 Open Access Article
Open Access ArticlePolydopamine as a versatile optical indicator for colorimetric and fluorescence-based biosensing
Jena Subhra Sulipta†
,
Haejin Jeong† and
Seonki Hong *
*
Department of Physics and Chemistry, Daegu Gyeongbuk Institute of Science and Technology (DGIST), Daegu, Republic of Korea. E-mail: seonkihong@dgist.ac.kr
First published on 20th March 2025
Abstract
Beyond their well-established adhesive properties, polydopamine (pDA) and pDA-like materials are emerging as superior alternatives to conventional optical indicators in biosensing applications due to their exceptional biocompatibility, tunable optical properties, and high sensitivity, arising from their eumelanin-like physicochemical characteristics. These materials attract significant attention for their ability to function as optical probes and transducers, enabling precise and sensitive detection in complex biological environments. This review highlights recent advancements in developing pDA-based optical probes, emphasizing strategies for fine-tuning synthetic parameters to optimize material properties, clarifying the fundamental sensing mechanisms underlying pDA-based systems, and exploring their potential roles in addressing global healthcare challenges. By facilitating early disease detection, real-time monitoring, and targeted therapeutic intervention, pDA-based optical probes offer transformative solutions to pressing biomedical needs. Through a comprehensive examination of cutting-edge research, this review aims to illuminate how the unique attributes of pDA materials drive innovation in biosensing technologies and contribute to improved healthcare outcomes.
1. Introduction
Biosensing techniques are crucial for identifying specific biomolecules from various specimens, serving a significant role in disease treatment, drug discovery, environmental monitoring, and other essential applications. In particular, in vitro diagnostics that detect biomarkers in biofluids, such as blood, saliva, sweat, and tears, have gained considerable attention in personalized healthcare. These techniques enable more targeted therapies and improve patient outcomes by facilitating early diagnosis, treatment monitoring, and prognosis.1,2Various detection principles have been developed to produce measurable signals upon biomarker recognition. In line with advancements in novel materials (such as MXene, graphene, and Borophene), which are known for their high surface reactivity as well as excellent thermal and electrical conductivity, bioassays based on electrochemical, magnetic, and mass-based methods have also seen significant progress.3,4 However, many of these detection techniques require expensive instruments and highly trained operators, limiting their accessibility for real-world applications. Optical sensing, in particular, has been extensively studied due to its convenience and rapid readout capabilities. Colorimetric detection is widely utilized among optical methods, particularly in home-based commercial kits, as exemplified by lateral-flow immunoassays (LFIA). This approach allows results to be visually interpreted without requiring specialized equipment, making it highly accessible for self-testing.5 Similarly, fluorescence-based sensing is a preferred strategy due to its high sensitivity for detecting low-abundance molecules, utilizing both commercially available devices and compact, custom-built equipment.6
In both colorimetric and fluorescence-based techniques, developing novel colorants and fluorophores is essential for enhancing sensitivity and specificity, which are critical to improving the overall accuracy of these assays. Small molecular chromophores have been extensively explored for biosensing due to their well-defined optical properties and the capacity to fine-tune spectral ranges through precise structure–property modifications.7,8 Despite their potential, challenges such as photobleaching and low aqueous solubility limit their use in complex biological and environmental contexts.9,10 Nanomaterials have emerged as a robust alternative, providing distinct advantages, including tunable spectral properties, enhanced optical stability, and improved robustness under diverse conditions.2,11,12 Examples include quantum dots, gold nanoparticles, and upconversion nanoparticles, each suited to biosensing applications in various physiological and environmental settings. In addition, their high surface area facilitates functionalization with bioreceptors, enabling target-specific recognition and expanding their utility.13 However, challenges persist. Variability in nanomaterial synthesis often results in batch-to-batch inconsistencies, adversely affecting assay reproducibility and reliability.14,15 In addition, concerns about the cytotoxicity of specific nanomaterials limit their direct application in biological systems, necessitating the development of safer, biocompatible alternatives.14,16 Large-scale production poses significant challenges, as scalability often compromises material quality and performance.15 Overcoming these issues requires innovative strategies to engineer materials with enhanced stability, biocompatibility, and scalability, ensuring they meet the practical demands of biosensing applications while addressing current limitations.
Polydopamine (pDA) and pDA-like materials have gained recognition as versatile materials for biosensing applications due to their unique physicochemical properties and biomimetic characteristics. A key feature of pDA in biosensing is its effectiveness as an interfacial layer for immobilizing bioreceptors onto transducers.17,18 The adhesive characteristics of pDA facilitate the robust attachment of proteins, nucleic acids, or small molecules, improving the stability and reproducibility of biosensor performance. Beyond its role as an adhesive layer, pDA's broadband light absorption, spanning the ultraviolet to near-infrared (NIR) regions and resembling natural eumelanin biopigments, unlocks novel optical signal transduction possibilities.19–21 For instance, its visible light absorption supports colorimetric sensing, enabling visually detectable biomarker quantification. Simultaneously, pDA's NIR absorption facilitates advanced techniques such as photoacoustic signal generation and photothermal therapies. In addition, its broadband absorption allows pDA to function as a universal fluorescent quencher, enhancing its utility in fluorescence-based biosensor platforms. Traditionally, pDA NPs have been considered non-fluorescent. However, recent advancements in synthetic methods have resulted in fluorescent pDA analogues, significantly expanding their application potential. These fluorescent derivatives have introduced new opportunities in biosensing, bioimaging, and in vitro diagnostics. These developments highlight the multifaceted nature of pDA and its derivatives, bridging material science and biomedical applications to address diverse biosensing requirements.
This review investigates advanced strategies utilizing pDA and pDA-like organic materials as optical probes in biosensing. The focus is on optimizing synthetic conditions to enhance material properties, clarifying the operational principles of pDA-based optical sensing, and examining their potential applications in healthcare. The review is structured into three main sections to provide a comprehensive analysis:
(1) pDA-based colorimetric sensing: Highlighting applications that use pDA's visible light absorption for simple and direct biomarker detection.
(2) Biosensing utilizing pDA as fluorescent quenchers: Exploring the utility of pDA in fluorescence-based platforms through its quenching capabilities.
(3) Fluorescent pDA analogues in biosensing: Discussing the emerging applications of fluorescent pDA derivatives in bioimaging and diagnostics.
The aim is to provide insights into how nature-inspired pDA materials can address critical challenges in biosensing by exploring these advancements. These innovations are poised to enable more accurate, cost-effective, and accessible healthcare solutions, with significant implications for the future of personalized medicine and diagnostics.
