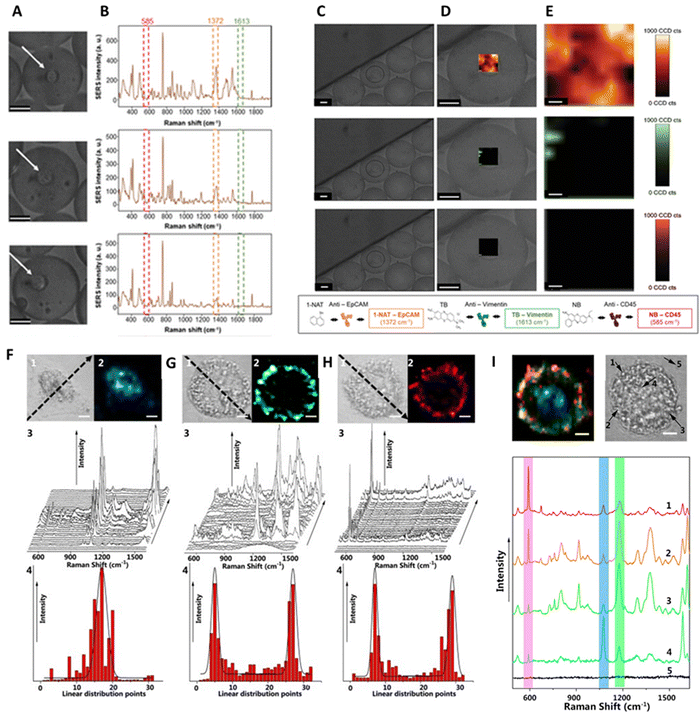 | ||
Fig. 14 (A) Three bright-field images of three different cell-containing microdroplets; (B) SERS spectra after measurements in each cell; (C)–(E) multiplex phenotypic characterization of MDA-MB-435 cells using three different SERS tags; (C) bright-field of an MDA-MB-435S cell labeled with Au NSs@RaR encapsulated in a microdroplet; (D) high magnification image and overlay of SERS mapping on top of a single cell; (E) respective SERS intensity maps for each RaR, and corresponding intensity colour scale (scale bar 3 μm). Inset: Pairing of RaRs with specific antibodies for recognition of cell membrane receptors. Reproduced from ref. 538 with permission from Wiley-VCH GmbH publisher, copyright 2021 (F)–(H) SERS imaging of a HeLa cell treated with MBA-coated Au NPs (F), CV-coated Au NPs (G), and CVa-coated Au NPs (H); (F1–CH1) Bright-field images and (F2-G2) SERS images corresponding to the detected HeLa cell. (F3–G3) SERS spectra were obtained from different positions within the cell via point-by-point detection. The histogram displays the Raman intensity at 1078![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cm−1 (MBA-coated, A4), 1175 cm−1 (MBA-coated, A4), 1175![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cm−1 (CV-coated, B4) and 595 cm−1 (CV-coated, B4) and 595![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cm−1 (CVa-coated, C4) obtained from the spectra in (A3–C3) along the dotted line in (F1–H1) (scale bar ≡ 4 cm−1 (CVa-coated, C4) obtained from the spectra in (A3–C3) along the dotted line in (F1–H1) (scale bar ≡ 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) μm); (I) Multi-targeting SERS imaging of a HeLa cell treated with CVa-coated, CV-coated and MBA-coated AuNPs; (I) left: overlap of SERS images of CVa-coated Au NPs (red), CV-coated Au NPs (green) and MBA-coated Au NPs (blue); right: bright-field image of the investigated HeLa cell; bottom: SERS spectra obtained from different positions in the cell (marked in bright-field cell image above by arrows (scale bar ≡ 4 μm); (I) Multi-targeting SERS imaging of a HeLa cell treated with CVa-coated, CV-coated and MBA-coated AuNPs; (I) left: overlap of SERS images of CVa-coated Au NPs (red), CV-coated Au NPs (green) and MBA-coated Au NPs (blue); right: bright-field image of the investigated HeLa cell; bottom: SERS spectra obtained from different positions in the cell (marked in bright-field cell image above by arrows (scale bar ≡ 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) μm)). Reproduced from ref. 544 with permission from Nature publisher, Copyright 2016. μm)). Reproduced from ref. 544 with permission from Nature publisher, Copyright 2016. | ||
The use of label-free methods for cell analysis using SERS offers advantages such as simplicity, lower costs and time savings, and with the advent of data analytics its potential is boosted with respect to codified SERS strategies, which were most pursued in the past decade.544–546 For example, Agnieszka Kamińska et al. in which they used a membrane containing plasmonic nanoparticles to capture and analyze CTCs.547 Unlike the previous works presented, in this case they present a label-free analysis of cancer cells (leucocytes, HeLa, and PC3 cells). A tailor-made membrane was used for both the capture of cells and their subsequent analysis. A combination of eletrospinning of polymer fibers followed by physical vapor deposition of AuAg alloy was the chosen strategy. After the acquisition of reference SERS spectra of the three cell lines, the authors show the successful classification of cell types by using PCA achieving an accuracy of 95% in 2D PCA to 98% in 3D PCA.
Besides the detection of whole cells, it is worth mentioning that SERS has proven to be quite successful in the development of the detection strategies for cell related intra-,548,549 and extracellular metabolites,550,551 extracellular vesicles (EVs),552,553 cell lysates,554 cells within tissues555 or cell imaging.556,557 For example, the work by Masson and co-workers described the monitoring of metabolic events of cells using SERS.558 In this work, in which the report a sensitivity down to the single molecule level, they used a set of borosilicate nanopipettes decorated with Au NPs as plasmonic sensor. The metabolites detected were the common suspects such as pyruvate, lactate, ATP, and urea simultaneously. In this approach, the detection was done by placing the SERS sensor at varying distances of living cells and after the application of a lysis process to boost cell secretion. In this way, the authors were able to monitor metabolite gradients and mixtures that could be related to disease stages.
As shown, a broad range of SERS applied to cell detection in different fashions is available in the literature. However, it is needed to consider the chosen SERS strategy taking into account the clinical need for a specific use case. As an example, in most cases for cancer diagnosis and monitoring, therapy guidance and clinical utility are key for the uptake of novel technologies. As such, guided therapy is based either on protein expression or on specific mutations. In this scenario, cell identification alone would not be sufficient to offer clinical utility, and the use of SERS tags to identify specific mutations, proteins or biomarkers would offer the relevant clinical information. However, some diseases can be diagnosed based on the presence/absence of aberrant cells, in which label-free approaches for rare cell detection could make a huge difference in clinical practice. Finally, with the advent of advanced data analytics, heavy SERS analytical data can be now interpreted and deconvoluted with much more efficiency and robustness, which should be exploited by the community to finally bring SERS to hospitals as diagnostic and clinically useful tools.559
3.4.4.4.4. Bacteria and virus sensing. Specific bacteria or viruses can cause infectious diseases with a significant impact on human health. For example, sepsis is a severe and life-threatening disease caused by bacteria or bacterial toxins in the bloodstream, triggering a severe inflammatory response.560 The respiratory disease SARS-CoV-2 has ravaged the world for the past four years, causing the rapid spread of the infectious disease and resulting in hundreds of millions of infected individuals worldwide, leading to the loss of lives of many elderly people.561,562 Therefore, the accurate and fast diagnosis and prevention of the spread of these infections are crucial. Reverse transcription-polymerase chain reaction (RT-PCR) is a widely used standard diagnostic method that extracts, amplifies, and detects RNA or DNA. However, it has limitations in detection time and requires experts who can effectively utilize it.563,564 The LFA strip, which extracts protein biomarkers instead of RNA/DNA and provides quick on-the-spot diagnosis, has been commercially successful in diagnosing SARS-CoV-2. However, the sensitivity is low, leading to a high false-negative diagnostic rate for early infected or asymptomatic infected individuals.565,566 Consequently, there is a need for a new high-sensitivity detection method that can significantly improve diagnostic sensitivity and address the problem of infection spread due to false-negative diagnoses.
SERS detection method has been recognized as a new diagnostic technique that can overcome the sensitivity limits of existing biomedical detection techniques in terms of absorbance, fluorescence, and chemiluminescence.567,568 In addition, SERS-LFA or SERS-ELISA technology, which combines SERS detection with an immunoassay platform, has been developed, which may overcome the sensitivity limit of immunoassays. The novel SERS-LFA diagnostic system was developed to diagnose infectious diseases caused by bacteria or viruses, such as Orientia tsutsugamushi (O. tsutsugamushi), a Gram-negative intracellular bacterium, or SARS-CoV-2 on site.569Fig. 15a shows a conceptual diagram of the system, which consists of a SERS-LFA strip, a portable Raman reader that can measure Raman signals of the test and control lines of the strip, and a lysis buffer used to extract the target protein (nucleocapsid protein) from clinical samples. The sample preparation process for the quantitative analysis of SARS-CoV-2 is similar to that of a commercial LFA strip, for which the result is interpreted as positive or negative by the eye.570 First, the clinical sample is placed in a running buffer, including a lysis buffer solution to lyse the virus. Then, the target protein is extracted and loaded into the inlet of the SERS-LFA strip along with the running buffer. This solution moves to the conjugate pad by capillary force and binds to antibody-conjugated SERS nanotags through antigen–antibody binding when the target protein is present. These target protein-SERS nanotag complexes continue to flow in the direction of the absorbent pad and are accumulated by combining with the antibodies immobilized on the test line. The SERS nanotags that did not bind to the antigen accumulate on the control line through antibody-antibody interactions. When the target protein is absent, the SERS nanotags do not bind to the test line and only bind to the control line through antibody-antibody interactions. Test and control lines that combine with SERS nanotags show a red color due to the surface plasmon effects of Au NPs, like commercial colorimetric LFA strips. However, due to the low sensitivity of these colorimetric LFA strips, it is difficult to detect the SARS-CoV-2 virus below 350 plaque-forming units (PFU) mL−1, resulting in false-negative diagnoses.
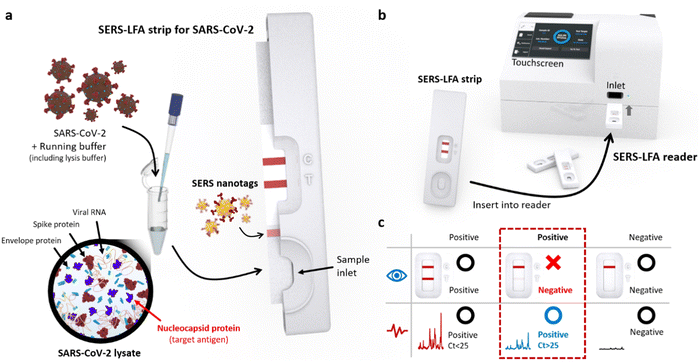 | ||
| Fig. 15 Illustration of the SERS-LFA system for on-site diagnosis of SARS-CoV-2. (a) Schematic of the SERS-LFA strip for SARS-CoV-2 antigen testing. SARS-CoV-2 is added to the running buffer, including a lysis buffer to lyse the virus. After mixing the SARS-CoV-2 with the running buffer, the solution is loaded into the inlet of the SERS-LFA strip. When this SARS-CoV-2 lysate reaches the conjugate pad, the target NC proteins and NC antibody-conjugated SERS nanotags form immunocomplexes through antibody–antigen interactions. The running buffer, including the NC protein-SERS nanotag complexes, moves towards the test and control lines. (b) After the running buffer reaches the absorption pad through the test and control lines, the SERS-LFA strip is inserted into the portable Raman reader. (c) Advantage of the SERS-LFA system over the commercial LFA strip. Both LFA and SERS-LFA show a negative result for SARS-CoV-2-negative patients. Both methods are positive for clinical samples with a high virus concentration (Ct < 25 in RT-PCR). The LFA strip usually shows a false-negative result for clinical samples with a relatively low virus concentration (25 < Ct in RT-PCR); however, the SERS-LFA system shows a true-positive result owing to its high sensitivity, Reproduced from ref. 570 with permission from ACS publisher, Copyright 2022. | ||
A portable Raman-LFA strip reader has been developed to solve sensitivity issues. Fig. 15b shows a schematic diagram of the SERS-LFA strip reader. A 632.8 nm diode laser serves as the light source, and the laser beam is focused through an objective lens on the test and control lines of the LFA strip. The laser beam is moved in 200 μm intervals in the x and y axes using a laser scanning module to obtain a reproducible Raman signal, and Raman mapping signals for 39 pixels for both test and control lines are obtained and averaged. For quantitative analysis, the target protein concentration is determined based on the Raman intensity ratio obtained for the test and control lines, which would indicate a positive or negative result. Fig. 15c compares the sensitivity of the on-site SERS-LFA system using a portable Raman strip reader with that of a commercial colorimetric LFA strip. Based on the cycle threshold (Ct) value of RT-PCR, when the concentration of the virus is very high (Ct < 25), both LFA and SERS-LFA would show a positive result, and both methods would show a negative result for non-infected samples. However, the false-negative diagnostic rate is expected to be significantly reduced when using the SERS-LFA system for early infected patients with a low virus concentration or asymptomatic infected patients (Ct > 25). After testing 54 clinical samples in the range of 0 < Ct < 35, a commercial LFA strip showed a sensitivity of approximately 35% for 31 clinical samples with 25 < Ct < 35. On the other hand, the sensitivity of the SERS-LFA system was 93%. Therefore, the SERS-LFA system could markedly improve the false-negative diagnostic rate by significantly increasing the diagnostic sensitivity for patients with a low concentration of the SARS-CoV-2 virus.570
ELISA techniques are widely used instead of LFA strips to perform protein assays in the laboratory. For SERS-ELISA, Au NPs are often used as SERS nanotags; however, signal enhancement is limited due to the lack of hotspots between particles in the nanogap. A 96-well plate coated with Au has been commercialized for nano-plasmonic immunoassay to induce hotspots between Au NPs and substrate surfaces. However, such a plate is expensive, and it is not easy to control hotspots accurately during the assay process. To address this problem, SiO2-coated core–satellite (CS) Au@Au SERS nanotags have been developed by assembling 32 nm and around 75 nm Au NPs.571 Multiple hotspots could be formed within individual CS NPs due to the abundance of nanogaps formed between the Au NP core and Au NPs satellite, resulting in greatly amplified EM signals. In addition, the stability of SERS nanotags could be improved by encapsulating SiO2 on the CS surface, improving antibody immobilization and assay effectiveness. Immunoassays were performed using SARS-CoV-2 lysates at the same concentration to evaluate the plasmonic coupling characteristics and stability of CS@SiO2 SERS nanotags. In comparison with conventional ELISA based on absorption/fluorescence detection methods or SERS-ELISA using Au NPs, the sensitivity and reproducibility of SERS-ELISA using CS@SiO2 SERS nanotags were significantly improved. Fig. 16 shows a schematic of SERS-ELISA using CS@SiO2 SERS nanotags. A 384-well plate was used in SERS-ELISA instead of the 96-well plate typically used in conventional ELISA to analyze as many clinical samples as possible in a high-throughput manner. In comparison with the 96-well plate, the 384-well plate could lower the assay cost by reducing the amount of expensive capture and detection antibodies required for the assay because the sample volume in each well is reduced from 400 μL to 130 μL. Additionally, the amount of target antigens can be quantified by measuring the Raman signal of SERS nanotags directly bound to them without an additional enzymatic reaction to induce a color change with a secondary antibody, which is required in conventional absorbance-based ELISA. This straightforward approach is advantageous for applying the SERS-ELISA technique to biomedical analysis (Fig. 16c). Fig. 16d shows the results of assays with SARS-CoV-2 lysates using three different SERS nanotags: CS@SiO2, silica-encapsulated hollow Au NP (HAuNP@SiO2), and silica-encapsulated AuNP (AuNP@SiO2). As expected, HAuNP@SiO2 and AuNP@SiO2 demonstrated poor sensitivity due to limited EM enhancement at the individual particle level. On the other hand, multiple hotspots could be created within one unit using CS@SiO2 SERS nanotags, resulting in significantly increased sensitivity. Therefore, the SERS-based assay platform may be a next-generation in vitro diagnostic technology that can overcome the sensitivity limits of existing on-site diagnostic methods for infectious diseases.
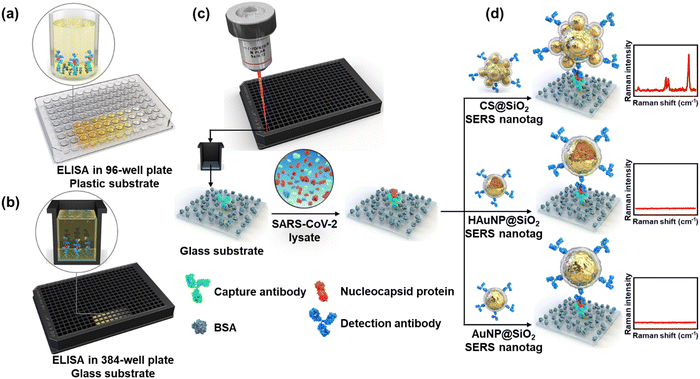 | ||
| Fig. 16 Schematic illustration of the SERS-ELISA method using CS@SiO2 (SiO2-coated core–satellite Au@Au NPs) SERS nanotags. Conventional ELISA using (a) a 96-well plastic plate and (b) a 384-well glass plate. (c) SERS-ELISA of SARS-CoV-2 in a 384-well glass plate using SERS nanotags. (d) Evaluation of SERS-ELISA for detecting SARS-CoV-2 using CS@SiO2 (top), HAuNP@SiO2 (middle), and AuNP@SiO2 (bottom) SERS nanotags. Reproduced from ref. 571 with permission from Elsevier publisher, Copyright 2023. | ||
3.5. SERS for monitoring reaction progress and intermediates
It is critical to analyze the intermediates of a reaction to understand their mechanisms. Traditionally, IR, NMR, or other spectroscopy techniques have been used to probe the reaction progress and intermediates. In the last decade, SERS has emerged as an alternative and noninvasive method to monitor reaction progress by in situ analysis of reaction products and intermediates because of the ability to provide molecular-level information about the chemical composition of molecules.572 It has been used to study a wide range of chemical reactions, including catalytic reactions, electrochemical reactions, and photochemical reactions because it provides important insights into the reaction mechanism and kinetics, and the nature of the reaction intermediates (Fig. 17). In this section, we will discuss the research progress on studying chemical transformation by SERS probing of reactants and products.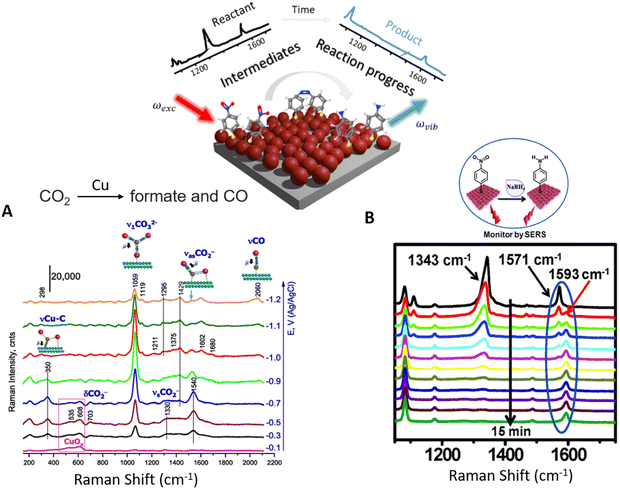 | ||
| Fig. 17 Schematic illustration of probing reaction progress and intermediates by SERS by collecting inelastic light scattering by molecules adsorbed on a metallic surface. (a) SERS spectra of intermediates of the transformation of CO2 into formate and CO on a Cu surface Reproduced from ref. 573 with permission from PNAS, Copyright 2018. (b) Time-dependent SERS spectra of the conversion of p-NP conversion into p-aminophenol. The reaction can be probed by the disappearance of p-NP bands and the evolution of peaks of the product (p-aminothiophenol) with time. Reproduced from ref. 574 with permission from RSC publisher, Copyright 2015. | ||
Besides, SERS has also been employed to study the reaction progress of photochemical reactions. For instance, one of the most important photochemical reactions is the reduction of CO2 into other organic species, which is critical to reducing greenhouse gases. Probing such a catalytic process and knowing intermediates could help to design better catalytic systems. With this aim, Devasia et al. have used SERS to determine a large number of C–C species formed by the photocatalyst reduction of CO2 in the presence of a plasmonic structure.576 Likewise, Kumari et al. studied the reaction progress of the photocatalytic reduction of CO2 on a plasmonic nanoparticle by SERS.577 They also found the formation of intermediate species, whose concentration increased at the early stage of the reaction and then decreased in later states, suggesting that these species were the intermediates in the reaction, thanks to the high specificity and sensitivity of SERS.
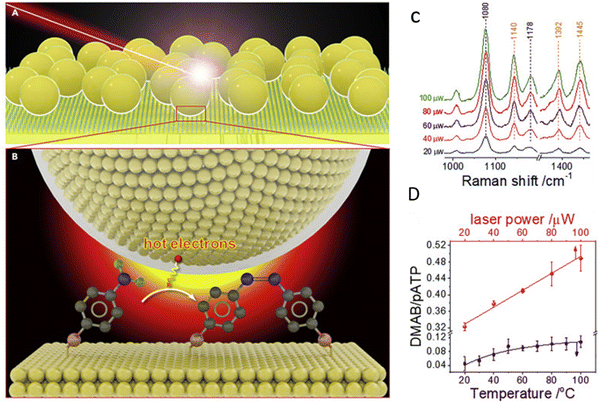 | ||
| Fig. 18 (a) Scheme of in situ SERS study on the interfacial hot electron transfer in the photocatalytic conversion of p-aminothiophenol (p-ATP) to dimercaptoazobenzene (DMAB). (b) SERS spectra of the conversion of p-ATP catalyzed by Au NPs at different laser power excitations. (c) The relative intensity of DMAB to p-ATP bands with different incident laser powers. The figure is adapted from ref. 582 with permission from Springer, Copyright 2020. | ||
Similarly, SERS has also been used to study the reaction intermediates in oxygen reduction reactions on a platinum surface.583 It was found that the type of intermediates depends on the pH or the crystalline structure of the surface. In this case, at high pH, only one intermediate was observed independent of the Pt crystalline structure. When the oxygen reduction reaction was performed at normal conditions, the intermediate 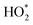 was adsorbed on the Pt (111) surface, while the Pt (110) and Pt (100) surfaces preferentially adsorb the intermediate HO*. In conclusion, SERS has evolved as a powerful analytical technique that can provide molecular-level information about the transformations and reaction kinetics. However, it is only limited to specific reactions where the analytes physically or chemically adsorb on plasmonic substrates. More studies are needed in the future to fully exploit the power of SERS in understanding reaction mechanisms.
was adsorbed on the Pt (111) surface, while the Pt (110) and Pt (100) surfaces preferentially adsorb the intermediate HO*. In conclusion, SERS has evolved as a powerful analytical technique that can provide molecular-level information about the transformations and reaction kinetics. However, it is only limited to specific reactions where the analytes physically or chemically adsorb on plasmonic substrates. More studies are needed in the future to fully exploit the power of SERS in understanding reaction mechanisms.
3.6. Tip-enhanced Raman scattering
As a powerful analytical technique, SERS can demonstrate detection sensitivity at the molecular scale under the plasmon enhancement effect. It can provide rich molecular structure information to analyze chemical composition and structure accurately.85,195,584 However, the enhancement effect of the SERS signal depends on the experimental conditions and the nanostructure of the sample surface, which is not stable enough in practical applications. Second, background interference significantly affects SERS signals, especially those from fluorescence and scattering, reducing the detection sensitivity of target substances.198 In addition, fabricating and controlling high-quality SERS-enhanced substrates remains challenging, especially for large-scale fabrication and practical applications. Tip-enhanced Raman scattering (TERS) is based on scanning electron/atomic force microscopy, which can achieve high-sensitivity Raman signal detection and perform high-precision morphology characterization of detection targets in real-time and realize detection at the single-molecule scale. SERS and TERS utilize the plasmon enhancement effect to enhance the Raman scattering signal. SERS focuses on nanostructured surfaces local EM field enhancement effect, while TERS achieves higher resolution and more sensitive analysis through the local enhancement effect of nanotip probes. TERS uses the local enhancement effect of tip probes (such as metal needle tips) to focus the enhanced electric field to the nanometer scale to achieve high-resolution Raman images and spectral information acquisition of samples.72,585–587 Compared with SERS, TERS has higher spatial resolution and sensitivity requirements, so it has a wide range of applications in nanomaterial characterization, surface catalysis, and biological analysis. TERS is an enhanced Raman technique based on metal tip probes, generally consisting of a tip (such as a metal tip) and a Raman spectrometer shown in Fig. 19a. | ||
| Fig. 19 (a) Schematic diagram of the TERS system. (b) TERS spectra of C60 and BCB samples and mapping of BCB, Reproduced from ref. 588 with permission from Elesvier publisher, Copyright 2000 (c) TERS and RRS of CN−1. Top panel: Ir tip/Au (111), bottom panel: Au tip/Pt (110) Reproduced from ref. 589 with permission from American Physical Society publisher, Copyright 2004 (d) Schematic and physical mechanism of HV-TERS, Reproduced from ref. 590 with permission from Nature publisher, Copyright 2012 (e) HV-TERS system structure (including analysis chamber, fast loading chamber, and molecular beam epitaxy chamber), Reproduced from ref. 591 with permission from AIP publisher, Copyright 2016. | ||
The initial development of TERS can be traced back to the early 1990s due to advances in atomic force microscopy (AFM).592 At that time, researchers began to try to use the tip to achieve high spatial resolution Raman spectroscopy measurements on samples. Initial studies focused on using the local electric field enhancement effect at the metal tip to enhance the intensity of the Raman signal. Deckert et al. found in 2000 that the Raman scattering signals of brilliant cresyl blue (BCB) molecules and C60 molecules could be enhanced in the nanometer range using the AFM-TERS system in an atmospheric environment, as shown in Fig. 19b.588 The spatial resolution of the sample is limited only by the size and shape of the apex of the metal tip and does not require special sample preparation. Moreover, the Raman signal enhancement uniformity at any sample position is excellent, so the quantitative measurement of the SERS spectrum can be performed. In addition, when the tip of the AFM-TERS system scans the surface of the sample, the morphology characterization results of the sample can be obtained simultaneously and correspond to the spectral data. Comparing the Raman signal when the metal tip is close to and away from the sample, it can be shown that the essence of the enhancement is due to the local EM field accumulated at the apex of the metal tip, which amplifies the Raman signal of nearby molecules. Moreover, using this method, the laser wavelength can be adjusted to precisely match the corresponding plasmon frequency to achieve Raman signal enhancement of a single molecule. For the TERS structure, the metal tip and substrate are the cores of the whole detection system. Choosing different materials to prepare metal tips and substrates with various shapes will also affect the detected Raman signal. Pettinger et al. used Ir and Au as other metal tip materials. They used Au (111) and Pt (110) single crystal surfaces as scanning tunneling microscopy (STM)-TERS substrates, respectively, to study the Raman signal enhancement effect of malachite CN−.589 As shown in Fig. 19c, the enhancement effect of the Ir tip on the CN− molecules on the Au (111) substrate is close to 4 × 105, and when the tip is far away, the resonance Raman scattering (RRS) on the Au (111) is also apparent. But for the Au tip and Pt (110) substrate, the TERS signal of the CN− molecule is gradually enhanced when the Au tip is close. However, when the Au tip moved away slowly, no RRS signal was found on the surface of the Pt (110) substrate. On the one hand, the interaction between Au and CN− molecules will make the Au substrate and CN− molecules form a stable coordination bond, while the adsorption of CN− molecules on the Pt surface is weak. On the other hand, the Ir tip has a higher local electric field enhancement effect than the Pt substrate, which can enhance the Raman scattering signal intensity of CN− molecules. However, the local electric field enhancement effect between the Au tip and the Pt substrate is weak.
Despite that, various gas molecules existing in the atmospheric environment may interact with the sample surface and interfere with the acquisition and analysis of Raman signals. In the atmospheric environment, the electrostatic effect on the sample surface will cause its charge distribution to change, thereby interfering with the Raman signal. Fortunately, these unfavorable factors can be perfectly solved in the high vacuum (HV)-TERS system, but the insufficient optical collection efficiency is a huge hindrance to designing HV-TERS systems. As shown in (Fig. 19d) Sun et al. designed an HV-TERS system based on STM equipment to meet the requirements of optical collection efficiency and used it to research the reduction reaction of 4-nitrobenzenethiol (4-NBT) to dimercaptoazobenzene (DMAB) catalyzed by local hot-electron induction.590 The plasmon intensity can control the chemical reactions in the high vacuum environment, while the local EM field intensity at the tip can be controlled by the incident laser intensity, tunneling current and bias voltage in the HV-TERS system. The high vacuum environment not only solves the problems in the measurement process, but also avoids the interference of molecules in the atmospheric environment on the Raman signal, and also prevents the interaction between the atmospheric environment and the sample, so that the monitoring of catalytic reactions and reduction reactions can be realized. The completion of this work provides a new way to realize efficient HV-TERS system, chemical catalysis and molecular synthesis. After that, Sun et al. further optimized the HV-TERS system. As shown in Fig. 19e, they placed the sample preparation, transfer, spectral measurement, and imaging in a high vacuum environment of 10−7 Pa, which can ensure the cleanliness of the sample and avoid material interference.591 This study enables the analysis of the nature of plasmon-driven reactions on metal surfaces in a pure environment.
After completing the design and practicality of the HV-TERS system, how to complete the imaging and detection of TERS spectra at the single-molecule spatial resolution scale has become a new direction for researchers. Fig. 20a shows that based on the spectral matching between plasmon resonance and molecular vibrational electronic transitions, Hou et al. have realized Raman spectral imaging with a spatial resolution below 1 nm and can resolve the internal structure and surface configuration of a single molecule. They exploited the extremely strong plasmon resonance effect between the Ag tip and the substrate to realize the detection of the single-molecule TERS spectrum of meso-tetrakis(3,5-di-tert-butylphenyl)porphyrin (H2TBPP) and study its dependence on the molecular orientation. The TERS mapping of H2TBPP single molecule on Ag (111) substrate was also completed. This work advances the detection and imaging technology of the HV-TERS system at the single-molecule scale with spatial resolution. Wu et al. designed a high-performance TERS structure based on the low-temperature HV-TERS system combined with the molecular beam epitaxy (MBE) system.593 In Fig. 20b, it can not only realize the preparation of 2D materials, but also complete in situ STM imaging and TERS detection. In an ultra-low temperature environment, the HV-STM-TERS system can achieve an enhancement factor of 109 and a spatial resolution of 0.5 nm for 2D silicene. If the TERS structure is combined with a femtosecond laser system, it is possible to realize femtosecond excitation TERS on a time-resolved scale. In previous work, they also used this system to identify different vibrational properties of silicene phases in different bending directions of Si–Si bonds. The local vibration signatures of silicene defects and edge positions can also be successfully identified.594 In TERS studies, the monitoring of catalytic reactions at time resolution enables the detection and recording of fast vibrations and dynamic changes in the sample. Molecular dynamics on nanosecond and sub-nanosecond time scales can be captured. Weckhuysen et al. used the TERS system to study time-resolved tip-enhanced Raman spectroscopy to monitor catalytic reactions at the nanoscale, which is shown in Fig. 20c.595 A silver tip to enhance the Raman signal and act as a catalyst is placed near the surface of the Au substrate with p-nitrothiophenol (pNPT). A 532 nm laser was used to induce the photocatalytic reduction process at the apex of the tip, and a 633 nm laser was used to monitor the conversion process during the reaction. This research, based on the high spatial resolution of TERS, shows great promise for studying time-resolved molecular dynamics processes on individual catalytic particles, especially for heterogeneous catalysis.
 | ||
| Fig. 20 (a) Single-molecule TERS spectra and their dependency on molecular orientations, and TERS mapping of a single H2TBPP molecule on Ag (111), Reproduced from ref. 596 with permission from Nature publishing group, Copyright 2013 (b) STM image of the Si (111) and in situ normal Raman spectrum of the Si (111) surface/coexisting phases, Reproduced from ref. 593 with permission from Elsevier publisher, Copyright 2018 (c) time-dependent TERS measurement before and after reaction, Reproduced from ref. 595 with permission from Elsevier publisher, Copyright 2012 (d) TERS-BL borophene system and plexciton traits, Reproduced from ref. 597 with permission from AIP publisher, Copyright 2023. | ||
The surface plasmon is a collective oscillation mode at the interface of a metal surface and a medium, resulting from the interaction of free electrons with an optical field. Excitons are quasiparticles in solids, bound states formed by Coulomb interactions between electrons and holes. When an LSPR is coupled to an exciton, the plexciton is produced, which has both LSPR and exciton characteristics. It leads to the rearrangement of their energy level structure, the energy exchange between photons and electron–hole pairs, and coupling together to form new excited states. The field localization makes plexciton have a strong localized EM field enhancement effect in the nanoscale range, which will enhance the absorption and scattering of light. Based on the characteristics of plexciton, Sun et al. constructed the TERS-BL borophene system with the Ag tip and the Ag substrate surface double-layer borophene, as shown in Fig. 20d.597 The negative real part of BL borophene in the strong coupling region indicates that it has the plasmonic properties of metals, leading to the Rabi splitting of plexcitons. The Fanoresonant propagating plexcitons appear in weakly coupled regions. The plasmonic properties of TERS and BL borophene around 488 nm originate from the coupled plasmon–exciton interaction, resulting in two plexciton peaks. This research demonstrates that plexciton-selectively enhanced spectroscopy is possible for the TERS-BL borophene system based on the plasmon–exciton coupling effect. Meanwhile, it provides a new idea for the future use of the TERS system in the study of the corners of 2D materials. Both SERS and TERS are enhanced Raman scattering techniques, which have critical applications in molecular-level characterization and analysis. SERS utilizes the localized EM field on the surface of metal nanostructures to enhance the Raman signal, while TERS further enhances the Raman signal by forming a nanoscale localized EM field between the metal tip and the sample. Therefore, TERS can be seen as an extension and improvement of SERS, achieving higher spatial/time resolution and sensitivity.
3.7. SERS data analysis
In-depth analysis of the inherently complex SERS data often holds the key to elucidating the wealth of chemical information that is embedded within.246,598–600 In recent decades, the progressive shift from traditional manual visual inspections of acquired SERS spectra to state-of-the-art chemometric and machine learning (ML) approaches has unlocked numerous potential applications of SERS-based nanosensors in the biomedical, environmental, and food industries.601–604 Fundamentally, these exciting achievements leverage advancements in both SERS-based sensing technologies as well as ML-driven data processing to accomplish breakthroughs in analyte identification and quantification. However, such success is heavily dependent on the ability to judiciously translate subtle differences present within high-dimensional SERS data into unbiased analyte identification and quantification outcomes through robust visualization or predictive models.The inherent complexity of SERS data stems from the representation of its fingerprint region as thousands of individual wavenumber-to-intensity values, depending on the spectral resolution. For chemometric and ML analysis, such a large number of input features can complicate the problem unnecessarily as (1) the algorithms struggle to find a generalizable function that describes the observations, (2) the signal-to-noise ratio may be lowered as a result of uninformative data, leading to model overfitting.605 Hence, a common and intuitive approach is to decompose SERS data with dimensionality reduction algorithms. Some ubiquitous examples include linear methods such as PCA, linear discriminant analysis (LDA), and singular value decomposition (SVD), as well as nonlinear methods such as isometric mapping (Isomap), t-distributed stochastic neighbor embedding (t-SNE) and uniform manifold approximation and projection (UMAP) (Fig. 21a).606,607 In this section, we will briefly discuss some applications using PCA, t-SNE and UMAP.
 | ||
| Fig. 21 Dimensionality reduction algorithms decompose and visualize high dimensional SERS data based on their most significant variations. (A) Scheme describing common dimensionality reduction techniques. (B) PCA score plot of R- and S-propranolol (PRO) based on their naphthyl vibrational modes with and without an applied voltage. Reproduced from ref. 608 with permission from ACS publisher, Copyright 2021. (C) (i) PCA score plot showing the relative flavor data cluster separation using the SERS taster strategy. (ii) Correlating PCA scores in terms of the relative flavor data cluster positions with its loadings, elucidating key receptor–flavor interactions and their corresponding vibrational peak assignment. Reproduced from ref. 409 with permission from ACS publisher, Copyright 2021. (D) t-SNE plot of the 256-feature space projected onto a 2D map extracted by the modified 1D convolutional neural network model (RamanNet). Reproduced from ref. 609 with permission from RSC publisher, Copyright 2022. (E) UMAP representation of phenotypic responses based on the whole Raman fingerprint region of all individual cells under ampicillin treatment where each point is a measurement of a single cell. Reproduced from ref. 610 with permission from Wiley-VCH GmbH publisher Copyright 2023. | ||
As a stand-alone technique, PCA is a simple unsupervised clustering algorithm that allows automated, objective, and concurrent comparison of numerous SERS spectra based on their most significant variations. For example, Leong et al. utilized PCA to distinguish the propranolol enantiomeric pair accurately highlighting visible differences in their naphthyl vibrational modes with electrochemical SERS (EC-SERS) sensing on nanoporous Au bowls (Fig. 21b).608 Even if the spectral differences appear subtle, the ability to interpret PCA scores through their component loadings enables retrospective correlations to attribute key vibrational modes. This can be seen in Leong et al.'s work, in which detailed analyses of the PCA loadings plot revealed crucial receptor-flavor compound SERS interactions arising from each receptor, which contributed to the distinct score clusters of five different flavor compounds (Fig. 21c).409
Apart from functioning as a stand-alone technique, PCA can also act as a preliminary decomposition step that is easily integrated into customized workflows. This is highly desirable to avoid classifier oversampling of SERS data, where the number of features often easily outnumbers the number of samples.611,612 In Safir et al., PCA was utilized to reduce a 508 wavenumber SERS spectrum to 24 principal components accounting for 90% of the overall variance captured.613 Subsequent input to a random forest classifier enabled ≥87% classification accuracy across acoustic bioprinted droplets containing Au NRs mixed with three single-cell-line and three bacteria mixtures. Despite gaining widespread popularity, one of the most critical drawbacks of PCA being a linear dimensionality reduction technique has become increasingly evident, especially for tasks where a nonlinear relationship between key components is expected. This motivated the venture towards nonlinear techniques such as t-SNE and UMAP, which are capable of preserving much of the local structure while also revealing global structure as clusters at various scales in high-dimensional SERS data.614,615 Briefly, t-SNE is primarily a visualization technique that evaluates pair-wise similarities and models them in a low-dimensional (2D/3D) scatter plot. For instance, a t-SNE plot was employed to illustrate maximal cluster separations attained with 256 features extracted by a modified convolutional neural network (CNN) (Fig. 21d).609 The high distinctiveness is a result of clear SERS spectral differences of 11 bacterial endotoxins acquired using an Ag NR array.
