Open Access Article
Open Access ArticleA comparison of the effects of polystyrene and polycaprolactone nanoplastics on macrophages†
Véronique Collin-Faurea,
Maeva Bouléeb,
Hélène Diemercd,
Daphna Fenele,
Christine Moriscote,
Sarah Cianféranicd,
Elisabeth Darrouzet a,
Marie Carrière
a,
Marie Carrière b and
Thierry Rabilloud
b and
Thierry Rabilloud *a
*a
aChemistry and Biology of Metals, Univ. Grenoble Alpes, CNRS UMR5249, CEA, IRIG-LCBM, F-38054 Grenoble, France. E-mail: thierry.rabilloud@cnrs.fr
bUniv. Grenoble-Alpes, CEA, CNRS, Grenoble-INP, IRIG, SyMMES, CIBEST, Grenoble 38000, France
cLaboratoire de Spectrométrie de Masse BioOrganique (LSMBO), IPHC UMR 7178, Université de Strasbourg, CNRS, 67087 Strasbourg, France
dInfrastructure Nationale de Protéomique ProFI – UAR2048, 67087 Strasbourg, France
eInstitut de Biologie Structurale, Université Grenoble Alpes, CEA, CNRS, Grenoble, France
First published on 19th June 2025
Abstract
Plastics are persistent in the environment, which suggests that they may induce adverse effects due to their progressive accumulation over time. This progressive accumulation is facilitated by the fact that macroplastics released into the environment progressively fragment into micro and nanoplastics, which are easily taken up by a wide range of living organisms. As these micro and nanoplastics are particulate materials, they are handled in these living organisms by specialized phagocytic cells, namely macrophages in vertebrates, opening the possibility that plastics accumulating in macrophages may elicit a variety of responses. Thus, one way of alleviating such accumulation in macrophages and in other cell types would be to use biodegradable plastics. Polycaprolactone is a biodegradable plastic showing favorable degradation characteristics in several environments. We thus investigated the responses of macrophages upon treatment with polycaprolactone nanobeads, using a combination of proteomics and validation experiments, and compared these results to the ones induced by polystyrene nanobeads. Many changes detected by proteomics, for example in the mitochondrial, lysosomal or reticulum proteins, did not induce detectable physiological consequences. A slight decrease in the phagocytic capacity of polycaprolactone-treated cells was detected, but not for polystyrene-treated cells. We also showed that polycaprolactone nanobeads degrade within a few days in macrophages, modulating the macrophage responses. The decrease in phagocytosis disappeared, while polystyrene induced a delayed surge in the phagocytic response. A delayed decrease in the secretion of pro-inflammatory cytokines was also observed in polycaprolactone-treated cells, which may be linked to the release of hydroxycaproic acid.
Environmental significanceBiodegradable plastics appear as a solution to the problems posed by classical plastics, which persist for a very long time and contaminate persistently living organisms. Within biodegradable plastics, polycaprolactone has attracted interest due to its rather easy degradation in many environments. However, this degradation is not immediate, meaning polycaprolactone nanoparticles can be expected to be found in the environment, and are likely to contaminate living organisms. We have thus studied the effect of polycaprolactone nanoparticles on macrophages, i.e. professional phagocytes conserved in evolution, by a combination of proteomic and targeted approaches. In addition to limited immediate effects, polycaprolactone nanoparticles degrade relatively slowly in macrophages and induce delayed effects that may alter the normal functions of phagocytes. |
1. Introduction
Plastics are now such an important part of our lifestyle, through their extremely diverse uses, that they are produced in gigantic amounts, i.e. close to 500 Mt per year.1 Unfortunately a huge proportion of these plastics, estimated at 10 Mt per year, is released into the oceans alone,2 where they accumulate over time, leading to estimates of 150 Mt of plastics in the oceans in 2025.3 This plastic waste has deleterious effects that are more and more documented in detail on a wide range of marine taxa,4 including sea birds.5 This pollution was first documented in aquatic marine environments,6–9 but is now found in marine sediments,10,11 freshwater environments,12–14 and also in terrestrial ones.15,16Part of the interest in plastics resides in their chemical resistance, which translates into very long half lives in the environment amounting to decades.17 However the plastics released into the environment do fragment, first into microplastics (less than 1 mm in size) and then into nanoplastics (less than 1 μm in size). It is anticipated that the smaller the plastic particles are, the more easily they will cross biological barriers and be able to disturb the homeostasis of living organisms. This hypothesis has prompted extensive research on the effects of micro and nanoplastics on a wide variety of living organisms from various phyla, ranging from worms18,19 to molluscs,20–22 crustaceans,23,24 insects,25,26 and then vertebrates such as fish27–29 and of course mammals30 including human models (e.g. in ref. 31–35).
Multicellular organisms defend themselves against particles, including plastic particles, by a series of mechanisms. The first line of defense is represented by biological barriers (e.g. intestinal or epidermal barriers) and research has been devoted to understanding how these barriers interact with plastic particles. When translocation across these barriers occurs,31 then a second line of barriers comes into play, and is represented by professional phagocytes. This cell type is encountered in invertebrates (annelids coelomocytes, insect hemocytes), as well as in vertebrates (macrophages, neutrophils). Indeed, it has been shown that this cell type does respond to plastic particles.19,26,36–42
In view of these deleterious effects, the development of biodegradable plastics has been investigated and is a very active area of research. There is however a compromise to be made between the technical properties of plastics and their biodegradability. Many applications, such as food packaging, require good resistance to water, and thus reasonable hydrophobicity, which decreases the interest in polysaccharide-based plastics, for example. In this respect, aliphatic polyesters represent an attractive choice. These plastics are rather hydrophobic, and yet the ester bond is relatively labile, ensuring a much better degradability than plastics where the backbone is a very long aliphatic chain without heteroatoms. Among the aliphatic polyesters, polycaprolactone (PCL) has retained attention because of its attractive properties, as reviewed in ref. 43. It uses a single monomer, ε-caprolactone, which polymerizes by ring opening, allowing an easy control of the polymerization reaction. This plastic, however, suffers from a relatively high price of the monomer, and hence of the polymer, which has led to its current use in niche applications, especially biomedical ones. Indeed, its slow but efficient degradation in mammals makes it a very good candidate for slow delivery drug release,44–47 or for building scaffolds, e.g. for orthopedics or various implants,48–50 which resorb slowly upon biocolonization by the host cells.51,52 In addition to these now well-established applications, there has been a renewal of interest in PCL for applications in membranes53 and packaging.54–60 This renewed interest is further supported by recent research showing that caprolactone can be obtained from bioresources,61 and not only from fossil oil as it is now.
This polymer seems to combine interesting technical properties and good degradability,62 e.g. through composting,63,64 with complete degradation between 100 and 300 days depending on the temperature63 and even in marine environments, with degradation of PCL films in a few weeks.65 Thus, there is a sharp contrast between the relatively easy degradation of PCL by microbes and its slow degradation in mammals. This poses in turn the problem of the effects that PCL particles may have on mammals. In biomedical applications, PCL is used in a kind of pulse-chase mode. The particles or the scaffolds represent a single and massive dose, which is slowly and completely eliminated. If PCL comes into everyday use products such as packaging films, we will then be subjected to a constant influx of PCL particles that, because of the slow degradation of this polymer, may lead to a constant loading of the cleaning system represented by phagocytes, or even to accumulation if the influx outweighs the efflux. Because of this potential caveat, we decided to investigate the effects of PCL nanoparticles on macrophages. As a positive control, we used the well-known polystyrene (PS) nanoparticles, which represent a non-degradable plastic. In order to obtain as much information as possible on the effects of these particles on macrophages, we decided to use a proteomic approach, as done earlier on other types of nanoparticles.66–68
2. Materials and methods
2.1. Plastic particles
Spherical pristine PCL particles (200 nm) were purchased from Phosphorex (Hopkinton, USA) or their retailer Millipore (#PCL200) and spherical pristine PS particles (200 nm) were from Polysciences (#07304-15). Deep red fluorescent labelled polystyrene (PS) particles (SkyBlue particles, 100–300 nm, catalog number #FP-0270-2, batch number #AL01) were purchased from Spherotech. The particles were used for both the proteomic experiments and the validation experiments. The nanoparticle suspensions were pasteurized overnight at 80 °C before use in cell culture.For hydrodynamic diameter and zeta potential measurements, PCL and PS particles were diluted to 100 μg mL−1 in ultrapure water, then transferred to a quartz spectroscopic cell. The hydrodynamic diameter was measured by DLS using a Litesizer 500 (Anton Paar, USA), and the surface charge was evaluated via zeta potential measurement using an Univette (Anton Paar, USA), with the same instrument. An average value was obtained from repeated measurements for each sample (n = 3) and analyzed with the instrument-associated Kaliope software.
PCL and PS particle primary diameters were assessed from transmission electron microscopy (TEM) images, obtained using negative staining on a grid. Samples of 10 mg mL−1 PCL or PS particles were deposited on the clean side of a carbon film on mica, stained with 2% uranyl acetate and 2% phosphotungstic acid, respectively, before being transferred onto a 400 mesh copper grid. Images were acquired using a Tecnai 12 LaB6 electron microscope equipped with a Gatan Orius 1000 CCD camera at an accelerating voltage of 120 kV, with defocus values ranging from 1.2 to 2.5 μm.
For FTIR analysis, 5 μL of 10 mg mL−1 particle suspensions were air-dried on a foil. Measurements were performed using a Nicolet iS50 (Thermo Scientific, USA) spectrometer equipped with a diamond ATR module. The number of scans was fixed to 32 with a resolution of 4 cm−1 between 4000 and 400 cm−1. FTIR spectra were acquired and analyzed using OMNIC software (Thermo Scientific, USA). Each spectrum corresponds to the average of three measurements taken on different zones on the sample, chosen manually in order to verify the absence of spectral variation through the sample. No variation was observed.
For some experiments requiring labelling of the particles, such as degradation in cells or internalization ratio determination, the PCL particles were labelled with the red fluorescent dye Disperse Blue 14 (1,4-bis(methylamino)anthraquinone, ABCR #AB177338, λex 640 nm, λem 685 nm) by the solvent swelling method.69 Briefly, a 10 mg ml−1 solution of Disperse Blue 14 in THF was prepared. 1% in volume of this solution and 10% THF were added to the bead suspension, and the mixture was agitated on a rotary wheel at room temperature for 30 minutes. Two volumes of water were then added and the particles collected by centrifugation (30 minutes at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g). The pellet was then resuspended in water and the beads recollected by centrifugation. The final pellet was then resuspended in 20% ethanol.
000g). The pellet was then resuspended in water and the beads recollected by centrifugation. The final pellet was then resuspended in 20% ethanol.
