Development of a xanthene-based NIR fluorescent probe for accurate and sensitive detection of γ-glutamyl transpeptidase in cancer diagnosis and treatment†
Chia-Kai
Lai
a,
Kuppan
Magesh
b,
Sivan
Velmathi
 b and
Shu-Pao
Wu
b and
Shu-Pao
Wu
 *a
*a
aDepartment of Applied Chemistry, National Yang Ming Chiao Tung University, Hsinchu, Taiwan, Republic of China. E-mail: spwu@nycu.edu.tw
bDepartment of Chemistry, National Institute of Technology, Tiruchirappalli, India
First published on 8th November 2024
Abstract
γ-Glutamyl transpeptidase (GGT) regulates glutathione (GSH), essential for cell functions and linked to cancer. High GGT levels in tumors make it a valuable cancer biomarker. Current GGT detection methods often lack sensitivity and specificity. To address this, we developed XM-Glu, a new near-infrared (NIR) fluorescent probe. XM-Glu features a xanthene-based structure with a hydroxy xanthene fluorophore and a malononitrile group for NIR emission and reduced background noise. It has a self-immolating linker masked with glutamate acid, which activates fluorescence when GGT is present. XM-Glu can detect GGT in the range of 1.0 to 20 mU with a low detection limit of 0.067 mU mL−1. It showed high specificity and minimal interference in cellular assays. In mice, XM-Glu effectively detected GGT in tumor, liver, and kidney tissues. Its NIR properties provide real-time insights into GGT activity, improving cancer diagnosis and monitoring. This new technology enhances cancer research and helps better understand GGT's role in cancer progression.
Introduction
γ-Glutamyl transpeptidase (GGT) plays a critical role in cellular physiology by regulating intracellular glutathione (GSH) levels, which are essential for maintaining redox balance and supporting various physiological processes.1,2 Abnormalities in GGT activity have been implicated in several pathological conditions, including cancer, where elevated GGT levels are frequently observed. This enzyme's involvement in cancer underscore its significance as a biomarker for disease diagnosis and therapeutic targeting.3–6Traditional methods for detecting GGT often struggle with sensitivity and specificity, limiting their clinical use. To improve this, advanced fluorescent probes have been developed for more precise and localized detection of GGT activity, which aids our understanding of its functions in normal cells and diseases, especially cancer.7–9 Recent advancements have focused on creating GGT-activated fluorescent probes, usually by linking glutamic acid to fluorophores using a self-immolative linker.10–13 Many existing probes use short wavelengths, which can lead to background noise and photodamage. To address this, near-infrared (NIR) fluorescent probes have been created. NIR probes penetrate tissues better and experience less background interference, making them especially useful for imaging in living organisms and deep tissues.14–38
This study introduces XM-Glu, a novel NIR fluorescent probe specifically designed to detect GGT activity with high sensitivity and selectivity. XM-Glu utilizes a xanthene-based structure incorporating a hydroxy xanthene fluorophore (XM-OH) coupled with a malononitrile group to enable NIR emission and minimize background signal. The probe features a self-immolating linker masked with glutamate acid, ensuring fluorescence activation upon enzymatic cleavage by GGT. In vitro studies have demonstrated XM-Glu's capability to detect GGT over a wide range of concentrations, with an impressive low detection limit of 0.067 mU mL−1. Cellular assays further confirmed XM-Glu's specificity and minimal cross-reactivity with other cellular components. Validation in murine models, including tumor, liver, and kidney tissues, underscored XM-Glu's effectiveness in real-time detection of GGT activity in complex biological environments.
Results and discussion
Design of the probe XM-Glu
Fig. 1 illustrates the molecular structure of XM-Glu, a near-infrared (NIR) fluorescent probe engineered for sensitive detection of γ-glutamyl transpeptidase (GGT). The probe is constructed around a xanthene-malononitrile framework, which serves as the NIR fluorophore, coupled with a glutamyl moiety that acts as the reactive site for GGT. These components are interconnected by a para-amino benzyl alcohol (PABA) linkage.23 The choice of the XM-OH fluorophore was motivated by its non-toxic profile and high quantum yield in fluorescence. The malononitrile group within XM-Glu possesses potent electron-withdrawing properties, essential for tuning the emission wavelength into the NIR-I window. Initially, XM-Glu remains non-fluorescent due to the presence of the PABA linker, which restricts electron transfer from the xanthene (donor) to the malononitrile (acceptor). Upon encountering GGT, the glutamyl group undergoes enzymatic cleavage, thereby removing the PABA linker and liberating the free fluorophore XM-OH. This enzymatic reaction restores near-infrared fluorescence, enabling precise detection of GGT activity in biological systems.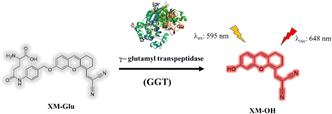 | ||
| Fig. 1 The molecular structure of XM-Glu and the predicted chemical process in response to GGT (PDB ID code 2DBU). | ||
General optical responses of the probe XM-Glu
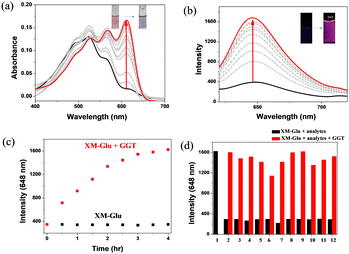
Initially, we investigated the optical response of XM-Glu upon interaction with γ-glutamyl transpeptidase (GGT) in a DMSO-H2O solution (v/v = 4/6, 6 mM PBS, pH 7.4). As shown in Fig. 2a, the color of XM-Glu shifted from red to blue, accompanied by a peak absorbance shift from 520 to 610 nm upon GGT interaction. Concurrently, fluorescence emission at 648 nm demonstrated a steady increase with increasing GGT concentration (Fig. 2b). The absorption at 610 nm and fluorescence intensity reached a plateau at 100 mU mL−1 GGT (see Fig. S1a and b in the ESI†). A linear relationship was observed between 0 and 20 mU mL−1 GGT (see Fig. S2 in the ESI†), with a low detection limit of 0.067 mU mL−1.The time-dependent fluorescence intensity of XM-Glu incubated with GGT revealed a gradual increase, reaching maximum fluorescence after 4 hours (Fig. 2c). In contrast, the fluorescence intensity remained stable in the absence of GGT, establishing a 4 hour incubation time for subsequent experiments. Enzymatic kinetic parameters between GGT and XM-Glu were also investigated, yielding KM, Vmax, and kcat values of 21.46 μM, 2.3 × 10−3 μM s−1, and 0.0094 s−1, respectively (Fig. S3 in ESI†), demonstrating XM-Glu's efficient enzymatic cleavage compared to previous probes (Table S1 in ESI†).
To evaluate XM-Glu's selectivity for GGT, we incubated the probe (20 μM) with various biologically relevant analytes in a DMSO-H2O solution (v/v = 4/6, 6 mM PBS, pH 7.4), including reactive oxygen species (H2O2, HOCl, 1O2, O2−), biothiols (Cys, Hcy, GSH), and enzymes (alkaline phosphatase, acetylcholinesterase, β-galactosidase, tyrosinase). Competitive studies further demonstrated minimal interference from other analytes, underscoring the probe's specificity (Fig. 2d).
The effect of pH on fluorescence was further investigated by monitoring the fluorescence response across a range of pH values (4.0 to 10.0). The results confirmed stable fluorescence within this range, with optimal signal observed at pH 8.0 (see Fig. S4 in the ESI†). These findings affirm XM-Glu's suitability for GGT detection under physiological conditions.
To validate the reaction mechanism, HPLC and ESI-MS analyses were performed on XM-Glu after treatment with GGT. The appearance of peaks corresponding to XM-Glu and XM-OH at 8 and 9 minutes, respectively, in HPLC (Fig. S5 in ESI†), and a mass spectrum (m/z 275.3) consistent with XM-OH in ESI-MS (Fig. S6 in ESI†) support the cleavage of XM-Glu's γ-glutamyl group by GGT, followed by 1,6-elimination to produce the fluorophore XM-OH (Scheme S2 in ESI†). These findings collectively underscore XM-Glu as a robust and selective probe for real-time detection of GGT activity in biological systems, offering insights into enzymatic kinetics and demonstrating potential for biomedical applications in cancer research and diagnosis.
Intracellular imaging experiment
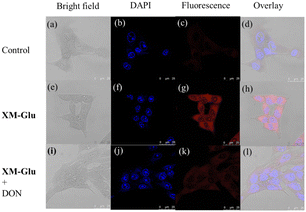
To assess XM-Glu's applicability for intracellular imaging, we initially conducted an MTT assay to evaluate its cytotoxic effects on HepG2 and HEK293 cells. The results indicated minimal cytotoxicity, with over 80% cell viability at 25 μM concentration of XM-Glu (Fig. S7 in ESI†). Subsequently, we selected HepG2 (human liver cancer), HeLa (cervical cancer), and HEK293 (human embryonic kidney) cell lines for further experiments based on their differential GGT activity levels.Treatment of HepG2, HeLa, and HEK293 cells with 10 μM XM-Glu for 2 hours resulted in robust red fluorescence, observed using confocal microscopy. As shown in Fig. 3g and Fig. S8 (ESI†), HepG2 and HeLa cells exhibited intense red fluorescence, indicative of high GGT levels. Conversely, HEK293 cells showed minimal red fluorescence under the same conditions, consistent with their lower GGT activity (Fig. S9 in ESI†). To validate that the observed fluorescence resulted from XM-Glu's interaction with GGT, we pre-treated cells with the GGT inhibitor 6-diazo-5-oxo-L-norleucine (DON, 1 mM) for 3 hours prior to exposure to XM-Glu (10 μM) for 2 hours. Fig. 3k illustrates reduced fluorescence in cells pre-treated with DON, confirming that XM-Glu specifically detects endogenous GGT in live cells. These results underscore XM-Glu's effectiveness in visualizing GGT activity with high specificity and minimal cytotoxicity, positioning it as a valuable tool for studying GGT-related processes in cellular contexts, particularly in cancer research and diagnostics.
In vivo imaging
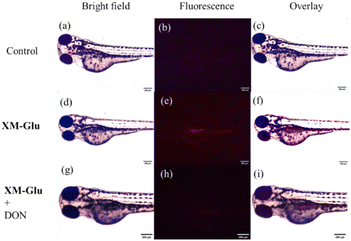
We utilized XM-Glu to investigate endogenous GGT activity in larval zebrafish and evaluated its efficacy for in vivo tracing of GGT. Newly hatched zebrafish were exposed to XM-Glu (50 μM) for two hours, and fluorescence microscopy captured red fluorescence in the larvae (Fig. 4f). Remarkably, fluorescence was predominantly localized in the liver of the larval zebrafish, consistent with previous findings indicating heightened GGT activity in zebrafish liver.34 To validate the specificity of XM-Glu for GGT detection in vivo, larval zebrafish were pre-treated with the GGT inhibitor 6-diazo-5-oxo-L-norleucine (DON, 1 mM) for three hours before incubation with XM-Glu. As depicted in Fig. 4i, pre-treatment with DON resulted in negligible red fluorescence, confirming that XM-Glu specifically detects endogenous GGT in larval zebrafish. These findings demonstrate XM-Glu's capability to visualize and trace GGT activity in vivo with high specificity, highlighting its potential as a valuable tool for studying GGT-related processes in complex biological environments.Building upon our success in cell and zebrafish imaging, we extended the application of our probe for detecting GGT in a living mouse model. A 5 week-old BALB/c nude mouse, bearing a HeLa tumor from subcutaneous injection of 2.5 × 107 cells into the left thigh, served as the experimental subject. Following five days of tumor growth, we administered XM-Glu (100 μM, 200 μL) via intratumoral injection. Fluorescence imaging of the mouse was conducted using an in vivo imaging system (IVIS). At the five-minute mark post-injection, a strong fluorescence signal was distinctly observed at the tumor site, as illustrated in Fig. 5b. Over time, the fluorescence signal diffused throughout the tumor and became detectable at the skin after one hour (Fig. 5c). By the two-hour mark post-injection, the fluorescence intensity began to diminish (Fig. S10 in the ESI†), indicating the metabolism of the probe within the mouse (Fig. 5d). These results highlight the effectiveness of XM-Glu in specifically imaging GGT activity in vivo within a living mouse model, showcasing its potential utility for non-invasive monitoring of GGT in tumor microenvironments.
Following the experiment, tissues from five organs (liver, kidney, gallbladder, heart, and lung) and from the HeLa tumor were harvested from the mouse model. Each tissue sample was exposed to XM-Glu (50 μM) for two hours, followed by fluorescence imaging using a fluorescence microscope. As depicted in Fig. S11 in the ESI,† strong red fluorescence signals were prominently observed in the tumor, liver, and kidney tissues. This observation aligns with previous research indicating elevated GGT activity in these organs and tumors. In contrast, minimal fluorescence was detected in the gallbladder, heart, and lung tissues. These findings underscore the effectiveness of XM-Glu in detecting endogenous GGT activity in tumor-bearing mice.
Experimental
Synthesis of XM-Glu
The synthesis of fluorophore XM-OH and compound 5 followed established procedures as previously reported (see the ESI†).23,39,40 In 5 mL of anhydrous DMF, a solution containing XM-OH (23 mg, 0.08 mmol), compound 5 (79 mg), and K2CO3 (23 mg) was prepared. The reaction proceeded for 20 hours at room temperature. Upon completion, the reaction mixture was extracted with dichloromethane. Compound 6 was purified as a red solid using column chromatography with a hexane/EtOAc mixture in a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) ratio (48 mg, 86%). 1H NMR (600 MHz, DMSO-d6) δ 9.96(s, 1H), 8.17 (s, 1H), 7.60 (d, J = 8.4 Hz, 2H), 7.41 (d, J = 8.6 Hz, 1H), 7.40 (d, J = 8.4 Hz, 2H), 7.35 (s, 1H), 7.33 (d, J = 2.4 Hz, 1H), 7.14 (d, J = 7.4 Hz, 1H), 6.94 (dd, J1 = 2.4 Hz, J2 = 8.6 Hz, 1H), 5.11 (s, 2H), 3.84 (m, 1H), 2.74 (d, J = 5.9 Hz, 2H), 2.61 (d, J = 5.9 Hz, 2H), 2.40 (m, 2H), 1.99 (m, 2H), 1.75 (m, 2H), 1.39 (s, 18H). 13C NMR (150 MHz, DMSO-d6) δ 172.0, 170.8, 161.5, 159.6, 155.95, 154.1, 150.3, 139.6, 132.7, 131.0, 129.1, 128.8, 126.2, 119.4, 118.1, 116.6, 115.2, 114.2, 109.7, 101.9, 80.8, 78.5, 70.3, 67.5, 54.3, 33.1, 28.6, 28.5, 28.1, 26.6, 24.7, 20.4. LRMS m/z (FD) calcd for C38H42N4O7
1 (v/v) ratio (48 mg, 86%). 1H NMR (600 MHz, DMSO-d6) δ 9.96(s, 1H), 8.17 (s, 1H), 7.60 (d, J = 8.4 Hz, 2H), 7.41 (d, J = 8.6 Hz, 1H), 7.40 (d, J = 8.4 Hz, 2H), 7.35 (s, 1H), 7.33 (d, J = 2.4 Hz, 1H), 7.14 (d, J = 7.4 Hz, 1H), 6.94 (dd, J1 = 2.4 Hz, J2 = 8.6 Hz, 1H), 5.11 (s, 2H), 3.84 (m, 1H), 2.74 (d, J = 5.9 Hz, 2H), 2.61 (d, J = 5.9 Hz, 2H), 2.40 (m, 2H), 1.99 (m, 2H), 1.75 (m, 2H), 1.39 (s, 18H). 13C NMR (150 MHz, DMSO-d6) δ 172.0, 170.8, 161.5, 159.6, 155.95, 154.1, 150.3, 139.6, 132.7, 131.0, 129.1, 128.8, 126.2, 119.4, 118.1, 116.6, 115.2, 114.2, 109.7, 101.9, 80.8, 78.5, 70.3, 67.5, 54.3, 33.1, 28.6, 28.5, 28.1, 26.6, 24.7, 20.4. LRMS m/z (FD) calcd for C38H42N4O7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 666.3; found for: 666.3. HRMS m/z (FD) calcd for C38H42N4O7
666.3; found for: 666.3. HRMS m/z (FD) calcd for C38H42N4O7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 666.3053; found for: 666.3048.
666.3053; found for: 666.3048.
Compound 6 (48 mg, 0.06 mmol) was dissolved in 3 mL of CH2Cl2, and 1 mL of trifluoroacetic acid was added dropwise with stirring at 0 °C. The reaction mixture was maintained at 0 °C for 2 hours. The resulting product was isolated through column chromatography using CH2Cl2/MeOH 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v). 1H NMR (400 MHz, DMSO-d6) δ 9.99(s, 1H), 8.17 (s, 1H), 7.61 (d, J = 7.88 Hz, 2H), 7.44 (d, J = 8.64 Hz, 1H), 7.38 (d, J = 7.88 Hz, 2H), 7.34 (m, 2H), 6.94 (d, J = 8.04 Hz, 1H), 5.11 (s, 2H), 3.23 (m, 1H), 2.74 (t, J = 5.56 Hz, 2H), 2.61 (t, J = 5.56 Hz, 2H), 2.41 (m, 2H), 1.89 (m, 2H), 1.75 (m, 2H). 13C NMR (150 MHz, DMSO-d6) δ 174.9, 171.3, 161.6, 159.6, 154.1, 150.3, 139.7, 132.6, 130.9, 128.9, 126.2, 119.4, 118.1, 116.6, 115.2, 114.2, 109.7, 101.9, 80.6, 70.3, 67.7, 54.4, 33.0, 30.3, 28.5, 24.8, 20.5 LRMS m/z (ESI) calcd for C29H27N4O5+: 511.2; found for: 511.4. HRMS m/z (ESI) calcd for C29H27N4O5+: 511.1976; found for: 511.1980.
1 (v/v). 1H NMR (400 MHz, DMSO-d6) δ 9.99(s, 1H), 8.17 (s, 1H), 7.61 (d, J = 7.88 Hz, 2H), 7.44 (d, J = 8.64 Hz, 1H), 7.38 (d, J = 7.88 Hz, 2H), 7.34 (m, 2H), 6.94 (d, J = 8.04 Hz, 1H), 5.11 (s, 2H), 3.23 (m, 1H), 2.74 (t, J = 5.56 Hz, 2H), 2.61 (t, J = 5.56 Hz, 2H), 2.41 (m, 2H), 1.89 (m, 2H), 1.75 (m, 2H). 13C NMR (150 MHz, DMSO-d6) δ 174.9, 171.3, 161.6, 159.6, 154.1, 150.3, 139.7, 132.6, 130.9, 128.9, 126.2, 119.4, 118.1, 116.6, 115.2, 114.2, 109.7, 101.9, 80.6, 70.3, 67.7, 54.4, 33.0, 30.3, 28.5, 24.8, 20.5 LRMS m/z (ESI) calcd for C29H27N4O5+: 511.2; found for: 511.4. HRMS m/z (ESI) calcd for C29H27N4O5+: 511.1976; found for: 511.1980.
Cell culture and imaging
At a temperature of 37 °C and with 5% CO2, HepG2 and HEK293 cells were cultured in 2 mL of DMEM supplemented with 10% FBS and 1% antibiotic solution. The cells were initially seeded in 6-well plates and allowed to grow for 24 hours to facilitate cell imaging. Subsequently, the media in the wells were replaced with 2 mL of DMEM containing the probe solution (10 μM) and placed in a CO2 incubator for 2 hours.For the inhibitory experiment, DON (1 mM) was pre-treated for 3 hours before incubating with XM-Glu (10 μM) for an additional 2 hours. Following the completion of the experiments, the media were aspirated, and the cells were washed twice with PBS. DAPI (4′,6-diamidino-2-phenylindole) was then added to the medium at a concentration of 0.02 mg mL−1. After washing with PBS, the cells were fixed for 15 minutes with 4% formaldehyde. Finally, confocal fluorescence microscopy was utilized to capture cell fluorescence images.
Zebrafish imaging
Newly hatched larval zebrafish were distributed into 6 wells, with 4–5 fish per well. Each zebrafish underwent a thorough wash with tap water before being incubated with XM-Glu (50 μM) for a duration of 2 hours. Following the probe incubation, the larval zebrafish underwent three washes with tap water to remove any excess probe. In the inhibition experiment, 6-diazo-5-oxo-L-norleucine (DON) (1 mM) was pre-treated for 3 hours and then incubated with XM-Glu (50 μM) for an additional 2 hours. Fluorescence images of the larval zebrafish were captured using a fluorescent microscope.In vivo imaging of mice
A 5 week-old BALB/c nude mouse, obtained from BioLASCO in Taiwan, underwent HeLa tumor implantation by subcutaneously injecting 2.5 × 107 HeLa cells into the left thigh. All animal experiments were approved by National Yang Ming Chiao Tung University and conducted according to the manual of the National Laboratory Animal Center in Taiwan. Five days post-implantation, the mouse received an intratumoral injection of 200 μL of XM-Glu (100 μM). Fluorescence images of the mice were captured using the Caliper IVIS Spectrum System with the XGI-8 Anesthesia System. Following the experiment, the mouse was euthanized using CO2. The tumor, along with five organs (liver, kidneys, gallbladder, heart, and lungs), was excised and preserved in 10% formaldehyde. The tissues were then incubated with XM-Glu (50 μM) for 2 hours, followed by three washes with PBS. Fluorescence images of the tissues were acquired using a fluorescence microscope.Conclusions
In conclusion, this study has comprehensively detailed the design and synthesis of the near-infrared (NIR)-based fluorescent probe XM-Glu, highlighting its structural components: the xanthene-malononitrile fluorophore, glutamyl reactive site, and para-amino benzyl alcohol (PABA) linker. The probe functions through enzymatic cleavage of the glutamyl group by GGT, liberating the free fluorophore XM-OH and restoring near-infrared fluorescence. Thorough in vitro investigations have validated XM-Glu's efficacy in selectively detecting GGT, characterized by a distinct spectral response including a color shift and enhanced fluorescence upon GGT interaction. Importantly, XM-Glu has demonstrated exceptional selectivity for GGT, showing minimal cross-reactivity with other analytes and enzymes, alongside impressive sensitivity with a low detection limit of 0.067 mU mL−1.Cellular studies further confirmed the biocompatibility of XM-Glu, revealing its ability to specifically detect endogenous GGT in various cell lines with low cytotoxicity. In vivo imaging experiments conducted in larval zebrafish and a live mouse model underscored XM-Glu's capability to accurately trace GGT activity within complex biological environments, particularly in tumors. Collectively, these findings position XM-Glu as a promising tool for sensitive and selective GGT detection across biological systems, with potential applications in live cell imaging and preclinical research. The robust performance of XM-Glu in diverse experimental settings highlights its utility for exploring GGT-related processes in both cellular and in vivo contexts.
Author contributions
Chia-Kai Lai: conceptualization, methodology, software, validation, formal analysis, investigation, writing – original draft preparation, writing – review & edition. Kuppan Magesh: investigation, writing – review & edition. Sivan Velmathi: writing – original draft preparation, writing – review & edition. Shu-Pao Wu: conceptualization, methodology, validation, formal analysis, writing – original draft preparation, writing – review & edition, supervision, project administration, funding acquisition.Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
We gratefully acknowledge the projects from the National Science and Technology Council (Taiwan) under a grant (NSTC 112-2113-M-A49-035).References
- R. D. Allison, Methods Enzymol., 1985, 113, 419–437 CrossRef CAS.
- S. Jiang, D. Jiang and Y. Tao, Exp Clin Cardiol., 2013, 18, 53–56 CAS.
- C. Grimm, G. Hofstetter, S. Aust, I. Mutz-Dehbalaie, M. Bruch, G. Heinze, J. Rahhal-Schupp, A. Reinthaller, N. Concin and S. Polterauer, Br. J. Cancer, 2013, 109, 610–614 CrossRef CAS PubMed.
- E. J. Cho, S. Jeong, G. E. Chung, J. Yoo, Y. Cho, K. Lee, D. W. Shin, Y. J. Kim, J. Yoon, K. Han and S. J. Yu, Scientific Reports, 2023, 13, 1751 CrossRef CAS PubMed.
- S. Su, L. Liu, C. Sun, Y. Nie, H. Guo, Y. Hu, S. Guo and S. Pang, Front. Oncol., 2021, 11, 648904 CrossRef CAS PubMed.
- M. V. Hemelrijck, W. Jassem, G. Walldius, I. S. Fentiman, N. Hammar, M. Lambe, H. Garmo, I. Jungner and L. Holmberg, Eur. J. Cancer, 2011, 47, 2033–2041 CrossRef PubMed.
- Z. Hu, X. Chen, Y. Yang, S. Wang, Z. Hu and K. Wang, Coord. Chem. Rev., 2024, 501, 215562 CrossRef CAS.
- Y. Zhang, Z. Zhang, M. Wu and R. Zhang, ACS Meas. Sci. Au, 2024, 4, 54–75 CrossRef CAS PubMed.
- Z. Luo, R. An and D. Ye, ChemBioChem, 2019, 20, 474–487 CrossRef CAS PubMed.
- Y. Li, C. Xue, Z. Fang, W. Xu and H. Xie, Anal. Chem., 2020, 92, 15017–15024 CrossRef CAS PubMed.
- Y. Urano, M. Sakabe, N. Kosaka, M. Ogawa, M. Mitsunaga, D. Asanuma, M. Kamiya, M. R. Young, T. Nagano, P. L. Choyke and H. Kobayashi, Sci. Transl. Med., 2011, 3, 110–119 Search PubMed.
- F. Wang, Y. Zhu, L. Zhou, L. Pan, Z. Cui, Q. Fei, S. Luo, D. Pan, Q. Huang, R. Wang, C. Zhao, H. Tian and C. Fan, Angew. Chem., Int. Ed., 2015, 54, 7349–7353 CrossRef CAS.
- R. Obara, M. Karmiya, Y. Tanaka, A. Abe, R. Kojima, T. Kawaguchi, M. Sugawara, A. Takahashi, T. Noda and Y. Urano, Angew. Chem., Int. Ed., 2021, 60, 2125–2129 CrossRef CAS.
- Y. J. Reo, M. Dai, Y. J. Yang and K. H. Ahn, Anal. Chem., 2020, 92, 12678–12685 CrossRef CAS.
- Y. J. Reo, Y. W. Jun, S. Sarkar, M. Dai and K. H. Ahn, Anal. Chem., 2019, 91, 14101–14108 CrossRef CAS PubMed.
- H. Liu, F. Liu, F. Wang, R. Yu and J. Jiang, Analyst, 2018, 143, 5530–5535 RSC.
- S. M. Usama, F. Inagaki, H. Kobayashi and M. J. Schnermann, J. Am. Chem. Soc., 2021, 143, 5674–5679 CrossRef CAS.
- W. Liu, B. Hung, Z.-X. Tong, S. Wang, Y.-J. Li and Y.-Y. Dai, New J. Chem., 2018, 42, 5403–5407 RSC.
- Y. Liu, B. Feng, X. Cao, G. Tang, H. Liu, F. Chen, M. Liu, Q. Chen, K. Yuan, Y. Gu, X. Feng and W. Zeng, Analyst, 2019, 144, 5136–5142 RSC.
- X. Hou, Q. Yu, F. Zeng, C. Yu and S. Wu, Chem. Commun., 2014, 50, 3417–3420 RSC.
- R. Huo, X. Zheng, W. Liu, L. Zhang, J. Wu, F. Li, W. Zhang, C. S. Lee and P. Wang, Chem. Commun., 2020, 56, 10902–10905 RSC.
- R. An, S. Wei, Z. Huang, F. Liu and D. Ye, Anal. Chem., 2019, 91, 13639–13646 CrossRef CAS PubMed.
- Z. Luo, L. Feng, R. An, G. Duan, R. Yan, H. Shi, J. He, Z. Zhou, C. Ji, H. Y. Chen and D. Ye, Chem. – Eur. J., 2017, 23, 14778–14785 CrossRef CAS PubMed.
- B. Bai, C. Yan, Y. Zhang, Z. Guo and W. H. Zhu, Chem. Commun., 2018, 54, 12393–12396 RSC.
- Z. Hai, J. Wu, L. Wang, J. Xu, H. Zhang and G. Liang, Anal. Chem., 2017, 89, 7017–7021 CrossRef CAS.
- L. Li, W. Shi, Z. Wang, Q. Gong and H. Ma, Anal. Chem., 2015, 87, 8353–8359 CrossRef CAS PubMed.
- J. Ou-Yang, Y. Li, W. L. Jiang, S. Y. He, H. W. Liu and C. Y. Li, Anal. Chem., 2019, 91, 1056–1063 CrossRef CAS PubMed.
- S. Park, S.-Y. Lim, S. M. Bae, S.-Y. Kim, S.-J. Myung and H.-J. Kim, ACS Sens., 2016, 1, 579–583 CrossRef CAS.
- H. Tong, Y. Zheng, L. Zhou, X. Li, R. Qian, R. Wang, J. Zhao, K. Lou and W. Wang, Anal. Chem., 2016, 88, 10816–10820 CrossRef CAS.
- R. J. Iwatate, M. Kamiya and Y. Urano, Chem. – Eur. J., 2016, 22, 1696–1703 CrossRef CAS PubMed.
- R. J. Iwatate, M. Kamiya, K. Umezawa, H. Kashima, M. Nakadate, R. Kojima and Y. Urano, Bioconjug. Chem., 2018, 29, 241–244 CrossRef CAS PubMed.
- X. Hou, F. Zeng and S. Wu, Biosens. Bioelectron., 2016, 85, 317–323 CrossRef CAS PubMed.
- F. Liu, Z. Wang, W. Wang, J. G. Luo and L. Kong, Anal. Chem., 2018, 90, 7467–7473 CrossRef CAS PubMed.
- P. Zhang, X. F. Jiang, X. Nie, Y. Huang, F. Zeng, X. Xia and S. Wu, Biomaterials, 2016, 80, 46–56 CrossRef CAS PubMed.
- Z. Luo, Z. Huang, K. Li, Y. Sun, J. Lin, D. Ye and H. Y. Chen, Anal. Chem., 2018, 90, 2875–2883 CrossRef CAS PubMed.
- L. Li, W. Shi, X. Wu, Q. Gong, X. Li and H. Ma, Biosens. Bioelectron., 2016, 81, 395–400 CrossRef CAS PubMed.
- J. Ou-Yang, Y. Li, P. Wu, W. Jiang, H. Liu and C. Li, ACS Sens., 2018, 3, 1354–1361 CrossRef CAS PubMed.
- F. Liu, D. Zhu, Y. Li, M. Kong, Y. Li, J. Luo and L. Kong, Sens. Actuators, B, 2022, 363, 131838 CrossRef CAS.
- Y. Qi, Y. Huang, B. Li, F. Zeng and S. Wu, Anal. Chem., 2018, 90, 1014–1020 CrossRef CAS.
- C. Wu, R. Xie, X. Pang, Y. Li, Z. Zhou and H. Li, Spectrochim. Acta, Part A, 2020, 243, 118764 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4tb01841a |
| This journal is © The Royal Society of Chemistry 2025 |