2. pDA as visible indicators
2.1. Fundamentals in the absorption spectrum of pDA and its connection to eumelanin
Eumelanin, a class of organic biopigments responsible for brown-to-black coloration in nature, exhibits broad light absorption across UV-Vis-NIR wavelengths, which monotonically increases toward high-energy regions. This unique property, rarely observed in natural organic chromophores, is often regarded as semiconductor-like behavior.22 Such behavior is attributed to eumelanin's highly complex and disordered chemical structure and hierarchical nanoscale assembly. In nature, eumelanin synthesis involves the enzymatic oxidation of tyrosine to 3,4-dihydroxyphenylalanine (DOPA), followed by the formation of intermediates such as DOPAchrome, DHI, and DHICA (Fig. 1A). These intermediates grow into heterogeneous oligomeric chromophores, commonly called protomolecules, which define the primary absorption characteristics across various wavelengths. However, this alone does not fully explain the monotonic increase in absorption across the wide spectral range extending from UV to NIR. Several studies indicate that excitonic and/or electronic coupling between protomolecules, facilitated by π-stacking and higher-order aggregation, broadens the absorption spectrum, introducing new transition energies distinct from those of individual chromophores (Fig. 1B).22–24 Partial proton transfer from catechol hydroxy groups to the solvent has also been proposed as a contributing factor.24 These molecular interactions enable eumelanin to dissipate absorbed energy as heat through rapid non-radiative relaxation processes, resulting in an extremely low fluorescence quantum yield (<0.05%).25,26 This mechanism is critical to the photoprotective function of eumelanin in nature.24 | ||
| Fig. 1 Fundamentals in eumelanin synthesis. (A) The initial stages of eumelanin synthesis begin with the oxidation of tyrosine to DOPA, which is further converted into the key intermediates DHICA and DHI. The DHI-involved pathways in eumelanin synthesis overlap with those of pDA synthesis. Adapted from ref. 27, Copyright 2014, ACS. (B) A proposed structure–property relationship of eumelanin, highlighting spectral broadening toward low energies observed during hierarchical assembly. Adapted from ref. 22, Copyright 2018, ACS. | ||
pDA is widely recognized as the most acceptable functional mimic of eumelanin due to its remarkable similarities in synthetic pathways and resulting material properties.19–21 The synthesis of pDA closely mirrors that of eumelanin, beginning with the oxidation of catechol-containing precursors, followed by the formation of protomolecules that further assemble into nanoscale structures. As a result, pDA forms brown-black, water-insoluble aggregates with broadband absorption spanning the UV to NIR regions, similar to eumelanin.28 Although the DA precursor used in pDA synthesis lacks the carboxyl group necessary for generating DHICA-based protomolecules, the inclusion of 5,6-dihydroxyindole (DHI), a major intermediate extensively discussed in prior eumelanin studies, strongly supports the designation of pDA as a functional mimic of eumelanin (Fig. 1A). These similarities highlight the potential of pDA to replicate the biological roles of eumelanin, such as photoprotection and antioxidation, while serving as an innovative material for optical and biomedical applications. In biosensing, the absorption properties of pDA in the visible range are particularly advantageous, facilitating the development of colorimetric and fluorescence-based readouts for point-of-care testing (POCT).
2.2. pDA NP-based colorimetric assays
Colorimetric bioassays represent one of the most accessible detection methods, allowing results to be directly observed with the naked eye without requiring specialized analytical equipment. This accessibility renders colorimetric assays particularly suitable for point-of-care (POC) applications, facilitating their use in real-world settings outside conventional laboratories.pDA nanoparticles (NPs) have been successfully integrated into LFIA for POC diagnostics. For instance, Tong et al. developed a colorimetric LFIA for COVID-19 detection, utilizing pDA NPs as visible indicators.29 Their study demonstrated that pDA NPs exhibit superior absorption in the visible range compared to AuNPs at the same mass fraction (4.5%), resulting in greater sensitivity for the pDA-based LFIA kits than commercial Au-based alternatives. Similarly, Zhang et al. synthesized pDA-coated metal–organic frameworks (MOFs) as visible probes in LFIA. Their system achieved a detection limit of 0.045 ng mL−1 for enrofloxacin, which improved upon the 0.095 ng mL−1 limit observed in conventional AuNP-incorporated systems.30 Xu et al. also reported that pDA coatings on AuNPs enhance color brightness, increasing the molar extinction coefficient by 1.68-fold and grayscale intensity by 1.87-fold.31 This enhancement improved the sensitivity of LFIA while providing high dispersibility across a wide pH range (6–14) and in NaCl-rich environments, broadening its compatibility with diverse bioassay conditions.
The versatility of pDA as a colorant extends beyond LFIA applications. For example, its application in enzyme-linked immunosorbent assays (ELISA) has shown considerable potential. Horseradish peroxidase (HRP)-accelerated pDA deposition significantly enhanced sensitivity, as demonstrated by Xu et al., who observed a 100-fold improvement in sensitivity compared to conventional enhanced chemiluminescence (ECL) kits.32
2.3. Spatial marking of biomarkers
Two primary strategies are employed to generate optical signals in bioassays. One involves using pre-prepared colorants, such as quantum dots and AuNPs, which bind specifically to target molecules (via antibody–antigen interactions), enabling visualization of biomarker distribution. However, this approach often suffers from limited signal amplification because the optical density of these materials is fixed during preparation. Alternatively, enzyme-incorporated strategies amplify signals by converting numerous substrate molecules into detectable indicators. Although this method provides substantial amplification, it is typically restricted to solution-based assays, as the generated colorants tend to diffuse in aqueous environments, making precise spatial localization challenging.In this context, pDA has emerged as an innovative adhesive colorant capable of addressing these limitations. Its local generation near biomarkers ensures precise spatial marking while enabling signal amplification. For example, Li et al. developed a pDA-based immunoassay named EASE, involving the in situ generation of pDA near biomarkers pre-labeled with HRP.33 This approach produced a detectable brownish color under bright-field imaging. In addition, the secondary immobilization of quantum dots (QDs) or HRP on the pDA layer further enhanced sensitivity (Fig. 2A). Using this method, an ELISA demonstrated remarkable sensitivity, detecting HIV antigens in blood samples at concentrations below 3 fg mL−1. Kim et al. employed in situ deposition of pyrocatechol-based adhesive colorants, showing the localized formation of pigments near HRP, in contrast to the diffusion of TMB in solution (Fig. 2B).34 Both studies were validated in cellular systems (Fig. 2C and D). In addition, Li et al. exploited the spectral shift in localized surface plasmon resonance (LSPR) of AuNPs induced by pDA coating as a visible marker (Fig. 2E). This method effectively detected food contaminants such as biogenic amines, with the pDA coating occurring significantly faster in the presence of organic amines.35 These advancements highlight the unique advantages of pDA and pDA-like materials in spatially marking biomarkers, providing precise localization and robust signal amplification for various biosensing applications.
 | ||
| Fig. 2 Colorimetric bioassays enabled by localized in situ generation of pDA and pDA-like materials. (A) A pDA-based immunoassay with enhanced sensitivity was achieved by secondary incorporating HRP (purple) onto pDA generated around biomarker-associated sites (brown). Adapted from ref. 33, Copyright 2017, Springer Nature. (B) A comparison of color localization between pDA-like adhesive colorants and conventional TMB activated by HRP. Adapted from ref. 34, Copyright 2020, Wiley. (C and D) Spatial marking of biomarkers in a cellular system. (C) was adapted from ref. 33, Copyright 2017, Springer Nature, and (D) was adapted from ref. 34, Copyright 2020, Wiley. (E) In situ pDA coating in the presence of food contaminant biogenic amines, tuning the LSPR of AuNPs as a visible indicator. Adapted from ref. 35, Copyright 2021, Elsevier. | ||
3. pDA-based fluorescent quenchers in bioassays
3.1. Fundamentals of pDA-mediated fluorescence quenching
pDA has been proposed as a multi-wavelength quencher for various fluorophores; however, its fluorescence quenching mechanisms remain not fully understood and appear to vary depending on the type of fluorophores involved and their proximity to pDA. Qiang et al. suggested that the quenching of small molecular organic dyes, such as Cy5, results from a combination of dynamic and static mechanisms, primarily involving fluorescence resonance energy transfer (FRET). This conclusion was supported by a decrease in fluorescence decay time and the observation of a nonlinear Stern–Volmer plot with an upward curvature (concave toward the y-axis).36 In addition, they noted that an electron transfer mechanism cannot be excluded, as monomeric dopamine (DA), which lacks spectral overlap with the fluorophore, also induced some fluorescence quenching. Medintz et al. previously confirmed the redox coupling between quantum dots and the quinone state of DA, demonstrating its applicability in pH sensing.37 Similarly, Wang et al. investigated the quenching mechanism of upconversion fluorescent nanoparticles (UCNPs) by pDA, identifying photoinduced electron transfer (PET) as the primary mechanism.38Chen et al. proposed that the primary quenching mechanism of carbon dots by pDA is the inner filter effect (IFE), based on their calculation of the contribution of IFE to quenching efficiency.39 They also suggested that neither dynamic nor static quenching mechanisms contributed to the quenching of carbon dots, as the fluorescence lifetime of carbon dots remained unchanged in the presence of pDA, and no evidence was found for forming non-fluorescent complexes between pDA and carbon dots. Similarly, Wang et al. argued that IFE, rather than FRET, is the more plausible quenching mechanism for pDA with three tested fluorescent particulate donors, aggregation-induced emission fluorescent microspheres, fluorescent microspheres, and quantum dot beads, based on the observation that the fluorescence lifetime decay showed no significant changes after mixing with pDA.40 However, Liu et al. reported that the direct coating of pDA on fluorescent carbon dots decreases fluorescence lifetime, indicating that FRET can still contribute to the quenching of carbon dots, mainly when they are in direct contact with pDA.41
3.2. Nucleic acid-based assays utilizing pDA NPs as fluorescent quenchers
Various fluorescence-based sensing schematics utilizing “turn-on” and “turn-off” mechanisms can be designed (Table 1). Due to the high affinity between nucleic acids and pDA, a common assay approach involves nucleic acid probes and/or aptamers. PDA quenches these probes in their single-stranded state, but fluorescence is restored when the probes form complexes with their targets and detach from pDA (Fig. 3A) or when fluorophores are released as free fluorophores from pDA due to probe degradation (Fig. 3B and C). Incorporating enzymes that selectively degrade nucleic acids further amplifies fluorescence signals, enhancing overall assay sensitivity. Qiang et al. demonstrated that pDA nanospheres serve as universal quenchers, achieving up to 97% quenching efficiency for four widely used fluorophores with emissions ranging from 400 to 700 nm, including aminomethylcoumarin acetate (AMCA), 6-carboxyfluorescein (FAM), 6-carboxytetramethylrhodamine (TAMRA), and Cy5.36 Building on this, they developed a platform for detecting single-base mismatches in DNA and thrombin. Qiang et al. enhanced assay sensitivity by incorporating exonuclease III, which degrades the probe–target duplex and releases free fluorophores from pDA.42 Including exonuclease III improved the detection limit by up to 20-fold, reducing it to 5 pM for DNA. Fan et al. validated the feasibility of an exonuclease III-incorporated system with pDA nanotubes as quenchers, developing a fluorescent detection platform for human immunodeficiency virus (HIV) DNA and ATP, achieving detection limits of 3.5 pM and 150 nM, respectively.43 Another platform, developed by Wang et al., utilized DNase I to regenerate target miRNA by selectively degrading DNA probes hybridized with the target miRNA.44 The regenerated miRNA triggered further degradation of DNA probes, increasing the concentration of free fluorophores in the solution. Deng et al. employed duplex-specific nuclease (DSN) to selectively degrade DNA probes on synthesized pDA-PEI nanodots, demonstrating effective miRNA sensing in serum.45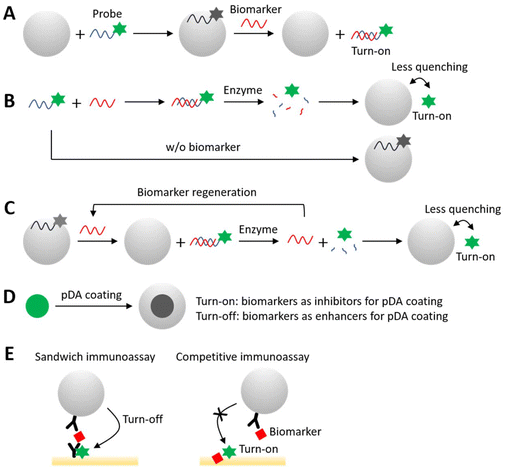 | ||
| Fig. 3 Various types of fluorescence-based sensing schematics are mediated by pDA, utilizing “turn-on” and “turn-off” mechanisms. | ||
| Year | Mechanism | Quencher | Fluorescent dye (used Ex/Em) | Target biomarker | Sensitivity | Ref. | |
|---|---|---|---|---|---|---|---|
| Nucleic acid probe | 2014 | Turn-on | Spherical pDA NPs (diameter: 336 nm) | AMCA (335/448 nm), FAM (470/518 nm), TAMRA (535/575 nm), Cy5 (625/660 nm) | Model DNA | Linear range: 0.78–25 nM, LOD: 0.1 nM | 36 |
| Thrombin | Linear range: 1.56–37.5 nM, LOD: 0.5 nM | ||||||
| 2015 | Turn-on (Exo III-assisted amplification) | Spherical pDA NPs with a diameter of 297.3 ± 20.4 nm | FAM (470/518 nm) | Model DNA | Linear range: 0.078–5 nM, LOD: 5 pM | 42 | |
| ATP | Linear range: logarithm of 0.39–1600 μM, LOD: 180 nM | ||||||
| 2016 | Turn-on (Exo III-assisted amplification) | pDA nanotubes | FAM | HIV DNA | Linear range: 10–200 pM, LOD: 3.5 pM | 43 | |
| ATP | Linear range: logarithm of 0.35–800 μM, LOD: 150 nM | ||||||
| 2017 | Turn-on (DNase I-assisted amplification) | Mesoporous pDA NPs (diameter: 70 nm) | FAM (480/525 nm) | Let-7a | LOD: 40 pM | 44 | |
| miRNA-21 | LOD: 32 pM | ||||||
| 2022 | Turn-on (DNase I-assisted amplification) | DA-PEI co-polymerized nanodots (diameter: 6.7 nm) | AMCA (365/452 nm) | miRNA-21 | Linear range: 0.8–50 nM, LOD: 0.52 nM | 45 | |
| In situ pDA generation | 2023 | Turn-off | In situ pDA coating | Fluorescent carbon dots (370/500 nm) | DA | Linear range: 0.1–15 μM, LOD: 37 nM | 41 |
| 2015 | Turn-on | In situ pDA coating | Upconversion fluorescent NPs (980/540 nm) | Total antioxidant capacity | Recoveries: 94.1%–100.1% | 38 | |
| 2019 | Turn-off | In situ pDA coating | Fluorescent carbon dots (350/452 nm) | Acid phosphatase | Linear range: 1–60 U L−1, LOD: 0.45 U L−1 | 39 | |
| Immunoassays | 2022 | Turn-on (LFIA) | Spherical pDA NPs (diameter: 205 nm) | AIEFM (320/505 nm), FM (495/530 nm), QB (354/615 nm) | Sulfamethazine | Linear range: 0.05–10 ng mL−1, LOD: 0.043 ng mL−1 | 40 |
| 2018 | Turn-off (sandwich) | pDA coated mesoporous SiO2 | ECL (luminol-O2 system) | Insulin | Linear range: 0.0001–50 ng mL−1, LOD: 26 fg mL−1 | 46 | |
| 2024 | Turn-on (competitive) | Hybrid pDA@ZIFs NPs | ECL (g-C3N4/K2S2O8 system) | Ochratoxin A | Linear range: 10.0 fg mL−1–1.0 ng mL−1, LOD: 4.8 fg mL−1 | 47 | |
| 2023 | Turn-on (LFIA) | Fe(III)-complexed pDA NPs | Fluorescent quantum dots (365/625 nm) | Enrofloxacin | LOD: 0.016 ng mL−1 | 48 |
Due to their biocompatibility and hydrophilicity, which are favorable for cellular environments, pDA-based fluorescence probes for nucleic acids are well-suited for both in vitro and in vivo imaging. These properties ensure that DNA probes physisorbed on pDA remain stable in biological systems and selectively detach during hybridization with target nucleic acids. Lin et al. synthesized multifunctional Fe3O4@pDA core–shell nanocomposites for intracellular mRNA detection and imaging-guided photothermal therapy.49 DNA probes physisorbed on the surface of these nanocomposites detached and emitted fluorescence upon hybridization with target mRNA, enabling the visualization of mRNA location and distribution within living cells. Mao et al. further demonstrated the feasibility of the pDA-DNA probe system for in vivo fluorescence sensing of miRNAs.50 They also proposed combining these probes with doxorubicin for potential applications in synergistic cancer therapy.