2.2. Cell culture
The J774A.1 cell line (mouse macrophages) was purchased from the European cell culture collection (Salisbury, UK). Cells were routinely propagated in DMEM supplemented with 10% fetal bovine serum (FBS) in non-adherent flasks (Cellstar flasks for suspension culture, Greiner Bio One, Les Ulis, France). For routine culture, the cells were seeded at 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per ml and passaged two days later, with a cell density ranging from 800
000 cells per ml and passaged two days later, with a cell density ranging from 800![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 1
000 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per ml. For exposure to plastic particles and to limit the effects of cell growth, cells were seeded at 500
000 cells per ml. For exposure to plastic particles and to limit the effects of cell growth, cells were seeded at 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per ml in 6 or 12 wells plates, let settle and recover for 24 hours, and then exposed to the particles at 80 μg ml−1 for 24 hours before harvesting for the experiments. Proteomic experiments were carried out in 6 well plates, and all the other experiments in 12 well plates. The medium volume was adjusted to keep the same height across all cell culture formats. Cells were used at passage numbers from 5 to 15 post-reception from the repository. Cell viability was measured by the propidium iodide method,70 or with the SytoxGreen probe (Thermofisher S7020) using the protocol provided by the supplier. Those viability tests were systematically performed along functional tests to check that cell viability was always correct. Thus, depending on the fluorophores used in the functional tests, different fluorophores had to be used for the viability tests in order to avoid interferences.
000 cells per ml in 6 or 12 wells plates, let settle and recover for 24 hours, and then exposed to the particles at 80 μg ml−1 for 24 hours before harvesting for the experiments. Proteomic experiments were carried out in 6 well plates, and all the other experiments in 12 well plates. The medium volume was adjusted to keep the same height across all cell culture formats. Cells were used at passage numbers from 5 to 15 post-reception from the repository. Cell viability was measured by the propidium iodide method,70 or with the SytoxGreen probe (Thermofisher S7020) using the protocol provided by the supplier. Those viability tests were systematically performed along functional tests to check that cell viability was always correct. Thus, depending on the fluorophores used in the functional tests, different fluorophores had to be used for the viability tests in order to avoid interferences.
For aging experiments, the cells were seeded at 300![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per ml in DMEM containing 1% horse serum, to limit cell proliferation.71 The medium was changed every two days and the cells kept in culture for up to one week.
000 cells per ml in DMEM containing 1% horse serum, to limit cell proliferation.71 The medium was changed every two days and the cells kept in culture for up to one week.
Prolonged cell cultures in 1% horse serum were used to determine particle degradation. To this purpose, cells cultured in 12 well adherent plates were exposed to fluorescent particles (PS or PCL) for 24 hours. The particle-containing medium was removed, the cells rinsed twice with sterile PBS, and cultured in DMEM with a medium change every two days. To measure the cell associated fluorescence, the medium was removed, and the cell layer was then rinsed once with 1 ml PBS and the PBS removed. The cell layer was then lysed by the addition of 400 μl of 10 mM Hepes pH 7.5, and the cell lysate was collected. Its fluorescence was the measured using a DeNovix QFX instrument (excitation at 635 nm, emission collected in the 665–740 nm window). Untreated cell cultures of matched age were used to determine the cell autofluorescence background. This protocol is by construction independent of cell division, as each well is considered as a whole.
2.3. Proteomics
Proteomics was carried out essentially as described previously.41 However, the experimental details are given here for the sake of consistency.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 15 minutes) to pellet the nucleic acids. The supernatants were then stored at −20 °C until use.
000g, 15 minutes) to pellet the nucleic acids. The supernatants were then stored at −20 °C until use.As mentioned above, methylene blue was included in the cell lysis buffer. The other initiator solutions included a 1 M solution of sodium toluene sulfinate in water and a saturated water solution of diphenyliodonium chloride. The ready-to-use polyacrylamide solution consisted of 1.2 ml of a commercial 40% acrylamide/bis solution (37.5/1) to which 100 μl of diphenyliodonium chloride solution, 100 μl of sodium toluene sulfinate solution and 100 μl of water were added.
To the protein samples (15 μl), 5 μl of acrylamide solution were added and mixed by pipetting in a 500 μl conical polypropylene microtube. 100 μl of water-saturated butanol were then layered on top of the samples, and polymerization was carried out under a 1500 lumen 2700 K LED lamp for 2 hours, during which the initially blue gel solution discolored. At the end of the polymerization period, butanol was removed, and the gel plugs were fixed for 1 h with 200 μl of 30% ethanol 2% phosphoric acid, followed by 3× 15 minutes washes in 20% ethanol. The fixed gel plugs were then stored at −20 °C until use.
Gel plug processing, digestion, peptide extraction and nano LC–MS/MS were performed as previously described,73 without the robotic protein handling system and using a Q-Exactive HF-X mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). Further details are available in Methods S1.†
For protein identification, the MS/MS data were interpreted using a local Mascot server with MASCOT 2.6.2 algorithm (Matrix Science, London, UK) against an in-house database containing all Mus musculus and Rattus norvegicus entries from UniProtKB/SwissProt (version 2019_10, 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 313 sequences) and the corresponding 50
313 sequences) and the corresponding 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 313 reverse entries. Spectra were searched with a mass tolerance of 10 ppm for MS and 0.07 Da for MS/MS data, allowing a maximum of one trypsin missed cleavage. Trypsin was specified as enzyme. Acetylation of protein N-termini, carbamidomethylation of cysteine residues and oxidation of methionine residues were specified as variable modifications. Identification results were imported into Proline software version 2.1 (https://proline.profiproteomics.fr/) for validation. Peptide spectrum matches (PSM) with pretty rank equal to one were retained. The false discovery rate was then optimized to be below 1% at the PSM level using the Mascot adjusted E-value and below 1% at the protein level using the Mascot Mudpit score.
313 reverse entries. Spectra were searched with a mass tolerance of 10 ppm for MS and 0.07 Da for MS/MS data, allowing a maximum of one trypsin missed cleavage. Trypsin was specified as enzyme. Acetylation of protein N-termini, carbamidomethylation of cysteine residues and oxidation of methionine residues were specified as variable modifications. Identification results were imported into Proline software version 2.1 (https://proline.profiproteomics.fr/) for validation. Peptide spectrum matches (PSM) with pretty rank equal to one were retained. The false discovery rate was then optimized to be below 1% at the PSM level using the Mascot adjusted E-value and below 1% at the protein level using the Mascot Mudpit score.
Mass spectrometry data are available via ProteomeXchange with the identifier PXD049997.
Proteins were considered as significantly different if their p value in the Mann–Whitney U-test against control values was inferior to 0.05. The selected proteins were then subjected to pathway analysis using the DAVID tool,76 with a cutoff value set at an FDR of 0.1.
2.4. Phagocytosis assay
For this assay, the cells were first exposed to deep red-labelled PCL or PS particles at 80 μg ml−1. After 24 hours of exposure, the cells were then exposed to 0.5 μm latex beads (carboxylated surface, yellow green-labelled, from Polysciences, excitation 488 nm, emission 527/32 nm) for 3 hours at a concentration of 5 μg ml−1. After this second exposure, the cells were collected, rinsed twice with PBS, and analyzed for the two types of fluorescence (green and red) on a Melody flow cytometer. Controls with no latex beads showed no green fluorescence.2.5. Mitochondrial transmembrane potential assay
The mitochondrial transmembrane potential assay was performed essentially as described previously.42 After exposure to plastic beads (either PCL or PS, 80 μg ml−1 for 24 hours), rhodamine 123 (Rh123) was added to the cultures at an 80 nM final concentration (to avoid quenching77), and the cultures were further incubated at 37 °C for 30 minutes. At the end of this period, the cells were collected, washed in cold PBS containing 0.1% glucose, resuspended in PBS glucose and analyzed for green fluorescence (excitation 488 nm emission 525 nm) on a Melody flow cytometer. As a positive control, butanedione monoxime (BDM) was added at a 30 mM final concentration together with Rh123.78 As a negative control, carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) was added at a 5 μM final concentration together with Rh123.78 Control cells unexposed to Rh123 did not show any green fluorescence.2.6. Lysosomal assay
For the lysosomal function assay, the Lysosensor method was used, as described previously.42 After exposure to plastic beads (either PCL or PS, 80 μg ml−1 for 24 hours), the medium was removed, the cell layer was rinsed with complete culture medium and incubated with 1 μM Lysosensor Green (Molecular Probes) diluted in warm (37 °C) complete culture medium for 1 hour at 37 °C. At the end of this period, the cells were collected, washed in cold PBS containing 0.1% glucose, resuspended in PBS glucose and analyzed for green fluorescence (excitation 488 nm, emission 540 nm) on a Melody flow cytometer. Control cells unexposed to Lysosensor Green did not show any green fluorescence.2.7. Cytokine release assays
Cells were first exposed to nanoplastics (80 μg ml−1) for 24 hours. At the end of this exposure period, the culture medium was removed, the cell layer was rinsed with culture medium and fresh medium was added to the wells. In half of the wells LPS (1 ng ml−1) was added. After another 24 hours or 5 days, the medium was collected and analyzed for proinflammatory cytokines. Tumor necrosis factor and interleukin 6 levels were measured using the Cytometric Bead Array Mouse Inflammation Kit (catalog numbers 558299, 558301 and 558266, BD Biosciences, Le Pont de Claix), and analyzed with FCAP Array software (3.0, BD Biosciences) according to the manufacturer's instructions.2.8. Assay for oxidative stress
For the oxidative stress assay, a protocol based on the oxidation of dihydrorhodamine 123 (DHR123) was used, essentially as described previously.42 After exposure to plastic beads (80 μg ml−1 for 24 hours), the cells were treated in PBS containing 500 ng ml−1 DHR123 for 20 minutes at 37 °C. The cells were then harvested, washed in cold PBS containing 0.1% glucose, resuspended in PBS glucose and analyzed for green fluorescence (the same parameters as rhodamine 123) on a Melody flow cytometer. Menadione (applied on the cells for 2 hours prior to treatment with DHR123) was used as a positive control in a concentration range of 25–50 μM. Control cells unexposed to DHR123 did not show any green fluorescence.3. Results
3.1. Plastic bead characterization
PCL and PS particles were both described by their supplier as being 200 nm in diameter. Regarding PCL particles, chemical composition was validated through FTIR analysis, which showed the typical features of PCL (Fig. 1A). These features were bands at 2944 cm−1 and 2867 cm−1, corresponding to asymmetric and symmetric stretching of CH2 groups, and typical bands of esters, notably at 1723 cm−1, which is attributed to the stretching of the ester carbonyl (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), then at 1240 cm−1 and 1186 cm−1, corresponding to asymmetric and symmetric stretching of the C–O–C group. TEM images showed that PCL particles exhibited spherical morphologies with heterogenous size distribution (204 ± 85 nm) (Fig. 1B). The polydispersity index from DLS analysis of size distribution confirmed the heterogeneous size distribution, i.e., 17 ± 5%, and a Z-average of 197 ± 2 nm.
O), then at 1240 cm−1 and 1186 cm−1, corresponding to asymmetric and symmetric stretching of the C–O–C group. TEM images showed that PCL particles exhibited spherical morphologies with heterogenous size distribution (204 ± 85 nm) (Fig. 1B). The polydispersity index from DLS analysis of size distribution confirmed the heterogeneous size distribution, i.e., 17 ± 5%, and a Z-average of 197 ± 2 nm.
PS particles were also confirmed to being composed of PS via FTIR (Fig. 1C), with typical absorption bands from the aromatic C–H stretching vibrations at 3081, 3059, and 3025 cm−1 and asymmetrical and symmetrical stretching vibrations of methylene (–CH2) groups at 2921 and 2849 cm−1. Carbon–carbon stretches in aromatic ring were observed at 1601 cm−1 and 1583 cm−1, as expected,79 as well as at 1493 cm−1 and 1452 cm−1. The peak at 755 cm−1 is attributed to out-of-plane C–H deformation, while the peak at 697 cm−1, also related to this deformation, indicates ring curvature. TEM images showed that PS particles exhibited spherical morphologies with a rather homogeneous size distribution (303 ± 41 nm) (Fig. 1D).
The parameters obtained in aqueous suspensions for the two beads by DLS are summarized in Table 1.
| Beads | Average primary diameter (TEM) | Average hydrodynamic diameter | Polydispersity index | Zeta potential |
|---|---|---|---|---|
| The results are expressed as mean ± standard deviation (N = 3). | ||||
| PCL | 204 ± 85 nm | 197 ± 2 nm | 17 ± 5% | −10.4 ± 1.1 mV |
| PS | 191 ± 12 nm | 182 ± 12 nm | 8 ± 4% | −43.9 ± 5.7 mV |
The PS beads proved to be within the range of the nominal diameter given by the supplier (0.1–0.3 μm, mean 0.26 μm), while the electron microscopy data were much closer to the nominal values. The observed sizes led to a bead number/cell ratio higher than 9000 beads per cell (at 80 μg ml−1) for the PS beads and higher than 6000 beads per cell for the PCL beads, when corrected for the internalization ratios determined for both particles (Experiments S1†).
3.2. Viability of the plastic-treated cells
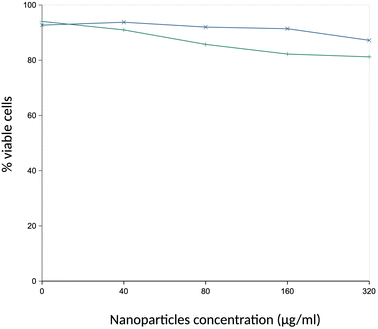
First, the toxic effects of the PCL or PS beads on J774A.1 cells were determined after a 24 hour exposure. The results, shown in Fig. 2, demonstrated a very low toxicity of the PCL and PS beads, with an LD50 that was greater than 300 μg ml−1. We decided to use an 80 μg ml−1 plastic concentration, i.e. the first concentration for which the viability was slightly but statistically significantly lower than the one of unexposed, control cells.3.3. Global analysis of the proteomic results
The shotgun proteomic analysis was able to detect and quantify 3402 proteins (Table S1†). As a first analysis, we analyzed the complete dataset via the PAST software, through a principal coordinates analysis. The results, displayed in Fig. 3, showed a good separation of all three groups (control, PS-treated and PCL-treated). The higher heterogeneity of the PS-treated group may be linked to the fact that some toxicity begins to appear, resulting in more divergent proteomes. The fact that the PCL and control groups were well separated in this global analysis also showed that PCL nanoparticles did modulate the cell proteome appreciably, although no toxicity is detected at the chosen concentration. The extent of the modulation is confirmed by the analysis of similarity test.80 This test showed a p-value (probability by random) of 0.0068 for the PCL vs. control comparison, 0.0071 for the PS vs. control comparison, and 0.0074 for the PCL vs. PS comparison. This indicated on the one hand significant proteome changes induced by the exposure to each plastic type, and on the other hand that the changes induced by PS and PCL are also different.As we have already published data on the proteomic response of macrophages to PS nanoparticles,41 we decided to focus the analysis on the cellular response of macrophages to PCL. Proteins modulated by the internalization of PCL particles were then selected on the basis of a Mann–Whitney U test in the comparison of plastic-treated cells to unexposed controls. This resulted in the selection of a total of 643 modulated proteins (Table S2†).
In order to gain further insight into the significance of the observed changes, this list of modulated proteins was used to perform pathways analyses by the DAVID software, and the results are shown in Table S3.† The pathways modulated by PS beads are shown in Table S4.† Regarding the pathways modulated by PCL, some indicated a global stress response, such as translation, nucleotide binding, nucleus, endoplasmic reticulum, mitochondria or carbon metabolism (Table S5†), which is expected for any cellular stress, while other pathways appeared more specific of cellular internalization of particles (e.g. lysosomes), and some specific for macrophages (e.g. innate immunity, hydrogen peroxide production). Compared to the PCL-modulated pathways, PS-modulated pathways also included mitochondria and lysosomes, but not endoplasmic reticulum. The proteasome-associated pathways were also highlighted for response to PS particles, as well as more metabolic pathways (e.g. pentose phosphate pathway, glycolysis). Macrophage-specific pathways did not appear as prominent in response to PS than in response to PCL.
This comprehensive analysis of the proteomic results led us to perform validation experiments on several functions.
3.4. Mitochondria and endoplasmic reticulum
Mitochondrial proteins represented an abundant class among the modulated proteins, with 80 proteins (Table S6†). Among those, 30 were increases and 50 decreases, suggesting a perturbation of the mitochondrial functions. In more detail, 10 were subunits of the respiratory chain (P19783, P62897, Q62425, Q91VR2, Q91WD5, Q9CQA3, Q9CR68, Q9CZ13, Q9D3D9, Q9DCX2) and one (Q9D1R1), a chaperone involved in the assembly of respiratory chain complex I. As the respiratory chain complexes I to III produce the proton gradient responsible for the mitochondrial transmembrane potential, which is in part dissipated by complex V, thereby ensuring the balance of the mitochondrial transmembrane potential, we decided to test this mitochondrial parameter. The results, displayed in Fig. 4A, showed no change of the mitochondrial transmembrane potential upon exposure of macrophages to PCL beads, and a slight and not statistically relevant decrease upon exposure of macrophages to PS beads. We did not check the effect of a longer exposure to PS beads. Interestingly, among the 32 proteins that show opposite modulations in response to PCL compared to PS, the three ATP synthase subunits selected in the proteomic screen show decreases in response to PCL particles and increases in response to PS particles. As ATP synthase dissipates the proton gradient to produce ATP, these changes in ATP synthase may be related to the differences in the mitochondrial transmembrane potential observed between the PCL-exposed cells and the PS-exposed cells.Endoplasmic reticulum proteins also represented an important class of modulated proteins in response to PCL particles (Table S7†), with 52 proteins (37 increases, 15 decreases). Among these 52 proteins, 5 were annotated directly or indirectly as related to the endoplasmic stress response (A1L1L2, Q5U2X6, Q5XI41, Q9QZN4, Q9Z2G6). This prompted us to test whether endoplasmic stress could be a determinant of toxicity for the PCL particles. To this purpose we used salubrinal, an inhibitor of the endoplasmic reticulum stress response,81 which is known to counteract the lethal effects of this stress on cells.82 The results, shown in Fig. 4B, indicated no effect of salubrinal on cell survival up to treatment with 320 μg ml−1 PCL particles for 24 hours.
3.5. Lysosomes and phagocytosis
In macrophages, phagocytosis is an integrated function coupling vesicle formation and transport to produce phagosomes, and finally fusion with lysosomes to produce phagolysosomes where the internalized material is subjected to degradation. Thus, in order to probe this function at the proteomic level, we integrated the proteins appearing with the keywords “lysosome” and “actin cytoskeleton” in a single protein list (Table S8†). We also added a protein appearing under the cluster “innate immune response”, but which is known to be implicated in the control of phagocytosis (STAP1, Q9JM90). This list encompassed 37 proteins (19 increases, 18 decreases). In the list of increased proteins were found two proteins participating in the acidification of the lysosomes, namely LAMP2 (O35114),83 and a catalytic subunit of the proton ATPase itself (P50516). This prompted us to test lysosomal acidification by the Lysosensor method. The results, displayed in Fig. 5A, showed a slight but not statistically significant (p = 0.1) increase in Lysosensor accumulation in response to PCL particles, and an even lower increase in response to PS. We then investigated how the uptake of PCL (or PS) particles influenced the residual phagocytic capability of macrophages. The results, displayed in Fig. 5B and C, showed a decrease in the phagocytic ability of PCL-treated cells, both in terms of percentage of phagocytic cells at 3 hours (Fig. 5B) and in terms of internalized beads' fluorescence (Fig. 5C), which was not observed for PS-treated cells.3.6. Immunity-related proteins and inflammation
The “innate immune response” keyword appeared in the pathway analysis, pointing to a subset of 47 modulated proteins (37 increases, 10 decreases, Table S9†). Among the increased proteins, we noticed both chains of cytochrome b-245, a critical component of NADPH oxidase, i.e. the enzyme producing superoxide in the phagosome. This suggested that the macrophages try to degrade the internalized plastic beads (both PCL and PS) by their classical defense mechanism.When examining in more detail the proteins present in this cluster, subgroups could be identified. One is related to proteins involved in antiviral responses (e.g. P0DOV2, P1103, P11928, Q3TL44, Q3UPF5, Q60710, Q64620, Q8BV66, Q8VCF0, Q8VI93, Q8VI94). The other important group included proteins that are known to modulate the inflammatory response. When trying to figure out the possible outcome of the modulations observed by proteomics in terms of cytokine production, we observed opposite trends. In response to the exposure to PCL particles, we observed increases in the abundances of negative regulators of inflammatory cytokine production (e.g. P25911,84 P54987,85 Q3UCV8,86 Q64281,87 Q9D8Y7,88 Q9R002 (ref. 89)), but also increases in positive regulators of inflammatory cytokine production (e.g. O88351,90 P56477,91 Q61107,92 Q9BDB7,93 Q9Z0E6 (ref. 92)) and decreases in negative regulators of cytokine production (Q9R007 (ref. 94)).
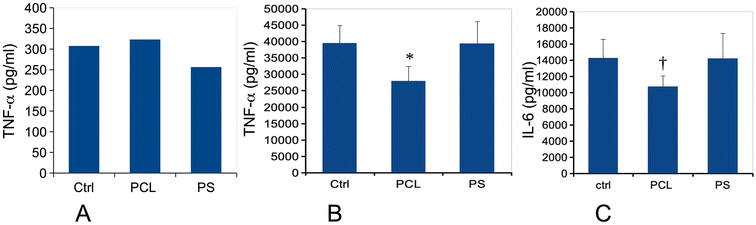
In view of these discrepant results, we tested the cytokine release of plastic-exposed macrophages under two conditions: either with plastic treatment only or in combination with a LPS stimulation. Responses to plastic alone allowed us to assess whether the plastics had an intrinsic pro-inflammatory effect or not. Responses to plastics followed by LPS allowed us to determine if the plastics could modulate further responses of the macrophages to bacteria. We tested two pro-inflammatory cytokines (TNF and IL-6). The results displayed in Fig. 6 showed first that PCL did not induce by itself any pro-inflammatory response. However, the combination of PCL-treatment with LPS stimulation resulted in a decrease of the magnitude of the response for both measured cytokines, although this decrease was at the limit of significance for IL-6. This effect was not observed in response to PS treatment.
3.7. Oxidative stress
In the course of the analysis of the proteomic data, we noticed that several proteins involved in the control of the cellular oxidative stress were modulated. These included the Cu–Zn superoxide dismutase (P08228, decreased), the mitochondrial Mn superoxide dismutase (P09671, increased) and peroxiredoxin 6 (O08709, decreased). Other proteins of this class (e.g. peroxiredoxins 1 to 5) did not show any significant modulation in response to treatment with PCL beads. These somewhat discrepant results, added to the induction of both cytochrome b245 subunits (ROS promoters), prompted us to study the level of cellular oxidative stress in response to PCL beads. The results, displayed in Fig. 7, showed a very slight but statistically significant decrease in the cellular oxidative stress in PCL-treated beads, while a slight but statistically significant increase was observed in PS-treated cells.3.8. Effect of lower particle concentration on cells
In order to determine to which extent the dose of particles is responsible for the effects that we could detect, we performed the same targeted experiments at a particle concentration of 10 μg ml−1, and the results are summarized in Table 2.| Parameter | Value | Value | Value | P-value T-test | P-value T-test |
|---|---|---|---|---|---|
| Control cells | PCL-treated cells | PS-treated cells | PCL vs. control | PS vs. control | |
| All results are expressed as mean ± standard deviation (N = 6). MFI: mean fluorescence index. | |||||
| Mitochondrial transmembrane potential (Rh123 fluorescence) | 57 ± 4.5 | 68.2 ± 3.4 | 58.2 ± 2.1 | 0.17 | 0.15 |
| Oxidative stress (DHR123 fluo.) | 23.8 ± 3.6 | 24.4 ± 1.4 | 17.5 ± 2.4 | 0.76 | 0.03 |
| Lysosensor signal | 174.3 ± 12.6 | 178.6 ± 14.2 | 159.2 ± 19.2 | 0.66 | 0.24 |
| % phagocytic cells | 90.1 ± 1.6 | 88.5 ± 0.5 | 91.1 ± 3.1 | 0.05 | 0.51 |
| MFI phagocytic cells | 250 ± 19 | 239 ± 13 | 263 ± 35 | 0.24 | 0.47 |
| Secreted IL-6 (pg ml−1) | 12.7 ± 2.9 | 10.7 ± 2.3 | 12.9 ± 3.1 | 0.21 | 0.90 |
| Secreted TNF (pg ml−1) | 1284 ± 280 | 1104 ± 258 | 1268 ± 289 | 0.27 | 0.93 |
| Secreted IL-6 with LPS stimul. (pg ml−1) | 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 270 ± 3510 270 ± 3510 |
16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 022 ± 3582 022 ± 3582 |
17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 070 ± 4296 070 ± 4296 |
0.55 | 0.93 |
| Secreted TNF with LPS stimul. (pg ml−1) | 105![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 343 ± 17 343 ± 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 530 530 |
118![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 843 ± 28 843 ± 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 666 666 |
106![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 908 ± 27 908 ± 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 445 445 |
0.35 | 0.91 |
These data showed that except for the level of oxidative stress, which is slightly but significantly decreased upon exposure to a low concentration of PS particles, this lower concentration of particles did not show any statistically significant changes in the measured parameters, which is consistent with dose-dependent affects.
3.9. Effect of particle aging in the cells
As PCL is known to be biodegradable,43 we first checked whether this was the case in our in vitro macrophage system. We thus used fluorescently labelled particles and used the fluorescence as a marker for the presence of the particles. Fluorescently-labelled PS particles were used as a low-degradation control. The results of these experiments are summarized in Table 3.| Bead type | Time in the presence of beads | ||
|---|---|---|---|
| 1 d | 4 d | 6 d | |
| The fluorescence intensities were normalized by the mean of the intensity observed after 1 day of loading. | |||
| PS beads | 100 ± 8 | 98 ± 4 | 66 ± 14 |
| PCL beads | 100 ± 17 | 99 ± 3 | 20 ± 3 |
The values obtained for polystyrene indicated the losses that are inherent to the experimental setup. The data, however, suggested an important degradation of the PCL beads at day 6, i.e. 1 day of exposure plus 5 days of recovery in a bead-free medium. We also showed that this degradation occurred in cells only, as PCL beads suspended in complete culture medium at 37 °C for 6 days did not show any fluorescence loss (Experiments S1†).
This prompted us to re-investigate the biological parameters already investigated just after a 24 hour exposure period. We did not detect any significant increase in oxidative stress levels in PCL-exposed cells compared to control cells after 5 days of recovery post-exposure. However, we detected significant changes in several other parameters, and the results are summarized in Table 4.
| Parameter | Value | Value | Value | P-value T-test | P-value T-test |
|---|---|---|---|---|---|
| Control cells | PCL-treated cells | PS-treated cells | PCL vs. control | PS vs. control | |
| All results are expressed as mean ± standard deviation (N = 6). MFI: mean fluorescence index. | |||||
| Mitochondrial transmembrane potential (Rh123 fluorescence) | 66.2 ± 7.4 | 81.4 ± 6.6 | 62.8 ± 4.5 | 0.02 | 0.46 |
| Oxidative stress (DHR123 fluo.) | 49.7 ± 1.8 | 49.4 ± 4.1 | 45.9 ± 3.4 | 0.88 | 0.11 |
| Lysosensor signal | 247.1 ± 52.6 | 289.6 ± 42.5 | 246.4 ± 36.8 | 0.26 | 0.98 |
| % phagocytic cells | 78.1 ± 1.5 | 80.9 ± 2.6 | 87.6 ± 1.7 | 0.05 | 0.000001 |
| MFI phagocytic cells | 445 ± 15 | 455 ± 39 | 598 ± 63 | 0.57 | 0.0014 |
| Secreted IL-6 (pg ml−1) | 119 ± 62 | 140 ± 72 | 274 ± 44 | 0.59 | 0.00075 |
| Secreted TNF (pg ml−1) | 1843 ± 268 | 1783 ± 217 | 2032 ± 227 | 0.68 | 0.22 |
| Secreted IL-6 with LPS stimul. (pg ml−1) | 1475 ± 284 | 844 ± 147 | 1324 ± 267 | 0.0015 | 0.4400 |
| Secreted TNF with LPS stimul. (pg ml−1) | 5361 ± 1570 | 3355 ± 670 | 5374 ± 1333 | 0.02 | 0.99 |
These results were in some cases consistent with the ones obtained immediately after exposure. A good example is represented by the lysosomal activity, which is increased by 12% immediately after exposure and by 17% after recovery. Other results are quite different from those obtained immediately after exposure. One striking example is the mitochondrial transmembrane potential, which is basically constant immediately after exposure and strongly increased after particle aging in the cells. Regarding phagocytosis, it was similar in PCL-treated and in unexposed cells, while PS-treated cells showed increased phagocytic indexes. These results were different from those obtained immediately after exposure. Oxidative stress levels were very similar in control, PCL-treated and PS-treated cells after aging of the particles in the cells. Regarding secretion of pro-inflammatory cytokines, a slight increase in the basal production of IL-6 (and less significantly for TNF) was observed in PS-treated cells where the NP stimulus persists, but not in PCL-treated cells. Oppositely, in the case of LPS-stimulated cells, a delayed anti-inflammatory effect was observed in PCL-treated cells, while the responses were similar to those of the control cells in PS-treated cells.
4. Discussion
Plastics are omnipresent in our daily lives because of their technical properties. They combine light weight to valuable mechanical properties, which range from deformability to mechanical strength depending on thickness and plastic type, and to good resistance to water and water-based environments, which makes them attractive for applications such as food packaging. However, the very same properties become a nuisance when degradation becomes required, i.e. at the end of life of the plastic items. This challenge is extremely important for single use plastics, where the end of life comes by definition quite early.A way to circumvent this degradation problem will be to use plastics that combine good technical properties with a chemical structure that renders them biodegradable. In this regard, purely aliphatic polyesters such as polylactide, polyhydroxybutyrate and polycaprolactone are promising, while mixed aromatic aliphatic polyesters such as poly-(ethylene terephthalate) are known to show poor biodegradability, albeit better than plastics with a pure polyolefinic backbone (e.g. polystyrene, polyacrylate, polyethylene).
Among this family of aliphatic polyesters, polycaprolactone (PCL) shows promising technical properties43,62 combined with good biodegradability under a variety of conditions.63,64,95 However, PCL is known to degrade very slowly in mammals, as confirmed by its medical uses.43,45,48 This poses the obvious question of its potential toxicological effects if particles of PCL cross the biological barriers and become internalized. To investigate this question we studied the effects of PCL nanoparticles on macrophages, i.e. the scavenger cell type in the body in charge of handling any particulates. As expected, the acute toxicity of PCL toward macrophages was very low, with the LD20 not reached at 300 μg ml−1. Indeed, the medical uses of PCL argue against any acute toxicity, but this does not mean that adverse effects will not exist. To investigate this question, we used a proteomic approach, which revealed wide changes in the proteome in response to the exposure to PCL (641 significant changes on 3402 quantified proteins). However, the changes observed at the proteome level may just reflect a homeostatic adaptation of the macrophages to the presence of the PCL particles in order to keep their full functions operational.
Indeed, homeostatic adaptations were observed for the endoplasmic reticulum (ER) and the mitochondria. Although numerous changes were observed for these two classes of proteins (80 for mitochondria, 52 for ER), we could not detect any implication of ER stress nor any mitochondrial depolarization or hyperpolarization in response to PCL after 24 hours. We also did not detect any damage to the lysosomes after 24 hours, opposite to what is observed with other nanomaterials such as silica.96
Interestingly, this absence of effects on mitochondria, while important changes in the mitochondrial proteome are observed, has also been demonstrated for macrophages treated with polystyrene particles41 or with polylactide particles.97 The same holds true for the endoplasmic reticulum. Regarding the lysosomes, we rather observed an increase in the lysosensor signal with the other plastics,97 which may be interpreted as an increase in the number of active lysosomes in response to plastic particle internalization. The fact that we did not observe this increase with PS particles at 80 μg ml−1 (this paper), compared to 50 μg ml−1,97 may be correlated with the onset of toxicity at 80 μg ml−1. It shall also be underlined that the absence of detected lysosomal damage with plastic particles may be due to their spherical shape, as sharp edges are known to induce lysosomal damage.98 This however shows that the polymers, including PCL, do not induce lysosomal damage per se.
While the general physiology of the macrophages was not affected by the treatment with PCL nanoparticles, the specialized macrophages functions were. We observed both a decrease in phagocytosis and a decrease of inflammatory cytokines in response to LPS-stimulation, suggesting that PCL-exposed cells will be less efficient than normal cells to respond to a bacterial infection. When comparing these results with those obtained on other plastics, polystyrene did not induce any decrease in the phagocytic activity but only a decrease in IL-6 release upon LPS stimulation. Polylactide particles, such as PCL, induced a decrease in the phagocytic activity and a decrease in IL-6 release.97 Once again, these results apply to the combined effects of the polymers and LPS on the cells. Regarding the intrinsic effects of the polymers, PCL did not induce any increase in TNF secretion, with polylactide and PS did.97 Interestingly, this increase in TNF secretion was observed at the nontoxic concentration of 50 μg ml−1,97 but not at the somewhat toxic dose of 80 μg ml−1 used here. Mechanical effects due to particles with sharp edges and inducing an inflammatory response98 cannot be excluded in a larger picture of the effects of plastic particles.
Oxidative stress was minimally impacted by treatment with the PCL nanoparticles, with a slight (−10%) but significant decrease in the level of oxidative stress. This is in contrast with the slight increase observed in response to PS particles, and the absence of oxidant response observed upon treatment with PLA nanoparticles. While oxidative stress is generally viewed as a deleterious signal, in macrophages it can be interpreted as a consequence of the oxidative burst that macrophages trigger upon ingestion of particulate material.99 Thus, the low level of oxidative stress observed in PCL-exposed cells, even below the control level, may well be another sign of the depression of the immune functions of the macrophages induced by PCL.
Quite interestingly, PCL beads were found to degrade in a few days within the macrophages, although more slowly than PLA beads.100 This reproduces the well-known behavior observed in medical nanoparticles.101 The degradation product of PCL is 6-hydroxyhexanoic acid,102 which is then oxidized in adipic acid.101 Adipic acid is known to have a complex metabolism including direct excretion and further degradation by beta-oxidation.103 However, its direct effects on mammalian cells are not known to date. Oppositely, 6-hydroxyhexanoic acid has been shown to exhibit a very low toxicity104 and to be the active component explaining the anti-inflammatory effects of Streptococcus gordonii supernatants.105,106 In line with the metabolic effects observed with 6-hydroxyhexanoic,106 the metabolism-associated proteins (Table S4†) that were most induced in response to PCL nanoparticles were the first enzymes of glycolysis (hexokinase, phosphofructokinase, glucokinase) and the fatty acid–CoA ligases, i.e. proteins implied in fatty acid metabolism.
This degradation of PCL nanoparticles within the macrophages prompted us to investigate the delayed effects on macrophages at this time point where the PCL nanoparticles have been extensively degraded, in comparison with PS nanoparticles which we know no to be degraded at this time point.42
Interestingly, we also detected an anti-inflammatory effect in our system, both immediately after exposure and a few days after exposure, when the PCL particles have been extensively degraded and the metabolite released. At this late time point, we also detected a metabolic effect in the form of mitochondrial hyperpolarization, which was neither observed immediately after exposure to PCL or PS nanoparticles, nor observed for PS beads in the delayed scheme. Oppositely, delayed effects, e.g. on phagocytosis and in basal pro-inflammatory cytokine production, were detected with PS particles but not with PCL particles, and contrast with the effects detected immediately after exposure. This may suggest that cellular stimulation occurs as a delayed response, which may explain why inflammatory responses are detected in vivo (e.g. in ref. 107), but not in vitro immediately after exposure. Overall, this further strengthens the interest in studies in which delayed effects are taken into account.
Last but certainly not least, it shall be kept in mind that the results described here have been obtained on nanospheres of pristine PCL. As such, this work describes the intrinsic effects of the pure polymer, i.e. the first, required step when evaluating the effects of micro and nanoplastics. While this work represents rather well PCL used in medical applications, it will not represent as well the complete effects of PCL micro and nanoplastics arising from other applications. This discrepancy arises from two main phenomena.
The first one is the presence of soluble chemicals in real-life particles, in addition to the polymer itself. These chemicals may be either additives used purposely for technological purposes, or environmental chemicals that adsorb on the plastic during its environmental weathering. Such eco-corona can be extremely complex,108 which will induce combinatorial explosion problems when trying to assess the toxicity of real-life plastic particles, which detailed chemical composition may be highly variable. Despite this complexity, scientists have tried to evaluate the potential synergies between plastics and their adsorbed chemicals. The results obtained so far show a variable response,109 and do not show any universal trend. In some cases a toxic synergy is observed,110 while in other cases plastics decrease the toxicity of chemicals,111,112 probably by a strong adsorption that prevents the release of the toxic chemicals, at least for the duration of the experiments.
The second one is the shape factor. In macrophages, it has been demonstrated on mineral nanomaterials such as silver nanowires113 or titanium dioxide,114 but also on cellulose,115,116 that a long aspect ratio is an intrinsic determinant for a pro-inflammatory effect. Sharp edges have also been described as a determinant for pro-inflammatory effects.117 Such effects have not been documented as well for microplastics so far, although some studies on fibers only are available.118 However, the fact that the only plastic particle for which we have observed a pro-inflammatory effect so far is PET nanopartcicles,119 which are not spherical and do show edges,120 argues in favor of the hypothesis that a spherical shape minimizes the effects of the nanomaterials on macrophages. Thus, our results may overall minimize the effects of real-life PCL plastics on macrophages.
5. Conclusions
The first important conclusion of this work on PCL particles is that they are slowly biodegradable within macrophages. However, this does not mean that they do not show immediate and delayed consequences on the physiology of macrophages. Comparison with PS nanoparticles shows that there are both common consequences and specific ones. Among the shared consequences, the induction of the fatty acyl CoA synthases can be noticed. As these enzymes are implied in both fatty acid synthesis and degradation, it can be suggested that they are implied in the biosynthesis of the lipids that are needed to build the vesicles containing the internalized particles. However, most of the parameters that we explored were altered differently by PCL and PS beads. This includes phagocytosis, cytokine production, and oxidative stress, and all these parameters suggested that PCL beads induced a reduced response of macrophages to further stimulation. Furthermore, delayed effects were observed after PCL internalization and degradation, but also for PS particles, which are not degraded. These results suggest that repeated exposure may be an interesting way to move forward, as responses to biodegradable plastics may differ even more from responses to non-biodegradable plastics.Data availability
The proteomic data have been deposited in the ProteomeXchange Consortium database121 and are available through the DOI: https://doi.org/10.6019/PXD049997. The non proteomic data have been deposited in BioStudies122 under the DOI: https://doi.org/10.6019/S-BSST1804.Author contributions
TR designed and supervised the study. MB, MC and DF carried out the characterization of the nanoparticles. VCF, HD and SC performed the proteomic experiments, which were then analyzed by TR and ED. VCF performed and interpreted the flow cytometry experiments. TR drafted the initial version of the manuscript, which was completed and amended by all co-authors, who approved the final version of the manuscript.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work used the flow cytometry facility supported by GRAL, a project of the University Grenoble Alpes graduate school (Ecoles Universitaires de Recherche) CBH-EUR-GS (ANR-17-EURE-0003), as well as the EM facilities at the Grenoble Instruct-ERIC Center (ISBG; UMS 3518 CNRS CEA-UGA-EMBL) with support from the French Infrastructure for Integrated Structural Biology (FRISBI; ANR-10-INSB-05-02) and GRAL, within the Grenoble Partnership for Structural Biology. The IBS Electron Microscope facility is supported by the Auvergne Rhône-Alpes Region, the Fonds Feder, the Fondation pour la Recherche Médicale and GIS-IBiSA. This work also used the platforms of the French Proteomic Infrastructure (ProFI) project (grant ANR-10-INBS-08-03). This work was carried out in the frame of the PlasticHeal project, which has received funding from the European Union's Horizon 2020 research and innovation programme under grant agreement No. 965196. This work was also supported by the ANR Plastox project (grant ANR-21-CE34-0028-04). We thank Guy Schoehn for the establishment of the IBS/ISBG EM facility.References
- R. Lehner, C. Weder, A. Petri-Fink and B. Rothen-Rutishauser, Emergence of Nanoplastic in the Environment and Possible Impact on Human Health, Environ. Sci. Technol., 2019, 53, 1748–1765 CrossRef CAS PubMed.
- P. J. Landrigan, J. J. Stegeman, L. E. Fleming, D. Allemand, D. M. Anderson, L. C. Backer, F. Brucker-Davis, N. Chevalier, L. Corra, D. Czerucka, M.-Y. D. Bottein, B. Demeneix, M. Depledge, D. D. Deheyn, C. J. Dorman, P. Fénichel, S. Fisher, F. Gaill, F. Galgani, W. H. Gaze, L. Giuliano, P. Grandjean, M. E. Hahn, A. Hamdoun, P. Hess, B. Judson, A. Laborde, J. McGlade, J. Mu, A. Mustapha, M. Neira, R. T. Noble, M. L. Pedrotti, C. Reddy, J. Rocklöv, U. M. Scharler, H. Shanmugam, G. Taghian, J. A. J. M. van de Water, L. Vezzulli, P. Weihe, A. Zeka, H. Raps and P. Rampal, Human Health and Ocean Pollution, Ann. Glob. Health, 2020, 86, 151 CrossRef PubMed.
- J. R. Jambeck, R. Geyer, C. Wilcox, T. R. Siegler, M. Perryman, A. Andrady, R. Narayan and K. L. Law, Plastic waste inputs from land into the ocean, Science, 2015, 347, 768–771 CrossRef CAS PubMed.
- J. G. B. Derraik, The pollution of the marine environment by plastic debris: a review, Mar. Pollut. Bull., 2002, 44, 842–852 CrossRef CAS PubMed.
- H. S. Charlton-Howard, A. L. Bond, J. Rivers-Auty and J. L. Lavers, ‘Plasticosis’: Characterising macro- and microplastic-associated fibrosis in seabird tissues, J. Hazard. Mater., 2023, 450, 131090 CrossRef CAS PubMed.
- K. Enders, R. Lenz, C. A. Stedmon and T. G. Nielsen, Abundance, size and polymer composition of marine microplastics ≥ 10 μm in the Atlantic Ocean and their modelled vertical distribution, Mar. Pollut. Bull., 2015, 100, 70–81 CrossRef CAS PubMed.
- A. Ter Halle, L. Jeanneau, M. Martignac, E. Jardé, B. Pedrono, L. Brach and J. Gigault, Nanoplastic in the North Atlantic Subtropical Gyre, Environ. Sci. Technol., 2017, 51, 13689–13697 CrossRef CAS PubMed.
- J. A. Brandon, A. Freibott and L. M. Sala, Patterns of suspended and salp-ingested microplastic debris in the North Pacific investigated with epifluorescence microscopy, Limnol. Oceanogr. Lett., 2020, 5, 46–53 CrossRef.
- M. Kedzierski, M. Palazot, L. Soccalingame, M. Falcou-Préfol, G. Gorsky, F. Galgani, S. Bruzaud and M. L. Pedrotti, Chemical composition of microplastics floating on the surface of the Mediterranean Sea, Mar. Pollut. Bull., 2022, 174, 113284 CrossRef CAS PubMed.
- E. S. Jones, S. W. Ross, C. M. Robertson and C. M. Young, Distributions of microplastics and larger anthropogenic debris in Norfolk Canyon, Baltimore Canyon, and the adjacent continental slope (Western North Atlantic Margin, U.S.A.), Mar. Pollut. Bull., 2021, 174, 113047 CrossRef PubMed.
- L. Cutroneo, M. Capello, A. Domi, S. Consani, P. Lamare, P. Coyle, V. Bertin, D. Dornic, A. Reboa, I. Geneselli and M. Anghinolfi, Microplastics in the abyss: a first investigation into sediments at 2443-m depth (Toulon, France), Environ. Sci. Pollut. Res., 2022, 29, 9375–9385 CrossRef CAS PubMed.
- T. Mani, A. Hauk, U. Walter and P. Burkhardt-Holm, Microplastics profile along the Rhine River, Sci. Rep., 2015, 5, 17988 CrossRef CAS PubMed.
- C. Scherer, A. Weber, F. Stock, S. Vurusic, H. Egerci, C. Kochleus, N. Arendt, C. Foeldi, G. Dierkes, M. Wagner, N. Brennholt and G. Reifferscheid, Comparative assessment of microplastics in water and sediment of a large European river, Sci. Total Environ., 2020, 738, 139866 CrossRef CAS PubMed.
- L. Weiss, W. Ludwig, S. Heussner, M. Canals, J.-F. Ghiglione, C. Estournel, M. Constant and P. Kerhervé, The missing ocean plastic sink: Gone with the rivers, Science, 2021, 373, 107–111 CrossRef CAS PubMed.
- C. J. Weber, C. Opp, J. A. Prume, M. Koch, T. J. Andersen and P. Chifflard, Deposition and in-situ translocation of microplastics in floodplain soils, Sci. Total Environ., 2022, 819, 152039 CrossRef CAS PubMed.
- H. Golwala, X. Zhang, S. M. Iskander and A. L. Smith, Solid waste: An overlooked source of microplastics to the environment, Sci. Total Environ., 2021, 769, 144581 CrossRef CAS PubMed.
- A. Chamas, H. Moon, J. Zheng, Y. Qiu, T. Tabassum, J. H. Jang, M. Abu-Omar, S. L. Scott and S. Suh, Degradation Rates of Plastics in the Environment, ACS Sustainable Chem. Eng., 2020, 8, 3494–3511 CrossRef CAS.
- L. Lei, M. Liu, Y. Song, S. Lu, J. Hu, C. Cao, B. Xie, H. Shi and D. He, Polystyrene (nano)microplastics cause size-dependent neurotoxicity, oxidative damage and other adverse effects in Caenorhabditis elegans, Environ. Sci.: Nano, 2018, 5, 2009–2020 RSC.
- X. Jiang, Y. Chang, T. Zhang, Y. Qiao, G. Klobučar and M. Li, Toxicological effects of polystyrene microplastics on earthworm (Eisenia fetida), Environ. Pollut., 2020, 259, 113896 CrossRef CAS PubMed.
- R. Sussarellu, M. Suquet, Y. Thomas, C. Lambert, C. Fabioux, M. E. J. Pernet, N. Le Goïc, V. Quillien, C. Mingant, Y. Epelboin, C. Corporeau, J. Guyomarch, J. Robbens, I. Paul-Pont, P. Soudant and A. Huvet, Oyster reproduction is affected by exposure to polystyrene microplastics, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 2430–2435 CrossRef CAS PubMed.
- Z. Liu, P. Yu, M. Cai, D. Wu, M. Zhang, Y. Huang and Y. Zhao, Polystyrene nanoplastic exposure induces immobilization, reproduction, and stress defense in the freshwater cladoceran Daphnia pulex, Chemosphere, 2019, 215, 74–81 CrossRef CAS PubMed.
- Y. Song, C. Cao, R. Qiu, J. Hu, M. Liu, S. Lu, H. Shi, K. M. Raley-Susman and D. He, Uptake and adverse effects of polyethylene terephthalate microplastics fibers on terrestrial snails (Achatina fulica) after soil exposure, Environ. Pollut., 2019, 250, 447–455 CrossRef CAS PubMed.
- A. H. D'Costa, Microplastics in decapod crustaceans: Accumulation, toxicity and impacts, a review, Sci. Total Environ., 2022, 832, 154963 CrossRef PubMed.
- O. Pikuda, E. Roubeau Dumont, Q. Chen, J.-R. Macairan, S. A. Robinson, D. Berk and N. Tufenkji, Toxicity of microplastics and nanoplastics to Daphnia magna: Current status, knowledge gaps and future directions, TrAC, Trends Anal. Chem., 2023, 167, 117208 CrossRef CAS.
- A. Muhammad, X. Zhou, J. He, N. Zhang, X. Shen, C. Sun, B. Yan and Y. Shao, Toxic effects of acute exposure to polystyrene microplastics and nanoplastics on the model insect, silkworm Bombyx mori, Environ. Pollut., 2021, 285, 117255 CrossRef CAS PubMed.
- M. Alaraby, D. Abass, J. Domenech, A. Hernández and R. Marcos, Hazard assessment of ingested polystyrene nanoplastics in Drosophila larvae, Environ. Sci.: Nano, 2022, 9, 1845–1857 RSC.
- N. R. Brun, P. van Hage, E. R. Hunting, A.-P. G. Haramis, S. C. Vink, M. G. Vijver, M. J. M. Schaaf and C. Tudorache, Polystyrene nanoplastics disrupt glucose metabolism and cortisol levels with a possible link to behavioural changes in larval zebrafish, Commun. Biol., 2019, 2, 382 CrossRef PubMed.
- I. Brandts, M. Garcia-Ordoñez, L. Tort, M. Teles and N. Roher, Polystyrene nanoplastics accumulate in ZFL cell lysosomes and in zebrafish larvae after acute exposure, inducing a synergistic immune response in vitro without affecting larval survival in vivo, Environ. Sci.: Nano, 2020, 7, 2410–2422 RSC.
- W. Gu, S. Liu, L. Chen, Y. Liu, C. Gu, H. Ren and B. Wu, Single-Cell RNA Sequencing Reveals Size-Dependent Effects of Polystyrene Microplastics on Immune and Secretory Cell Populations from Zebrafish Intestines, Environ. Sci. Technol., 2020, 54, 3417–3427 CrossRef CAS PubMed.
- X. Li, T. Zhang, W. Lv, H. Wang, H. Chen, Q. Xu, H. Cai and J. Dai, Intratracheal administration of polystyrene microplastics induces pulmonary fibrosis by activating oxidative stress and Wnt/β-catenin signaling pathway in mice, Ecotoxicol. Environ. Saf., 2022, 232, 113238 CrossRef CAS PubMed.
- V. Stock, L. Böhmert, E. Lisicki, R. Block, J. Cara-Carmona, L. K. Pack, R. Selb, D. Lichtenstein, L. Voss, C. J. Henderson, E. Zabinsky, H. Sieg, A. Braeuning and A. Lampen, Uptake and effects of orally ingested polystyrene microplastic particles in vitro and in vivo, Arch. Toxicol., 2019, 93, 1817–1833 CrossRef CAS PubMed.
- J. Domenech, A. Hernández, L. Rubio, R. Marcos and C. Cortés, Interactions of polystyrene nanoplastics with in vitro models of the human intestinal barrier, Arch. Toxicol., 2020, 94, 2997–3012 CrossRef CAS PubMed.
- S. Ballesteros, J. Domenech, I. Barguilla, C. Cortés, R. Marcos and A. Hernández, Genotoxic and immunomodulatory effects in human white blood cells after ex vivo exposure to polystyrene nanoplastics, Environ. Sci.: Nano, 2020, 7, 3431–3446 RSC.
- C. Meindl, K. Öhlinger, V. Zrim, T. Steinkogler and E. Fröhlich, Screening for Effects of Inhaled Nanoparticles in Cell Culture Models for Prolonged Exposure, Nanomaterials, 2021, 11, 606 CrossRef CAS PubMed.
- J. Antunes, P. Sobral, M. Martins and V. Branco, Nanoplastics activate a TLR4/p38-mediated pro-inflammatory response in human intestinal and mouse microglia cells, Environ. Toxicol. Pharmacol., 2023, 104, 104298 CrossRef CAS PubMed.
- M. Zhang, J. Li, G. Xing, R. He, W. Li, Y. Song and H. Guo, Variation in the internalization of differently sized nanoparticles induces different DNA-damaging effects on a macrophage cell line, Arch. Toxicol., 2011, 85, 1575–1588 CrossRef CAS PubMed.
- B. Prietl, C. Meindl, E. Roblegg, T. R. Pieber, G. Lanzer and E. Fröhlich, Nano-sized and micro-sized polystyrene particles affect phagocyte function, Cell Biol. Toxicol., 2014, 30, 1–16 CrossRef CAS PubMed.
- V. Paget, S. Dekali, T. Kortulewski, R. Grall, C. Gamez, K. Blazy, O. Aguerre-Chariol, S. Chevillard, A. Braun, P. Rat and G. Lacroix, Specific Uptake and Genotoxicity Induced by Polystyrene Nanobeads with Distinct Surface Chemistry on Human Lung Epithelial Cells and Macrophages, PLoS One, 2015, 10, e0123297 CrossRef PubMed.
- I. Florance, S. Ramasubbu, A. Mukherjee and N. Chandrasekaran, Polystyrene nanoplastics dysregulate lipid metabolism in murine macrophages in vitro, Toxicology, 2021, 458, 152850 CrossRef CAS PubMed.
- Q. Hu, H. Wang, C. He, Y. Jin and Z. Fu, Polystyrene nanoparticles trigger the activation of p38 MAPK and apoptosis via inducing oxidative stress in zebrafish and macrophage cells, Environ. Pollut., 2021, 269, 116075 CrossRef CAS PubMed.
- V. Collin-Faure, B. Dalzon, J. Devcic, H. Diemer, S. Cianférani and T. Rabilloud, Does size matter? A proteomics-informed comparison of the effects of polystyrene beads of different sizes on macrophages, Environ. Sci.: Nano, 2022, 9, 2827–2840 RSC.
- V. Collin-Faure, M. Vitipon, A. Torres, O. Tanyeres, B. Dalzon and T. Rabilloud, The internal dose makes the poison: higher internalization of polystyrene particles induce increased perturbation of macrophages, Front. Immunol., 2023, 14, 1092743 CrossRef CAS PubMed.
- M. A. Woodruff and D. W. Hutmacher, The return of a forgotten polymer—Polycaprolactone in the 21st century, Prog. Polym. Sci., 2010, 35, 1217–1256 CrossRef CAS.
- T. K. Dash and V. B. Konkimalla, Poly-ε-caprolactone based formulations for drug delivery and tissue engineering: A review, J. Controlled Release, 2012, 158, 15–33 CrossRef CAS PubMed.
- B. Nottelet, V. Darcos and J. Coudane, Aliphatic polyesters for medical imaging and theranostic applications, Eur. J. Pharm. Biopharm., 2015, 97, 350–370 CrossRef CAS PubMed.
- P. Grossen, D. Witzigmann, S. Sieber and J. Huwyler, PEG-PCL-based nanomedicines: A biodegradable drug delivery system and its application, J. Controlled Release, 2017, 260, 46–60 CrossRef CAS PubMed.
- N. Amiryaghoubi, M. Fathi, J. Barar, H. Omidian and Y. Omidi, Advanced nanoscale drug delivery systems for bone cancer therapy, Biochim. Biophys. Acta, Mol. Basis Dis., 2023, 1869, 166739 CrossRef CAS PubMed.
- J. C. Middleton and A. J. Tipton, Synthetic biodegradable polymers as orthopedic devices, Biomaterials, 2000, 21, 2335–2346 CrossRef CAS PubMed.
- V. P. Nirwan, T. Kowalczyk, J. Bar, M. Buzgo, E. Filová and A. Fahmi, Advances in Electrospun Hybrid Nanofibers for Biomedical Applications, Nanomaterials, 2022, 12, 1829 CrossRef CAS PubMed.
- D. Ege and V. Hasirci, Is 3D Printing Promising for Osteochondral Tissue Regeneration?, ACS Appl. Bio Mater., 2023, 6, 1431–1444 CrossRef CAS PubMed.
- H. Sun, L. Mei, C. Song, X. Cui and P. Wang, The in vivo degradation, absorption and excretion of PCL-based implant, Biomaterials, 2006, 27, 1735–1740 CrossRef CAS PubMed.
- C. X. F. Lam, D. W. Hutmacher, J. Schantz, M. A. Woodruff and S. H. Teoh, Evaluation of polycaprolactone scaffold degradation for 6 months in vitro and in vivo, J. Biomed. Mater. Res., 2009, 90A, 906–919 CrossRef CAS PubMed.
- A. Sadeghi, S. M. Mousavi, E. Saljoughi and S. Kiani, Biodegradable membrane based on polycaprolactone/polybutylene succinate: Characterization and performance evaluation in wastewater treatment, J. Appl. Polym. Sci., 2021, 138, 50332 CrossRef CAS.
- S. Khalid, L. Yu, M. Feng, L. Meng, Y. Bai, A. Ali, H. Liu and L. Chen, Development and characterization of biodegradable antimicrobial packaging films based on polycaprolactone, starch and pomegranate rind hybrids, Food Packag. Shelf Life, 2018, 18, 71–79 CrossRef.
- J. S. Lyu, J.-S. Lee and J. Han, Development of a biodegradable polycaprolactone film incorporated with an antimicrobial agent via an extrusion process, Sci. Rep., 2019, 9, 20236 CrossRef CAS PubMed.
- T.-T. Li, H. Zhang, S.-Y. Huang, X. Pei, Q. Lin, S. Tian, Z. Ma and J.-H. Lin, Preparation and property evaluations of PCL/PLA composite films, J. Polym. Res., 2021, 28, 156 CrossRef CAS.
- M. Thakur, I. Majid, S. Hussain and V. Nanda, Poly(ε-caprolactone): A potential polymer for biodegradable food packaging applications, Packag. Technol. Sci., 2021, 34, 449–461 CrossRef CAS.
- W. Kim, T. Han, Y. Gwon, S. Park, H. Kim and J. Kim, Biodegradable and Flexible Nanoporous Films for Design and Fabrication of Active Food Packaging Systems, Nano Lett., 2022, 22, 3480–3487 CrossRef CAS PubMed.
- K. İlaslan, F. Tornuk and M. Z. Durak, Development of polycaprolactone biodegradable films reinforced with silver-doped organoclay and effect on the microbiological quality of ground beef meat, J. Food Process. Preserv., 2022, 46, e16862 Search PubMed.
- E. Drago, R. Campardelli, A. Barbucci and P. Perego, Polycaprolactone sub-micrometric fibers optimization for primary packaging loaded with fatty acids as natural phase change materials, J. Food Eng., 2023, 358, 111680 CrossRef CAS.
- S.-H. Pyo, J. H. Park, V. Srebny and R. Hatti-Kaul, A sustainable synthetic route for biobased 6-hydroxyhexanoic acid, adipic acid and ε-caprolactone by integrating bio- and chemical catalysis, Green Chem., 2020, 22, 4450–4455 RSC.
- V. Massardier-Nageotte, C. Pestre, T. Cruard-Pradet and R. Bayard, Aerobic and anaerobic biodegradability of polymer films and physico-chemical characterization, Polym. Degrad. Stab., 2006, 91, 620–627 CrossRef CAS.
- A. S. Al Hosni, J. K. Pittman and G. D. Robson, Microbial degradation of four biodegradable polymers in soil and compost demonstrating polycaprolactone as an ideal compostable plastic, Waste Manage., 2019, 97, 105–114 CrossRef CAS PubMed.
- A. Richert and G. B. Dąbrowska, Enzymatic degradation and biofilm formation during biodegradation of polylactide and polycaprolactone polymers in various environments, Int. J. Biol. Macromol., 2021, 176, 226–232 CrossRef CAS PubMed.
- M. Rutkowska, M. Jastrzębska and H. Janik, Biodegradation of polycaprolactone in sea water, React. Funct. Polym., 1998, 38, 27–30 CrossRef CAS.
- C. Aude-Garcia, B. Dalzon, J. L. Ravanat, V. Collin-Faure, H. Diemer, J. M. Strub, S. Cianferani, A. Van Dorsselaer, M. Carriere and T. Rabilloud, A combined proteomic and targeted analysis unravels new toxic mechanisms for zinc oxide nanoparticles in macrophages, J. Proteomics, 2016, 134, 174–185 CrossRef CAS PubMed.
- B. Dalzon, C. Aude-Garcia, V. Collin-Faure, H. Diemer, D. Béal, F. Dussert, D. Fenel, G. Schoehn, S. Cianférani, M. Carrière and T. Rabilloud, Differential proteomics highlights macrophage-specific responses to amorphous silica nanoparticles, Nanoscale, 2017, 9, 9641–9658 RSC.
- A. Torres, V. Collin-Faure, H. Diemer, C. Moriscot, D. Fenel, B. Gallet, S. Cianférani, J.-A. Sergent and T. Rabilloud, Repeated Exposure of Macrophages to Synthetic Amorphous Silica Induces Adaptive Proteome Changes and a Moderate Cell Activation, Nanomaterials, 2022, 12, 1424 CrossRef CAS PubMed.
- T. Behnke, C. Würth, K. Hoffmann, M. Hübner, U. Panne and U. Resch-Genger, Encapsulation of Hydrophobic Dyes in Polystyrene Micro- and Nanoparticles via Swelling Procedures, J. Fluoresc., 2011, 21, 937–944 CrossRef CAS PubMed.
- A. Torres, V. Collin-Faure, D. Fenel, J.-A. Sergent and T. Rabilloud, About the Transient Effects of Synthetic Amorphous Silica: An In Vitro Study on Macrophages, Int. J. Mol. Sci., 2022, 24, 220 CrossRef PubMed.
- B. Dalzon, A. Torres, J. Devcic, D. Fenel, J.-A. Sergent and T. Rabilloud, A Low-Serum Culture System for Prolonged in Vitro Toxicology Experiments on a Macrophage System, Front. Toxicol., 2021, 3, 780778 CrossRef PubMed.
- L. Muller, L. Fornecker, M. Chion, A. Van Dorsselaer, S. Cianférani, T. Rabilloud and C. Carapito, Extended investigation of tube-gel sample preparation: a versatile and simple choice for high throughput quantitative proteomics, Sci. Rep., 2018, 8, 8260 CrossRef PubMed.
- C. Cavazza, V. Collin-Faure, J. Pérard, H. Diemer, S. Cianférani, T. Rabilloud and E. Darrouzet, Proteomic analysis of Rhodospirillum rubrum after carbon monoxide exposure reveals an important effect on metallic cofactor biosynthesis, J. Proteomics, 2022, 250, 104389 CrossRef CAS PubMed.
- T. Lyubimova, S. Caglio, C. Gelfi, P. G. Righetti and T. Rabilloud, Photopolymerization of polyacrylamide gels with methylene blue, Electrophoresis, 1993, 14, 40–50 CrossRef CAS PubMed.
- O. Hammer, D. A. T. Harper and P. D. Ryan, Paleontological statistics software package for education and data analysis, Palaeontol. Electronica, 2001, 4, 9 Search PubMed.
- D. W. Huang, B. T. Sherman and R. A. Lempicki, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources, Nat. Protoc., 2009, 4, 44–57 CrossRef CAS PubMed.
- S. W. Perry, J. P. Norman, J. Barbieri, E. B. Brown and H. A. Gelbard, Mitochondrial membrane potential probes and the proton gradient: a practical usage guide, BioTechniques, 2011, 50, 98–115 CrossRef CAS PubMed.
- J. F. Keij, C. Bell-Prince and J. A. Steinkamp, Staining of mitochondrial membranes with 10-nonyl acridine orange, MitoFluor Green, and MitoTracker Green is affected by mitochondrial membrane potential altering drugs, Cytometry, 2000, 39, 203–210 CrossRef CAS PubMed.
- D. Olmos, E. V. Martín and J. González-Benito, New molecular-scale information on polystyrene dynamics in PS and PS–BaTiO 3 composites from FTIR spectroscopy, Phys. Chem. Chem. Phys., 2014, 16, 24339–24349 RSC.
- K. R. Clarke, Non-parametric multivariate analyses of changes in community structure, Aust. J. Ecol., 1993, 18, 117–143 CrossRef.
- M. Boyce, K. F. Bryant, C. Jousse, K. Long, H. P. Harding, D. Scheuner, R. J. Kaufman, D. Ma, D. M. Coen, D. Ron and J. Yuan, A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress, Science, 2005, 307, 935–939 CrossRef CAS PubMed.
- T. L. Pan, P. W. Wang, Y. C. Hung, C. H. Huang and K. M. Rau, Proteomic analysis reveals tanshinone IIA enhances apoptosis of advanced cervix carcinoma CaSki cells through mitochondria intrinsic and endoplasmic reticulum stress pathways, Proteomics, 2013, 13, 3411–3423 CrossRef CAS PubMed.
- J. Zhang, W. Zeng, Y. Han, W.-R. Lee, J. Liou and Y. Jiang, Lysosomal LAMP proteins regulate lysosomal pH by direct inhibition of the TMEM175 channel, Mol. Cell, 2023, 83, 2524–2539 CrossRef CAS PubMed.
- S. Keck, M. Freudenberg and M. Huber, Activation of murine macrophages via TLR2 and TLR4 is negatively regulated by a Lyn/PI3K module and promoted by SHIP1, J. Immunol., 2010, 184, 5809–5818 CrossRef CAS PubMed.
- Y. Li, P. Zhang, C. Wang, C. Han, J. Meng, X. Liu, S. Xu, N. Li, Q. Wang, X. Shi and X. Cao, Immune responsive gene 1 (IRG1) promotes endotoxin tolerance by increasing A20 expression in macrophages through reactive oxygen species, J. Biol. Chem., 2013, 288, 16225–16234 CrossRef CAS PubMed.
- R. B. Damgaard, J. A. Walker, P. Marco-Casanova, N. V. Morgan, H. L. Titheradge, P. R. Elliott, D. McHale, E. R. Maher, A. N. J. McKenzie and D. Komander, The Deubiquitinase OTULIN Is an Essential Negative Regulator of Inflammation and Autoimmunity, Cell, 2016, 166, 1215–1230 CrossRef CAS PubMed.
- Z. Jiang, J.-J. Qin, Y. Zhang, W.-L. Cheng, Y.-X. Ji, F.-H. Gong, X.-Y. Zhu, Y. Zhang, Z.-G. She, Z. Huang and H. Li, LILRB4 deficiency aggravates the development of atherosclerosis and plaque instability by increasing the macrophage inflammatory response via NF-κB signaling, Clin. Sci., 2017, 131, 2275–2288 CrossRef CAS PubMed.
- H. Sun, S. Gong, R. J. Carmody, A. Hilliard, L. Li, J. Sun, L. Kong, L. Xu, B. Hilliard, S. Hu, H. Shen, X. Yang and Y. H. Chen, TIPE2, a negative regulator of innate and adaptive immunity that maintains immune homeostasis, Cell, 2008, 133, 415–426 CrossRef CAS PubMed.
- Q. Yin, D. P. Sester, Y. Tian, Y.-S. Hsiao, A. Lu, J. A. Cridland, V. Sagulenko, S. J. Thygesen, D. Choubey, V. Hornung, T. Walz, K. J. Stacey and H. Wu, Molecular mechanism for p202-mediated specific inhibition of AIM2 inflammasome activation, Cell Rep., 2013, 4, 327–339 CrossRef CAS PubMed.
- Y. Tsuchiya, T. Asano, K. Nakayama, T. Kato, M. Karin and H. Kamata, Nuclear IKKbeta is an adaptor protein for IkappaBalpha ubiquitination and degradation in UV-induced NF-kappaB activation, Mol. Cell, 2010, 39, 570–582 CrossRef CAS PubMed.
- A. Takaoka, H. Yanai, S. Kondo, G. Duncan, H. Negishi, T. Mizutani, S.-I. Kano, K. Honda, Y. Ohba, T. W. Mak and T. Taniguchi, Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors, Nature, 2005, 434, 243–249 CrossRef CAS PubMed.
- D. M. Pilla, J. A. Hagar, A. K. Haldar, A. K. Mason, D. Degrandi, K. Pfeffer, R. K. Ernst, M. Yamamoto, E. A. Miao and J. Coers, Guanylate binding proteins promote caspase-11-dependent pyroptosis in response to cytoplasmic LPS, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 6046–6051 CrossRef CAS PubMed.
- H. Jiang, L. Tsang, H. Wang and C. Liu, IFI44L as a Forward Regulator Enhancing Host Antituberculosis Responses, J. Immunol. Res., 2021, 2021, 5599408 Search PubMed.
- B. Joyce-Shaikh, M. E. Bigler, C.-C. Chao, E. E. Murphy, W. M. Blumenschein, I. E. Adamopoulos, P. G. Heyworth, S. Antonenko, E. P. Bowman, T. K. McClanahan, J. H. Phillips and D. J. Cua, Myeloid DAP12-associating lectin (MDL)-1 regulates synovial inflammation and bone erosion associated with autoimmune arthritis, J. Exp. Med., 2010, 207, 579–589 CrossRef CAS PubMed.
- M. Suzuki, Y. Tachibana, K. Oba, R. Takizawa and K. Kasuya, Microbial degradation of poly(ε-caprolactone) in a coastal environment, Polym. Degrad. Stab., 2018, 149, 1–8 CrossRef CAS.
- L. M. Costantini, R. M. Gilberti and D. A. Knecht, The Phagocytosis and Toxicity of Amorphous Silica, PLoS One, 2011, 6, e14647 CrossRef CAS PubMed.
- V. Collin-Faure, M. Vitipon, H. Diemer, S. Cianferani, E. Darrouzet and T. Rabilloud, Biobased, Biodegradable but not bio-neutral: about the effects of polylactic acid nanoparticles on macrophages, Environ. Sci.: Nano, 2024, 11, 4102–4116 RSC.
- V. Hornung, F. Bauernfeind, A. Halle, E. O. Samstad, H. Kono, K. L. Rock, K. A. Fitzgerald and E. Latz, Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization, Nat. Immunol., 2008, 9, 847–856 CrossRef CAS PubMed.
- R. B. Johnston, C. A. Godzik and Z. A. Cohn, Increased superoxide anion production by immunologically activated and chemically elicited macrophages, J. Exp. Med., 1978, 148, 115–127 CrossRef CAS PubMed.
- V. Collin-Faure, M. Vitipon, H. Diemer, S. Cianferani, E. Darrouzet and T. Rabilloud, Biobased, Biodegradable but not bio-neutral: about the effects of polylactic acid nanoparticles on macrophages, bioRxiv, 2024, preprint, DOI:10.1101/2024.07.15.603484.
- U. Edlund and A.-C. Albertsson, in Degradable Aliphatic Polyesters, Springer Berlin Heidelberg, Berlin, Heidelberg, 2002, vol. 157, pp. 67–112 Search PubMed.
- T. Douki, V. Bard, M. Boulée and M. Carrière, Extensive HPLC-tandem mass spectrometry characterization of soluble degradation products of biodegradable nanoplastics under environmentally relevant temperature and irradiation conditions, Environ. Sci.: Nano, 2024, 11, 3956–3965 RSC.
- I. I. Rusoff, R. R. Baldwin, F. J. Domingues, C. Monder, W. J. Ohan and R. Thiessen, Intermediary metabolism of adipic acid, Toxicol. Appl. Pharmacol., 1960, 2, 316–330 CrossRef CAS PubMed.
- A. Orchel, K. Jelonek, J. Kasperczyk and Z. Dzierzewicz, Growth of human fibroblasts in the presence of 6-hydroxyhexanoic acid, Acta Pol. Pharm., 2010, 67, 710–712 CAS.
- Y. Shu, C. Upara, Q. Ding, M. Zhu, E. Zeng, J. A. Banas and L. Hong, Spent culture supernatant of Streptococcus gordonii mitigates inflammation of human periodontal cells and inhibits proliferation of pathogenic oral microbes, J. Periodontol., 2023, 94, 575–585 CrossRef CAS PubMed.
- S. C. Sebag, M. Hao, Q. Qian, C. Upara, Q. Ding, M. Zhu, J. A. Banas, H. Cao, L. Hong and L. Yang, A medium chain fatty acid, 6-hydroxyhexanoic acid (6-HHA), protects against obesity and insulin resistance, Acta Pharm. Sin. B, 2024, 14, 1892–1894 CrossRef CAS PubMed.
- N. Ali, J. Katsouli, E. L. Marczylo, T. W. Gant, S. Wright and J. Bernardino de la Serna, The potential impacts of micro-and-nano plastics on various organ systems in humans, EBioMedicine, 2024, 99, 104901 CrossRef CAS PubMed.
- J. Cao, Q. Yang, J. Jiang, T. Dalu, A. Kadushkin, J. Singh, R. Fakhrullin, F. Wang, X. Cai and R. Li, Coronas of micro/nano plastics: a key determinant in their risk assessments, Part. Fibre Toxicol., 2022, 19, 55 CrossRef PubMed.
- L. Zimmermann, S. Göttlich, J. Oehlmann, M. Wagner and C. Völker, What are the drivers of microplastic toxicity? Comparing the toxicity of plastic chemicals and particles to Daphnia magna, Environ. Pollut., 2020, 267, 115392 CrossRef CAS PubMed.
- I. Barguilla, J. Domenech, L. Rubio, R. Marcos and A. Hernández, Nanoplastics and Arsenic Co-Exposures Exacerbate Oncogenic Biomarkers under an In Vitro Long-Term Exposure Scenario, Int. J. Mol. Sci., 2022, 23, 2958 CrossRef CAS PubMed.
- R. Trevisan, C. Voy, S. Chen and R. T. Di Giulio, Nanoplastics Decrease the Toxicity of a Complex PAH Mixture but Impair Mitochondrial Energy Production in Developing Zebrafish, Environ. Sci. Technol., 2019, 53, 8405–8415 CrossRef CAS PubMed.
- B. Arikan, C. Ozfidan-Konakci, E. Yildiztugay, M. Turan and H. Cavusoglu, Polystyrene nanoplastic contamination mixed with polycyclic aromatic hydrocarbons: Alleviation on gas exchange, water management, chlorophyll fluorescence and antioxidant capacity in wheat, Environ. Pollut., 2022, 311, 119851 CrossRef CAS PubMed.
- D. Toybou, C. Celle, C. Aude-Garcia, T. Rabilloud and J.-P. Simonato, A toxicology-informed, safer by design approach for the fabrication of transparent electrodes based on silver nanowires, Environ. Sci.: Nano, 2019, 6, 684–694 RSC.
- M. Allegri, M. G. Bianchi, M. Chiu, J. Varet, A. L. Costa, S. Ortelli, M. Blosi, O. Bussolati, C. A. Poland and E. Bergamaschi, Shape-Related Toxicity of Titanium Dioxide Nanofibres, PLoS One, 2016, 11, e0151365 CrossRef PubMed.
- M. Ilves, S. Vilske, K. Aimonen, H. K. Lindberg, S. Pesonen, I. Wedin, M. Nuopponen, E. Vanhala, C. Højgaard, J. R. Winther, M. Willemoës, U. Vogel, H. Wolff, H. Norppa, K. Savolainen and H. Alenius, Nanofibrillated cellulose causes acute pulmonary inflammation that subsides within a month, Nanotoxicology, 2018, 12, 729–746 CrossRef CAS PubMed.
- D. Musino, J. Devcic, C. Lelong, S. Luche, C. Rivard, B. Dalzon, G. Landrot, T. Rabilloud and I. Capron, Impact of Physico-Chemical Properties of Cellulose Nanocrystal/Silver Nanoparticle Hybrid Suspensions on Their Biocidal and Toxicological Effects, Nanomaterials, 2021, 11, 1862 CrossRef CAS PubMed.
- M. Ghiazza, M. Polimeni, I. Fenoglio, E. Gazzano, D. Ghigo and B. Fubini, Does vitreous silica contradict the toxicity of the crystalline silica paradigm?, Chem. Res. Toxicol., 2010, 23, 620–629 Search PubMed.
- J. S. Choi, K. Kim, S. H. Hong, K.-I. Park and J.-W. Park, Impact of polyethylene terephthalate microfiber length on cellular responses in the Mediterranean mussel Mytilus galloprovincialis, Mar. Environ. Res., 2021, 168, 105320 CrossRef CAS PubMed.
- V. Collin-Faure, A. Villacorta, M. Vitipon, H. Diemer, S. Cianférani, R. Marcos, E. Darrouzet, A. Hernandez and T. Rabilloud, About the effects of true-to-life polyethylene terephthalate nanoparticles on macrophages, Environ. Sci.: Nano, 2025, 12, 2799–2814 RSC.
- A. Villacorta, L. Rubio, M. Alaraby, M. López-Mesas, V. Fuentes-Cebrian, O. H. Moriones, R. Marcos and A. Hernández, A new source of representative secondary PET nanoplastics. Obtention, characterization, and hazard evaluation, J. Hazard. Mater., 2022, 439, 129593 CrossRef CAS PubMed.
- J. A. Vizcaino, E. W. Deutsch, R. Wang, A. Csordas, F. Reisinger, D. Rios, J. A. Dianes, Z. Sun, T. Farrah, N. Bandeira, P. A. Binz, I. Xenarios, M. Eisenacher, G. Mayer, L. Gatto, A. Campos, R. J. Chalkley, H. J. Kraus, J. P. Albar, S. Martinez-Bartolome, R. Apweiler, G. S. Omenn, L. Martens, A. R. Jones and H. Hermjakob, ProteomeXchange provides globally coordinated proteomics data submission and dissemination, Nat. Biotechnol., 2014, 32, 223–226 CrossRef CAS PubMed.
- U. Sarkans, M. Gostev, A. Athar, E. Behrangi, O. Melnichuk, A. Ali, J. Minguet, J. C. Rada, C. Snow, A. Tikhonov, A. Brazma and J. McEntyre, The BioStudies database—one stop shop for all data supporting a life sciences study, Nucleic Acids Res., 2018, 46, D1266–D1270 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5en00074b |
| This journal is © The Royal Society of Chemistry 2025 |