In contrast, UMAP is a generalizable dimensionality reduction technique like PCA, based upon the well-established manifold approximation theory. Moreover, UMAP offers comparable visualization quality to t-SNE, albeit being a relatively new technique. Yang et al. exemplified the capability of UMAP in revealing four bacterial subpopulations with distinct phenotypic responses to increased antibiotic stress during resistance evolution through 1250 single-cell SERS spectra.610 Importantly, these algorithms are built upon robust mathematical frameworks and are applicable for large datasets with complex underlying correlations, making them invaluable to numerous applications in SERS and beyond.
Besides trying to reduce the complexity of SERS data, it is also imperative to pick up even the most minute spectral variance to accurately predict the type and amount of an analyte present. To date, a whole suite of algorithms including but not limited to regression-based algorithms like partial least-squares (PLS), support vector machines (SVM), tree-based frameworks like random forest (RF) and neural networks like CNN exist for this purpose. In general, they traverse different routes to reach the same goal – that is to establish a universal set of mathematical functions to describe the observed relationship between SERS variations and assigned data classes. When adeptly deployed, these powerful algorithms can swiftly highlight subtle spectral variances almost invisible to the naked eye and provide new chemical insights that may otherwise be overlooked. This is apparent in Nguyen et al., where the use of SVM to construct calibration curves was instrumental in enabling multiplex trace quantification of small gaseous molecular targets (sulfur and nitrogen dioxides) (Fig. 22a).392 They justified that a ring complexation strategy involving two SERS-based molecular receptors was crucial to induce key SERS spectral variances, which in turn allowed the SVM model to achieve a 91.7% quantification accuracy. Similarly, Shin et al. employed a CNN model to attain a diagnostic sensitivity of 90.2% and specificity of 94.4% for six types of early-stage cancers (lung, breast, colon, liver, pancreas, and stomach) by analyzing the SERS profiles of exosomes (Fig. 22b).616 Importantly, they found that the first convolutional layer was particularly influential in elucidating the complex exosomal Raman fingerprints contributed by numerous protein constituents.
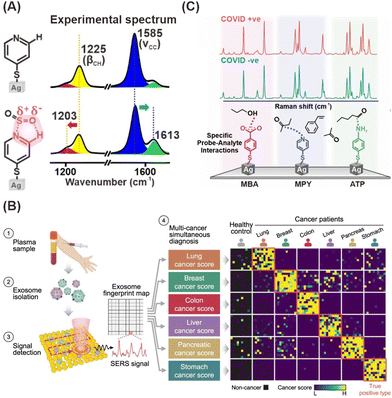 | ||
| Fig. 22 ML-driven classification and regression models constructed based on subtle SERS spectral differences. (A) Formation of a five-membered ring complex with sulfur dioxide inducing spectral variations that are visually subtle. Reproduced from ref. 392 with permission from Wiley-VCH GmbH publisher, Copyright 2024. (B) Obtaining exosome SERS fingerprints from plasma samples of patients with six different types of cancer. The CNN model outputs predictions about cancer presence and tissue of origin. Reproduced from ref. 616 with permission from Springer Nature publisher, Copyright 2023. (C) Combining three receptor SERS spectra as a “SERS super-profile” to discriminate the breath profiles of COVID-positive and healthy individuals. Reproduced from ref. 393 with permission from ACS publisher, Copyright 2022. | ||
It is also interesting to note that, while seemingly contradictory, deliberate amplification of SERS data dimensionality has been proven as a sound strategy specifically for molecular receptor-based SERS sensing. This is because hosting a series of nonspecific noncovalent interactions can effectively probe different facets of target analytes and provide meaningful information about complex sample matrices.617 This concept was highlighted by Leong et al., where the combination of spectral information arising from three molecular receptors allowed ≥ 95% sensitivity and specificity in the SERS-based distinction of healthy breath volatile organic compound (BVOC) profiles and individuals infected with coronavirus disease 2019 (COVID-19) (Fig. 22c).393 Such diverse strategic choices demonstrate the immense potential and versatility of ML algorithms in providing tailored solutions when analyzing complex SERS data.
While sophisticated algorithms can deliver unprecedented results, it is pertinent to ensure that a constructed predictive model is interpretable and remains grounded in strong scientific principles. In fact, the loss in model interpretability often results in the “black box” phenomena or the generation of grossly overfit models that crumble when exposed to unseen data. To avoid this pitfall, a straightforward approach is to extract feature importance and corroborate important features to known Raman vibrational modes, thereby deducing molecular-level insights. For algorithms that do not possess inherent interpretability, framework-agnostic methods such as local interpretable model-agnostic explanations (LIME), Shapley additive explanations (SHAP), and contrastive explanation method (CEM) aim to provide a generalized and comprehensive set of feature rankings.618–620 Alternatively, Živanović et al. evaluated specialized approaches like the minimal depth variable selection (MD) and surrogate minimal depth (SMD) technique in ranking important SERS bands found with a RF model.621 These bands elucidated the presence of cholesterol amongst sphingomyelin and other lipid entities in the analysis of lipid accumulation in lysozymes after treatment with three antidepressants. Naturally, such understanding will also guide downstream feature selection and engineering protocols which can convert wavenumber-based data into more chemically meaningful representations such as peak positions and intensities.
Overall, we have provided a concise overview of the application of state-of-the-art artificial intelligence-driven ML algorithms in SERS data analytics, including dimensionality reduction and visualization algorithms, classification and regression predictive modeling and interpretability techniques. Notably, this overview precludes other expansive topics such as feature selection and engineering as well as data augmentation methods which should be considered of equivalent importance to those covered. Nonetheless, the inherently challenging nature of high-dimensional SERS data presents an interesting conundrum that is likely to motivate further explorations and fuel more exciting discoveries.
4. Surface-enhanced fluorescence (SEF) plasmonic sensors
Various techniques, such as multiple-fluorophore labels,622 rolling circle amplification,623,624 and photonic crystal enhancement,625 have been explored to improve the signal-to-noise ratio of fluorescence-based sensing and imaging techniques. Despite the improved sensitivity, these technologies are not widely adopted in biomedical research or clinical settings. This is because most of these technologies require significant modifications to the existing practices, such as additional steps that significantly prolong the assay time, specialized and expensive read-out systems, non-traditional data processing and analysis, or the use of temperature-sensitive reagents, which usually require tightly controlled transport and storage conditions. Yet another attractive approach for fluorescence enhancement is metal- or plasmon-enhanced fluorescence. Enhancement in the emission of fluorophores in close vicinity to plasmonic nanostructures is attributed to the enhanced EM field at the surface of the plasmonic nanostructures and a decrease in the fluorescence lifetime due to the coupling between excited fluorophores and surface plasmons of the nanostructures.626–635Plasmon-enhanced fluorescence (PEF) biosensors harness the large enhancement of fluorescence near metal nanostructures to enhance the sensitivity of various bioassays. Depending on the integration strategy between the plasmonic nanostructures and the bioassay, PEF biosensors can be broadly categorized into three types: (i) In the first method, PEF biosensors involve metal nanostructures adsorbed or grown on rigid substrates such as glass, silicon, and plastics. This configuration fixes the plasmonic nanostructures to enhance the emission from molecular fluorophores typically used as labels in bioassays. (ii) In the second method, PEF biosensors use an “add-on” plasmonic nanostructure/surface that can be applied to completed fluoroimmunoassays. This method relaxes the reliance on plasmonically-active substrate and makes the PEF biosensor platform more versatile. (iii) Finally, PEF biosensors use ultrabright fluorescent nanolabels that directly replace conventional fluorophores. This strategy integrates the plasmonic nanostructure and the fluorophore into a single nanoscale label for maximal sensitivity, ease-of-use, and ultimate versatility of PEF-based biosensors. Below we briefly describe these methods by highlighting a few specific examples.
4.1. Plasmonically active substrates for enhancing the sensitivity of bioassays
There have been numerous reports that harness plasmonic nanostructures adsorbed on a surface to amplify the fluorescence signals from a bioassay implemented on the surface.636–640 Among the wide variety of metal nanostructures explored for PEF biosensors, gold nanoisland films known as plasmonic gold chips (pGOLD) have gained extensive attention, pioneered by Dai's group.629 Fabricated through a gold seeding process followed by the growth of winding and elongated gold islands across the surface, pGOLD substrates contain abundant nanogaps on the order of 10 nm between adjacent islands (Fig. 23a). These hotspot-rich films effectively couple incident light to generate greatly amplified NIR fluorescence, enhancing detection sensitivity up to 100-fold,630 Owing to the substantial fluorescence enhancement, high reproducibility, and small sample volumes, pGOLD assays have demonstrated immense potential for various bioassays and have been widely adopted across diverse diagnostic settings over the past decade. One implementation in lung cancer profiling utilized multiplexed pGOLD microarrays to simultaneously measure clinically relevant protein biomarkers carcinoembryonic antigen (CEA), Cyfra21-1, and neuron-specific enolase (NSE) in patient blood, achieving much higher sensitivity and specificity over standard bead-based immunoassays.632 More recently, by integrating with portable automated microfluidics, Zhang's group employed pGOLD chips as substrates for a platform termed FEMMAN (Fig. 23b) to perform rapid single nucleotide-level discrimination among 12 SARS-CoV-2 variants directly from patient samples, with performance rivaling tedious genome sequencing.641 DNA probes complementary to sequences unique to each viral lineage were immobilized on the plasmonically-active gold nanoislands for this nucleic acid-based assay. Patient RNA samples underwent an asymmetric amplification step in which 5’ biotinylated amplicons were generated, enabling specific capture at perfectly matched DNA probes to generate plasmon-enhanced fluorescence signal. Only 1 viral RNA copy was required without extraction or purification, with 98.8% sensitivity and 100% specificity metrics that showed 97.6% agreement with gold standard genome testing. | ||
| Fig. 23 (A) SEM image showing the plasmonically-active gold nanoisland film, Reproduced from ref. 630 with permission from Nature publishing group, Copyright 2014 (B) Schematic of the FEMMAN workflow. SARS-CoV-2 related DNA probe targeting different variants’ sites, a terminal biotin-labelled DNA probe as positive control and a DNA probe unrelated to SARS-CoV-2 as negative control are arrayed on the plasmonic gold substrate, Reproduced from ref. 641 with permission from Nature publishing group, Copyright 2023 (C) SEM image of SIF shows the heterogeneity of the particles’ shapes and sizes. The inset is a higher magnification of the SIF, Reproduced from ref. 642, 643 with permission from Elsevier publisher, Copyright 2006 and 2011 (D) Fluorescence image of labeled oligonucleotide targets hybridized to MEF and glass DNA arrays, Reproduced from ref. 642 with permission from Oxford University Press publisher, Copyright 2006 (E) SEM images of ZnO nanorods grown on glass Reproduced from ref. 643 with permission from Elsevier publishing, Copyright 2011 (F) Schematic of the fabrication of ZnO-based antibody microarray, Reproduced from ref. 643 with permission from Elsevier publisher, Copyright 2011. | ||
Beyond gold nanoislands, other nanostructure architectures, including silver island films (SIFs) developed by Lakowicz's group644 and zinc oxide nanorod substrates created by Li's group,643 have also enabled highly sensitive plasmon-enhanced fluorescence biosensing. Lakowicz's lab pioneered the concept of SIFs (Fig. 23c), comprised of silver island films grown at controlled rates on glass substrates using vapor deposition techniques. By immobilizing rabbit IgG on SIFs and capturing fluorescently labeled anti-rabbit IgG antibodies, they systematically studied the dependence of PEF on emission wavelength. Their results revealed more efficient fluorescence enhancement in the near-infrared (NIR) wavelengths compared to the visible wavelengths. In a follow-on work, they demonstrated up to a 28-fold increase in fluorescence signal for DNA microarrays by leveraging near-infrared Cy5 dye and SIFs, highlighting the potential to significantly improve nucleic acid detection sensitivity using far-red fluorophores642 (Fig. 23d). SIFs were also shown to provide 10–15× amplification for sandwich immunoassays detecting cardiac biomarker myoglobin, lowering detection limits below 50 ng mL−1 concentrations and underscoring the broader applicability to enhance the sensitivity of surface-based bioassays.645 Beyond metal films, Li's group introduced zinc oxide nanorod substrates (Fig. 23e) as an alternative PEF transducer architecture. By growing aligned zinc oxide nanorod arrays, precise sub-wavelength spacing between nanorods yielded exceptional optical properties that could be harnessed for surface plasmon-coupled fluorescence enhancement646,647 (Fig. 23f). Using this platform for cancer biomarker microarrays, they demonstrated detection sensitivity improvements down to 1 pg mL−1 for protein biomarkers such as carcinoembryonic antigen (CEA) and α-fetoprotein (AFP) in serum.643 Collectively, these examples highlight the versatility of nanostructure-enabled PEF for ultrasensitive bioanalysis across various assay formats beyond gold nanoisland films.
As described above, various plasmonic substrates such as periodic gold arrays648,649 and metal nano-islands629–632 have been shown to result in a large fluorescence enhancement. Although these plasmonic surfaces are highly attractive, their application in routine biomedical research and clinical settings has been limited. The limited application of plasmon-enhanced fluoroimmunoassays in research and clinical settings is due to: (i) stringent requirement for the fluoroimmunoassay to be implemented on pre-fabricated substrates, typically a rigid glass slide deposited with metal nanostructures, instead of standard or sometimes irreplaceable bioanalytical platforms (e.g., 96-well plates and nitrocellulose membranes), which significantly limits the broad applicability of the technique; More importantly, the requirement of special substrates limits cross-platform and cross-laboratory consistency and seamless integration with widely employed bioanalytical procedures, which represents a major bottleneck of conventional plasmon-enhanced fluorescence techniques; (ii) non-traditional bioconjugation procedures, complex interaction between biomolecules and the metal nanostructures, and poor stability of biomolecules (e.g., antibodies) immobilized on metal surfaces not only complicate the assay procedures but also impose further technical challenges in their widespread application;650 and (iii) fluorescence quenching due to non-radiative energy transfer, which is difficult to control when the immunoassay is performed on a metal surface. Thus, it is imperative to address these challenges to propel the plasmon-enhanced fluorescence techniques to real-world applications.
Overcoming these limitations, add-on plasmonic architectures that can be interfaced with completed bioassays provide a more flexible and easy-to-implement route for integrating PEF. An illustrative example of successfully transitioning to an add-on PEF biosensor is the plasmonic nanogap cavity assay developed by Mikkelsen and Chilkoti's groups.651 They fabricated the base platform by grafting a polymer brush coating of poly(oligo(ethylene glycol) methyl ether methacrylate) (POEGMA) chains onto a gold film-sputtered glass surface, which enabled inkjet printing of both capture antibodies and fluorescently-labeled detection antibodies (with Alexa Fluor 647 dye) as microarray spots. Two spot types were printed on the POEGMA layer – “stable” spots containing surface-immobilized capture antibodies, and “soluble” spots where detection antibodies were mixed with an excipient that dissolved upon sample addition to facilitate antibody release. After completing the immunoassay, Ag NCs were appended above each printed spot using a poly(allylamine hydrochloride)(PAH) intermediate layer or conjugating the NCs to a secondary antibody. This formed nanogap cavities with a plasmonic metal–dielectric–metal geometry structured to amplify fluorescence emission. They termed the resulting architecture the plasmonically enhanced D4 (PED4; D4 stands for Dispense, Dissolve, Diffuse, Detect) assay (Fig. 24a). Using cardiac biomarker B-type natriuretic peptide (BNP) as a model analyte, optimization of POEGMA brush thickness and NC parameters enabled ∼150-fold fluorescence enhancement along with 14-fold improvement in detection limits compared to standard assays (Fig. 24b). The combination of the versatile D4 assay format652 with the add-on nanostructure PEF amplifier imparts both excellent sensitivity and field-deployable ease-of-use relevant for point-of-care applications.
 | ||
| Fig. 24 (A) Schematic of workflow for D4 assay, Reproduced from ref. 651 with permission from the American Chemical Society, Copyright 2020 (B) comparison of fluorescence images for PED4 assay pre and post cubes addition, Reproduced from ref. 651 with permission from the American Chemical Society, Copyright 2020 (C) representative visual photo and SEM images of plasmonic patch. TEM images of nanorods used to modify the plasmonic patch, Reproduced from ref. 653 with permission from Nature publishing group, Copyright 2018 (D) fluorescence scanning images of fluorobioassay performed with and without the enhancement of plasmonic patch, Reproduced from ref. 653 with permission from Nature publishing group, Copyright 2018. | ||
Among other prominent efforts to develop add-on plasmonic amplifiers, Singamaneni's group has introduced a flexible “plasmonic patch”. Their key innovation involves adsorbing tunable plasmonic nanostructures onto an elastomeric polymer film to create a conformal nanoplasmonic interface653,654 (Fig. 24c). When interfaced with completed fluoroimmunoassays in microwell plates or microarrays, this plasmonic patch can provide up to 100-fold fluorescence enhancement. Critically, the LSPR wavelength of the embedded nanostructures can be tuned to match the absorption/emission profile of the fluorescent reporter for maximal amplification.654 Moreover, they systematically investigated the distance-dependent fluorescence enhancement to determine optimal spacer layer thickness. Assays for two key renal health indicators were evaluated to demonstrate the broad applicability across diverse protein biomarkers: kidney injury molecule-1 (KIM1) and neutrophil gelatinase-associated lipocalin (NGAL). Both biomarker immunoassays were performed using standard ELISA protocols, substituting streptavidin-fluorophore conjugates for streptavidin-horseradish peroxidase (HRP). By interfacing the completed assay with an LSPR-matched plasmonic patch, fluorescence intensity increased 36-fold, and detection limits improved 280-fold for KIM1 (Fig. 24d). Similarly, NGAL fluorescence boosted 103-fold while detection limits enhanced 100-fold. Furthermore, the plasmonic patch successfully enabled quantification of biomarker concentrations in patient samples that were undetectable without signal amplification. This highlights the versatility of the add-on nanostructure approach to harness plasmon-enhanced fluorescence for ultrasensitive clinically viable assays.
4.2. Ultrabright fluorescent nanolabels based on PEF
Building on their plasmonic patch concept, Singamaneni's group continued advancing integrations of plasmonic nanostructures with bioassays to harness sensitivity gains from PEF. By realizing optimized plasmonic nanostructure structures conjugated to fluorescent reporters, they developed exceptionally bright nanolabels, termed “plasmonic-fluors” (PFs), that are nearly 7000-fold brighter than their corresponding molecular fluorophores655 (Fig. 25a). Streptavidin modification enables the PFs to substitute for streptavidin-fluorophore or streptavidin-HRP conjugates in common assay formats like FLISA and ELISA, providing built-in signal amplification tailored to each biomarker system (Fig. 25b). Using the model cytokine IL-6, they demonstrated plasmon-enhanced fluorescence-linked immunosorbent assay termed p-FLISA that lowered detection limits 1440-fold versus standard FLISA and 189-fold below conventional ELISA, highlighting the ultrasensitive quantification (Fig. 25c). Moreover, all previously demonstrated PEF biosensors have relied on modifying the conventional substrates or assay procedures, constraining the real-world utility. The nanolabel architecture of the PFs liberates plasmonic signal enhancement for integration into diverse analytical formats, including competitive and sandwich immunoassays,656–658 CRISPR-based RNA detection,659 serological assays,660 flow cytometry,655 microneedle-based non-invasive sampling,661 LFA,662 and measurements of live cell protein secretion.663–665 In one demonstration, by integrating PFs into lateral flow immunoassays (p-LFAs) (Fig. 25d), detection sensitivity improved 1785-fold over conventional colorimetric LFAs (Fig. 25e) and 30-fold over standard clinical lab tests such as ELISA. Additionally, p-LFA successfully identified 34 out 35 positive COVID patient samples based on SARS-CoV-2 antigen levels, significantly outperforming colorimetric LFA. Coupled with a portable fluorescence scanner, the versatile plasmonic-fluor nanoparticle labels provide a broadly applicable solution to harness PEF for point-of-care testing applications spanning nucleic acids, antigens, and other biomarkers. | ||
| Fig. 25 (A) Schematic and TEM image of plasmonic-fluor structure, Reproduced from ref. 655 with permission from Nature publishing group, Copyright 2020 (B) schematic of workflow for plasmonic-fluor enhanced fluorobioassay, Reproduced from ref. 655 with permission from Nature publishing group, Copyright 2020 (C) comparison of ELISA and p-FLISA regarding the performance of human IL-6 detection Reproduced from ref. 655 with permission from Nature publishing group, Copyright 2020 (D) Schematic of construct and workflow of p-LFA in detection of SARS-CoV-2 N protein, Reproduced from ref. 662 with permission from Nature publishing group, Copyright 2023 (E) comparison of Au NP-based colorimetric and plasmonic-fluor-based fluorometric LFA performance of N protein detection, Reproduced from ref. 662 with permission from Nature publishing group, Copyright 2023. | ||
In a recent work, Tian's lab has demonstrated a buoyant biosensor that enabled multiplexed, in situ detection and quantification of attomolar cytokine concentrations in cell culture media with 15 min temporal resolution,666 Plasmonic-fluors combined with digital counting further enhanced the detection limit to 14 ag mL−1 for IL-6, preserving viable macrophages while quantifying dynamically secreted cytokines.667 The facile sensor demonstrates strong promise for quantifying the temporal protein biomarker concentration changes in biological systems without perturbation. Plasmonic-fluors have also been employed as ultrabright nanolabels for probing the interaction of charge-controlled nanostructures with neurons and for demonstrating plasmon-enhanced expansion microscopy.668,669 The emission from PFs was further amplified by coupling with resonant modes of a photonic crystal.670 Cunningham and co-workers demonstrated the synergistic coupling of plasmonic and photonic modes to significantly boost the emission from plasmonic-fluors. Optimal match of the resonant modes of a plasmonic fluor and a photonic crystal resulted in a 52-fold improvement in signal intensity. This enhanced emission was employed to achieve an ultrasensitive digital immunoassay on photonic crystal surface.
Overall, PEF has been extensively explored in the past two decades as a powerful tool to boost the sensitivity of various fluorescence-based bioanalytical techniques. Tremendous progress has been made in the last five years in overcoming the challenges associated with translation of these technologies to routine real-world use. We can expect a widespread use of these technologies in the near future based on the extensive commercialization interest in these technologies.
5. Localized surface plasmon resonance (LSPR) sensing
5.1. Fundamentals of LSPR sensing
As mentioned before, metal NPs support LSPR which are responsible for an intense EM field enhancement very close to their surfaces.671–673 Optically, these plasmonic NPs exhibit wavelength light absorption and scattering which is largely dependent on the NP dimensions, geometry, chemical nature, interparticle interactions, and the surrounding medium refractive index (RI).674–677 The latter is the reason why they have been extensively used for sensing applications, and by precisely tuning the former parameters it is possible to maximize their high refractive index sensitivities (RIS).678 For spheroidal NPs Mie–Gans theory can directly deduce the RIS for a NP with a given aspect-ratio (AR) as follows.:678,679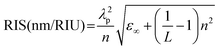 | (9) |
In the context of plasmonic sensors, in general, NPs exhibit smaller RIS compared to their thin film counterparts.685,687 Nevertheless, their usage can be advantageous in certain scenarios due to the absence of phase-matching conditions, which significantly reduces the costs associated with the interrogation setup.688 Moreover, the distribution and uniformity of NPs immobilization, particularly in the case of colloidal NPs, can be more easily achieved, especially when dealing with complex optical platforms where physical vapor deposition techniques may be challenging or impractical, as with micro-structured optical fibers.689,690 Also, plasmonic sensors based on colloidal NPs present far greater scalability potential and lower-cost setups for synthesis and immobilization, two crucial factors for the widespread implementation of any sensing technology. In the next sections, the interaction between NPs and optical platforms is explored, with particular interest in the use of optical fibers, recent trends in NP arrangement for enhanced RIS, and finally, providing an outlook on the development of optical fiber-NP systems for improved sensors.
5.2. Optical platform and sensor configurations
The linear light dispersion of plasmonic NPs contrasts with the strong light dispersion observed in thin film-based SPR. However, when NPs are near or in contact with substrates, strong polarizability effects emerge, causing their optical response to be significantly influenced by the NP-substrate distance, as well as the substrate material itself.673,685,691 This is caused by the EM-field enhancement being partly buried in the substrate (Fig. 26a), as so the effective RI around the NP gets modified, causing wavelength shifts and intensity extinction variations (Fig. 26b).692 Also, this presents relevant consequences regarding their RIS, where the RIS gets diminished by the immobilization over a dielectric substrate, when compared to the same particles in colloidal solution (Fig. 26c).691 However, a different trend is observed when the substrate material gets replaced by a metal. This optical response has been explored in recent years, showing that strong redshifts can be observed without significant band widening (Fig. 26d).693 The presence of the metal layer acts as a charge mirror, effectively mimicking the presence of another NP in its vicinity. Thus, by changing the dielectric spacer length, it is possible to tune the strength of the plasmon hybridization (Fig. 26e). Further tuning can be achieved by changing the thickness of the metal layer,694 or substrate materials, as the case of TiO2.695 Also, modifying the NP geometry, e.g. using nanorods, can produce even stronger optical variations with interesting results, as higher order plasmonic modes respond differently to polarization changes (Fig. 26f).695,696 These later configurations are commonly known as nanoparticle on film (NPoF) structures and can behave as light waveguiding structures, composing a metal–insulator–metal (MIM) configuration, where the metallic top layer is replaced by the NPs (Fig. 26g).697 Recently, this kind of structure has received a lot of attention and even other nanostructures have been explored. Nevertheless, for sensing purposes the main tuning parameter still lies within the nanocavity. This is based on the principle of achieving a large effective RI in the spacing layer with the decrease of its dimensions. Thus, the ability to decrease this spacing will greatly impact the structure performance (Fig. 26g). However, despite the recent progress observed with these MIM wave-guiding structures, the EM-field shows strong confinement between the NP and thin film layer, which is not ideal for sensing purposes, as the field enhancement should extend towards the surrounding medium. Accordingly, to increase the sensing performance of these structures, this issue should be addressed. Nevertheless, several high RIS were reported using these NPoF configurations, as shown in Table 2. To the moment, the highest reported RIS was presented by Xia et al., where an experimental RIS of 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 747 and 25
747 and 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 642 nm RIU−1 for Au NS and Au NR, respectively, was attained on a photonic crystal fiber (PCF) coated with an Au thin film.698
642 nm RIU−1 for Au NS and Au NR, respectively, was attained on a photonic crystal fiber (PCF) coated with an Au thin film.698
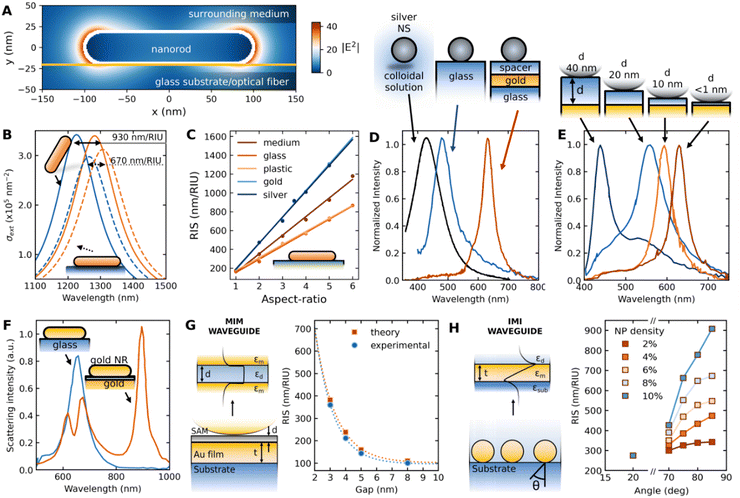 | ||
| Fig. 26 (A) Near field simulation of an Ag nanorod (NR) immobilized on a glass substrate using boundary elements method at the peak of the longitudinal mode LSPR band reproduced from ref. 692 with permission from Optica publisher, Copyright 2010 (B) spectra comparison upon change in the medium RI for an Ag nanorod immobilized on a glass substrate and on colloidal suspension. (C) Impact of the substrate material on the RI sensitivity (RIS). (D) Spectra of an Ag nanosphere (NS) on colloidal solution, over a glass substrate and at proximity (<1 nm) to an Au thin film layer. Reproduced from ref. 693 with permission from ACS publisher, Copyright 2010 (E) impact on the optical behavior on the changing of the spacer thickness, showing a strong red shift with decreased spacing. Reproduced from ref. 693 with permission from ACS publisher, Copyright 2010 (F) Au NR immobilized over an Au thin film and over a glass substrate, showing the appearance of higher order modes due to plasmon hybridization. Reproduced from ref. 699 with permission from ACS publishing, Copyright 2018 (G) metal–insulator–metal (MIM) waveguide structure composed of an Au thin film and Au NS along the RIS dependency on gap length. Reproduced from ref. 697 with permission ACS publisher Copyright 2021 and RIS results Reproduced from ref. 700 with permission from RSC publisher, Copyright 2022 (H) insulator–metal–insulator (IMI) waveguide structure representation, along the RIS dependence on launching angle. Reproduced from ref. 701 with permission from ACS publisher, Copyright 2011. | ||
| Optical platform | RIS (nm RIU−1) | Ref. |
|---|---|---|
| Photonic crystal fiber with Au thin film and Au NSs | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 747 747 |
698 |
| Photonic crystal fiber with Au thin film and Au NRs | 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 642 642 |
698 |
| Side polished multimode fiber with Au thin film and Au NSs | 3074 | 702 |
| Multimode fiber tip coated with Au thin film and carboxylated multi-walled carbon nanotubes | 2524 | 703 |
| Multimode fiber tip coated with Au thin film and carboxylated multi-walled carbon nanotubes, and Pt NPs | 5923 | 703 |
| Multimode fiber coated with SnO2 thin layer and 50 nm SnO2 NPs | 5334 | 704 |
| Multimode fiber coated with 9 PSS-SnO2 bilayers and 50 nm SnO2 NPs | 4704 | 704 |
| Photonic crystal fiber section between two MMF with Au film and Au NRs | 3915 | 705 |
Other waveguiding structures have also been proposed, showing better prospects for effective implementation of high-performance NP-based plasmonic structures. Namely, the insulator–metal–insulator (IMI) waveguide scheme (Fig. 26h). In this case, randomly assembled NPs over a glass substrate are seen as an effective RI layer between the substrate and the external medium. This metal layer can be modeled using effective medium theories such as Bruggeman or the Maxwell-Garnett effective index approximations,706 and it will present high effective indices enabling it to guide light. The light will be highly compressed according to λeff = λ/neff. Consequently, as the neff increases, the more efficient waveguiding becomes, which can be done by increasing the NP density. In addition, a stronger field in the meta-layer will lead to a stronger interaction with the surrounding medium, causing an increase in RIS (See right panel of Fig. 26h).701 As this is a waveguiding mechanism light dispersion is expected, so it will become dependent on the wave-vector of the excitation light, causing a dependence on the angle of incidence.
As the light illumination conditions can determine the sensing system performance, it is relevant to explore not only the optical behavior of NPs under colloidal suspension but also when immobilized over planar substrates or optical fibers and discuss the benefits of either situation. First, using the NPs in colloidal solutions allows for the simpler usage of all, as it can just be placed inside a lab bench spectrometer and measure the extinction coefficient. Nevertheless, it was seen that by placing them on a substrate, they can enhance the NPs RIS. In this situation, to control excitation conditions, a more complex optical setup such as the Kretschmann or Otto configuration is required, allowing for precise wavevector conditions control. However, the costs associated with these setups can rapidly increase and require a sampling system or microfluidic channel to transport the analyte into contact with the immobilized NPs. Another option is the use of optical fibers, as they allow for easy multiplexing, long-range sensing, potentially lower costs, and material choice, as well as a broad choice of sensing configurations such as sensing tips.707–710
Among the multitude of optical fiber sensing configurations available the most common are based on exposing the fiber core and immobilizing the NPs on that section, by side polishing, cladding removal on MMF, or bending a fiber to such a small curvature radius that causes an evanescent field to interact with the immobilized NPs.689,708,711–715 Another approach is structural modification via grating inscription to partially redirect the light to the fiber cladding allowing for evanescent interaction with the NPs, as in the case of the long-period fiber gratings (LPFG) or multi-single-multimode (SMS) fiber configuration (Fig. 27a).716,717 When NPs are assembled on optical fibers, the reports on RIS don’t match with the ones of the same kind of NPs immobilized on glass slides, indicating that not only polarizability effects are of concern (Fig. 27b). Effectively, several reports show RIS higher than 1000 nm RIU−1 for simple gold nanospheres (NS) when immobilized over optical fiber, contrasting to the case of glass slides and colloidal solutions. To the best of the author's knowledge, the highest reported RIS with gold NSs are 2016, 350, and 180 nm RIU−1 on optical fibers, glass slides, and colloidal dispersion, respectively.718,719 The large discrepancies observed between the optical fiber and the other two can be attributed more to the light excitation conditions than the material specificities. To the moment no consensus exists to explain the observations, whereas some authors as Otte et al.,701 consider that the higher sensitivities are related to wave-guiding phenomena, while others attribute to the creation of hotspots due to interparticle-coupling or lossy-mode resonances.718,720 The same trend was observed on other NP shapes and materials.721–738 Focusing on a better performant spheroidal NP shape than NS, i.e., Au NRs, by comparing their performance on different optical platforms (Fig. 27c), it is possible to see that fibers show the better overall performance with a RIS/Aspect-Ratio of 304 nm/RIU/Aspect-Ratio, contrasting to a ratio of 113 and 33 nm/RIU/Aspect-Ratio for the NRs on glass slides and colloidal solutions, respectively. Thus, suggesting that better performance can be accomplished with the conjunction of higher AR NRs and optical fibers. To the best of the authors knowledge there is no RIS data reported for NRs with AR larger than 5 on optical fibers. Thus, causing a low R2 value in the linear regression analysis. Nevertheless, as the NR synthesis protocols become more mature, resulting in higher yields, NP monodispersity, and larger AR, it is expected that more focus will be given to such high AR NRs – optical fiber plasmonic sensor configurations.
 | ||
| Fig. 27 (A) Typical nanoparticle-based plasmonic optical fiber sensor configurations (B) Refractive index sensitivity (RIS) comparison between Au nanospheres (NSs) immobilized on optical fiber sensor configurations, glass slides, or in colloidal solutions. The data labels correspond to the following works: A1-A8;718 B1,B2;739 C;716 D;736 E1,E2;734 F;740 G;741 H;742 H1-H9;719 I1-I12;720 J;743 (C) RIS for gold nanorods (NRs) comparison for the same two optical platforms and on colloidal solution. Data were reproduced from ref. 678, 724, 732, 736, 738, 740, 742, 744–748 with permission from ACS publication, Springer New York publisher, ACS publication Elsevier publisher, MDPI publisher, Copyright 2007, 2022, 2016, 2008, 2022, 2007, 2009, 2021, 2018 respectively. | ||
The recent advancements in synthesis methods have led to increased yields, enhanced NP monodispersity, improved colloidal stability, and enhanced biocompatibility.749 Consequently, the widespread implementation of sensing applications utilizing plasmonic NPs may be on the horizon. Recent trends of NPs-based plasmonic sensors were found to rely on optical fibers as the optical platform. As well, NPs stacking and even immobilization over or in between metallic thin films have been proven to unravel new paths towards unprecedented RIS and overall optical performance.711,750–754 Enhanced chemical and biochemical sensing applications have been recently reported, showing the feasibility of high-performance NP-based plasmonic sensing. In the future, more works in NIR wavelengths with NPs and optical fibers are expected, which can greatly advance the field and produce even higher RIS and overall chemical/biochemical sensors.
5.3. Colorimetric sensing
 | ||
| Fig. 28 Colorimetric sensing based on plasmonic nanoparticles categorized according to the type of analyte. (A) Refractive index sensors: (i) measurement of the RI change (1 < n > 1.6) through the shift of the change of hue of Al nanodisk array substrates. (ii) Lineal hue changes of a substrate comprising gold nanoparticles because of sucrose concentrations from reflectance and transmission images. (iii) Variation of hue of Ag nanodome arrays by changing the surrounding medium from water to ethylene glycol. (B) Colorimetric measure of temperature by the plasmon coupling of Au-coated pNIPAM microgels at different salt additives, where the color change is monitored through the chromaticity coordinates x and y. (C) Humidity sensor measured by Al NPs on Al mirror with pNIPAM as a spacer in which the color change is monitored in the CIE L*a*b* space. (D) Biosensor of volatile organic compounds detected by using Ag NCs on Ag mirror with a bacteriophage spacer as a transducer. Color difference (ΔE) is quantified for a wide range of ethanol concentrations. (a-i) and (a-ii) Reprinted from ref. 757, 758 with permission from ACS publisher, Copyright 2015, 2020. (a-iii) Reprinted from ref. 759 with permission from Wiley-VCH Verlag GmbH & Co. KGaA publisher, Copyright 2020 (b) Reprinted from ref. 760 with permission from Springer publisher, Copyright 2018. (c) Adapted with permission from the Authors of ref. 761. (d) Reprinted from ref. 762 with permission from ACS publisher, Copyright 2022. | ||
Traditionally, colorimetric plasmonic sensing has emerged from the LSPR spectral sensors, where the plasmon modes in the visible spectral range shift by the alteration of the refractive index (RI) of the surrounding medium. The initial attempt to transition from spectral to colorimetric sensing was limited to a qualitative readout, e.g., the presence or absence of an analyte. Over the past few years, the advancement in the design and fabrication of dynamic plasmonic systems enabled the implementation of standardized mathematical descriptions for color change due to RI variation. King et al.757 designed a 2D Fanoresonant plasmonic system consisting of a centered Al disk with N number of satellite planar Al nanodisks (Fig. 28a-i) that scattered light in the visible spectral range allowing apparent vibrant color (pure saturated). The authors introduced a Figure of Merit (FOM), |ΔHue/Δn|, a measure to quantify the change of hue in the polar coordinate of the CIE lightness-chroma-hue color space (CIE LCHab). The value of FOM correlated linearly with RI in the range from 1 to 1.6, achieving the Δhue as large as 216°. In a similar approach, Reinhard et al.758 designed a RI sensor comprising Au NPs (18–115 nm) attached to a solid substrate to detect sucrose solutions (Fig. 28a-ii). The authors found the hue was the most sensitive color parameter among 15 other parameters tested (e.g., hue, r, g, and b chromaticities). Through the ratio Δy/RIU, where y is the color parameter and RIU is the change of RI, the value of Δhue equal to 0.07 (or 26.5°) was obtained for 1.33 < RI < 1.42 (RIU = 0.09), exceeding by nearly 6-fold the sensitivity of the r chromaticity. The authors concluded that the sensitivity to RI increases with the particle size and that a larger color difference is achieved in transmission rather than reflection. In a more recent publication, Toma et al.759 designed a RI sensor consisting of polystyrene bead monolayers (200–300 nm) coated with Ag thin films to form Ag nanodomes whose surface colors were optimized by changing the size of the polystyrene beads (Fig. 28a-iii). The sensitivity was evaluated through the hue parameter for water and ethylene glycol at different volume ratios and three Ag nano-dome diameters. In the best-performing architecture, Δhue of 59.6° for a RIU = 0.1 was obtained. The authors demonstrated that the resolution of the colorimetric detection (5.0 × 10−5 RIU) was higher than spectral detection (10−4 RIU).
With the integration of plasmonic nanostructures alongside other advanced materials, colorimetric sensors have expanded beyond detecting changes in RI. Choe et al.760 have developed plasmonic colorimetric patches to sense temperature. The authors used Au-coated poly (N-isopropyl acrylamide) (pNIPAM) microgels embedded within a hydrogel matrix (Fig. 28b). The pNIPAM exhibits lower critical solution temperature (LCST) at 32 °C. Below LCST, the microgel particles gain volume, increasing the separation of Au NPs, hence the color transition from blue to red. Above LCST, the pNIPAM chains collapse, reducing the specific volume of microgels and thus turning red to blue due to plasmon coupling. To correlate the color change and temperature, the authors employed the RGB color space and varied the transition temperature by adding different salts to the hydrogel films in which the Au NPs pNIPAM microgels were incorporated: N1-0.2 M NaCl, N2-0.1 M NaCl, S1-1 mM SDS, S2-2 mM SDS, and control-no additives. The authors found that the red component exhibits high sensitivity for S1, ΔRS1 = −33, and S2, ΔRS2 = −61, in temperature ranges of 29–33 °C and 33–40 °C, respectively. Interestingly, the authors achieved a temperature resolution of 0.2 °C, allowing precise monitoring of temperature variations on various substrates. By exploiting the concept of particles on the mirror, Cencillo-Abad et al.761 have developed a humidity sensor using Al NPs (3 nm to 5.5 nm) on Al mirror (Fig. 28c). These nanoparticles were separated by a pNIPAM layer of 35.5 nm at relative humidity (RH) of 50% (at 20 °C). At low RH levels, the desorption of water caused shrinkage of the pNIPAM layer, resulting in the optical coupling of NPs with mirror, in consequence, an enhancement of the red component of the reflected light. At high RH levels, the pNIPAM layer expanded, reducing the coupling and enhancing the blue component in the reflected light. The color of the sensor switched from magenta to blue that could be tracked with the change of the component a* and b* of the CIE L*a*b* color space (L*-lightness-, a*-red to violet-, and b*-yellow to blue-coordinates, Fig. 28c). The device effectively transformed changes in relative humidity into observable color changes.
Recent advancements in biotechnology enabled the fabrication of biological constructs of synthetic and natural origin that can serve as biomarkers or signal transducers in colorimetric sensing involving plasmonic nanoparticles. Nguyen et al.762 have employed genetically engineered bacteriophage as a spacer between Ag NCs and silver film (gap plasmonic color film, GPCF) to detect volatile organic compounds (VOCs) as biomarkers of lung cancer (Fig. 28d). The bacteriophage layer worked as an actuator, adjusting the gap in response to the concentration of VOC. The researchers selected over 43 VOC biomarkers and tested the biosensor's performance with five gases: acetone, ethanol, isopropyl alcohol, diethyl ether, and hexane. The analyte changed the gap and thus the color difference (Fig. 28d shows the ΔE for ethanol). The GPCF-based biosensor demonstrated exceptional selectivity, and ease of fabrication, making it a promising platform for developing novel diagnostic devices. The colorimetric sensing of biomolecules was proposed by Sanromán-Iglesias et al.,763 where aggregating DNA-coated Au nanoparticles were used as an optical signal transduction to detect bacterial nucleases – a biomarker of Salmonella spp. In the presence of nucleases, the aggregation of nanoparticles was inhibited (red color) due to enzymatic cleavage of double-stranded probe sequences (positive readout). Without nucleases, the nanoparticles were allowed to aggregate (red-to-blue color transition, negative readout) due to the selective binding of complementary DNA strands. Through naked-eye inspection, the method detected living bacteria down to 1 CFU mL−1 in naturally contaminated food samples. Unlike substrate-based plasmonic sensors, colorimetric detection based on nanoparticle aggregation suffers from a poor quantitative description of color change. This is because many geometrical parameters (mutual particle orientation or interparticle-gap) condition the reproducibility of color difference. Recently, Montaño-Priede et al.764 have evaluated the effect of geometrical descriptors (particle shape and size, number of particles per cluster, and interparticle distance) on the resulting change in Hue during the aggregation of gold nanoparticles. Through the comparison of experimental and theoretical data, the authors found that gold decahedra with an edge length of 30 nm outperform other shapes, showing ΔH = 72% (ΔH = ΔHue/max(ΔHue)). The proposed methodology for calculating hue values and associated FOM ΔH provided new universal and quantitative tools to monitor color transitions during the aggregation of nanoparticles induced by any molecule.
To close, plasmonic colorimetric sensing offers a quantitative measure of color change in the presence of biomolecules and environmental stressors. This type of sensor is expected to experience substantial improvements in forthcoming years. The main driving force is the progress in bottom-up fabrication that allows for obtaining structural diversity, reproducibility, and switchable optical outcome that can be quantified through standardized color spaces (RGB, CIE XYZ, or HSV). Further integration of active plasmonic sensors into substrates obtained by the top-down approach will offer portability, cost-effectiveness, and suitability for point-of-care applications by reducing the need for expensive, specialized instruments. Still, the performance of a plasmonic colorimetric sensor depends on the selection of suitable color space, requiring a trial-and-error approach. Thus, further standardization of the current methods is needed to ultimately render high-performance and hyperchromatic sensing.765
In this section, we will highlight some examples to illustrate these mechanisms. Growth-based plasmonic sensors have been developed to detect various analytes, including metabolites,774 DNA,775 enzymes,775 protein biomarkers,776 bacteria,777 and viruses.778 The growth of plasmonic nanoparticles can be modulated by enzymes. Common precursors include Au3+ and Ag+ for gold and silver nanostructure growth. For example, a plasmonic ELISA based on Au NP growth modulated by enzymatic decomposition of a reducing agent was reported by the Stevens group.776 Following a standard ELISA procedure, the reducing agent, hydrogen peroxide (H2O2), and gold ion precursor solution were introduced sequentially (Fig. 29a). In the presence of targets, the enzymes linked to detection antibodies consume H2O2, slow down the gold reduction in 2-(N-morpholino)ethanesulphonic acid (MES) buffer and yield aggregated gold nanoparticles, which result in blue color solution. In the absence of targets, the high concentration of H2O2 leads to a fast reduction and the Au NPs are not aggregated, which yields red color solution. A distinguishable blue to red color transition happens in a narrow H2O2 concentration range between 100 and 120 μM. The method enabled the detection of prostate specific antigen (PSA) and human immunodeficiency virus HIV-1 capsid antigen p24 in whole serum at an ultralow concentration of 1× 10−18 g mL−1. Detecting p24 at such a low concentrations using a standard ELISA format without the need for expensive nucleic acid-based tests could offer a robust alternative for diagnosing HIV infection through naked-eye observation. Based on a similar approach, Huang et al. reported naked-eye detection of Gram-negative Escherichia coli (E. coli) and Klebsiella pneumoniae (K. pn) in urine samples.777 These bacteria produce catalase that decomposes H2O2, which modulates the growth of Au NPs to yield different colors. This approach takes 85–100 minutes from sample collection to analysis and provides a limit of detection of ∼106 CFU mL−1. Many enzymes, such as glucose oxidase,774 tyrosinase,779 and alkaline phosphatase,778 can catalyze the decomposition of substrates into reducing agents which induce nanoparticle growth. A simple example is an enzymatic glucose sensor, in which H2O2 was generated in the presence of glucose and oxygen.774 The glucose concentration is proportional to the amount of H2O2 produced to modulate the NP growth. The same growth mechanism can also expand to the detection of enzyme775 and reducing agent780 targets. In addition to self-nucleation, Au and Ag growth can occur on many predefined nanostructures, such as Au NSs,774 Au NRs,780 Au NBPs,778 and Au NSTs.775
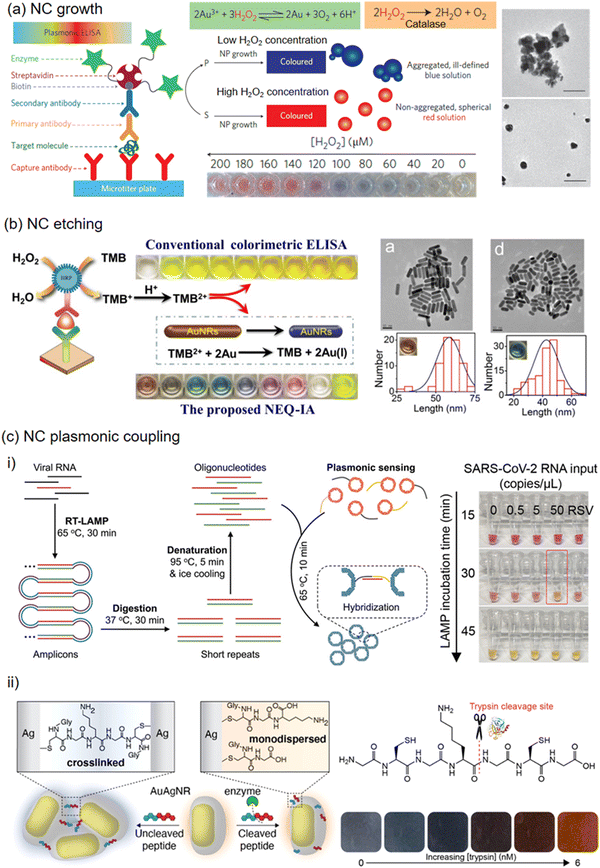 | ||
| Fig. 29 Colloidal NP-based colorimetric sensing examples categorized according to signal generation mechanisms. (A) NP growth: ultrasensitive detection of prostate specific antigen and HIV-1 capsid antigen p24 based on the growth of gold NPs band reproduced with permission from ref. 776 Copyright Nature publisher (B) NP etching: multichromatic semiquantitative immunoassay enabled by TMB2+etched Au NRs band reproduced from ref. 781 with permission from Elsevier publisher, Copyright 2017 (C) NC plasmonic coupling: (i) specific and sensitive detection of SARS-CoV-2 RNA through the agglomeration of gold-silver alloy nanoshells involving loop-mediated isothermal amplification reproduced from ref. 782 with permission from Wiley-VCH publisher, Copyright 2022 (ii) Trypsin detection by modulating the aggregation of Ag-coated Au NRs reproduced from ref. 783 with permission from ACS publisher, Copyright 2022. | ||
Etching-based colorimetric sensing involves the etching of predefined NPs by the targets or the intermediates produced following the presence of targets. The predefined NCs include Au NSs,784 Au NRs,781 triangular Ag NPts,785 Au NBPs,786 and Ag-coated Au NSs.787 The etching-based colorimetric sensors have been reported to detect various targets, including metal ions,784 anions,788 metabolites,785 DNA,789 proteins,781 and exosomes.790 The presence of various ions, such as Cl−, Br−, and CN−, can reduce the redox potential of gold and silver and induce the etching under certain conditions.787,788,791 This can be used to detect these ions in the presence of oxidizing agents, including oxygen,787 hydrogen peroxide,792 and nitric acid.791 Similarly, targets with oxidizing ability, such as NO2−, Cu2+, and Cr6+, can be detected after the redox potential of Au and Ag decreases.788,793,794 The formation of Pb–Au alloys can accelerate the etching of gold nanospheres in the presence of 2-mercaptoethanol and thiosulfate (S2O32−), which was used to detect Pb2+.784 Enzymatic reactions that produce H2O2 to etch triangular Ag NPts785 or Au NRs795 can be used to detect glucose. The H2O2 produced in ELISA can be used to detect DNA based on color changes in the etched silver triangular nanoplates.789 Another universal way was based on the TMB2+ produced in the HRP-3,3′,5,5′-tetramethylbenzidine (TMB) ELISA, which etches gold nanorods to yield vivid color responses (Fig. 29b).781 TMB2+ shortened Au NRs in the presence of cetyltrimethyl ammonium bromide (CTAB) within 90 seconds. This led to a blue shift of LSPR peak and a colorful transition from brown (Au NRs) to pink (Au NSs). The colorless solution indicates the thorough oxidization of Au NRs into AuBr2− and the yellow color at even higher TMB2+ concentrations is due to excess TMB2+. The multichromatic response allows for semi-quantitative detection of carcinoembryonic antigen and PSA with naked eyes with low LODs at ∼2.5 ng mL−1 and ∼75 pg mL−1, respectively. The approach can be adapted to other biomarkers with such a simple add-on step, which changes a monochromic response from the traditional colorimetric assay to a multicolor display.
In plasmonic-coupling-based colorimetric sensors, analytes can modulate aggregation or dispersion of plasmonic NPs, generating a colorimetric response. The reported plasmonic NPs include Au NSs,796 Au NRs,797 AuAg nanoshells,782 and Ag-coated Au NRs.783 The plasmonic-coupling-based colorimeter sensors have been reported to detect various targets, including metal ions,798 proteins,796 DNA,799 bacteria,800 and viruses. A common approach is to modulate the weak interaction between high-affinity ligand-modified nanoparticles. For example, an HRP-mediated, iodide-catalyzed cascade reaction was reported to control the oxidation of cysteine to cystine and the following dispersion/aggregation of Au NPs for the detection of proteins.796 An enzyme-free colorimetric sensor was based on the release of cysteine from liposomes and subsequent aggregations of Au NPs.800 This approach enabled the visual detection of single-digit bacteria and target antibodies at an attomolar concentration. Plasmonic NPs can be functionalized with an aptamer, DNAzyme, or probe DNA, which changes color after complementary DNA is added to initiate DNA hybridization and NP aggregation.799,801 Ye et al. demonstrated specific and sensitive naked-eye detection of SARS-CoV-2 RNA with a low limit of detection at 10 copies per reaction and a ∼75-min assay time (Fig. 29c, i).782 The RNA was reverse transcribed and amplified with loop-mediated isothermal amplification. The amplicons were then prepared into short repeats in oligonucleotides. For colorimetric readout, the oligonucleotide-functionalized AuAg alloy nanoshells were introduced, and a color change from red to yellow was induced by the agglomeration of the plasmonic nanoshells via DNA hybridization within 10 min at 65 °C. The AuAg nanoshells as plasmonic sensors show 4 times stronger extinction and 20 times lower limit of detection compared to gold counterparts. Creyer et al. constructed a colorimetric assay using core–shell AuAg NPs for trypsin detection.783 In this application, the authors investigated three types of nanostructures, including Ag-coated Au NRs with orange slice shape, Au-coated Ag NSs with spherical shape, and Ag-coated Au NRs with nanocuboid shape. Cysteine-containing peptides (Gly-Cys-Gly-Lys-Gly-Cys-Gly) were used to mediate the aggregation of NPs, producing a color response from orange to blueish grey (Fig. 29c, ii). In the presence of trypsin, the crosslinker is cleaved, and the aggregation of the NPs is attenuated. Among the three nanostructures, the orange slice-like Ag-coated Au NRs showed the strongest colorimetric response and provided a low LOD of trypsin at ∼0.5 nM.
Plasmonic colorimetric sensors have the potential to detect a wide range of analytes, including ions, metabolites, proteins, bacteria, and viruses, at point-of-care and resource-limited settings. Colloidal colorimetric sensors rely on chemically synthesized Au and Ag NPs, which are low-cost and easy to scale up compared to lithography-fabricated plasmonic NPs on substrates. To further reduce material cost, plasmonic NPs made of other materials, such as copper, aluminum, and magnesium, can be considered.802 Surface functionalization plays an important role in achieving the desired sensitivity, specificity, and stability of colorimetric sensors. The growth-based and etching-based colorimetric sensors offer better stability and reproducibility than the plasmonic coupling-based approach. Further improvement can be realized by immobilizing nanomaterials on low-cost substrates, such as paper and foam, which enables highly uniform nanostructure immobilization and easy integration with a microfluidic platform.803–811 The strategy to enable multiplexed detection with a small volume of samples is useful for many applications, such as disease diagnosis and health monitoring.811–813 The specificity of colorimetric sensors needs vigorous testing against potential interferents, considering the complexity of real samples in targeted applications. To achieve specificity, many sensors utilize antibodies and enzymes, which have limited stability at room temperature and under harsh environmental conditions. Stable biorecognition elements, such as molecular imprinted polymers and aptamers, can be utilized to address this challenge.814–816 Alternatively, preservation materials, such as glycerol, organosiloxane, silk, and metal–organic framework, can be deployed to enhance biomolecule stability and prolong sensor shelf life.666,817–819 Among these, the best-performing materials require removal to make biomolecules accessible for specific interaction with targets, which complicates the implementation in colorimetric sensor operation. There is still a need to develop superior materials to provide robust preservation capability, which can be easily implemented in high throughput and low cost. Another challenge is to detect analytes by differentiating the changes in color wavelength and intensity with the naked eye. A promising solution is to use a smartphone camera, which is widely available, with image-processing algorithms to achieve robust detection and quantification of targeted analytes.
5.4. Lateral flow assays (LFAs)
LFAs are another type of plasmonic NP-based sensor whose response is also based on colorimetry.820–822 LFAs are paper-based biosensors in which the sample flows by capillarity, dragging with it the nanoparticles to the areas where the recognition takes place, the test and control lines (TL, CL). In fact, these types of biosensors are an evolution of paper chromatography823 and the technology behind well-known products as the pregnancy test. LFAs are ideal for point-of-care (POC) testing,74 where the patient is located, either in the hospital (diagnostics), at home (it grants the patient a plus of privacy) or even in the field (environmental sensing). The qualities that make LFAs so suitable for POC are their portability, easiness of use (no special training is required, and any person can use them) and the fact that they are completely battery and equipment-free. In addition, LFAs are very affordable and sustainable, since the main component is, as we said, paper (cellulose and nitrocellulose). The common design of a LFA is a paper-strip composed of 4 different pads,824,825 as illustrated in Fig. 30. A sample pad is generally composed of cotton fiber (cellulose), although other materials such as polyester, polypropylene and glass fiber can be also employed, and it is where the sample is dropped (Fig. 30a).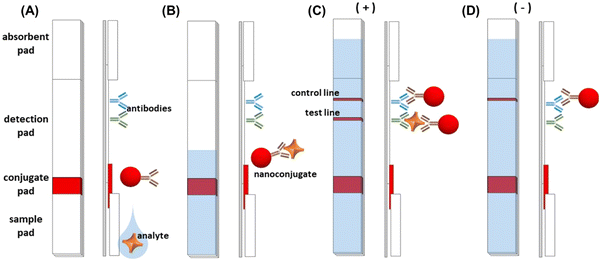 | ||
| Fig. 30 Illustration of a LFA and its working principle. (A) Components of a LFA paper strip (front and side views). (B) Sample flowing across the paper strip. (C) Positive response, nanoconjugate reaches TL and CL. (D) Negative response, in absence of the analyte, only CL is visible. Reprinted from ref. 822 with permission from https://www.sensor100.com/Sens_Tech_Dir/lateral-flow/, copyright 2023. | ||
A sample pad is normally pre-treated with salts, which will act as buffer for all the reactions across the strip, and other compounds such as surfactants, that may contribute the fluidic or releasing the sample (e.g. by breaking a viral capsid). The sample will then flow to the conjugate pad (Fig. 30b), commonly made of glass fiber, polyester, or polypropylene, although hybrid materials have been emerging during the last years (e.g. Fusion 5).826 In the conjugate pad there is a nanoconjugate (NP conjugated with a bioreceptor, e.g. an antibody or a DNA strain, that specifically recognizes the target analyte) dry stored that will be rehydrated and dragged by the flow. The nanoconjugate captures the target analyte, if present in the sample, and flow together throught the detection pad. The detection pad is made of nitrocellulose on a plastic support. The two parameters of this material that are key for the assay are: the pore size of the paper, which will control the flow speed (being s/4 cm a common flow unit given by nitrocellulose manufacturers), and the affinity for the proteins (related to the amount of nitro groups on the nitrocellulose structure) which can be tuned by pre-treating the material with surfactants or proteins. In the detection pad there are the TL and CL are in the detection pad (Fig. 30c). When color is visible in the TL the response is positive (the analyte is present on the sample). TL consists of a bioreceptor able to capture the analyte, which at the same time is attached to the nanoconjugate, thus in a positive sample a “sandwich” recognition takes place (TL bioreceptor – analyte – nanoconjugate bioreceptor – nanomaterial). CL indicates if the assay is working properly. Thus, CL presents a printed bioreceptor that targets the bioreceptor of the nanoconjugate. The absence of CL may indicate a failure in the assay therefore considering the result null. If the sample is negative (Fig. 30d) there will be no recognition on TL and thus only CL will be visible. The methodology described follows the “standard” or “direct” LFA mechanism, however, other systems employ “competitive” or “indirect” strategies, not discussed within this review, which are less reported but useful for cases in which the analyte is too small and the “sandwich” recognition may not work due steric hindrances between the bioreceptors.820
LFAs can be employed for the recognition of different types of analytes, from proteins and hormones,827 to allergens in food,828 or dust,829 bacteria,830 DNA,831 extracellular vesicles,832 and even heavy metals;833 practically any type of analyte as long as there is a bioreceptor able to carry on the recognition. Actually, LFAs can be used for multiplexing, detecting several analytes at the time by placing several TL on the strip, or a dots array instead.834,835
There exist several strategies that can be carried out using nanomaterials to enhance the colorimetric signal,842,843 such as increasing the ratio of nanoparticles per unit of analyte (which can be done by combining nanoparticles together to increase the plasmonic intensiry,844,845 or by reducing the number of bioreceptors on the surface of non-spheroidal nanoparticles,846 so each nanoparticle captures a minimum amount of target analyte units), exploring novel nanomaterials with stronger plasmonic response (e.g. C dots847 or IrO2 NPs848) and even fluorescent properties,849 using nanomaterials to modify the paper substrate properties,850,851 relying on secondary reactions to enhance the colorimetric signal (e.g. nanozymes: mimicking enzymatic reactions using NPs),852,853 modifying paper architecture (altering the flow across the strip)854,855 or seeking for novel sensing techniques, as thermochromism.856 Some selected examples are explained below with more detail.
NP clusters (nanoclusters) exhibit higher sensitivity levels than single particles on LFAs,848 however working with them can be hard since they easily tend to aggregate. Oh et al. synthetized colour-preserved Au NPs clusters stable against aggregation for LFAs.857 In their work, they compared different clusters selecting those of 40 nm diameter Au NPs, in which the particles remained stably separated with a gap around 15 nm thanks to streptavidin and biotinylated antibodies (Fig. 31a). Their LFA exhibited a LoD of 0.038 ng mL−1 for the SARS-CoV-2 nucleocapsid protein; this is around 4000 times lower than a non-enhanced LFA for a model protein (e.g. human IgG).850 It must be noted that authors may use different methods to calculate LOD values, so the comparison between systems is not always “precise”. We recommend calculating the LOD from the equation of the calibrate, e.g. f(x) = a + b·log(x), considering as “f(x)” the value of blank plus 3.3 times its standard deviation and then isolating from the equation the concentration parameter “x” as the LoD value.858
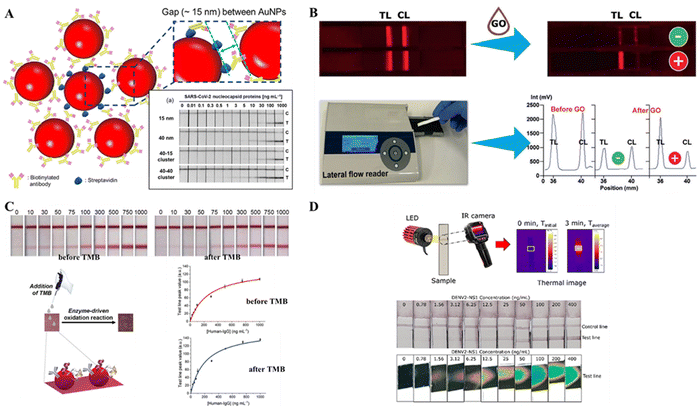 | ||
| Fig. 31 (A) Streptavidin-stabilized Au NPs nanoclusters for LFAs. Reproduced from ref. 857 with permission from Elsevier, copyright 2022 (B) Fluorescent response observed on QDs-based LFAs in which the signal is quenched by graphene. Reproduced from ref. 859 with permission from Elsevier publisher, Copyright 2019. (C) Enzymatic reaction using an artificial enzyme to enhance LFAs response. Reproduced from ref. 852 with permission from Wiley-VCH publisher, Copyright 2023 (D) Au NPs being heated on a LFAs and thermochromically measured. Reproduced from ref. 856 with permission from RSC publisher, Copyright 2023. | ||
Graphene has the property to quench (turn-off) the fluorescence of several nanomaterials (e.g. nanoclusters860 and quantum dots,861 QDs), and this property can be also applied inside the LFAs.859 Morales-Narváez et al.861 and Hassan et al.859 took advantage of this effect and designed a system very different from conventional LFAs: they immobilized QDs on TL and CL. QDs were conjugated to the antibodies recognizing the target (the bacterium Escherichia coli in both cases), and bare QDs were immobilized in CL. After the bacteria has been added to the strip, the graphene solution is dropped and quenches the fluorescence of QDs, except if bacteria are there (in the TL) since it generates a gap between QDs and graphene, impeding their quenching. Thus, if the sample is negative, all the fluorescence, both TL and CL, are quenched. If the sample is positive, after adding graphene only the TL should be visible (Fig. 31b). The response can be quantified with a table fluorescence reader,862 reaching a LOD as low as 105 CFU mL−1, only 100 times greater than commercial ELISA kits.863
As previously mentioned, another way to enhance the signal on LFAs is by carrying out enzymatic colorimetric reactions on the nanomaterials, which may act as nanozymes.853 However, in the work of Renzi et al. they immobilized an artificial metalloenzyme (a.k.a. mimochrome) called “FeMC6*a” on Au NPs,852 claiming its advantages over nanozymes. In the assays, nanozymes may lead to some irreproducibility issues due to little variations in NPs shape and size can provoke variations in the catalytic reactions. Instead, metalloenzyme behaviour imitates the molecular level behaviour of enzymes, being able to replicate native enzyme functions with greater reproducibility.864 So, in the work of Renzi et al., FeMC6*a was immobilized on Au NPs and, after running the assay as a standard LFA, a substrate (3,3′,5,5′-tetramethylbenzidine, TMB) was added to the strip that, when oxidized by the metalloenzyme, increased the colour intensity of the TL (Fig. 31c). With this method the LOD of the assay was decreased from 36.4 to 8.2 ng mL−1.
As a last example, Trakoolwilaiwan et al. explored different nanomaterials for their application in thermochromic LFAs, as a new signal quantification method.856 It must be highlighted that the orientation of plasmonic NPs has a great impact on the ability to be heated, since the closer the particles can be, the greater the heat transmission. Then, as they found, spherical Au NPs exhibited greater response when heated by LED light and measured by an IR camera (Fig. 31d), compared to other structures such as NRs, which difficult the contact among the nanomaterials. They observed that the working range of their LFAs did not differ too much from conventional optical LFAs. However, the sensitivity of the assay (the capacity to discern between close values, i.e. the slope of the calibration plot) was greater, allowing a better quantification. Thermal measurements on LFAs have reported up to an 8-fold increase in the sensitivity of the assays.865
6. Chiroptical sensing
Chirality is a universal phenomenon observed at various length scales ranging from small molecules, NPs, and large structures.876,877 Of special interest has been the chiral signatures in amino acid units, which are the building blocks of proteins. Protein chirality can further extend to complex structural units which in turn has resulted in the induction of optical activity into other biomolecular systems such as DNA and RNA. Due to the rich abundance of optically active species in living systems, the development of sensing platforms dependent on chiroptical spectroscopy can be a powerful tool for the detection of biologically relevant molecules.878 Realizing these facts, different research groups have focused their attention towards the development of chiroptical sensors that are relevant to the detection of various diseases and biomarkers.879,880Among different classes of biosensors that rely on chiroptical spectroscopy, the ones based on chiral plasmonic nanomaterials, owing to their ability to focus light onto nanoscale dimensions,881,882 have emerged as promising candidates for applications in the field of biomedical research.883,884 Most sensors rely on circular dichroism (CD) which measures the differential absorption of left- and right-circularly polarized light. In this regard, plasmonic nanostructures, due to their large absorption and scattering cross sections, can generate intense CD signals. Recent advances in synthetic protocols have offered versatile avenues for the fabrication of a wide range of nanomaterials.885 Efforts in this direction have resulted in the fabrication of chiral nanostructures with enhanced sensing capabilities.884 Detection limits as low as attomolar and zeptomolar concentrations have been achieved rendering this class of sensors extremely relevant in the field of forensic science, drug industry, and biomedical applications, areas that demand very high sensitivities.879,880 Moreover, these nanomaterials generate chiral plasmonic signals in the visible and NIR regions of the EM spectrum, a spectral range that is clear from the interference of biomolecule and solvent signals.
The interaction of plasmonic NPs with chiral analytes can generate plasmonic CD signals that can be used as a biosensing platform. Depending on the nature of interactions and the mechanism of chiral induction, plasmonic chirality for biosensing applications can be broadly divided into two classes; (i) assembly of achiral NPs on the chiral templates leading to the generation of coupled plasmonic CD signals137,886–891 and (ii) generation of induced plasmonic CD through the interaction of chiral molecules with the plasmonic field of achiral NPs.892–898 The following section will discuss a few examples wherein plasmonic CD signals generated through these two approaches are used for sensing of biologically relevant analytes.
6.1. Template-assisted NP assemblies
Plasmonic NPs, due to their unique LSPR properties that enable the trapping of light into nano-dimensions, experience larger chiroptical responses compared to their molecular counterparts. However, self-assembly of these NPs into chiral geometries, results in plasmonic coupling between adjacent NPs leading to intense chiroptical responses that aid sensing of the analytes at extremely low concentrations.137,880 One of the common techniques to fabricate chiral-shaped nanostructures and their assemblies is electron-beam lithography. However, the technique is limited by scalability and high cost, and hence we will focus our discussion on the various solution-processed bottom-up approaches developed for the synthesis of chiral NP assemblies. DNA, RNA, peptides, proteins, and other biomolecular assemblies act as robust templates for the assembly of NPs, and some of the relevant examples are discussed in the following section.137,886–890Initial studies on the generation of plasmonic chirality relied on the use DNA and peptides, even though the objective then was not on the biosensing.886,887 Thomas and co-workers, in one of the initial reports, used self-assembled peptides to obtain mirror image plasmonic CD signals from Au NPs attached to opposite isomers of the peptide surface.887 Since then, there has a large number of reports on generation and tuning of plasmonic chirality using a variety of templates. Recent efforts in this direction have focused on finding applications for nanohybrid systems, with biosensing is at the forefront of all of these endeavors. Liz-Marzan and co-workers utilized the assembly of NRs on the protein fibrils formed from α-synuclein for the detection of Parkinson's disease (Fig. 32a).137 The chiral nature of the fibrils facilitated helical assembly of NRs which in turn generated plasmonic CD signals in the visible and NIR region (Fig. 32b). Fibrillation of protein being a hallmark of neurodegenerative diseases, the chiral plasmonic signals could be used for the detection of Parkinson's disease. Kuang and co-workers developed a DNA-based detection platform for the sensing of mycotoxin, namely, ochratoxin A (OTA).888 Chiral satellite superstructures synthesized using core–shell Ag@Au NPs (functionalized with OTA aptamers) and Au NSs (functionalized with complementary sequence of OTA aptamers) exhibiting intense negative CD signal at 530 nm was employed as the plasmonic platform. Upon addition of OTA, the superstructures disassembled leading to a gradual reduction in the CD signals as a function of OTA concentration. The linear fit could be used for the qualitative and quantitative detection of OTA. In different approach, Liedl and co-workers used DNA origami template to generate switchable plasmonic chirality that could detect specific RNA at picomolar concentrations.889
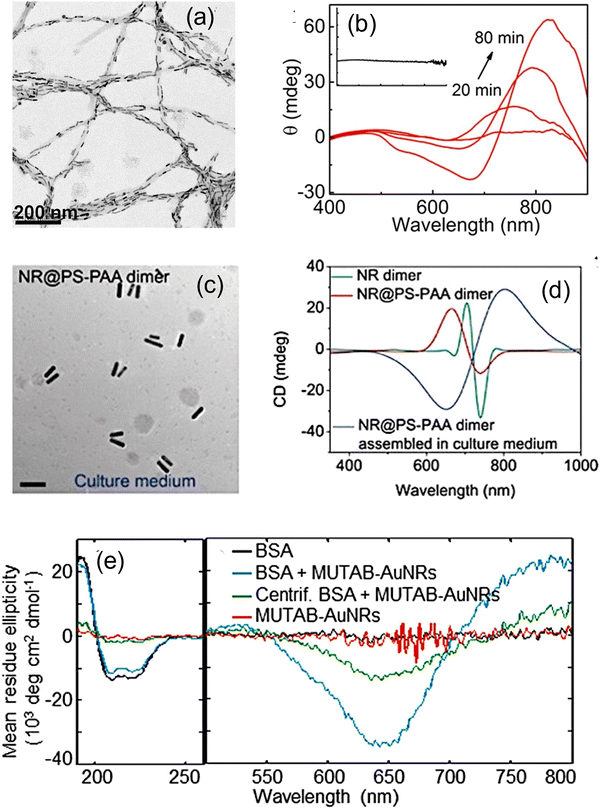 | ||
| Fig. 32 Detection of Parkinson's disease. (A) TEM image depicting the assembly of Au NRs on protein fibrils and (B) the corresponding CD spectra showing an enhancement as the assembly progresses. Spectra in the inset shows no CD for NRs in the native protein sample. Reproduced from ref. 137 with permission from PANAS publisher, Copyright 2018. (c,d) Detection of miRNA. (C) TEM image of NRs encapsulated by PS-PAA in culture medium (scale bar = 100 nm). (D) CD spectra side-by-side assembled dimers of bare Au NRs (green trace) and NRs encapsulated by PS-PAA in PBS buffer (red trace) and culture medium in the presence of 100 pM of synthetic microRNA-21 (right). Reproduced from ref. 890 with permission from Wiley publisher, Copyright 2018 (E) Unfolding of BSA upon adsorption to MUTAB-AuNRs. CD spectra of BSA (black trace), MUTAB-AuNRs alone (red trace), BSA/MUTAB-AuNRs (blue trace), centrifuged BSA/MUTAB-AuNRs (green trace) Reproduced from ref. 891 with permission from ACS publisher, Copyright 2016. | ||
Xu and co-workers used the side-by-side assembly of Au NRs for the detection of microRNA (miRNA). DNA functionalized Au NR dimers were further stabilized with poly (styrene-b-acrylic acid) (PS-PAA) for intracellular stability and biocompatibility.890 miRNA driven side-by-side self-assembly of the PS-PAA-coated Au NRs in living cells generated distinct plasmonic CD signals that helped the in vivo detection of miRNA in picomolar concentrations (Fig. 32c and d). BSA detection through the aggregation of mercaptoundecyltrimethylammonium bromide (MUTAB)-coated Au NRs was demonstrated by Link and co-workers.891 Adsorption of BSA on the NP surface led to the unfolding of the protein. Subsequent interactions between the unfolded proteins drive the NP aggregation resulting in plasmonic CD signals (Fig. 32e). Au NR aggregation at nanomolar concentrations of BSA facilitates the use of plasmonic platforms for biosensing applications. Kotov and co-workers studied the cell internalization of NPs using plasmonic CD signals from assembled NR dimers.892 DNA-bridged chiral NP dimers upon introduction to mammalian cell media showed a variation in the magnitude and sign of CD signal for NPs internalized in the cell. The CD reversal was attributed to the spontaneous twisting motion around the DNA bridge due to changes in the electrostatic repulsion between NP in a dimer during the transmembrane transport. There are several reports on similar NP assemblies being used for applications such as drug screening, and detection of other biologically relevant analytes. The enhancement in signal intensity during the assembly process makes this approach an attractive tool for the highly sensitive detection of analytes.
6.2 Induced plasmonic chirality as a biosensing tool
Another approach for the generation of plasmonic CD signals is through the Coulombic interaction between a chiral molecule and an achiral NP. While there are different classifications to such an effect, we restrict our discussions to the plasmon-coupled circular dichroism phenomenon which deals with the chiral induction into nanoantenna by a chiral molecule through dipolar or multipolar interactions. Govorov et al. framed detailed theoretical formulations for these effects and attributed the origin of plasmonic chirality to different mechanisms such as far-field EM coupling, near-field interactions, and orbital hybridization.893 The initial investigations in this direction were carried out by Naik and co-workers who created optically active Au NPs through their interaction with random coil and α-helix peptides as chiral analytes.894 The peptide–Au NP interactions produced plasmonic CD at the LSPR of Au NPs. Later, the same effect was widely used for the generation of plasmonic CD signals in achiral NPs that were coupled with a wide variety of chiral molecules, such as DNA, peptides, proteins, and dye molecules.Markovich and co-workers used a chiral molecule riboflavin to induce chiral plasmonic signals into Au islands deposited on a glass slide.895 While the Au islands coated with the achiral polymer, poly (methyl methacrylate) (PMMA), did not exhibit any CD signals, riboflavin-embedded PMMA when coated onto the Au islands showed CD peaks in the plasmonic region (Fig. 33a). They could further demonstrate the distance dependence of the same effect by coating an achiral polyelectrolyte spacer between the Au island and the chiral layer. Enhanced plasmonic CD signal in achiral core–shell Au@Ag NCs was achieved through DNA attachment by Gang and co-workers.899 The greater sensitivity was attributed to the lower plasmon loss and stronger LSPR enhancement of Ag compared to Au in the visible region of the spectrum. There have been several attempts to obtain higher sensitivity by modifying the composition as well as the morphological features of the synthesized nanostructures. Attempts in this direction have led to a unique phenomenon termed as hotspot enhanced plasmonic chirality.
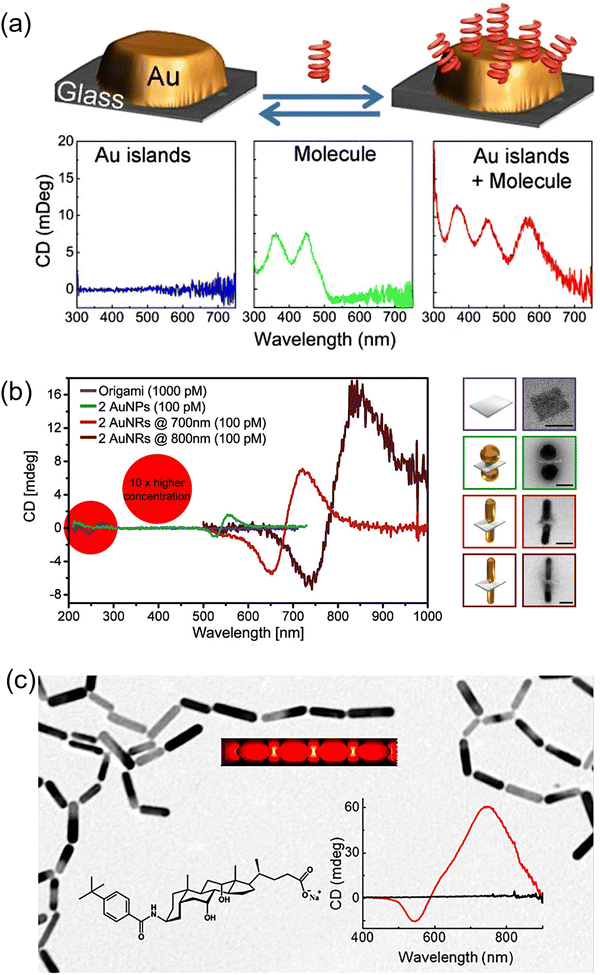 | ||
| Fig. 33 (A) Induced chirality in Au islands. CD spectra of bare Au islands (blue trace), riboflavin molecule (green trace), and Au Island with riboflavin embedded in PMMA (red trace). Reproduced from ref. 895 with permission from ACS publisher, Copyright 2013. (B) Hotspot-enhanced CD in DNA origami-assembled Au NP dimers. Compared to the bare DNA origami template (purple), 30-fold and 300-fold amplified CD is observed for Au NP dimers (green trace) and Au NR dimers (red and brown traces). Scale bars = 40 nm. Reproduced from ref. 897 with permission from ACS publisher, Copyright 2018 (C) Hotspot enhanced CD induction in linearly assembled Au NRs. Bile salt derivative acts as the linker for the linear assembly of NRs Reproduced from ref. 898 with permission from ACS publisher, Copyright 2020. | ||
The theoretical formulations for hotspot-enhanced plasmonic chirality were first provided by Zhang and Govorov on Ag nanosphere dimer with varying gap space.896 Later, Liedl and co-workers experimentally verified this effect on Au NP dimers assembled using DNA-origami.897 They could observe orders of magnitude CD enhancement which could be attributed to the high field created at the hotspots of NPs (Fig. 33b). Recently, Severoni et al. demonstrated the hotspot-driven chiral enhancement in linearly assembled Au NRs wherein chiral bile salt derivative was used as the linker (Fig. 33c).898 This peculiar effect has the potential to develop into an efficient platform reaching up to single-molecule detection. However, the major challenge is to precisely position the analytes at the plasmonic hotspots. In short, it can be summarized that the advancement in the field of NP synthesis has boosted the potential of chiral plasmonic detection platforms to a great extent. However, a lot needs to be explored in understanding the analyte structural/conformational changes and their interaction with the NPs at the molecular level. Further experimental and theoretical investigations can help understand these fundamental effects better. Future challenges also lie in demonstration in in vivo experiments and transitioning the detection tool for application in the market.
7. Outlook
Plasmonic NP-based sensors have emerged as formidable tools across a diverse array of applications. Even before a comprehensive understanding of their colorimetric properties, these NPs were used as colorimetric sensors, gradually evolving into practical applications such as LFIAsLFIAs, now commonplace in-home diagnostics. Subsequent comprehension of the principles governing their colorimetric behavior, coupled with advancements in SPR technologies, opened new vistas for plasmonic sensors. However, the discovery of SERS catalyzed an exponential surge in the development of new plasmonic nanoparticle sensors.Despite the significant advantages offered by surface-enhanced characterization techniques, including theirtheir non-invasive nature, real-time monitoring capabilities, and multiplexing potential, theirtheir adoption outside academia has been relatively limited, failing to transition seamlessly into industrial settings. Numerous studies showcasee the exceptional detection limits and selectivity of plasmonic NP-basedbased sensors across various applications, yet translating this technology into practical use faces several hurdles.
Foremost among these challenges is the development of commercial plasmonic NP-based sensors sensors that meet industrial requirements: reproducibility, stability, and cost-effectiveness. Recent strides in this area have been promising, with notable progress achieved in fabricating nanostructures withwith excellent quality and tailored morphologies. In some instances, these advancements have led to the creation of nanostructures with smaller, more precisely engineered 2D and 3D nanogaps. Such innovative fabrication strategies and techniques have facilitated the rapid, cost-effective, and reproducible large-scale production of substrates and nanotags exhibiting robust signal responses, suitable for both general-purpose and target-specific sensing applications. However, further investigation is needed in areas such as the development of flexible plasmonic nanoparticle substrates that facilitate on-site analyte collection and transportation for later characterization, as well as their integration into small devices or even wearable devices. Another necessary improvement is finding a way to reuse the plasmonic substrates without a loss in efficiency. Significant advances have been made by applying UV light or plasma etching, but further investigation is required.
For plasmonic SERS sensors, a significant obstacle lies in the interpretation of SERS spectra. Despite theirtheir immense potential, the complex and intricate nature of SERS spectra often poses challenges in accurately deciphering and extracting meaningful information. The inherent variability in spectra associated with substrate fabrication arisesarises from factors such as nanoparticle size, shape, composition, and hotspot distribution, as well as the influence of the surrounding environment, complicatinging spectral analysis. Furthermore, the presence of spectral features originating from multiple analytes,contaminants, or plasmonic NP ligands further exacerbates the issue, confounding the identification and quantification of target molecules. Addressing these challenges requires innovative approaches, and one promising avenue is the integration of artificial intelligence or machine learning protocols. Machine learning algorithms offer a powerful means to navigate the complexity of SERS spectra, enabling the extraction of valuable insights and facilitating the identification of relevant spectral features. By leveraging advanced data processing techniques, machine learning algorithms can discern subtle patterns within spectra, discriminate between different analytes, and mitigate the effects of spectral variability. Moreover, machine learning models can adapt and improve over time, refining their predictive capabilities and enhancing the accuracy of spectral interpretation. Integrating machine learning protocols into SERS-based sensing platforms holds immense potential for enhancing analytical performance and expanding the scope of applications. By harnessing the complementary strengths of machine learning and SERS spectroscopy, researchers can unlock new opportunities for sensitive, selective, and reliable detection across diverse fields, ranging from biomedical diagnostics to environmental monitoring and beyond. Moreover, the synergistic combination of these technologies paves the way for developingdeveloping autonomous sensing systems capable of real-time, on-site analysis, thereby revolutionizing traditional analytical workflows and empowering users with actionable insights in various settings.
On the other hand, SEF applications also benefit from the implementation of machine learning algorithms to filter background noise and autofluorescence as well as to separate overlapping spectral features from the fluorophores and the metallic nanostructures, leading to clearer and more accurate fluorescence signals. However, the major limitation in this case is controlling the interaction between plasmonic nanoparticles and fluorophores to overcome the quenching effect. Controlling the distance between the fluorophore and the metal surface requires precise control and still needs further investigation.
Plasmonic colorimetric sensors require NP surface functionalization to achieve the desired sensitivity, specificity, and stability. Their major limitation is specificity since, considering the complexity of real samples in targeted applications, they can suffer from potential interferents. To avoid this limitation, many sensors utilize antibodies and enzymes, which have limited stability at room temperature and under harsh environmental conditions. However, this challenge continues to need addressing through new and more complex functionalization options. In the particular case of LFA, apart from finding new possible functionalizations for the plasmonic NPs, it is necessary to search for new bioreceptors such as DNA aptamers that could allow the integration of DNA amplification techniques into the LFA protocol. Also, another key parameter for the future improvement of LFAs is the employment of more precise quantification techniques besides the naked eye, such as the combination with SERS spectroscopy.
Chiral plasmonic detection platforms are a relatively new characterization technique that still requires further exploration to understand the analyte structural/conformational changes and their interaction with the NPs at the molecular level. Further experimental and theoretical investigations can help understand these fundamental effects better. In general, one major area for future development is the integration of plasmonic NP-based sensors with modern digital and communication technologies. Combining plasmonic sensors with smartphones, portable devices, and Internet of Things (IoT) platforms could enable real-time, remote, and user-friendly diagnostics. For example, the integration of LFA sensors with smartphones will allow the recording of the test result, guaranteeing its correct interpretation, and even sharing the response with a clinician or specialist if necessary. This integration requires advancements in miniaturization and the development of robust, cost-effective, and easy-to-use sensor kits.
In conclusion, plasmonic NP-based sensors have a bright future with the potential to revolutionize various fields, including medical diagnostics, environmental monitoring, and security. By addressing the current challenges and leveraging technological advancements, these sensors can become indispensable tools for real-time, highly sensitive, and specific detection of a wide range of analytes.
Author contributions:
K. K., L. P., and I. P. S. initiated and coordinated the review. L. P. and I. P. S. edited the manuscript. The manuscript was written through the contributions of all authors. L. P. contributed to the introduction; X. X. and J. W. contributed in different type of plasmonic metal nano crystal; L. P. contributed to the Methods of plasmonic sensing; R. A. A. contributed to the Basics of SERS; T. L. contributed to the Single molecule SERS; K. K. and D. R. contributed to the SESR Sensing platforms; V. R. S., S. S. B. M., and R. B., contributed to the Chemical Sensing Medium; L. G. C. and D. G. L. contributed to the Ionic Species sensing; M. L., R. M., and M. P. P. contributed to the Gas Sensing; P. W., S. M. and V. B., contributed to the SERS-based Nucleic acid detection; B. R., and S. K., contributed to the SERS Protein detection; S. A. C. contributed to the SESR cell Detection; Y. J., Q. Y., and J. C., contributed to the SERS-based bacterial/viral sensors; P. H., and S. G.-G, contributed to the Sensing Reaction progress and intermediates; Y. C., and M. S., contributed to the SERS + Other technologies; S. S., Y. L., and Y. L. contributed to the Surface enhanced fluorescence based plasmonic sensors; X. Y. L., contributed to the SERS Data Analysis; P. S. S., P. L. C., and J. M. M. M. A., contributed to the LSPR; J. L. M. P., and M. G., contributed to the colorimetric sensing; H. G., and L. T., contributed to the colloidal colorimetric sensing; D. Q. G., and A. M., contributed to the LFAs; J. K., and S. M., contributed to the Chiroptical; J. P. J and I. P. S., contributed to the Outlook. All authors read the manuscript and have approved the final version of the manuscript.Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Conflicts of interest
The authors declare the following competing financial interest(s): Srikanth Singamaneni is an inventor on a pending patent related to plasmonic-fluor technology, and the technology has been licensed by the Office of Technology Management at Washington University in St. Louis to Auragent Bioscience LLC. He is a co-founder/shareholder of Auragent Bioscience LLC. He along with Washington University, may have financial gain through Auragent Bioscience, LLC through this licensing agreement.Acknowledgements
Krishna Kant acknowledge European Union's Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement no. 894227. Lara González-Cabaleiro acknowledge Xunta de Galicia for a predoctoral scholarship (Programa de axudas á etapa predoutoral da Consellería de Cultura, Educación e Universidades da Xunta de Galicia, reference number: 2022/294). Heng Guo and Limei Tian acknowledge the funding from the National Science Foundation (Grant No: 1648451), and the National Institutes of Health (Grant No: R21EB029064 and R35 GM147568). Ramon A. Alvarez-Puebla acknowledges support by the projects PID2020-120306RB-I00 (funded by MCIN/AEI/10.13039/501100011033), PDC2021-121787-I00 (funded by MCIN/AEI/10.13039/501100011033 and European Union Next Generation EU/PRTR), 2020SGR00166 (funded by Generalitat de Cataluña) and 2021PFR-URV-B2-02 (funded by Universitat Rovira i Virgili). Maria P. Pina acknowledges support from: the European Union's Horizon 2020 research and innovation program under grant agreements No. 883390 (H2020-SU-SECU-2019 SERSing Project), No. 823895 (H2020-MSCA-RISE-2018 SENSOFT), AEI Spain (PID2019-108660RB-I00). Marta Lafuente, thanks Ministerio de Universidades (Spain) and Next Generation EU for the postdoctoral grant “Margarita Salas”. Rizia Bardhana and Siddhant Kothadiya acknowledge support from the National Institutes of Health (NIH) award R01EB029756-01A1 and congressionally directed medical research program (CDMRP) award W81XWH-20-1-0620. Jaebum Choo thanks the financial support from the National Research Foundation of Korea (Grant numbers 2020R1A5A1018052 and RS-2024-00352256). Daniel Quesada-González and Arben Merkoçi acknowledge that the ICN2 is funded by the CERCA programme/Generalitat de Catalunya. The ICN2 is supported by the Severo Ochoa Centres of Excellence programme, Grant CEX2021-001214-S, funded by MCIN/AEI/10.13039.501100011033 and Grant PID2021-124795NB-I00 funded by MICIU/AEI/10.13039/501100011033 and by “ERDF/EU” and Departament de Recerca i Universitats of Generalitat de Catalunya for the grant 2021 SGR 01464. Sergio Gómez-Graña acknowledges support by the projects PID2020-117371RA-I00, TED 2021-131628A-I00 and CNS2022-135531 funded by MCIN/AEI/10.13039/501100011033. Soma Venugopal Raoexpresses thanks to Defence Research and Development Organization, India, for the financial support received through ACRHEM [#ERIP/ER/1501138/M/01/319/D(R&D)] and the financial support from the Institute of Eminence (IoE) [ref. No. UOH/IOE/RC1/RC1-20-016]. The IoE project was awarded to the University of Hyderabad by the Ministry of Education, Government of India, per the MHRD notification F11/9/2019-U3(A). Sara Abalde-Cela acknowledges supported by the 3DSecret project, funded by the EU under the program HORIZON-EIC-2022-PATHFINDEROPEN-01-01 (ga 101099066) and by the UK Research and Innovation (UKRI) under the UK government's Horizon Europe funding guarantee (ga 10063360). José Luis Montaño-Priede acknowledges the financial support received from the IKUR Strategy under the collaboration agreement between the Ikerbasque Foundation and Materials Physics Center on behalf of the Department of Education of the Basque Government. Marek Grzelczak acknowledges financial support from EITB Maratoia. Srikanth Singamaneni acknowledges National Science Foundation (CBET-2316285, CBET-2224610) and National Institutes of Health (R21AI178217). L. P. acknowledges support from the Spanish Ministerio de Ciencia e Innovación through Ramón y Cajal grant (grant no. RYC2018-026103-I), the Spanish State Research Agency (grant no. PID2020-117371RA-I00 and TED2021-131628A-I00), and a grant from the Xunta de Galicia (grant no. ED431F2021/05). The authors acknowledge the Universidad de Vigo/CSIUG for open access funding. Jorge Pérez-Juste and Isabel Pastoriza-Santos acknowledge support from MICIU/AEI/10.13039/501100011033 and ERDF/EU (Grant Number: PID2022-138724NB-I00) and the European Innovation Council (Horizon 2020 Project Number: 965018—BIOCELLPHE).References
- C. R. Lowe, Trends Biotechnol., 1984, 2, 59–65 CrossRef CAS.
- S. M. Borisov and O. S. Wolfbeis, Chem. Rev., 2008, 108, 423–461 CrossRef CAS PubMed.
- N. J. Ronkainen, H. B. Halsall and W. R. Heineman, Chem. Soc. Rev., 2010, 39, 1747 RSC.
- L. Su, W. Jia, C. Hou and Y. Lei, Biosens. Bioelectron., 2011, 26, 1788–1799 CrossRef CAS PubMed.
- S. Balbinot, A. M. Srivastav, J. Vidic, I. Abdulhalim and M. Manzano, Trends Food Sci. Technol., 2021, 111, 128–140 CrossRef CAS.
- J. R. Mejía-Salazar and O. N. Oliveira, Chem. Rev., 2018, 118, 10617–10625 CrossRef PubMed.
- M. Ahmed, M. O. Mavukkandy, A. Giwa, M. Elektorowicz, E. Katsou, O. Khelifi, V. Naddeo and S. W. Hasan, npj Clean Water, 2022, 5, 12 CrossRef CAS.
- B. Brunekreef and S. T. Holgate, Lancet, 2002, 360, 1233–1242 CrossRef CAS PubMed.
- J. Homola, Chem. Rev., 2008, 108, 462–493 CrossRef CAS PubMed.
- K. M. Mayer and J. H. Hafner, Chem. Rev., 2011, 111, 3828–3857 CrossRef CAS PubMed.
- O. Tokel, F. Inci and U. Demirci, Chem. Rev., 2014, 114, 5728–5752 CrossRef CAS PubMed.
- L. Song, J. Chen, B. B. Xu and Y. Huang, ACS Nano, 2021, 15, 18822–18847 CrossRef CAS PubMed.
- J. Langer, D. Jimenez de Aberasturi, J. Aizpurua, R. A. Alvarez-Puebla, B. Auguié, J. J. Baumberg, G. C. Bazan, S. E. J. Bell, A. Boisen, A. G. Brolo, J. Choo, D. Cialla-May, V. Deckert, L. Fabris, K. Faulds, F. J. García de Abajo, R. Goodacre, D. Graham, A. J. Haes, C. L. Haynes, C. Huck, T. Itoh, M. Käll, J. Kneipp, N. A. Kotov, H. Kuang, E. C. Le Ru, H. K. Lee, J.-F. Li, X. Y. Ling, S. A. Maier, T. Mayerhöfer, M. Moskovits, K. Murakoshi, J.-M. Nam, S. Nie, Y. Ozaki, I. Pastoriza-Santos, J. Perez-Juste, J. Popp, A. Pucci, S. Reich, B. Ren, G. C. Schatz, T. Shegai, S. Schlücker, L.-L. Tay, K. G. Thomas, Z.-Q. Tian, R. P. Van Duyne, T. Vo-Dinh, Y. Wang, K. A. Willets, C. Xu, H. Xu, Y. Xu, Y. S. Yamamoto, B. Zhao and L. M. Liz-Marzán, ACS Nano, 2020, 14, 28–117 CrossRef CAS PubMed.
- L. Polavarapu and L. M. Liz-Marzán, Phys. Chem. Chem. Phys., 2013, 15, 5288 RSC.
- L. Polavarapu, J. Pérez-Juste, Q.-H. Xu and L. M. Liz-Marzán, J. Mater. Chem. C, 2014, 2, 7460 RSC.
- A. M. Shrivastav, U. Cvelbar and I. Abdulhalim, Commun. Biol., 2021, 4, 70 CrossRef CAS PubMed.
- R. Rahad, A. K. M. Rakib, M. A. Haque, S. S. Sharar and R. H. Sagor, Results Phys., 2023, 49, 106478 CrossRef.
- T. Liyanage, B. Alharbi, L. Quan, A. Esquela-Kerscher and G. Slaughter, ACS Omega, 2022, 7, 2411–2418 CrossRef CAS PubMed.
- F. Jafrasteh, A. Farmani and J. Mohamadi, Sci. Rep., 2023, 13, 15349 CrossRef CAS PubMed.
- M. Beheshti Asl, J. Karamdel, M. Khoshbaten and A. Rostami, Opt. Continuum, 2022, 1, 2043 CrossRef CAS.
- L. Wang, Sensors, 2017, 17, 1572 CrossRef PubMed.
- I. Chirisa, T. Mutambisi, M. Chivenge, E. Mabaso, A. R. Matamanda and R. Ncube, Geochem. J., 2022, 87, 815–828 Search PubMed.
- B. Giri, S. Pandey, R. Shrestha, K. Pokharel, F. S. Ligler and B. B. Neupane, Anal. Bioanal. Chem., 2021, 413, 35–48 CrossRef CAS PubMed.
- D. Ndwandwe and C. S. Wiysonge, Curr. Opin. Immunol., 2021, 71, 111–116 CrossRef CAS PubMed.
- N. W. Schluger, D. Kinney, T. J. Harkin and W. N. Rom, Chest, 1994, 105, 1116–1121 CrossRef CAS PubMed.
- P. Hornbeck, Curr. Protoc. Immunol., 2015, 110, 2.1.1–2.1.23 Search PubMed.
- A. Francisco-Cruz, E. R. Parra, M. T. Tetzlaff and I. I. Wistuba, Biomarkers Immunother., Cancer: Methods Protoc., 2020, 467–495 CAS.
- S. D. Richardson, Anal. Chem., 2009, 81, 4645–4677 CrossRef CAS PubMed.
- X. Huang, Y. Zhu and E. Kianfar, J. Mater. Res. Technol., 2021, 12, 1649–1672 CrossRef CAS.
- A. Chamorro-Garcia and A. Merkoçi, Nanobiomedicine, 2016, 3, 184954351666357 CrossRef PubMed.
- A. K. Yadav, N. Basavegowda, S. Shirin, S. Raju, R. Sekar, P. Somu, U. T. Uthappa and G. Abdi, Mol. Biotechnol., 2024 DOI:10.1007/s12033-024-01157-y.
- T. S. Dhahi, A. K. Y. Dafhalla, S. A. Saad, D. M. I. Zayan, A. E. T. Ahmed, M. E. Elobaid, T. Adam and S. C. B. Gopinath, Biotechnol. Appl. Biochem., 2024, 71, 429–445 CrossRef CAS PubMed.
- F. Achi, A. M. Attar and A. A. Lahcen, TrAC, Trends Anal. Chem., 2024, 170, 117423 CrossRef CAS.
- E. Priyadarshini and N. Pradhan, Sens. Actuators, B, 2017, 238, 888–902 CrossRef CAS.
- K. Saha, S. S. Agasti, C. Kim, X. Li and V. M. Rotello, Chem. Rev., 2012, 112, 2739–2779 CrossRef CAS PubMed.
- J. R. Mejía-Salazar and O. N. Oliveira, Chem. Rev., 2018, 118, 10617–10625 CrossRef PubMed.
- J. Zhao, X. Zhang, C. R. Yonzon, A. J. Haes and R. P. Van Duyne, Nanomedicine, 2006, 1, 219–228 CrossRef CAS PubMed.
- E. Petryayeva and U. J. Krull, Anal. Chim. Acta, 2011, 706, 8–24 CrossRef CAS PubMed.
- K. A. Willets and R. P. Van Duyne, Annu. Rev. Phys. Chem., 2007, 58, 267–297 CrossRef CAS PubMed.
- L. M. Liz-Marzán, Langmuir, 2006, 22, 32–41 CrossRef PubMed.
- N. Zhou, V. López-Puente, Q. Wang, L. Polavarapu, I. Pastoriza-Santos and Q.-H. Xu, RSC Adv., 2015, 5, 29076–29097 RSC.
- X. Lu, M. Rycenga, S. E. Skrabalak, B. Wiley and Y. Xia, Annu. Rev. Phys. Chem., 2009, 60, 167–192 CrossRef CAS PubMed.
- C. M. Cobley, S. E. Skrabalak, D. J. Campbell and Y. Xia, Plasmonics, 2009, 4, 171–179 CrossRef CAS.
- Y. Xia and N. J. Halas, MRS Bull., 2005, 30, 338–348 CrossRef CAS.
- M. Rycenga, C. M. Cobley, J. Zeng, W. Li, C. H. Moran, Q. Zhang, D. Qin and Y. Xia, Chem. Rev., 2011, 111, 3669–3712 CrossRef CAS PubMed.
- J. Kimling, M. Maier, B. Okenve, V. Kotaidis, H. Ballot and A. Plech, J. Phys. Chem. B, 2006, 110, 15700–15707 CrossRef CAS PubMed.
- J. Turkevich, Gold Bull., 1985, 18, 86–91 CrossRef CAS.
- N. R. Jana, L. Gearheart and C. J. Murphy, Chem. Commun., 2001, 617–618 RSC.
- B. Nikoobakht and M. A. El-Sayed, Chem. Mater., 2003, 15, 1957–1962 CrossRef CAS.
- Y. Sun and Y. Xia, Science, 2002, 298, 2176–2179 CrossRef CAS PubMed.
- J. E. Millstone, S. J. Hurst, G. S. Métraux, J. I. Cutler and C. A. Mirkin, Small, 2009, 5, 646–664 CrossRef CAS PubMed.
- L. R. Hirsch, A. M. Gobin, A. R. Lowery, F. Tam, R. A. Drezek, N. J. Halas and J. L. West, Ann. Biomed. Eng., 2006, 34, 15–22 CrossRef PubMed.
- A. Guerrero-Martínez, S. Barbosa, I. Pastoriza-Santos and L. M. Liz-Marzán, Curr. Opin. Colloid Interface Sci., 2011, 16, 118–127 CrossRef.
- I. B. Becerril-Castro, I. Calderon, N. Pazos-Perez, L. Guerrini, F. Schulz, N. Feliu, I. Chakraborty, V. Giannini, W. J. Parak and R. A. Alvarez-Puebla, Analysis Sensing, 2022, 2, e202200005 CrossRef CAS.
- Y. Luo, C. Chi, M. Jiang, R. Li, S. Zu, Y. Li and Z. Fang, Adv. Opt. Mater., 2017, 5(16), 1700040 CrossRef.
- V. K. Valev, J. J. Baumberg, C. Sibilia and T. Verbiest, Adv. Mater., 2013, 25, 2517–2534 CrossRef CAS PubMed.
- J. Dostálek and W. Knoll, Biointerphases, 2008, 3, FD12–FD22 CrossRef PubMed.
- M. Cottat, N. Thioune, A.-M. Gabudean, N. Lidgi-Guigui, M. Focsan, S. Astilean and M. Lamy de la Chapelle, Plasmonics, 2013, 8, 699–704 CrossRef CAS.
- C. A. Mirkin, R. L. Letsinger, R. C. Mucic and J. J. Storhoff, Nature, 1996, 382, 607–609 CrossRef CAS PubMed.
- A. P. Alivisatos, K. P. Johnsson, X. Peng, T. E. Wilson, C. J. Loweth, M. P. Bruchez and P. G. Schultz, Nature, 1996, 382, 609–611 CrossRef CAS PubMed.
- L. Polavarapu, J. Pérez-Juste, Q.-H. Xu and L. M. Liz-Marzán, J. Mater. Chem. C, 2014, 2, 7460 RSC.
- M. Moskovits, Rev. Mod. Phys., 1985, 57, 783–826 CrossRef CAS.
- M. Moskovits, Rev. Mod. Phys., 1985, 57, 783–826 CrossRef CAS.
- R. Pilot, R. Signorini, C. Durante, L. Orian, M. Bhamidipati and L. Fabris, Biosensors, 2019, 9, 57 CrossRef CAS PubMed.
- D.-K. Lim, K.-S. Jeon, H. M. Kim, J.-M. Nam and Y. D. Suh, Nat. Mater., 2010, 9, 60–67 CrossRef CAS PubMed.
- A. B. Zrimsek, N. Chiang, M. Mattei, S. Zaleski, M. O. McAnally, C. T. Chapman, A.-I. Henry, G. C. Schatz and R. P. Van Duyne, Chem. Rev., 2017, 117, 7583–7613 CrossRef CAS PubMed.
- P. Zhan, T. Wen, Z. Wang, Y. He, J. Shi, T. Wang, X. Liu, G. Lu and B. Ding, Angew. Chem., Int. Ed., 2018, 57, 2846–2850 CrossRef CAS PubMed.
- S. Nie and S. R. Emory, Science, 1997, 275, 1102–1106 CrossRef CAS PubMed.
- R. Gupta, P. Gupta, S. Wang, A. Melnykov, Q. Jiang, A. Seth, Z. Wang, J. J. Morrissey, I. George, S. Gandra, P. Sinha, G. A. Storch, B. A. Parikh, G. M. Genin and S. Singamaneni, Nat. Biomed. Eng., 2023, 7, 1556–1570 CrossRef CAS PubMed.
- M. Fan, G. F. S. Andrade and A. G. Brolo, Anal. Chim. Acta, 2020, 1097, 1–29 CrossRef CAS PubMed.
- Y. Yuan, N. Panwar, S. H. K. Yap, Q. Wu, S. Zeng, J. Xu, S. C. Tjin, J. Song, J. Qu and K.-T. Yong, Coord. Chem. Rev., 2017, 337, 1–33 CrossRef CAS.
- T. Itoh, M. Procházka, Z.-C. Dong, W. Ji, Y. S. Yamamoto, Y. Zhang and Y. Ozaki, Chem. Rev., 2023, 123, 1552–1634 CrossRef CAS PubMed.
- H. K. Lee, Y. H. Lee, C. S. L. Koh, G. C. Phan-Quang, X. Han, C. L. Lay, H. Y. F. Sim, Y.-C. Kao, Q. An and X. Y. Ling, Chem. Soc. Rev., 2019, 48, 731–756 RSC.
- D. Quesada-González and A. Merkoçi, Chem. Soc. Rev., 2018, 47, 4697–4709 RSC.
- E. Garcia-Rico, R. A. Alvarez-Puebla and L. Guerrini, Chem. Soc. Rev., 2018, 47, 4909–4923 RSC.
- A. I. Pérez-Jiménez, D. Lyu, Z. Lu, G. Liu and B. Ren, Chem. Sci., 2020, 11, 4563–4577 RSC.
- X. X. Han, R. S. Rodriguez, C. L. Haynes, Y. Ozaki and B. Zhao, Nat. Rev. Methods Primers, 2022, 1, 87 CrossRef.
- M. Fleischmann, P. J. Hendra and A. J. McQuillan, Chem. Phys. Lett., 1974, 26, 163–166 CrossRef CAS.
- M. G. Albrecht and J. A. Creighton, J. Am. Chem. Soc., 1977, 99, 5215–5217 CrossRef CAS.
- D. L. Jeanmaire and R. P. Van Duyne, J. Electroanal. Chem. Interfacial Electrochem., 1977, 84, 1–20 CrossRef CAS.
- M. Moskovits and B. D. Piorek, J. Raman Spectrosc., 2021, 52, 279–284 CrossRef CAS.
- B. Sharma, R. R. Frontiera, A.-I. Henry, E. Ringe and R. P. Van Duyne, Mater. Today, 2012, 15, 16–25 CrossRef CAS.
- R. A. Álvarez-Puebla, J. Phys. Chem. Lett., 2012, 3, 857–866 CrossRef PubMed.
- K. Kneipp, Y. Wang, H. Kneipp, L. T. Perelman, I. Itzkan, R. R. Dasari and M. S. Feld, Phys. Rev. Lett., 1997, 78, 1667–1670 CrossRef CAS.
- K. Kneipp, Y. Wang, H. Kneipp, L. T. Perelman, I. Itzkan, R. R. Dasari and M. S. Feld, Phys. Rev. Lett., 1997, 78, 1667–1670 CrossRef CAS.
- A. Otto, I. Mrozek, H. Grabhorn and W. Akemann, J. Phys.: Condens. Matter, 1992, 4, 1143–1212 CrossRef CAS.
- C. Liang, J. Luan, Z. Wang, Q. Jiang, R. Gupta, S. Cao, K.-K. Liu, J. J. Morrissey, E. D. Kharasch, R. R. Naik and S. Singamaneni, ACS Appl. Mater. Interfaces, 2021, 13, 11414–11423 CrossRef CAS PubMed.
- R. Goodrum and H. Li, Biotechnol. J., 2024, 19, 2300519 CrossRef CAS PubMed.
- A. Sultangaziyev and R. Bukasov, Sens Biosensing Res, 2020, 30, 100382 CrossRef PubMed.
- M. Bauch, K. Toma, M. Toma, Q. Zhang and J. Dostalek, Plasmonics, 2014, 9, 781–799 CrossRef CAS PubMed.
- C. D. Geddes and J. R. Lakowicz, J. Fluoresc., 2002, 12, 121–129 CrossRef.
- J. R. Lakowicz, Plasmonics, 2006, 1, 5–33 CrossRef CAS PubMed.
- K. Aslan, I. Gryczynski, J. Malicka, E. Matveeva, J. R. Lakowicz and C. D. Geddes, Curr. Opin. Biotechnol, 2005, 16, 55–62 CrossRef CAS PubMed.
- J. R. Lakowicz, Anal. Biochem., 2005, 337, 171–194 CrossRef CAS PubMed.
- A. Seth, Y. Liu, R. Gupta, Z. Wang, E. Mittal, S. Kolla, P. Rathi, P. Gupta, B. A. Parikh, G. M. Genin, S. Gandra, G. A. Storch, J. A. Philips, I. A. George and S. Singamaneni, Nano Lett., 2024, 24, 229–237 CrossRef CAS PubMed.
- J. R. Lakowicz, Anal. Biochem., 2005, 337, 171–194 CrossRef CAS PubMed.
- K. Aslan, I. Gryczynski, J. Malicka, E. Matveeva, J. R. Lakowicz and C. D. Geddes, Curr. Opin. Biotechnol, 2005, 16, 55–62 CrossRef CAS PubMed.
- C. D. Geddes and J. R. Lakowicz, J. Fluoresc., 2002, 12, 121–129 CrossRef.
- J. R. Lakowicz, Plasmonics, 2006, 1, 5–33 CrossRef CAS PubMed.
- D. Gontero, A. V. Veglia and A. G. Bracamonte, Photochem. Photobiol. Sci., 2020, 19, 1168–1188 CrossRef CAS PubMed.
- S. M. Fothergill, C. Joyce and F. Xie, Nanoscale, 2018, 10, 20914–20929 RSC.
- M. Zheng, Y. Li, L. Zhang, C. Li, M. Liu and H. Tang, Anal. Methods, 2024, 16, 3099–3108 RSC.
- J.-H. Choi and J.-W. Choi, Nano Lett., 2020, 20, 7100–7107 CrossRef CAS PubMed.
- Y. Jeong, Y.-M. Kook, K. Lee and W.-G. Koh, Biosens. Bioelectron., 2018, 111, 102–116 CrossRef CAS PubMed.
- C. Joyce, S. M. Fothergill and F. Xie, Mater. Today Adv., 2020, 7, 100073 CrossRef.
- W. P. Hall, S. N. Ngatia and R. P. Van Duyne, J. Phys. Chem. C, 2011, 115, 1410–1414 CrossRef CAS PubMed.
- M. P. Raphael, J. A. Christodoulides, S. P. Mulvaney, M. M. Miller, J. P. Long and J. M. Byers, Anal. Chem., 2012, 84, 1367–1373 CrossRef CAS PubMed.
- B. Sepúlveda, P. C. Angelomé, L. M. Lechuga and L. M. Liz-Marzán, Nano Today, 2009, 4, 244–251 CrossRef.
- S. Unser, I. Bruzas, J. He and L. Sagle, Sensors, 2015, 15, 15684–15716 CrossRef PubMed.
- A. Vestri, M. Rippa, V. Marchesano, D. Sagnelli, G. Margheri, J. Zhou and L. Petti, J. Mater. Chem. B, 2021, 9, 9153–9161 RSC.
- R. Wang, L. Schirmer, T. Wieduwilt, R. Förster, M. A. Schmidt, U. Freudenberg, C. Werner, A. Fery and C. Rossner, Langmuir, 2022, 38, 12325–12332 CrossRef CAS PubMed.
- S. Qian, Y. Cui, Z. Cai and L. Li, Biosens. Bioelectron.: X, 2022, 11, 100173 CAS.
- B. Liu, J. Zhuang and G. Wei, Environ. Sci.: Nano, 2020, 7, 2195–2213 RSC.
- H. Aldewachi, T. Chalati, M. N. Woodroofe, N. Bricklebank, B. Sharrack and P. Gardiner, Nanoscale, 2018, 10, 18–33 RSC.
- V. X. T. Zhao, T. I. Wong, X. T. Zheng, Y. N. Tan and X. Zhou, Mater. Sci. Energy Technol., 2020, 3, 237–249 CAS.
- X. Huang and M. A. El-Sayed, J. Adv. Res., 2010, 1, 13–28 CrossRef.
- M. Song, D. Wang, S. Peana, S. Choudhury, P. Nyga, Z. A. Kudyshev, H. Yu, A. Boltasseva, V. M. Shalaev and A. V. Kildishev, Appl. Phys. Rev., 2019, 6(4), 41308 CAS.
- J.-S. Lee, P. A. Ulmann, M. S. Han and C. A. Mirkin, Nano Lett., 2008, 8, 529–533 CrossRef CAS PubMed.
- R. Kanjanawarut and X. Su, Anal. Chem., 2009, 81, 6122–6129 CrossRef CAS PubMed.
- M. H. Jazayeri, T. Aghaie, A. Avan, A. Vatankhah and M. R. S. Ghaffari, Sens. Biosens. Res., 2018, 20, 1–8 Search PubMed.
- E. Celikbas, E. Guler Celik and S. Timur, Anal. Chem., 2018, 90, 12325–12333 CrossRef CAS PubMed.
- X. Xue, F. Wang and X. Liu, J. Am. Chem. Soc., 2008, 130, 3244–3245 CrossRef CAS PubMed.
- J. Du, L. Jiang, Q. Shao, X. Liu, R. S. Marks, J. Ma and X. Chen, Small, 2013, 9, 1467–1481 CrossRef CAS PubMed.
- R. Cao, B. Li, Y. Zhang and Z. Zhang, Chem. Commun., 2011, 47, 12301 RSC.
- B. Li, X. Li, Y. Dong, B. Wang, D. Li, Y. Shi and Y. Wu, Anal. Chem., 2017, 89, 10639–10643 CrossRef CAS PubMed.
- X. Xie, W. Xu and X. Liu, Acc. Chem. Res., 2012, 45, 1511–1520 CrossRef CAS PubMed.
- F. Zhang and J. Liu, Anal. Sens., 2021, 1, 30–43 CAS.
- S. Mason, Trends Pharmacol. Sci., 1986, 7, 20–23 CrossRef CAS.
- F. Wang, X. Yue, Q. Ding, H. Lin, C. Xu and S. Li, Nanoscale, 2023, 15, 2541–2552 RSC.
- N. H. Cho, H. Kim, J. W. Kim, Y.-C. Lim, R. M. Kim, Y. H. Lee and K. T. Nam, Chemistry, 2024, 10, 1052–1070 CrossRef CAS.
- W. Ma, L. Xu, A. F. de Moura, X. Wu, H. Kuang, C. Xu and N. A. Kotov, Chem. Rev., 2017, 117, 8041–8093 CrossRef CAS PubMed.
- H.-E. Lee, H.-Y. Ahn, J. Mun, Y. Y. Lee, M. Kim, N. H. Cho, K. Chang, W. S. Kim, J. Rho and K. T. Nam, Nature, 2018, 556, 360–365 CrossRef CAS PubMed.
- L. Zhang, Y. Chen, J. Zheng, G. R. Lewis, X. Xia, E. Ringe, W. Zhang and J. Wang, Angew. Chem., Int. Ed., 2023, 135(52), e202312615 CrossRef.
- G. González-Rubio, J. Mosquera, V. Kumar, A. Pedrazo-Tardajos, P. Llombart, D. M. Solís, I. Lobato, E. G. Noya, A. Guerrero-Martínez, J. M. Taboada, F. Obelleiro, L. G. MacDowell, S. Bals and L. M. Liz-Marzán, Science, 2020, 368, 1472–1477 CrossRef PubMed.
- D. Vila-Liarte, N. A. Kotov and L. M. Liz-Marzán, Chem. Sci., 2022, 13, 595–610 RSC.
- C. Song, M. G. Blaber, G. Zhao, P. Zhang, H. C. Fry, G. C. Schatz and N. L. Rosi, Nano Lett., 2013, 13, 3256–3261 CrossRef CAS PubMed.
- J. Kumar, H. Eraña, E. López-Martínez, N. Claes, V. F. Martín, D. M. Solís, S. Bals, A. L. Cortajarena, J. Castilla and L. M. Liz-Marzán, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 3225–3230 CrossRef CAS PubMed.
- X. Wu, L. Xu, L. Liu, W. Ma, H. Yin, H. Kuang, L. Wang, C. Xu and N. A. Kotov, J. Am. Chem. Soc., 2013, 135, 18629–18636 CrossRef CAS PubMed.
- Y. Xia, K. D. Gilroy, H. Peng and X. Xia, Angew. Chem., Int. Ed., 2017, 56, 60–95 CrossRef CAS PubMed.
- Y. Xia, Y. Xiong, B. Lim and S. E. Skrabalak, Angew. Chem., Int. Ed., 2009, 48, 60–103 CrossRef CAS PubMed.
- E. Carbó-Argibay and B. Rodríguez-González, Isr. J. Chem., 2016, 56, 214–226 CrossRef.
- N. G. Bastús, J. Comenge and V. Puntes, Langmuir, 2011, 27, 11098–11105 CrossRef PubMed.
- Y.-N. Wang, W.-T. Wei, C.-W. Yang and M. H. Huang, Langmuir, 2013, 29, 10491–10497 CrossRef CAS PubMed.
- F. Qin, T. Zhao, R. Jiang, N. Jiang, Q. Ruan, J. Wang, L. Sun, C. Yan and H. Lin, Adv. Opt. Mater., 2016, 4, 76–85 CrossRef CAS.
- J. Zheng, X. Cheng, H. Zhang, X. Bai, R. Ai, L. Shao and J. Wang, Chem. Rev., 2021, 121, 13342–13453 CrossRef CAS PubMed.
- Q. Ruan, L. Shao, Y. Shu, J. Wang and H. Wu, Adv. Opt. Mater., 2014, 2, 65–73 CrossRef.
- Y. Zheng, X. Zhong, Z. Li and Y. Xia, Part. Part. Syst. Charact., 2014, 31, 266–273 CrossRef CAS.
- F. Zhou, Z.-Y. Li, Y. Liu and Y. Xia, J. Phys. Chem. C, 2008, 112, 20233–20240 CrossRef CAS.
- S. Mazzucco, N. Geuquet, J. Ye, O. Stéphan, W. Van Roy, P. Van Dorpe, L. Henrard and M. Kociak, Nano Lett., 2012, 12, 1288–1294 CrossRef CAS PubMed.
- L. Zhang, Y. Zhang, J. Ahn, X. Wang and D. Qin, Chem. Mater., 2019, 31, 1057–1065 CrossRef CAS.
- L. Scarabelli, A. Sánchez-Iglesias, J. Pérez-Juste and L. M. Liz-Marzán, J. Phys. Chem. Lett., 2015, 6, 4270–4279 CrossRef CAS PubMed.
- X. Zhuo, X. Zhu, Q. Li, Z. Yang and J. Wang, ACS Nano, 2015, 9, 7523–7535 CrossRef CAS PubMed.
- J.-H. Lee, K. J. Gibson, G. Chen and Y. Weizmann, Nat. Commun., 2015, 6, 7571 CrossRef CAS PubMed.
- S. Atta, M. Beetz and L. Fabris, Nanoscale, 2019, 11, 2946–2958 RSC.
- X. Cui, F. Qin, Q. Ruan, X. Zhuo and J. Wang, Adv. Funct. Mater., 2018, 28, 1705516 CrossRef.
- S. Yoo, S. Go, J. Son, J. Kim, S. Lee, M. Haddadnezhad, H. Hilal, J.-M. Kim, J.-M. Nam and S. Park, J. Am. Chem. Soc., 2021, 143, 15113–15119 CrossRef CAS PubMed.
- Q. Li, X. Zhuo, S. Li, Q. Ruan, Q. Xu and J. Wang, Adv. Opt. Mater., 2015, 3, 801–812 CrossRef CAS.
- X. Kou, S. Zhang, C.-K. Tsung, M. H. Yeung, Q. Shi, G. D. Stucky, L. Sun, J. Wang and C. Yan, J. Phys. Chem. B, 2006, 110, 16377–16383 CrossRef CAS PubMed.
- X. Zhuo, H. K. Yip, X. Cui, J. Wang and H.-Q. Lin, Light: Sci. Appl., 2019, 8, 39 CrossRef PubMed.
- S. Li, R. Ai, K. K. Chui, Y. Fang, Y. Lai, X. Zhuo, L. Shao, J. Wang and H.-Q. Lin, Nano Lett., 2023, 23, 4183–4190 CrossRef CAS PubMed.
- J. Lee, P. Lee, H. Lee, D. Lee, S. S. Lee and S. H. Ko, Nanoscale, 2012, 4, 6408 RSC.
- T. H. Chow, N. Li, X. Bai, X. Zhuo, L. Shao and J. Wang, Acc. Chem. Res., 2019, 52, 2136–2146 CrossRef CAS PubMed.
- H. Chen, L. Shao, T. Ming, Z. Sun, C. Zhao, B. Yang and J. Wang, Small, 2010, 6, 2272–2280 CrossRef CAS PubMed.
- X. Yang, Y. Liu, S. H. Lam, J. Wang, S. Wen, C. Yam, L. Shao and J. Wang, Nano Lett., 2021, 21, 8205–8212 CrossRef CAS PubMed.
- L. Peng, H. Chan, P. Choo, T. W. Odom, S. K. R. S. Sankaranarayanan and X. Ma, Nano Lett., 2020, 20, 5866–5872 CrossRef CAS PubMed.
- X. Cui, Y. Lai, R. Ai, H. Wang, L. Shao, H. Chen, W. Zhang and J. Wang, Adv. Opt. Mater., 2020, 8, 2001173 CrossRef CAS.
- H. Huang, H. Wang, S. Li, J. Jiang, Y. Liu, M. Cai, L. Shao, H. Chen and J. Wang, ACS Nano, 2022, 16, 14874–14884 CrossRef CAS PubMed.
- S. Schlücker, Angew. Chem., Int. Ed., 2014, 53, 4756–4795 CrossRef PubMed.
- M. Moskovits, J. Chem. Phys., 1978, 69, 4159–4161 CrossRef CAS.
- D. Pines and D. Bohm, Phys. Rev., 1952, 85, 338–353 CrossRef CAS.
- J. A. Creighton, C. G. Blatchford and M. G. Albrecht, J. Chem. Soc., Faraday Trans. 2, 1979, 75, 790 RSC.
- J. Billmann, G. Kovacs and A. Otto, Surf. Sci., 1980, 92, 153–173 CrossRef CAS.
- J. A. Creighton, Surf. Sci., 1983, 124, 209–219 CrossRef CAS.
- M. Moskovits and J. S. Suh, J. Phys. Chem., 1984, 88, 5526–5530 CrossRef CAS.
- H. Metiu, Prog. Surf. Sci., 1984, 17, 153–320 CrossRef CAS.
- M. Kerker, Acc. Chem. Res., 1984, 17, 271–277 CrossRef CAS.
- A. Wokaun, Mol. Phys., 1985, 56, 1–33 CrossRef CAS.
- M. Kerker, (No Title).
- G. C. Schatz and R. P. Van Duyne, in Handbook of Vibrational Spectroscopy, ed. P. R. Griffiths, Wiley, 2001 Search PubMed.
- P. Etchegoin, L. F. Cohen, H. Hartigan, R. J. C. Brown, M. J. T. Milton and J. C. Gallop, J. Chem. Phys., 2003, 119, 5281–5289 CrossRef CAS.
- R. Aroca, Surface-Enhanced Vibrational Spectroscopy, Wiley, 2006 Search PubMed.
- C. F. Bohren and D. R. Huffman, Absorption and Scattering of Light by Small Particles, Wiley, 1998 Search PubMed.
- M. Kerker, Science, 1969, 666 Search PubMed.
- K. L. Kelly, E. Coronado, L. L. Zhao and G. C. Schatz, J. Phys. Chem. B, 2003, 107, 668–677 CrossRef CAS.
- R. Alvarez-Puebla, L. M. Liz-Marzán and F. J. García de Abajo, J. Phys. Chem. Lett., 2010, 1, 2428–2434 CrossRef CAS.
- L. Rodríguez-Lorenzo, R. A. Álvarez-Puebla, I. Pastoriza-Santos, S. Mazzucco, O. Stéphan, M. Kociak, L. M. Liz-Marzán and F. J. García de Abajo, J. Am. Chem. Soc., 2009, 131, 4616–4618 CrossRef PubMed.
- E. Hao and G. C. Schatz, J. Chem. Phys., 2004, 120, 357–366 CrossRef CAS PubMed.
- A. Kudelski and J. Bukowska, Surf. Sci., 1996, 368, 396–400 CrossRef CAS.
- K. Kneipp, H. Kneipp, V. B. Kartha, R. Manoharan, G. Deinum, I. Itzkan, R. R. Dasari and M. S. Feld, Phys. Rev. E, 1998, 57, R6281–R6284 CrossRef CAS.
- E. Pazos, M. Garcia-Algar, C. Penas, M. Nazarenus, A. Torruella, N. Pazos-Perez, L. Guerrini, M. E. Vázquez, E. Garcia-Rico, J. L. Mascareñas and R. A. Alvarez-Puebla, J. Am. Chem. Soc., 2016, 138, 14206–14209 CrossRef CAS PubMed.
- K. Ataka, T. Yotsuyanagi and M. Osawa, J. Phys. Chem., 1996, 100, 10664–10672 CrossRef CAS.
- D. L. Allara and R. G. Nuzzo, Langmuir, 1985, 1, 52–66 CrossRef CAS.
- Handbook of Vibrational Spectroscopy, ed. J. M. Chalmers and P. R. Griffiths, Wiley, 2001 Search PubMed.
- S. Fornasaro, F. Alsamad, M. Baia, L. A. E. Batista de Carvalho, C. Beleites, H. J. Byrne, A. Chiadò, M. Chis, M. Chisanga, A. Daniel, J. Dybas, G. Eppe, G. Falgayrac, K. Faulds, H. Gebavi, F. Giorgis, R. Goodacre, D. Graham, P. La Manna, S. Laing, L. Litti, F. M. Lyng, K. Malek, C. Malherbe, M. P. M. Marques, M. Meneghetti, E. Mitri, V. Mohaček-Grošev, C. Morasso, H. Muhamadali, P. Musto, C. Novara, M. Pannico, G. Penel, O. Piot, T. Rindzevicius, E. A. Rusu, M. S. Schmidt, V. Sergo, G. D. Sockalingum, V. Untereiner, R. Vanna, E. Wiercigroch and A. Bonifacio, Anal. Chem., 2020, 92, 4053–4064 CrossRef CAS PubMed.
- B. Sharma, R. R. Frontiera, A.-I. Henry, E. Ringe and R. P. Van Duyne, Mater. Today, 2012, 15, 16–25 CrossRef CAS.
- C. E. Ott and L. E. Arroyo, WIREs Forensic Sci., 2023, 5, e1483 CrossRef CAS.
- W. R. Premasiri, Y. Chen, J. Fore, A. Brodeur and L. D. Ziegler, Frontiers and Advances in Molecular Spectroscopy, Elsevier, 2018, pp. 327–367 Search PubMed.
- L. Jiang, M. M. Hassan, S. Ali, H. Li, R. Sheng and Q. Chen, Trends Food Sci. Technol., 2021, 112, 225–240 CrossRef CAS.
- Podagatlapalli Gopala Krishna, Hamad Syed and V. R. Soma, J. Phys. Chem. C, 2015, 119, 16972–16983 CrossRef.
- H. Liu, X. Gao, C. Xu and D. Liu, Theranostics, 2022, 12, 1870–1903 CrossRef CAS PubMed.
- Y. Liu, Y. Zhang, M. Tardivel, M. Lequeux, X. Chen, W. Liu, J. Huang, H. Tian, Q. Liu, G. Huang, R. Gillibert, M. L. de la Chapelle and W. Fu, Plasmonics, 2020, 15, 743–752 CrossRef CAS.
- M. Richard-Lacroix and V. Deckert, Light: Sci. Appl., 2020, 9, 35 CrossRef CAS PubMed.
- L. A. Lane, X. Qian and S. Nie, Chem. Rev., 2015, 115, 10489–10529 CrossRef CAS PubMed.
- Y. Wang, B. Yan and L. Chen, Chem. Rev., 2013, 113, 1391–1428 CrossRef CAS PubMed.
- Z. Tao, W. Zhao, S. Wang, B. Zhao, R. Hua, J. Qin and Z. Xu, Nanotechnol. Precis. Eng., 2021, 4(4), 043004 CrossRef CAS.
- Y. Wu, Y. Jiang, X. Zheng, S. Jia, Z. Zhu, B. Ren and H. Ma, R. Soc. Open Sci., 2018, 5, 172034 CrossRef PubMed.
- L. Petti, R. Capasso, M. Rippa, M. Pannico, P. La Manna, G. Peluso, A. Calarco, E. Bobeico and P. Musto, Vib. Spectrosc., 2016, 82, 22–30 CrossRef CAS.
- M. Li, J. Lu, J. Qi, F. Zhao, J. Zeng, J. C.-C. Yu and W.-C. Shih, J. Biomed. Opt., 2014, 19, 050501 CrossRef PubMed.
- S. Yan, H. Tang, J. Sun, C. Zhu, Q. Pan, B. Chen and G. Meng, Adv. Opt. Mater., 2024, 12, 2302010 CrossRef CAS.
- A. Pandya, J. C. Kumaradas and A. Douplik, in Novel Biophotonics Techniques and Applications V, ed. A. Amelink and S. K. Nadkarni, SPIE, 2019, vol. Part F142-ECBO, pp. 5 Search PubMed.
- C. Hamon and L. M. Liz-Marzán, J. Colloid Interface Sci., 2018, 512, 834–843 CrossRef CAS PubMed.
- A. Marques, B. Veigas, A. Araújo, B. Pagará, P. V. Baptista, H. Águas, R. Martins and E. Fortunato, Sci. Rep., 2019, 9, 17922 CrossRef PubMed.
- J. Choi, J. Lee and J. H. Jung, Biosens. Bioelectron., 2020, 169, 112611 CrossRef CAS PubMed.
- H. Pu, W. Xiao and D.-W. Sun, Trends Food Sci. Technol., 2017, 70, 114–126 CrossRef CAS.
- B. L. Scott and K. T. Carron, Anal. Chem., 2012, 84, 8448–8451 CrossRef CAS PubMed.
- J. P. Camden, J. A. Dieringer, J. Zhao and R. P. Van Duyne, Acc. Chem. Res., 2008, 41, 1653–1661 CrossRef CAS PubMed.
- P. L. Stiles, J. A. Dieringer, N. C. Shah and R. P. Van Duyne, Annu. Rev. Anal. Chem., 2008, 1, 601–626 CrossRef CAS PubMed.
- E. C. Le Ru and P. G. Etchegoin, Chem. Phys. Lett., 2006, 423, 63–66 CrossRef CAS.
- E. J. Blackie, E. C. Le Ru and P. G. Etchegoin, J. Am. Chem. Soc., 2009, 131, 14466–14472 CrossRef CAS PubMed.
- J. A. Dieringer, R. B. Lettan, K. A. Scheidt and R. P. Van Duyne, J. Am. Chem. Soc., 2007, 129, 16249–16256 CrossRef CAS PubMed.
- D.-K. Lim, K.-S. Jeon, H. M. Kim, J.-M. Nam and Y. D. Suh, Nat. Mater., 2010, 9, 60–67 CrossRef CAS PubMed.
- J. H. Yoon, F. Selbach, L. Langolf and S. Schlücker, Small, 2018, 14, 1870018 CrossRef.
- J. H. Yoon, F. Selbach, L. Schumacher, J. Jose and S. Schlücker, ACS Photonics, 2019, 6, 642–648 CrossRef CAS.
- H. Cha, J. H. Yoon and S. Yoon, ACS Nano, 2014, 8, 8554–8563 CrossRef CAS PubMed.
- J.-H. Lee, J.-M. Nam, K.-S. Jeon, D.-K. Lim, H. Kim, S. Kwon, H. Lee and Y. D. Suh, ACS Nano, 2012, 6, 9574–9584 CrossRef CAS PubMed.
- P. W. K. Rothemund, Nature, 2006, 440, 297–302 CrossRef CAS PubMed.
- W. Fang, S. Jia, J. Chao, L. Wang, X. Duan, H. Liu, Q. Li, X. Zuo, L. Wang, L. Wang, N. Liu and C. Fan, Sci. Adv., 2019, 5, eaau4506 CrossRef PubMed.
- J. Prinz, B. Schreiber, L. Olejko, J. Oertel, J. Rackwitz, A. Keller and I. Bald, J. Phys. Chem. Lett., 2013, 4, 4140–4145 CrossRef CAS.
- V. V. Thacker, L. O. Herrmann, D. O. Sigle, T. Zhang, T. Liedl, J. J. Baumberg and U. F. Keyser, Nat. Commun., 2014, 5, 3448 CrossRef PubMed.
- P. Kühler, E.-M. Roller, R. Schreiber, T. Liedl, T. Lohmüller and J. Feldmann, Nano Lett., 2014, 14, 2914–2919 CrossRef PubMed.
- M. Pilo-Pais, A. Watson, S. Demers, T. H. LaBean and G. Finkelstein, Nano Lett., 2014, 14, 2099–2104 CrossRef CAS PubMed.
- K. Tapio, A. Mostafa, Y. Kanehira, A. Suma, A. Dutta and I. Bald, ACS Nano, 2021, 15, 7065–7077 CrossRef CAS PubMed.
- S. Simoncelli, E.-M. Roller, P. Urban, R. Schreiber, A. J. Turberfield, T. Liedl and T. Lohmüller, ACS Nano, 2016, 10, 9809–9815 CrossRef CAS PubMed.
- A. Otto, J. Raman Spectrosc., 2002, 33, 593–598 CrossRef CAS.
- J. Szczerbiński, L. Gyr, J. Kaeslin and R. Zenobi, Nano Lett., 2018, 18, 6740–6749 CrossRef PubMed.
- C. Heck, Y. Kanehira, J. Kneipp and I. Bald, Molecules, 2019, 24 Search PubMed.
- E. J. Bjerneld, F. Svedberg, P. Johansson and M. Käll, J. Phys. Chem. A, 2004, 108, 4187–4193 CrossRef CAS.
- P. Senthil Kumar, I. Pastoriza-Santos, B. Rodríguez-González, F. Javier García de Abajo and L. M. Liz-Marzán, Nanotechnology, 2008, 19, 15606 CrossRef PubMed.
- Y. Kanehira, K. Tapio, G. Wegner, S. Kogikoski Jr., S. Rüstig, C. Prietzel, K. Busch and I. Bald, ACS Nano, 2023, 17, 21227–21239 CrossRef PubMed.
- S. Tanwar, V. Kaur, G. Kaur and T. Sen, J. Phys. Chem. Lett., 2021, 12, 8141–8150 CrossRef CAS PubMed.
- C. Heck, Y. Kanehira, J. Kneipp and I. Bald, Angew. Chem., Int. Ed., 2018, 57, 7444–7447 CrossRef CAS PubMed.
- F. Schuknecht, K. Kołątaj, M. Steinberger, T. Liedl and T. Lohmueller, Nat. Commun., 2023, 14, 7192 CrossRef CAS PubMed.
- F. Schuknecht, K. Kołątaj, M. Steinberger, T. Liedl and T. Lohmueller, Nat. Commun., 2023, 14, 7192 CrossRef CAS PubMed.
- M. Sharma, C. Kaur, P. Singhmar, S. Rai and T. Sen, Nanoscale, 2024, 16, 15128–15140 RSC.
- L. M. Wassermann, M. Scheckenbach, A. V. Baptist, V. Glembockyte and A. Heuer-Jungemann, Adv. Mater., 2023, 35, 2212024 CrossRef CAS PubMed.
- F. Lussier, V. Thibault, B. Charron, G. Q. Wallace and J.-F. Masson, TrAC, Trends Anal. Chem., 2020, 124, 115796 CrossRef CAS.
- A. B. Zrimsek, N. Chiang, M. Mattei, S. Zaleski, M. O. McAnally, C. T. Chapman, A.-I. Henry, G. C. Schatz and R. P. Van Duyne, Chem. Rev., 2017, 117, 7583–7613 CrossRef CAS PubMed.
- J.-A. Huang, M. Z. Mousavi, Y. Zhao, A. Hubarevich, F. Omeis, G. Giovannini, M. Schütte, D. Garoli and F. De Angelis, Nat. Commun., 2019, 10, 5321 CrossRef PubMed.
- Y. Qiu, C. Kuang, X. Liu and L. Tang, Sensors, 2022, 22, 4889 CrossRef CAS PubMed.
- Z. Zhang, S. Sheng, R. Wang and M. Sun, Anal. Chem., 2016, 88, 9328–9346 CrossRef CAS PubMed.
- J. Zhou, P.-L. Zhou, Q. Shen, S. A. Ahmed, X.-T. Pan, H.-L. Liu, X.-L. Ding, J. Li, K. Wang and X.-H. Xia, Anal. Chem., 2021, 93, 11679–11685 CrossRef CAS PubMed.
- S. Jiang, Y. Zhang, R. Zhang, C. Hu, M. Liao, Y. Luo, J. Yang, Z. Dong and J. G. Hou, Nat. Nanotechnol., 2015, 10, 865–869 CrossRef CAS PubMed.
- Z. Liu, S.-Y. Ding, Z.-B. Chen, X. Wang, J.-H. Tian, J. R. Anema, X.-S. Zhou, D.-Y. Wu, B.-W. Mao, X. Xu, B. Ren and Z.-Q. Tian, Nat. Commun., 2011, 2, 305 CrossRef PubMed.
- J. Qi, J. Zeng, F. Zhao, S. H. Lin, B. Raja, U. Strych, R. C. Willson and W.-C. Shih, Nanoscale, 2014, 6, 8521–8526 RSC.
- B. Guven, M. Eryilmaz, A. Üzer, I. H. Boyaci, U. Tamer and R. Apak, RSC Adv., 2017, 7, 37039–37047 RSC.
- B. Soundiraraju and B. K. George, ACS Nano, 2017, 11, 8892–8900 CrossRef CAS PubMed.
- P. Garg, Bharti, R. K. Soni and R. Raman, J. Mater. Sci.: Mater. Electron., 2020, 31, 1094–1104 CrossRef CAS.
- N. Singh, T. K. Naqvi, P. Awasthi, A. M. Siddiqui, A. K. Srivastava and P. K. Dwivedi, Sens Actuators, A, 2022, 347, 113915 CrossRef CAS.
- T. K. Naqvi, M. Sree Satya Bharati, A. K. Srivastava, M. M. Kulkarni, A. M. Siddiqui, S. V. Rao and P. K. Dwivedi, ACS Omega, 2019, 4, 17691–17701 CrossRef CAS PubMed.
- V. R. Kahkhaie, M. H. Yousefi, S. M. R. Darbani and A. Mobashery, Photonics Nanostruct. Fundam. Appl., 2020, 41, 100801 CrossRef.
- T. Liyanage, A. Rael, S. Shaffer, S. Zaidi, J. V. Goodpaster and R. Sardar, Analyst, 2018, 143, 2012–2022 RSC.
- C. Wang, B. Liu and X. Dou, Sens. Actuators, B, 2016, 231, 357–364 CrossRef CAS.
- C. Byram, J. Rathod, S. S. B. Moram, A. Mangababu and V. R. Soma, Nanomaterials, 2022, 12, 2150 CrossRef CAS PubMed.
- W. Fan, S. Yang, Y. Zhang, B. Huang, Z. Gong, D. Wang and M. Fan, ACS Sens., 2020, 5, 3599–3606 CrossRef CAS PubMed.
- R. Gao, X. Song, C. Zhan, C. Weng, S. Cheng, K. Guo, N. Ma, H. Chang, Z. Guo, L.-B. Luo and L. Yu, Sens. Actuators, B, 2020, 314, 128081 CrossRef CAS.
- R. Gao, H. Qian, C. Weng, X. Wang, C. Xie, K. Guo, S. Zhang, S. Xuan, Z. Guo and L.-B. Luo, Sens. Actuators, B, 2020, 321, 128543 CrossRef CAS.
- S. S. B. Moram, C. Byram, S. N. Shibu, B. M. Chilukamarri and V. R. Soma, ACS Omega, 2018, 3, 8190–8201 CrossRef CAS PubMed.
- S. Ben-Jaber, W. J. Peveler, R. Quesada-Cabrera, C. W. O. Sol, I. Papakonstantinou and I. P. Parkin, Nanoscale, 2017, 9, 16459–16466 RSC.
- V. Heleg-Shabtai, A. Zaltsman, M. Sharon, H. Sharabi, I. Nir, D. Marder, G. Cohen, I. Ron and A. Pevzner, RSC Adv., 2021, 11, 26029–26036 RSC.
- K. J. Squire, K. Sivashanmugan, B. Zhang, J. Kraai, G. Rorrer and A. X. Wang, ACS Appl. Nano Mater., 2020, 3, 1656–1665 CrossRef CAS.
- A.-M. Dowgiallo, A. Branham and D. Guenther, Spectroscopyonline, 2017, 32, 8–17 Search PubMed.
- V. S. Vendamani, S. V. S. N. Rao, A. P. Pathak and V. R. Soma, RSC Adv., 2020, 10, 44747–44755 RSC.
- V. S. Vendamani, R. Beeram, M. M. Neethish, S. V. S. N. Rao and S. V. Rao, iScience, 2023, 25, 104849 CrossRef PubMed.
- S. S. B. Moram, A. K. Shaik, C. Byram, S. Hamad and V. R. Soma, Anal. Chim. Acta, 2020, 1101, 157–168 CrossRef CAS PubMed.
- S. Adhikari, E. K. Ampadu, M. Kim, D. Noh, E. Oh and D. Lee, Sensors, 2021, 21, 5567 CrossRef CAS PubMed.
- D. Banerjee, M. Akkanaboina, R. K. Kanaka and V. R. Soma, Appl. Surf. Sci., 2023, 616, 156561 CrossRef CAS.
- G. K. Podagatlapalli, S. Hamad, M. A. Mohiddon and S. V. Rao, Laser Phys. Lett., 2015, 12, 36003 CrossRef.
- R. K. Avasarala, T. Jena, S. K. Balivada, C. Angani, H. Syed, V. R. Soma and G. K. Podagatlapalli, Results Opt., 2021, 5, 100153 CrossRef.
- C. Byram, S. S. B. Moram and V. R. Soma, Analyst, 2019, 144, 2327–2336 RSC.
- S. V. Rao, G. K. Podagatlapalli and S. Hamad, International Conference on Fibre Optics and Photonics, 2016, Th4D.2.
- X. He, Y. Liu, Y. Liu, S. Cui, W. Liu and Z. Li, CrystEngComm, 2020, 22, 776–785 RSC.
- U. P. Shaik, S. Hamad, M. Ahamad Mohiddon, V. R. Soma and M. Ghanashyam Krishna, J. Appl. Phys., 2016, 119, 093013 CrossRef.
- H. Syed, G. Krishna Podagatlapalli, M. A. Mohiddon and V. Rao Soma, Adv. Mater. Lett., 2015, 6, 1073–1080 CrossRef CAS.
- S. Hamad, S. S. Bharati Moram, B. Yendeti, G. K. Podagatlapalli, S. V. S. Nageswara Rao, A. P. Pathak, M. A. Mohiddon and V. R. Soma, ACS Omega, 2018, 3, 18420–18432 CrossRef CAS PubMed.
- J. Rathod, C. Byram, R. K. Kanaka, M. Sree Satya Bharati, D. Banerjee, M. Akkanaboina and V. R. Soma, ACS Omega, 2022, 7, 15969–15981 CrossRef CAS PubMed.
- S. S. B. Moram, C. Byram and V. R. Soma, RSC Adv., 2023, 13, 2620–2630 RSC.
- M. S. S. Bharati, B. Chandu and S. V. Rao, RSC Adv., 2019, 9, 1517–1525 RSC.
- A. K. Verma and R. K. Soni, Opt. Laser Technol., 2023, 163, 109429 CrossRef CAS.
- C. Xiao, Z. Chen, M. Qin, D. Zhang and L. Fan, Photonic Sens., 2018, 8, 278–288 CrossRef CAS.
- B. Samransuksamer, M. Horprathum, T. Jutarosaga, A. Kopwitthaya, S. Limwichean, N. Nuntawong, C. Chananonnawathorn, V. Patthanasettakul, P. Muthitamongkol and A. Treetong, Sens. Actuators, B, 2018, 277, 102–113 CrossRef CAS.
- M. Ge, W. Zhao, Y. Han, H. Gai and C. Zong, Front. Chem., 2023, 11, 1–9 Search PubMed.
- N. Bi, Y.-H. Zhang, M.-H. Hu, J. Xu, W. Song, J. Gou, Y.-X. Li and L. Jia, Spectrochim. Acta, Part A, 2023, 284, 121777 CrossRef CAS PubMed.
- C. Chin, C. Chen, X. Chen, H. Yen, H. Hsien, J. Young and Y. Chen, Sens. Actuators, B, 2021, 347, 130614 CrossRef CAS.
- T. K. Naqvi, A. Bajpai, M. S. S. Bharati, M. M. Kulkarni, A. M. Siddiqui, V. R. Soma and P. K. Dwivedi, J. Hazard. Mater., 2021, 407, 124353 CrossRef CAS PubMed.
- X.-R. Bai, Y. Zeng, X.-D. Zhou, X.-H. Wang, A.-G. Shen and J.-M. Hu, Anal. Chem., 2017, 89, 10335–10342 CrossRef CAS PubMed.
- N. Singh, A. M. Shrivastav, N. Vashistha and I. Abdulhalim, Sens. Actuators, B, 2023, 374, 132813 CrossRef CAS.
- X. Kong, Y. Xi, P. Le Duff, X. Chong, E. Li, F. Ren, G. L. Rorrer and A. X. Wang, Biosens. Bioelectron., 2017, 88, 63–70 CrossRef CAS PubMed.
- W. Liu, Z. Wang, Z. Liu, J. Chen, L. Shi, L. Huang, Y. Liu, S. Cui and X. He, ACS Sens., 2023, 8, 1733–1741 CrossRef CAS PubMed.
- X. He, Y. Liu, X. Xue, J. Liu, Y. Liu and Z. Li, J. Mater. Chem. C, 2017, 5, 12384–12392 RSC.
- Y. Shi, W. Wang and J. Zhan, Nano Res., 2016, 9, 2487–2497 CrossRef CAS.
- D. Lin, R. Dong, P. Li, S. Li, M. Ge, Y. Zhang, L. Yang and W. Xu, Talanta, 2020, 218, 121157 CrossRef CAS PubMed.
- J. Wu, Y. Feng, L. Zhang and W. Wu, Carbohydr. Polym., 2020, 248, 116766 CrossRef CAS PubMed.
- D. Chen, X. Zhu, J. Huang, G. Wang, Y. Zhao, F. Chen, J. Wei, Z. Song and Y. Zhao, Anal. Chem., 2018, 90, 9048–9054 CrossRef CAS PubMed.
- K. Milligan, N. C. Shand, D. Graham and K. Faulds, Anal. Chem., 2020, 92, 3253–3261 CrossRef CAS PubMed.
- R. Beeram, V. S. Vendamani and V. R. Soma, Spectrochim. Acta, Part A, 2023, 289, 122218 CrossRef CAS PubMed.
- R. Beeram, D. Banerjee, L. M. Narlagiri and V. R. Soma, Anal. Methods, 2022, 14, 1788–1796 RSC.
- R. Beeram and V. R. Soma, Opt. Mater., 2023, 137, 113615 CrossRef CAS.
- F. Wan, Q. Liu, W.-P. Kong, Z.-Y. Luo, S.-F. Gao, Y.-Y. Wang and W.-G. Chen, IEEE Sens. J., 2023, 23, 6849–6856 CAS.
- L. Juhlin, T. Mikaelsson, A. Hakonen, M. S. Schmidt, T. Rindzevicius, A. Boisen, M. Käll and P. O. Andersson, Talanta, 2020, 211, 120721 CrossRef CAS PubMed.
- M. Lafuente, I. Pellejero, V. Sebastián, M. A. Urbiztondo, R. Mallada, M. P. Pina and J. Santamaría, Sens. Actuators, B, 2018, 267, 457–466 CrossRef CAS.
- T. Li, B. Wen, Y. Zhang, L. Zhang and J. Li, J. Raman Spectrosc., 2022, 53, 1386–1393 CrossRef CAS.
- A. Hakonen, T. Rindzevicius, M. S. Schmidt, P. O. Andersson, L. Juhlin, M. Svedendahl, A. Boisen and M. Käll, Nanoscale, 2016, 8, 1305–1308 RSC.
- M. Lafuente, D. Sanz, M. Urbiztondo, J. Santamaría, M. P. Pina and R. Mallada, J. Hazard. Mater., 2020, 384, 121279 CrossRef CAS PubMed.
- W.-C. Huang and H.-R. Chen, Molecules, 2023, 28, 520 CrossRef CAS PubMed.
- N. Taranenko, J. Alarie, D. L. Stokes and T. Vo-Dinh, J. Raman Spectrosc., 1996, 27, 379–384 CrossRef CAS.
- J. Wang, G. Duan, G. Liu, Y. Li, Z. Chen, L. Xu and W. Cai, J. Hazard. Mater., 2016, 303, 94–100 CrossRef CAS PubMed.
- Z. Wang, Y. Dai, X. Zhou, Z. Liu, W. Liu, L. Huang, M. Yuan, S. Cui and X. He, Talanta, 2023, 258, 124460 CrossRef CAS PubMed.
- J. Wu, L. Zhang, F. Huang, X. Ji, H. Dai and W. Wu, J. Hazard. Mater., 2020, 387, 121714 CrossRef CAS PubMed.
- D. J. Klapec, G. Czarnopys and J. Pannuto, Forensic Sci. Int., 2023, 6, 100298 Search PubMed.
- K. C. To, S. Ben-Jaber and I. P. Parkin, ACS Nano, 2020, 14, 10804–10833 CrossRef CAS PubMed.
- F. Zapata, M. López-López and C. García-Ruiz, Appl. Spectrosc. Rev., 2016, 51, 227–262 CrossRef CAS.
- S. Botti, S. Almaviva, L. Cantarini, A. Palucci, A. Puiu and A. Rufoloni, J. Raman Spectrosc., 2013, 44, 463–468 CrossRef CAS.
- A. P. Lister, W. J. Sellors, C. R. Howle and S. Mahajan, Anal. Chem., 2020, 93, 417–429 CrossRef PubMed.
- A. W. Fountain III, S. D. Christesen, R. P. Moon, J. A. Guicheteau and E. D. Emmons, Appl. Spectrosc., 2014, 68, 795–811 CrossRef PubMed.
- C. Byram, M. Sree Satya Bharati, B. Dipanjan, B. Reshma, R. Jagannath and S. Venugopal Rao, J. Opt., 2023, 25, 043001 CrossRef.
- R. Beeram, K. R. Vepa and V. R. Soma, Biosensors, 2023, 13, 328 CrossRef CAS PubMed.
- S. Adhikari, D. Noh, M. Kim, D. Ahn, Y. Jang, E. Oh and D. Lee, Spectrochim. Acta, Part A, 2024, 123996 CrossRef CAS PubMed.
- S. Adhikari, D. Noh, M. Kim, D. Ahn, Y. Jang, E. Oh and D. Lee, Available at SSRN 4258144.
- V. Heleg-Shabtai, H. Sharabi, A. Zaltsman, I. Ron and A. Pevzner, Analyst, 2020, 145, 6334–6341 RSC.
- G. C. Phan-Quang, H. K. Lee, H. W. Teng, C. S. L. Koh, B. Q. Yim, E. K. M. Tan, W. L. Tok, I. Y. Phang and X. Y. Ling, Angew. Chem., Int. Ed., 2018, 57, 5792–5796 CrossRef CAS PubMed.
- X. Zhang, K. Zhang, H. von Bredow, C. Metting, G. Atanasoff, R. M. Briber and O. Rabin, Front. Phys., 2022, 9, 752943 CrossRef.
- Y. Huang, W. Liu, Z. Gong, W. Wu, M. Fan, D. Wang and A. G. Brolo, ACS Sens., 2020, 5, 2933–2939 CrossRef CAS PubMed.
- K. R. Kumar, D. Banerjee, A. Mangababu, R. S. P. Goud, A. P. Pathak, V. R. Soma and S. V. S. N. Rao, J. Phys. D: Appl. Phys., 2022, 55, 405103 CrossRef CAS.
- D. Wang, Z. Gong, M. Tang, W. Fan, B. Huang and M. Fan, Anal. Methods, 2022, 14, 3798–3801 RSC.
- D. Banerjee, M. Akkanaboina, S. Ghosh and V. R. Soma, Materials, 2022, 15, 4155 CrossRef CAS.
- G. A. Khan, Ö. Demirtaş, A. K. Demir, Ö. Aytekin, A. Bek, A. S. Bhatti and W. Ahmed, Colloids Surf., A, 2021, 619, 126542 CrossRef CAS.
- A. K. Verma and R. K. Soni, Opt. Mater., 2023, 139, 113820 CrossRef CAS.
- M. S. S. Bharati and V. R. Soma, Opto-Electron. Adv., 2021, 4, 210048 CAS.
- S. Kalasung, K. Aiempanakit, I. Chatnuntawech, N. Limsuwan, K. Lertborworn, V. Patthanasettakul, M. Horprathum, N. Nuntawong and P. Eiamchai, Sens Actuators, B, 2022, 366, 131986 CrossRef CAS.
- M. Akkanaboina, D. Banerjee, K. R. Kumar, R. S. P. Goud, V. R. Soma and S. V. S. N. Rao, Opt. Lett., 2023, 48, 5539–5542 CrossRef CAS PubMed.
- D. Banerjee, M. Akkanaboina, R. K. Kanaka, B. Ghose and V. R. Soma, J. Phys. Chem. C, 2024, 128, 4655–4665 CrossRef CAS.
- H. Marrapu, R. Avasarala, V. R. Soma, S. K. Balivada and G. K. Podagatlapalli, RSC Adv., 2020, 10, 41217–41228 RSC.
- M. Akkanaboina, D. Banerjee, K. R. Kumar, R. S. P. Goud, V. R. Soma and S. V. S. N. Rao, Surf. Interfaces, 2023, 36, 102563 CrossRef CAS.
- J. Yin, T. Wu, J. Song, Q. Zhang, S. Liu, R. Xu and H. Duan, Chem. Mater., 2011, 23, 4756–4764 CrossRef CAS.
- V. M. Zamarion, R. A. Timm, K. Araki and H. E. Toma, Inorg. Chem., 2008, 47, 2934–2936 CrossRef CAS PubMed.
- J. Li, L. Chen, T. Lou and Y. Wang, ACS Appl. Mater. Interfaces, 2011, 3, 3936–3941 CrossRef CAS PubMed.
- J. Li, B. Zheng, Z. Zheng, Y. Li and J. Wang, Sens. Actuators Rep., 2020, 2(1), 100013 CrossRef.
- X. Ding, L. Kong, J. Wang, F. Fang, D. Li and J. Liu, ACS Appl. Mater. Interfaces, 2013, 5, 7072–7078 CrossRef CAS.
- C. Song, B. Yang, Y. Yang and L. Wang, Sci. China: Chem., 2016, 59, 16–29 CrossRef CAS.
- Q. Zou, X. Li, T. Xue, J. Zheng and Q. Su, Talanta, 2019, 195, 497–505 CrossRef CAS PubMed.
- Y. Wang and J. Irudayaraj, Chem. Commun., 2011, 47, 4394–4396 RSC.
- Y. X. Yuan, L. Ling, X. Y. Wang, M. Wang, R. A. Gu and J. L. Yao, J. Raman Spectrosc., 2007, 38, 1280–1287 CrossRef CAS.
- L. Polavarapu, J. Pérez-Juste, Q. H. Xu and L. M. Liz-Marzán, J. Mater. Chem. C, 2014, 2, 7460–7476 RSC.
- G. Wang, C. Lim, L. Chen, H. Chon, J. Choo, J. Hong and A. J. deMello, Anal. Bioanal. Chem., 2009, 394, 1827–1832 CrossRef CAS PubMed.
- S. De Marchi, D. García-Lojo, G. Bodelón, J. Pérez-Juste and I. Pastoriza-Santos, ACS Appl. Mater. Interfaces, 2021, 13, 61587–61597 CrossRef CAS PubMed.
- D. Han, S. Y. Lim, B. J. Kim, L. Piao and T. D. Chung, Chem. Commun., 2010, 46, 5587 RSC.
- S. Tan, M. Erol, S. Sukhishvili and H. Du, Langmuir, 2008, 24, 4765–4771 CrossRef CAS PubMed.
- S. W. Bishnoi, C. J. Rozell, C. S. Levin, M. K. Gheith, B. R. Johnson, D. H. Johnson and N. J. Halas, Nano Lett., 2006, 6, 1687–1692 CrossRef CAS PubMed.
- L. S. Lawson, J. W. Chan and T. Huser, Nanoscale, 2014, 6, 7971–7980 RSC.
- L. Bi, Y. Wang, Y. Yang, Y. Li, S. Mo, Q. Zheng and L. Chen, ACS Appl. Mater. Interfaces, 2018, 10, 15381–15387 CrossRef CAS PubMed.
- K. V. Kong, U. S. Dinish, W. K. O. Lau and M. Olivo, Biosens. Bioelectron., 2014, 54, 135–140 CrossRef CAS PubMed.
- B. Liu, Y. Huang, W. Zheng, D. Wang and M. Fan, Anal. Methods, 2022, 14, 1856–1861 RSC.
- X. S. Zheng, P. Hu, Y. Cui, C. Zong, J. M. Feng, X. Wang and B. Ren, Anal. Chem., 2014, 86, 12250–12257 CrossRef CAS.
- C. S. L. Koh, H. K. Lee, X. Han, H. Y. F. Sim and X. Y. Ling, Chem. Commun., 2018, 54, 2546–2549 RSC.
- J. Prakash, P. R. de Oliveira, H. C. Swart, M. Rumyantseva, M. Packirisamy, B. C. Janegitz and X. Li, Sens. Diagn., 2022, 1, 1143–1164 RSC.
- M. Lafuente, S. De Marchi, M. Urbiztondo, I. Pastoriza-Santos, I. Pérez-Juste, J. Santamaría, R. Mallada and M. Pina, ACS Sens., 2021, 6, 2241–2251 CrossRef CAS PubMed.
- M. Lafuente, D. Sanz, M. Urbiztondo, J. Santamaría, M. P. Pina and R. Mallada, J. Hazard. Mater., 2020, 384, 121279 CrossRef CAS PubMed.
- A. Hakonen, P. O. Andersson, M. Stenbæk Schmidt, T. Rindzevicius and M. Käll, Anal. Chim. Acta, 2015, 893, 1–13 CrossRef CAS PubMed.
- Y. Chen, Y. Zhang, F. Pan, J. Liu, K. Wang, C. Zhang, S. Cheng, L. Lu, W. Zhang, Z. Zhang, X. Zhi, Q. Zhang, G. Alfranca, J. M. de la Fuente, D. Chen and D. Cui, ACS Nano, 2016, 10, 8169–8179 CrossRef CAS PubMed.
- J. Fu, Z. Zhong, D. Xie, Y. Guo, D. Kong, Z. Zhao, Z. Zhao and M. Li, Angew. Chem., Int. Ed., 2020, 59, 20489–20498 CrossRef CAS.
- X. Qiao, B. Su, C. Liu, Q. Song, D. Luo, G. Mo and T. Wang, Adv. Mater., 2018, 30, 1–8 Search PubMed.
- K. Chang, Y. Zhao, M. Wang, Z. Xu, L. Zhu, L. Xu and Q. Wang, Chem. Eng. J., 2023, 459, 141539 CrossRef CAS.
- N. Taranenko, J.-P. Alarie, D. L. Stokes and T. Vo-Dinh, J. Raman Spectrosc., 1996, 27, 379–384 CrossRef CAS.
- D. A. Stuart, K. B. Biggs and R. P. Van Duyne, Analyst, 2006, 131, 568 RSC.
- R. K. Lauridsen, P. B. Skou, T. Rindzevicius, K. Wu, S. Molin, S. B. Engelsen, K. G. Nielsen, H. K. Johansen and A. Boisen, Anal. Methods, 2017, 9, 5757–5762 RSC.
- C. L. Wong, U. S. Dinish, M. S. Schmidt and M. Olivo, Anal. Chim. Acta, 2014, 844, 54–60 CrossRef CAS PubMed.
- X. He, H. Wang, Z. Li, D. Chen, J. Liu and Q. Zhang, Nanoscale, 2015, 7, 8619–8626 RSC.
- S. Kim, D.-H. Kim and S.-G. Park, Analyst, 2018, 143, 3006–3010 RSC.
- S. Kim, H. S. Jung, D.-H. Kim, S.-H. Kim and S.-G. Park, Analyst, 2019, 144, 7162–7167 RSC.
- S. D. Christesen, Appl. Spectrosc., 1988, 42, 318–321 CrossRef CAS.
- Y. Liu, M. Kim, S. H. Cho and Y. S. Jung, Nano Today, 2021, 37, 101063 CrossRef CAS.
- V. M. Szlag, R. S. Rodriguez, J. He, N. Hudson-Smith, H. Kang, N. Le, T. M. Reineke and C. L. Haynes, ACS Appl. Mater. Interfaces, 2018, 10, 31825–31844 CrossRef CAS.
- M. Oh, R. De and S. Yim, J. Raman Spectrosc., 2018, 49, 800–809 CrossRef CAS.
- E.-C. Lin, J. Fang, S.-C. Park, F. W. Johnson and H. O. Jacobs, Nat. Commun., 2013, 4, 1636 CrossRef PubMed.
- M. Lafuente, E. Berenschot, R. Tiggelaar, R. Mallada, N. Tas and M. Pina, Micromachines, 2018, 9, 60 CrossRef PubMed.
- A. Chauvin, M. Lafuente, J. Y. Mevellec, R. Mallada, B. Humbert, M. P. Pina, P.-Y. Tessier and A. El Mel, Nanoscale, 2020, 12, 12602–12612 RSC.
- K. J. Squire, K. Sivashanmugan, B. Zhang, J. Kraai, G. Rorrer and A. X. Wang, ACS Appl. Nano Mater., 2020, 3, 1656–1665 CrossRef CAS.
- K. Huang, S. Gong, L. Zhang, H. Zhang, S. Li, G. Ye and F. Huang, Chem. Commun., 2021, 57, 2144–2147 RSC.
- K. Yang, K. Zhu, Y. Wang, Z. Qian, Y. Zhang, Z. Yang, Z. Wang, L. Wu, S. Zong and Y. Cui, ACS Nano, 2021, 15, 12996–13006 CrossRef CAS PubMed.
- K. Yang, C. Zhang, K. Zhu, Z. Qian, Z. Yang, L. Wu, S. Zong, Y. Cui and Z. Wang, ACS Nano, 2022, 16, 19335–19345 CrossRef CAS PubMed.
- M. Mueller, M. Tebbe, D. V. Andreeva, M. Karg, R. A. Alvarez Puebla, N. Pazos Perez and A. Fery, Langmuir, 2012, 28, 9168–9173 CrossRef CAS PubMed.
- L. B. T. Nguyen, Y. X. Leong, C. S. L. Koh, S. X. Leong, S. K. Boong, H. Y. F. Sim, G. C. Phan-Quang, I. Y. Phang and X. Y. Ling, Angew. Chem., 2022, 61(33), e202207447 CrossRef CAS PubMed.
- S. X. Leong, Y. X. Leong, E. X. Tan, H. Y. F. Sim, C. S. L. Koh, Y. H. Lee, C. Chong, L. S. Ng, J. R. T. Chen, D. W. C. Pang, L. B. T. Nguyen, S. K. Boong, X. Han, Y.-C. Kao, Y. H. Chua, G. C. Phan-Quang, I. Y. Phang, H. K. Lee, M. Y. Abdad, N. S. Tan and X. Y. Ling, ACS Nano, 2022, 16, 2629–2639 CrossRef CAS PubMed.
- H. Bao, H. Zhang, H. Fu, L. Zhou, P. Zhang, Y. Li and W. Cai, Nanoscale Horiz., 2020, 5, 739–746 RSC.
- W. Xu, H. Bao, H. Zhang, H. Fu, Q. Zhao, Y. Li and W. Cai, J. Hazard. Mater., 2021, 420, 126668 CrossRef CAS PubMed.
- Z. Zhao, H. Bao, Q. Zhao, H. Fu, L. Zhou, H. Zhang, Y. Li and W. Cai, ACS Appl. Mater. Interfaces, 2022, 14, 47999–48010 CrossRef CAS PubMed.
- H. Bao, Y. Guo, T. Zhang, H. Fu, S. Zhu, L. Zhou, H. Zhang, Y. Li and W. Cai, Adv. Mater. Interfaces, 2022, 9, 1–10 Search PubMed.
- M. Lafuente, F. Almazan, E. Bernad, I. Florea, R. Arenal, M. A. Urbiztondo, R. Mallada and M. P. Pina, Lab Chip, 2023, 3160 RSC.
- L. He, Y. Liu, J. Liu, Y. Xiong, J. Zheng, Y. Liu and Z. Tang, Angew. Chem., Int. Ed., 2013, 52, 3741–3745 CrossRef CAS PubMed.
- L. E. Kreno, N. G. Greeneltch, O. K. Farha, J. T. Hupp and R. P. Van Duyne, Analyst, 2014, 139, 4073 RSC.
- J. Chen, L. Guo, L. Chen, B. Qiu, G. Hong and Z. Lin, ACS Sens., 2020, 5, 3964–3970 CrossRef CAS PubMed.
- C. Huang, A. Li, X. Chen and T. Wang, Small, 2020, 16, 1–17 Search PubMed.
- T. Zorlu, M. A. Correa-Duarte and R. A. Alvarez-Puebla, J. Chem. Phys., 2023, 158, 171001 CrossRef CAS PubMed.
- G. C. Phan-Quang, N. Yang, H. K. Lee, H. Y. F. Sim, C. S. L. Koh, Y.-C. Kao, Z. C. Wong, E. K. M. Tan, Y.-E. Miao, W. Fan, T. Liu, I. Y. Phang and X. Y. Ling, ACS Nano, 2019, 13, 12090–12099 CrossRef CAS PubMed.
- X. Qiao, B. Su, C. Liu, Q. Song, D. Luo, G. Mo and T. Wang, Adv. Mater., 2018, 30, 1–8 Search PubMed.
- Q.-Q. Chen, R.-N. Hou, Y.-Z. Zhu, X.-T. Wang, H. Zhang, Y.-J. Zhang, L. Zhang, Z.-Q. Tian and J.-F. Li, Anal. Chem., 2021, 93, 7188–7195 CrossRef CAS PubMed.
- K. Yang, S. Zong, Y. Zhang, Z. Qian, Y. Liu, K. Zhu, L. Li, N. Li, Z. Wang and Y. Cui, ACS Appl. Mater. Interfaces, 2020, 12, 1395–1403 CrossRef CAS PubMed.
- A. Li, X. Qiao, K. Liu, W. Bai and T. Wang, Adv. Funct. Mater., 2022, 32, 1–7 Search PubMed.
- Y. X. Leong, Y. H. Lee, C. S. L. Koh, G. C. Phan-Quang, X. Han, I. Y. Phang and X. Y. Ling, Nano Lett., 2021, 21, 2642–2649 CrossRef CAS PubMed.
- K. Nemciauskas, L. Traksele, A. Salaseviciene and V. Snitka, Microelectron. Eng., 2020, 225, 111282 CrossRef CAS.
- J. Plou, M. Charconnet, I. García, J. Calvo and L. M. Liz-Marzán, ACS Nano, 2021, 15, 8984–8995 CrossRef CAS PubMed.
- L. Ma, H. Wu, Y. Huang, S. Zou, J. Li and Z. Zhang, ACS Appl. Mater. Interfaces, 2016, 8, 27162–27168 CrossRef CAS PubMed.
- E. Pyrak, J. Krajczewski, A. Kowalik, A. Kudelski and A. Jaworska, Molecules, 2019, 24, 4423 CrossRef CAS PubMed.
- C. Chen, W. Liu, S. Tian and T. Hong, Sensors, 2019, 19, 1712 CrossRef CAS PubMed.
- T. K. Sharma, R. Ramanathan, R. Rakwal, G. K. Agrawal and V. Bansal, Proteomics, 2015, 15, 1680–1692 CrossRef CAS PubMed.
- G. K. Agrawal, A. M. Timperio, L. Zolla, V. Bansal, R. Shukla and R. Rakwal, J. Proteomics, 2013, 93, 74–92 CrossRef CAS PubMed.
- D. J. Sarkar, B. K. Behera, P. K. Parida, V. K. Aralappanavar, S. Mondal, J. Dei, B. K. Das, S. Mukherjee, S. Pal, P. Weerathunge, R. Ramanathan and V. Bansal, Biosens. Bioelectron., 2023, 219, 114771 CrossRef CAS PubMed.
- F. Arshad, M. Deliorman, P. Sukumar, M. A. Qasaimeh, J. S. Olarve, G. N. Santos, V. Bansal and M. U. Ahmed, Trends Food Sci. Technol., 2023, 136, 145–158 CrossRef CAS.
- E. Pyrak, A. Kowalczyk, J. L. Weyher, A. M. Nowicka and A. Kudelski, Spectrochim. Acta, Part A, 2023, 295, 122606 CrossRef CAS PubMed.
- A. Szaniawska and A. Kudelski, Front. Chem., 2021, 9, 664134 CrossRef CAS PubMed.
- B. K. Behera, B. Dehury, A. K. Rout, B. Patra, N. Mantri, H. J. Chakraborty, D. J. Sarkar, N. K. Kaushik, V. Bansal, I. Singh, B. K. Das, A. R. Rao and A. Rai, Gen. Rep., 2021, 25, 101372 CrossRef CAS.
- X. Wei, S. Su, Y. Guo, X. Jiang, Y. Zhong, Y. Su, C. Fan, S. Lee and Y. He, Small, 2013, 9, 2493–2499 CrossRef CAS PubMed.
- Y. He, S. Su, T. Xu, Y. Zhong, J. A. Zapien, J. Li, C. Fan and S.-T. Lee, Nano Today, 2011, 6, 122–130 CrossRef CAS.
- L.-J. Xu, Z.-C. Lei, J. Li, C. Zong, C. J. Yang and B. Ren, J. Am. Chem. Soc., 2015, 137, 5149–5154 CrossRef CAS PubMed.
- E. Papadopoulou and S. E. J. Bell, Chem. Commun., 2011, 47, 10966 RSC.
- A. Barhoumi, D. Zhang, F. Tam and N. J. Halas, J. Am. Chem. Soc., 2008, 130, 5523–5529 CrossRef CAS PubMed.
- E. Papadopoulou and S. E. J. Bell, Angew. Chem., Int. Ed., 2011, 50, 9058–9061 CrossRef CAS PubMed.
- E. Papadopoulou and S. E. J. Bell, Chem. – Eur. J., 2012, 18, 5394–5400 CrossRef CAS PubMed.
- Y. Li, X. Han, Y. Yan, Y. Cao, X. Xiang, S. Wang, B. Zhao and X. Guo, Anal. Chem., 2018, 90, 2996–3000 CrossRef CAS PubMed.
- Y. Li, X. Han, S. Zhou, Y. Yan, X. Xiang, B. Zhao and X. Guo, J. Phys. Chem. Lett., 2018, 9, 3245–3252 CrossRef CAS PubMed.
- T. Zhang, X. Quan, N. Cao, Z. Zhang and Y. Li, Nanomaterials, 2022, 12, 3119 CrossRef CAS PubMed.
- D. Li, L. Xia, Q. Zhou, L. Wang, D. Chen, X. Gao and Y. Li, Anal. Chem., 2020, 92, 12769–12773 CrossRef CAS PubMed.
- Y. Li, T. Gao, G. Xu, X. Xiang, B. Zhao, X. X. Han and X. Guo, Anal. Chem., 2019, 91, 7980–7984 CrossRef CAS PubMed.
- E. Prado, A. Colin, L. Servant and S. Lecomte, J. Phys. Chem. C, 2014, 118, 13965–13971 CrossRef CAS.
- L. Guerrini, Ž. Krpetić, D. van Lierop, R. A. Alvarez-Puebla and D. Graham, Angew. Chem., Int. Ed., 2015, 54, 1144–1148 CrossRef CAS PubMed.
- J. Morla-Folch, H. Xie, P. Gisbert-Quilis, S. G. Pedro, N. Pazos-Perez, R. A. Alvarez-Puebla and L. Guerrini, Angew. Chem., Int. Ed., 2015, 54, 13650–13654 CrossRef CAS PubMed.
- J. Morla-Folch, R. A. Alvarez-Puebla and L. Guerrini, J. Phys. Chem. Lett., 2016, 7, 3037–3041 CrossRef CAS PubMed.
- A. Barhoumi and N. J. Halas, J. Am. Chem. Soc., 2010, 132, 12792–12793 CrossRef CAS PubMed.
- Y. Qian, T. Fan, Y. Yao, X. Shi, X. Liao, F. Zhou and F. Gao, Sens. Actuators, B, 2018, 254, 483–489 CrossRef CAS.
- Y. Yao, H. Zhang, T. Tian, Y. Liu, R. Zhu, J. Ji and B. Liu, Talanta, 2021, 235, 122728 CrossRef CAS PubMed.
- E. Bailo and V. Deckert, Angew. Chem., Int. Ed., 2008, 47, 1658–1661 CrossRef CAS PubMed.
- Y. C. Cao, R. Jin and C. A. Mirkin, Science, 2002, 297, 1536–1540 CrossRef CAS PubMed.
- R. Jin, Y. C. Cao, C. S. Thaxton and C. A. Mirkin, Small, 2006, 2, 375–380 CrossRef CAS PubMed.
- Z. Y. Jiang, X. X. Jiang, S. Su, X. P. Wei, S. T. Lee and Y. He, Appl. Phys. Lett., 2012, 100(20), 203104 CrossRef.
- T. Kang, S. M. Yoo, I. Yoon, S. Y. Lee and B. Kim, Nano Lett., 2010, 10, 1189–1193 CrossRef CAS PubMed.
- G. Braun, S. J. Lee, M. Dante, T.-Q. Nguyen, M. Moskovits and N. Reich, J. Am. Chem. Soc., 2007, 129, 6378–6379 CrossRef CAS PubMed.
- J.-M. Li, C. Wei, W.-F. Ma, Q. An, J. Guo, J. Hu and C.-C. Wang, J. Mater. Chem., 2012, 22, 12100 RSC.
- S. He, K.-K. Liu, S. Su, J. Yan, X. Mao, D. Wang, Y. He, L.-J. Li, S. Song and C. Fan, Anal. Chem., 2012, 84, 4622–4627 CrossRef CAS PubMed.
- D. Graham, D. G. Thompson, W. E. Smith and K. Faulds, Nat. Nanotechnol., 2008, 3, 548–551 CrossRef CAS PubMed.
- X. Qian, X. Zhou and S. Nie, J. Am. Chem. Soc., 2008, 130, 14934–14935 CrossRef PubMed.
- T. Donnelly, W. E. Smith, K. Faulds and D. Graham, Chem. Commun., 2014, 50, 12907–12910 RSC.
- J.-M. Li, W.-F. Ma, L.-J. You, J. Guo, J. Hu and C.-C. Wang, Langmuir, 2013, 29, 6147–6155 CrossRef CAS PubMed.
- H. Zhang, M. H. Harpster, W. C. Wilson and P. A. Johnson, Langmuir, 2012, 28, 4030–4037 CrossRef CAS PubMed.
- H. Zhang, M. H. Harpster, H. J. Park, P. A. Johnson and W. C. Wilson, Anal. Chem., 2011, 83, 254–260 CrossRef CAS PubMed.
- X. Fu, Z. Cheng, J. Yu, P. Choo, L. Chen and J. Choo, Biosens. Bioelectron., 2016, 78, 530–537 CrossRef CAS PubMed.
- X. Wang, N. Choi, Z. Cheng, J. Ko, L. Chen and J. Choo, Anal. Chem., 2017, 89, 1163–1169 CrossRef CAS PubMed.
- A. MacAskill, D. Crawford, D. Graham and K. Faulds, Anal. Chem., 2009, 81, 8134–8140 CrossRef CAS PubMed.
- L. Fabris, M. Dante, G. Braun, S. J. Lee, N. O. Reich, M. Moskovits, T.-Q. Nguyen and G. C. Bazan, J. Am. Chem. Soc., 2007, 129, 6086–6087 CrossRef CAS PubMed.
- C. Yuan, J. Fang, M. L. de la Chapelle, Y. Zhang, X. Zeng, G. Huang, X. Yang and W. Fu, TrAC, Trends Anal. Chem., 2021, 143, 116401 CrossRef CAS.
- D. Ma, C. Huang, J. Zheng, J. Tang, J. Li, J. Yang and R. Yang, Biosens. Bioelectron., 2018, 101, 167–173 CrossRef CAS PubMed.
- W. Xu, A. Zhao, F. Zuo, R. Khan, H. M. J. Hussain and J. Chang, Microchim. Acta, 2020, 187, 384 CrossRef CAS PubMed.
- Y. Pang, C. Wang, J. Wang, Z. Sun, R. Xiao and S. Wang, Biosens. Bioelectron., 2016, 79, 574–580 CrossRef CAS PubMed.
- Y. Pang, C. Wang, L. Lu, C. Wang, Z. Sun and R. Xiao, Biosens. Bioelectron., 2019, 130, 204–213 CrossRef CAS PubMed.
- X. Yang, S. Wang, Y. Wang, Y. He, Y. Chai and R. Yuan, ACS Appl. Mater. Interfaces, 2018, 10, 12491–12496 CrossRef CAS PubMed.
- Y. He, X. Yang, R. Yuan and Y. Chai, Anal. Chem., 2017, 89, 8538–8544 CrossRef CAS PubMed.
- J. Hu and C. Zhang, Anal. Chem., 2010, 82, 8991–8997 CrossRef CAS PubMed.
- B. Guven, I. H. Boyaci, U. Tamer, E. Acar-Soykut and U. Dogan, Talanta, 2015, 136, 68–74 CrossRef CAS PubMed.
- Y. He, X. Yang, R. Yuan and Y. Chai, Anal. Chem., 2017, 89, 2866–2872 CrossRef CAS PubMed.
- C. Liu, C. Chen, S. Li, H. Dong, W. Dai, T. Xu, Y. Liu, F. Yang and X. Zhang, Anal. Chem., 2018, 90, 10591–10599 CrossRef CAS PubMed.
- Y. Si, L. Xu, T. Deng, J. Zheng and J. Li, ACS Sens., 2020, 5, 4009–4016 CrossRef CAS PubMed.
- W. Wang, Y. Li, A. Nie, G.-C. Fan and H. Han, Analyst, 2021, 146, 848–854 RSC.
- Y. Sun and T. Li, Anal. Chem., 2018, 90, 11614–11621 CrossRef CAS PubMed.
- Y. Mao, Y. Sun, J. Xue, W. Lu and X. Cao, Anal. Chim. Acta, 2021, 1178, 338800 CrossRef CAS PubMed.
- J. Chen, Y. Wu, C. Fu, H. Cao, X. Tan, W. Shi and Z. Wu, Biosens. Bioelectron., 2019, 143, 111619 CrossRef CAS PubMed.
- J. Zhang, X. Miao, C. Song, N. Chen, J. Xiong, H. Gan, J. Ni, Y. Zhu, K. Cheng and L. Wang, Biosens. Bioelectron., 2022, 212, 114379 CrossRef CAS PubMed.
- S. Ye, Y. Wu, X. Zhai and B. Tang, Anal. Chem., 2015, 87, 8242–8249 CrossRef CAS PubMed.
- F. Gao, J. Lei and H. Ju, Anal. Chem., 2013, 85, 11788–11793 CrossRef CAS PubMed.
- Y. Zhao, L. Liu, H. Kuang, L. Wang and C. Xu, RSC Adv., 2014, 4, 56052–56056 RSC.
- B. Yin, Q. Zhang, X. Xia, C. Li, W. K. H. Ho, J. Yan, Y. Huang, H. Wu, P. Wang, C. Yi, J. Hao, J. Wang, H. Chen, S. H. D. Wong and M. Yang, Theranostics, 2022, 12, 5914–5930 CrossRef CAS PubMed.
- Y. Pang, Q. Li, C. Wang, S. Zhen, Z. Sun and R. Xiao, Chem. Eng. J., 2022, 429, 132109 CrossRef CAS.
- J. Liang, P. Teng, W. Xiao, G. He, Q. Song, Y. Zhang, B. Peng, G. Li, L. Hu, D. Cao and Y. Tang, J. Nanobiotechnology, 2021, 19, 273 CrossRef CAS PubMed.
- C. Lin, S. Liang, Y. Peng, L. Long, Y. Li, Z. Huang, N. V. Long, X. Luo, J. Liu, Z. Li and Y. Yang, Nanomicro Lett., 2022, 14, 75 CAS.
- P. Senthil Kumar, I. Pastoriza-Santos, B. Rodríguez-González, F. Javier García de Abajo and L. M. Liz-Marzán, Nanotechnology, 2008, 19, 015606 CrossRef PubMed.
- A. Guerrero-Martínez, S. Barbosa, I. Pastoriza-Santos and L. M. Liz-Marzán, Curr. Opin. Colloid Interface Sci., 2011, 16, 118–127 CrossRef.
- R. A. Alvarez-Puebla, A. Agarwal, P. Manna, B. P. Khanal, P. Aldeanueva-Potel, E. Carbó-Argibay, N. Pazos-Pérez, L. Vigderman, E. R. Zubarev, N. A. Kotov and L. M. Liz-Marzán, Proc. Natl. Acad. Sci., 2011, 108, 8157–8161 CrossRef CAS PubMed.
- P. Kusch, S. Mastel, N. S. Mueller, N. Morquillas Azpiazu, S. Heeg, R. Gorbachev, F. Schedin, U. Hübner, J. I. Pascual, S. Reich and R. Hillenbrand, Nano Lett., 2017, 17, 2667–2673 CrossRef CAS PubMed.
- Y. Xu, M. P. Konrad, W. W. Y. Lee, Z. Ye and S. E. J. Bell, Nano Lett., 2016, 16, 5255–5260 CrossRef CAS PubMed.
- M. P. Konrad, A. P. Doherty and S. E. J. Bell, Anal. Chem., 2013, 85, 6783–6789 CrossRef CAS PubMed.
- Y. He, X. Yang, R. Yuan and Y. Chai, J. Mater. Chem. B, 2019, 7, 2643–2647 RSC.
- A. Woo, K. Lim, B. H. Cho, H. S. Jung and M. Lee, Anal. Sci. Ad., 2021, 2, 397–407 CrossRef CAS PubMed.
- M. Kim, S. M. Ko, C. Lee, J. Son, J. Kim, J.-M. Kim and J.-M. Nam, Anal. Chem., 2019, 91, 10467–10476 CrossRef CAS PubMed.
- D.-K. Lim, K.-S. Jeon, J.-H. Hwang, H. Kim, S. Kwon, Y. D. Suh and J.-M. Nam, Nat. Nanotechnol., 2011, 6, 452–460 CrossRef CAS PubMed.
- J.-E. Shim, Y. J. Kim, J.-H. Choe, T. G. Lee and E.-A. You, ACS Appl. Mater. Interfaces, 2022, 14, 38459–38470 CrossRef CAS PubMed.
- M. Kim, S. M. Ko, J.-M. Kim, J. Son, C. Lee, W.-K. Rhim and J.-M. Nam, ACS Cent. Sci., 2018, 4, 277–287 CrossRef CAS PubMed.
- O. Guselnikova, A. Trelin, A. Skvortsova, P. Ulbrich, P. Postnikov, A. Pershina, D. Sykora, V. Svorcik and O. Lyutakov, Biosens. Bioelectron., 2019, 145, 111718 CrossRef CAS PubMed.
- Y. Yang, H. Li, L. Jones, J. Murray, J. Haverstick, H. K. Naikare, Y.-Y. C. Mosley, R. A. Tripp, B. Ai and Y. Zhao, ACS Sens., 2023, 8, 297–307 CrossRef CAS PubMed.
- M. Prochazka, Surface-Enhanced Raman Spectroscopy: Bioanalytical, Biomolecular and Medical Applications, Springer International Publishing, Cham, 2016, pp. 149–211 Search PubMed.
- Y. Wu, M. R. K. Ali, K. Chen, N. Fang and M. A. El-Sayed, Nano Today, 2019, 24, 120–140 CrossRef CAS.
- S. E. J. Bell, G. Charron, E. Cortés, J. Kneipp, M. L. de la Chapelle, J. Langer, M. Procházka, V. Tran and S. Schlücker, Angew. Chem., Int. Ed., 2020, 59, 5454–5462 CrossRef CAS PubMed.
- Y. Zhang, H. Hong, D. V. Myklejord and W. Cai, Small, 2011, 7, 3261–3269 CrossRef CAS PubMed.
- Y. Li, Z. Wang, X. Mu, A. Ma and S. Guo, Biotechnol. Adv., 2017, 35, 168–177 CrossRef CAS PubMed.
- R. M. Crist, S. S. K. Dasa, C. H. Liu, J. D. Clogston, M. A. Dobrovolskaia and S. T. Stern, WIREs Nanomed. Nanobiotechnol., 2021, 13, e1665 CrossRef CAS PubMed.
- B. Andreiuk, F. Nicolson, L. M. Clark, S. R. Panikkanvalappil, M. Rashidian, S. Harmsen and M. F. Kircher, Nanotheranostics, 2022, 6, 10–30 CrossRef PubMed.
- B. Tim, M. Rojewska and K. Prochaska, Int. J. Mol. Sci., 2022, 23, 12822 CrossRef CAS PubMed.
- Y.-C. Ou, J. A. Webb, C. M. O’Brien, I. J. Pence, E. C. Lin, E. P. Paul, D. Cole, S.-H. Ou, M. Lapierre-Landry, R. C. DeLapp, E. S. Lippmann, A. Mahadevan-Jansen and R. Bardhan, Nanoscale, 2018, 10, 13092–13105 RSC.
- C. L. Zavaleta, B. R. Smith, I. Walton, W. Doering, G. Davis, B. Shojaei, M. J. Natan and S. S. Gambhir, Proc. Natl. Acad. Sci., 2009, 106, 13511–13516 CrossRef CAS PubMed.
- J. H. Yu, I. Steinberg, R. M. Davis, A. V. Malkovskiy, A. Zlitni, R. K. Radzyminski, K. O. Jung, D. T. Chung, L. D. Curet, A. L. D’Souza, E. Chang, J. Rosenberg, J. Campbell, H. Frostig, S. Park, G. Pratx, C. Levin and S. S. Gambhir, ACS Nano, 2021, 15, 19956–19969 CrossRef CAS PubMed.
- S. Bock, Y.-S. Choi, M. Kim, Y. Yun, X.-H. Pham, J. Kim, B. Seong, W. Kim, A. Jo, K.-M. Ham, S. G. Lee, S. H. Lee, H. Kang, H. S. Choi, D. H. Jeong, H. Chang, D.-E. Kim and B.-H. Jun, J. Nanobiotechnol., 2022, 20, 130 CrossRef CAS PubMed.
- O. E. Eremina, A. T. Czaja, A. Fernando, A. Aron, D. B. Eremin and C. Zavaleta, ACS Nano, 2022, 16, 10341–10353 CrossRef CAS PubMed.
- G. Cutshaw, S. Uthaman, N. Hassan, S. Kothadiya, X. Wen and R. Bardhan, Chem. Rev., 2023, 123, 8297–8346 CrossRef CAS PubMed.
- J. Deal, S. Mayes, C. Browning, S. Hill, P. Rider, C. Boudreaux, T. C. Rich and S. J. Leavesley, J. Biomed. Opt., 2018, 24, 1 Search PubMed.
- J. Wang, D. Liang, J. Feng and X. Tang, Anal. Chem., 2019, 91, 11045–11054 CrossRef CAS PubMed.
- Y. Zou, S. Huang, Y. Liao, X. Zhu, Y. Chen, L. Chen, F. Liu, X. Hu, H. Tu, L. Zhang, Z. Liu, Z. Chen and W. Tan, Chem. Sci., 2018, 9, 2842–2849 RSC.
- F. Nicolson, M. F. Kircher, N. Stone and P. Matousek, Chem. Soc. Rev., 2021, 50, 556–568 RSC.
- F. Nicolson, B. Andreiuk, C. Andreou, H.-T. Hsu, S. Rudder and M. F. Kircher, Theranostics, 2019, 9, 5899–5913 CrossRef CAS PubMed.
- P. Strobbia, V. Cupil-Garcia, B. M. Crawford, A. M. Fales, T. J. Pfefer, Y. Liu, M. Maiwald, B. Sumpf and T. Vo-Dinh, Theranostics, 2021, 11, 4090–4102 CrossRef CAS PubMed.
- J. L. Rutter, D. B. Goldstein, A. Philippakis, J. W. Smoller, G. Jenkins, E. Dishman and J. C. Denny, N. Engl. J. Med., 2019, 381, 668 CrossRef PubMed.
- A. P. Rao, N. Bokde and S. Sinha, Appl. Sci., 2020, 10, 767 CrossRef CAS.
- C. Zhang, Z. Xu, H. Di, E. Zeng, Y. Jiang and D. Liu, Theranostics, 2020, 10, 6061–6071 CrossRef CAS PubMed.
- M. A. Wall, T. M. Shaffer, S. Harmsen, D.-F. Tschaharganeh, C.-H. Huang, S. W. Lowe, C. M. Drain and M. F. Kircher, Theranostics, 2017, 7, 3068–3077 CrossRef CAS PubMed.
- B. Shi, D. Li, W. Yao, W. Wang, J. Jiang, R. Wang, F. Yan, H. Liu, H. Zhang and J. Ye, Biomater. Sci., 2022, 10, 2577–2589 RSC.
- Y.-C. Ou, X. Wen, C. A. Johnson, D. Shae, O. D. Ayala, J. A. Webb, E. C. Lin, R. C. DeLapp, K. L. Boyd, A. Richmond, A. Mahadevan-Jansen, M. Rafat, J. T. Wilson, J. M. Balko, M. N. Tantawy, A. E. Vilgelm and R. Bardhan, ACS Nano, 2020, 14, 651–663 CrossRef CAS PubMed.
- X. Chen, Z. Wu, Y. He, Z. Hao, Q. Wang, K. Zhou, W. Zhou, P. Wang, F. Shan, Z. Li, J. Ji, Y. Fan, Z. Li and S. Yue, Adv. Sci., 2023, 10(21), e2300961 CrossRef PubMed.
- H. G. Lee, W. Choi, S. Y. Yang, D.-H. Kim, S.-G. Park, M.-Y. Lee and H. S. Jung, Sens. Actuators, B, 2021, 326, 128802 CrossRef CAS.
- J. C. Denny, N. Engl. J. Med., 2019, 381, 668–676 CrossRef PubMed.
- R. T. Busch, F. Karim, J. Weis, Y. Sun, C. Zhao and E. S. Vasquez, ACS Omega, 2019, 4, 15269–15279 CrossRef CAS PubMed.
- Y. Liu, L. Zhan, Z. Qin, J. Sackrison and J. C. Bischof, ACS Nano, 2021, 15, 3593–3611 CrossRef CAS PubMed.
- S. Yadav, M. A. Sadique, P. Ranjan, N. Kumar, A. Singhal, A. K. Srivastava and R. Khan, ACS Appl. Bio Mater., 2021, 4, 2974–2995 CrossRef CAS PubMed.
- M. Muhammad, C. Shao and Q. Huang, Spectrochim. Acta, Part A, 2019, 223, 117282 CrossRef CAS PubMed.
- M. R. Willner, K. S. McMillan, D. Graham, P. J. Vikesland and M. Zagnoni, Anal. Chem., 2018, 90, 12004–12010 CrossRef CAS PubMed.
- R. Chen, X. Du, Y. Cui, X. Zhang, Q. Ge, J. Dong and X. Zhao, Small, 2020, 16, 2002801 CrossRef CAS PubMed.
- X. Wen, Y. Ou, H. F. Zarick, X. Zhang, A. B. Hmelo, Q. J. Victor, E. P. Paul, J. M. Slocik, R. R. Naik, L. M. Bellan, E. C. Lin and R. Bardhan, Bioeng. Transl. Med., 2020, 5, e10165 CrossRef CAS PubMed.
- Y. Su, S. Xu, J. Zhang, X. Chen, L.-P. Jiang, T. Zheng and J.-J. Zhu, Anal. Chem., 2019, 91, 864–872 CrossRef CAS PubMed.
- J. Wang, A. Wuethrich, A. A. I. Sina, R. E. Lane, L. L. Lin, Y. Wang, J. Cebon, A. Behren and M. Trau, Sci. Adv., 2020, 6, eaax3223 CrossRef CAS PubMed.
- U. Mogera, H. Guo, M. Namkoong, M. S. Rahman, T. Nguyen and L. Tian, Sci. Adv., 2022, 8, 1736 CrossRef PubMed.
- R. Alder, L. Xiao and S. Fu, Drug Test. Anal., 2021, 13, 944–952 CrossRef CAS PubMed.
- A. Azziz, W. Safar, Y. Xiang, M. Edely and M. Lamy de la Chapelle, J. Mol. Struct., 2022, 1248, 131519 CrossRef CAS.
- S. Abalde-Cela, L. Wu, A. Teixeira, K. Oliveira and L. Diéguez, Adv. Opt. Mater., 2021, 9, 2001171 CrossRef CAS.
- A. N. Ramya, M. M. Joseph, J. B. Nair, V. Karunakaran, N. Narayanan and K. K. Maiti, ACS Appl. Mater. Interfaces, 2016, 8, 10220–10225 CrossRef CAS PubMed.
- Y. Pang, C. Wang, R. Xiao and Z. Sun, Chem. – Eur. J., 2018, 24, 7060–7067 CrossRef CAS PubMed.
- X. Wu, Y. Xia, Y. Huang, J. Li, H. Ruan, T. Chen, L. Luo, Z. Shen and A. Wu, ACS Appl. Mater. Interfaces, 2016, 8, 19928–19938 CrossRef CAS PubMed.
- J. Wang, R. Zhang, X. Ji, P. Wang and C. Ding, Anal. Chim. Acta, 2021, 1141, 206–213 CrossRef CAS PubMed.
- K. Oliveira, A. Teixeira, J. M. Fernandes, C. Lopes, A. Chícharo, P. Piairo, L. Wu, L. Rodríguez-Lorenzo, L. Diéguez and S. Abalde-Cela, Adv. Opt. Mater., 2023, 11, 2201500 CrossRef CAS.
- Y. Chen, X. Bai, L. Su, Z. Du, A. Shen, A. Materny and J. Hu, Sci. Rep., 2016, 6, 19173 CrossRef CAS PubMed.
- W. Nam, X. Ren, S. A. S. Tali, P. Ghassemi, I. Kim, M. Agah and W. Zhou, Nano Lett., 2019, 19, 7273–7281 CrossRef CAS PubMed.
- G. Kuku, M. Altunbek and M. Culha, Anal. Chem., 2017, 89, 11160–11166 CrossRef CAS PubMed.
- A. Kamińska, T. Szymborski, E. Witkowska, E. Kijeńska-Gawrońska, W. Świeszkowski, K. Niciński, J. Trzcińska-Danielewicz and A. Girstun, Nanomaterials, 2019, 9, 366 CrossRef PubMed.
- D. Wu, Y. Chen, S. Hou, W. Fang and H. Duan, ChemBioChem, 2019, 20, 2432–2441 CrossRef CAS PubMed.
- Y. Zhang, X. Ye, G. Xu, X. Jin, M. Luan, J. Lou, L. Wang, C. Huang and J. Ye, RSC Adv., 2016, 6, 5401–5407 RSC.
- J. Plou, I. García, M. Charconnet, I. Astobiza, C. García-Astrain, C. Matricardi, A. Mihi, A. Carracedo and L. M. Liz-Marzán, Adv. Funct. Mater., 2020, 30(17), 1910335 CrossRef CAS.
- J. Plou, P. S. Valera, I. García, D. Vila-Liarte, C. Renero-Lecuna, J. Ruiz-Cabello, A. Carracedo and L. M. Liz-Marzán, Small, 2023, 2207658 CrossRef CAS PubMed.
- M. Tavakkoli Yaraki, A. Tukova and Y. Wang, Nanoscale, 2022, 14, 15242–15268 RSC.
- J. Wang, Y.-C. Kao, Q. Zhou, A. Wuethrich, M. S. Stark, H. Schaider, H. P. Soyer, L. L. Lin and M. Trau, Adv. Funct. Mater., 2022, 32, 2010296 CrossRef CAS.
- A. Teixeira, J. Hernández-Rodríguez, L. Wu, K. Oliveira, K. Kant, P. Piairo, L. Diéguez and S. Abalde-Cela, Appl. Sci., 2019, 9, 1387 CrossRef CAS.
- Z. Du, Y. Qi, J. He, D. Zhong and M. Zhou, WIREs Nanomed. Nanobiotechnol., 2021, 13, e1672 CrossRef CAS.
- A. F. Palonpon, J. Ando, H. Yamakoshi, K. Dodo, M. Sodeoka, S. Kawata and K. Fujita, Nat. Protoc., 2013, 8, 677 CrossRef CAS.
- E. Lenzi, D. Jimenez de Aberasturi and L. M. Liz-Marzán, ACS Sens., 2019, 4, 1126–1137 CrossRef CAS.
- F. Lussier, T. Brulé,Brulé, M. Vishwakarma, T. Das, J. P. Spatz and J.-F. O. Masson, Nano Lett., 2016, 16(6), 3866–3871 CrossRef CAS PubMed.
- A. Y. F. Mahmoud, A. Teixeira, M. Aranda, M. S. Relvas, S. Quintero, M. Sousa-Silva, A. Chícharo, M. Chen, M. Hashemi, J. B. King, J. W. Tunnell, C. Morasso, F. Piccotti, F. Corsi, M. Henriksen-Lacey, D. J. de Aberasturi, D. M. Merino, A. Rodríguez-Patón, S. Abalde-Cela and L. Diéguez, TrAC, Trends Anal. Chem., 2023, 117311 CrossRef CAS.
- G. Ramachandran, Virulence, 2014, 5, 213–218 CrossRef PubMed.
- M. McKee and D. Stuckler, Nat. Med., 2020, 26, 640–642 CrossRef CAS PubMed.
- G. Bonaccorsi, F. Pierri, M. Cinelli, A. Flori, A. Galeazzi, F. Porcelli, A. L. Schmidt, C. M. Valensise, A. Scala, W. Quattrociocchi and F. Pammolli, Proc. Natl. Acad. Sci., 2020, 117, 15530–15535 CrossRef CAS PubMed.
- V. M. Corman, O. Landt, M. Kaiser, R. Molenkamp, A. Meijer, D. K. Chu, T. Bleicker, S. Brünink, J. Schneider, M. L. Schmidt, D. G. Mulders, B. L. Haagmans, B. van der Veer, S. van den Brink, L. Wijsman, G. Goderski, J.-L. Romette, J. Ellis, M. Zambon, M. Peiris, H. Goossens, C. Reusken, M. P. Koopmans and C. Drosten, Eurosurveillance, 2020, 25, 2000045 Search PubMed.
- M. Shen, Y. Zhou, J. Ye, A. A. Abdullah AL-maskri, Y. Kang, S. Zeng and S. Cai, J. Pharm. Anal., 2020, 10, 97–101 CrossRef PubMed.
- K. Dighe, P. Moitra, M. Alafeef, N. Gunaseelan and D. Pan, Biosens. Bioelectron., 2022, 200, 113900 CrossRef CAS PubMed.
- H. Harpaldas, S. Arumugam, C. Campillo Rodriguez, B. A. Kumar, V. Shi and S. K. Sia, Lab Chip, 2021, 21, 4517–4548 RSC.
- Z. Cheng, N. Choi, R. Wang, S. Lee, K. C. Moon, S.-Y. Yoon, L. Chen and J. Choo, ACS Nano, 2017, 11, 4926–4933 CrossRef CAS PubMed.
- X. Wang, S. Park, J. Ko, X. Xiao, V. Giannini, S. A. Maier, D. Kim and J. Choo, Small, 2018, 14, 1801623 CrossRef PubMed.
- S. H. Lee, J. Hwang, K. Kim, J. Jeon, S. Lee, J. Ko, J. Lee, M. Kang, D. R. Chung and J. Choo, Anal. Chem., 2019, 91, 12275–12282 CrossRef CAS PubMed.
- Y. Joung, K. Kim, S. Lee, B.-S. Chun, S. Lee, J. Hwang, S. Choi, T. Kang, M.-K. Lee, L. Chen and J. Choo, ACS Sens., 2022, 7, 3470–3480 CrossRef CAS PubMed.
- Q. Yu, H. D. Trinh, Y. Lee, T. Kang, L. Chen, S. Yoon and J. Choo, Sens. Actuators, B, 2023, 382, 133521 CrossRef CAS.
- P. Mandal and B. S. Tewari, Surf. Interfaces, 2022, 28, 101655 CrossRef CAS.
- I. V. Chernyshova, P. Somasundaran and S. Ponnurangam, Proc. Natl. Acad. Sci., 2018, 115, E9261–E9270 CrossRef CAS PubMed.
- G. Zheng, L. Polavarapu, L. M. Liz-Marzán, I. Pastoriza-Santos and J. Pérez-Juste, Chem. Commun., 2015, 51, 4572–4575 RSC.
- P. Taladriz-Blanco, L. Rodríguez-Lorenzo, M. Sanles-Sobrido, P. Hervés, M. A. Correa-Duarte, R. A. Álvarez-Puebla and L. M. Liz-Marzán, ACS Appl. Mater. Interfaces, 2009, 1, 56–59 CrossRef CAS PubMed.
- D. Devasia, A. J. Wilson, J. Heo, V. Mohan and P. K. Jain, Nat. Commun., 2021, 12, 2612 CrossRef CAS PubMed.
- G. Kumari, X. Zhang, D. Devasia, J. Heo and P. K. Jain, ACS Nano, 2018, 12, 8330–8340 CrossRef CAS PubMed.
- S. Rodal-Cedeira, V. Montes-García, L. Polavarapu, D. M. Solís, H. Heidari, A. La Porta, M. Angiola, A. Martucci, J. M. Taboada, F. Obelleiro, S. Bals, J. Pérez-Juste and I. Pastoriza-Santos, Chem. Mater., 2016, 28, 9169–9180 CrossRef CAS.
- J. Li, J. Liu, Y. Yang and D. Qin, J. Am. Chem. Soc., 2015, 137, 7039–7042 CrossRef CAS PubMed.
- P. Li, B. Ma, L. Yang and J. Liu, Chem. Commun., 2015, 51, 11394–11397 RSC.
- W. Xie, R. Grzeschik and S. Schlücker, Angew. Chem., Int. Ed., 2016, 55, 13729–13733 CrossRef CAS PubMed.
- H. Zhang, J. Wei, X.-G. Zhang, Y.-J. Zhang, P. M. Radjenovica, D.-Y. Wu, F. Pan, Z.-Q. Tian and J.-F. Li, Chemistry, 2020, 6, 689–702 CrossRef CAS.
- J.-C. Dong, X.-G. Zhang, V. Briega-Martos, X. Jin, J. Yang, S. Chen, Z.-L. Yang, D.-Y. Wu, J. M. Feliu, C. T. Williams, Z.-Q. Tian and J.-F. Li, Nat. Energy, 2018, 4, 60–67 CrossRef.
- Samriti, V. Rajput, R. K. Gupta and J. Prakash, J. Mater. Chem. C, 2022, 10, 73–95 RSC.
- Y. Cao and M. Sun, Rev. Phys., 2022, 8, 100067 CrossRef.
- J. Ma, Y. Cheng and M. Sun, Nanoscale, 2021, 13, 10712–10725 RSC.
- N. Kumar, B. M. Weckhuysen, A. J. Wain and A. J. Pollard, Nat. Protoc., 2019, 14, 1169–1193 CrossRef CAS PubMed.
- R. M. Stöckle, Y. D. Suh, V. Deckert and R. Zenobi, Chem. Phys. Lett., 2000, 318, 131–136 CrossRef.
- B. Pettinger, B. Ren, G. Picardi, R. Schuster and G. Ertl, Phys. Rev. Lett., 2004, 92, 096101 CrossRef PubMed.
- M. Sun, Z. Zhang, H. Zheng and H. Xu, Sci. Rep., 2012, 2, 647 CrossRef PubMed.
- Y. Fang, Z. Zhang and M. Sun, Rev. Sci. Instrum., 2022, 7(1), 1165–1172 Search PubMed.
- G. Binnig, C. F. Quate and C. Gerber, Phys. Rev. Lett., 1986, 56, 930–933 CrossRef PubMed.
- S. Sheng, W. Li, J. Gou, P. Cheng, L. Chen and K. Wu, Rev. Sci. Instrum., 2018, 89, 053107 CrossRef PubMed.
- S. Sheng, J. Wu, X. Cong, W. Li, J. Gou, Q. Zhong, P. Cheng, P. Tan, L. Chen and K. Wu, Phys. Rev. Lett., 2017, 119, 196803 CrossRef PubMed.
- E. M. van Schrojenstein Lantman, T. Deckert-Gaudig, A. J. G. Mank, V. Deckert and B. M. Weckhuysen, Nat. Nanotechnol., 2012, 7, 583–586 CrossRef CAS PubMed.
- R. Zhang, Y. Zhang, Z. C. Dong, S. Jiang, C. Zhang, L. G. Chen, L. Zhang, Y. Liao, J. Aizpurua, Y. Luo, J. L. Yang and J. G. Hou, Nature, 2013, 498, 82–86 CrossRef CAS PubMed.
- Y. Cao, Y. Feng, Y. Cheng, L. Meng and M. Sun, Appl. Phys. Lett., 2023, 122, 231601 CrossRef CAS.
- J.-F. Masson, J. S. Biggins and E. Ringe, Nat. Nanotechnol., 2023, 18, 111–123 CrossRef CAS PubMed.
- Y. X. Leong, E. X. Tan, S. X. Leong, C. S. Lin Koh, L. B. Thanh Nguyen, J. R. Ting Chen, K. Xia and X. Y. Ling, ACS Nano, 2022, 16, 13279–13293 CrossRef CAS PubMed.
- F. Cui, Y. Yue, Y. Zhang, Z. Zhang and H. S. Zhou, ACS Sens., 2020, 5, 3346–3364 CrossRef CAS PubMed.
- R. Beeram, K. R. Vepa and V. R. Soma, Biosensors, 2023, 13, 328 CrossRef CAS PubMed.
- C. Liu, D. Xu, X. Dong and Q. Huang, Trends Food Sci. Technol., 2022, 128, 90–101 CrossRef CAS.
- S. X. Leong, Y. X. Leong, C. S. L. Koh, E. X. Tan, L. B. T. Nguyen, J. R. T. Chen, C. Chong, D. W. C. Pang, H. Y. F. Sim, X. Liang, N. S. Tan and X. Y. Ling, Chem. Sci., 2022, 13, 11009–11029 RSC.
- J. Perumal, Y. Wang, A. B. E. Attia, U. S. Dinish and M. Olivo, Nanoscale, 2021, 13, 553–580 RSC.
- J. Hua, Z. Xiong, J. Lowey, E. Suh and E. R. Dougherty, Bioinformatics, 2005, 21, 1509–1515 CrossRef CAS PubMed.
- R. Gautam, S. Vanga, F. Ariese and S. Umapathy, EPJ Tech. Instrum., 2015, 2, 8 CrossRef.
- S. Nanga, A. T. Bawah, B. A. Acquaye, M.-I. Billa, F. D. Baeta, N. A. Odai, S. K. Obeng and A. D. Nsiah, J. Data Anal. Inform. Process., 2021, 09, 189–231 Search PubMed.
- S. X. Leong, C. S. L. Koh, H. Y. F. Sim, Y. H. Lee, X. Han, G. C. Phan-Quang and X. Y. Ling, ACS Nano, 2021, 15, 1817–1825 CrossRef CAS PubMed.
- Y. Yang, B. Xu, J. Haverstick, N. Ibtehaz, A. Muszyński, X. Chen, M. E. H. Chowdhury, S. M. Zughaier and Y. Zhao, Nanoscale, 2022, 14, 8806–8817 RSC.
- K. Yang, F. Xu, L. Zhu, H. Li, Q. Sun, A. Yan, B. Ren, Y. Zhu and L. Cui, Angew. Chem., Int. Ed., 2023, 62, e202217412 CrossRef CAS PubMed.
- M. Sugiyama, Statistical Reinforcement Learning, Chapman and Hall/CRC, 2015 Search PubMed.
- A. L. Tarca, V. J. Carey, X. Chen, R. Romero and S. Drăghici, PLoS Comput. Biol., 2007, 3, e116 CrossRef PubMed.
- F. Safir, N. Vu, L. F. Tadesse, K. Firouzi, N. Banaei, S. S. Jeffrey, A. A. E. Saleh, B. P. T. Khuri-Yakub and J. A. Dionne, Nano Lett., 2023, 23, 2065–2073 CrossRef CAS PubMed.
- L. J. P. van der Maaten and G. E. Hinton, J. Mach. Learn. Res., 2008, 9, 2579–2605 Search PubMed.
- L. McInnes, J. Healy and J. Melville.
- H. Shin, B. H. Choi, O. Shim, J. Kim, Y. Park, S. K. Cho, H. K. Kim and Y. Choi, Nat. Commun., 2023, 14, 1644 CrossRef CAS PubMed.
- H. K. Lee, Y. H. Lee, C. S. L. Koh, G. C. Phan-Quang, X. Han, C. L. Lay, H. Y. F. Sim, Y.-C. Kao, Q. An and X. Y. Ling, Chem. Soc. Rev., 2019, 48, 731–756 RSC.
- M. T. Ribeiro, S. Singh and C. Guestrin, Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, ACM, New York, NY, USA, 2016, vol. 13–17, 1135–1144.
- S. M. Lundberg and S.-I. Lee, Adv. Neural Inf. Process Syst., 2017, 30, 3149–3157 Search PubMed.
- A. Dhurandhar, P.-Y. Chen, R. Luss, C.-C. Tu, P. Ting, K. Shanmugam and P. Das, Adv. Neural Inf. Process Syst., 2018, 590–601 Search PubMed.
- V. Živanović, S. Seifert, D. Drescher, P. Schrade, S. Werner, P. Guttmann, G. P. Szekeres, S. Bachmann, G. Schneider, C. Arenz and J. Kneipp, ACS Nano, 2019, 13, 9363–9375 CrossRef PubMed.
- R. L. Woodbury, S. M. Varnum and R. C. Zangar, J. Proteome Res., 2002, 1, 233–237 CrossRef CAS PubMed.
- B. Schweitzer, S. Roberts, B. Grimwade, W. Shao, M. Wang, Q. Fu, Q. Shu, I. Laroche, Z. Zhou and V. T. Tchernev, Nat. Biotechnol., 2002, 20, 359–365 CrossRef CAS PubMed.
- M. M. Ali, F. Li, Z. Zhang, K. Zhang, D.-K. Kang, J. A. Ankrum, X. C. Le and W. Zhao, Chem. Soc. Rev., 2014, 43, 3324–3341 RSC.
- N. Ganesh, W. Zhang, P. C. Mathias, E. Chow, J. Soares, V. Malyarchuk, A. D. Smith and B. T. Cunningham, Nat. Nanotechnol., 2007, 2, 515–520 CrossRef PubMed.
- V. Flauraud, R. Regmi, P. M. Winkler, D. T. L. Alexander, H. Rigneault, N. F. Van Hulst, M. F. García-Parajo, J. Wenger and J. Brugger, Nano Lett., 2017, 17, 1703–1710 CrossRef CAS PubMed.
- F. Tam, G. P. Goodrich, B. R. Johnson and N. J. Halas, Nano Lett., 2007, 7, 496–501 CrossRef CAS PubMed.
- A. Kinkhabwala, Z. Yu, S. Fan, Y. Avlasevich, K. Müllen and W. E. Moerner, Nat. Photonics, 2009, 3, 654–657 CrossRef CAS.
- S. M. Tabakman, L. Lau, J. T. Robinson, J. Price, S. P. Sherlock, H. Wang, B. Zhang, Z. Chen, S. Tangsombatvisit, J. A. Jarrell, P. J. Utz and H. Dai, Nat. Commun., 2011, 2, 466 CrossRef PubMed.
- B. Zhang, R. B. Kumar, H. Dai and B. J. Feldman, Nat. Med., 2014, 20, 948–953 CrossRef CAS PubMed.
- B. Zhang, B. A. Pinsky, J. S. Ananta, S. Zhao, S. Arulkumar, H. Wan, M. K. Sahoo, J. Abeynayake, J. J. Waggoner and C. Hopes, Nat. Med., 2017, 23, 548–550 CrossRef CAS PubMed.
- B. Liu, Y. Li, H. Wan, L. Wang, W. Xu, S. Zhu, Y. Liang, B. Zhang, J. Lou, H. Dai and K. Qian, Adv. Funct. Mater., 2016, 26, 7994–8002 CrossRef CAS.
- K. M. Mayer and J. H. Hafner, Chem. Rev., 2011, 111, 3828–3857 CrossRef CAS PubMed.
- A. J. Haes, L. Chang, W. L. Klein and R. P. Van Duyne, J. Am. Chem. Soc., 2005, 127, 2264–2271 CrossRef CAS PubMed.
- R. Bardhan, N. K. Grady, J. R. Cole, A. Joshi and N. J. Halas, ACS Nano, 2009, 3, 744–752 CrossRef CAS PubMed.
- H. Li, C.-Y. Chen, X. Wei, W. Qiang, Z. Li, Q. Cheng and D. Xu, Anal. Chem., 2012, 84, 8656–8662 CrossRef CAS PubMed.
- K.-X. Xie, S.-H. Cao, Z.-C. Wang, Y.-H. Weng, S.-X. Huo, Y.-Y. Zhai, M. Chen, X.-H. Pan and Y.-Q. Li, Sens. Actuators, B, 2017, 253, 804–808 CrossRef CAS.
- Z. A. R. Jawad, I. G. Theodorou, L. R. Jiao and F. Xie, Sci. Rep., 2017, 7, 14309 CrossRef PubMed.
- J. Lee, S. R. Ahmed, S. Oh, J. Kim, T. Suzuki, K. Parmar, S. S. Park, J. Lee and E. Y. Park, Biosens. Bioelectron., 2015, 64, 311–317 CrossRef CAS PubMed.
- K. Ray, H. Szmacinski and J. R. Lakowicz, Anal. Chem., 2009, 81, 6049–6054 CrossRef CAS PubMed.
- Y. Liu, Y. Yang, G. Wang, D. Wang, P.-L. Shao, J. Tang, T. He, J. Zheng, R. Hu, Y. Liu, Z. Xu, D. Niu, J. Lv, J. Yang, H. Xiao, S. Wu, S. He, Z. Tang, Y. Liu, M. Tang, X. Jiang, J. Yuan, H. Dai and B. Zhang, Nat. Biomed. Eng., 2023, 7(12), 1636–1648 CrossRef CAS PubMed.
- C. R. Sabanayagam and J. R. Lakowicz, Nucleic Acids Res., 2006, 35, e13–e13 CrossRef PubMed.
- W. Hu, Y. Liu, H. Yang, X. Zhou and C. M. Li, Biosens. Bioelectron., 2011, 26, 3683–3687 CrossRef CAS PubMed.
- J. Zhang, E. Matveeva, I. Gryczynski, Z. Leonenko and J. R. Lakowicz, J. Phys. Chem. B, 2005, 109, 7969–7975 CrossRef CAS PubMed.
- E. G. Matveeva, I. Gryczynski, A. Barnett, Z. Leonenko, J. R. Lakowicz and Z. Gryczynski, Anal. Biochem., 2007, 363, 239–245 CrossRef CAS PubMed.
- T. W. Ebbesen, H. J. Lezec, H. F. Ghaemi, T. Thio and P. A. Wolff, Nature, 1998, 391, 667–669 CrossRef CAS.
- G. Hawa, L. Sonnleitner, A. Missbichler, A. Prinz, G. Bauer and C. Mauracher, Anal. Biochem., 2018, 549, 39–44 CrossRef CAS PubMed.
- P. P. Pompa, L. Martiradonna, A. Della Torre, F. Della Sala, L. Manna, M. De Vittorio, F. Calabi, R. Cingolani and R. Rinaldi, Nat. Nanotechnol., 2006, 1, 126–130 CrossRef CAS PubMed.
- L. Zhou, F. Ding, H. Chen, W. Ding, W. Zhang and S. Y. Chou, Anal. Chem., 2012, 84, 4489–4495 CrossRef CAS PubMed.
- C. Wang, S. Tadepalli, J. Luan, K. Liu, J. J. Morrissey, E. D. Kharasch, R. R. Naik and S. Singamaneni, Adv. Mater., 2018, 3(2), 342–351 CAS.
- D. F. Cruz, C. M. Fontes, D. Semeniak, J. Huang, A. Hucknall, A. Chilkoti and M. H. Mikkelsen, Nano Lett., 2020, 20, 4330–4336 CrossRef CAS PubMed.
- D. Y. Joh, A. M. Hucknall, Q. Wei, K. A. Mason, M. L. Lund, C. M. Fontes, R. T. Hill, R. Blair, Z. Zimmers, R. K. Achar, D. Tseng, R. Gordan, M. Freemark, A. Ozcan and A. Chilkoti, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E7054–e7062 CrossRef CAS PubMed.
- J. Luan, J. J. Morrissey, Z. Wang, H. G. Derami, K.-K. Liu, S. Cao, Q. Jiang, C. Wang, E. D. Kharasch, R. R. Naik and S. Singamaneni, Light: Sci. Appl., 2018, 7, 29 CrossRef PubMed.
- C. Liang, J. Luan, Z. Wang, Q. Jiang, R. Gupta, S. Cao, K.-K. Liu, J. J. Morrissey, E. D. Kharasch, R. R. Naik and S. Singamaneni, ACS Appl. Mater. Interfaces, 2021, 13, 11414–11423 CrossRef CAS PubMed.
- J. Luan, A. Seth, R. Gupta, Z. Wang, P. Rathi, S. Cao, H. Gholami Derami, R. Tang, B. Xu, S. Achilefu, J. J. Morrissey and S. Singamaneni, Nat. Biomed. Eng., 2020, 4, 518–530 CrossRef CAS PubMed.
- Z. Wang, Q. Zhou, A. Seth, S. Kolla, J. Luan, Q. Jiang, P. Rathi, P. Gupta, J. J. Morrissey, R. R. Naik and S. Singamaneni, Biosens. Bioelectron., 2022, 200, 113918 CrossRef CAS PubMed.
- Y. Kim, Z. Wang, C. Li, K. Kidd, Y. Wang, B. G. Johnson, S. Kmoch, J. J. Morrissey, A. J. Bleyer, J. S. Duffield, S. Singamaneni and Y. M. Chen, Am. J. Physiol.: Renal Physiol., 2021, 321, F236–F244 CrossRef CAS PubMed.
- Y. Wang, J. J. Morrissey, P. Gupta, P. Chauhan, R. K. Pachynski, P. K. Harris, A. Chaudhuri and S. Singamaneni, ACS Appl. Mater. Interfaces, 2023, 15, 18598–18607 CrossRef CAS PubMed.
- L. Liu, Z. Wang, Y. Wang, J. Luan, J. J. Morrissey, R. R. Naik and S. Singamaneni, Adv. Healthcare Mater., 2021, 10, 2100956 CrossRef CAS PubMed.
- Z. Wang, J. J. Morrissey, L. Liu, Y. Wang, Q. Zhou, R. R. Naik and S. Singamaneni, Anal. Chem., 2022, 94, 909–917 CrossRef CAS PubMed.
- Z. Wang, J. Luan, A. Seth, L. Liu, M. You, P. Gupta, P. Rathi, Y. Wang, S. Cao, Q. Jiang, X. Zhang, R. Gupta, Q. Zhou, J. J. Morrissey, E. L. Scheller, J. S. Rudra and S. Singamaneni, Nat. Biomed. Eng., 2021, 5, 64–76 CrossRef CAS PubMed.
- R. Gupta, P. Gupta, S. Wang, A. Melnykov, Q. Jiang, A. Seth, Z. Wang, J. J. Morrissey, I. George, S. Gandra, P. Sinha, G. A. Storch, B. A. Parikh, G. M. Genin and S. Singamaneni, Nat. Biomed. Eng., 2023, 7, 1556–1570 CrossRef CAS PubMed.
- A. Seth, E. Mittal, J. Luan, S. Kolla, M. B. Mazer, H. Joshi, R. Gupta, P. Rathi, Z. Wang, J. J. Morrissey, J. D. Ernst, C. Portal-Celhay, S. C. Morley, J. A. Philips and S. Singamaneni, Cells Rep. Methods, 2022, 2, 100267 CrossRef CAS PubMed.
- E. Mittal, A. T. Roth, A. Seth, S. Singamaneni, W. Beatty and J. A. Philips, eLife, 2023, 12, e85416 CrossRef CAS PubMed.
- H. Joshi, A. Almgren-Bell, E. P. Anaya, E. M. Todd, S. J. Van Dyken, A. Seth, K. M. McIntire, S. Singamaneni, F. Sutterwala and S. C. Morley, Cell Rep., 2022, 38, 110507 CrossRef CAS PubMed.
- H. Guo, R. Gupta, D. Sharma, E. Zhanov, C. Malone, R. Jada, Y. Liu, M. Garg, S. Singamaneni, F. Zhao and L. Tian, Nano Lett., 2023, 23, 10171–10178 CrossRef CAS PubMed.
- A. Seth, Y. Liu, R. Gupta, Z. Wang, E. Mittal, S. Kolla, P. Rathi, P. Gupta, B. A. Parikh, G. M. Genin, S. Gandra, G. A. Storch, J. A. Philips, I. A. George and S. Singamaneni, Nano Lett., 2024, 24(1), 229–237 CrossRef CAS PubMed.
- P. Gupta, P. Rathi, R. Gupta, H. Baldi, Q. Coquerel, A. Debnath, H. G. Derami, B. Raman and S. Singamaneni, Nanoscale Horiz., 2023, 8, 1537–1555 RSC.
- P. Rathi, P. Gupta, A. Debnath, H. Baldi, Y. Wang, R. Gupta, B. Raman and S. Singamaneni, Nano Lett., 2023, 23, 5654–5662 CrossRef CAS PubMed.
- P. Barya, Y. Xiong, S. Shepherd, R. Gupta, L. D. Akin, J. Tibbs, H. Lee, S. Singamaneni and B. T. Cunningham, Small, 2023, 19, 2207239 CrossRef CAS PubMed.
- N. G. Khlebtsov and L. A. Dykman, J. Quant. Spectrosc. Radiat. Transfer, 2010, 111, 1–35 CrossRef CAS.
- U. Guler, V. M. Shalaev and A. Boltasseva, Mater. Today, 2015, 18, 227–237 CrossRef CAS.
- K. M. Mayer and J. H. Hafner, Chem. Rev., 2011, 111(6), 3828–3857 CrossRef CAS PubMed.
- J. Katyal and R. K. Soni, Plasmonics, 2015, 10, 1729–1740 CrossRef CAS.
- V. Devaraj, J. M. Lee and J. W. Oh, Curr. Appl. Phys., 2020, 20, 1335–1341 CrossRef.
- A. Yuksel, M. Cullinan, E. T. Yu and J. Murthy, J. Quant. Spectrosc. Radiat. Transfer, 2020, 254, 107207 CrossRef CAS.
- E. R. Encina and E. A. Coronado, J. Phys. Chem. C, 2007, 111, 16796–16801 CrossRef CAS.
- K. Ueno, S. Juodkazis, M. Mino, V. Mizeikis and H. Misawa, J. Phys. Chem. C, 2007, 111, 4180–4184 CrossRef CAS.
- J. Jatschka, A. Dathe, A. Csáki, W. Fritzsche and O. Stranik, Sens. Biosens. Res., 2016, 7, 62–70 Search PubMed.
- W. Cui, A. Xiao, J. Zheng, P. Chen, J. Liang, Y. Huang and B. O. Guan, ACS Appl. Nano Mater., 2023, 6, 6345–6353 CrossRef CAS.
- C. Goldmann, M. De Frutos, E. H. Hill, D. Constantin and C. Hamon, Chem. Mater., 2021, 33, 2948–2956 CrossRef CAS.
- A. Sa, X. Zhuo, W. Albrecht, S. Bals and L. M. Liz-Marza, ACS Mater. Lett., 2020, 2(9), 1246–1250 CrossRef.
- Y. Koike and K. Koike, J. Polym. Sci., Part B: Polym. Phys., 2011, 49, 2–17 CrossRef CAS.
- M. Arumugam, Pramana, 2001, 57, 849–869 CrossRef.
- M. Chauhan and V. K. Singh, Opt. Fiber Technol., 2021, 64, 102580 CrossRef CAS.
- L. Tian, E. Chen, N. Gandra, A. Abbas and S. Singamaneni, Langmuir, 2012, 28, 17435–17442 CrossRef CAS PubMed.
- J. Cao, E. K. Galbraith, T. Sun and K. T. V. Grattan, IEEE Sens. J., 2012, 12(7), 2355–2361 CAS.
- B. Kaur and A. K. Sharma II, Proc. SPIE, 2019, 11028, 405–410 Search PubMed.
- C. Christopher, A. Subrahmanyam and V. V. R. Sai, Plasmonics, 2018, 13, 493–502 CrossRef CAS.
- J. Boehm, A. François, H. Ebendorff-Heidepriem and T. M. Monro, Plasmonics, 2011, 6, 133–136 CrossRef CAS.
- V. Myroshnychenko, J. Rodríguez-Fernández, I. Pastoriza-Santos, A. M. Funston, C. Novo, P. Mulvaney, L. M. Liz-Marzán and F. J. García de Abajo, Chem. Soc. Rev., 2008, 37, 1792–1805 RSC.
- R. K. Harrison and A. Ben-Yakar, Opt. Express, 2010, 18, 22556–22571 CrossRef CAS PubMed.
- M. Hu, A. Ghoshal, M. Marquez and P. G. Kik, J. Phys. Chem. C, 2010, 114, 7509–7514 CrossRef CAS.
- R. E. Armstrong, J. C. Van Liempt and P. Zijlstra, J. Phys. Chem. C, 2019, 123, 25801–25808 CrossRef CAS.
- K. Hei Willis Ho, A. Shang, F. Shi, T. Wing Lo, P. Hong Yeung, Y. Sing Yu, X. Zhang, K. Wong, D. Yuan Lei, K. H. W. Ho, A. Shang, F. Shi, T. W. Lo, P. H. Yeung, Y. S. Yu, X. Zhang, D. Y. Lei and K. Wong, Adv. Funct. Mater., 2018, 28, 1800383 CrossRef.
- H. Sugimoto, S. Yashima and M. Fujii, ACS Photonics, 2018, 5, 3421–3427 CrossRef CAS.
- R. Chikkaraddy and J. J. Baumberg, ACS Photonics, 2021, 8, 2811–2817 CrossRef CAS PubMed.
- F. Xia, H. Song, Y. Zhao, W. M. Zhao, Q. Wang, X. Z. Wang, B. T. Wang and Z. X. Dai, Measurement, 2020, 164, 108083 CrossRef.
- H. Sugimoto, S. Yashima and M. Fujii, ACS Photonics, 2018, 5, 3421–3427 CrossRef CAS.
- Q. Wang, L. Hou, C. Li, H. Zhou, X. Gan, K. Liu, F. Xiao and J. Zhao, Nanoscale, 2022, 14, 10773–10779 RSC.
- M. A. Otte, M. C. Estévez, D. Regatos, L. M. Lechuga and B. Sepúlveda, ACS Nano, 2011, 5, 9179–9186 CrossRef CAS PubMed.
- L. Y. Niu, Q. Wang, J. Y. Jing and W. M. Zhao, Opt. Commun., 2019, 450, 287–295 CrossRef CAS.
- X. Jiang and Q. Wang, Opt. Fiber Technol., 2019, 51, 118–124 CrossRef CAS.
- Q. Wang, X. Li, W. M. Zhao and S. Jin, Appl. Surf. Sci., 2019, 492, 374–381 CrossRef CAS.
- B. T. Wang and Q. Wang, IEEE Sens. J., 2018, 18, 8303–8310 CAS.
- V. A. Markel, J. Opt. Soc. Am. A, 2016, 33, 1244 CrossRef PubMed.
- Z. Wang, X. Jia, H. Wu, F. Peng, Y. Fu and Y. Rao, 25th International Conference, Opt. Fiber Sens., 2017, 10323, 103230 Search PubMed.
- L. Liu, S. Deng, J. Zheng, L. Yuan, H. Deng and C. Teng, Sensors, 2021, 21, 1516 CrossRef CAS PubMed.
- C. Caucheteur, T. Guo and J. Albert, Anal. Bioanal. Chem., 2015, 407, 3883–3897 CrossRef CAS PubMed.
- J. H. Qu, B. Peeters, F. Delport, K. Vanhoorelbeke, J. Lammertyn and D. Spasic, Biosens. Bioelectron., 2021, 192, 113549 CrossRef CAS PubMed.
- S. Jiang, Z. Li, C. Zhang, S. Gao, Z. Li, H. Qiu, C. Li, C. Yang, M. Liu and Y. Liu, J. Phys. D: Appl. Phys., 2017, 50, 165105 CrossRef.
- T. J. Ray, A. Wang, X. Jia, W. Zhou and Y. Zhu, Virgina Tech., 2019 Search PubMed.
- H. Dong, L. Chen, J. Zhou, J. Yu, H. Guan, W. Qiu, J. Dong, H. Lu, J. Tang, W. Zhu, Z. Cai, Y. I. Xiao, J. Zhang and Z. Chen, Opt. Express, 2017, 25(5), 5352–5365 CrossRef PubMed.
- L. Zhuo, J. Tang, W. Zhu, H. Zheng, H. Guan, H. Lu, Y. Chen, Y. Luo, J. Zhang, Y. Zhong, J. Yu and Z. Chen, Photonic Sens., 2022, 13, 1–24 Search PubMed.
- W. Udos, K. S. Lim, C. L. Tan, M. N. S. M. Ismail, C. W. Ooi, R. Zakaria and H. Ahmad, Optics, 2020, 219, 164970 CAS.
- J. A. García, D. Monzón-Hernández, J. Manríquez and E. Bustos, Opt. Mater., 2016, 51, 208–212 CrossRef.
- J. L. Tang, S. F. Cheng, W. T. Hsu, T. Y. Chiang and L. K. Chau, Sens. Actuators, B, 2006, 119, 105–109 CrossRef CAS.
- S. Jia, C. Bian, J. Sun, J. Tong and S. Xia, Biosens. Bioelectron., 2018, 114, 15–21 CrossRef CAS PubMed.
- E. Martinsson, B. Sepulveda, P. Chen, A. Elfwing, B. Liedberg and D. Aili, Plasmonics, 2014, 9, 773–780 CrossRef CAS.
- M. Lin, M. Lu, Y. Liang, L. Yu and W. Peng, ACS Appl. Nano Mater., 2022, 5, 6171–6180 CrossRef CAS.
- K. C. Chen, Y. Le Li, C. W. Wu and C. C. Chiang, Sensors, 2018, 18, 1217 CrossRef PubMed.
- S. Jiang, Z. Li, C. Zhang, S. Gao, Z. Li, H. Qiu, C. Li, C. Yang, M. Liu and Y. Liu, J. Phys. D: Appl. Phys., 2017, 50, 165105 CrossRef.
- J. Cao, M. H. Tu, T. Sun and K. T. V. Grattan, Sens. Actuators, B, 2013, 181, 611–619 CrossRef CAS.
- J. Wang, H. Zhang, Y. Tang, M. Wen, B. Yao, S. Yuan, W. Zhang and H. Lei, ACS Biomater. Sci. Eng., 2022, 1060–1066 CrossRef CAS PubMed.
- J. Feng, J. Gao, W. Yang, R. Liu, M. Shafi, Z. Zha, C. Liu, S. Xu, T. Ning, T. Ning, S. Jiang and S. Jiang, Opt. Express, 2022, 30, 10187–10198 CrossRef CAS PubMed.
- M. H. Tu, T. Sun and K. T. V. Grattan, Sens. Actuators, B, 2014, 191, 37–44 CrossRef CAS.
- N. Cennamo, G. D’Agostino, A. Donà, G. Dacarro, P. Pallavicini, M. Pesavento and L. Zeni, Sensors, 2013, 13, 14676–14686 CrossRef PubMed.
- C. Zhang, Z. Li, S. Z. Jiang, C. H. Li, S. C. Xu, J. Yu, Z. Li, M. H. Wang, A. H. Liu and B. Y. Man, Sens. Actuators, B, 2017, 251, 127–133 CrossRef CAS.
- B. Sciacca and T. M. Monro, Langmuir, 2014, 30, 946–954 CrossRef CAS PubMed.
- J. Chen, S. Shi, R. Su, W. Qi, R. Huang, M. Wang, L. Wang and Z. He, Sensors, 2015, 15, 12205–12217 CrossRef CAS PubMed.
- C. Zhou, Opt. Lasers Eng., 2012, 50, 1592–1595 CrossRef.
- S. Rodal-Cedeira, V. Montes-García, L. Polavarapu, D. M. Solís, H. Heidari, A. La Porta, M. Angiola, A. Martucci, J. M. Taboada, F. Obelleiro, S. Bals, J. Pérez-Juste and I. Pastoriza-Santos, Chem. Mater., 2016, 28, 9169–9180 CrossRef CAS.
- S. Barbosa, A. Agrawal, L. Rodríguez-Lorenzo, I. Pastoriza-Santos, R. A. Alvarez-Puebla, A. Kornowski, H. Weller and L. M. Liz-Marzán, Langmuir, 2010, 26, 14943–14950 CrossRef CAS PubMed.
- E. Martinsson, M. A. Otte, M. M. Shahjamali, B. Sepulveda and D. Aili, J. Phys. Chem. C, 2014, 118, 24680–24687 CrossRef CAS.
- C. L. Nehl, H. Liao and J. H. Hafner, Nano Lett., 2006, 6, 683–688 CrossRef CAS PubMed.
- H. Chen, X. Kou, Z. Yang, W. Ni and J. Wang, Langmuir, 2008, 24, 5233–5237 CrossRef CAS PubMed.
- R. Bukasov and J. S. Shumaker-Parry, Nano Lett., 2007, 7, 1113–1118 CrossRef CAS PubMed.
- H. R. Hegde, S. Chidangil and R. K. Sinha, J. Mater. Sci.: Mater. Electron., 2022, 33, 4011–4024 CrossRef CAS.
- W. Te Wu, C. H. Chen, C. Y. Chiang and L. K. Chau, Sensors, 2018, 18, 23–30 Search PubMed.
- S. M. Marinakos, S. Chen and A. Chilkoti, Anal. Chem., 2007, 79, 5278–5283 CrossRef CAS PubMed.
- C. A. Barrios, T. Mirea and M. H. Represa, Sensors, 2023, 1, 66 Search PubMed.
- Y. Khalavka, J. Becker and C. Sönnichsen, J. Am. Chem. Soc., 2009, 131, 1871–1875 CrossRef CAS PubMed.
- N. Nath and A. Chilkoti, Anal. Chem., 2002, 74, 504–509 CrossRef CAS PubMed.
- M. Lu, H. Zhu, M. Lin, F. Wang, L. Hong, J. F. Masson and W. Peng, Sens. Actuators, B, 2021, 329, 129094 CrossRef CAS.
- X. Zhuo, X. Zhu, Q. Li, Z. Yang and J. Wang, ACS Nano, 2015, 9(7), 7523–7535 CrossRef CAS PubMed.
- C. D. Chen, S. F. Cheng, L. K. Chau and C. R. C. Wang, Biosens. Bioelectron., 2007, 22, 926–932 CrossRef CAS PubMed.
- J. Cao, M. H. Tu, T. Sun and K. T. V. Grattan, Sens. Actuators, B, 2013, 181, 611–619 CrossRef CAS.
- C. Zhuang, Y. Xu, N. Xu, J. Wen, H. Chen and S. Deng, Sensors, 2018, 18(10), 3458 CrossRef PubMed.
- A. Sánchez-Iglesias, N. Winckelmans, T. Altantzis, S. Bals, M. Grzelczak and L. M. Liz-Marzán, J. Am. Chem. Soc., 2016, 139, 107–110 CrossRef PubMed.
- C. Zhang, B. Man, Z. Li, X. Xiu, L. Hou, J. Yu, S. Jiang, X. Zhao, Q. Peng, S. Qiu and C. Li, Nanophotonics, 2021, 10, 1529–1540 CrossRef.
- J. K. Nayak and R. Jha, IEEE Photonics Technol. Lett., 2018, 30, 1667–1670 CAS.
- G. C. Li, Q. Zhang, S. A. Maier and D. Lei, Nanophotonics, 2018, 7, 1865–1889 CrossRef CAS.
- A. Moreau, C. Ciracì, J. J. Mock, D. R. Smith, R. T. Hill, A. Chilkoti, Q. Wang and B. J. Wiley, Nature, 2012, 492(7427), 86–89 CrossRef CAS PubMed.
- J. Feng, J. Gao, W. Yang, R. Liu, M. Shafi, Z. Zha, C. Liu, S. Xu, T. Ning, T. Ning, S. Jiang and S. Jiang, Opt. Express, 2022, 30(6), 10187–10198 CrossRef CAS PubMed.
- W. S. Mokrzycki and M. Tatol, Mach. Graphics Vision Int. J., 2011, 20, 383–411 Search PubMed.
- G. Sharma, W. Wu and E. N. Dalal, Color Res. Appl., 2005, 30, 21–30 CrossRef.
- N. S. King, L. Liu, X. Yang, B. Cerjan, H. O. Everitt, P. Nordlander and N. J. Halas, ACS Nano, 2015, 9, 10628–10636 CrossRef CAS PubMed.
- I. Reinhard, K. Miller, G. Diepenheim, K. Cantrell and W. P. Hall, ACS Appl. Nano Mater., 2020, 3, 4342–4350 CrossRef CAS.
- M. Toma, Y. Itakura, S. Namihara and K. Kajikawa, Adv. Eng. Mater., 2023, 25, 2200912 CrossRef CAS.
- A. Choe, J. Yeom, R. Shanker, M. P. Kim, S. Kang and H. Ko, NPG Asia Mater., 2018, 10, 912–922 CrossRef CAS.
- P. Cencillo-Abad, P. Mastranzo-Ortega, D. Appavoo, T. Guo, L. Zhai, J. Sanchez-Mondragon and D. Chanda, Adv. Opt. Mater., 2023, 11, 2300300 CrossRef CAS.
- T. M. Nguyen, J. H. Chung, G.-H. Bak, Y. H. Kim, M. Kim, Y.-J. Kim, R. J. Kwon, E.-J. Choi, K. H. Kim, Y. S. Kim and J.-W. Oh, ACS Sens., 2023, 8, 167–175 CrossRef CAS PubMed.
- M. Sanromán-Iglesias, V. Garrido, Y. Gil-Ramírez, J. Aizpurua, M. Grzelczak and M.-J. Grilló, Sens. Actuators, B, 2021, 349, 130780 CrossRef.
- J. L. Montaño-Priede, M. Sanromán-Iglesias, N. Zabala, M. Grzelczak and J. Aizpurua, ACS Sens., 2023, 8, 1827–1834 CrossRef PubMed.
- T. H. Talukdar, B. McCoy, S. K. Timmins, T. Khan and J. D. Ryckman, Proc. Natl. Acad. Sci., 2020, 117, 30107–30117 CrossRef CAS PubMed.
- K. Saha, S. S. Agasti, C. Kim, X. Li and V. M. Rotello, Chem. Rev., 2012, 112, 2739–2779 CrossRef CAS PubMed.
- W. Zhou, X. Gao, D. Liu and X. Chen, Chem. Rev., 2015, 115, 10575–10636 CrossRef CAS PubMed.
- S. K. Ghosh and T. Pal, Chem. Rev., 2007, 107, 4797–4862 CrossRef CAS PubMed.
- L. Tang and J. Li, ACS Sens., 2017, 2, 857–875 CrossRef CAS PubMed.
- Z. Zhang, H. Wang, Z. Chen, X. Wang, J. Choo and L. Chen, Biosens. Bioelectron., 2018, 114, 52–65 CrossRef CAS PubMed.
- E. Mauriz, Sensors, 2020, 20(21), 6214 CrossRef CAS PubMed.
- N. Gandra, A. Abbas, L. Tian and S. Singamaneni, Nano Lett., 2012, 12, 2645–2651 CrossRef CAS PubMed.
- A. Abbas, R. Kattumenu, L. Tian and S. Singamaneni, Langmuir, 2013, 29, 56–64 CrossRef CAS PubMed.
- M. Zayats, R. Baron, I. Popov and I. Willner, Nano Lett., 2005, 5, 21–25 CrossRef CAS PubMed.
- Y. Guo, J. Wu, J. Li and H. Ju, Biosens. Bioelectron., 2016, 78, 267–273 CrossRef CAS PubMed.
- R. de la Rica and M. M. Stevens, Nat. Nanotechnol., 2012, 7, 821–824 CrossRef CAS PubMed.
- R. Huang, X. Cai, J. Du, J. Lian, P. Hui, M. Gu, F. Li, J. Wang and W. Chen, ACS Nano, 2022, 16, 19229–19239 CrossRef CAS PubMed.
- S. Xu, W. Ouyang, P. Xie, Y. Lin, B. Qiu, Z. Lin, G. Chen and L. Guo, Anal. Chem., 2017, 89, 1617–1623 CrossRef CAS PubMed.
- R. Baron, M. Zayats and I. Willner, Anal. Chem., 2005, 77, 1566–1571 CrossRef CAS PubMed.
- G. Wang, Z. Chen and L. Chen, Nanoscale, 2011, 3, 1756–1759 RSC.
- X. Ma, Y. Lin, L. Guo, B. Qiu, G. Chen, H. H. Yang and Z. Lin, Biosens. Bioelectron., 2017, 87, 122–128 CrossRef CAS PubMed.
- H. Ye, C. Nowak, Y. Liu, Y. Li, T. Zhang, L. Bleris and Z. Qin, Small, 2022, 18, e2107832 CrossRef PubMed.
- M. N. Creyer, Z. Jin, M. Retout, W. Yim, J. Zhou and J. V. Jokerst, Langmuir, 2022, 38, 14200–14207 CrossRef CAS PubMed.
- Y.-Y. Chen, H.-T. Chang, Y.-C. Shiang, Y.-L. Hung, C.-K. Chiang and C.-C. Huang, Anal. Chem., 2009, 81, 9433–9439 CrossRef CAS PubMed.
- Y. Xia, J. Ye, K. Tan, J. Wang and G. Yang, Anal. Chem., 2013, 85, 6241–6247 CrossRef CAS PubMed.
- D. Wang, Y. Zhang, X. Zhao and Z. Xu, Sens. Actuators, B, 2019, 296, 126646 CrossRef CAS.
- J. Zeng, Y. Cao, J. Chen, X. Wang, J. Yu, B. Yu, Z. Yan and X. Chen, Nanoscale, 2014, 6, 9939–9943 RSC.
- Z. Chen, Z. Zhang, C. Qu, D. Pan and L. Chen, Analyst, 2012, 137, 5197–5200 RSC.
- X. Yang, Y. Yu and Z. Gao, ACS Nano, 2014, 8, 4902–4907 CrossRef CAS PubMed.
- Y. Zhang, J. Jiao, Y. Wei, D. Wang, C. Yang and Z. Xu, Anal. Chem., 2020, 92, 15244–15252 CrossRef CAS PubMed.
- S. K. Tripathy, J. Y. Woo and C.-S. Han, Anal. Chem., 2011, 83, 9206–9212 CrossRef CAS PubMed.
- L. Tian, E. Chen, N. Gandra, A. Abbas and S. Singamaneni, Langmuir, 2012, 28, 17435–17442 CrossRef CAS PubMed.
- F.-M. Li, J.-M. Liu, X.-X. Wang, L.-P. Lin, W.-L. Cai, X. Lin, Y.-N. Zeng, Z.-M. Li and S.-Q. Lin, Sens. Actuators, B, 2011, 155, 817–822 CrossRef CAS.
- Z. Zhang, Z. Chen, C. Qu and L. Chen, Langmuir, 2014, 30, 3625–3630 CrossRef CAS PubMed.
- Z. Zhang, Z. Chen, F. Cheng, Y. Zhang and L. Chen, Biosens. Bioelectron., 2017, 89, 932–936 CrossRef CAS PubMed.
- Y. Xianyu, Y. Chen and X. Jiang, Anal. Chem., 2015, 87, 10688–10692 CrossRef CAS PubMed.
- L. Lu and Y. Xia, Anal. Chem., 2015, 87, 8584–8591 CrossRef CAS PubMed.
- C.-H. Lu, Y.-W. Wang, S.-L. Ye, G.-N. Chen and H.-H. Yang, NPG Asia Mater., 2012, 4, e10–e10 CrossRef.
- P. Valentini and P. P. Pompa, Angew. Chem., Int. Ed., 2016, 55, 2157–2160 CrossRef CAS PubMed.
- M.-P. N. Bui, S. Ahmed and A. Abbas, Nano Lett., 2015, 15, 6239–6246 CrossRef CAS PubMed.
- K. Zagorovsky and W. C. W. Chan, Angew. Chem., Int. Ed., 2013, 52, 3168–3171 CrossRef CAS PubMed.
- S. Kim, J.-M. Kim, J.-E. Park and J.-M. Nam, Adv. Mater., 2018, 30, 1704528 CrossRef PubMed.
- C. H. Lee, L. Tian and S. Singamaneni, ACS Appl. Mater. Interfaces, 2010, 2, 3429–3435 CrossRef CAS PubMed.
- C. H. Lee, M. E. Hankus, L. Tian, P. M. Pellegrino and S. Singamaneni, Anal. Chem., 2011, 83, 8953–8958 CrossRef CAS PubMed.
- L. Tian, J. Luan, K.-K. Liu, Q. Jiang, S. Tadepalli, M. K. Gupta, R. R. Naik and S. Singamaneni, Nano Lett., 2016, 16, 609–616 CrossRef CAS PubMed.
- L. Tian, J. J. Morrissey, R. Kattumenu, N. Gandra, E. D. Kharasch and S. Singamaneni, Anal. Chem., 2012, 84, 9928–9934 CrossRef CAS PubMed.
- L. Tian, Q. Jiang, K.-K. Liu, J. Luan, R. R. Naik and S. Singamaneni, Adv. Mater. Interfaces, 2016, 3, 1600214 CrossRef.
- U. Mogera, H. Guo, M. Namkoong, M. S. Rahman, T. Nguyen and L. Tian, Sci. Adv., 2022, 8(12), eabn1736 CrossRef CAS PubMed.
- S. Z. Nergiz, N. Gandra, M. E. Farrell, L. M. Tian, P. M. Pellegrino and S. Singamaneni, J. Mater. Chem. A, 2013, 1, 6543–6549 RSC.
- L. M. Tian, S. Tadepalli, M. E. Farrell, K. K. Liu, N. Gandra, P. M. Pellegrino and S. Singamaneni, J. Mater. Chem. C, 2014, 2, 5438–5446 RSC.
- L. Tian, S. Tadepalli, S. Hyun Park, K.-K. Liu, J. J. Morrissey, E. D. Kharasch, R. R. Naik and S. Singamaneni, Biosens. Bioelectron., 2014, 59, 208–215 CrossRef CAS PubMed.
- D. Tu, A. Holderby, H. Guo, S. Mabbott, L. Tian and G. L. Coté, Anal. Chim. Acta, 2022, 1198, 339562 CrossRef CAS PubMed.
- H. Guo, Z. Yin, M. Namkoong, Y. Li, T. Nguyen, E. Salcedo, I. Arizpe and L. Tian, ACS Appl. Mater. Interfaces, 2022, 14, 10729–10737 CrossRef CAS PubMed.
- S. Song, L. Wang, J. Li, C. Fan and J. Zhao, TrAC, Trends Anal. Chem., 2008, 27, 108–117 CrossRef CAS.
- A. Abbas, L. Tian, J. J. Morrissey, E. D. Kharasch and S. Singamaneni, Adv. Funct. Mater., 2013, 23, 1789–1797 CrossRef CAS PubMed.
- L. M. Tian, K. K. Liu, J. J. Morrissey, N. Gandra, E. D. Kharasch and S. Singamaneni, J. Mater. Chem. B, 2014, 2, 167–170 RSC.
- Z. Yin, H. Guo, Y. Li, J. Chiu and L. Tian, ACS Appl. Mater. Interfaces, 2020, 12, 35977–35985 CrossRef CAS PubMed.
- Y. Li, H. Guo, Z. Yin, K. Lyle and L. Tian, ACS Appl. Mater. Interfaces, 2021, 13, 5564–5573 CrossRef CAS PubMed.
- C. Wang, J. Luan, S. Tadepalli, K.-K. Liu, J. J. Morrissey, E. D. Kharasch, R. R. Naik and S. Singamaneni, ACS Appl. Mater. Interfaces, 2016, 8, 26493–26500 CrossRef CAS PubMed.
- D. Quesada-González and A. Merkoçi, Biosens. Bioelectron., 2015, 73, 47–63 CrossRef PubMed.
- V.-T. Nguyen, S. Song, S. Park and C. Joo, Biosens. Bioelectron., 2020, 152, 112015 CrossRef CAS PubMed.
- D. Quesada-González and A. Merkoçi, Lateral Flow Biosensors, Sensor Technology Directory (eBook), Sensor100, 2023 Search PubMed.
- David E. Charlton, US Pat., US6485982B1, 1988 Search PubMed.
- Lateral Flow Immunoassay, ed. R. Wong and H. Tse, Humana Press, Totowa, NJ, 2009 Search PubMed.
- C. Parolo, A. Sena-Torralba, J. F. Bergua, E. Calucho, C. Fuentes-Chust, L. Hu, L. Rivas, R. Álvarez-Diduk, E. P. Nguyen, S. Cinti, D. Quesada-González and A. Merkoçi, Nat. Protoc., 2020, 15, 3788–3816 CrossRef CAS PubMed.
- Whatman Fusion 5|Cytiva, https://www.cytivalifesciences.com/en/es/shop/lab-filtration/immunodiagnostics/lateral-flow-pads/fusion-5-p-00787, (accessed 22 May 2024).
- A. Chamorro-Garcia, A. de la Escosura-Muñiz, M. Espinoza-Castañeda, C. J. Rodriguez-Hernandez, C. de Torres and A. Merkoçi, Nanomedicine, 2016, 12, 53–61 CrossRef CAS PubMed.
- A. Sena-Torralba, J. Gabaldón-Atienza, A. Cubells-Gómez, P. Casino, Á. Maquieira and S. Morais, Biosensors, 2022, 12, 980 CrossRef CAS PubMed.
- M. A. V. Ngu, J. H. Bergantin and J. D. A. Ramos, Protein Pept. Lett., 2019, 26, 357–363 CrossRef CAS PubMed.
- A. Rubio-Monterde, A. Génova-Tesoro, B. Contreras-Repiso, R. Monaghan, R. Merriman, B. Teixeira-Dias, L. Rivas, J. Escudero-Garcia, M. Gallegos, L. Ainsworth, S. Daniels and D. Quesada-González, A. Rubio-Monterde, 31st ECCMID Search PubMed.
- T. Wang, L. Chen, A. Chikkanna, S. Chen, I. Brusius, N. Sbuh and R. N. Veedu, Theranostics, 2021, 11, 5174–5196 CrossRef PubMed.
- M. Oliveira-Rodríguez, S. López-Cobo, H. T. Reyburn, A. Costa-García, S. López-Martín, M. Yáñez-Mó, E. Cernuda-Morollón, A. Paschen, M. Valés-Gómez and M. C. Blanco-López, J. Extracell. Vesicles, 2016, 5(1), 31803 CrossRef PubMed.
- D. Quesada-González, G. A. Jairo, R. C. Blake, D. A. Blake and A. Merkoçi, Sci. Rep., 2018, 8, 16157 CrossRef PubMed.
- N. A. Taranova, N. A. Byzova, V. V. Zaiko, T. A. Starovoitova, Y. Y. Vengerov, A. V. Zherdev and B. B. Dzantiev, Microchim. Acta, 2013, 180, 1165–1172 CrossRef CAS.
- L. Anfossi, F. Di Nardo, S. Cavalera, C. Giovannoli and C. Baggiani, Biosensors, 2018, 9, 2 CrossRef PubMed.
- D. Quesada-González and A. Merkoçi, Biosens. Bioelectron., 2017, 92, 549–562 CrossRef PubMed.
- Using Portable SPR to Speed Up Your Lateral Flow Assay Development, https://www.affiniteinstruments.com/post/spr-in-aiding-the-development-of-lateral-flow-assays, (accessed 12 October 2023).
- J. Kimling, M. Maier, B. Okenve, V. Kotaidis, H. Ballot and A. Plech, J. Phys. Chem. B, 2006, 110, 15700–15707 CrossRef CAS PubMed.
- M. H. Jazayeri, H. Amani, A. A. Pourfatollah, H. Pazoki-Toroudi and B. Sedighimoghaddam, Sens. Biosens. Res., 2016, 9, 17–22 Search PubMed.
- K. Tripathi and J. D. Driskell, ACS Omega, 2018, 3, 8253–8259 CrossRef CAS PubMed.
- Gold Nanoparticle Covalent Conjugation—Fortis Life Sciences, https://www.fortislife.com/lateral-flow-covalent-conjugation, (accessed 12 October 2023).
- Y. Deng, H. Jiang, X. Li and X. Lv, Microchim. Acta, 2021, 188, 379 CrossRef CAS PubMed.
- D. Quesada González, Universitat Autònoma de Barcelona. Departament de Química., TDX (Tesis Doctorals en Xarxa) Search PubMed.
- Y. Shen and G. Shen, ACS Omega, 2019, 4, 5083–5087 CrossRef CAS.
- E. Garrido, E. Climent, M. D. Marcos, F. Sancenón, K. Rurack and R. Martínez-Máñez, Nanoscale, 2022, 14, 13505–13513 RSC.
- D. Quesada and M. Gallegos, EU Pat., EP4070105A1/WO2021110958, 2020 Search PubMed.
- G. Ross, M. Bremer, J. Wichers, A. van Amerongen and M. Nielen, Biosensors, 2018, 8, 130 CrossRef CAS PubMed.
- D. Quesada-González, A. Sena-Torralba, W. P. Wicaksono, A. de la Escosura-Muñiz, T. A. Ivandini and A. Merkoçi, Biosens. Bioelectron., 2019, 132, 132–135 CrossRef PubMed.
- B. Zhang, W. Ma, F. Li, W. Gao, Q. Zhao, W. Peng, J. Piao, X. Wu, H. Wang, X. Gong and J. Chang, Nanoscale, 2017, 9, 18711–18722 RSC.
- D. Quesada-González, C. Stefani, I. González, A. de la Escosura-Muñiz, N. Domingo, P. Mutjé and A. Merkoçi, Biosens. Bioelectron., 2019, 141, 111407 CrossRef PubMed.
- T. You, W. Jeong, H. Lee, Y. S. Huh, S. M. Kim and T.-J. Jeon, Microchim. Acta, 2021, 188, 364 CrossRef CAS PubMed.
- E. Renzi, A. Piper, F. Nastri, A. Merkoçi and A. Lombardi, Small, 2023, 19, 2207949 CrossRef CAS PubMed.
- A. Scarsi, D. Pedone and P. P. Pompa, Nanoscale Adv., 2023, 5, 2167–2174 RSC.
- I. N. Katis, P. J. W. He, R. W. Eason and C. L. Sones, Biosens. Bioelectron., 2018, 113, 95–100 CrossRef CAS PubMed.
- M. Díaz-González and A. de la Escosura-Muñiz, Anal. Bioanal. Chem., 2021, 413, 4111–4117 CrossRef PubMed.
- T. Trakoolwilaiwan, Y. Takeuchi, T. S. Leung, M. Sebek, L. Storozhuk, L. Nguyen, L. D. Tung and N. T. K. Thanh, Nanoscale, 2023, 15, 12915–12925 RSC.
- H.-K. Oh, K. Kim, J. Park, H. Im, S. Maher and M.-G. Kim, Biosens. Bioelectron., 2022, 205, 114094 CrossRef CAS PubMed.
- D. A. Armbruster and T. Pry, Clin. Biochem. Rev., 2008, 29(Suppl 1), S49–S52 Search PubMed.
- A. H. A. Hassan, J. F. Bergua, E. Morales-Narváez and A. Mekoçi, Food Chem., 2019, 297, 124965 CrossRef CAS PubMed.
- X. Y. Wong, D. Quesada-González, S. Manickam and K. Muthoosamy, Anal. Chim. Acta, 2021, 1175, 338745 CrossRef CAS PubMed.
- E. Morales-Narváez, T. Naghdi, E. Zor and A. Merkoçi, Anal. Chem., 2015, 87, 8573–8577 CrossRef PubMed.
- ESEQuant Lateral Flow System-QIAGEN, https://www.qiagen.com/us/resources/resourcedetail?id=60ed2e10-7c42-464b-a805-08a6f9eacc22&lang=en, (accessed 12 October 2023).
- E. coli E. coli O157:H7 ELISA Kit (ABIN5650391), https://www.antibodies-online.com/kit/5650391/Escherichia+Coli,+O157H7+ELISA+Kit/, (accessed 12 October 2023).
- A. M. Ashrafi, Z. Bytesnikova, J. Barek, L. Richtera and V. Adam, Biosens. Bioelectron., 2021, 192, 113494 CrossRef CAS PubMed.
- Y. Wang, Z. Qin, D. R. Boulware, B. S. Pritt, L. M. Sloan, I. J. González, D. Bell, R. R. Rees-Channer, P. Chiodini, W. C. W. Chan and J. C. Bischof, Anal. Chem., 2016, 88, 11774–11782 CrossRef CAS PubMed.
- N. K. Bakirhan, B. D. Topal, G. Ozcelikay, L. Karadurmus and S. A. Ozkan, Crit. Rev. Anal. Chem., 2022, 52, 519–534 CrossRef CAS PubMed.
- J. Cheng, G. Yang, J. Guo, S. Liu and J. Guo, Analyst, 2022, 147, 554–570 RSC.
- A. Sena-Torralba, R. Álvarez-Diduk, C. Parolo, A. Piper and A. Merkoçi, Chem. Rev., 2022, 122, 14881–14910 CrossRef CAS PubMed.
- A. Sena-Torralba, Y. D. Banguera-Ordoñez, L. Mira-Pascual, Á. Maquieira and S. Morais, Trends Biotechnol., 2023, 41, 1299–1313 CrossRef CAS PubMed.
- Z. Wang, Z. Zhang, W. Luo, L. Wang, X. Han, R. Zhao, X. Liu, J. Zhang, W. Yu, J. Li, Y. Yang, C. Zuo and G. Xie, Nanoscale, 2023, 15, 12660–12669 RSC.
- A. Rubio-Monterde, D. Quesada-González and A. Merkoçi, Anal. Chem., 2023, 95, 468–489 CrossRef CAS PubMed.
- Y. Wang, J. Niu, M. Sun, Z. Li, X. Wang, Y. He and J. Qi, Int. J. Mol. Sci., 2023, 24, 7733 CrossRef CAS PubMed.
- M. S. Cordray and R. R. Richards-Kortum, Malar. J., 2015, 14, 472 CrossRef PubMed.
- A. Salazar, F. M. Ochoa-Corona, J. L. Talley and B. H. Noden, Sci. Rep., 2021, 11, 15962 CrossRef CAS PubMed.
- M. Jauset-Rubio, M. Svobodová, T. Mairal, C. McNeil, N. Keegan, M. S. El-Shahawi, A. S. Bashammakh, A. O. Alyoubi and C. K. O’Sullivan, Anal. Chem., 2016, 88, 10701–10709 CrossRef CAS PubMed.
- J. Kumar, K. G. Thomas and L. M. Liz-Marzán, Chem. Commun., 2016, 52, 12555–12569 RSC.
- J. Kumar and L. M. Liz-Marzán, Bull. Chem. Soc. Jpn., 2019, 92, 30–37 CrossRef CAS.
- Q. Zhang, T. Hernandez, K. W. Smith, S. A. H. Jebeli, A. X. Dai, L. Warning, R. Baiyasi, L. A. McCarthy, H. Guo, D. H. Chen, J. A. Dionne, C. F. Landes and S. Link, Science, 2019, 365, 1475–1478 CrossRef CAS PubMed.
- W. Ma, H. Kuang, L. Xu, L. Ding, C. Xu, L. Wang and N. A. Kotov, Nat. Commun., 2013, 4, 2689 CrossRef PubMed.
- Y. Zhao, A. N. Askarpour, L. Sun, J. Shi, X. Li and A. Alù, Nat. Commun., 2017, 8, 6–13 CrossRef PubMed.
- P. Mulvaney, Langmuir, 1996, 12, 788–800 CrossRef CAS.
- R. Thomas, J. Kumar, R. S. Swathi and K. G. George Thomas, Curr. Sci., 2012, 102, 85–96 CAS.
- L. A. Warning, A. R. Miandashti, L. A. McCarthy, Q. Zhang, C. F. Landes and S. Link, ACS Nano, 2021, 15, 15538–15566 CrossRef CAS PubMed.
- W. Ma, L. Xu, A. F. De Moura, X. Wu, H. Kuang, C. Xu and N. A. Kotov, Chem. Rev., 2017, 117, 8041–8093 CrossRef CAS PubMed.
- S. Maniappan, C. Dutta, D. M. Solís, J. M. Taboada and J. Kumar, Angew. Chem., Int. Ed., 2023, 62, e202300461 CrossRef CAS PubMed.
- G. Shemer, O. Krichevski, G. Markovich, T. Molotsky, I. Lubitz and A. B. Kotlyar, J. Am. Chem. Soc., 2006, 128, 11006–11007 CrossRef CAS PubMed.
- J. George and K. G. Thomas, J. Am. Chem. Soc., 2010, 132, 2502–2503 CrossRef CAS PubMed.
- J. Cai, C. Hao, M. Sun, W. Ma, C. Xu and H. Kuang, Small, 2018, 14, 1–8 Search PubMed.
- T. Funck, F. Nicoli, A. Kuzyk and T. Liedl, Angew. Chem., Int. Ed., 2018, 57, 13495–13498 CrossRef CAS PubMed.
- L. Xu, Y. Gao, H. Kuang, L. M. Liz-Marzán and C. Xu, Angew. Chem., Int. Ed., 2018, 57, 10544–10548 CrossRef CAS PubMed.
- S. Dominguez-Medina, L. Kisley, L. J. Tauzin, A. Hoggard, B. Shuang, A. S. D. S. Indrasekara, S. Chen, L. Y. Wang, P. J. Derry, A. Liopo, E. R. Zubarev, C. F. Landes and S. Link, ACS Nano, 2016, 10, 2103–2112 CrossRef CAS PubMed.
- M. Sun, L. Xu, J. H. Banhg, H. Kuang, S. Alben, N. A. Kotov and C. Xu, Nat. Commun., 2017, 8, 1–10 CrossRef CAS PubMed.
- X. T. Kong, L. V. Besteiro, Z. Wang and A. O. Govorov, Adv. Mater., 2020, 32, 1–14 Search PubMed.
- A. O. Govorov, Z. Fan, P. Hernandez, J. M. Slocik and R. R. Naik, Nano Lett., 2010, 10, 1374–1382 CrossRef CAS PubMed.
- B. M. Maoz, Y. Chaikin, A. B. Tesler, O. Bar Elli, Z. Fan, A. O. Govorov and G. Markovich, Nano Lett., 2013, 13, 1203–1209 CrossRef CAS PubMed.
- H. Zhang and A. O. Govorov, Phys. Rev. B: Condens. Matter Mater. Phys., 2013, 87, 1–8 Search PubMed.
- L. M. Kneer, E. M. Roller, L. V. Besteiro, R. Schreiber, A. O. Govorov and T. Liedl, ACS Nano, 2018, 12, 9110–9115 CrossRef CAS PubMed.
- E. Severoni, S. Maniappan, L. M. Liz-Marzán, J. Kumar, F. J. Garciá De Abajo and L. Galantini, ACS Nano, 2020, 14, 16712–16722 CrossRef CAS PubMed.
- F. Lu, Y. Tian, M. Liu, D. Su, H. Zhang, A. O. Govorov and O. Gang, Nano Lett., 2013, 13, 3145–3151 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |