 Open Access Article
Open Access ArticleAntimicrobial cobaltocenium copolymers: tuning amphiphilicity against NDM-1 bacteria†
Md Waliullah
Hossain
a,
Ian
Brand
a,
Swagatam
Barman
a,
Alimi
Abidoun
a,
JiHyeon
Hwang
a,
Adam
Parris
a,
Xiaoming
Yang
b,
Prakash
Nagarkatti
b,
Mitzi
Nagarkatti
b and
Chuanbing
Tang
 *a
*a
aDepartment of Chemistry and Biochemistry, University of South Carolina, Columbia, SC 29208, USA. E-mail: tang4@mailbox.sc.edu
bDepartment of Pathology, Microbiology and Immunology, University of South Carolina, School of Medicine, Columbia, SC 29209, USA
First published on 11th June 2025
Abstract
The emergence of Gram-negative superbugs coupled with a steep decline in antibiotic pipelines has imposed a serious threat to global public health. Cationic metallopolymers have gained significant attention due to their antimicrobial efficacy. In this work, we developed a range of broad-spectrum antimicrobial cobaltocenium and ammonium containing copolymers with different compositions, which attain the amphiphilic balance without compromising the total charges for enhanced interaction with bacterial membranes. The copolymers showed high antimicrobial efficacy with greater selectivity than the corresponding ammonium-containing methacrylate polymers. The mechanistic investigations of the lead polymer using bacterial strains harboring the New Delhi metallo-β-lactamase (NDM-1) enzyme revealed its membrane-active nature. The copolymer with 69% dimethyl cobaltocenium showed a minimal increase in the minimal inhibitory concentration over 14 passages, whereas polymyxin-B showed a 256-fold increase. These findings provided insights into metallopolymers with optimal amphiphilicity as potent antimicrobial agents to tackle Gram-negative superbugs.
1. Introduction
Antimicrobial resistance (AMR) is no longer a future predicament but one of the greatest challenges to public health worldwide.1,2 Antibiotics are arguably among the greatest discoveries ever for treating bacterial infections.3 However, there is a significant reduction in new antibiotic discovery compared to the growing antibiotic resistance. Between 1962 and 2000, there were no reports of discovering a new class of antibiotics that can treat common and deadly infections associated with Gram-negative bacteria.4 Bacterial resistance to antibiotics arises through various mechanisms.5,6 Gram-positive bacteria possess a thick, net negatively-charged peptidoglycan layer, while Gram-negative bacteria have an asymmetric lipid bilayer, which contains a significant amount of negatively-charged lipopolysaccharides (LPS).7 Many of the latter, listed in the World Health Organization's ‘critical group’ of priority pathogens, present a greater threat due to the emergence of extended-spectrum serine β-lactamase (ESBL) and New Delhi metallo-β-lactamase (NDM) containing strains.8 In particular, treatment options for NDM-1-producing bacteria remain scarce.9,10 To bypass these resistance mechanisms, developing antimicrobial agents that leverage amphiphilicity to disrupt bacterial membranes offers a promising universal strategy.11Antimicrobial peptides (AMPs) are part of the innate immune system of multicellular eukaryotes that can prevent infectious diseases by combating pathogenic bacteria, viruses, and fungi.12 AMPs are composed of varying combinations of cationic, hydrophilic, and hydrophobic groups, possessing overall positive charges, which allow them to selectively target negatively charged bacterial membranes.13–17 Their membrane-disrupting properties make them effective against multidrug-resistant (MDR) bacteria.18,19 However, clinical use is limited due to low bioavailability, poor stability and high manufacturing cost. Synthetic polymers mimicking AMPs offer a scalable, cost-effective alternative, retaining cationic charges and amphiphilicity for antibacterial activity.20–24 Ongoing research aims to optimize these AMP-mimicking polymers for enhanced efficacy and broader clinical application.25–34
Metallopolymers have emerged as a unique class of polymers, which combine an organic polymeric framework with functional inorganic metal centers to synergistically tune the properties of the combined framework.35–38 Although the broad impacts of this emerging class of polymers remain underexplored in the field of antimicrobial therapeutics,39–44 cobaltocenium-containing homopolymers have been reported to form bioconjugates with antibiotics and used as adjuvants to resensitize antibiotics.44–47 However, most of these polymers alone exhibit poor antimicrobial activity. While adjuvants help revive antibiotics, developing new antimicrobial agents is crucial for preventing resistance and strengthening the therapeutic arsenal. Ammonium-containing polymers have been studied extensively as antimicrobial agents,48,49 employing binary and ternary systems to generate libraries of copolymers with various compositions to achieve significant efficacy.50,51
Herein, we report cobaltocenium-containing methacrylate copolymers as novel antimicrobial agents that combine with primary ammonium charges to tune the amphiphilicity without altering the overall charge of the polymers. In addition, cobaltocenium and dimethyl substituted cobaltocenium were compared to elucidate their effect on antimicrobial efficacy. The introduction of the latter moiety is considered as an additional handle to alter the hydrophobicity of the cobaltocenium moieties. The antimicrobial activity of these copolymers was tested against methicillin-resistant S. aureus (MRSA) strains and Gram-negative bacteria, including NDM-1 strains, and their structure–activity relationship was evaluated. Killing kinetics was performed against both planktonic and stationary bacterial cells harboring NDM-1. Mechanistic investigations, including outer membrane permeability, membrane depolarization, LPS inhibition assay, and alteration of bacterial surface potential, revealed membrane-perturbing properties of these cobaltocenium-containing copolymers. Furthermore, the resistance profile of a lead copolymer was assessed for 14 passages.