3.3. In situ pDA generation for fluorescence quenching-based assays
In another approach, the in situ generation of pDA as a fluorescence quencher has proven effective for detecting DA as a biomarker and identifying factors involved in pDA generation (Fig. 3D). DA, along with initiators or accelerators of pDA production, results in a “turn-off” sensing of co-existing fluorophores, while inhibitors of pDA generation maintain fluorophore fluorescence, resulting in a “turn-on” effect. Liu et al. developed a fluorescence quenching-based assay for DA detection by monitoring in situ pDA coating on fluorescent carbon dots, which induces fluorescence quenching.41 The system exhibited minimal interference from common biomolecules in biofluids, such as salts, amino acids, ascorbic acid, and glutathione, achieving good recoveries ranging from 94.0 to 106.7% of DA spiked in human serum. In another study, Wang et al. evaluated the inhibition of pDA formation by antioxidants by measuring fluorescence quenching of upconversion NPs during pDA coating (Fig. 4A).38 Various antioxidants, including biothiols, vitamin C, and Trolox, were specifically detected even in the presence of 100-fold higher concentrations of other common biofluid components. The feasibility of this system was validated for monitoring the antioxidant capacity of cell extracts and human plasma. Chen et al. utilized fluorescence quenching induced by in situ pDA formation to indicate acid phosphatase (ACP) activity under neutral conditions.39 They highlighted that ACP's ability to generate pDA using DA as a substrate distinguishes it from alkaline phosphatase (ALP) under neutral conditions, addressing a challenge in previously reported methods. This system showed recoveries ranging from 93.7% to 109.1% for detecting ACP in 20-times diluted human serum. Lastly, Lee et al. developed a pDA-based sensor platform to detect bacterial hypoxia using the requirement of oxygen for pDA synthesis (Fig. 4B).51 During the exponential growth phase of Escherichia coli, pDA synthesis was significantly inhibited due to bacterial oxygen consumption. This inhibition was monitored through the quenching of co-existing fluorescent dextran NPs. This system monitored bacterial growth in the presence of antibiotics, enabling a rapid antibiotic susceptibility test within 5 h, much faster than traditional culture-based assays that typically require 2 to 7 days. | ||
| Fig. 4 In situ pDA generation for fluorescence quenching-based assays. (A) pDA quencher coupled to upconversion nanoparticles (UCNPs) for NIR-excited sensing of in vitro antioxidant capacity. Adapted from ref. 38, Copyright 2015, ACS. (B) pDA-based sensor platform for detecting bacterial hypoxia. Adapted from ref. 51, Copyright 2021, Wiley. | ||
3.4. Immunoassays
pDA NPs can function as fluorescence quenchers in immunoassays, such as LFIA and ELISA. Both “turn-on” and “turn-off” sensing mechanisms are applicable in these systems (Fig. 3E). Xing et al. developed a sandwich assay for insulin detection utilizing pDA-coated silica as quencher probes for the ECL signals of a luminol-O2 system.46 The target antigen bridges luminol and pDA in this approach, weakening ECL signals. This method enabled reliable insulin detection with a linear range of 0.0001–50 ng mL−1 and a 26 fg mL−1 detection limit. Similarly, Yang et al. employed pDA-incorporated ZIFs as detection probes for g-C3N4/K2S2O8 ECL in a sandwich assay for mycotoxins, achieving a linear range of 10.0 fg mL−1–1.0 ng mL−1 and a detection limit of 4.8 fg mL−1.47 In addition, Lai et al. established an LFIA using iron-doped pDA (Fe-pDA) submicrobeads as quencher probes for pre-loaded quantum dots on the strip.48 The incorporation of iron into pDA resulted in a 14.9-fold higher fluorescence quenching constant than regular pDA due to the synergistic effects of FRET and the IFE. Based on this, a competitive assay was demonstrated in which antibody-decorated Fe-pDA beads were selectively bound to the pre-loaded target molecules near the quantum dots on the surface without the target in the medium, resulting in a negative signal. This approach achieved a detection limit of 0.016 ng mL−1.4. Fluorescent analogues of pDA
4.1. Synthesis of fluorescent pDA analogues
pDA NPs are typically synthesized as non-fluorescent spheres with sizes ranging from a few hundred nanometers. However, reports exist on synthesizing pDA analogues exhibiting fluorescence, making them valuable for biosensing and bioimaging applications. Understanding the mechanism behind this fluorescence emission remains challenging due to the complex chemical structure of pDA. Nevertheless, based on the non-radiative energy dissipation via electronic and/or excitonic coupling between stacked protomolecules, it is plausible that aggregation-caused quenching (ACQ) is linked to the growth of pDA NPs derived from intrinsically fluorescent protomolecules. Therefore, the synthesis of fluorescent pDA analogues (F-pDA) primarily focuses on reducing the degree of assembly and/or weakening the non-covalent interactions between protomolecules. This can be achieved using either a bottom-up approach, which involves enhanced oxidative conditions or the inclusion of additive molecules during synthesis, or a top-down approach, where pre-synthesized NPs are chemically degraded.Non-fluorescent pDA is typically synthesized under mildly basic pH in aerobic aqueous conditions, where dissolved oxygen serves as the oxidant, eliminating the need for additional chemical oxidants. Enhanced oxidative conditions have been reported to facilitate the formation of F-pDA analogues. For example, Zhang et al. demonstrated that introducing H2O2 into a 15-minute pre-reacted pDA synthesis batch led to worm-like structured F-pDA analogues.52 This process required a relatively high concentration of H2O2, with 10 mL of concentrated H2O2 (30% w/w) added to 40 mL of the synthetic mixture. Then, Liu et al. utilized iron oxide NPs (Fe3O4) with peroxidase-like activity to significantly reduce the required H2O2 concentration for F-pDA synthesis to 5 mM (Fig. 5A),53 while Li et al. employed the Fenton reaction, using Fe2+ and H2O2 to generate ˙OH, achieving a reduction in the required H2O2 concentration to as low as 2 mM.54 Other inorganic oxidants, such as KMnO4, MnO2, and CoOOH, have also been employed to generate F-pDA, which can be further applied in various bioassays based on in situ F-pDA generation (section 4.3). Liu et al. reported a rapid and additive-free synthesis method using nonthermal air plasma treatment.55 This method involved applying a 1 minute plasma treatment at 270 W to a neutral DA solution, generating various reactive species, such as persistent reactive oxygen/nitrogen species (H2O2 and NO3−), short-lived radical/atomic species (O, ˙NO, and ˙OH), and non-radical chemical compounds (1O2).
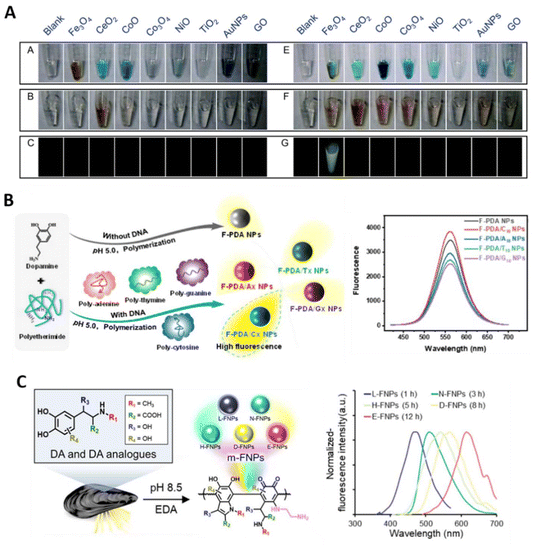 | ||
| Fig. 5 Fluorescent pDA analogues synthesized through various strategies. (A) Images of oxidized TMB (top), oxidized DA (middle), and fluorescence of oxidized DA under 470 nm excitation (bottom row) after reaction with each nanomaterial in the presence of H2O2. Adapted from ref. 53, Copyright 2016, RSC. (B) Sequence-specific enhancement of fluorescence in PEI-incorporated pDA dots. Adapted from ref. 56, Copyright 2022, ACS. (C) NPs emitting distinct fluorescence spectra ranging from 410 to 680 nm, synthesized by reacting DA and DA analogues as precursors with ethylenediamine (EDA). Reproduced under the terms of the CC-BY license,57 Copyright 2024, The Authors, Published by Elsevier. | ||
Another strategy for synthesizing F-pDA involves incorporating additives such as polyethyleneimine (PEI), ethylenediamine (EDA), and folic acid during synthesis. These additives chemically modify the structure via Michael addition and Schiff-base formation or disrupt non-covalent interactions, reducing assembly and altering the electronic states of the resulting NPs. Interestingly, Liu et al. reported that the fluorescent intensity of F-pDA NPs synthesized with PEI can be further enhanced by co-synthesizing them with polycytosine, among the homopolymers composed of DNA bases (polyadenine, polythymine, polyguanine, and polycytosine) (Fig. 5B).56
In addition to DA, its analogues can also serve as precursors for synthesizing fluorescent carbon dots. For example, Lee et al. utilized four different DA analogues, levodopa (LVD), norepinephrine (NPP), 6-hydroxydopamine (HDA), and epinephrine (EPP), as precursors reacted with EDA, producing NPs that emitted distinct fluorescence spectra ranging from 410 to 680 nm (Fig. 5C).57 Variations in fluorescence were attributed to substituents at different positions on the DA molecules, which influenced the π–π* electronic transitions (260–320 nm) and n–π* electronic transitions (320–440 nm) of the synthesized NPs.
Top-down approaches involving the chemical degradation of pre-synthesized pDA NPs are also utilized to synthesize F-pDA analogues. For instance, Yin et al. demonstrated that reducing pDA NPs with sodium borohydride resulted in blue-shifted fluorescence emission, enhancing the quantum yield to 5.1%.58 This enhancement was attributed to converting carbonyl and carboxyl groups into electron-donating hydroxyl groups. Similarly, Quignard et al. reported the photo-oxidation of pDA-coated oil droplets under UVA illumination, indicating that the partial dissolution of pDA degradation products into the hydrophilic oil matrix contributed to fluorescence development, likely by excluding water, which acts as a fluorescence quencher.59
Table 2 indicates that the size and fluorescence characteristics of synthesized F-pDA NPs vary depending on the synthetic conditions. Given the focus on applying pDA in biosensing, sections 4.2 and 4.3 were organized based on the working principles of biomarker detection rather than solely comparing synthetic strategies and material properties. Categorizing the content based on detection principles clarifies how pDA analogues are specifically designed for various biosensing applications.
| Category | Year | F-pDA synthesis | Size of F-pDAs | Ex/Em for sensing | Sensing mechanism | Target biomarker | Sensitivity | Ref. |
|---|---|---|---|---|---|---|---|---|
| As-prepared F-pDA NPs | 2016 | Bottom-up (oxidant: iron oxide nanozyme/H2O2) | A few to 15 nm | 360/500 nm | Turn-on | Zn2+ | LOD: 60 nM (linear: up to 5 μM) | 53 |
| 2018 | Top-down (NaBH4-induced reduction) | 34.83 nm | 335/423 nm | Turn-off | Fe3+ | LOD: 0.15 μM (linear: 0.5–20 μM) | 58 | |
| 2022 | Bottom-up (one-pot, polycytosine & PEI as additives) | 25 ± 6 nm | 400/550 nm | Turn-off | Cu2+ | LOD: 0.03 μM (linear: 0–10 μM) | 56 | |
| 2023 | Bottom-up (one-pot, folic acid as an additive) | 1.9 ± 0.3 nm | 360/around 440–450 nm (not specified) | Turn-off | Hg2+ | LOD: 0.18 μM (linear: 0–18 μM) | 60 | |
| 2015 | Top-down (degradation using hydroxyl radicals) | 330/440 nm | Turn-off | Fe3+ | LOD: 0.3 μM (linear: 1–50 μM) | 61 | ||
| 2018 | Bottom-up (GSH as an additive) | 2.7–3.28 nm | 360/450 nm | Turn-off | Fe3+ | LOD: 30 nM (linear: 0.1–3 μM) | 62 | |
| Turn-on (by removing Fe3+ from F-pDA) | Pyrophosphate | LOD: 0.6 μM (linear: 2–12 μM) | ||||||
| Pre-quenched F-pDA NPs by protamine | 2021 | Top-down (degradation using hydroxyl radicals) | Diameter: 4.8 nm | 340/435 nm | Turn-on (by degrading protamine) | Trypsin | LOD: 6.7 ng mL−1 (linear: 0.01–0.1 μg mL−1) | 63 |
| In situ F-pDA generation | 2021 | Bottom-up (Fe2+/H2O2 as oxidants) | Monodispersed, about 8 nm | 380/522 nm | Turn-on | Fe2+ | LOD: 0.09 μM (linear: 0.5–10 μM) | 54 |
| DA | LOD: 0.07 μM (linear: 0–300 μM) | |||||||
| H2O2 | LOD: 0.49 μM (linear: 1 to 60 μM) | |||||||
| Glucose (GOx-incorporated) | LOD: 1.61 μM (linear: 0–100 μM) | |||||||
| 2018 | Bottom-up (Ficin/H2O2 as oxidants) | Few tens to hundreds of nanometers | 400/476 nm | Turn-on | DA | LOD: 5.5 nM (linear: 10 nM–5.0 μM) | 64 | |
| 2023 | Bottom-up (MnO2 as an oxidant) | Spherical, less than 50 nm | 420/503 nm | Turn-on | Quercetin | LOD: 0.1 μM (linear: 0.5–250 μM) | 65 | |
| 2016 | Bottom-up (MnO2 as an oxidant) | Irregular, from several to tens of nanometers | 400/485 nm | Turn-off (by consuming the oxidant) | Glutathione | LOD: 1.5 μM (linear: 0–350 μM) | 66 | |
| 2022 | Bottom-up (KMnO4 as an oxidant) | Spherical, 4 nm | 400/480 nm | Butyrylcholinesterase | LOD: 0.047 U L−1 (linear: 0.5–200 U L−1) | 67 | ||
| 2017 | Bottom-up (CoOOH as an oxidant) | Around 10–50 nm | 400/495 nm | Ascorbic acid | LOD: 4.8 μM (linear: 0–500 μM) | 68 | ||
| 2021 | About 5 nm with a wide distribution | 400/490 nm | Alkaline phosphatase | LOD: 0.1 U L−1 (linear: 0.5–300 U L−1) | 69 | |||
| 2020 | Irregular, about 10–50 nm | 390/490 nm | Glutathione | LOD: 1.93 μM (linear: 10–100 μM) | 70 |
4.2. Bioassays utilizing as-prepared fluorescent pDA NPs
The complexation of F-pDA NPs with metal ions can quench or enhance their fluorescence, governed by mechanisms similar to those described earlier for the fluorescence quenching of dyes by non-fluorescent pDA NPs in section 3.1. For instance, Tian et al. reported F-pDA NPs with an excitation-independent emission wavelength of approximately 466 nm (spanning an excitation range of 368 to 428 nm), which were effectively quenched upon complexation with Hg2+ ions.71 This property was further adapted for detecting alkaline phosphatase, an enzyme that hydrolyzes L-ascorbic acid-2-phosphate (AA2P) to L-ascorbic acid (AA), reducing Hg2+ to Hg0. Similarly, F-pDA analogues synthesized with folic acid exhibited selective fluorescence quenching upon complexation with Hg2+, demonstrating minimal interference from other metal ions, anions, and amino acids.60 In addition, F-pDA NPs synthesized by degrading non-fluorescent pDA NPs via refluxing with H2O2 at alkaline pH61 and those with fluorescence enhanced through reduction with sodium borohydride58 both exhibited selective fluorescence quenching by Fe3+. The Fe3+-mediated quenching mechanism can be exploited for pyrophosphate (PPi) detection, as PPi binds with Fe3+, removing it from the F-pDA NPs and restoring their fluorescence.62 In contrast, Liu et al. synthesized F-pDA analogues with PEI and polycytosine as additives, which were selectively quenched upon Cu2+ chemisorption, making these materials suitable for detecting and removing Cu2+ ions.56 Interestingly, unlike most studies reporting fluorescence quenching of F-pDA NPs upon metal complexation, Liu et al. demonstrated a unique system where F-pDA NPs synthesized via peroxidase-mimicking Fe3O4 NPs exhibited enhanced fluorescence upon binding with Zn2+ under 360 nm excitation.53 This enabled a “light-up” sensing system for Zn2+ with a detection limit of 60 nM.Beyond metal ion detection, F-pDA NPs provide opportunities for designing rapid and convenient bioassays for various biomarkers. For example, Li et al. developed a novel trypsin assay using aggregated F-pDA NPs.63 They demonstrated that the fluorescence of F-pDA NPs was quenched upon aggregation induced by protamine and recovered when trypsin digested the protamine, releasing free F-pDA NPs. This assay exhibited a linear detection range of 0.01–0.1 mg mL−1, with a 6.7 ng mL−1 detection limit.