2. Results
2.1. Synthesis of cobaltocenium-containing copolymers
Cobaltocenium is a unique metallocene cation, which has previously shown electrostatic interaction with anions,52,53 anionic polyelectrolytes,54,55 and negatively charged bacterial membranes.45,46 The balance of cationic charges and amphiphilicity plays a crucial role in designing an effective antimicrobial polymer.2 To tune amphiphilicity, cobaltocenium was quantitatively incorporated into methacrylate copolymers, as shown in Scheme 1. A homopolymer (PAEMA) of 2-aminoethyl methacrylate with a molecular weight of 4.0 kDa was first synthesized via Reversible Addition Fragmentation Transfer (RAFT) polymerization using 2-cyano-2-propyl benzodithioate (CPB) as a chain transfer agent, and 2,2′-azobisisobutyronitrile (AIBN) as an initiator (Scheme S1†), according to a previously reported literature study.56 Cobaltocenium carboxylic acid and dimethyl substituted cobaltocenium carboxylic acid were synthesized according to prior work.57 Attachment of cobaltocenium to PAEMA was achieved using an amide coupling reaction. Successful integration of cobaltocenium was indicated by the presence of aromatic cyclopentadienyl (Cp) protons at 6.1–6.4 ppm and 5.7–6.0 ppm from 1H NMR (Fig. 1). For dimethyl substituted cobaltocenium, the Cp proton peaks appear at 5.5–6.2 ppm and the two methyl peaks appear approximately at 2.0–2.2 ppm and 2.3–2.5 ppm (Fig. 1). By tuning the compositions, we could control the percentages of cobaltocenium (CC) and dimethyl cobaltocenium (dmCC) and obtain six different random copolymers (Table 1). Finally, ion exchange was performed to change the counter ion from PF6− to Cl− for much enhanced solubility in the culture media and to prevent aggregation in aqueous media.58 All polymers were water soluble and were tested up to 20 mg mL−1. Polymer size and zeta potential were determined at different concentrations (0.5, 1, and 2 mg mL−1) using dynamic light scattering (DLS) (see ESI Table S1†).59 At these concentrations, the homopolymer P1 did not show any aggregation. However, CC-substituted copolymers (P2–P4) observed the hydrodynamic diameter DH ranging from 6.9 nm to 22.4 nm, while dmCC-substituted copolymers showed a range of 40.9–70.9 nm. This indicated that the more hydrophobic dmCC likely induced the aggregation of copolymers. Zeta potential analysis revealed that the homopolymer P1 has the lowest zeta potential of <10 mV possibly due to the absence of aggregation, whereas the CC-substituted copolymers increased in the zeta potential ranging from ∼+14 mV to ∼+25 mV and dmCC-substituted copolymers showed an even higher zeta potential ranging from ∼+29 mV to ∼+47 mV.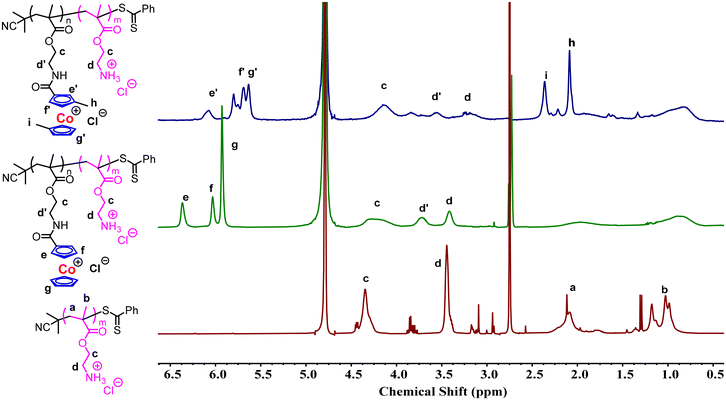 | ||
| Fig. 1 1H NMR spectra of the PAEMA homopolymer (P1), the cobaltocenium containing copolymer (P2) and the dimethyl substituted cobaltocenium containing copolymer (P5) (deuterated solvent: D2O). | ||
2.2. Antimicrobial activity of cobaltocenium-containing copolymers
The antimicrobial activity of CC- or dmCC-containing copolymers (P2–P7) was determined against two strains of Gram-positive methicillin-resistant S. aureus (MRSA) and three Gram-negative bacterial strains: E. coli, P. aeruginosa, and E. cloacae. It was carried out through a broth microdilution method following a well-established protocol adopted by the Clinical Laboratory Standards Institute (CLSI) and described as the minimum inhibitory concentration (MIC).60 Polymers were tested at concentrations ranging from 256 μg mL−1 to 0.5 μg mL−1. The results are included in Table 2.| Bacterial strain | P1 | P2 (CC42%) | P3 (CC50%) | P4 (CC81%) | P5 (dmCC54%) | P6 (dmCC58%) | P7 (dmCC69%) |
|---|---|---|---|---|---|---|---|
| E. coli | 256 | 128 | 128 | 32 | 32 | 16 | 16 |
| P. aeruginosa | 256 | 128 | 128 | 128 | 64 | 32 | 32 |
| E. cloacae | 256 | 256 | 128 | 4 | 64 | 16 | 8 |
| MRSA-1 | 32 | 16 | 16 | 4 | 4 | 16 | 2 |
| MRSA-2 | 32 | 16 | 8 | 2 | 4 | 16 | 4 |
While P1 showed no activity against Gram-negative bacteria, it was moderately active against both MRSA strains with a MIC90 value of 32 μg mL−1. For cobaltocenium copolymers, we noticed that with an increasing percentage of cobaltocenium, the copolymers exhibited higher efficacy against Gram-negative strains. The dmCC-containing copolymers showed higher activity compared to their unsubstituted counterparts. To further investigate the effect of cobaltocenium, we tested the copolymers against MDR Gram-negative strains including NDM-1 bearing strains, as shown in Table 3. We found a more prominent effect of dmCC in these strains where copolymers with moderate (P5: PAEMA-dmCC54%, P6: PAEMA-dmCC58%) to higher (P7: PAEMA-dmCC69%) fractions exhibited very high activities with MIC90 values as low as 2 μg mL−1, whereas CC-containing copolymers showed a gradual enhancement in activities from lower (P2: PAEMA-CC42%, P3: PAEMA-CC50%) to higher fractions (P4: PAEMA-CC80%), reaching a MIC90 value of 4 μg mL−1.
| Bacterial strains | P1 | P2 (CC42%) | P3 (CC50%) | P4 (CC81%) | P5 (dmCC54%) | P6 (dmCC58%) | P7 (dmCC69%) |
|---|---|---|---|---|---|---|---|
| E. coli-1 | 128 | 32 | 16 | 4 | 8 | 4 | 4 |
| E. coli-2 | >256 | 64 | 32 | 4 | 16 | 8 | 8 |
| E. hormaechei | 256 | 128 | 32 | 4 | 4 | 32 | 4 |
| K. pneumoniae | >256 | 128 | 32 | 4 | 16 | 2 | 4 |
| P. aeruginosa | 64 | 64 | 32 | 8 | 8 | 32 | 2 |
2.3 Bactericidal kinetics against planktonic and stationary bacteria
Carbapenem-resistant Enterobacteriaceae (CRE) are among the urgent threat pathogens. These bacteria can render antibiotics ineffective by producing NDM-1 enzyme to hydrolyze β-lactam antibiotics.6 Thus, we intended to perform killing kinetics of cobaltocenium copolymers against two NDM-1 containing Gram-negative strains: MDR E. coli (BAA-2471), MDR K. pneumoniae (BAA-2473). Based on the growth conditions, bacteria can multiply in different sub-populations: planktonic and stationary phases. Planktonic bacterial cells grow under favorable conditions, while, in contrast, stationary bacterial cells grow under nutrient-starved conditions. Antibiotics that can inhibit planktonic bacteria (metabolically active) often fail in treating stationary bacteria (metabolically inactive) because of the difference in cellular metabolic processes.2First, we treated planktonic bacterial cells of both strains and observed that it took 8 hours for the copolymers (P4, P6, and P7) to kill the bacteria completely. During treatment against MDR E. coli, all copolymers at different concentrations (2×, 4×, and 8× MIC90) showed a significant reduction of bacteria within 4 hours (∼4 log reduction) (Fig. 2A). For MDR K. pneumoniae, an even faster reduction of bacterial colonies was noticed. Within 2 hours, P6 achieved ∼2.5 log reduction, whereas P4 and P7 achieved ∼4 log reduction, indicating more efficient inhibition of bacteria with higher cobaltocenium contents in the copolymers (Fig. 2B).
The better performing copolymers P4 and P7 were selected to treat stationary phase bacteria. Two β-lactam antibiotics, Aztreonam and Imipenem, were used as antibiotic controls at concentrations not lower than the copolymers. At 2 hours, the control (without any treatment) and antibiotics did not induce reduction, while all concentrations (4× and 8× MIC90) of the copolymers displayed significant reduction. P7 was able to eliminate MDR E. coli completely within 4 hours, whereas the antibiotics displayed only ∼2 log reduction up to 12 hours (Fig. 2C). Against MDR K. pneumoniae, similar results were observed for copolymer P7 and antibiotic treatment (Fig. 2D). These outcomes indicated significant killing efficiency of the copolymers against both planktonic and stationary cells of Gram-negative bacteria.
2.4. Toxicity and selectivity
It is crucial to evaluate the biocompatibility of antimicrobial polymers. Hemolysis of mouse red blood cells (RBCs) was assessed and is summarized in Fig. 2E. All polymers exhibited a low hemolysis rate at various concentrations, and the HC50 (concentration of the test drug required for 50% lysis of RBCs) was found to be >1000 μg mL−1. It is noteworthy that dmCC-containing copolymers (P5, P6, and P7) showed lower hemolysis than the precursor polymer (P1).The therapeutic index is an indicator of the applicability of a new antimicrobial agent. A suitable antimicrobial agent should display selective antimicrobial characteristics towards pathogenic microbes over mammalian cells. The selectivity of polymers is defined as the ratio between the concentrations leading to 50% hemolysis (HC50) and the minimum inhibitory concentration (MIC90) of polymer required to inhibit bacterial growth (selectivity = HC50/MIC90). Higher selectivity indicates selective killing of pathogens over mammalian cells. Since all polymers have HC50 over 1000 μg mL−1, the selectivity index depends on the MICs of the polymers. Table 4 shows the selectivity index of the copolymers (P3–P7) against Gram-negative bacterial strains, excluding precursor homopolymer P1 and copolymer P2 (PAEMA-CC42%) as they have high MIC values against bacterial strains. A higher selectivity was obtained for copolymers with higher cobaltocenium contents. The selectivity index is in the range of >31 to >500, which manifests excellent selectivity toward bacterial cells over mammalian cells.
| Bacterial strains | P3 (CC50%) | P4 (CC81%) | P5 (dmCC54%) | P6 (dmCC58%) | P7 (dmCC69%) |
|---|---|---|---|---|---|
| E. coli-1 | >62.5 | >250 | >125 | >250 | >250 |
| E. coli-2 | >31.25 | >250 | >62.5 | >125 | >125 |
| E. hormaechei | >31.25 | >250 | >250 | >31.25 | >250 |
| K. pneumoniae | >31.25 | >250 | >62.5 | >500 | >250 |
| P. aeruginosa | >31.25 | >125 | >125 | >31.25 | >500 |
2.5. Mechanisms of action
Both planktonic and stationary bacterial cells were treated separately with P7 at different concentrations (1×, 2×, and 4× MIC90). In the case of MDR E. coli, increased fluorescence was observed with higher doses of the copolymer, indicating dose-dependent damage to the outer membrane of planktonic bacteria (Fig. 3A). For stationary bacteria, the fluorescence intensity also increased but was not as prominent as planktonic bacteria, indicating acquired resistance by the bacteria though not enough to prevent membrane rupture (Fig. 3C). Similar observations were found against MDR K. pneumoniae, where lower doses of the polymer caused less damage to the outer membrane and higher doses showed enhanced perturbation for both planktonic and stationary bacteria (Fig. 3B and D).
2.6. Propensity to resistance
One major cause of drug resistance in antibiotics is sub-lethal and/or repeated dosing of antibiotics, which can lead to the formation of multidrug resistant mutant strains. AMP-mimicking polymers usually have an advantage over conventional small molecular antibiotics due to their membrane targeting properties. To investigate the propensity to resistance, NDM-1 containing MDR K. pneumoniae was treated with the lead polymer P7 and polymyxin-B. The bacteria were exposed to 14 serial passages. The polymer exhibited only a 2-fold increase in the MIC by passage 11, in contrast to polymyxin B, which showed a 256-fold MIC increase by passage 7 and maintained at that level throughout the remainder of the treatment period (Fig. 4E). The experiment suggested that the polymer exhibited a markedly low propensity for resistance development compared to conventional antibiotics, likely due to its membrane-targeting mechanism.3. Discussion
Gram-negative bacteria, especially NDM-1 producing strains, are of urgent concern with no efficient treatment strategy currently in existence. Therefore, new antimicrobial agent discovery besides conventional antibiotics can work as a standalone or combined synergistic treatment method. In this work, we aim to combat Gram-negative bacteria by developing a library of metallopolymers with two charged species to unveil a new approach towards AMP-mimicking polymers. Combining cobaltocenium with the primary ammonium containing methacrylate polymer and carefully controlling the incorporation of a cobaltocenium moiety, an effective amphiphilic balance was obtained without compromising the overall cationic nature of the polymer. In particular, copolymers P4 (CC81%) and P7 (dmCC69%) with a higher percentage of cobaltocenium stood out with excellent antimicrobial activity against Gram-negative bacteria compared to the rest of the polymers. The bactericidal kinetics revealed complete eradication of MDR bacterial strains (E. coli and K. pneumoniae) within a short time with the copolymer treatment, reflecting the fast-acting properties of AMP mimicking agents. Another significant feature of antimicrobial agents is their high selectivity towards bacterial cells. The copolymers showed hemocompatibility up to >1000 μg mL−1 and were highly selective towards Gram-negative bacterial cells.Furthermore, we performed an in-depth analysis of the mechanistic action of the most active copolymer P7, with an emphasis on NDM-1 containing bacteria. The outer membrane and proton motive force (PMF) of Gram-negative bacteria make multiple treatments fail, thus resulting in MDR bacteria. The copolymer was able to disrupt the outer membrane and caused membrane depolarization showing the dose-dependent killing efficiency of MDR bacteria. Another crucial aspect of Gram-negative bacteria is the negatively charged LPS, an essential molecule integrated throughout the outer membrane. An increase in the MIC90 of copolymer P7 in the presence of LPS molecules revealed strong electrostatic interactions. This was further evidenced by observing the change in the surface charge of the bacterial solution from negative to positive zeta potentials with the addition of copolymer P7. Finally, the resistance profile of the copolymer was measured for a period of two weeks against MDR K. Pneumoniae (BAA-2473), which showed a minimal increase in the MIC90 for our copolymer treatment, whereas Polymyxin-B, a last-resort antibiotic, failed to inhibit resistance. These investigations revealed that cobaltocenium in combination with the primary ammonium-containing methacrylate polymer yields an effective antimicrobial AMP mimicking metallopolymer and to our knowledge this is the first cobaltocenium copolymer that can act as a standalone therapeutic agent without the use of antibiotics.
4. Conclusions
In summary, we synthesized a new library of copolymers by incorporating two distinct cationic species to maintain amphiphilic balance. The inclusion of cobaltocenium endowed the polymers with broad-spectrum antimicrobial activity and enabled rapid eradication of NDM-1 producing bacteria without altering the overall cationic charge density. Mechanistic investigations revealed membrane-targeting properties via outer membrane permeabilization and membrane depolarization. The copolymers’ high selectivity and low propensity to resistance can be ascribed to their membrane-perturbing mechanism and strong electrostatic interaction with LPS molecules, also demonstrated via bacterial surface charge alteration. Notably, the lead polymer was immune to bacterial resistance development. These findings establish metallopolymers with tunable amphiphilicity as promising therapeutic candidates to combat the escalating antibiotic crisis while unveiling a new frontier in AMP-mimicking polymer design.5. Experimental section
5.1. Materials
All reagents were purchased from commercial sources and used as received unless stated otherwise. 2-Aminoethyl methacrylate hydrochloride (AEMA·HCl) was purchased from Aaron Chemicals. 2-Cyano-2-propyl benzodithioate (CPB) was purchased from Boron Molecular. N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC·HCl) was purchased from Oakwood Chemicals. Sulfo-NHS (N-hydroxysulfosuccinimide sodium salt 97%) was purchased from Ambeed, Inc. 2,2′-Azobis(2-methylpropionitrile) (AIBN, Sigma-Aldrich, 98%) and solvents such as dimethylformamide (DMF), dimethyl sulfoxide (DMSO), and dichloromethane (DCM) were purified by standard procedures. Deionized water was purified using a Millipore water purification system with a minimum resistivity of 18.2 megaohms per cm.5.2. Polymer synthesis and characterization
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)–), δ = 3.96–4.50 (2H, –O-CH2-CH2-NH2–), δ = 5.8–5.97 (5H, Cp), δ = 5.97–6.10 (2H, Cp), δ = 6.25–6.46 (2H, Cp). The 1H NMR of P3 is presented in Fig. S3.†1H NMR (400 MHz, D2O): δ = 0.50–1.30 (3H, –CH2-C-CH3), δ = 1.50–2.26 (2H, –CH2-C-CH3), δ = 3.22–3.43 (2H, –CH2-CH2-NH2·HCl), δ = 3.50–3.75 (2H, –CH2-CH2-NH–(C
O)–), δ = 3.96–4.50 (2H, –O-CH2-CH2-NH2–), δ = 5.8–5.97 (5H, Cp), δ = 5.97–6.10 (2H, Cp), δ = 6.25–6.46 (2H, Cp). The 1H NMR of P3 is presented in Fig. S3.†1H NMR (400 MHz, D2O): δ = 0.50–1.30 (3H, –CH2-C-CH3), δ = 1.50–2.26 (2H, –CH2-C-CH3), δ = 3.22–3.43 (2H, –CH2-CH2-NH2·HCl), δ = 3.50–3.75 (2H, –CH2-CH2-NH–(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)–), δ = 3.85–4.36 (2H, –O-CH2-CH2-NH2–), δ = 5.75–5.86 (5H, Cp), δ = 5.86–5.97 (2H, Cp), δ = 6.15–6.32 (2H, Cp). The 1H NMR of P4 is presented in Fig. S4.†1H NMR (400 MHz, D2O): δ = 0.36–1.1 (3H, –CH2-C-CH3), δ = 1.38–2.18 (2H, –CH2-C-CH3), δ = 3.16–3.34 (2H, –CH2-CH2-NH2·HCl), δ = 3.39–3.70 (2H, –CH2-CH2-NH–(C
O)–), δ = 3.85–4.36 (2H, –O-CH2-CH2-NH2–), δ = 5.75–5.86 (5H, Cp), δ = 5.86–5.97 (2H, Cp), δ = 6.15–6.32 (2H, Cp). The 1H NMR of P4 is presented in Fig. S4.†1H NMR (400 MHz, D2O): δ = 0.36–1.1 (3H, –CH2-C-CH3), δ = 1.38–2.18 (2H, –CH2-C-CH3), δ = 3.16–3.34 (2H, –CH2-CH2-NH2·HCl), δ = 3.39–3.70 (2H, –CH2-CH2-NH–(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)–), δ = 3.74–4.22 (2H, –O-CH2-CH2-NH2–), δ = 5.67–5.80 (5H, Cp), δ = 5.80–5.93 (2H, Cp), δ = 6.08–6.27 (2H, Cp).
O)–), δ = 3.74–4.22 (2H, –O-CH2-CH2-NH2–), δ = 5.67–5.80 (5H, Cp), δ = 5.80–5.93 (2H, Cp), δ = 6.08–6.27 (2H, Cp).
For the dimethyl substituted cobaltocenium-containing polymer, a similar protocol was used. The 1H NMR of P5 is presented in Fig. S6.†1H NMR (400 MHz, D2O): δ = 0.60–1.36 (3H, –CH2-C-CH3), δ = 1.57–2.50 (2H, –CH2-C-CH3), δ = 1.99–2.17 (3H, Cp-CH3), δ = 2.27–2.47 (3H, Cp-CH3), δ = 3.25–3.45 (2H, –CH2-CH2-NH2·HCl), δ = 3.45–3.91 (2H, –CH2-CH2-NH–(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)–), δ = 3.94–4.38 (2H, –O-CH2-CH2-NH2–), δ = 5.55–5.86 (7H, Cp), δ = 5.96–6.18 (2H, Cp). The 1H NMR of P6 is presented in Fig. S7.†1H NMR (400 MHz, D2O): δ = 0.58–1.40 (3H, –CH2-C-CH3), δ = 1.55–2.60 (2H, –CH2-C-CH3), δ = 1.99–2.16 (3H, Cp-CH3), δ = 2.26–2.46 (3H, Cp-CH3), δ = 3.00–3.38 (2H, –CH2-CH2-NH2·HCl), δ = 3.43–3.92 (2H, –CH2-CH2-NH–(C
O)–), δ = 3.94–4.38 (2H, –O-CH2-CH2-NH2–), δ = 5.55–5.86 (7H, Cp), δ = 5.96–6.18 (2H, Cp). The 1H NMR of P6 is presented in Fig. S7.†1H NMR (400 MHz, D2O): δ = 0.58–1.40 (3H, –CH2-C-CH3), δ = 1.55–2.60 (2H, –CH2-C-CH3), δ = 1.99–2.16 (3H, Cp-CH3), δ = 2.26–2.46 (3H, Cp-CH3), δ = 3.00–3.38 (2H, –CH2-CH2-NH2·HCl), δ = 3.43–3.92 (2H, –CH2-CH2-NH–(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)–), δ = 3.93–4.41 (2H, –O-CH2-CH2-NH2–), δ = 5.51–5.87 (7H, Cp), δ = 5.97–6.20 (2H, Cp). The 1H NMR of P7 is presented in Fig. S8.†1H NMR (400 MHz, D2O): δ = 0.50–1.30 (3H, –CH2-C-CH3), δ = 1.53–2.46 (2H, –CH2-C-CH3), δ = 1.93–2.12 (3H, Cp-CH3), δ = 2.22–2.40 (3H, Cp-CH3), δ = 3.00–3.38 (2H, –CH2-CH2-NH2·HCl), δ = 3.38–3.89 (2H, –CH2-CH2-NH–(C
O)–), δ = 3.93–4.41 (2H, –O-CH2-CH2-NH2–), δ = 5.51–5.87 (7H, Cp), δ = 5.97–6.20 (2H, Cp). The 1H NMR of P7 is presented in Fig. S8.†1H NMR (400 MHz, D2O): δ = 0.50–1.30 (3H, –CH2-C-CH3), δ = 1.53–2.46 (2H, –CH2-C-CH3), δ = 1.93–2.12 (3H, Cp-CH3), δ = 2.22–2.40 (3H, Cp-CH3), δ = 3.00–3.38 (2H, –CH2-CH2-NH2·HCl), δ = 3.38–3.89 (2H, –CH2-CH2-NH–(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)–), δ = 3.89–4.41 (2H, –O-CH2-CH2-NH2–), δ = 5.48–5.90 (7H, Cp), δ = 5.92–6.20 (2H, Cp).
O)–), δ = 3.89–4.41 (2H, –O-CH2-CH2-NH2–), δ = 5.48–5.90 (7H, Cp), δ = 5.92–6.20 (2H, Cp).
5.3. Measurement of antibacterial activity
Escherichia coli (ATCC-11775), Pseudomonas aeruginosa (ATCC-10145), Enterobacter cloacae (ATCC-13047), Acinetobacter baumannii (ATCC-19606), MRSA-1 (BAA-1717), MRSA-2 (BAA-44), MDR E. coli-1 (BAA-2452), MDR E. coli-2 (BAA-2471), MDR E. hormaechei (BAA-2468), MDR K. pneumoniae (BAA-2473), and MDR P. aeruginosa (BAA-2108) were purchased from ATCC. Bacteria were streaked on tryptic soy agar (TSA) plates from their primary glycerol stock. After incubation at 37 °C for 24 h, a single colony was inoculated in 3 mL of tryptic soy broth (TSB) at 37 °C for 6 h under constant shaking at 190 rpm to reach the mid-log phase. Specific antibiotics were used in both TSA plates and TSB for specific bacteria: imipenem (25 μg mL−1) for MDR E. coli-2 (BAA-2471), MDR E. cloacae (BAA-2468), and MDR K. pneumoniae (BAA-2473). All bacteria were adjusted to an optical density of 0.5 McFarland standard (OD600 = 0.70) for further use.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 in nutrient broth (NB) and incubated at 37 °C with shaking at 190 rpm for 16 hours. Following incubation, the culture was centrifuged at 3500g for 5 minutes, and the supernatant was removed. The bacterial pellet was resuspended in normal saline. The stationary cells were then used for kinetic experiments, following the same procedure as time-kill kinetics against planktonic bacterial cells. All experiments were performed in duplicate. Bacteria were treated with P4 and P7, while water was used as a negative control.
1000 in nutrient broth (NB) and incubated at 37 °C with shaking at 190 rpm for 16 hours. Following incubation, the culture was centrifuged at 3500g for 5 minutes, and the supernatant was removed. The bacterial pellet was resuspended in normal saline. The stationary cells were then used for kinetic experiments, following the same procedure as time-kill kinetics against planktonic bacterial cells. All experiments were performed in duplicate. Bacteria were treated with P4 and P7, while water was used as a negative control.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of 5 mM HEPES and 5 mM glucose buffer. N-Phenyl naphthylamine (NPN) dye was added to the bacterial suspension at a final concentration of 10 μM. A Corning black and clear bottom 96-well plate was prepared, with 180 μL of the prepared bacterial suspension added to each well. The fluorescence intensity of the NPN dye was measured at an excitation wavelength of 350 nm and an emission wavelength of 420 nm every 2 minutes. After 4 minutes, 20 μL of the test compound in water was added to the wells, with the final concentration adjusted using the microdilution method. The fluorescence intensity was recorded for an additional 24 minutes. Wells containing 20 μL of water instead of the compound served as negative controls. All experiments were performed in triplicate.60
1 mixture of 5 mM HEPES and 5 mM glucose buffer. N-Phenyl naphthylamine (NPN) dye was added to the bacterial suspension at a final concentration of 10 μM. A Corning black and clear bottom 96-well plate was prepared, with 180 μL of the prepared bacterial suspension added to each well. The fluorescence intensity of the NPN dye was measured at an excitation wavelength of 350 nm and an emission wavelength of 420 nm every 2 minutes. After 4 minutes, 20 μL of the test compound in water was added to the wells, with the final concentration adjusted using the microdilution method. The fluorescence intensity was recorded for an additional 24 minutes. Wells containing 20 μL of water instead of the compound served as negative controls. All experiments were performed in triplicate.60
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of 5 mM HEPES buffer, 5 mM glucose, and 100 mM KCl solution and 0.2 mM ethylenediaminetetraacetic acid (EDTA) was added additionally for Gram-negative bacteria. DiSC3 (3,3′-dipropylthiadicarbocyanine iodide) was added to the bacterial suspension to a final concentration of 2 μM, and the mixture was incubated in darkness for 1 hour. A Corning black and clear bottom 96-well plate was prepared with 180 μL of the bacterial suspension per well. The fluorescence intensity of the DiSC3 dye was measured at an excitation wavelength of 622 nm and an emission wavelength of 670 nm every 2 minutes. After 4 minutes, 20 μL of the test compound in Millipore water was added, with the final concentration adjusted using the microdilution method. The fluorescence intensity was recorded for an additional 24 minutes. Wells containing 20 μL of Millipore water instead of the compound served as negative controls. All experiments were performed in triplicate.44
1 mixture of 5 mM HEPES buffer, 5 mM glucose, and 100 mM KCl solution and 0.2 mM ethylenediaminetetraacetic acid (EDTA) was added additionally for Gram-negative bacteria. DiSC3 (3,3′-dipropylthiadicarbocyanine iodide) was added to the bacterial suspension to a final concentration of 2 μM, and the mixture was incubated in darkness for 1 hour. A Corning black and clear bottom 96-well plate was prepared with 180 μL of the bacterial suspension per well. The fluorescence intensity of the DiSC3 dye was measured at an excitation wavelength of 622 nm and an emission wavelength of 670 nm every 2 minutes. After 4 minutes, 20 μL of the test compound in Millipore water was added, with the final concentration adjusted using the microdilution method. The fluorescence intensity was recorded for an additional 24 minutes. Wells containing 20 μL of Millipore water instead of the compound served as negative controls. All experiments were performed in triplicate.44
Author contributions
M. W. H: writing – original draft, investigation, formal analysis, and methodology; I. B: writing – review & editing, formal analysis, and methodology; S. B: writing – review & editing, formal analysis, and methodology; A. A: formal analysis; A. P: formal analysis; X. Y: formal analysis and methodology; P. N: writing – review & editing; M. N: writing – review & editing; C. T: writing – original draft, conceptualization, supervision, investigation, formal analysis, resources, and funding acquisition. All authors approved the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the ESI.†Acknowledgements
The authors acknowledge the University of South Carolina for financial support. The partial funding support from the National Institutes of Health (R01AI149810) is also acknowledged. We would also like to acknowledge BioRender for the TOC image (created in BioRender by Hossain, M. W. (2025); https://BioRender.com/nax4lzk).References
- I. N. Okeke, M. E. A. de Kraker, T. P. Van Boeckel, C. K. Kumar, H. Schmitt, A. C. Gales, S. Bertagnolio, M. Sharland and R. Laxminarayan, Lancet, 2024, 403, 2426–2438 CrossRef CAS PubMed.
- S. Barman, L. B. Kurnaz, R. Leighton, M. W. Hossain, A. W. Decho and C. Tang, Biomaterials, 2024, 122690 CrossRef CAS PubMed.
- K. Lewis, Cell, 2020, 181, 29–45 CrossRef CAS PubMed.
- M. I. Hutchings, A. W. Truman and B. Wilkinson, Curr. Opin. Microbiol., 2019, 51, 72–80 CrossRef CAS PubMed.
- G. D. Wright, Chem. Commun., 2011, 47, 4055 RSC.
- G. D. Wright, Trends Microbiol., 2016, 24, 862–871 CrossRef CAS PubMed.
- H. I. Zgurskaya, C. A. Lopez and S. Gnanakaran, ACS Infect. Dis., 2015, 1, 512–522 CrossRef CAS PubMed.
- WHO bacterial priority pathogens list, 2024: bacterial pathogens of public health importance, to guide research, development, and strategies to prevent and control antimicrobial resistance, World Health Organization, 2024 Search PubMed.
- L. Maryam, S. Khalid, A. Ali and A. U. Khan, Future Microbiol., 2019, 14, 671–689 CrossRef CAS PubMed.
- M. Shahid, N. Ahmad, N. K. Saeed, M. Shadab, R. M. Joji, A. Al-Mahmeed, K. M. Bindayna, K. S. Tabbara and F. K. Dar, Front. Cell. Infect. Microbiol., 2022, 12, 1033305 CrossRef CAS PubMed.
- P. Pham, S. Oliver and C. Boyer, Macromol. Chem. Phys., 2023, 224, 2200226 CrossRef CAS.
- M. Zasloff, Nature, 2002, 415, 389–395 CrossRef CAS PubMed.
- Z. Song, Z. Han, S. Lv, C. Chen, L. Chen, L. Yin and J. Cheng, Chem. Soc. Rev., 2017, 46, 6570–6599 RSC.
- M. Xiong, Z. Han, Z. Song, J. Yu, H. Ying, L. Yin and J. Cheng, Angew. Chem., Int. Ed., 2017, 56, 10826–10829 CrossRef CAS PubMed.
- A. A. Bahar and D. Ren, Pharmaceuticals, 2013, 6, 1543–1575 CrossRef PubMed.
- R. E. Hancock, M. A. Alford and E. F. Haney, Nat. Rev. Microbiol., 2021, 19, 786–797 CrossRef CAS PubMed.
- S. Barman, L. B. Kurnaz, X. Yang, M. Nagarkatti, P. Nagarkatti, A. W. Decho and C. Tang, ACS Infect. Dis., 2023, 9, 1769–1782 CrossRef CAS PubMed.
- Y. Chen, L. Yu, B. Zhang, W. Feng, M. Xu, L. Gao, N. Liu, Q. Wang, X. Huang, P. Li and W. Huang, Biomacromolecules, 2019, 20, 2230–2240 CrossRef CAS PubMed.
- C. H. Chen and T. K. Lu, Antibiotics, 2020, 9, 24 CrossRef CAS PubMed.
- G. N. Tew, R. W. Scott, M. L. Klein and W. F. Degrado, Acc. Chem. Res., 2010, 43, 30–39 CrossRef CAS PubMed.
- E. F. Palermo and K. Kuroda, Appl. Microbiol. Biotechnol., 2010, 87, 1605–1615 CrossRef CAS PubMed.
- T. Zhu, Y. Sha, J. Yan, P. Pageni, M. A. Rahman, Y. Yan and C. Tang, Nat. Commun., 2018, 9, 4329 CrossRef PubMed.
- M. Zhou, Y. Qian, J. Xie, W. Zhang, W. Jiang, X. Xiao, S. Chen, C. Dai, Z. Cong, Z. Ji, N. Shao, L. Liu, Y. Wu and R. Liu, Angew. Chem., Int. Ed., 2020, 59, 6412–6419 CrossRef CAS PubMed.
- H. Takahashi, G. A. Caputo and K. Kuroda, Biomater. Sci., 2021, 9, 2758–2767 RSC.
- K. Kuroda and W. F. Degrado, J. Am. Chem. Soc., 2005, 127, 4128–4129 CrossRef CAS PubMed.
- M. F. Ilker, K. Nüsslein, G. N. Tew and E. B. Coughlin, J. Am. Chem. Soc., 2004, 126, 15870–15875 CrossRef CAS PubMed.
- K. Lienkamp, K. N. Kumar, A. Som, K. Nüsslein and G. N. Tew, Chem. – Eur. J., 2009, 15, 11710–11714 CrossRef CAS PubMed.
- E. F. Palermo and K. Kuroda, Biomacromolecules, 2009, 10, 1416–1428 CrossRef CAS PubMed.
- E. H. Wong, M. M. Khin, V. Ravikumar, Z. Si, S. A. Rice and M. B. Chan-Park, Biomacromolecules, 2016, 17, 1170–1178 CrossRef CAS PubMed.
- P. R. Judzewitsch, T. K. Nguyen, S. Shanmugam, E. H. H. Wong and C. Boyer, Angew. Chem., Int. Ed., 2018, 57, 4559–4564 CrossRef CAS PubMed.
- P. R. Judzewitsch, L. Zhao, E. H. H. Wong and C. Boyer, Macromolecules, 2019, 52, 3975–3986 CrossRef CAS.
- J. Xie, M. Zhou, Y. Qian, Z. Cong, S. Chen, W. Zhang, W. Jiang, C. Dai, N. Shao, Z. Ji, J. Zou, X. Xiao, L. Liu, M. Chen, J. Li and R. Liu, Nat. Commun., 2021, 12, 5898 CrossRef CAS PubMed.
- A. Vishwakarma, F. Dang, A. Ferrell, H. A. Barton and A. Joy, J. Am. Chem. Soc., 2021, 143, 9440–9449 CrossRef CAS PubMed.
- J. Yan, X. Zhang, P. Xu, J. Zhang, X. Li, F. Zhong, Y. Xu, Q. Zhang, Y. Zhu and Y. Yan, Adv. Mater. Technol., 2025, 10, 2401559 CrossRef CAS.
- G. R. Whittell, M. D. Hager, U. S. Schubert and I. Manners, Nat. Mater., 2011, 10, 176–188 CrossRef CAS PubMed.
- C. G. Hardy, L. Ren, J. Zhang and C. Tang, Isr. J. Chem., 2012, 52, 230–245 CrossRef CAS.
- L. Zhao, X. Liu, L. Zhang, G. Qiu, D. Astruc and H. Gu, Coord. Chem. Rev., 2017, 337, 34–79 CrossRef CAS.
- Y. Wang, D. Astruc and A. S. Abd-El-Aziz, Chem. Soc. Rev., 2019, 48, 558–636 RSC.
- M. R. Roner, C. E. Carraher Jr, K. Shahi and G. Barot, Materials, 2011, 4, 991–1012 CrossRef CAS PubMed.
- M. Patra, G. Gasser and N. Metzler-Nolte, Dalton Trans., 2012, 41, 6350–6358 RSC.
- G. Gasser and N. Metzler-Nolte, Curr. Opin. Chem. Biol., 2012, 16, 84–91 CrossRef CAS PubMed.
- F. Li, J. G. Collins and F. R. Keene, Chem. Soc. Rev., 2015, 44, 2529–2542 RSC.
- A. S. Abd-El-Aziz, C. Agatemor and N. Etkin, Biomaterials, 2017, 118, 27–50 CrossRef CAS PubMed.
- J. Hwang, S. Barman, R. Gao, X. Yang, A. O'Malley, P. Nagarkatti, M. Nagarkatti, M. Chruszcz and C. Tang, Adv. Healthcare Mater., 2023, 12, e2301764 CrossRef PubMed.
- J. Zhang, Y. P. Chen, K. P. Miller, M. S. Ganewatta, M. Bam, Y. Yan, M. Nagarkatti, A. W. Decho and C. Tang, J. Am. Chem. Soc., 2014, 136, 4873–4876 CrossRef CAS PubMed.
- P. Yang, M. Bam, P. Pageni, T. Zhu, Y. P. Chen, M. Nagarkatti, A. W. Decho and C. Tang, ACS Infect. Dis., 2017, 3, 845–853 CrossRef CAS PubMed.
- Z. Si, W. Zheng, D. Prananty, J. Li, C. H. Koh, E. T. Kang, K. Pethe and M. B. Chan-Park, Chem. Sci., 2022, 13, 345–364 RSC.
- S. Hong, H. Takahashi, E. T. Nadres, H. Mortazavian, G. A. Caputo, J. G. Younger and K. Kuroda, PLoS One, 2017, 12, e0169262 CrossRef PubMed.
- Q. Zhou, K. Li, K. Wang, W. Hong, J. Chen, J. Chai, L. Yu, Z. Si and P. Li, Sci. Adv., 2024, 10, eadp6604 CrossRef CAS PubMed.
- R. Namivandi-Zangeneh, Z. Sadrearhami, D. Dutta, M. Willcox, E. H. H. Wong and C. Boyer, ACS Infect. Dis., 2019, 5, 1357–1365 CrossRef CAS PubMed.
- S. C. Williams, M. B. Chosy, C. K. Jons, C. Dong, A. N. Prossnitz, X. Liu, H. L. Hernandez, L. Cegelski and E. A. Appel, ACS Cent. Sci., 2025, 11, 486–496 CrossRef CAS PubMed.
- J. Zhang, Y. Yan, M. W. Chance, J. Chen, J. Hayat, S. Ma and C. Tang, Angew. Chem., Int. Ed., 2013, 52, 13387–13391 CrossRef CAS PubMed.
- S. Wickramasinghe, A. Hoehn, S. T. Wetthasinghe, H. Lin, Q. Wang, J. Jakowski, V. Rassolov, C. Tang and S. Garashchuk, J. Phys. Chem. B, 2023, 127, 10129–10141 CrossRef CAS PubMed.
- T. Zhu, Y. Sha, H. A. Firouzjaie, X. Peng, Y. Cha, D. M. M. M. Dissanayake, M. D. Smith, A. K. Vannucci, W. E. Mustain and C. Tang, J. Am. Chem. Soc., 2020, 142, 1083–1089 CrossRef CAS PubMed.
- H. Lin, L. Ramos, J. Hwang, T. Zhu, M. W. Hossain, Q. Wang, S. Garashchuk and C. Tang, Macromolecules, 2023, 56, 6375–6384 CrossRef CAS.
- L. He, E. S. Read, S. P. Armes and D. J. Adams, Macromolecules, 2007, 40, 4429–4438 CrossRef CAS.
- L. Ren, C. G. Hardy, S. Tang, D. B. Doxie, N. Hamidi and C. Tang, Macromolecules, 2010, 43, 9304–9310 CrossRef CAS.
- Y. Lou, J. Gaitor, M. Treichel, K. J. T. Noonan and E. F. Palermo, ACS Macro Lett., 2023, 12, 215–220 CrossRef CAS PubMed.
- M. A. Rahman, Y. Cha, L. Yuan, P. Pageni, T. Zhu, M. S. Jui and C. Tang, J. Polym. Sci., 2020, 58, 77–83 CrossRef CAS PubMed.
- L. B. Kurnaz, S. Barman, X. Yang, C. Fisher, F. W. Outten, P. Nagarkatti, M. Nagarkatti and C. Tang, Biomaterials, 2023, 301, 122275 CrossRef CAS PubMed.
- A. H. Delcour, Biochim. Biophys. Acta, Proteins Proteomics, 2009, 1794, 808–816 CrossRef CAS PubMed.
- M. A. Farha, C. P. Verschoor, D. Bowdish and E. D. Brown, Chem. Biol., 2013, 20, 1168–1178 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5bm00497g |
| This journal is © The Royal Society of Chemistry 2025 |