4.3. Synthetic parameters as biomarkers
Similar to the in situ generation of pDA as a quencher for co-incorporated fluorophores discussed in section 3.3, the in situ generation of F-pDA presents valuable opportunities for designing bioassays, particularly for identifying factors associated with F-pDA synthesis. Li et al. achieved green-emitting F-pDA dots with a long-wavelength emission at 522 nm and a large Stokes shift of 142 nm using a synthesis involving the Fenton reaction.54 Various metal ions, including Fe2+, Fe3+, Cu2+, Ag+, Zn2+, Mn2+, Ni2+, Al3+, Co2+, and Mg2+, were tested; however, fluorescent dots were obtained only with Fe2+ and Fe3+ in the presence of H2O2. The fluorescence intensity of the synthesized NPs depended significantly on the concentrations of synthetic components, including Fe2+, H2O2, and DA, enabling the development of quantitative assays for these components. In addition, this system was extended to glucose sensing by incorporating glucose oxidase (GOx), which produces H2O2 in the presence of glucose. Similarly, Pang et al. reported that peroxidase-like ficin mediated the synthesis of F-pDA NPs in the presence of H2O2, facilitating DA quantification as the DA concentration directly influenced the fluorescence intensity of the resulting NPs.64 Momeni et al. observed that the fluorescence intensity of F-pDA increased significantly with higher concentrations of co-polymerized quercetin, making this system effective for quercetin quantification.65 The detection limit of the fluorescent assay was 0.3 μM, with a dynamic range of 0.5 to 250 μM. The system also effectively assessed quercetin content in urine and food samples.Regulatory inorganic oxidants have also been employed in F-pDA generation for ‘turn-off’ biosensing applications. For example, Kong et al. utilized MnO2-mediated F-pDA synthesis to detect reduced glutathione (GSH) based on a mechanism where GSH reduces MnO2 to Mn2+, preventing F-pDA synthesis.66 Although other reducing agents such as cysteine, homocysteine, and vitamin C can also reduce MnO2, their typically lower concentrations enabled the system to validate specificity for GSH in human whole blood, with recovery rates ranging from 97.2% to 103.2% and relative standard deviations between 3.2% and 5.7%. In another study, Li et al. used KMnO4 as an oxidant to synthesize F-pDA NPs, which were utilized to detect butyrylcholinesterase activity, as the enzymatic product, thiocholine, reduces KMnO4 to Mn2+.67 Zhao et al. were the first to report that CoOOH nanosheets can function as oxidants for synthesizing F-pDA NPs.68 Since CoOOH can be converted to Co2+ by ascorbic acid and GSH, CoOOH-based F-pDA synthesis serves as an effective indicator for detecting these molecules and enzymes associated with them.68–70
5. Future directions and challenges
pDA and pDA-like materials have firmly established themselves as versatile optical probes and transducers for biosensing applications due to their unique physicochemical properties, strong adhesive nature, and adaptable optical characteristics. These materials have been successfully employed in colorimetric and fluorescence-based detection systems, enabling highly sensitive and specific assays for healthcare diagnostics, environmental monitoring, and various other fields. However, several challenges remain, as outlined in sections 5.1 to 5.3. Addressing these issues requires innovative synthetic strategies, rigorous validation of materials properties, and the integration of pDA into diverse sensing platforms. Through continued interdisciplinary collaboration and technological advancements, pDA-based materials have the potential to play a transformative role in the future of biosensing.5.1. Commercialization challenges 1: synthesis of pDA-based materials
The commercialization of pDA-based biosensing systems necessitates overcoming key hurdles related to reproducibility, large-scale production, standardization, and market integration of pDA-based materials.72 Most current studies remain confined to laboratory settings, which prolongs the transition to commercial applications. Progress in synthetic strategies is essential to translating pDA technologies into viable market solutions.A deeper understanding of pDA's chemical structure and polymerization pathways is critical for refining synthesis processes and mitigating the inherent heterogeneity that currently undermines batch-to-batch consistency. This refinement will facilitate precise tuning of optical properties and the development of tools to improve structural uniformity. For example, techniques such as additive-assisted growth control or substituting pDA with well-defined catechol-grafted or catechol-conjugated polymeric materials provide viable pathways to enhanced reproducibility and reliability. In addition, creating stimuli-responsive pDA materials capable of reacting to environmental triggers such as pH or temperature shifts can enable innovative “smart” biosensing systems that dynamically adapt to changing conditions.
Optimizing large-scale synthesis methods and cost-effective production strategies is another crucial step toward achieving standardization in manufacturing processes. Advances in automation and AI-driven quality control could further enhance production scalability and reliability.
5.2. Commercialization challenges 2: long-term sensing performance validation
For pDA-adapted biosensing platforms, validating long-term performance in terms of stability and reliability is critical. Many currently developed sensors suffer from time-dependent performance degradation, primarily due to limited durability in real-world conditions, such as exposure to atmospheric elements or biofluids.72 To mitigate this issue, improving the robustness of sensing components and packaging systems is essential to ensure consistent performance over time. Additionally, minimizing interfacial mismatches between sensing components with different chemical compositions is crucial for further enhancing long-term stability and reliability.5.3. Expansion of application scope 1: in vivo adaptation
Future developments in pDA-based sensing systems are anticipated to explore new frontiers in applications and mechanisms. A particularly promising direction involves using pDA's optical properties for theragnostic applications, where it can perform dual roles in vivo imaging and photothermal therapy. However, achieving this potential requires further studies to validate pDA's biocompatibility and in vivo clearance to alleviate concerns about toxicity. Recent advancements in proteomic technologies enable the study of complex molecular interactions at the surface of pDA nanoparticles in biological fluids.73,74 These studies will provide deeper insights into the toxicity of pDA-based materials and create opportunities to fine-tune biological responses through chemical modifications, ultimately improving their suitability for clinical applications.5.4. Expansion of application scope 2: integration with other signal transduction systems
Investigating pDA's capabilities in alternative signal transduction mechanisms can further expand its application scope. Recent research on the optoelectronic properties of natural melanin and pDA highlights their potential for next-generation materials and devices capable of biological signaling in a controlled, spatiotemporal manner. In addition, pDA's inherent redox activity presents opportunities for its integration into electrochemical transduction platforms, which require highly sensitive sensing capabilities. Interdisciplinary collaborations will be essential in driving the widespread adoption of pDA-based biosensing technologies and ensuring their successful market integration.Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grant (2018R1A5A1025511) and the DGIST R&D program (24-SENS2-04) funded by the Korean Government (MSIT).References
- H. L. Shao, H. Im, C. M. Castro, X. Breakefield, R. Weissleder and H. H. Lee, Chem. Rev., 2018, 118, 1917–1950 CrossRef CAS PubMed.
- W. Zhou, X. Gao, D. B. Liu and X. Y. Chen, Chem. Rev., 2015, 115, 10575–10636 CrossRef CAS PubMed.
- V. Chaudhary, S. Sonu, B. A. Taha, P. Raizada, S. Rustagi, S. Chahal, P. Singh, A. Khosla and V. Nguyen, J. Mater. Sci. Technol., 2025, 218, 236–262 CrossRef CAS.
- V. Chaudhary, B. A. Taha, B. R. Lucky, S. Rustagi, A. Khosla, P. Papakonstantinou and N. Bhalla, ACS Sens., 2024, 9, 4469–4494 CrossRef CAS PubMed.
- A. Sena-Torralba, R. Alvarez-Diduk, C. Parolo, A. Piper and A. Merkoci, Chem. Rev., 2022, 122, 14881–14910 CrossRef CAS PubMed.
- D. K. Mal, H. Pal and G. Chakraborty, TrAC, Trends Anal. Chem., 2024, 171, 117493 CrossRef CAS.
- K. Yan, Z. B. Hu, P. Yu, Z. Y. He, Y. Chen, J. J. Chen, H. T. Sun, S. F. Wang and F. Zhang, Nat. Commun., 2024, 15, 2593 CrossRef CAS PubMed.
- Z. Thiel, J. Nguyen and P. Rivera-Fuentes, Angew. Chem., Int. Ed., 2020, 59, 7669–7677 CrossRef CAS PubMed.
- X. Q. Jiang, L. F. Wang, S. L. Carroll, J. W. Chen, M. C. Wang and J. Wang, Antioxid. Redox Signal., 2018, 29, 518–540 CrossRef CAS PubMed.
- M. C. Leake and S. D. Quinn, Chem. Phys. Rev., 2023, 4, 011302 CrossRef CAS.
- S. Jampasa, W. Khamcharoen, S. Wirojsaengthong, A. Suea-Ngam, S. Traipop, T. Ozer, F. Unob, P. Puthongkham and O. Chailapakul, TrAC, Trends Anal. Chem., 2024, 180, 117914 CrossRef CAS.
- A. T. Krasley, E. G. E. Li, J. M. Galeana, C. Bulumulla, A. G. Beyene and G. S. Demirer, Chem. Rev., 2024, 124, 3085–3185 CrossRef CAS PubMed.
- A. M. Wagner, J. M. Knipe, G. Orive and N. A. Peppas, Acta Biomater., 2019, 94, 44–63 CrossRef CAS PubMed.
- S. Dordevic, M. M. Gonzalez, I. Conejos-Sánchez, B. Carreira, S. Pozzi, R. C. Acúrcio, R. Satchi-Fainaro, H. F. Florindo and M. J. Vicent, Drug Delivery Transl. Res., 2022, 12, 500–525 CrossRef PubMed.
- R. M. Crist, S. S. K. Dasa, C. H. Liu, J. D. Clogston, M. A. Dobrovolskaia and S. T. Stern, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2021, 13, e1665 CAS.
- L. J. Johnston, N. Gonzalez-Rojano, K. J. Wilkinson and B. S. Xing, NanoImpact, 2020, 18, 100219 CrossRef.
- M. L. Alfieri, T. Weil, D. Y. W. Ng and V. Ball, Adv. Colloid Interface Sci., 2022, 305, 102689 CrossRef CAS PubMed.
- S. P. Gao, D. Zhang, M. Pedrero, Z. M. Guo, J. M. Pingarrón, S. Campuzano and X. B. Zou, Coord. Chem. Rev., 2024, 501, 215564 CrossRef CAS.
- W. J. Bai, P. Yang, H. J. Liu, Y. Zou, X. H. Wang, Y. Yang, Z. P. Gu and Y. W. Li, Macromolecules, 2022, 55, 3493–3501 CrossRef CAS.
- M. Xiao, M. D. Shawkey and A. Dhinojwala, Adv. Opt. Mater., 2020, 8, 2000932 CrossRef CAS.
- M. Xiao and W. Li, Melanins: Functions, Biotechnological Production, and Applications, 2023, pp. 127–146 Search PubMed.
- K. Y. Ju, M. C. Fischer and W. S. Warren, ACS Nano, 2018, 12, 12050–12061 CrossRef CAS PubMed.
- C. T. Chen, C. Chuang, J. S. Cao, V. Ball, D. Ruch and M. J. Buehler, Nat. Commun., 2014, 5, 3859 CrossRef CAS PubMed.
- A. Ilina, K. E. Thorn, P. A. Hume, I. Wagner, R. R. Tamming, J. J. Sutton, K. C. Gordon, S. K. Andreassend, K. Chen and J. M. Hodgkiss, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2212343119 CrossRef CAS PubMed.
- C. Grieco, F. R. Kohl, A. T. Hanes and B. Kohler, Nat. Commun., 2020, 11, 4569 CrossRef CAS PubMed.
- J. B. Nofsinger, T. Ye and J. D. Simon, J. Phys. Chem. B, 2001, 105, 2864–2866 CrossRef CAS.
- M. d'Ischia, A. Napolitano, V. Ball, C. T. Chen and M. J. Buehler, Acc. Chem. Res., 2014, 47, 3541–3550 CrossRef PubMed.
- S. Hong, Y. Wang, S. Y. Park and H. Lee, Sci. Adv., 2018, 4, eaat7457 CrossRef PubMed.
- H. Tong, C. Cao, M. You, S. Han, Z. Liu, Y. Xiao, W. He, C. Liu, P. Peng, Z. Xue, Y. Gong, C. Yao and F. Xu, Biosens. Bioelectron., 2022, 213, 114449 CrossRef CAS PubMed.
- G. Zhang, S. Deng, B. Fang, G. Zhang, X. Lai, L. Su, W. He and W. Lai, Anal. Bioanal. Chem., 2022, 414, 7315–7323 CrossRef CAS PubMed.
- S. Xu, G. Zhang, B. Fang, Q. Xiong, H. Duan and W. Lai, ACS Appl. Mater. Interfaces, 2019, 11, 31283–31290 CrossRef CAS PubMed.
- L. Xu, R. Chopdat, D. Li and K. T. Al-Jamal, Biosens. Bioelectron., 2020, 169, 112576 CrossRef CAS PubMed.
- J. Li, M. A. Baird, M. A. Davis, W. Tai, L. S. Zweifel, K. M. Adams Waldorf, M. Gale Jr., L. Rajagopal, R. H. Pierce and X. Gao, Nat. Biomed. Eng., 2017, 0082, DOI:10.1038/s41551-017-0082.
- S. Kim and S. Hong, Adv. Healthcare Mater., 2020, 9, e2000540 CrossRef PubMed.
- H. Li, J. Gan, Q. Yang, L. Fu and Y. Wang, Talanta, 2021, 234, 122706 CrossRef CAS PubMed.
- W. Qiang, W. Li, X. Li, X. Chen and D. Xu, Chem. Sci., 2014, 5, 3018–3024 RSC.
- I. L. Medintz, M. H. Stewart, S. A. Trammell, K. Susumu, J. B. Delehanty, B. C. Mei, J. S. Melinger, J. B. Blanco-Canosa, P. E. Dawson and H. Mattoussi, Nat. Mater., 2010, 9, 676–684 CrossRef CAS PubMed.
- D. Wang, C. Chen, X. Ke, N. Kang, Y. Shen, Y. Liu, X. Zhou, H. Wang, C. Chen and L. Ren, ACS Appl. Mater. Interfaces, 2015, 7, 3030–3040 CrossRef CAS PubMed.
- Y. Chen, Z. Wang, X. Hao, F. Li, Y. Zheng, J. Zhang, X. Lin and S. Weng, Sens. Actuators, B, 2019, 297, 126784 CrossRef CAS.
- Z. Wang, K. Xing, N. Ding, S. Wang, G. Zhang and W. Lai, J. Hazard. Mater., 2022, 423, 127204 CrossRef CAS PubMed.
- X. Liu, W. Yu, X. Mu, W. Zhang, X. Wang and Q. Gu, Spectrochim. Acta, Part A, 2023, 287, 122112 CrossRef CAS PubMed.
- W. Qiang, X. Wang, W. Li, X. Chen, H. Li and D. Xu, Biosens. Bioelectron., 2015, 71, 143–149 CrossRef CAS PubMed.
- D. Fan, X. Zhu, Q. Zhai, E. Wang and S. Dong, Anal. Chem., 2016, 88, 9158–9165 CrossRef CAS PubMed.
- Z. Wang, J. Zhang, F. Chen and K. Cai, Analyst, 2017, 142, 2796–2804 RSC.
- X. Deng, S. Wu, S. Zang, X. Liu and Y. Ma, Anal. Chem., 2022, 94, 14546–14553 CrossRef CAS PubMed.
- B. Xing, W. Zhu, X. Zheng, Y. Zhu, Q. Wei and D. Wu, Sens. Actuators, B, 2018, 265, 403–411 CrossRef CAS.
- Q. Yang, J. Xiong, L. Duan, S. Chen, Z. Peng, X. Liao, Z. Ning and D. Wang, Food Chem., 2024, 439, 138058 CrossRef CAS PubMed.
- X. Lai, G. Zhang, S. Deng, Z. Huang, J. Peng, G. Zhang, L. Su, W. He, Y. Wu, N. Ding, Z. Zhang and W. Lai, Chem. Eng. J., 2023, 454, 140444 Search PubMed.
- L. S. Lin, Z. X. Cong, J. B. Cao, K. M. Ke, Q. L. Peng, J. Gao, H. H. Yang, G. Liu and X. Chen, ACS Nano, 2014, 8, 3876–3883 Search PubMed.
- W. Mao, C. Hu, H. Zheng, J. Xie, X. Shi, Y. Du and F. Wang, Mol. Ther. – Nucleic Acids, 2020, 22, 27–37 CrossRef CAS PubMed.
- J. H. Lee, J. S. Ryu, Y. K. Kang, H. Lee and H. J. Chung, Adv. Funct. Mater., 2021, 31, 2007993 CrossRef CAS.
- X. Zhang, S. Wang, L. Xu, L. Feng, Y. Ji, L. Tao, S. Li and Y. Wei, Nanoscale, 2012, 4, 5581–5584 Search PubMed.
- B. Liu, X. Han and J. Liu, Nanoscale, 2016, 8, 13620–13626 Search PubMed.
- Q. Li, T. Zhang, J. Chen, W. Ji and Y. Wei, J. Mater. Chem. B, 2021, 9, 5503–5513 RSC.
- Y. Liu, M. Yang, J. Li, W. Zhang and X. Jiang, Anal. Chem., 2019, 91, 6754–6760 CrossRef CAS PubMed.
- X. Liu, M. Jin, L. Zou, W. Mei, X. Yang, Q. Wang, H. Wang, Q. Zou and K. Wang, ACS Appl. Nano Mater., 2022, 5, 2038–2047 CrossRef CAS.
- K. K. Lee, J. Y. Shin, S. C. Lee and C.-S. Lee, Chem. Eng. J., 2024, 495, 152739 CrossRef CAS.
- H. Yin, K. Zhang, L. Wang, K. Zhou, J. Zeng, D. Gao, Z. Xia and Q. Fu, Nanoscale, 2018, 10, 18064–18073 RSC.
- S. Quignard, M. d'Ischia, Y. Chen and J. Fattaccioli, ChemPlusChem, 2014, 79, 1254–1257 CrossRef CAS.
- L. Chen, C. Chen, Y. Yan, L. Yang, R. Liu, J. Zhang, X. Zhang and C. Xie, Polymers, 2023, 15, 1892 CrossRef CAS PubMed.
- J. H. Lin, C. J. Yu, Y. C. Yang and W. L. Tseng, Phys. Chem. Chem. Phys., 2015, 17, 15124–15130 RSC.
- L. Tang, S. Mo, S. G. Liu, L. L. Liao, N. B. Li and H. Q. Luo, Sens. Actuators, B, 2018, 255, 754–762 CrossRef CAS.
- F. Li, Y. Chen, R. Lin, C. Miao, J. Ye, Q. Cai, Z. Huang, Y. Zheng, X. Lin, Z. Zheng and S. Weng, Anal. Chim. Acta, 2021, 1148, 338201 CrossRef CAS PubMed.
- Y. Pang, Y. Shi, Y. Pan, Y. Yang, Y. Long and H. Zheng, Sens. Actuators, B, 2018, 263, 177–182 CrossRef CAS.
- S. Momeni, A. M. Ramezani, S. Talebi and I. Nabipour, J. Food Compos. Anal., 2023, 120, 105296 CrossRef CAS.
- X. J. Kong, S. Wu, T. T. Chen, R. Q. Yu and X. Chu, Nanoscale, 2016, 8, 15604–15610 RSC.
- Q. Li, Y. M. Guo and G. L. Li, Spectrochim. Acta, Part A, 2022, 274, 121097 CrossRef CAS PubMed.
- Y.-Y. Zhao, L. Li, R.-Q. Yu, T.-T. Chen and X. Chu, Anal. Methods, 2017, 9, 5518–5524 RSC.
- Q. Li, Y.-M. Guo, Z. Wu and G.-L. Li, Dyes Pigm., 2021, 194, 109616 CrossRef CAS.
- Q. Yu, Y. Zhao, W. Deng, T. Chen and X. Chu, Anal. Sci., 2020, 36, 347–352 CrossRef CAS PubMed.
- J. X. Tian, Y. Z. Fang, R. Yu, Z. Y. Zhang, Y. T. Zhuo, J. Y. He, S. Wu, Q. Xiao and X. J. Kong, Anal. Methods, 2021, 13, 322–326 RSC.
- R. K. Talreja, H. Sable, V. Chaudhary, S. Kadian, M. Singh, M. Kumar, J. Kishore, V. Chaudhary and A. Khosla, ECS Sens. Plus, 2024, 3, 041602 CrossRef CAS.
- G. Emilsson, K. Liu, F. Hook, L. Svensson, L. Rosengren, L. Lindfors and K. Sigfridsson, ACS Nano, 2023, 17, 24725–24742 CrossRef CAS PubMed.
- M. Battaglini, A. Carmignani, D. Z. Ciobanu, A. Marino, F. Catalano, A. Armirotti and G. Ciofani, ACS Appl. Mater. Interfaces, 2025, 17, 10485–10498 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |
