 Open Access Article
Open Access ArticleDigital PCR: from early developments to its future application in clinics
Amandine Troucheta,
Guillaume Gines b,
Leonor Benhaimac and
Valerie Taly
b,
Leonor Benhaimac and
Valerie Taly *ad
*ad
aCentre de Recherche des Cordeliers, UMRS1138, INSERM, CNRS, Sorbonne Université, USPC, Université Paris Cité, Equipe Labellisée Ligue Nationale Contre le Cancer, CNRS SNC 5096, Paris, France. E-mail: valerie.taly@parisdescartes.fr
bLaboratoire Gulliver, UMR7083 CNRS/ESPCI Paris-PSL Research University, 10 rue Vauquelin, 75005, Paris, France
cDepartment of Oncologic Surgery, Gustave Roussy Cancer Campus, Paris-Saclay University, 114 rue Edouard Vaillant, Villejuif, France
dMETHYS Dx, 67 rue Saint-Jacques, Paris, France
First published on 21st July 2025
Abstract
Digital PCR (dPCR) is the third generation of PCR technology, after conventional PCR and real-time quantitative PCR. It is based on the partitioning of a PCR mixture supplemented with the sample to analyse into a large number of parallel reactions, so that each partition contains either 0, 1 or a few nucleic acid targets, according to a Poisson distribution. Following PCR amplification, the fraction of positive partitions is extracted from an end-point measurement, allowing the computation of the target concentration. This calibration-free technology presents powerful advantages including high sensitivity, absolute quantification, high accuracy and reproducibility as well as rapid turnaround time and has therefore rapidly spread. Digital PCR offers a wide range of applications in research, clinical diagnostics, and biotechnology. Among the first clinically relevant applications of dPCR was its ability to detect rare genetic mutations within a background of wild-type genes. This breakthrough paved the way to tumour heterogeneity analysis in oncology and enabled liquid biopsy applications, such as the monitoring of treatment response. The scope of dPCR applications has since rapidly extended to include prenatal diagnosis through the detection of aneuploidy or inherited mutations, as well as pathogen identification via the detection of virus-specific genes or antibiotic-resistance genes in bacteria. This review focuses on the clinical applications of dPCR, highlighting its advantages over existing technologies and providing an outlook on future developments.
I. Introduction to dPCR
Modern medicine requires precise and sensitive techniques for disease diagnosis and patient follow-up. The pathologies should be detected and identified at the earliest to increase the chances for finding a cure. Historically, infectious diseases were diagnosed with serological tests for antibody or antigen detection, or with sample culture for bacteria identification. Although they are easy to perform, widely standardised and inexpensive, these tests can be time consuming and exhibit low sensitivity. The COVID-19 pandemic has emphasised the urgent need for highly sensitive and accurate detection methods.11. History and principle of dPCR
In 1986, Karry Mullis invented the polymerase chain reaction (PCR), a technique that would become the gold standard for nucleic acid detection.2 This molecular biology method enables the exponential replication of specific DNA sequences, through a mix of – at least – two synthetic target-specific oligonucleotides (primers), a thermostable DNA-replicative enzyme (DNA polymerase) and deoxyribonucleotide triphosphate monomers (dNTP).2 In its initial development, the product of the amplification reaction was analysed by gel electrophoresis, providing semi-quantitative information based on band intensity. In 1992, Russel Higuchi developed the second-generation PCR, the quantitative PCR (qPCR, also known as real-time PCR), where the amplification reaction is monitored in real-time using for example a fluorescent DNA-intercalating dye or specific fluorescent probes (TaqMan probes or molecular beacons).3 From the fluorescence signal, the amplification time (i.e. the cycle at which the fluorescence crosses a given threshold) is extracted and compared to standard samples of known concentration, allowing for a relative quantification.In a precursor work from 1989, Peter Simmonds used limiting dilution PCR to detect single copies of HIV provirus in infected cells and concluded that the disease stage correlates with the proportion of infected Peripheral Blood Mononuclear Cells (PBMC, ranging from 1 per 5000 to 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells for asymptomatic patients to 1 per 700 to 3300 cells for late stage/stage IV patients).4 Three years later, Morley and Sykes combined limiting dilution PCR with Poisson statistics to isolate, detect and quantify single nucleic acid molecules, laying the foundations of digital PCR:5 in their study, sample dilutions were replicated, PCR-amplified and analysed by gel electrophoresis, enabling an accurate count of target molecules based on the fraction of negative partitions. The authors successfully detected, within bone marrow samples of leukemia patients, mutated IgH rearranged heavy chain gene as low as 2 targets in 160
000 cells for asymptomatic patients to 1 per 700 to 3300 cells for late stage/stage IV patients).4 Three years later, Morley and Sykes combined limiting dilution PCR with Poisson statistics to isolate, detect and quantify single nucleic acid molecules, laying the foundations of digital PCR:5 in their study, sample dilutions were replicated, PCR-amplified and analysed by gel electrophoresis, enabling an accurate count of target molecules based on the fraction of negative partitions. The authors successfully detected, within bone marrow samples of leukemia patients, mutated IgH rearranged heavy chain gene as low as 2 targets in 160![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 wild-type sequences. In 1999, the term digital PCR, the third and latest PCR generation, was coined by Bert Vogelstein and collaborators (see Fig. 1), who developed a workflow involving limiting dilution distributed on 96-well plates combined with a fluorescence readout to detect mutations of RAS oncogene in the stools of patients with colorectal cancer.6
000 wild-type sequences. In 1999, the term digital PCR, the third and latest PCR generation, was coined by Bert Vogelstein and collaborators (see Fig. 1), who developed a workflow involving limiting dilution distributed on 96-well plates combined with a fluorescence readout to detect mutations of RAS oncogene in the stools of patients with colorectal cancer.6
 | ||
| Fig. 1 Schematic chronology of dPCR focused on historical works and commercial developments. Created with https://Biorender.com. References cited are Mullis et al.,2 Higuchi et al.,3 Vogelstein et al.,6 Dressman et al.,8 Huggett et al.390 | ||
The technology of dPCR was born, but the need for microtiter plates limited its practicability and some improvements were therefore needed. In 1997, Olga Kalinina and collaborators introduced volume miniaturisation by using microcapillaries (∼10 nL) for the partition process, which reduced the cost of reagents and improved the amplification efficiency.7 In 2003, Bert Vogelstein et al. reported the BEAMing (beads, emulsion, amplification and magnetics) technology,8–10 further simplifying the compartmentalisation process by utilising water-in-oil droplets parallelising PCR. The method involved encapsulating individual DNA molecules with magnetic beads coated with primers, permitting PCR amplification within the droplet. The amplified products were then recovered magnetically and analysed by flow cytometry using DNA probes and/or immunostaining. Some derived protocols of BEAMing replaced the flow cytometry analysis by the imaging of planar arrays of hydrogel beads.11 This adaptation has been used to detect early-stage colorectal cancer by assessing oncogene expression in tissue and stool samples.12
Modern dPCR protocols are built upon those foundational principles and generally follow four key steps: i) partitioning the PCR mixture that contains the sample into thousands to millions of compartments. This step implies the random distribution of the targets among the partitions; ii) amplifying individual target-containing partitions; iii) performing end-point fluorescence analysis of the partitions; iv) computing the target concentration using Poisson statistics, based on the fraction of positive and negative partitions (see Fig. 2). This provides PCR with high sensitivity and calibration-free absolute quantification13 owing to the single-molecule detection attribute.13 For the past decades, two major types of partitioning methods have emerged: water-in-oil droplet emulsification and microchambers.
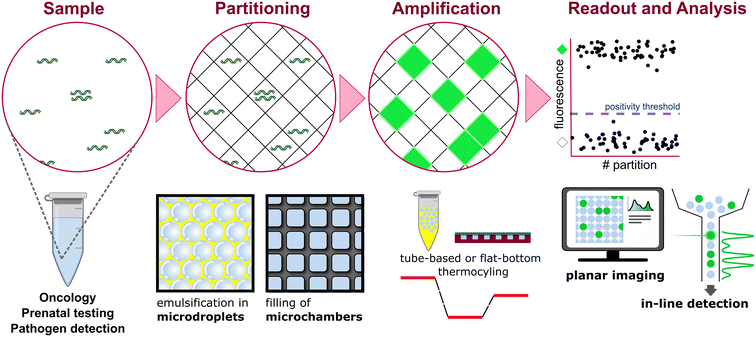 | ||
| Fig. 2 Principle of dPCR based on limited dilution, distribution in partitions, amplification, fluorescence detection and data analysis. | ||
In droplet digital PCR (ddPCR), the sample is dispersed into tiny (pL to nL) droplets within an immiscible oil phase. Monodisperse droplets can be generated at high speed (typically 1–100 kHz) using a microfluidic chip leveraging passive forces or actively breaking the aqueous/oil interface (for an exhaustive review on microfluidic designs for droplet generation, see Xu et al.14). It is to be noted that water-in-oil droplets are prone to coalescence (especially during the harsh temperature variation of the PCR protocol) and their stabilisation with an appropriate surfactant is of prime importance.15
Microchamber-based dPCR uses an array of thousands of microscopic wells or chambers embedded in a solid chip. While ddPCR offers greater scalability and cost-effectiveness, it requires precise emulsification and droplet stability. On the other hand, microchamber dPCR provides higher reproducibility and ease of automation but is limited by the fixed number of partitions and typically higher costs.
As for the droplet-signal reading technology, again, two primary readout methods are available: in-line detection and planar imaging. In in-line detection, commonly used in ddPCR, the droplets are flowed through a microfluidic channel or capillary and their fluorescence is measured one by one using a light source coupled to detectors. This allows the analysis of a large number of droplets but requires precise control of the flow. In contrast, planar arrays of microchambers or microdroplets can be imaged using a fluorescence microscope or scanner and provide a static snapshot of the partitions. Note that 3D imaging16 and analysis17 techniques have been developed to assay in a shorter time a larger number of droplets.
2. A path towards commercialisation of dPCR platforms
The rise of dPCR has been driven by significant advances in microfabrication and microfluidics, expanding the possibilities for volume miniaturisation.18,19 This progress has led to the development of various dPCR techniques, ranging from 96-well plate-compatible protocols20 to sophisticated lab-on-chip prototypes, potentially suitable for commercialisation.One notable example is Slip Chip, a microfabricated chip composed of a bottom plate with microchambers filled with PCR solution. This chip slides under a top plate which contains the samples, enabling interaction and amplification with an end-point analysis under a fluorescence microscope.21 Another innovative system, the spinning disk, uses centrifugation to separate the sample into nanoliter wells for an end-point fluorescent analysis.22 Although these systems are technologically advanced, they remain mainly used as laboratory prototypes.
In contrast, the first compartment-based dPCR nanofluidic platform was commercialised by Fluidigm in 2006. It is composed of an integrated fluidic controller (IFC) that loads the samples automatically into microchambers using on-chip valves; a fluorescence analyser with or without an integrated thermocycler – that allows real-time PCR (Biomark) or endpoint (EP1) analysis respectively. Although no longer commercially available, this platform was proven to be efficient for the detection of bacterial signatures,23 for the measurement of gene expression in tissues,24 or gene copy numbers in breast cancer samples.25 The next significant commercial dPCR system was the Quantstudio 3D (QS3D), marketed by Applied Biosystem in 2013. Originally developed as the Open Array Platform by BioTrove, it was acquired by Life Technologies in 2009 and replaced by Absolute Q in 2022. In 2013, Formulatrix introduced its Constellation dPCR instrument. The company was bought by Qiagen in 2019 and the same instrument was renamed QIAcuity in 2020. The system developed by Roche (Digital LightCycler) followed in 2022 (see Table 1).
| INSTRUMENT characteristics | CHIP/ARRAY characteristics | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Brand | Instrument | Launch Date | Integration/number of machines | Type of analysis | Nb of chips or plates per run | Nb. of analysis channels and dyes associated | Chip, samples and nb. of partitions | Type of partitions | Volume of partition | Real-time option |
| Abbreviations: nb.: number,a not commercialized anymore.b In-line: droplets flow in front of a detector; planar: partitions are analyzed by 2D-scanning.c OpenArray was initially commercialized by BioTrove; QX100 was developed following Quantalife acquisition; Qiacuity suite was developed based on the Formulatrix Constellation system | ||||||||||
| Thermofisher Scientific | Quantstudio Absolute Q | 2022 | 1 instrument for partitioning, thermocycling and data acquisition | Planar | 1 plate: up to 16 samples (4, 8, 12 or 16)90 min per run (depend on thermocycling conditions) | 5 channels4 for sample analysis: FAM, HEX/VIC, TAMRA/Atto550, Cy5 1 used as a reference/QC: ROX/Atto590 (589 nm/625 nm) |
Microfluidic array plate (MAP), MAP16 plate for 16 samples, each divided into 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 480 partitions 480 partitions |
Micro-chambers | ∼0.4 nL | No |
| Applied Biosystems QuantStudio 12K Flex with the OpenArrayc platform | 2009 | 2 instruments for:- Partitioning: manually (+ a sealing device) or Accu Fill system for automated partionning of 4 arrays - Thermocycling integrated with data acquisition |
Planar | Up to 4 chips in 4 hours | 6 channels:FAM, HEX/VIC, TAMRA, ROX, Cy5, Cy5.5 | 1 chip Open Array for 1 sample, divided into 3072 partitions | Micro-chambers | ∼33 nL | Yes | |
| Quantstudio 3Da (QS3D) | 2013(discontinued in 2023) | 3 instruments for:- Partitioning: Chip loader for 1 chip - Thermocycling: need for the additional thermocycler Dual Flat block PCR system with adapters - Data acquisition |
Planar | Up to 24 chips per run of thermocycling (2.5 h)1 chip per run of data acquisition (30 s) | 3 channels:FAM, HEX/VIC, ROX | 1 chip for 1 sample, divided into 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 partitions 000 partitions |
Micro-chambers | ∼0.8 nL | No | |
| Qiagen | QIAcuityc One | 2020 | 1 instrument for partitioning, thermocycling and data acquisition | Planar | 1 nanoplate per run | Qiacuity one 2 plex: 2Qiacuity one 5 plex: up to 8 (6 plus 2 hybrid (e.g. for long Stokes shift (LSS) probes) | Nanoplates:24 samples in 8500 partitions 96 samples in 8500 partitions24 samples in 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 partitions 000 partitions8 samples in 26 |
Micro-chambers | ∼1.5 nL | No |
| QIAcuityc Four | up to 4 nanoplate per run | 8 channels: 6 plus 2 hybrid (e.g. for long Stokes shift (LSS) probes) | ||||||||
| QIAcuityc Eight | up to 8 nanoplate per run | 8 channels: 6 plus 2 hybrid (e.g. for long Stokes shift (LSS) probes) | ||||||||
| Roche | Digital LightCycler system | 2022 | 2 instruments for:- Partitioning: partitioning engine for 1 plate - Thermocycling integrated with data acquisition |
Planar | Up to 12 plates per run (8–96 samples) | 6 channels:Cyan500/Atto425, FAM, HEX/VIC, LC610/Texas Red, CY5/LC640, Cy5.5 | 1 plate for 8 samples3 types of plates:high resolution plate: 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 partitionsUniversal plate: 28 000 partitionsUniversal plate: 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 partitions High sensitivity plate: 20 000 partitions High sensitivity plate: 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 partitions 000 partitions |
Micro-chambers | ∼0.15 nL∼1 nL∼2.5 nL | No |
| Bio-Rad | QX100a,a | 2011 | 3 instruments for:- Partitioning: QX Droplet Generator (1-8 samples per partitioning) or AutoDG for automatic generation (up to 96 samples per partitioning) + plate sealer - Thermocycling: need for the additional thermocycler BioC1000 Touch or PTC tempo - Data acquisition: QX Droplet reader |
In-lineb | Up to 96 samples in a 96-well plate | 2 channels: FAM, HEX/VIC | DG8 cartridge for 8 samples, each divided into 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 partitionsDG32 Automated Droplet Generator Cartridges: 4 × 8 samples, each divided into 20 000 partitionsDG32 Automated Droplet Generator Cartridges: 4 × 8 samples, each divided into 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 partitions 000 partitions |
Droplets | ∼nL | No |
| QX200 | 2013 | 2 channels: FAM, HEX/VIC | ||||||||
| QX600 | 2022 | 6 channels:FAM, HEX/VIC, Cy5, Cy5.5, ROX, and Atto590 | ||||||||
| QX ONE | 2019 | 1 instrument for partitioning, thermocycling and data acquisition | 5 plates = 480 samples | 4 channels:FAM, HEX/VIC, Cy5, Cy5.5 | GCR96 cartdriges: 96 samples, each divided into 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 partitions 000 partitions |
Droplets | ∼nL | No | ||
| Raindrop dPCRa | 2012 | 3 instruments for:- Partitioning: Raindrop Source, 1 chip for 8 samples - Thermocycling: need for an additional thermocycler - Data acquisition: Raindrop Sense |
In-lineb | 1 chip per run (8 samples) | 2 channels: FAM, HEX/VIC | 1 chip for 8 samples, each divided into 1 million to 10 million partitions | Droplets | ∼pL | No | |
| Stilla Technologies | NAICA 3 | 2016 | 2 instruments for:- Partitioning and thermocycling: Geode - Data acquisition: PRISM3 or 6 |
Planar | 3 chips per run (for both instruments) | Prism 3 with 3 channels:FAM, HEX/VIC, Cy5 Prism 6 with 6 channels: FAM, YY, Atto550, ROX, Cy5, Cy5.5 |
Sapphire Chip: 4 samples per chip, each divided into 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 partitionsRuby chip: 16 samples per chip, each divided into 17 000 partitionsRuby chip: 16 samples per chip, each divided into 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 partitions 000 partitions |
droplets | ∼0.8 nL∼0.3 nL | No |
| NAICA 6 | 2020 | |||||||||
| Nio™ E | 2023 | 1 instrument for partitioning, thermocycling and data acquisitionNio e: 3 PCR programs per run Nio: 12 PCR programs per run Nio+: 2 integrated thermocyclers = 24 PCR programs per run |
Planar | 3 chips per run(48 samples) | 7 channels:FAM, YY, Atto550, ROX, Cy5, Cy5.5, DY-521-XL | Ruby chip: 16 samples per chip, each divided into 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 partitions 000 partitions |
Droplets | ∼0.3 nL | No | |
| Nio™ | 12 chips per run(192 samples) | |||||||||
| Nio™+ | 24 chips per run(384 samples) | |||||||||
| Fluidigm | Biomark HDa | 2006 | 2 instruments for:- Partitioning: 3 devices (MX, RX or Juno) - Thermocycling integrated with data acquisition |
Planar | 1 | 2 channels: FAM, HEX/VIC | Integrated microfluidic circuits (IFC) allowing the partitioning of 12 to 192 samples into a range of partition numbers from 12 to 770 partitions | Micro-chambers | ∼nL | Yes |
| JN MedSys | Claritya | 2016 | 3 instruments for:- Partitioning in tube: Clarity Auto-Loader + Clarity Sealing Enhancer, 8 tubes/ partitioning run - Thermocycling: need for an additional thermocycler with adjustable ramp and 0.2 mL tube - Data acquisition: Clarity Plus Reader |
Planar | 32 samples per thermocycling and data acquisition runs | 2 channels: FAM, HEX/VIC | 1 sample per tube, divided into 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 partitions 000 partitions |
Micro-chambers | ∼1.5 nL | No |
| Clarity+ | 2020 | Planar | Up to 96 samples per thermocycling and data acquisition runs | 6 channels: FAM, HEX/VIC, Atto550, Texas Red, Cy5, Cy5.5 | 1 sample per tube, divided into 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 partitions 000 partitions |
Micro-chambers | ∼0.3 nL | No | ||
| Optolane | Genotizer™/Dr. PCR™ | 2019 | 2 instruments for:- Partitioning: POSTMAN (sample loader) for 1 chip - Thermocycling and data acquisition: LOAA analyzer |
Planar | 1 | 2 channels: FAM, HEX/VIC | 1 sample per chip, divided into 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 163 partitions 163 partitions |
Micro-chambers | ∼33 nL | Yes |
In droplet-based dPCR, laboratory prototypes tend towards fully integrated on chip systems. They usually contain microfluidic valves26 and/or electrodes and magnets27 to generate the droplets that are thermocycled in either a chamber28 or in a microchannel that traverses alternating temperatures areas.27,29 However, at the present time, the use of a separated 3-step protocol for on-chip droplet generation, off-chip in-tube thermocycling and on-chip droplet fluorescence analysis still presents clear advantages such as reliability and flexibility. Moreover, it permits compliance with clinical constraints which could imply separate rooms for pre-PCR, PCR and post-PCR with the aim of avoiding cross-contaminations. It is thus central to most commercial systems and has been used in research and clinical laboratories, for example, to analyse rare mutations of the KRAS gene.13 An optimised multiplex with fluorescence intensity encoding (using different green and red probe concentrations, see also section 2.a) led to a 5-plex assay capable of the simultaneous identification of the c815A>G mutation and copy number variation of genes implicated in spinal muscular atrophy.30 In the same article, the authors mentioned the achievement of a 10-plex assay. Concurrent developments focused on using a 96-well plate for droplet collection to parallelise sample thermocycling. It allowed processing of 8 samples simultaneously and led to the first droplet-based dPCR commercialised instrument by Quantalife.31 The American company Bio-Rad bought Quantalife in 2011 and its other competitor Raindance in 2017, making it the global leader in droplet dPCR.
Since 2011, Bio-Rad has commercialised a 2-color system based on the 3-step workflow, namely the QX100™ Droplet Digital™ PCR System and its next version the QX200™ followed by the QX600 6-color version, and the all-in-one fully automated QX1 system (see Table 1 for characteristics). In 2016, the French company Stilla Technologies commercialised a 3-color system (NAICA 3), replaced by a 6-color system (NAICA 6) and the brand-new all in one Nio+ system series (7 colors ddPCR). In 2025, Bio-Rad entered the process of acquiring Stilla Technologies. From 2019, other competitors emerged including Rainsure, Targeting One, Forevergen, Sniper or Pilot Gene Tech, diversifying the market of ddPCR platforms. Important studies that compared these droplet dPCR platforms confirmed a high degree of consistency, as shown in the case of SARS-CoV-2 gene-associated detection.32,33 The calibration of droplet volume remains recommended to maintain the consistency between platforms.33–35
Many studies have compared dPCR instruments based on microchambers versus droplets with the ultimate goal of calibrating differences (see Table 1) and standardising dPCR for clinical use. The evaluation of the QX100/QX200 and QS3D instruments, with different types of partition and readout, was conducted to assess their ability to detect mutations in samples in various situations including prenatal non-invasive testing,36 lung cancer follow-up37 and HIV follow-up.38 It was demonstrated that both platforms achieved comparable results with similar sensitivity. The QX200 platform was also compared to the Absolute Q for the detection of early-stage breast cancer. These platforms displayed >90% concordance in ctDNA positivity within 46 plasma samples.39 Further work also compared QX200 and QIAcuity platforms for the detection of specific mutations,40,41 and both allowed the detection of DNA quantities as low as 9 picograms,40 although a moderate agreement was found due to the sampling effect and threshold settings.41 To assess the impact of partition number, the QX200, QS3D and Raindrop (a system with 250-fold more partitions, from Raindance Technologies) systems were compared for the detection of the BCR-ABL1 fusion gene (leukemia biomarker) and were found to have a common 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log dynamic range and to correlate only for frequency >0.1%.42 These platforms were also compared to the QX100, the Constellation, and the Biomark systems for the detection of mutated KRAS oncogene, taking plasma mass spectrometry as a reference. It showed a variability in concentration values less than 1.3-fold.43 The QX100, the Biomark and the Raindrop systems have also been compared to the Quantstudio 12K Flex. The analysis required partition volume correction and indicated comparable effectiveness for the quantification of a certified plasmid reference material.44
log dynamic range and to correlate only for frequency >0.1%.42 These platforms were also compared to the QX100, the Constellation, and the Biomark systems for the detection of mutated KRAS oncogene, taking plasma mass spectrometry as a reference. It showed a variability in concentration values less than 1.3-fold.43 The QX100, the Biomark and the Raindrop systems have also been compared to the Quantstudio 12K Flex. The analysis required partition volume correction and indicated comparable effectiveness for the quantification of a certified plasmid reference material.44
The numerous advantages of dPCR (high sensitivity45 and reproducibility, absolute quantification, less competition between DNA targets and so less bias due to PCR efficiency differences, less sensitivity to PCR inhibitors,46 easy analysis,47 lower volumes and turnaround times) explain its rapid expansion, in the last years (see Fig. 3a), as a powerful tool for potential clinical applications. This is, all the more true, knowing that the clinical implementation of liquid biopsies is becoming a standard of care. Liquid biopsy is the act of sampling biological fluids for the analysis of nucleic acids, circulating cells or subcellular structures as exosomes. It is mostly known in the field of oncology, but it is also used for other disease diagnoses. Despite the minimally invasive character of liquid biopsy, the minute amount of material available is still a real challenge.
 | ||
| Fig. 3 a) Number of yearly publications on digital PCR and digital PCR in clinics. b) Applications of dPCR in different fields in general and in clinics. Source: Scopus [data assessed: 27/11/2024]. | ||
However, very few applications are, at the present time, FDA-approved in clinics. Indeed, the FDA validated the use of dPCR in 3 particular cases: SARS-CoV-2 detection,48 BCR::ABL1 detection to follow up patients with chronic myelogenous leukemia49 and residual host cell DNA detection in biologic drugs produced in E. coli.50 It's worth noting that the FDA has so far decided not to regulate laboratory-developed tests, such as non-invasive prenatal testing.51 In order to pave the way of dPCR for patient diagnosis and follow up, a large range of clinical trials are comparing its performance to the gold standard methods currently used in clinics, particularly in oncology (see Fig. 3b). This review will present chosen examples of these studies, referencing advantages and drawbacks of dPCR. Finally, an overview of promising improvements will be proposed, although the list is not exhaustive.14
II. Applications of dPCR in oncology
In 2022, cancer was responsible for approximately 9.7 million deaths worldwide and remains the second leading cause of mortality.52 Among the estimated 20 million new cases per year, the most common are breast cancer (BC), colorectal cancer (CRC) and lung cancer (LC). According to the National Cancer Institute, cancer is defined as a disease “in which some of the body's cells grow uncontrollably and spread to other parts of the body”.53 This abnormal cell proliferation is triggered by the accumulation of alterations within cells implying changes at different key levels including genomic, transcriptional or epigenomic. All these alterations constitute potential cancer biomarkers54 useful for cancer detection, disease prognosis, treatment selection or analysis of response to treatment.55 At the genomic level, somatic molecular alterations can be fusion gene, point mutations or copy number variation (CNV) of specific oncogenes or tumour suppressor genes such as HER2, PIK3CA, KRAS, BRAF, EGFR, and TP53 (ref. 56) (see Table 2).| Gene of interest | Type of alteration | Biomarker type | Type of cancer | References |
|---|---|---|---|---|
| HER2 | CNV | Tissue, ctDNA | Breast cancer, gastric cancer, NSCLC | 12, 61, 79, 96, 97 |
| RAS | Point mutation | Tissue, ctDNA, CTC | CRC, pancreatic cancer, melanoma | 41, 55, 72–75, 82, 86, 100, 102, 109, 157, 341–343, 347, 348, 392–394 |
| BRAF | Point mutation | Tissue, ctDNA | CRC, melanoma | 55, 68, 73, 75, 85, 86, 95, 99, 101, 102, 341, 347, 393, 394 |
| EGFR | Point mutation | Tissue, ctDNA, CTC | NSCLC | 37, 41, 55, 68, 70, 71, 78, 84, 88, 90–94, 106, 125, 150, 151, 153, 158, 395–397 |
| TP53 | Point mutation | Tissue, ctDNA | Ovarian cancer, breast cancer, NSCLC, pancreatic cancer, hepatocellular carcinoma, melanoma | 39, 65, 80, 106, 109, 122, 398 |
| PIK3CA | Point mutation | Tissue, ctDNA, CTC | Breast cancer, CRC, NSCLC | 39, 75, 81, 100, 102, 106, 110, 147, 148, 347, 399, 400 |
| TERT | CNV, point mutation | Tissue, ctDNA | Lung cancer, bladder cancer, hepatocellular carcinoma | 58, 120, 122–124, 398, 401 |
| MYC | CNV | Tissue, ctDNA | Lung cancer | 58, 402 |
| ESR1 | Point mutation | ctDNA, CTC, EV | Breast cancer | 147, 149, 346 |
| BCR::ABL1 | Fusion gene | ctDNA | Leukemia | 34, 42, 49, 87, 113–117 |
| ALK | Fusion gene | Tissue, ctDNA, CTC | NSCLC | 63, 119, 165, 166, 169 |
| MET | CNV, point mutation | Tissue, ctDNA, CTC | Lung cancer, ovarian cancer | 55, 59, 62, 150, 169 |
| PD-L1 | Transcript | CTC | Head and neck squamous cell carcinoma | 154 |
| WIF1 | Hypermethylation | ctDNA | CRC | 127 |
| NPY | Hypermethylation | ctDNA | CRC | 127 |
1. Solid tissue analysis
Tissue biopsies are used by the pathologist to confirm the diagnosis of cancer by direct observation of the cells and tissues' morphologic features. This observation is combined with other analyses such as immunohistochemistry (IHC) and/or fluorescent in situ hybridisation (FISH) and/or traditional molecular analysis. These methods are both expensive and time-consuming and could introduce a subjective dimension into the diagnosis.54 Complementary analysis by dPCR can deliver interesting information on DNA extracted from solid tumour tissue. As an example, it is possible to analyse the CNV of genes of interest (using a gene of reference) to discriminate the tumour from the normal tissue. For instance, the matrix metalloproteinase-9 gene (MMP-9) CNV was found only in tumour tissue and not in adjacent tissues. Coupled to mRNA expression analysis, it showed a potential value as a diagnostic biomarker in hepatocellular carcinoma (HCC) samples (P < 0.0001, AUC = 0.76).57 Similarly, the CNV analysis of tumour and non-tumour tissues has enabled the diagnosis of lung cancers with a pre-defined specificity of 99% and a sensitivity of 41% for MYC and 51% for TERT individually, whereas the combination of both genes gave an improved sensitivity of 60%,58 highlighting that targeting several genes can improve clinical sensitivity. The MET polysomy detection by dPCR indicated a 100% concordance with FISH in non-small cell lung cancer (NSCLC) samples.59 The determination of HER2 CNV is particularly interesting: if upregulated, HER2 is a treatment target in several solid tumours including breast and gastric cancers. The traditional analysis of HER2 CNV by FISH uses a reference gene, often CEP17 (centromere of chromosome 17). Up to 20% of false negative results have been reported with FISH, especially in the case of polysomy of chromosome 17.60 In BC,61 a strategy based on a 3 duplex dPCR with 3 different reference genes has been developed, resulting in the same HER2 CNV detected as with IHC and FISH combined, without the use of CEP17. Thereby using several reference genes can significantly improve the sensitivity, making it competitive with standard methods. The CNV determination by sequencing analysis is also feasible, but dPCR has been suggested as a pertinent validation for unambiguous results.62For the analysis of point mutations, dPCR is often challenged against sequencing methods (Sanger or next-generation sequencing).63–67 Indeed, dPCR had a similar sensitivity and a faster turnaround time than NGS for testing DNA and RNA biomarker panels in samples from patients with NSCLC68 or acute myeloid leukemia (AML).69 In NSCLC, the treatment with EGFR (endothelial growth factor receptor) tyrosine kinase inhibitor (TKI) is inefficient in patients with EGFR resistance mutations. It has been shown that the presence of T790M EGFR mutation correlates with a faster rate of disease progression in the first five months.70 In contrast, dPCR offers quantitative analysis of the EGFR mutation and revealed that low-level T790M does not impact the treatment response or the survival, indicating that a threshold is needed to determine who will benefit from EGFR-TKI.70 Thus, the impact of mutations on treatment resistance can be studied by dPCR.71 In the case of CRC, resistance to anti-EGFR therapies arises from mutations of the RAS gene and BRAF. As for NSCLC, highly sensitive dPCR revealed the need for a threshold in the mutant allele frequency (MAF) detection for prognosis. The clinically relevant threshold analysed by extended pathway genotyping of RAS and BRAF in large patient populations seemed comprised between 1% (ref. 72–74) and 5% of RAS/BRAF mutant.75 Unfortunately, solid biopsies are invasive and therefore not suitable for regular patient monitoring. Moreover, solid biopsies only provide information from a limited area of the tumour in a disease known to be highly heterogeneous.
2. Liquid biopsy analysis
In recent years, liquid biopsy in oncology, consisting of the analysis of tumour-specific components released in bodily fluids such as blood, urine, saliva, pleural, peritoneal or cerebrospinal fluids, has shown to be pertinent to overcome the limitations of tissue biopsy. The concept of liquid biopsy was first coined by Catherine Alix-Panabières and Klaus Pantel by detecting, in blood, circulating tumour cells (CTCs) – intact cells that detached from a primary tumour and entered the bloodstream.76 Liquid biopsies are non or minimally invasive and their analysis offers several benefits in comparison with solid biopsies, including real-time analysis and reflection of tumour heterogeneity. Moreover, liquid biopsy analyses present applications for detection of early cancer, cancer progression and minimal residual disease (MRD) as well as for the real-time monitoring of treatment response. The FDA approved the Cell Search CTC system, for monitoring breast cancer via CTC isolation/enumeration in 2004, metastatic CRC in 2007 and metastatic prostate cancer in 2008. Many other biomarkers can be analysed in liquid biopsies including exosomes, microRNAs (miRNAs), and cell-free circulating tumour DNA (ctDNA). ctDNA consists of fragments of DNA released into bloodstream by tumour cells, allowing the search for tumour specific molecular changes in real-time. | ||
| Fig. 4 Multiplex assay with two fluorescent probes labelling wild-type and mutations of KRAS gene for ctDNA analysis in CRC patients. a) Multiplexing strategy for a 5-plex assay and b) representative data from 2 available panels including the 7 frequently seen mutations in KRAS codon 12 and 13. Reproduced from ref. 82 with permission from Oxford University Press, copyright 2013. | ||
These comparative studies encourage considering the use of molecular analysis in liquid biopsy as companion diagnostic for routine analysis.
As already discussed, the performances of various commercially available dPCR platforms are regularly analysed. A conclusion would be that dPCR is generally more sensitive than the fully automated qPCR-based method.86,87 Nevertheless, some levels of standardisation have become mandatory, particularly when so many dPCR platforms are available. In the case of NSCLC, the ctDNA detection of EGFR mutations was first FDA-approved in 2016 with a qPCR assay for clinical use (COBAS assay).88,89 Indeed, the efficacy of the EGFR inhibitor depends on the EGFR-signalling activation with EGFR-sensitising mutations (exon 19 and 21) and EGFR-resistant mutation (T790M substitution in exon 20).90,91 Provencio et al. described that patients with sensitising mutations detected by dPCR at a mutant allele frequency (MAF) <7% had lower risk of death (60%). During the follow-up, plasma samples, taken while under EGFR inhibition treatment, revealed the emergence of T790M in 52.8% of patients subjected to disease progression.92 Another study that used dPCR correlates the absence of mutated EGFR, at baseline and/or at 4 weeks of iconitib therapy (TKI), with longer progression-free survival (PFS).93 In a different study, undetectable levels of sensitising mutation, during osimertinib therapy (TKI), were associated with higher PFS, whereas its re-emergence alone or together with p.T790M was associated with shorter PFS. Surprisingly, patients with the triplet molecular pattern (sensitising+/T790M+/C797S+) had 12.3 months of median time to progression compared to 4.9 months for patients with sensitising mutation only and 2.17 months for patients also presenting p.T790M mutation.94 Similarly, the FIRE-4.5 study concluded that liquid biopsy evaluating ctDNA is informative and relevant to guide treatment choices in patients with BRAF V600E-mutated metastatic CRC,95 in which case a clear superiority of FOLFOXIRI plus bevacizumab was demonstrated. During the follow-up of patients with HER2+ BC, a decrease of HER2 CNV (15% (ref. 96)) in plasma was correlated with better prognostics and could predict clinical benefit.96,97 In AML, dPCR has proven to be suitable for FLT3-TKD mutation detection, but more clinical studies are needed to conclude about a clinical value.98 These discoveries highlight the importance of treatment response monitoring and the pertinence of the use of liquid biopsy for this purpose. Moreover, the rapid turnaround time and the quantitative character of dPCR associated with the low invasiveness of liquid biopsy make it an outstanding tool for therapy monitoring for daily clinical practice.
Apart from the metastatic setting, dPCR has also been evaluated in the perioperative period, particularly for MRD detection. After surgery of patients with CRC, the ctDNA monitoring for BRAFV600E mutation by dPCR revealed a correlation of the detected MAF with tumour diameter but not with tumour recurrence.99 In contrast, after liver resection in patients with CRC liver metastases, KRAS and PIK3CA mutations were associated with a shorter overall survival (OS).100 Similarly, in patients with resected cutaneous melanoma under therapy, the detection of BRAF-mutated ctDNA was associated with significantly worse OS.101
Furthermore, tumour-informed strategies are studied using the combination of NGS analysis of primary tumour and ctDNA monitoring by dPCR for NGS-identified mutations after surgery and during chemotherapy. They allowed the prediction of early relapse, 3 to 6 months ahead of conventional imaging examinations in the case of CRC102,103 or ahead of the serum biomarker (cancer antigen 125) rise in the case of gynaecological cancers.104 This combination of NGS for mutation identification followed by dPCR enables custom adjustments during long-term treatment response monitoring.105,106 It can also be called personalised dPCR,107 a first step into personalised medicine (see Fig. 5). Many studies using such tumour-informed approaches have shown a good early prediction of patient relapse, after surgery or chemotherapy, in various cancers and settings.66,77,107–112
 | ||
| Fig. 5 Workflow of generating customised dual-color digital PCR assays for routine and extended longitudinal monitoring of circulating tumour DNA throughout treatment. Reproduced from ref. 107 with permission from Elsevier, copyright 2020. | ||
Fusion gene also represents a highly interesting cancer biomarker, particularly in the case of hematopoietic and lymphoid malignancies. Although the current gold standard method for treatment monitoring is reverse transcriptase qPCR (RT-qPCR) and flow cytometry, it is actively challenged by dPCR, particularly for MRD detection that requires high sensitivity.113 Indeed, in 2019, the FDA approved the dPCR assay, from Bio-Rad, aiming to detect BCR::ABL1 in samples of patients with chronic myeloid leukemia.49 Since then, it exhibited good performance: the fusion gene was detected in 63% (ref. 114) and 68% (ref. 115) of patient samples initially negative by RT-qPCR. These results suggest that dPCR could help the early selection of patients admissible for treatment discontinuation.116,117 Other fusion genes were targeted for MRD detection, such as Ig::TCR gene in acute lymphoblastic leukemia (ALL), which was detected by dPCR in 83% (29/35) of the ambiguous qPCR cases.118 Similarly, dPCR has proved its ability to identify accurately patients with high relapse risk via NPM::ALK gene detection in anaplastic large cell lymphoma (ALCL).119
Even though liquid biopsy on blood samples is widespread, other biofluids could be used. For example, in the case of urothelial bladder cancer, the liquid in continuous contact with the tumour surface is urine. The high diversity of genetic alterations that can be found in this cancer (including mutations in genes TERT, FGFR3, PIK3CA, ERBB2, HRAS and GPR126)120 requires the use of tumour-informed dPCR (combined to NGS analysis of tumour tissues) for each patient.121,122 These studies revealed the potential of urinary ctDNA variant allele frequency as a molecular biomarker of recurrence after surgery. In particular, the detection of TERT mutations in urine was proven to be of particular interest: studies of bladder cancer urine samples have highlighted a superior sensitivity of dPCR (79.7%) compared to cytology (59.5%) and uromonitor (56.8%)123 or similar performances to qPCR.124 On the other hand, in lung adenocarcinoma, a comparison between blood and other biofluid samples (pleural effusion, cerebrospinal fluid, ascites and pericardial effusion) from EGFR-positive patients concluded that cell-free DNA (cfDNA) was less abundant in blood and that sensitising mutations were detected in 16 vs. 21 samples respectively.125 In the case of central nervous system tumours, dPCR successfully detected the H3K27 variant in cerebrospinal fluids and was in accordance with tissue analysis.40
Recently, a large body of literature has shown that ctDNA could be detected thanks to the detection of tumour specific methylation dysregulation.126 Indeed, the search for such tumour-specific methylation markers has shown them to act as universal markers of cancer that do not require previous analysis of tumour tissue. Generally, dPCR is associated with the bisulfite conversion to detect and quantify cancer specific markers such as the hypermethylated genes WIF1 and NPY in CRC,127 RASSF1A and GSTP1 in prostate cancer,128,129 HOXD8 and POU4F1 in metastatic pancreatic cancer,130 SEPT9 in gastrointestinal tumours131 and the hypermethylated promoters of SOX17, CDO1, TAC1 and HOXA7 in NSCLC132,133 and of OXT/ZSCQN12 in endometrial carcinoma.134 Similarly, this strategy has been used to target biomarkers in genes EMX1, Chr5q14.1 and NXPH1 for multi-cancer detection (AUC = 0.948).135 These studies highlighted that hypermethylated ctDNA was highly correlated to ctDNA and thus to tumour burden, making it a good tool for patient monitoring, treatment management and even timing of intervention.136 The study of methylations by dPCR revealed the possibility to differentiate liver metastases originated from colorectal or pancreatic ductal adenocarcinoma cancers, and liver cancer types, such as liver adenocarcinoma.137 Indeed, some methylations remain unaltered between primary tumours and liver metastases, whereas some others change and could potentially be drivers of the metastatic cascade.137,138 Another study developed the Methyl-BEAMing technology, combining bisulfite conversion and BEAMing: the bisulfite conversion is followed by a first round of amplification of methylated DNA and reference DNA, then a second round of emulsion PCR on magnetic beads enables the analysis by flow cytometry thanks to fluorescent methylation-specific probes. This method showed a higher sensitivity than dPCR for low DNA quantities.139
Dysregulated methylation could also be assessed by using methylation sensitive restriction enzyme (MSRE) prior to dPCR analysis.140 Based on this technology, the study of hypo- and hypermethylation of promoters demonstrated their value as potential biomarkers for detection of oral cavity cancer.141 Moreover, the team of Takahiro Yamasaki included MSRE-dPCR in predictive models for cancer diagnosis. For example, measuring methylated somatostatin (SST) coupled to fecal immunochemical test and age (FAMS) allowed for the efficient detection of CRC and advanced colorectal adenocarcinoma (AUC = 0.90).142 Evaluation of hTERT and methylated RUNX3 coupled to age and sex (ASTEm-R3) allowed the detection of early gastric cancer (AUC = 0.93, sensitivity 79.7%, specificity 91.1%).143 Assessment of methylated HOXA1 coupled to classical markers (AFP, DCP) as well as age and sex (ASDAm-H1) permitted the accurate detection of hepatocellular carcinoma (AUC = 0.96, sensitivity 86.2%, specificity 93.9%).144 However, in another study, this strategy has shown some discordance with the gold standard, the OSNA method (one-step nucleic acid amplification), to detect RASSF1A methylated.145
Studies on non-coding RNA biomarkers have also been published.159 For example, miRNA 320a expression levels were able to differentiate patients with ovarian cancer from healthy donors by RT-dPCR more reliably than by RT-qPCR.160 Similarly, miRNA 181a appeared as a promising biomarker (ROC = 0.849) in cerebrospinal fluid for detection of central nervous system leukemia and for identification of therapy-admissible patients.161 Also, the prognosis value of long non-coding RNA (lncRNA) MYU in prostate cancer was demonstrated by RT-dPCR on urine samples.162 However, a study compared the biomarker value of several sorts of RNAs in urine samples from prostate cancer patients, and it concluded that miRNAs (miR-27b-3p, miR-574-3p and miR-125b-5p) are more efficient biomarkers than lncRNAs or mRNAs (PCA3, PCAT18 and KLK13).163 Moreover, observations of expression changes of miR-205-5p, miR-222-3p and SNORD48 in a cohort of patients with endometrial cancer suggested their implication in cancer development.164 On the other hand, RNAs can be carried in extracellular vesicles (EV), where they are protected from degradation by RNases. It has been demonstrated that non-coding RNAs contained in extracellular vesicles (EV) are involved in regulation of transcription and post-transcription and thus are an efficient biomarker to monitor cancer progression.165 Mutated tumoral RNA from EVs has been detected in small amounts by RT-dPCR in the case of ovarian cancer45 and of NSCLC.166,167 Furthermore, the combination of circulating and vesicle-associated miRNAs showed potential clinical significance for the identification of pancreatic cancer patients.168 The EV study remains for now limited by the efficiency of the step of EV isolation/enrichment,169 as for CTCs.
3. Other applications of dPCR in oncology
In addition to potential applications in diagnostic of solid and liquid biopsies, dPCR is a powerful tool for analysing bone marrow aspirates and/or peripheral blood, where the entire peripheral blood DNA is studied without distinguishing between cell-associated and cfDNA.Such analyses are conducted to detect mixed chimerism (MC) following allogeneic stem cell transplantation (HSCT). In patients with haematological disorders, surgery is not an option as they do not present any solid tumour. However, HSCT offers a curative treatment, along with cellular therapies like virus-specific T cells.170 HSCT involves replacing the patient stem cells with haematopoietic stem cells from a compatible donor, which can lead to MC. Prolonged MC is undesirable, as it is often linked to disease recurrence.171 The detection by dPCR of MRD and MC after transplantation has shown to be competitive compared to the gold standard methods, the short tandem repeat amplification by PCR (STR-PCR) with a good correlation and a shorter turnaround time,170,172 allowing for a more effective monitoring of remission and adjustment of treatment.170,171 When combined to multiparameter flow cytometry, dPCR has also permitted the precise identification of patients with high risk of relapse from bone marrow aspirates after HSCT.173
dPCR can also be used for quality control of biotherapies. For instance, in the chimeric antigen receptor (CAR) T-cell therapy, an emerging and highly personalised immunotherapy: it consists in genetically modifying ex vivo the T cells of the patient. The transduced T cells will express CAR on their surface enabling the specific recognition of tumour cells by the immune system. This genetic modification approach has been declared as potentially oncogenic and toxic necessitating quality and safety controls. Indeed, the FDA requires a maximum of 5 vector copies per transduced cell, which is enough to be efficient while minimising the oncogenic risk.174 dPCR allowed for the precise quantification of vector copy number in CAR T-cells expressing both anti-CD19 and anti-CD22 receptors, called AUTO3.175 Similarly, a triplex dPCR was demonstrated to be as efficient as two duplex dPCR, to quantify 3 targets in AUTO6NG T-cells, an improvement of AUTO6 (anti-GD2 and anti-RQR8 CARs) against neuroblastoma.176 Furthermore, clinical reports have testified that the continuous proliferation of CAR-T cells in vivo is a key factor to ensure the therapeutic effects.177,178 Thus CAR-T cell monitoring became crucial to follow treatment response. dPCR has shown stable results in quantifying the CAR transgene after CAR-T cell infusion in peripheral blood samples179,180 and other sample types such as bone marrow and lymph node material.181 The limit of detection (LoD) was 20 copies per μg DNA.181
In conclusion, these studies suggest that dPCR is a powerful tool for clinical applications in cancer medicine. Its performances in terms of sensitivity and specificity are mostly similar to the current gold standard methods, such as qPCR. Its high reproducibility to detect oncogene mutations, CNV or fusion gene is due to its ability to perform absolute quantification not relying on standards. It makes it reliable and suitable for patient monitoring during the perioperative period, during and after treatment, for therapy response study or minimal residual disease detection. Also, in the cases of cancers where solid biopsy is not an option (haematological disorders), the detection of biomarkers in liquid biopsy by dPCR allows diagnostic and patient follow up. However, the small quantity of ctDNA or CTCs shed into bloodstream still represents a real technological challenge for dPCR to be used in clinics, particularly for early-stage diagnostic. From a non-clinical, fundamental research perspective, dPCR has facilitated the study of complex cancer mechanisms, notably enabling biomarker discovery.182–185
III. Prenatal testing
The emergence of cfDNA as a tool in cancer medicine has inspired researchers in the field of prenatal testing. Indeed, in 1997, a simple PCR targeting DYS14 gene on the Y chromosome highlighted the presence of cell-free fetal DNA (cffDNA) in plasma of pregnant women bearing male foetuses.186 Since this discovery, invasive procedures such as amniocentesis and chorionic villus sampling (CVS), generally associated with up to 1% risk of miscarriage,187 could be avoided. Non-invasive prenatal testing (NIPT) has become a clinical reality to evaluate numerous genetic disorders. Although rising during gestation, cffDNA represents a low fraction of cfDNA in maternal plasma ranging from 0.5 to ≈30%.188,189 For this reason, dPCR seems a more suitable method for NIPT than the current gold standard methods, namely qPCR or NGS in the case of genetic disorders. Moreover, the targeted nature of dPCR avoids ethical questions rising from NGS screening of an unborn child genome and from pregnancy choices. For inherited diseases, when the presence of an allele variant cannot conclude on the affected foetal status, genotyping is needed and dPCR can be coupled with the digital relative mutation dosage (RMD).189,190 Indeed, dPCR permits precise allele quantification and RMD determines if the dosages of the mutant and wild-type alleles of a disease-causing gene are balanced or unbalanced in maternal plasma.191 Knowing the parental genotypes, it enables to deduce the foetus status (see Fig. 6b). | ||
| Fig. 6 a) Example of a workflow for implementation of NIPT in the case of paternally inherited monogenic disorder or in the case of de novo mutations in clinical practice as a first step into personalised medicine. Reproduced from ref. 211 with permission from John Wiley and Sons, copyright 2022; b) the principle of digital relative mutation dosage. It allows one to deduce the foetus status from the parental genotypes and from the amount of mutant allele (M) and wild-type allele (W) in maternal plasma. For instance, if both parents are heterozygous, M = W if the foetus is heterozygous, whereas W > M or W < M, if the foetus is homozygous for the wild-type or the mutant allele, respectively. When the mother is heterozygous and the father is homozygous and mutated, M = W if the foetus is heterozygous and W < M if the foetus is homozygous. When the mother is homozygous and mutated and the father is heterozygous, W < M if the foetus is heterozygous and without wild-type allele if the foetus is homozygous. | ||
Evaluation of the cffDNA fraction is the first step of NIPT, playing a crucial role in determining sample quality and test reliability. For a male foetus, dPCR has shown to be efficient and reliable by targeting the SRY gene on the Y chromosome as early as 7 weeks of pregnancy.192 For a female foetus, a positive test is preferred to the assessment of the absence of result for the SRY gene. Thereby, it is possible to examine the paternal X-chromosomal alleles for multiple insertion/deletion polymorphisms by dPCR, and it allowed the detection of 42/63 patients bearing a female foetus.193 In addition to evaluating the cffDNA fraction, these methods enable sex determination, which can lead to further analysis, for example in the case of X-linked inherited human disorder such as haemophilia, adrenal hypoplasia or muscular dystrophy. In haemophilia, only a male foetus will suffer from bleeding disorders, caused by mutations in the coagulation factor genes F8 and F9. With the study of 15 male cases, dPCR has proved to be an affordable method to directly detect these variants in samples with cffDNA ranging from 3% to 33%,194 enabling an adaptive intervention, like a caesarean to reduce the risk of intracranial haemorrhage during birth.195 Other strategies based on MSRE and dPCR successfully estimated the cffDNA fraction via seven fetal-specific differentially methylated regions.196
Although autosomal monogenic diseases are well understood due to their simple inheritance patterns (dominant or recessive), their detection through NIPT has only recently begun. In the case of dominant allele inheritance, the presence of paternal mutation in cffDNA will directly conclude an affected foetus,197 whereas a maternal mutation will need RMD to determine the foetal genotype.191 For example, achondroplasia is an autosomal dominant genetic disease caused by mutations in the FGFR3 gene, leading to dwarfism or skeletal dysplasia. It is usually detected during routine ultrasound in the 3rd trimester of pregnancy and confirmed by molecular testing on foetal genomic DNA obtained by an invasive procedure. In a study on 25 women carrying a foetus at risk of achondroplasia according to ultrasound results, dPCR was compared to mini-sequencing on plasmas and to conventional Sanger sequencing on foetal DNA obtained by amniocentesis. dPCR and mini-sequencing were both concordant with traditional testing, detecting 4/4 cases of achondroplasia.198 Likewise, a case study of a man affected by an autosomal dominant disease (MEN1) used NGS analysis to identify and reclassify the MEN1 c.654G>T mutation as a pathogenic variant. In this study, dPCR has been performed as a personalised medicine service with a specific design of primers and probe, on the cffDNA of his pregnant wife. It resulted in the absence of the mutated variant, excluding the risk of disease for the foetus.199 dPCR also enabled the detection of neurofibromatosis, another autosomal dominant disease, due to mutations in the NF1 gene, at the early late trimester by targeting the paternal NF1 variant in 3 out of 4 couples and thus correlating with the results from foetal genotyping by invasive sampling.200 In parallel, this study investigated the CFTR mutations causing an autosomal recessive disease namely cystic fibrosis, but necessitated invasive testing to conclude.200,201 Indeed, in the case of autosomal recessive disease, the presence of the variant alone is inconclusive with regards to the affected status. The use of RMD associated with the highly sensitive allelic quantification of dPCR allowed the foetal genotyping in the case of phenylketonuria due to mutations in the PAH gene,202 in the case of spinal muscular atrophy due to deletion of SMN1 gene203 or in the case of diabetes associated with GCK or HNF4A variants.204 Thalassemia is another autosomal-recessive inherited disease, resulting from abnormal haemoglobin chain synthesis and leading to blood disorders. The thalassemia type, called α-thallassemia, is caused by the deletion of the α-globin gene, and is seen mainly in Southeast Asia. This variant CNV was detected accurately by dPCR in at least 90% of cases,205,206 but the detection of the second variant, β-thalassemia, by dPCR was not conclusive.206 Indeed, β-thalassemia is caused by many mutations in the β-globin HBB genes. The most frequent mutations in the Mediterranean area are β+IVSI-110 G>A207 and β039,208 whereas in Asia it's a 4-base pair deletion (-CTTT) at codon 41/42.206 dPCR coupled to RMD and Z-score analysis has permitted the identification of almost all homozygous mutated cases, which correspond to the real case in which the foetus could become a β-thalassemia patient independently of the mutation origin,208,209 avoiding the need for invasive obstetrical procedures. Moreover, allelic ratios of the heterozygous and wild-type homozygous foetuses were clearly distinguishable without overlapping, permitting correct genotyping as early as the seventh week of gestation.208 However, inconclusive or misclassified cases may occur from either an insufficient foetal fraction or excessively fragmented cffDNA,208 highlighting the importance of quality control in cffDNA studies. Thanks to dPCR, it is now possible to screen for multiple disorders with reasonable quantity of maternal blood.210 Moreover, it is progressively leading to personalised analysis, with the target mutations deduced directly from the parent genotyping211 (see Fig. 6a).
Other haemoglobinopathies can benefit from dPCR advantages, such as alloimmunisation disorders. Indeed, a pregnant woman presenting an antibody for a blood group antigen requires intensive monitoring to prevent risks of haemolytic disease of the foetus or newborn (HDFN). In the case of Rh blood group antigen D, RHD genotyping done in parallel of sex determination by dPCR was found to be much more sensitive than qPCR (sensitivity of 100% for dPCR vs. 83% for qPCR), allowing the RHD-negative women to be administrated prophylactic anti-D treatment.212 In late first semester samples, dPCR has been demonstrated to be highly reliable in the genotyping of other blood groups, such as in the Kell and Duffy systems, by detecting single nucleotide variants (SNVs) in the D or Fya and Fyb antigens respectively.36,213,214 This contrasts with RhD genotyping, which relies on detecting a gene deletion rather than SNVs. Alloimmunised antibodies can also recognise human platelet antigens (HPA) and generate foetal and neonatal alloimmune thrombocytopenia (FNAIT). The most common antigens are HPA-1a, HPA-5b, HPA-3a and HPA-15b detectable via mono or biallelic polymorphisms classified in the Immuno Polymorphism Database. Here again, dPCR has shown to be efficient for the early identification of pregnancy at high risk of FNAIT,188 with an LoD as low as 0.05% for HPA-1a and non-ambiguous results on the 13 pregnant women tested.213
Historically, one of the first targets of prenatal diagnosis was chromosomal aneuploidies, as trisomy diseases originate from copy number aberrations of chromosomes 13, 18 and 21 (for example). The test employed in clinical practice is FISH, a labor-intensive, long (overnight hybridisation is generally needed) and costly technique, or qPCR. In 2019, a combination of duplex dPCRs helped to identify cases of CNV of the chromosomes 13, 18, 21 and Y or X by targeting respectively the genes MBNL2, EHZF, PRDM15 and SRY, and non-coding region on chromosome X, in a cohort of 133 prenatal CVS samples.215 It has proved the rapidity, the simplicity and the cost-effectiveness of dPCR as a tool for NIPT. Moreover, in the development of less invasive procedures targeting cffDNA in maternal plasma, the real challenge comes from the low cffDNA concentration. In order to meet the challenge, a proof-of-concept study on trisomy 21 increased the number of targets in a two color 8-plex ddPCR, with 4 FAM-probes targeting genes on chromosome 21 (BRWD1, LTN1, NCAM2, RUNX1) and 4 VIC-probes targeting genes on chromosome 18 (CTIF, RIT2, SMAD4, TCF4) as a reference, with the aim of increasing positive droplets. This test succeeded to detect trisomic DNA content with a sensitivity of 94% and a specificity of 98% and revealed 16/21 cases of trisomy 21 on a large cohort of 213 pregnant women already screened with an invasive procedure to have foetal karyotype.47 A study added an enrichment step of cffDNA by size selection to a ddPCR targeting 4 genes on chromosome 21 with FAM-probes (SETD4, CRB1, UBE2G2, CLDN14) with references (VIC probes) on chromosome 1 and 2. This method showed an improved sensitivity of 100% for the 50 positive samples and 3 false positive results for the 827 negative samples, giving an overall accuracy of 99.66% on 877 pregnant women plasma samples.216 Another study identified cases of trisomy 21, 18 and 13 in 283 clinical samples with a sensitivity of 100% and a specificity of 95.12%.217
Although not yet in NIPT, de novo mutations are another process for disease apparition in newborns. However, it has been reported that such mutations can actually come from parental mosaicism,218,219 a condition in which cells within the same person possess more than one genetic line. Indeed, in a study on alternating hemiplegia of childhood, dPCR results revealed that 7.5% (6/80) of cases classified by sequencing as de novo were actually linked to parental mosaicism220 and they correlated the MAF of mosaicism with phenotype severity. Many other dPCR-based studies revealed mosaicism from the mother221,222 or father,223,224 whereas newborns were initially classified as presenting de novo mutations with asymptomatic parents. Such studies highlight the importance of mosaicism identification in both parents and newborns, to provide supportive genetic counselling and guidance on fertility choices. Moreover, aneuploidy has been shown to be detectable by dPCR even with high maternal mosaic contamination.225
In conclusion, the high sensitivity of dPCR enables the analysis of the foetal DNA fraction in the maternal blood sample, making non-invasive prenatal diagnosis a reality. Genetic aberrations such as monogenic disorders, alloimmunisation, aneuploidy and even parental mosaicism can be efficiently identified. Moreover, the targeted nature of dPCR is an advantage in NIPT, compared to non-targeted NGS methods, as it decreases the costs, and it avoids rising questions on ethics from accessing the constitutive genomic sequences of an unborn child.47
IV. Pathogen detection
1. Viral infection
Viruses are found in almost every ecosystem on Earth and are the most abundant type of biological entities. They need a host living cell of other organisms to enter and replicate in. For an early detection, very sensitive assays are needed. The gold standard method for detecting viruses is qPCR after an RNA/DNA extraction. But the high sensitivity of dPCR makes it very attractive for an earlier diagnosis.A recent example of a disease that welcomed dPCR for a more accurate diagnosis is the coronavirus disease 2019 (COVID-19). The COVID-19 outbreak, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), triggered a worldwide public health problem, declared as a pandemic by the World Health Organisation (WHO). Despite the development of antigen and antibody testing kits for rapid diagnosis, the World Health Organisation (WHO) recommends the use of a nucleic acid test as a standard method of confirmation of SARS-CoV-2 infection.1 Detection protocols by RT-qPCR usually target at least 2 independent genes of the virus genome among parts of the open frame reading gene (ORF1), the spike gene (S), the envelope gene (E) or the nucleocapsid gene (N).1,226,227 Although RT-qPCR is high-throughput, compatible with automation and sensitive,228,229 in some cases the clinical symptoms were not in correlation with the nucleic acid test results (false negatives),229,230 leading to time and material consuming repeated swab tests. These false negative results could be explained by an insufficient viral load, by experimental errors or by the presence of inhibitors in the swabs that are known to reduce RT-qPCR efficiency. Unfortunately, they could lead to a delay in infection confirmation, an incorrect diagnosis of treated patients in recovery and a relapse after discharge, leading to disease spread. In contrast, dPCR improves sensitivity and accuracy of the diagnosis in clinical samples, particularly in low viral samples.231 Tao Suo and collaborators demonstrated that, with an LoD of 2.1 and 1.8 copies per reaction for ORF1ab and N, respectively, ddPCR is 500 times more sensitive than qPCR (LoD of 1039 and 873.2 copies per reaction for the same genes).232 From qPCR to dPCR, the sensitivity rose from 40% to 96% and 26/77 patients were detected negative by qPCR but positive by dPCR. Similarly, Paolo Poggio and collaborators found that 11 (61%) out of 18 qPCR negative patients were positive by dPCR in a cohort of 64 patients, increasing the sensitivity to 89% compared to qPCR (72%).233 Finally, Chong Liu et al. studied only recovering hospitalised patients (43) and determined a cut-off value of 0.6 copy per reaction. On the 9 discharged patients by qPCR, 8 turned out to be positive by dPCR.234 These results clearly indicate that dPCR drastically reduced the number of false negatives, which makes it especially suited to study asymptomatic and suspected patients or close contacts. Moreover, the reproducibility of dPCR is much better than that of qPCR. Indeed, where qPCR requires calibration curves for quantification, dPCR allows an absolute quantification of RNA by counting the positive reactions. It shows a high degree of consistency by avoiding the variations coming from experimental conditions (analytical protocols, instruments, operators or laboratories) and from the references needed to produce calibration curves.32,33,235,236 A study recently reported the successful use of RT-dPCR, compared to RT-qPCR, as a reference measurement procedure to perform external quality assessment for molecular diagnostic testing of SARS-CoV-2. While, among three institutes, 61 laboratories observed a good agreement of median values between both technologies, only a <2-fold difference between laboratories was demonstrated for RT-dPCR, whereas RT-qPCR differences were generally between 10 and 50-fold.237 The superior accuracy and reproducibility of dPCR make it suitable for long-time monitoring of viral load in convalescent patients but also for monitoring the influence of treatment or vaccination.238 Indeed, the promising drug azvudine (FNC) has been tested on a 281-patient cohort, and the results indicated that it permits a faster virus elimination and a reduced time of treatment.239 Also, as dPCR is highly resistant to inhibitors, it enables the detection of viral RNA in complex body fluids such as blood. It has also been shown that the quantitative detection of SARS-CoV-2 (RNAemia) in blood is highly correlated to disease severity.228,240 This prognostic biomarker could be a crucial asset to predict clinical deteriorations. The inhibitor resistance also led to the development of more direct quantification by shortening the protocols typically with a 1-step RT-dPCR. But the efficiency of this method had questionable sensitivity compared to RNA extracted and analysed by dPCR in 2 steps.228,241,242 Finally, the potential drawback of diagnosis by acid nucleic testing is the impossibility to distinguish infectious viral particles from non-infectious RNA.242 However, in the epidemiological context of COVID-19, dPCR presents several advantages, such as rapidity and safety, over the classical culture-based method, which is a labor-intensive and time-consuming (3–4 days) protocol, potentially risky due to required manipulations in high biosafety level settings (BSL3 out of 4) and prone to significant variability from non-standardised protocols and operator errors.
The use of dPCR technology has also shown great interest for the detection and quantification of human immunodeficiency virus (HIV). Although HIV appeared in the 80s, it is still a major issue for global health.243 It causes the acquired immunodeficiency syndrome (AIDS), which induces a progressive failure of the immune system through the infection of macrophages, dendritic cells and helper T cells (particularly the CD4+ T cells).244 During the primary infection, the symptoms are not worse than the ones of a general influenza, but in time, the immune system becomes vulnerable to life-threatening opportunistic infections and cancers. The current treatment of HIV consists in the use of antiretroviral therapies (ART) that block different steps of the HIV transcriptional cycle.245 Despite an effective suppression of plasma viremia by ART,246–248 the virus remains present in the so-called latent reservoir of infected cells249 harboring replication competent proviral HIV DNA in their genome, allowing its persistence and rebirth as soon as ART is stopped. It has been reported that HIV DNA, as well as HIV RNA, before and during treatment, has prognostic significance and can predict treatment efficacy.249,250 As early as 2012, dPCR was used to monitor levels of total HIV DNA in patients on ART.246 Compared to the gold standard qPCR methods, the dPCR superiority in terms of sensitivity has been questionable. Semi-nested qPCR was shown to be more sensitive than ddPCR in samples from patients on ART, particularly for low viral charge samples.251,252 In contrast, similar sensitivity between these methods has been demonstrated by others.38,253 False positive signals were also described to affect the detection power of dPCR,38,246,251,254,255 and the threshold between positive and negative partitions is a real challenge to determine.254,255 On the other hand, dPCR exhibited a better accuracy and reproducibility.252,256,257 Moreover, dPCR absolute quantification enabled one to highlight the progressive loss during culture of HIV from 8E5 cells, the cell line used as a classic standard for qPCR calibration. A deviation of the number of HIV DNA contained per 8E5 cell from 1 DNA copy initially to 0.73–0.43 copy per cell depending on sources has been demonstrated.252,256 Such results imply an overestimation of the DNA copy number detected by qPCR and then of the latent viral reservoir. It could thus lead to incorrect patient monitoring that would have consequences on patient health. Follow up of treatment response by dPCR is also possible.258 For example, studies revealed how important are the timing of treatment initiation and the treatment itself (regimen and exposure) to affect the HIV reservoir.259,260 Furthermore, dPCR multiplexing and robustness to target sequence variations turned out to be an important feature in the detection and study of HIV. Indeed, the HIV genome often contains defects such as hypermutations or deletions and might not be efficiently transcribed after latency reversal.251,261 Therefore, the study of intact proviruses is crucial. Some dPCR methods such as Rainbow 5-plex dPCR262 or an intact proviral DNA assay (IPDA)234,236 differentiated and quantified intact proviruses (<10% of the total proviruses) from replication-defective ones and thus studied their dynamics. For these studies, dPCR presents the advantage of being faster, more accurate and less time and reagent consuming than culture methods.263 Also, this sequence tolerance allowed the development of two HIV assays by ddPCR for the detection of the worldwide most HIV prevalent subtypes.250,264 Adaptation of dPCR to RNA detection has also been useful to study HIV transcription mechanisms.245,265,266 Not only for patients on ART, dPCR has been used for the monitoring of patients, who underwent allogeneic stem-cell transplantation, with genetically modified cells, in remission at 18 months after ART interruption.267
The high sensitivity of dPCR was also demonstrated to be pertinent for the detection of the hepatitis B virus (HBV). Indeed, similar to HIV, HBV DNA is inserted into the nucleus of infected cells, in a more stable converted form, a covalently closed circular DNA (cccDNA).268 The persistence of cccDNA in infected hepatocytes is a major obstacle to curing chronic hepatitis B. Thus, dPCR methods to detect HBV and monitor patients under treatment have been developed.269 dPCR has been massively compared for cccDNA detection to classical serological tests270–272 or to more sensitive qPCR assays.273,274 Over these routine tests, dPCR demonstrated superior sensitivity and accuracy. Indeed, dPCR's LoD was evaluated at 8 copies per mL in plasma samples,273 100 copies per mL in serum samples271 and 1 copy/20 ng in liver tissue samples.274 Moreover, the correlation between the tumour stage of HCC and HBV was demonstrated by dPCR, whereas serological tests presented 18.3% of false negative results for HBV DNA detection.270 Similarly, the integration rate has been correlated by dPCR to the natural clearance of chronic HBV infection.275 dPCR also allowed the study of occult hepatitis B infection, that is transmitted usually during liver transplantation or blood transfusion. Indeed, as it is characterised by very low concentrations of serum HBV DNA, dPCR provides an added value in the optimisation of its diagnosis276 but also in the improvement of the patient therapeutic management before or after a liver transplantation.277
The multiplexing capacity of dPCR is highly attractive for the identification of other viruses. For example, it is useful for detecting the four serotypes of the dengue virus278 or for identifying high-risk human papillomavirus (HPV) serotypes, such as HPV16/18/11/45.279,280 Considering that HPV infections can increase the risk of developing cancer, it has been shown that ctHPV-DNA is highly correlated to tumour viral load in HPV-associated cancers, such as oropharyngeal squamous cell carcinoma,281 cervical cancer282 or anal cancer.283 These studies demonstrated the potential value of ctHPV-DNA as a biomarker at baseline and during and/or after treatment, highlighting its value for treatment response monitoring,284–286 as well as for diagnosis.287
The use of dPCR was also described as a quality control of adeno-associated virus vectors for HIV immunisation by neutralising antibodies,288 but also for other viruses like dengue289,290 or Ebola291 viruses to determine the ratio of particles to infectious units requested by the WHO for vaccine manufacturing.
In conclusion, dPCR offers exceptional sensitivity and accuracy for viral diagnostics, especially in cases of low viral loads or complex infections. Its success in COVID-19 detection has paved the way for its use in monitoring chronic infections like HIV and hepatitis B, and its multiplexing ability enhances detection of various viruses. Despite some challenges, dPCR shows great potential for improving early diagnosis, treatment monitoring, and standardising viral testing globally.292
2. Non-viral infection
Non-viral infections can be caused by bacteria, fungi or parasites. The methods usually used for their detection are culture-based. But inevitably, they are time-consuming and labour-intensive, which inflates the costs, making them unaffordable in some countries. Nucleic acid testing such as dPCR can provide a solution to these problems. Here we present specific examples of diseases that benefited from dPCR's advantages.With 10.6 million infected people and 1.3 million deaths, tuberculosis, caused by Mycobacterium tuberculosis (MTB), was the second highest killer worldwide in 2022, after COVID-19.293 MTB usually infects the lungs, but it can also disseminate in extrapulmonary organs,286 complicating its detection: indeed, collecting samples from these distant sites often requires invasive procedures like surgery. It has been demonstrated by dPCR that MTB can be detected through circulating DNA in plasma.294,295 Particularly in comorbidity situations, like the case reported by the team of Yamamoto, the non-invasiveness of dPCR is crucial: dPCR successfully detected MTB in plasma, where urine, sputum and blood samples all tested negative using non-dPCR based commercial tests, except in liver tissues after autopsy.296 Moreover, for an earlier diagnosis of MTB, it has been shown that the detection rate in low concentration samples is higher when using exosomal DNA than cfDNA:297 the clinical sensitivity was estimated to 75% and 61.4%, respectively. Otherwise, most of the time, tuberculosis is latent and cannot be easily detected. Thanks to the multiplexing character of RT-dPCR, differentiation between MTB, latent MTB and other diseases has been proven feasible by targeting different transcriptional signatures.298 Furthermore, a dPCR-based study in CD34-positive PBMCs has reported that MTB DNA is a good biomarker of latent MTB.299 On the other hand, MTB is usually treated with antibiotics, but multi-drug resistance emerges when inappropriate health care is provided. Thus, precise strain identification and drug susceptibility testing (DST) are needed. Unfortunately, usual culture assays are not suitable as their turnaround times are too long (5–24 days for identification and DST), the results are not reliable, and they require a lot of material which increases the costs.300 For these reasons, new methods have been developed for DST. For example, the combination of culture and ddPCR enabled the detection in 5 hours and the DST within 4 days directly from sputum.301 Also, a drop-off triplex ddPCR assay targeting all the mutations on the major resistant genes for isoniazid has been optimised allowing patient monitoring during treatment (see Fig. 7a). It revealed a correlation between bacterial load and symptoms, an interference of hyperglycaemia with drug efficacy and a slower decrease of bacterial load in the case of multi-drug resistance.302
 | ||
| Fig. 7 Schematic illustration of the drop off dPCR strategy to target all mutations in antimicrobial resistance (AMR) genes: the reaction contains a FAM-labelled drop-off probe targeting katG 315 and a HEX-labelled reference probe spanning an adjacent invariable region. Mutations in katG 315 were detected as FAMlow/HEXhigh with a vertical shift, which could be distinguished from the FAMhigh/HEXhigh double positive droplets of the wild-type sequence. Reproduced from ref. 302 with permission from the American Society for Microbiology, copyright 2023; b) schematic overview of the mediator probe PCR principle, showing the separation between the target detection and fluorescence signal generation steps. Reproduced from ref. 355 with permission from MDPI, copyright 2024. | ||
Bloodstream infections (BSI) are another important public health threat worldwide with high mortality and morbidity, particularly those leading to sepsis.303 The definition of sepsis was adjusted in 2016 as “a life-threatening organ dysfunction caused by a dysregulated host response to infection”.304 25–30% of sepsis cases are due to bloodstream infections.305 These diseases can be due to diverse pathogens as fungi or bacteria, but the key ones are Klebsiella spp. (species), Escherichia coli, Staphylococcus aureus, Pseudomonas aeruginosa, Enterococcus spp., Streptococcus spp. and coagulase-negative staphylococci.306 They are treated with antibiotics, and it has been shown that the sooner the antimicrobial therapy starts the better the chances of survival. Indeed, each hour of delay between hospital registration and antibiotics administration is associated with a 9% increase in the odds of mortality and the median time is 2.1 h.307 Thereby, patients are rapidly treated with broad-spectrum antibiotic, which can be inadequate and can result in drug toxicity, antimicrobial drug resistance and an increase in hospital readmissions and health costs. Indeed, the current testing assays are culture-based, but their turnaround time is too long.308,309 It is thus urgent to develop fast and accurate assays for identification and DST. An infection is classically detected via the host immune response, i.e. through the white blood cell count, C-reactive protein or procalcitonin (PCT) levels for example. It has been shown that dPCR targeting HLA-DRA RNA (coding for the alpha protein of the MHCII complex) used in combination with PCT had a better predictive ability than PCT only to detect sepsis.310 Also, the IgM response detected at a transcriptomic level by dPCR has been proved to be a promising approach for an early diagnosis.311 On the other hand, the direct detection of species-specific signature by dPCR has been demonstrated to detect rapidly in approximately 4 hours,312–314 with a high sensitivity and specificity, the key bacteria responsible for BSI in blood,312,313,315 enabling an early detection of sepsis.316 Indeed, the multiplexing capability of dPCR has been a major asset for targeting both specific signatures and antibiotic resistance gene,317–319 allowing guidance for antimicrobial therapy. To enlarge the BSI cause identification, fungi-specific genes can be added as targets in addition or not to bacterial genes and antimicrobial resistance (AMR) genes in blood samples,320–322 but also in fecal samples323 or pleural and peritoneal fluids.324 One of these studies highlighted that dPCR is more efficient to detect polymicrobial infections than culture-based assays.321 The highly conserved bacterial 16S rRNA and fungal 28S rRNA can also be targeted by duplex dPCR, for the differentiation between bacterial and fungal BSI.325 It's worth noting that reagents used for bacterial lysis are generally incompatible with dPCR buffers or droplets,326 implying an indirect detection of DNA released by bacteria in bloodstream or other biofluids. Isothermal amplification-based technologies, less prone to detergent inhibition, could be investigated for direct detection.327,328 However, because of their low multiplexing and specificity, they require further development in order to be considered for the clinic.
Finally, in a non-clinical application but non negligeable topic, dPCR is very useful for sensitive detection of foodborne pathogens such as Shiga toxin-producing Escherichia coli329,330 or of biothreat bacteria such as Bacillus anthracis or Yersinia pestis.331
In conclusion, dPCR is highly efficient for the precise identification of pathogens without the need for enrichment steps, considerably reducing turnaround time to approximately 4 hours. This enables timely patient management, reduces morbidity and mortality and helps prevent the escalation of antimicrobial resistance.
V. Perspectives of dPCR
1. Integration of dPCR with other methods of detection
Besides nucleic acids, proteins are another important class of biomarkers closely linked to each individual phenotype. Traditional protein detection methods, such as western blot or ELISA bioassays, often lack sensitivity and accuracy. The nascent field of digital protein detection has nonetheless rapidly expanded, offering the same benefits as digital PCR over qPCR. We will focus on the technologies that bridges these two fields (for a comprehensive review on digital detection of proteins, the reader may refer to D. Duffy's article332). In fact, PCR has for a long time been employed to enhance the sensitivity of protein detection. Notable examples include heterogeneous formats like immuno-PCR and other adaptations of the microplate ELISA protocol with isothermal nucleic acid amplification methods such as immuno-RCA or immuno-LAMP. Additionally, homogenous assays like proximity extension assays (PEA) or proximity ligation assays (PLA) have been developed. In these approaches, two DNA-labelled monoclonal antibodies bind to separate epitopes of the same target protein, enabling the DNA tags to come into close proximity and be either extended or ligated. The resulting duplex can then be quantified by qPCR. These assays have since been adapted to a digital format to answer the need for accurate quantification.Schröder et al. modified the standard microplate immuno-PCR protocol to release the DNA-tagged antibody following the formation of the complex with the target protein (IL-2 or IL-6), allowing for its subsequent quantification by ddPCR.333 Another approach involves grafting the capture antibody on nano/microparticles instead of on a microplate. At high particle concentration, the formation of the sandwich [capture antibody/target/DNA-tagged detection antibody] is governed by the Poisson law, which allows for digital readout after isolating the particles followed by PCR amplification of single DNA tags. Zhang et al. employed magnetic nanoparticles combined with droplet PCR for the quantification of α-synuclein in serum;334 Vanness et al. used fluorescently-encoded microparticles isolated in microwells for the multiplex and multimodal detection of miRNA let-7a and cytokine IL-6.335 Li et al. introduced another multimodal approach, termed digital simultaneous cross-dimensional output and unified tracking (dSCOUT), which integrates CTC enrichment and DNA–antibody conjugate tagging of surface markers. This approach enabled the simultaneous analysis of three proteins (including the tumour specific-marker EpCAM) and three mRNAs in CTCs, demonstrating its diagnostic potential for HCC.336
Extracellular vesicles and exosomes can similarly benefit from ddPCR quantification by being labelled with DNA-tagged antibodies. Ko et al. applied direct labelling of EVs with orthogonal antibody–DNA conjugates, which allows single EV phenotyping for EGFR and EpCAM markers.337 Lin et al. adapted the PLA approach to analyse the PD-L1 status of tumour-derived exosomes;405 in their method, two aptamers recognised EpCAM and PD-L1, producing a ligated product only if the exosome displays both markers, eliminating the interference of non-tumour-derived exosomes and soluble proteins.
Abasıyanık et al. adapted the PLA protocol to the detection of both nucleic acids (bacterial DNA) and proteins (IL-6 and TNA-α) for the prediction of septic shock outcome in patients.338 Byrnes et al. developed a simplified protocol, which does not require washing the excess of antibody and uses a polydisperse emulsion.339
In summary, these new technologies are paving the way to diversify the field of application of dPCR, beyond genomic or transcriptomic, to proteomic.
2. Remaining challenges of dPCR & potential ways of improvement
Despite its undeniable strengths (calibration-free, high sensitivity, absolute quantification), some technical points remain to be optimised in order to reach better performances (Table 3).| Challenge | Technology | Principle | References |
|---|---|---|---|
| Multiplexing | Lock nucleic acid-modified probes | LNA makes the probes thermally more stable and more specific | 341, 343–346 |
| Drop-off probes | Fig. 7a | 81, 302, 347 | |
| Melting curves analysis coupled to dPCR | Melting curve at the end of the real-time dPCR: amplicons discrimination by their different Tm | 348–352 | |
| Photobleaching probes | See article | 353 | |
| Mediator probes | Separate the DNA detection from the fluorescent signal generation | 354, 355 | |
| Dynamic range | 3D analysis dPCR | 1-Million bilayer droplet array | 28 |
| dPCR coupled to light-sheet microscopy | 16, 356, 403 | ||
| Multivolume droplet-dPCR | Smaller droplets used for a better upper limit of quantification (LQ), larger ones are used to decline the lower LQ | 358, 361, 362, 404 | |
| Virtual partitioning | In a high target concentration regime: intensity-encoding with multicolor probes | 364 | |
| Threshold determination | Computation | Automatic thresholding | 364, 365 |
| Computation | Automatic cluster labelling | 366, 367 | |
| Portability | Microfluidic-free partitioning | Polydisperse emulsion with analog readout | 370 |
| Particle-templated emulsification system | 368 | ||
| Pipette-based droplets microprinting | 352 | ||
| Centrifuge-based partitioning | Dual flow-focusing function; lab-on-a-disc | 371, 372 (respectively) | |
| Smartphone-based POCT | Integration of dPCR on a smartphone for point-of-care testing | 373–378 | |
| Handling-free protocol | Droplet microfluidic (DMF) system | EWOD-controlled droplet movement | 386, 387 |
| DMF and centrifugal microfluidic | POCT dPCR (portable and all-integrated protocol) | 388 |
The use of the locked nucleic acid (LNA) in the detection probes has been considerably used and reported in dPCR. Thanks to a 2′-O,4′-C methylene bridge in the ribose moiety, these optimised probes have a higher affinity to the complementary DNA, which increases both duplex stability and mismatch discrimination. Their use has been suggested to reduce the rain droplets.341 The high stability and specificity of LNA-modified probes allowed the detection of driver mutations at early stage342 or in metastatic343 pancreatic cancers and to target more mutations, up to 40 biomarkers, in the case of foetal aneuploidy for trisomy 21 detection.344,345 Moreover, Hashimoto et al. developed an LNA-clamp ddPCR strategy, where LNA-modified oligonucleotides bind to the wild-type sequence in order to inhibit its amplification. This study highlighted the presence of minor mutated clones of ESR1 with a MAF of <0.1% in fresh frozen tissues of patients with BC.346
Another strategy to increase the number of biomarkers targeted is the drop-off dPCR (see Fig. 7a), where a reference probe targets an invariable region in the vicinity of the mutational hotspot, whereas a drop off probe targets a wild-type sequence, leading to an absence of a double positive signal in the case of a mutated allele. It has been successful in the detection of drug-resistance mutations in MTB302 and of cancer mutations81,347 improving the number of targets up to 69 hotspot mutations, but not permitting the identification of these mutations.
From a technological point of view, multiplexing is the ability to detect and identify simultaneously multiple targets using orthogonal signals. dPCR displayed a relatively low multiplexing capability, often limited to less than a dozen targets, as opposed to microarrays that can measure hundreds to thousands of targets simultaneously. This limitation arises primarily from the restricted number of available fluorescence channels, and the spectral overlap that occurs as more fluorophores are added. In addition, increasing the numbers of primer pairs and probes may affect the amplification efficiency and raises the risk of artefactual reactions such as primer dimers.
Multiplex dPCR typically relies on three main strategies: spectral-encoding, intensity encoding or combinatorial-encoding, or a combination of these. Spectral encoding utilises distinct fluorescence channels with orthogonal probes (up to 7-color for the latest Nio system). Intensity-encoding differentiates targets based on varying probe concentrations,30 producing distinct fluorescence clusters. Alternatively, an intercalating dye can also achieve intensity encoding given that the amplicons are of different sizes. Combinatorial encoding expands multiplexing further by assigning unique targets to specific combinations of fluorophores, effectively increasing the number of detectable targets beyond the available spectral channels.
Planar array imaging integrated with a thermal control enables the recording of melting curves for individual partitions, which has been leveraged for discriminating different amplicons from single-color probes. Applied in microchambers, this strategy has shown promising results for KRAS genotyping in pancreatic cancer samples,348,349 where the amplicons from different mutants display distinguishable Tm. To implement this idea to droplet-based dPCR, an algorithm has been developed to correct the droplet displacement during thermocycling.350 In a study on AMR-bacterial infection, this technology was coupled to a machine learning algorithm, enabling the discrimination of two single-color probes with very similar signals from the same droplets, via the probe differences on the entire kinetic profile during amplification, not only their differences in Tm.351
In another study, Li et al. developed a sophisticated strategy to exploit the difference in the melting temperature between the target and probes with no need to record the complete temperature profile.352 In this case, the non-hydrolytic probe is composed of a universal forward primer anchor domain, a signal domain modified with a fluorophore and a quencher and multiple barcode domains that are complementary to different reverse primers. In an asymmetric PCR regime (with an excess of forward primers that bear the probe binding domain), the single-stranded amplicon can anneal to the probe forming a loop with the universal forward primer binding domain and the target-specific reverse primer barcode domain that display differential melting temperature. As these complexes display differential melting temperature, the target amplicon can be identified by measuring the droplet fluorescence at different temperatures. Using four probes of different colours and two imaging temperatures, the authors successfully demonstrated an 8-plex, that includes a reference sequence and seven EGFR mutation variants encompassing 35 subtypes.
The differential photobleaching property of fluorochromes was also leveraged in a multiplex configuration. In this approach, two probes are used: one labeled with a photosensitive dye and the other with a photostable fluorophore. Droplet imaging is performed before and after photobleaching, causing the signal from the photosensitive dye to extinguish. This enables unambiguous indexing of the targets using a single fluorescence channel. Using the same strategy with three channels, the authors developed a 6-plex compatible with different target panels that gave promising results still to be tested on non-synthetic samples.353 One may imagine that recording the entire photobleaching kinetics would allow the creation of more than two virtual channels, although this has not yet been demonstrated, to the best of our knowledge.
On the other hand, besides the physical limitations of fluorescence, a higher multiplexing capacity implies a cumbersome molecular optimisation coming from increasing the numbers of primer pairs and probes. To overcome this limitation, a strategy consists in separating the DNA detection from the fluorescent signal generation, by using a mediator probe (see Fig. 7b). Compared to LNA-modified probes, this technology gave similar performances for the detection of KRAS and BRAF mutations (4-plex) on samples from patients with CRC.354 Based on this technology, the same team elaborated a generic fluorogenic 6-plex reporter set compatible with different target panels that gave promising results still to be tested on non-synthetic samples.355
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log dynamic range. In addition, in both CLEAR-PCR and UltraPCR, every ddPCR step happened in the same tube: compartmentalisation in droplets is done in a centrifuged tube that is then closed, thermocycled and analysed by 3D imaging. The advantage of such a process is to avoid sample loss issue and operational contamination.
log dynamic range. In addition, in both CLEAR-PCR and UltraPCR, every ddPCR step happened in the same tube: compartmentalisation in droplets is done in a centrifuged tube that is then closed, thermocycled and analysed by 3D imaging. The advantage of such a process is to avoid sample loss issue and operational contamination.
 | ||
| Fig. 8 a) Schematic illustration of the CLEAR-dPCR process with (A) the droplet generation by centrifugation, (B) the high-throughput readout of bulk PCR droplets by 3D light-sheet microscopy, (C) the dual-channel light-sheet fluorescence image sequences, and (D) the volumetric reconstruction of dual-channel images. Reproduced from ref. 16 with permission from United States National Academy of Sciences, copyright 2020; b) example of a fully-integrated smartphone-based device of dPCR for point-of-care testing allowing an isothermal amplification. Reproduced from ref. 373 with permission from the American Chemical Society, copyright 2021. | ||
The dynamic range may also be artificially enlarged simply by testing samples at different dilutions, which mathematically brings a concentrated sample that exceeds the dynamic range (by partition saturation) within the quantification region.357 Alternatively, the sample may be split into partitions of different sizes, effectively associating target concentrations with multiple Poisson parameters (λ). This approach is compatible with multiple partitioning techniques such as surface-assisted droplet printing,358 centrifugal step emulsification359 or microchambers.360 The Ismagilov group adapted the SlipChip design to accommodate wells ranging from 1 to 125 nL, achieving a dynamic range of 5–6 OoM for the quantification of viral RNA associated with HIV viral load.361 In addition to being cost and time-effective, polydisperse emulsions naturally generate a wide range of droplet sizes, which might be leveraged to extend the dynamic range, given a proper droplet analysis framework is provided.
To extend the upper limit of detection beyond the partition saturation point, several methods have been proposed to estimate the number of targets per partition. Luo et al. combined the SlipChip multivolume design with real-time partition monitoring: at partition saturation, where the digital information is lost, the Cq extraction from the analog PCR regime can be used to infer the average target concentration.362 Similarly, Jacky et al. introduced virtual partition dPCR, an intensity-encoding scheme with multicolour probes, coupled with a mathematical model, known as high-definition PCR,363 to estimate the number of target copies per partition.364 At high target concentrations, where negative partitions are nearly absent, quantitative information can still be retrieved by analysing the target distribution across all positive droplets. However, this strategy is only applicable in a multiplex context, as it relies on detecting different target combinations within individual droplets, each yielding a unique signal (Fig. 9).
 | ||
| Fig. 9 Summary of challenges in dPCR, as ways of improvement of dPCR technology. Beyond spectral-encoding, intensity encoding and combinatorial-encoding can be used to index the positive partitions to their target. Other methods such as melt-curve analysis or the use of photo-sensitive/resistant dyes in PCR probes may be used in standalone or in combination with the above-mentioned strategies. The dynamic range may be extended using multi-volume partitioning. When close to saturation, strategies to infer the number of copies per partition such as switching to the analog mode of qPCR or virtual partitioning364 may further increase the higher limit of quantification. Alternative partitioning strategies have been proposed to simplify the process. These include microfluidic-free techniques such as polydisperse emulsification, particle-templated emulsion or the OsciDrop technology (image adapted from ref. 391. The Slip-Chip technology has also been developed to democratize dPCR by proposing a versatile and easy-to-use microchamber-based chip. | ||
A strategy to facilitate the accessibility to ddPCR is to develop microfluidic-free partitioning techniques. In a seminal work, the Abate group reported the use of a particle-templated emulsification system. Monodisperse hydrogel (agarose or acrylamide) particles are mixed with the PCR mix and the sample, before being vigorously agitated with the immiscible oil phase. This results in the isolation of the particles in water-in-oil droplet with similar monodispersity.368 Along the same line, Heinrich et al. used agarose particles covered with a chitosan layer that non-specifically binds to the DNA fragment, allowing the enrichment of the target from a large sample volume, prior to templated emulsification.369 The Abate group also proposed to convert the digital information, obtained by single molecule isolation in a polydisperse emulsion, into an analog readout:370 following the dPCR amplification, the emulsion is broken and the enriched amplicon population quantified by a simple qPCR protocol, eliminating the need for a complex droplet analysis technique. The author reported similar analytical performance to a standard dPCR workflow, but this system reintroduces the need for calibration and raised the question of contamination due to the breaking of the post-PCR emulsion.
Another idea was suggested to eliminate microfabricated chips by using pipette-based droplet microprinting.352 This OsciDrop system allowed a multiplex dPCR to detect EGFR mutations or HER2 CNV in LC or BC FFPE samples, respectively. Despite reduced costs and a rapid process, the machine remained bulky and not portable. Centrifugal microfluidics, cleared from pressure control, has emerged as a simpler and promising solution to address the portability limitation of ddPCR. The team of Shuwen Zeng has developed a lab-on-a-disc (LOAD) device enabling, on the same chip, the generation, thermocycling and analysis (by an external microscope) of droplets. Good performance was demonstrated for screening viruses in clinical samples, but the process still required an external microscope.371 To fully integrate the analysis, the team of Gangyin Luo has proposed a fully automated instrument with a microfluidic chip, where a rotary valve allows the flow-focusing design (with pressure control) to both generate and analyse droplets. This in-line readout based instrument showed good results for the quantification of the HER2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CEP17 ratio in cell lines, and tests on human samples could validate this promising technology for clinical diagnosis.372 In the case of microchamber-based dPCR, smartphone-adapted dPCR devices have been designed, offering on chip compartmentalisation, thermocycling, data acquisition via a smartphone camera and image analysis (see Fig. 8b). A lot of research has been conducted on the optimisation of an embedded heating/cooling system, which requires both flexibility and accuracy to support various protocols, from isothermal373,374 to thermal cycling amplification.374–378 Another challenge of miniaturised dPCR devices is designing chips compatible with industrial scale production374 and that can accommodate as many wells as possible to reach the best sensitivity possible with a smartphone camera imager. Most of the devices developed are still at the level of proof of concept with tests on plasmid DNA, but good performances have been proven with 45 cycles in 49 minutes tested for cancer, aneuploidy and COVID-19 detection377 or with an LoD of 1 copy per μL in the range of 2–1000 copies per μL in the detection of EGFR L858R gene mutation in NSCLC.373
CEP17 ratio in cell lines, and tests on human samples could validate this promising technology for clinical diagnosis.372 In the case of microchamber-based dPCR, smartphone-adapted dPCR devices have been designed, offering on chip compartmentalisation, thermocycling, data acquisition via a smartphone camera and image analysis (see Fig. 8b). A lot of research has been conducted on the optimisation of an embedded heating/cooling system, which requires both flexibility and accuracy to support various protocols, from isothermal373,374 to thermal cycling amplification.374–378 Another challenge of miniaturised dPCR devices is designing chips compatible with industrial scale production374 and that can accommodate as many wells as possible to reach the best sensitivity possible with a smartphone camera imager. Most of the devices developed are still at the level of proof of concept with tests on plasmid DNA, but good performances have been proven with 45 cycles in 49 minutes tested for cancer, aneuploidy and COVID-19 detection377 or with an LoD of 1 copy per μL in the range of 2–1000 copies per μL in the detection of EGFR L858R gene mutation in NSCLC.373
To improve the ease of operation, the next level of optimisation is the integration of all the protocol steps in a single automated apparatus. In addition to the fully integrated commercial instruments such as NIo, QX ONE, QIAcuity and Absolute Q systems, new platforms are currently under development. For example, an integrated platform based on microfluidic array partitioning has been tested to quantify EGFR T790M in NSCLC samples and BCR::ABL1 fusion gene in chronic myeloid leukemia, detecting MAF as low as 0.01%.379 Although these platforms are integrated in a single apparatus, they still require a professional to handle the various protocol steps. An emerging technology, digital microfluidics (DMF), could address this problem. DMF is based on electrowetting-on-dielectric (EWOD), and it allows the precise handling of picoliter-to-nanoliter-sized droplets on an array with microelectrodes. Over the past decade, DMF has found extensive applications in molecular diagnostics.380–384 The GenMark ePlex system, a DMF-based platform for syndromic pathogen detection, has even been approved by the FDA.385 A DMF-Bimol system has been developed to couple PLA and RT-ddPCR for the analysis of CD147 protein and its transcript, once again revealing a poor correlation between the two.386 One of the main challenges of an automatic sample preparation for dPCR arises from the wide variety of contaminants depending on the type of sample (sputum, blood, saliva…). Walter Hu's team has developed a system, using magnetic bead-based nucleic acid extraction coupled to DMF qPCR for the detection of 15 pathogens in nasopharyngeal swabs, oropharyngeal swabs, bronchoalveolar lavage fluid and sputum.387 A handling time of 1 minute could be achieved, as well as a total sample-to-answer detection within 80 minutes with good sensitivity (200–628 copies per mL). The strategy of DNA extraction by magnetic beads could easily be adapted to dPCR.
Another system took advantage of both centrifugal microfluidic and DMF, to propose a sample-to-answer quantification of viral load by a fully-integrated RT-ddPCR. It uses the centrifuge force to move the sample from chambers, each associated with one step of the protocol, namely the lysis, RNA extraction, wash and elution, mixing with PCR reagents, droplet generation and finally droplet imaging.388 Its applications to SARS-CoV-2 detection demonstrated an LoD of 0.1 copies per μL, as well as 100% accuracy compared to qPCR on 14 nasopharyngeal swab samples. This very promising system allows a handling-free workflow for sample preparation thanks to a centrifugal platform with pneumatic pumping, as well as portability with on-chip droplet generation, thermocycling and analysis thanks to an embedded heater and epi-fluorescence imaging modules.
Conclusion
Digital PCR has the potential to become a major technology in the diagnostic field. The demonstration of its high sensitivity and specificity compared to other techniques, along with its accuracy and reproducibility, has made dPCR a powerful tool for interrogating genetic or epigenetic information of individual patients – particularly in the context of the shift towards personalised and non-invasive medicine. The samples for dPCR can range from DNA extracted from tissue samples (FFPE, frozen) to cfDNA, ctDNA, CTCs or EVs recovered from body fluids such as blood, urine, saliva, pleural or cerebrospinal fluids. The use of dPCR has been validated for a wide range of clinical applications in many medical fields including oncology, prenatal testing, and pathogen detection. However, this technology still presents limitations that hinder its widespread clinical implementation. For example, detection of mutations in ctDNA at early cancer stages remains challenging, and complementary systems are often needed alongside dPCR. Moreover, despite the wide choice in commercialised dPCR platforms, they are still cost intensive (as compared to qPCR for example) and are not yet suitable for point-of-care applications.Nevertheless, further developments in dPCR are being fuelled by technological advancements, including isothermal amplification,327,328,389 novel imaging and scanning technologies, and algorithm optimisation for automated thresholding and analysis. These may lead to even better performance in terms of multiplexing, sensitivity and dynamic range, as well as reduced costs. In the field of personalised medicine, dPCR and NGS could represent complementary technologies. An example can be found in oncology, where tumour-informed strategies may involve initial tumour characterisation by NGS, followed by the design of patient-tailored assays for follow-up using dPCR. Furthermore, the increasing development of fully automated dPCR instruments, coupled to the decreasing costs of NGS, may accelerate the spread of personalised medicine, often described as the future of healthcare. Moreover, portable, hands-free, all-integrated processes are central to ongoing development efforts, paving the way for point-of-care applications, such as pathogen detection in resource-limited areas.
Data availability
No primary research results, software, or code have been included and no new data were generated or analysed as part of this review.Author contributions
Supervision: VT; conceptualization: AT, GG, VT; investigation (analysis of >300 publications) and writing of original draft: AT; writing – review & editing: VT, GG, AT, LB.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge the support of the Paris Sciences et Lettres Research University, ESPCI-Paris, the Université Paris Cité, the Centre National de la Recherche Scientifique (CNRS), the Institut National de la Santé et de la Recherche Médicale (IN-SERM), the Ligue Nationale Contre le Cancer (LNCC, Program “Equipe labelisée LIGUE”; no. EL2016.LNCC/VaT). This work was supported by the European Research Council under the framework program H2020 for research and innovation (grant project MoP-MiP, No. 949493), ITMO cancer re-search program PCSI (grant M2DIA), and the Agence Nationale de la recherche (grant PROJECT SNAP-Seq, ANR-21-CE19-0019).References
- World Health Organization, Diagnostic testing for SARS-CoV-2 [Internet], 2020, Sep [cited 2024 Apr 1], Available from: https://www.who.int/publications/i/item/WHO-2019-nCoV-SurveillanceGuidance-2022.2.
- K. Mullis, F. Faloona, S. Scharf, R. Saiki, G. Horn and H. Erlich, Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction, Cold Spring Harbor Symp. Quant. Biol., 1986, Pt1, 263–273, DOI:10.1101/sqb.1986.051.01.032.
- R. Higuchi, G. Dollinger, P. S. Walsh and R. Griffith, Simultaneous amplification and detection of specific DNA sequences, Biotechnology, 1992, 10(4), 413–417 CrossRef CAS PubMed , Available from: http://www.nature.com/naturebiotechnology.
- P. Simmonds, P. Balfe, J. F. Peutherer, C. A. Ludlam, J. O. Bishop and A. J. Brown, Human immunodeficiency virus-infected individuals contain provirus in small numbers of peripheral mononuclear cells and at low copy numbers, J. Virol., 1990, 64(2), 864–872 CrossRef CAS PubMed , Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=249182&tool=pmcentrez&rendertype=abstract.
- P. J. Sykes and A. A. Morley, Quantitation of targets for PCR by use of limiting dilution, BioTechniques, 1992, 13(3), 444–449 CAS.
- B. Vogelstein and K. W. Kinzler, Digital PCR, Proc. Natl. Acad. Sci. U. S. A., 1999, 96(16), 9236–9241 CrossRef CAS PubMed , Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=17763&tool=pmcentrez&rendertype=abstract.
- O. Kalinina, I. Lebedeva, J. Brown and J. Silver, Nanoliter scale PCR with TaqMan detection, Nucleic Acids Res., 1997, 25(10), 1999–2004 CrossRef CAS PubMed.
- D. Dressman, H. Yan, G. Traverso, K. W. Kinzler and B. Vogelstein, Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations, Proc. Natl. Acad. Sci. U. S. A., 2003, 100(15), 8817–8822 CrossRef CAS PubMed.
- F. Diehl, M. Li, Y. He, K. W. Kinzler, B. Vogelstein and D. Dressman, BEAMing: Single-molecule PCR on microparticles in water-in-oil emulsions, Nat. Methods, 2006, 3(7), 551–559 CrossRef CAS PubMed.
- M. Li, F. Diehl, D. Dressman, B. Vogelstein and K. W. Kinzler, BEAMing up for detection and quantification of rare sequence variants, Nat. Methods, 2006, 3(2), 95–97 CrossRef CAS PubMed.
- H. Huang, Z. Qi, L. Deng, G. Zhou, T. Kajiyama and H. Kambara, Highly sensitive mutation detection based on digital amplification coupled with hydrogel bead-array, Chem. Commun., 2009, 4094–4096 RSC.
- Z. Qi, Y. Ma, L. Deng, H. Wu, G. Zhou and T. Kajiyama, et al., Digital analysis of the expression levels of multiple colorectal cancer-related genes by Multiplexed Digital-PCR coupled with Hydrogel Bead-array, Analyst, 2011, 136(11), 2252–2259 RSC.
- D. Pekin, Y. Skhiri, J. C. Baret, D. Le Corre, L. Mazutis and C. Ben Salem, et al., Quantitative and sensitive detection of rare mutations using droplet-based microfluidics, Lab Chip, 2011, 11(13), 2156–2166 RSC.
- D. Xu, W. Zhang, H. Li, N. Li and J. M. Lin, Advances in droplet digital polymerase chain reaction on microfluidic chips, Lab Chip, 2023, 23(5), 1258–1278 RSC.
- J. C. Baret, F. Kleinschmidt, A. E. Harrak and A. D. Griffiths, Kinetic aspects of emulsion stabilization by surfactants: A microfluidic analysis, Langmuir, 2009, 25(11), 6088–6093 CrossRef CAS PubMed.
- P. Liao, M. Jiang, Z. Chen, F. Zhang, Y. Sun and J. Nie, et al., Three-dimensional digital PCR through light-sheet imaging of optically cleared emulsion, Proc. Natl. Acad. Sci. U. S. A., 2020, 117(41), 25628–25633 CrossRef CAS PubMed , Available from: https://www.pnas.org/lookup/suppl/.
- O. Chen-Yin, T. Vu, J. T. Grunwald, M. Toledano, J. Zimak and M. Toosky, et al., An Ultrasensitive Test for Profiling Circulating Tumor DNA using Integrated Comprehensive Droplet Digital Detection, Lab Chip, 2019, 19(6), 993–1005 RSC.
- X. Casadevall i Solvas and A. J. DeMello, Droplet microfluidics: recent developments and future applications, Chem. Commun., 2011, 47(7), 1936–1942 RSC , Available from: http://www.ncbi.nlm.nih.gov/pubmed/20967373.
- A. Huebner, S. Sharma, M. Srisa-Art, F. Hollfelder, J. B. Edel and A. J. DeMello, Microdroplets: A sea of applications?, Lab Chip, 2008, 8(8), 1244 RSC , Available from: http://xlink.rsc.org/?DOI=b806405a.
- N. B. Y. Tsui, R. Akolekar, R. W. K. Chiu, K. C. K. Chow, T. Y. Leung and T. K. Lau, et al., Synergy of total PLAC4 RNA concentration and measurement of the RNA single-nucleotide polymorphism allelic ratio for the noninvasive prenatal detection of trisomy 21, Clin. Chem., 2010, 56(1), 73–81 CrossRef CAS PubMed.
- F. Shen, W. Du, J. E. Kreutz, A. Fok and R. F. Ismagilov, Digital PCR on a SlipChip, Lab Chip, 2010, 10(20), 2666–2672 RSC.
- S. O. Sundberg, C. T. Wittwer, C. Gao and B. K. Gale, Spinning disk platform for microfluidic digital polymerase chain reaction, Anal. Chem., 2010, 82(4), 1546–1550 CrossRef CAS PubMed.
- E. A. Ottesen, J. W. Hong, S. R. Quake and J. R. Leadbetter, Microfluidic Digital PCR Enables Multigene Analysis of Individual Environmental Bacteria, PLoS One, 2006, 314(5804), 1464–1467 CAS , Available from: https://www.science.org.
- S. L. Spurgeon, R. C. Jones and R. Ramakrishnan, High throughput gene expression measurement with real time PCR in a microfluidic dynamic array, PLoS One, 2008, 3(2), 1662–1667 CrossRef PubMed.
- J. Qin, R. C. Jones and R. Ramakrishnan, Studying copy number variations using a nanofluidic platform, Nucleic Acids Res., 2008, 36(18), 116–118 CrossRef PubMed.
- K. A. Heyries, C. Tropini, M. Vaninsberghe, C. Doolin, O. I. Petriv and A. Singhal, et al., Megapixel digital PCR, Nat. Methods, 2011, 8(8), 649–651, DOI:10.1038/nmeth.1640.
- Z. Hua, J. L. Rouse, A. E. Eckhardt, V. Srinivasan, V. K. Pamula and W. A. Schell, Multiplexed real-time polymerase chain reaction on a digital microfluidic platform, Anal. Chem., 2010, 82(6), 2310–2316 CrossRef CAS PubMed.
- A. C. Hatch, J. S. Fisher, A. R. Tovar, A. T. Hsieh, R. Lin and S. L. Pentoney, et al., 1-Million droplet array with wide-field fluorescence imaging for digital PCR, Lab Chip, 2011, 11(22), 3838–3845 RSC.
- N. R. Beer, B. J. Hindson, E. K. Wheeler, S. B. Hall, K. A. Rose and I. M. Kennedy, et al., On-chip, real-time, single-copy polymerase chain reaction in picoliter droplets, Anal. Chem., 2007, 79(22), 8471–8475 CrossRef CAS PubMed.
- Q. Zhong, S. Bhattacharya, S. Kotsopoulos, J. Olson, V. Taly and A. D. Griffiths, et al., Multiplex digital PCR: Breaking the one target per color barrier of quantitative PCR, Lab Chip, 2011, 11(13), 2167–2174 RSC.
- B. J. Hindson, K. D. Ness, D. A. Masquelier, P. Belgrader, N. J. Heredia and A. J. Makarewicz, et al., High-throughput droplet digital PCR system for absolute quantitation of DNA copy number, Anal. Chem., 2011, 83(22), 8604–8610 CrossRef CAS PubMed.
- H. Zhou, D. Liu, L. Ma, T. Ma, T. Xu and L. Ren, et al., A SARS-CoV-2 Reference Standard Quantified by Multiple Digital PCR Platforms for Quality Assessment of Molecular Tests, Anal. Chem., 2021, 93(2), 715–721 CrossRef CAS PubMed.
- C. Niu, X. Wang, Y. Zhang, L. Lu, D. Wang and Y. Gao, et al., Interlaboratory assessment of quantification of SARS-CoV-2 RNA by reverse transcription digital PCR, Anal. Bioanal. Chem., 2021, 413(29), 7195–7204, DOI:10.1007/s00216-021-03680-2.
- W. Saisaard and W. Owattanapanich, Comparative analysis of BCR::ABL1 p210 mRNA transcript quantification and ratio to ABL1 control gene converted to the International Scale by chip digital PCR and droplet digital PCR for monitoring patients with chronic myeloid leukemia, Clin. Chem. Lab. Med., 2024, 63(2), 279–290 CrossRef PubMed.
- K. R. Emslie, J. L. H. McLaughlin, K. Griffiths, M. Forbes-Smith, L. B. Pinheiro and D. G. Burke, Droplet volume variability and impact on digital pcr copy number concentration measurements, Anal. Chem., 2019, 91(6), 4124–4131 CrossRef CAS PubMed.
- A. Orzińska, M. Krzemienowska, S. Purchla-Szepioła, I. Kopeć and K. Guz, Noninvasive diagnostics of fetal KEL*01.01 allele from maternal plasma of immunized women using digital PCR protocols, Transfusion, 2022, 62(4), 863–870 CrossRef PubMed.
- Q. Guo, L. Wang, X. Liang, M. Zhao, X. Huang and W. Xu, et al., Comparative analysis of QS3D versus droplet digital PCR for quantitative measures of EGFR T790M mutation from identical plasma, Heliyon, 2022, 8(11), e1339 Search PubMed.
- K. J. Bosman, M. Nijhuis, P. M. Van Ham, A. M. J. Wensing, K. Vervisch and L. Vandekerckhove, et al., Comparison of digital PCR platforms and semi-nested qPCR as a tool to determine the size of the HIV reservoir, Sci. Rep., 2015, 5, 13811 CrossRef CAS PubMed.
- V. Sánchez-Martín, E. López-López, D. Reguero-Paredes, A. Godoy-Ortiz, M. E. Domínguez-Recio and B. Jiménez-Rodríguez, et al., Comparative study of droplet-digital PCR and absolute Q digital PCR for ctDNA detection in early-stage breast cancer patients, Clin. Chim. Acta, 2024, 552, 117673 CrossRef PubMed.
- M. Zaytseva, N. Usman, E. Salnikova, A. Sanakoeva, A. Valiakhmetova and A. Chervova, et al., Methodological Challenges of Digital PCR Detection of the Histone H3 K27M Somatic Variant in Cerebrospinal Fluid, Pathol. Oncol. Res., 2022, 28, 1610024 CrossRef CAS PubMed.
- S. Crucitta, M. Ruglioni, C. Novi, M. Manganiello, R. Arici and I. Petrini, et al., Comparison of digital PCR systems for the analysis of liquid biopsy samples of patients affected by lung and colorectal cancer, Clin. Chim. Acta, 2023, 541, 117239 CrossRef CAS PubMed.
- M. Alikian, A. S. Whale, S. Akiki, K. Piechocki, C. Torrado and T. Myint, et al., RT-qPCR and RT-digital PCR: A comparison of different platforms for the evaluation of residual disease in chronic myeloid leukemia, Clin. Chem., 2017, 63(2), 525–531 CrossRef CAS PubMed.
- A. S. Whale, G. M. Jones, J. Pavšič, T. Dreo, N. Redshaw and S. Akyürek, et al., Assessment of digital PCR as a primary reference measurement procedure to support advances in precision medicine, Clin. Chem., 2018, 64(9), 1296–1307 CrossRef CAS PubMed.
- L. Dong, Y. Meng, Z. Sui, J. Wang, L. Wu and B. Fu, Comparison of four digital PCR platforms for accurate quantification of DNA copy number of a certified plasmid DNA reference material, Sci. Rep., 2015, 5, 13174 CrossRef CAS PubMed.
- C. Y. Sung, C. C. Huang, Y. S. Chen, K. F. Hsu and G. B. Lee, Isolation and quantification of extracellular vesicle-encapsulated microRNA on an integrated microfluidic platform, Lab Chip, 2021, 21(23), 4660–4671 RSC.
- T. C. Dingle, R. H. Sedlak, L. Cook and K. R. Jerome, Tolerance of droplet-digital PCR vs real-time quantitative PCR to inhibitory substances, Clin. Chem., 2013, 59, 1670–1672 CrossRef CAS PubMed.
- L. A. El Khattabi, C. Rouillac-Le Sciellour, D. Le Tessier, A. Luscan, A. Coustier and R. Porcher, et al., Could digital PCR be an alternative as a non-invasive prenatal test for trisomy 21: A proof of concept study, PLoS One, 2016, 11(5), e0155009 CrossRef PubMed.
- FDA, COVID-19 RT-Digital PCR Detection Kit Instructions for Use For Emergency Use Authorization Only [Internet], 2020, [cited 2025 Apr 1], Available from: https://www.fda.gov/media/136738/download.
- D. N. Shelton, P. Bhagavatula, N. Sepulveda, L. Beppu, S. Gandhi and D. Qin, et al., Performance characteristics of the first Food and Drug Administration (FDA)-cleared digital droplet PCR (ddPCR) assay for BCR::ABL1 monitoring in chronic myelogenous leukemia, PLoS One, 2022, 17(3), e0265278 CrossRef CAS PubMed.
- T. Azam, C. Sommers, J. Rodriguez, D. Keire and K. Yang, Direct analysis of residual host cell DNA by droplet digital PCR (ddPCR) in biologic drugs produced in E. Coli, 2021.
- Nuffield Council on Bioethics, Non-invasive prenatal testing: ethical issues, Nuffield Council on Bioethics, 2017, p. 149.
- J. Ferlay, M. Colombet, I. Soerjomataram, D. M. Parkin, M. Piñeros and A. Znaor, et al., Cancer statistics for the year 2020: An overview, Int. J. Cancer, 2021, 149(4), 778–789 CrossRef CAS PubMed.
- National Cancer Institute, What Is Cancer? [Internet], 2021, Available from: https://www.cancer.gov/about-cancer/understanding/what-is-cancer.
- V. K. Sarhadi and G. Armengol, Molecular Biomarkers in Cancer, Biomolecules, 2022, 12, 1021 CrossRef CAS PubMed.
- J. Simon, D. Reita, E. Guerin, B. Lhermitte, N. Weingertner and F. Lefebvre, et al., Clinical impact of large genomic explorations at diagnosis in 198 pediatric solid tumors: a monocentric study aiming practical feasibility of precision oncology, BMC Cancer, 2024, 24(1), 1296 CrossRef CAS PubMed.
- U. Gezer, A. J. Bronkhorst and S. Holdenrieder, The Clinical Utility of Droplet Digital PCR for Profiling Circulating Tumor DNA in Breast Cancer Patients, Diagnostics, 2022, 12, 3042 CrossRef CAS PubMed.
- X. Yu, J. Huang, S. Wu, Y. Huang, Y. Shan and C. Lu, Copy number variations of mmp-9 are prognostic biomarkers for hepatocellular carcinoma, Transl. Cancer Res., 2020, 9(2), 698–706 CrossRef CAS PubMed.
- A. Brik, K. Wichert, D. G. Weber, K. Szafranski, P. Rozynek and S. Meier, et al., Assessment of MYC and TERT copy number variations in lung cancer using digital PCR, BMC Res. Notes, 2023, 16(1), 279 CrossRef CAS PubMed.
- R. C. M. Shek, P. S. N. Li, S. C. M. Leung, H. T. Chu, F. Hioe and V. W. L. Tang, et al., A Novel Digital PCR Assay for Accurate Detection and Differentiation of Focal and Non-Focal Subtypes of Mesenchymal–Epithelial Transition (MET) Gene Amplification in Lung Cancer, Cancers, 2025, 17(5), 811 CrossRef CAS PubMed.
- A. C. Wolff, M. Elizabeth, H. Hammond, J. N. Schwartz, K. L. Hagerty and D. C. Allred, et al., Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer [Internet], Arch. Pathol. Lab. Med., 2007, 131(1), 18–43 CrossRef CAS PubMed , Available from: http://arpa.allenpress.com.
- X. Wang, D. Xing, Z. Liu, Y. Zhang, B. Cheng and S. Sun, et al., Establishment and evaluation of digital PCR methods for HER2 copy number variation in breast cancer, Anal. Bioanal. Chem., 2023, 415(4), 725–733 CrossRef CAS PubMed.
- P. A. Grebnev, I. O. Meshkov, P. V. Ershov, A. V. Makhotenko, V. B. Azarian and M. V. Erokhina, et al., Benchmarking of Approaches for Gene Copy-Number Variation Analysis and Its Utility for Genetic Aberration Detection in High-Grade Serous Ovarian Carcinomas, Cancers, 2024, 16(19), 3252 CrossRef CAS PubMed.
- E. Sánchez-Herrero, M. B. Clemente, V. Calvo, M. Provencio and A. Romero, Next-generation sequencing to dynamically detect mechanisms of resistance to ALK inhibitors in ALK-positive NSCLC patients: A case report, Transl. Lung Cancer Res., 2020, 9(2), 366–372 CrossRef PubMed.
- L. M. Wainman, G. Huang, D. C. Green, G. J. Tsongalis, L. J. Tafe and W. A. Khan, et al., Diagnostic next-generation sequencing to detect MYD88 L265P in Lymphoplasmacytic lymphoma compared to ddPCR, Exp. Mol. Pathol., 2025, 141, 104956 CrossRef CAS PubMed.
- S. R. Vitale, F. H. Groenendijk, R. van Marion, C. M. Beaufort, J. C. Helmijr and H. J. Dubbink, et al., TP53 mutations in serum circulating cell-free tumor DNA as longitudinal biomarker for high-grade serous ovarian cancer, Biomolecules, 2020, 10(3), 415 CrossRef CAS PubMed.
- P. Darville-O'Quinn, N. Gokgoz, K. M. Tsoi, I. L. Andrulis and J. S. Wunder, Investigating the Use of Circulating Tumor DNA for Sarcoma Management, J. Clin. Med., 2024, 13(21), 6539 CrossRef PubMed.
- K. Li, N. Zhang, B. Xu, Z. Liu, D. Zhao and Y. Dong, et al., Utility of Circulating Tumor DNA Assay in Identifying Mutations and Guiding Matched Targeted Therapy in Lung Cancers. Clin Med Insights, Onco Targets Ther, 2024, 18, 1–12 Search PubMed.
- K. Cabrera, J. Gole, B. Leatham, M. J. Springer, M. Smith and L. Herdt, et al., Analytical Performance and Concordance with Next-Generation Sequencing of a Rapid, Multiplexed dPCR Panel for the Detection of DNA and RNA Biomarkers in Non-Small-Cell Lung Cancer, Diagnostics., 2023, 13(21), 3299 CrossRef CAS PubMed.
- J. Di, T. Sheng, R. Arora, J. Stocks-Candelaria, S. Wei and C. Lutz, et al., The Validation of Digital PCR–Based Minimal Residual Disease Detection for the Common Mutations in IDH1 and IDH2 Genes in Patients with Acute Myeloid Leukemia, J. Mol. Diagn., 2024, 100–108 Search PubMed.
- L. Ye, N. Mesbah Ardakani, C. Thomas, K. Spilsbury, C. Leslie and B. Amanuel, et al., Detection of Low-level EGFR c.2369 C > T (p.Thr790Met) Resistance Mutation in Pre-treatment Non-small Cell Lung Carcinomas Harboring Activating EGFR Mutations and Correlation with Clinical Outcomes, Pathol. Oncol. Res., 2020, 26(4), 2371–2379 CrossRef CAS PubMed.
- T. M. Carvalho, R. M. Dourado, S. M. Nakatani, C. A. Barros Duarte, S. O. Ioshii and I. N. Riediger, et al., Digital PCR detection of EGFR somatic mutations in non-small-cell lung cancer formalin fixed paraffin embedded samples, Mol. Cell. Probes, 2021, 58, 101745 CrossRef CAS PubMed.
- P. Laurent-Puig, D. Pekin, C. Normand, S. K. Kotsopoulos, P. Nizard and K. Perez-Toralla, et al., Clinical relevance of KRAS-mutated subclones detected with picodroplet digital PCR in advanced colorectal cancer treated with Anti-EGFR therapy, Clin. Cancer Res., 2015, 21(5), 1087–1097 CrossRef CAS PubMed.
- D. Azuara, C. Santos, A. Lopez-Doriga, J. Grasselli, M. Nadal and X. Sanjuan, et al., Nanofluidic digital PCR and extended genotyping of RAS and BRAF for improved selection of metastatic colorectal cancer patients for anti-EGFR therapies, Mol. Cancer Ther., 2016, 15(5), 1106–1112 CrossRef CAS PubMed.
- C. Santos, D. Azuara, R. Garcia-Carbonero, P. G. Alfonso, A. Carrato and M. E. Elez, et al., Optimization of RAS/BRAF mutational analysis confirms improvement in patient selection for clinical benefit to anti-EGFR treatment in metastatic colorectal cancer, Mol. Cancer Ther., 2017, 16(9), 1999–2007 CrossRef CAS PubMed.
- C. Santos, D. Azuara, J. M. Viéitez, D. Páez, E. Falcó and E. Élez, et al., Phase II study of high-sensitivity genotyping of KRAS, NRAS, BRAF and PIK3CA to ultra-select metastatic colorectal cancer patients for panitumumab plus FOLFIRI: The ULTRA trial, Ann. Oncol., 2019, 30(5), 796–803 CrossRef CAS PubMed.
- K. Pantel and C. Alix-Panabières, The clinical significance of circulating tumor cells, Nat. Clin. Pract. Oncol., 2007, 4, 62–63 CrossRef PubMed.
- M. Allegretti, G. Cottone, F. Carboni, E. Cotroneo, B. Casini and E. Giordani, et al., Cross-sectional analysis of circulating tumor DNA in primary colorectal cancer at surgery and during post-surgery follow-up by liquid biopsy, J. Exp. Clin. Cancer Res., 2020, 39(1), 69 CrossRef CAS PubMed.
- L. Y. W. Shong, J. Y. Deng, H. H. Kwok, N. C. M. Lee, S. C. Z. Tseng and L. Y. Ng, et al., Detection of EGFR mutations in patients with suspected lung cancer using paired tissue-plasma testing: a prospective comparative study with plasma ddPCR assay, Sci. Rep., 2024, 14(1), 25701 CrossRef CAS PubMed.
- S. Xie, Y. Wang, Z. Gong, Y. Li, W. Yang and G. Liu, et al., Liquid Biopsy and Tissue Biopsy Comparison with Digital PCR and IHC/FISH for HER2 Amplification Detection in Breast Cancer Patients, J. Cancer, 2022, 13(3), 744–751 CrossRef CAS PubMed.
- A. Dobilas, Y. Chen, C. Brueffer, P. Leandersson, L. H. Saal and C. Borgfeldt, Preoperative ctDNA Levels Are Associated With Poor Overall Survival in Patients With Ovarian Cancer, Cancer Genomics Proteomics, 2023, 20(6), 763–770 CrossRef CAS PubMed.
- J. Corné, F. Le Du, V. Quillien, F. Godey, L. Robert and H. Bourien, et al., Development of multiplex digital PCR assays for the detection of PIK3CA mutations in the plasma of metastatic breast cancer patients, Sci. Rep., 2021, 11(1), 17316 CrossRef PubMed.
- V. Taly, D. Pekin, L. Benhaim, S. K. Kotsopoulos, D. Le Corre and X. Li, et al., Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients, Clin. Chem., 2013, 59(12), 1722–1731 CrossRef CAS PubMed.
- H. Qi, A. Xiong, L. Jiang, H. Van, J. Xu and J. Wu, et al., Blood digital polymerase chain reaction as a potential method to detect human epidermal growth factor receptor 2 amplification in non-small cell lung cancer, Transl. Lung Cancer Res., 2021, 10(11), 4235–4249 CrossRef CAS PubMed.
- J. Xu, W. Wu, C. Wu, Y. Mao, X. Qi and L. Guo, et al., A large-scale, multicentered trial evaluating the sensitivity and specificity of digital PCR versus ARMS-PCR for detecting ctDNA-based EGFR p.T790M in non-small-cell lung cancer patients. Transl Lung, Cancer Res., 2021, 10(10), 3888–3901 CAS.
- S. L. Csoma, K. Madarász, Y. C. Chang Chien, G. Emri, J. Bedekovics and G. Méhes, et al., Correlation Analyses between Histological Staging and Molecular Alterations in Tumor-Derived and Cell-Free DNA of Early-Stage Primary Cutaneous Melanoma, Cancers, 2023, 15(21), 5141 CrossRef CAS PubMed.
- C. Franczak, A. Witz, K. Geoffroy, J. Demange, M. Rouyer and M. Husson, et al., Evaluation of KRAS, NRAS and BRAF mutations detection in plasma using an automated system for patients with metastatic colorectal cancer, PLoS One, 2020, 15(1), e0227294 CrossRef CAS PubMed.
- D. Smitalova, D. Dvorakova, Z. Racil and M. Romzova, Digital PCR can provide improved BCR-ABL1 detection in chronic myeloid leukemia patients in deep molecular response and sensitivity of standard quantitative methods using EAC assays, Pract. Lab. Med., 2021, 25, e00210 CrossRef CAS PubMed.
- S. Jenkins, J. C. H. Yang, S. S. Ramalingam, K. Yu, S. Patel and S. Weston, et al., Plasma ctDNA Analysis for Detection of the EGFR T790M Mutation in Patients with Advanced Non–Small Cell Lung Cancer, J. Thorac. Oncol., 2017, 12(7), 1061–1070 CrossRef PubMed.
- J. Claus, D. De Smet, J. Breyne, J. Wesolowski, U. Himpe and I. Demedts, et al., Patient-centric thresholding of Cobas® EGFR mutation Test v2 for surveillance of EGFR-mutated metastatic non-small cell lung cancer, Sci. Rep., 2024, 14(1), 18191 CrossRef CAS PubMed.
- M. Riudavets, V. Lamberts, E. Auclin, M. Aldea, D. Vasseur and C. Jovelet, et al., 22P Clinical utility of ddPCR for detection of sensitizing and resistance EGFRm in pts with advanced NSCLC, J. Thorac. Oncol., 2021, 16(4), S707–S708 CrossRef.
- C. Silveira, A. C. Sousa, A. Janeiro, S. Malveiro, E. Teixeira and E. Brysch, et al., Detection and quantification of EGFR T790M mutation in liquid biopsies by droplet digital PCR, Transl. Lung Cancer Res., 2021, 10(3), 1200–1208 CrossRef CAS PubMed.
- M. Provencio, R. Serna-Blasco, F. Franco, V. Calvo, A. Royuela and M. Auglytė, et al., Analysis of circulating tumour DNA to identify patients with epidermal growth factor receptor–positive non-small cell lung cancer who might benefit from sequential tyrosine kinase inhibitor treatment, Eur. J. Cancer, 2021, 149, 61–72 CrossRef CAS PubMed.
- Y. Hong, W. Zhuang, J. Lai, H. Xu, Y. He and J. Lin, et al., Plasma EGFR mutation ctDNA dynamics in patients with advanced EGFR-mutated NSCLC treated with Icotinib: phase 2 multicenter trial result, Sci. Rep., 2024, 14(1), 23115 CrossRef CAS PubMed , Available from: http://www.ncbi.nlm.nih.gov/pubmed/39367090.
- A. Romero, R. Serna-Blasco, C. Alfaro, E. Sánchez-Herrero, M. Barquín and M. C. Turpin, et al., ctDNA analysis reveals different molecular patterns upon disease progression in patients treated with osimertinib, Transl. Lung Cancer Res., 2020, 9(3), 532–540 CrossRef CAS PubMed.
- S. Klein-Scory, A. Baraniskin, W. Schmiegel, T. Mika, R. Schroers and S. Held, et al., Evaluation of circulating tumor DNA as a prognostic and predictive biomarker in BRAF V600E mutated colorectal cancer—results from the FIRE-4.5 study, Mol. Oncol., 2024, 19(2), 344–356 CrossRef PubMed.
- Y. K. Kang, Y. R. Si, J. Ju, Z. Q. Jia, N. L. Hu and H. Dong, et al., Assessing early changes in plasma HER2 levels is useful for predicting therapeutic response in advanced breast cancer: A multicenter, prospective, noninterventional clinical study, Cancer Med., 2023, 12(5), 5323–5333 CrossRef CAS PubMed.
- E. Giordani, M. Allegretti, A. Sinibaldi, F. Michelotti, G. Ferretti and E. Ricciardi, et al., Monitoring changing patterns in HER2 addiction by liquid biopsy in advanced breast cancer patients, J. Exp. Clin. Cancer Res., 2024, 43(1), 182 CrossRef CAS PubMed.
- E. W. Li, N. Y. K. Tran, D. McCulloch, M. Krigstein, A. Catalano and J. Othman, et al., FLT3-TKD Measurable Residual Disease Detection Using Droplet Digital PCR and Clinical Applications in Acute Myeloid Leukemia, Int. J. Mol. Sci., 2024, 25(11), 5771 CrossRef CAS PubMed.
- K. Tanaka, Y. Yoshida, T. Yamada, T. Hayashi, H. Shimaoka and F. Yoshimura, et al., Oncological evaluation in the perioperative period using cfDNA with BRAF V600E mutation in patients with colorectal cancer, Sci. Rep., 2021, 11(1), 13263 CrossRef CAS PubMed.
- J. A. Kalil, L. Krzywon, S. K. Petrillo, M. Tsamchoe, O. Zlotnik and A. Lazaris, et al., Feasibility of ctDNA in detecting minimal residual disease and predicting recurrence for colorectal cancer liver metastases. Front, Onco Targets Ther, 2024, 14, 1418696 CAS.
- S. Marchisio, A. A. Ricci, G. Roccuzzo, E. Bongiovanni, E. Ortolan and L. Bertero, et al., Monitoring circulating tumor DNA liquid biopsy in stage III BRAF-mutant melanoma patients undergoing adjuvant treatment, J. Transl. Med., 2024, 22(1), 1074 CrossRef CAS PubMed.
- M. Yaegashi, T. Iwaya, N. Sasaki, M. Fujita, Z. Ju and D. Siwak, et al., Frequent post-operative monitoring of colorectal cancer using individualised ctDNA validated by multiregional molecular profiling, Br. J. Cancer, 2021, 124(9), 1556–1565 CrossRef CAS PubMed.
- T. V. Henriksen, C. Demuth, A. Frydendahl, J. Nors, M. Nesic and M. H. Rasmussen, et al., Unraveling the potential clinical utility of circulating tumor DNA detection in colorectal cancer—evaluation in a nationwide Danish cohort, Ann. Oncol., 2024, 35(2), 229–239 CrossRef CAS PubMed.
- N. Balasan, F. Kharrat, G. Di Lorenzo, E. Athanasakis, A. M. Bianco and A. Conti, et al., Sensitive Detection of Gynecological Cancer Recurrence Using Circulating Tumor DNA and Digital PCR: A Comparative Study with Serum Biochemical Markers, Int. J. Mol. Sci., 2024, 25(22), 11997 CrossRef CAS PubMed , Available from: http://www.ncbi.nlm.nih.gov/pubmed/39596073.
- T. Sasaki, H. Hiraki, A. Yashima-Abo, H. Nagashima, F. Endo and M. Yaegashi, et al., Comprehensive Genome Profiling-Initiated Tumor-Informed Circulating Tumor DNA Monitoring for Patients With Advanced Cancer, Cancer Sci., 2025, 116, 764–774 CrossRef CAS PubMed.
- H. Kaneda, H. Daga, A. Okada, Y. Nakatani, Y. Tani and T. Oka, et al., Efficacy of ramucirumab combined with erlotinib or osimertinib in untreated EGFR-mutated NSCLC patients with asymptomatic brain metastases: insights from molecular biomarkers in the RELAY-brain trial, Invest. New Drugs, 2025, 23, 147–156, DOI:10.1007/s10637-025-01505-y.
- C. M. Wood-Bouwens, D. Haslem, B. Moulton, A. F. Almeda, H. Lee and G. M. Heestand, et al., Therapeutic Monitoring of Circulating DNA Mutations in Metastatic Cancer with Personalized Digital PCR, J. Mol. Diagn., 2020, 22(2), 247–261 CrossRef CAS PubMed.
- R. Kogo, T. Manako, T. Iwaya, S. Nishizuka, H. Hiraki and Y. Sasaki, et al., Individualized circulating tumor DNA monitoring in head and neck squamous cell carcinoma, Cancer Med., 2022, 11(21), 3960–3968 CrossRef CAS PubMed.
- C. Maulat, C. Canivet, B. Cabarrou, A. Pradines, J. Selves and A. Casanova, et al., Prognostic impact of circulating tumor DNA detection in portal and peripheral blood in resected pancreatic ductal adenocarcinoma patients, Sci. Rep., 2024, 14(1), 27296 CrossRef CAS PubMed.
- G. A. Martens, J. Demol, F. Dedeurwaerdere, J. Breyne, K. De Smet and P. De Jaeger, et al., Rational thresholding of circulating tumor DNA concentration for improved surveillance of metastatic breast cancer, ESMO Open, 2024, 9(2), 102235 CrossRef CAS PubMed.
- K. B. Lygre, R. B. Forthun, T. Høysæter, S. M. Hjelle, G. E. Eide and B. T. Gjertsen, et al., Assessment of postoperative circulating tumour DNA to predict early recurrence in patients with stage I–III right-sided colon cancer: prospective observational study, BJS Open, 2024, 8(1), zrad146 CrossRef PubMed.
- M. Coakley, G. Villacampa, P. Sritharan, C. Swift, K. Dunne and L. Kilburn, et al., Comparison of Circulating Tumor DNA Assays for Molecular Residual Disease Detection in Early-Stage Triple-Negative Breast Cancer, Clin. Cancer Res., 2024, 30(4), 895–903 CrossRef CAS PubMed.
- X. Tang, S. Liu, Y. Hu, F. Chen, L. Wang and T. Li, et al., Clearing MRD positivity with blinatumomab in pediatric B-cell acute lymphoblastic leukemia: insights from droplet digital PCR and flow cytometry, Ann. Hematol., 2024, 104, 559–564 Search PubMed.
- E. Abruzzese, M. Bocchia, M. M. Trawinska, D. Raspadori, F. Bondanini and A. Sicuranza, et al., Minimal Residual Disease Detection at RNA and Leukemic Stem Cell (LSC) Levels: Comparison of RT-qPCR, d-PCR and CD26+ Stem Cell Measurements in Chronic Myeloid Leukemia (CML) Patients in Deep Molecular Response (DMR), Cancers, 2023, 15(16), 4112 CrossRef CAS PubMed.
- S. Bernardi, A. Cavalleri, S. Mutti, L. Garuffo, M. Farina and A. Leoni, et al., Digital PCR (dPCR) is able to anticipate the achievement of stable deep molecular response in adult chronic myeloid leukemia patients: results of the DEMONSTRATE study, Ann. Hematol., 2024, 104, 207–217 Search PubMed.
- C. Kockerols, P. J. M. Valk, P. Hogenbirk, J. J. Cornelissen and P. E. Westerweel, BCR::ABL1 Deep Molecular Response Quantification and Transcript Type Identification in Chronic Myeloid Leukemia Using a US Food and Drug Administration–Approved Droplet-Based Digital PCR Assay, J. Mol. Diagn., 2025, 27(2), 109–118 CrossRef CAS PubMed.
- C. Kockerols, P. J. M. Valk, P. Hogenbirk, I. Geelen, N. M. A. Blijlevens and J. J. W. M. Janssen, et al., Treatment-Free Remission Outcomes in a BCR::ABL1 Digital PCR Selected Clinical Cohort of CML Patients, Eur. J. Haematol., 2025, 114, 900–907 CrossRef CAS PubMed.
- Y. Lu, Z. Li, E. H. Lim, P. T. Huan, S. K. Y. Kham and A. E. J. Yeoh, Digital PCR for Minimal Residual Disease Quantitation Using Immunoglobulin/T-Cell Receptor Gene Rearrangements in Acute Lymphoblastic Leukemia: A Proposed Analytic Algorithm, J. Mol. Diagn., 2022, 24(6), 655–665 CrossRef CAS PubMed.
- C. Damm-Welk, F. Lovisa, G. Contarini, J. Lüdersen, E. Carraro and F. Knörr, et al., Quantification of Minimal Disease by Digital PCR in ALK-Positive Anaplastic Large Cell Lymphoma: A Step towards Risk Stratification in International Trials?, Cancers, 2022, 14(7), 1703 CrossRef CAS PubMed.
- M. Jain, A. Tivtikyan, D. Kamalov, S. Avdonin, T. Rakhmatullin and E. Pisarev, et al., Development of a Sensitive Digital Droplet PCR Screening Assay for the Detection of GPR126 Non-Coding Mutations in Bladder Cancer Urine Liquid Biopsies, Biomedicines, 2023, 11(2), 495 CrossRef CAS PubMed.
- M. Abe, H. Hiraki, T. Tsuyukubo, S. Ono, S. Maekawa and D. Tamura, et al., The Clinical Validity of Urinary Pellet DNA Monitoring for the Diagnosis of Recurrent Bladder Cancer, J. Mol. Diagn., 2024, 26(4), 278–291 CrossRef CAS PubMed.
- D. Tamura, M. Abe, H. Hiraki, N. Sasaki, A. Yashima-Abo and D. Ikarashi, et al., Postoperative recurrence detection using individualized circulating tumor DNA in upper tract urothelial carcinoma, Cancer Sci., 2024, 115(2), 529–539 CrossRef CAS PubMed.
- A. Rabien, D. Rong, S. Rabenhorst, T. Schlomm, F. Labonté and S. Hofbauer, et al., Diagnostic performance of Uromonitor and TERTpm ddPCR urine tests for the non-invasive detection of bladder cancer, Sci. Rep., 2024, 14(1), 30616 CrossRef PubMed.
- M. Bessa-Gonçalves, J. P. Brás, T. T. Jesus, H. Prazeres, P. Soares and J. Vinagre, TERTmonitor Efficacy and Performance in Detecting Mutations by Droplet Digital PCR, Genes, 2024, 15(11), 1424 CrossRef PubMed.
- C. Pérez-Barrios, E. Sánchez-Herrero, N. Garcia-Simón, M. Barquín, M. B. Clemente and M. Provencio, et al., CtDNA from body fluids is an adequate source for EGFR biomarker testing in advanced lung adenocarcinoma, Clin. Chem. Lab. Med., 2021, 59(7), 1221–1229 CrossRef PubMed.
- L. Li and Y. Sun, Circulating tumor DNA methylation detection as biomarker and its application in tumor liquid biopsy: advances and challenges, MedComm, 2024, 5, e766 CrossRef CAS PubMed.
- S. Garrigou, G. Perkins, F. Garlan, C. Normand, A. Didelot and D. Le Corre, et al., A study of hypermethylated circulating tumor DNA as a universal colorectal cancer biomarker, Clin. Chem., 2016, 62(8), 1129–1139 CrossRef CAS PubMed.
- M. Friedemann, F. Horn, K. Gutewort, L. Tautz, C. Jandeck and N. Bechmann, et al., Increased sensitivity of detection of rassf1a and gstp1 dna fragments in serum of prostate cancer patients: Optimisation of diagnostics using obbpa-ddpcr, Cancers, 2021, 13(17), 4459 CrossRef CAS PubMed.
- M. Friedemann, C. Jandeck, L. Tautz, K. Gutewort, L. von Rein and O. Sukocheva, et al., Blood-Based DNA Methylation Analysis by Multiplexed OBBPA-ddPCR to Verify Indications for Prostate Biopsies in Suspected Prostate Cancer Patients, Cancers, 2024, 16(7), 1324 CrossRef CAS PubMed.
- D. Pietrasz, S. Wang-Renault, J. Taieb, L. Dahan, M. Postel and J. Durand-Labrunie, et al., Prognostic value of circulating tumour DNA in metastatic pancreatic cancer patients: post-hoc analyses of two clinical trials, Br. J. Cancer, 2021, 126, 440–448 CrossRef PubMed.
- R. Olivera-Salazar, G. Salcedo Cabañas, L. Vega-Clemente, D. Alonso-Martín, V. M. Castellano Megías and P. Volward, et al., Pilot Study by Liquid Biopsy in Gastrointestinal Stromal Tumors: Analysis of PDGFRA D842V Mutation and Hypermethylation of SEPT9 Presence by Digital Droplet PCR, Int. J. Mol. Sci., 2024, 25(12), 6783 CrossRef CAS PubMed.
- Y. Zhao, C. M. O'Keefe, K. Hsieh, L. Cope, S. C. Joyce and T. R. Pisanic, et al., Multiplex Digital Methylation-Specific PCR for Noninvasive Screening of Lung Cancer. Advanced, Science, 2023, 10(16), e2206518 Search PubMed.
- A. L. S. A. Vicente, F. A. de Souza Santos, W. Y. Hirai, D. Lissa, R. de Oliveira Cavagna and A. L. V. da Silva, et al., HOXA9 methylation is not associated with survival in Brazilian patients with lung adenocarcinoma, Clin. Epigenet., 2025, 17(1), 25 CrossRef CAS PubMed.
- G. Beinse, B. Borghese, M. Métairie, P. A. Just, G. Poulet and S. Garinet, et al., Highly Specific Droplet-Digital PCR Detection of Universally Methylated Circulating Tumor DNA in Endometrial Carcinoma, Clin. Chem., 2022, 68(6), 782–793 CrossRef PubMed.
- I. Neefs, N. De Meulenaere, T. Vanpoucke, J. Vandenhoeck, D. Peeters and M. Peeters, et al., Simultaneous detection of eight cancer types using a multiplex droplet digital PCR assay, Mol. Oncol., 2024, 19(1), 188–203 CrossRef PubMed.
- L. Raunkilde, R. F. Andersen, C. B. Thomsen, T. F. Hansen and L. H. Jensen, A prospective study of methylated ctDNA in patients undergoing treatment for liver metastases from colorectal cancer, Eur J Surg Oncol, 2025, 51(5), 109586 CrossRef PubMed.
- T. Draškovič, B. Ranković, N. Zidar and N. Hauptman, DNA methylation biomarker panels for differentiating various liver adenocarcinomas, including hepatocellular carcinoma, cholangiocarcinoma, colorectal liver metastases and pancreatic adenocarcinoma liver metastases, Clin. Epigenet., 2024, 16(1), 153 CrossRef PubMed.
- E. J. Rodger, G. Gimenez, P. Ajithkumar, P. A. Stockwell, S. Almomani and S. A. Bowden, et al., An epigenetic signature of advanced colorectal cancer metastasis, iScience, 2023, 26(6), 106986 CrossRef CAS PubMed.
- M. Macagno, V. Pessei, N. Congiusta, L. Lazzari, S. E. Bellomo and F. Idrees, et al., A Comparative Study of Methyl-BEAMing and Droplet Digital PCR for MGMT Gene Promoter Hypermethylation Detection, Diagnostics, 2024, 14(22), 2467 CrossRef CAS PubMed.
- N. Redshaw, J. F. Huggett, M. S. Taylor, C. A. Foy and A. S. Devonshire, Quantification of epigenetic biomarkers: An evaluation of established and emerging methods for DNA methylation analysis, BMC Genomics, 2014, 15(1), 1174 CrossRef PubMed.
- S. Das, S. Karuri, J. Chakraborty, B. Basu, A. Chandra and S. Aravindan, et al., Universal penalized regression (Elastic-net) model with differentially methylated promoters for oral cancer prediction, Eur. J. Med. Res., 2024, 29(1), 458, DOI:10.1186/s40001-024-02047-4.
- Y. Inoue, A. Ishiguro, Y. Suehiro, Y. Kunimune, Y. Yamaoka and S. Hashimoto, et al., A novel index combining fecal immunochemical test, DNA test, and age improves detection of advanced colorectal adenoma, Cancer Sci., 2024, 115(11), 3682–3694 CrossRef CAS PubMed.
- K. Nakamura, Y. Suehiro, K. Hamabe, A. Goto, S. Hashimoto and Y. Kunimune, et al., A Novel Index Including Age, Sex, hTERT, and Methylated RUNX3 Is Useful for Diagnosing Early Gastric Cancer, Onco Targets Ther, 2025, 103(4), 320–326 CAS.
- Y. Kunimune, Y. Suehiro, I. Saeki, Y. Yamauchi, N. Tanabe and T. Matsumoto, et al., Combination Assay of Methylated HOXA1 with Tumor Markers Shows High Sensitivity for Detection of Early-Stage Hepatocellular Carcinoma, Liver Cancer, 2024, 13(5), 487–497 CrossRef CAS PubMed.
- S. A. Park, N. Masunaga, N. Kagara, Y. Ohi, N. Gondo and K. Abe, et al., Evaluation of RASSF1A methylation in the lysate of sentinel lymph nodes for detecting breast cancer metastasis: A diagnostic accuracy study, Oncol. Lett., 2023, 26(5), 475 CrossRef CAS PubMed.
- A. J. de Melo-Silva, J. P. Lucena and T. Hueneburg, The evolution of molecular diagnosis using digital polymerase chain reaction to detect cancer via cell-free DNA and circulating tumor cells, Cell Biol. Int., 2020, 44(3), 735–743 CrossRef PubMed.
- M. Szostakowska-Rodzos, E. A. Grzybowska, I. Mysliwy, R. Zub, A. Jagiello-Gruszfeld and M. Rubach, et al., The Combined Assessment of CTC and ESR1 Status in Liquid Biopsy Samples Enhances the Clinical Value of Prediction in Metastatic Breast Cancer, Int. J. Mol. Sci., 2025, 26(5), 2038 CrossRef CAS PubMed.
- S. Smilkou, L. Kaklamanis, I. Balgouranidou, H. Linardou, A. M. Papatheodoridi and F. Zagouri, et al., Direct comparison of an ultrasensitive real-time PCR assay with droplet digital PCR for the detection of PIK3CA hotspot mutations in primary tumors, plasma cell-free DNA and paired CTC-derived gDNAs. Front, Onco Targets Ther, 2024, 14, 1435559 CAS.
- S. Smilkou, A. Ntzifa, V. Tserpeli, I. Balgkouranidou, A. Papatheodoridi and E. Razis, et al., Detection rate for ESR1 mutations is higher in circulating-tumor-cell-derived genomic DNA than in paired plasma cell-free DNA samples as revealed by ddPCR, Mol. Oncol., 2025, 1–11 Search PubMed , Available from: http://www.ncbi.nlm.nih.gov/pubmed/39754401.
- M. Vismara, C. Reduzzi, M. Silvestri, F. Murianni, G. Lo Russo and O. Fortunato, et al., Single-Cell Phenotypic and Molecular Characterization of Circulating Tumor Cells Isolated from Cryopreserved Peripheral Blood Mononuclear Cells of Patients with Lung Cancer and Sarcoma, Clin. Chem., 2022, 68(5), 691–701 CrossRef PubMed.
- W. Gao, T. Huang, H. Yuan, J. Yang, Q. Jin and C. Jia, et al., Highly sensitive detection and mutational analysis of lung cancer circulating tumor cells using integrated combined immunomagnetic beads with a droplet digital PCR chip, Talanta, 2018, 185, 229–236 CrossRef CAS PubMed.
- A. Alba-Bernal, A. Godoy-Ortiz, M. E. Domínguez-Recio, E. López-López, M. E. Quirós-Ortega and V. Sánchez-Martín, et al., Increased blood draws for ultrasensitive ctDNA and CTCs detection in early breast cancer patients. NPJ, Breast Cancer, 2024, 10(1), 36 CAS.
- A. Ntzifa, A. Kotsakis, V. Georgoulias and E. Lianidou, Detection of EGFR Mutations in Plasma cfDNA and Paired CTCs of NSCLC Patients before and after Osimertinib Therapy Using Crystal Digital PCR, Cancer, 2021, 13, 2736, DOI:10.3390/cancers13112736.
- A. Strati, M. Zavridou, P. Economopoulou, S. Gkolfinopoulos, A. Psyrri and E. Lianidou, Development and Analytical Validation of a Reverse Transcription Droplet Digital PCR (RT-ddPCR) Assay for PD-L1 Transcripts in Circulating Tumor Cells, Clin. Chem., 2021, 67(4), 642–652 CrossRef PubMed.
- M. Winter, Z. Cai, K. Winkler, K. Georgiou, D. Inglis and T. Lavranos, et al., Circulating tumour cell RNA characterisation from colorectal cancer patient blood after inertial microfluidic enrichment, MethodsX, 2019, 6, 1512–1520 CrossRef PubMed.
- M. Zavridou, S. Smilkou, V. Tserpeli, A. Sfika, E. Bournakis and A. Strati, et al., Development and Analytical Validation of a 6-Plex Reverse Transcription Droplet Digital PCR Assay for the Absolute Quantification of Prostate Cancer Biomarkers in Circulating Tumor Cells of Patients with Metastatic Castration-Resistant Prostate Cancer, Clin. Chem., 2022, 68(10), 1323–1335 CrossRef PubMed.
- J. A. Denis, A. Patroni, E. Guillerm, D. Pépin, N. Benali-Furet and J. Wechsler, et al., Droplet digital PCR of circulating tumor cells from colorectal cancer patients can predict KRAS mutations before surgery, Mol. Oncol., 2016, 10(8), 1221–1231 CrossRef CAS PubMed.
- H. Dejima, H. Nakanishi, R. Takeyama, T. Nishida, Y. Yamauchi and Y. Saito, et al., Detection of Circulating Tumor Cells and EGFR Mutation in Pulmonary Vein and Arterial Blood of Lung Cancer Patients Using a Newly Developed Immunocytology-Based Platform, Diagnostics, 2024, 14(18), 2064 CrossRef CAS PubMed.
- B. Hossam Abdelmonem, L. T. Kamal, L. W. Wardy, M. Ragheb, M. M. Hanna and M. Elsharkawy, et al., Non-coding RNAs: emerging biomarkers and therapeutic targets in cancer and inflammatory diseases, Front. Oncol., 2025, 15, 1534862 CrossRef PubMed.
- P. D. R. Cirillo, K. Margiotti, A. Mesoraca and C. Giorlandino, Quantification of circulating microRNAs by droplet digital PCR for cancer detection, BMC Res. Notes, 2020, 13(1), 28556 Search PubMed.
- B. Péterffy, T. J. Nádasi, S. Krizsán, A. Horváth, Á. Márk and G. Barna, et al., Digital PCR-based quantification of miR-181a in the cerebrospinal fluid aids patient stratification in pediatric acute lymphoblastic leukemia, Sci. Rep., 2024, 14(1), 28556 CrossRef PubMed.
- D. Liu, H. Yin, Y. Wang, Y. Cao, J. Yin and J. Zhang, et al., Development of a highly sensitive digital PCR assay to quantify long non-coding RNA MYU in urine samples which exhibited great potential as an alternative diagnostic biomarker for prostate cancer, Transl. Androl. Urol., 2021, 10(10), 3815–3825 CrossRef PubMed.
- M. Stella, G. I. Russo, R. Leonardi, D. Carcò, G. Gattuso and L. Falzone, et al., Extracellular RNAs from Whole Urine to Distinguish Prostate Cancer from Benign Prostatic Hyperplasia, Int. J. Mol. Sci., 2024, 25(18), 10079 CrossRef CAS PubMed , Available from: http://www.ncbi.nlm.nih.gov/pubmed/39337566.
- A. Bogaczyk, N. Potocka, S. Paszek, M. Skrzypa, A. Zuchowska and M. Kośny, et al., Absolute Quantification of Selected microRNAs Expression in Endometrial Cancer by Digital PCR, Int. J. Mol. Sci., 2024, 25(6), 3286 CrossRef CAS PubMed.
- S. R. Vitale, J. A. Helmijr, M. Gerritsen, H. Coban, L. F. van Dessel and N. Beije, et al., Detection of tumor-derived extracellular vesicles in plasma from patients with solid cancer, BMC Cancer, 2021, 21(1), 315 CrossRef CAS PubMed.
- E. Sánchez-Herrero, C. Campos-Silva, Y. Cáceres-Martell, L. Robado De Lope, S. Sanz-Moreno and R. Serna-Blasco, et al., ALK-Fusion Transcripts Can Be Detected in Extracellular Vesicles (EVs) from Nonsmall Cell Lung Cancer Cell Lines and Patient Plasma: Toward EV-Based Noninvasive Testing, Clin. Chem., 2022, 68(5), 668–679 CrossRef PubMed.
- C. Genova, S. Marconi, G. Chiorino, F. Guana, P. Ostano and S. Santamaria, et al., Extracellular vesicles miR-574-5p and miR-181a-5p as prognostic markers in NSCLC patients treated with nivolumab, Clin. Exp. Med., 2024, 24(1), 182 CrossRef CAS PubMed.
- G. Girolimetti, I. A. Pelisenco, L. H. Eusebi, C. Ricci, B. Cavina and I. Kurelac, et al., Dysregulation of a Subset of Circulating and Vesicle-Associated miRNA in Pancreatic Cancer, Noncoding RNAs, 2024, 10(3), 29 CAS.
- A. Giménez-Capitán, E. Sánchez-Herrero, L. Robado de Lope, A. Aguilar-Hernández, I. Sullivan and V. Calvo, et al., Detecting ALK, ROS1, and RET fusions and the METΔex14 splicing variant in liquid biopsies of non-small-cell lung cancer patients using RNA-based techniques, Mol. Oncol., 2023, 17(9), 1884–1897 CrossRef PubMed.
- D. Kliman, G. Castellano-Gonzalez, B. Withers, J. Street, E. Tegg and O. Mirochnik, et al., Ultra-Sensitive Droplet Digital PCR for the Assessment of Microchimerism in Cellular Therapies, Biol. Blood Marrow Transplant., 2018, 24(5), 1069–1078 CrossRef CAS PubMed.
- M. Waterhouse, D. Pfeifer, J. Duque-Afonso, M. Follo, J. Duyster and M. Depner, et al., Droplet digital PCR for the simultaneous analysis of minimal residual disease and hematopoietic chimerism after allogeneic cell transplantation, Clin. Chem. Lab. Med., 2019, 57(5), 641–647 CAS.
- T. Stahl, C. Rothe, M. U. Böhme, A. Kohl, N. Kröger and B. Fehse, Digital PCR panel for sensitive hematopoietic chimerism quantification after allogeneic stem cell transplantation, Int. J. Mol. Sci., 2016, 17(9), 1515 CrossRef PubMed.
- W. Chen, J. Huang, Y. Zhao, L. Huang, Z. Yuan and M. Gu, et al., Measurable residual disease monitoring by ddPCR in the early posttransplant period complements the traditional MFC method to predict relapse after HSCT in AML/MDS: a multicenter retrospective study, J. Transl. Med., 2024, 22(1), 410 CrossRef CAS PubMed.
- Fda Cber, Long Term Follow-Up After Administration of Human Gene Therapy Products, Guidance for Industry [Internet], 2020, Available from: https://www.fda.gov/vaccines-blood-biologics/guidance-compliance-.
- L. A. Murphy, R. C. Marians, K. Miller, M. D. Brenton, R. L. V. Mallo and M. E. Kohler, et al., Digital polymerase chain reaction strategies for accurate and precise detection of vector copy number in chimeric antigen receptor T-cell products, Cytotherapy, 2023, 25(1), 94–102 CrossRef CAS PubMed.
- W. Wang, M. Al-Hajj and A. S. Alavi, Detection and quantification of integrated vector copy number by multiplex droplet digital PCR in dual-transduced CAR T cells, Mol. Ther.--Methods Clin. Dev., 2023, 30, 403–410 CrossRef CAS PubMed.
- S. L. Maude, T. W. Laetsch, J. Buechner, S. Rives, M. Boyer and H. Bittencourt, et al., Tisagenlecleucel in Children and Young Adults with B-Cell Lymphoblastic Leukemia, N. Engl. J. Med., 2018, 378(5), 439–448 CrossRef CAS PubMed.
- I. de la Iglesia-San Sebastián, M. López-Esteban, M. Bastos-Oreiro, S. Fernández de Córdoba-Oñate, M. Gutierrez and D. Carbonell, et al., Chimeric antigen receptor copies in cell-free DNA predict relapse in aggressive B-cell lymphoma patients treated with CAR T-cell therapy, Br. J. Haematol., 2025, 206(1), 195–203 CrossRef PubMed.
- Y. Lou, C. Chen, X. Long, J. Gu, M. Xiao and D. Wang, et al., Detection and Quantification of Chimeric Antigen Receptor Transgene Copy Number by Droplet Digital PCR versus Real-Time PCR, J. Mol. Diagn., 2020, 22(5), 699–707 CrossRef CAS PubMed.
- M. L. Schubert, C. Berger, A. Kunz, A. Schmitt, A. Badbaran and B. Neuber, et al., Comparison of single copy gene-based duplex quantitative PCR and digital droplet PCR for monitoring of expansion of CD19-directed CAR T cells in treated patients, Int. J. Oncol., 2022, 60(5), 48 CrossRef CAS PubMed.
- G. Wiedemann, U. Bacher, R. Joncourt, F. Solly, C. C. Widmer and S. Zeerleder, et al., A Comprehensive ddPCR Strategy for Sensitive and Reliable Monitoring of CAR-T Cell Kinetics in Clinical Applications, Int. J. Mol. Sci., 2024, 25(16), 8556 CrossRef CAS PubMed.
- Y. Cui, L. J. Zhang, J. Li, Y. J. Xu and M. Y. Liu, Diagnostic value of circular free DNA for colorectal cancer detection, World J Gastrointest Oncol, 2023, 15(6), 1086–1095 CrossRef PubMed.
- A. Murillo Carrasco, O. Acosta, J. Ponce, J. Cotrina, A. Aguilar and J. Araujo, et al., PUM1 and RNase P genes as potential cell-free DNA markers in breast cancer, J. Clin. Lab. Anal., 2021, 35(4), e23720 CrossRef CAS PubMed.
- M. Du, C. C. Huang, W. Tan, M. Kohli and L. Wang, Multiplex digital pcr to detect amplifications of specific androgen receptor loci in cell-free DNA for prognosis of metastatic castration-resistant prostate cancer, Cancers, 2020, 12(8), 1–13 CrossRef PubMed.
- Y. Zhang, Y. Bae, S. Shibayama, X. Wang, M. Kato and L. Dong, International co-validation on absolute quantification of single nucleotide variants of KRAS by digital PCR, Anal. Bioanal. Chem., 2022, 414(19), 5899–5906 CrossRef CAS PubMed.
- Y. M. Dennis Lo, N. Corbetta, P. F. Chamberlain, V. Rai, I. L. Sargent and G. Redman, et al., Presence of fetal DNA in maternal plasma and serum, Lancet, 1997, 350, 485–487 CrossRef PubMed.
- A. Tabor, M. Madsen, E. B. Obel, P. John, B. Jens and N.-P. Bent, Randomised controlled trial of genetic amniocentesis in 4606 low-risk women, Lancet, 1986, 1287–1293 CrossRef CAS PubMed.
- Y. Ouzegdouh Mammasse, C. Chenet, D. Drubay, C. Martageix, J. P. Cartron and W. Vainchenker, et al., A new efficient tool for non-invasive diagnosis of fetomaternal platelet antigen incompatibility, Br. J. Haematol., 2020, 190(5), 787–798 CrossRef CAS PubMed.
- J. Shaw, E. Scotchman, B. Paternoster, M. Ramos, S. Nesbitt and S. Sheppard, et al., Non-invasive fetal genotyping for maternal alleles with droplet digital PCR: A comparative study of analytical approaches, Prenatal Diagn., 2023, 43(4), 477–488 CrossRef CAS PubMed.
- M. Y. Chang, S. Ahn, M. Y. Kim, J. H. Han, H. R. Park and H. K. Seo, et al., One-step noninvasive prenatal testing (NIPT) for autosomal recessive homozygous point mutations using digital PCR, Sci. Rep., 2018, 8(1), 2877 CrossRef PubMed.
- S. Perlado, A. Bustamante-Aragonés, M. Donas, I. Lorda-Sánchez, J. Plaza and M. R. De Alba, Fetal genotyping in maternal blood by digital PCR: Towards NIPD of monogenic disorders independently of parental origin, PLoS One, 2016, 11(4), e0153258 CrossRef PubMed.
- E. D'Aversa, G. Breveglieri, P. Pellegatti, G. Guerra, R. Gambari and M. Borgatti, Non-invasive fetal sex diagnosis in plasma of early weeks pregnants using droplet digital PCR, Mol. Med., 2018, 24(1), 14 Search PubMed.
- I. Zednikova, E. Pazourkova, S. Lassakova, B. Vesela and M. Korabecna, Detection of cell-free foetal DNA fraction in female-foetus bearing pregnancies using X-chromosomal insertion/deletion polymorphisms examined by digital droplet PCR, Sci. Rep., 2020, 10(1), 20036 CrossRef CAS PubMed.
- I. Hudecova, P. Jiang, J. Davies, Y. M. D. Lo, R. A. Kadir and R. W. K. Chiu, Noninvasive detection of F8 int22h-related inversions and sequence variants in maternal plasma of hemophilia carriers, Blood, 2017, 130(3), 340–347 CrossRef CAS PubMed.
- A. J. Johnson, Might broader be better?, Blood, 2017, 130, 239–240 CrossRef CAS PubMed.
- M. Ioannides, A. Achilleos, S. Kyriakou, E. Kypri, C. Loizides and K. Tsangaras, et al., Development of a new methylation-based fetal fraction estimation assay using multiplex ddPCR, Mol. Genet. Genomic Med., 2020, 8(2), e1094 CrossRef CAS PubMed.
- J. Wang, W. Wang, W. Zhou, Y. Zhou, L. Zhou and X. Wang, et al., Preliminary study of noninvasive prenatal screening for 22q11.2 deletion/duplication syndrome using multiplex dPCR assay, Orphanet J. Rare Dis., 2023, 18(1), 278 CrossRef PubMed.
- L. Orhant, O. Anselem, M. Fradin, P. H. Becker, C. Beugnet and N. Deburgrave, et al., Droplet digital PCR combined with minisequencing, a new approach to analyze fetal DNA from maternal blood: Application to the non-invasive prenatal diagnosis of achondroplasia, Prenatal Diagn., 2016, 36(5), 397–406 CrossRef CAS PubMed.
- T. Huby, E. Le Guillou, C. Burin Des Roziers, L. Pacot, A. Briand-Suleau and A. Chansavang, et al., Noninvasive Prenatal Diagnosis of a Paternally Inherited MEN1 Pathogenic Splicing Variant, J. Clin. Endocrinol. Metab., 2022, 107(4), E1367–E1373 CrossRef PubMed.
- A. Gruber, M. Pacault, L. A. El Khattabi, N. Vaucouleur, L. Orhant and T. Bienvenu, et al., Non-invasive prenatal diagnosis of paternally inherited disorders from maternal plasma: Detection of NF1 and CFTR mutations using droplet digital PCR, Clin. Chem. Lab. Med., 2018, 56(5), 728–738 CrossRef CAS PubMed.
- E. Debrand, A. Lykoudi, E. Bradshaw and S. K. Allen, A non-invasive droplet digital PCR (ddPCR) assay to detect paternal CFTR mutations in the cell-free fetal DNA (cffDNA) of three pregnancies at risk of cystic fibrosis via compound heterozygosity, PLoS One, 2015, 10(11), e0142729 CrossRef PubMed.
- Y. Yan, F. Wang, C. Zhang, X. Jin, Q. Zhang and X. Feng, et al., Evaluation of droplet digital PCR for non-invasive prenatal diagnosis of phenylketonuria, Anal. Bioanal. Chem., 2019, 411(27), 7115–7126 CrossRef CAS PubMed.
- X. Wei, W. Lv, H. Tan, D. Liang and L. Wu, Development and validation of a haplotype-free technique for non-invasive prenatal diagnosis of spinal muscular atrophy, J. Clin. Lab. Anal., 2020, 34(2), e23046 CrossRef CAS PubMed.
- R. C. Caswell, T. Snowsill, J. A. L. Houghton, A. J. Chakera, M. H. Shepherd and T. W. Laver, et al., Noninvasive Fetal Genotyping by Droplet Digital PCR to Identify Maternally Inherited Monogenic Diabetes Variants, Clin. Chem., 2020, 66(7), 958–965 CrossRef PubMed.
- Y. Li, J. Ye, L. Liang, X. Tan, L. Zheng and T. Qin, et al., Detection of α-thalassemia South-East Asian deletion based on a fully integrated digital polymerase chain reaction system DropXpert S6, Hematology, 2024, 29(1), 2365596 CrossRef PubMed.
- K. Sawakwongpra, K. Tangmansakulchai, W. Ngonsawan, S. Promwan, S. Chanchamroen and W. Quangkananurug, et al., Droplet-based digital PCR for non-invasive prenatgenetic diagnosis of α and β-thalassemia, Biomed Rep., 2021, 15(4), 82 CrossRef CAS PubMed.
- C. G. Constantinou, E. Karitzi, S. Byrou, C. Stephanou, K. Michailidou and C. Makariou, et al., Optimized Droplet Digital PCR Assay on Cell-Free DNA Samples for Non-Invasive Prenatal Diagnosis: Application to Beta-Thalassemia, Clin. Chem., 2022, 68(8), 1053–1063 CrossRef PubMed.
- E. D'aversa, G. Breveglieri, E. Boutou, A. Balassopoulou, E. Voskaridou and P. Pellegatti, et al., Droplet Digital PCR for Non-Invasive Prenatal Detection of Fetal Single-Gene Point Mutations in Maternal Plasma, Int. J. Mol. Sci., 2022, 23(5), 2819 CrossRef PubMed.
- P. Charoenkwan, K. Traisrisilp, S. Sirichotiyakul, A. Phusua, T. Sanguansermsri and T. Tongsong, Noninvasive Prenatal Diagnosis of Beta-Thalassemia Disease by Using Digital PCR Analysis of Cell-Free Fetal DNA in Maternal Plasma, Fetal Diagn Ther., 2023, 49(11–12), 468–478 Search PubMed.
- J. Camunas-Soler, H. Lee, L. Hudgins, S. R. Hintz, Y. J. Blumenfeld and Y. Y. El-Sayed, et al., Noninvasive prenatal diagnosis of single-gene disorders by use of droplet digital PCR, Clin. Chem., 2018, 64(2), 336–345 CrossRef CAS PubMed.
- M. Pacault, C. Verebi, M. Lopez, N. Vaucouleur, L. Orhant and N. Deburgrave, et al., Non-invasive prenatal diagnosis of single gene disorders by paternal mutation exclusion: 3 years of clinical experience, BJOG, 2022, 129(11), 1879–1886 CrossRef CAS PubMed.
- K. A. Sillence, L. A. Roberts, H. J. Hollands, H. P. Thompson, M. Kiernan and T. E. Madgett, et al., Fetal sex and RHD genotyping with digital PCR demonstrates greater sensitivity than real-time PCR, Clin. Chem., 2015, 61(11), 1399–1407 CrossRef CAS PubMed.
- M. Eryilmaz, D. Müller, G. Rink, H. Klüter and P. Bugert, Introduction of Noninvasive Prenatal Testing for Blood Group and Platelet Antigens from Cell-Free Plasma DNA Using Digital PCR, Transfus Med Hemother, 2020, 47(4), 292–301 CrossRef PubMed.
- O.' B. Helen, H. Catherine, S. Elizna, F. Robert, D. James and G. Glenn, Non-invasive prenatal testing (NIPT) for fetal Kell, Duffy and Rh blood group antigen prediction in alloimmunised pregnant women: power of droplet digital PCR, Br. J. Haematol., 2020, 189, e64–e118 Search PubMed.
- A. Nykel, M. Kaszkowiak, W. Fendler and A. Gach, Chip-based digital PCR approach provides a sensitive and cost-effective single-day screening tool for common fetal aneuploidies-A proof of concept study, Int. J. Mol. Sci., 2019, 20(21), 5486 CrossRef CAS PubMed.
- S. Y. Lee, S. J. Kim, S. H. Han, J. S. Park, H. J. Choi and J. J. Ahn, et al., A new approach of digital PCR system for non-invasive prenatal screening of trisomy 21, Clin. Chim. Acta, 2018, 476, 75–80 CrossRef CAS PubMed.
- P. Dai, Y. Yang, G. Zhao, Z. Gu, H. Ren and S. Hu, et al., A dPCR-NIPT assay for detections of trisomies 21, 18 and 13 in a single-tube reaction-could it replace serum biochemical tests as a primary maternal plasma screening tool?, J. Transl. Med., 2022, 20(1), 269 CrossRef CAS PubMed.
- M. Lee, A. C. Y. Lui, J. C. K. Chan, P. H. L. Doong, A. K. Y. Kwong and C. C. Y. Mak, et al., Revealing parental mosaicism: the hidden answer to the recurrence of apparent de novo variants, Hum. Genomics, 2023, 17(1), 91 CrossRef CAS PubMed.
- E. Ehn, J. Eisfeldt, J. M. Laffita-Mesa, H. Thonberg, J. Schoumans and A. M. Portaankorva, et al., A de novo, mosaic and complex chromosome 21 rearrangement causes APP triplication and familial autosomal dominant early onset Alzheimer disease, Sci. Rep., 2025, 15(1), 2912 CrossRef CAS PubMed.
- X. Yang, X. Yang, J. Chen, S. Li, Q. Zeng and A. Y. Huang, et al., ATP1A3 mosaicism in families with alternating hemiplegia of childhood, Clin. Genet., 2019, 96(1), 43–52 CrossRef CAS PubMed.
- B. Liu, M. Chen, Y. Yang, Y. Huang, Y. Qian and M. Dong, Identification of of a PAX2 mutation from maternal mosaicism causes recurrent renal disorder in siblings, Clin. Chim. Acta, 2022, 525, 23–28 CrossRef CAS PubMed.
- F. Zhang, Z. Wang, Q. Meng, J. Song, S. Yang and X. Tang, et al., Disparate phenotypes in two unfavorable pregnancies due to maternal mosaicism of a novel RET gene mutation, Clin. Chim. Acta, 2022, 531, 84–90 CrossRef CAS PubMed.
- P. Arts, J. Garland, A. B. Byrne, T. S. E. Hardy, M. Babic and J. Feng, et al., Paternal mosaicism for a novel PBX1 mutation associated with recurrent perinatal death: Phenotypic expansion of the PBX1-related syndrome, Am. J. Med. Genet., Part A, 2020, 182(5), 1273–1277 CrossRef CAS PubMed.
- Y. Qian, J. Liu, Y. Yang, M. Chen, C. Jin and P. Chen, et al., Paternal low-level mosaicism-caused SATB2-associated syndrome, Front. Genet., 2019, 10, 630 CrossRef CAS PubMed.
- A. Nykel, R. Wo, A. Gach, R. Kimata Pooh and E. J. Pavlik, The Influence of Maternal Cell Contamination on Fetal Aneuploidy Detection Using Chip-Based Digital PCR Testing, Diagnostics, 2021, 11(9), 1607, DOI:10.3390/diagnostics11091607.
- V. Corman, T. Bleicker, S. Brünink, C. Drosten, O. Landt and M. Koopmans, et al., Users looking for a workflow protocol consult the last three pages of this document [Internet], 2020, Available from: https://virologie-ccm.charite.de/en/.
- Public Health HKU- LKS Faculty of Medecine. Detection of 2019 novel coronavirus (2019-nCoV) in suspected human cases by RT-PCR.
- L. Dong, J. Zhou, C. Niu, Q. Wang, Y. Pan and S. Sheng, et al., Highly accurate and sensitive diagnostic detection of SARS-CoV-2 by digital PCR, Talanta, 2021, 224, 121726 CrossRef CAS PubMed.
- Z. Wu and J. M. McGoogan, Characteristics of and Important Lessons from the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72314 Cases from the Chinese Center for Disease Control and Prevention, JAMA, J. Am. Med. Assoc., 2020, 323, 1239–1242 CrossRef CAS PubMed.
- X. Xie, Z. Zhong, W. Zhao, C. Zheng, F. Wang and J. Liu, Chest CT for Typical Coronavirus Disease 2019 (COVID-19) Pneumonia: Relationship to Negative RT-PCR Testing, Radiology, 2020, 296(2), E41–E45 CrossRef PubMed.
- J. Zhang, J. Han, Y. Liang, C. Bai, W. Liu and X. Wang, et al., The development of a droplet digital PCR for accurate detection of SARS-CoV- 2 by simultaneous determination of dual gene targets, Research Square, 2022, preprint, DOI:10.21203/rs.3.rs-2214895/v1.
- T. Suo, X. Liu, J. Feng, M. Guo, W. Hu and D. Guo, et al., ddPCR: a more accurate tool for SARS-CoV-2 detection in low viral load specimens, Emerging Microbes Infect., 2020, 9(1), 1259–1268 CrossRef CAS PubMed.
- P. Poggio, P. Songia, C. Vavassori, V. Ricci, C. Banfi and S. S. Barbieri, et al., Digital PCR for high sensitivity viral detection in false-negative SARS-CoV-2 patients, Sci. Rep., 2021, 11(1), 4310 CrossRef CAS PubMed.
- C. Liu, Q. Shi, M. Peng, L. Ruyue, H. Li and Y. Cai, et al., Evaluation of droplet digital PCR for quantification of SARS-CoV-2 Virus in discharged COVID-19 patients, Aging., 2020, 20997–21003 CrossRef CAS PubMed.
- C. Niu, L. Dong, Y. Gao, Y. Zhang, X. Wang and J. Wang, Quantitative analysis of RNA by HPLC and evaluation of RT-dPCR for coronavirus RNA quantification, Talanta, 2021, 228, 122227 CrossRef CAS PubMed.
- S. S. Lee, S. Kim, H. M. Yoo, D. H. Lee and Y. K. Bae, Development of SARS-CoV-2 packaged RNA reference material for nucleic acid testing, Anal. Bioanal. Chem., 2022, 414(5), 1773–1785 CrossRef CAS PubMed.
- S. Falak, D. M. O'sullivan, M. H. Cleveland, S. Cowen, E. J. Busby and A. S. Devonshire, et al., The Application of Digital PCR as a Reference Measurement Procedure to Support the Accuracy of Quality Assurance for Infectious Disease Molecular Diagnostic Testing, Clin. Chem., 2025, 71(3), 378–386 CrossRef CAS PubMed.
- A. Badbaran, R. K. Mailer, C. Dahlke, J. Woens, A. Fathi and S. C. Mellinghoff, et al., Digital PCR to quantify ChAdOx1 nCoV-19 copies in blood and tissues, Mol. Ther.--Methods Clin. Dev., 2021, 23, 418–423 CrossRef CAS PubMed.
- R. M. da Silva, P. Gebe Abreu Cabral, S. B. de Souza, R. F. Arruda, S. P. de F Cabral and A. L. E. M. de Assis, et al., Serial viral load analysis by DDPCR to evaluate FNC efficacy and safety in the treatment of mild cases of COVID-19, Front. Med., 2023, 10, 1143485 CrossRef PubMed.
- D. Veyer, S. Kernéis, G. Poulet, M. Wack, N. Robillard and V. Taly, et al., Highly Sensitive Quantification of Plasma Severe Acute Respiratory Syndrome Coronavirus 2 RNA Sheds Light on its Potential Clinical Value, Clinical Infectious Diseases., 2021, 73(9), E2890–E2897 CrossRef CAS PubMed.
- M. Deiana, A. Mori, C. Piubelli, S. Scarso, M. Favarato and E. Pomari, Assessment of the direct quantitation of SARS-CoV-2 by droplet digital PCR, Sci. Rep., 2020, 10(1), 18764 CrossRef CAS PubMed.
- M. Brandolini, F. Taddei, M. M. Marino, L. Grumiro, A. Scalcione and M. E. Turba, et al., Correlating qrt-pcr, dpcr and viral titration for the identification and quantification of sars-cov-2: A new approach for infection management, Viruses., 2021, 13(6), 1022 CrossRef CAS PubMed.
- World Health Organisation, What's New in TreatmentMonitoring: Viral Load and CD4 Testing [Internet], 2017, Available from: https://www.who.int/hiv.
- D. A. Bejarano, M. C. Puertas, K. Börner, J. Martinez-Picado, B. Müller and H. G. Kräusslich, Detailed characterization of early HIV-1 replication dynamics in primary human macrophages, Viruses, 2018, 10(11), 620 CrossRef CAS PubMed.
- S. A. Yukl, P. Kaiser, P. Kim, S. Telwatte, S. K. Joshi and M. Vu, et al., HIV latency in isolated patient CD4+ T cells may be due to blocks in HIV transcriptional elongation, completion, and splicing, Sci. Transl. Med., 2018, 10(430), eaap9927 CrossRef PubMed.
- T. J. Henrich, S. Gallien, J. Z. Li, F. Pereyra and D. R. Kuritzkes, Low-level detection and quantitation of cellular HIV-1 DNA and 2-LTR circles using droplet digital PCR, J. Virol. Methods, 2012, 186(1–2), 68–72 CrossRef CAS PubMed.
- E. M. Anderson, F. R. Simonetti, R. J. Gorelick, S. Hill, M. A. Gouzoulis and J. Bell, et al., Dynamic shifts in the HIV proviral landscape during long term combination antiretroviral therapy: Implications for persistence and control of HIV infections, Viruses, 2020, 12(2), 136 CrossRef CAS PubMed.
- E. Bruzzesi, A. Gabrieli, D. Bernasconi, G. Marchetti, A. Calcagno and D. Ripamonti, et al., HIV-DNA decrease during treatment in primary HIV-1 infection with three different drug regimens: Italian Network of Acute HIV Infection (INACTION) clinical trial, J. Med. Virol., 2023, 95(9), e29114 CrossRef CAS PubMed.
- E. Muñoz-Muela, M. Trujillo-Rodríguez, A. Serna-Gallego, A. Saborido-Alconchel, C. Gasca-Capote and A. Álvarez-Ríos, et al., HIV-1-DNA/RNA and immunometabolism in monocytes: contribution to the chronic immune activation and inflammation in people with HIV-1, Lancet, 2024, 105338 Search PubMed , Available from: https://www.thelancet.com.
- C. Alteri, R. Scutari, C. Stingone, G. Maffongelli, M. Brugneti and F. Falasca, et al., Quantification of HIV-DNA and residual viremia in patients starting ART by droplet digital PCR: Their dynamic decay and correlations with immunological parameters and virological success, J. Clin. Virol., 2019, 117, 61–67 CrossRef CAS PubMed.
- M. Kiselinova, A. O. Pasternak, W. De Spiegelaere, D. Vogelaers, B. Berkhout and L. Vandekerckhove, Comparison of droplet digital PCR and seminested real-time PCR for quantification of cell-associated HIV-1 RNA, PLoS One, 2014, 9(1), e85999 CrossRef PubMed.
- E. Busby, A. S. Whale, R. Bridget Ferns, P. R. Grant, G. Morley and J. Campbell, et al., Instability of 8E5 calibration standard revealed by digital PCR risks inaccurate quantification of HIV DNA in clinical samples by qPCR, Sci. Rep., 2017, 7(1), 1209 CrossRef PubMed.
- I. Falces-Romero, I. García-Pérez, L. Martín-Carbonero, J. García-Rodríguez and J. Mingorance, Quantification of human immunodeficiency virus type 2 (HIV-2) viral load in plasma: Comparison of three commercial assays, J. Clin. Virol., 2024, 175, 105745 CrossRef CAS PubMed.
- M. C. Strain, S. M. Lada, T. Luong, S. E. Rought, S. Gianella and V. H. Terry, et al., Highly Precise Measurement of HIV DNA by Droplet Digital PCR, PLoS One, 2013, 8(4), e55943 CrossRef CAS PubMed.
- S. Rutsaert, K. Bosman, W. Trypsteen, M. Nijhuis and L. Vandekerckhove, Digital PCR as a tool to measure HIV persistence, Retrovirology, 2018, 15, 16 CrossRef PubMed.
- C. Renault, K. Bolloré, A. Pisoni, C. Motto-Ros, P. Van de Perre and J. Reynes, et al., Accuracy of real-time PCR and digital PCR for the monitoring of total HIV DNA under prolonged antiretroviral therapy, Sci. Rep., 2022, 12(1), 9323 CrossRef CAS PubMed.
- W. van Snippenberg, D. Gleerup, S. Rutsaert, L. Vandekerckhove, W. De Spiegelaere and W. Trypsteen, Triplex digital PCR assays for the quantification of intact proviral HIV-1 DNA, Methods, 2022, 201, 41–48 CrossRef CAS PubMed.
- A. Saborido-Alconchel, A. Serna-Gallego, M. Trujillo-Rodriguez, E. Muñoz-Muela, A. I. Álvarez-Ríos and C. Lozano, et al., Long-term effects on immunological, inflammatory markers, and HIV-1 reservoir after switching to a two-drug versus maintaining a three-drug regimen based on integrase inhibitors, Front. Immunol., 2024, 15, 1423734 CrossRef CAS PubMed.
- A. De Nicolò, A. Palermiti, S. Dispinseri, G. Marchetti, M. Trunfio and E. De Vivo, et al., Plasma, Intracellular and Lymph Node Antiretroviral Concentrations and HIV DNA Change During Primary HIV Infection: Results from the INACTION P25 Study, Int. J. Antimicrob. Agents, 2024, 64(2), 107200 CrossRef PubMed.
- I. González-Navarro, V. Urrea, C. Gálvez, M. del C, S. Morón-López and M. C. Puertas, et al., Assessing advances in three decades of clinical antiretroviral therapy on the HIV-1 reservoir, J. Clin. Invest., 2025, 135(2), e183952 CrossRef PubMed , Available from: https://www.jci.org/articles/view/183952.
- K. M. Bruner, Z. Wang, F. R. Simonetti, A. M. Bender, K. J. Kwon and S. Sengupta, et al., A quantitative approach for measuring the reservoir of latent HIV-1 proviruses, Nature, 2019, 566(7742), 120–125 CrossRef CAS PubMed.
- M. Delporte, L. Lambrechts, E. E. Blomme, W. van Snippenberg, S. Rutsaert and M. Verschoore, et al., Integrative Assessment of Total and Intact HIV-1 Reservoir by a 5-Region Multiplexed Rainbow DNA Digital PCR Assay, Clin. Chem., 2025, 71(1), 203–214 CrossRef CAS PubMed.
- A. Myerski, A. Siegel, J. Engstrom, I. McGowan and R. M. Brand, The Use of Droplet Digital PCR to Quantify HIV-1 Replication in the Colorectal Explant Model, AIDS Res. Hum. Retroviruses, 2019, 35(3), 326–334 CrossRef CAS PubMed.
- K. J. Bosman, A. M. Wensing, A. E. Pijning, W. J. van Snippenberg, P. M. van Ham and D. M. de Jong, et al., Development of sensitive ddPCR assays to reliably quantify the proviral DNA reservoir in all common circulating HIV subtypes and recombinant forms, J. Int. AIDS Soc., 2018, 21, e25185, DOI:10.1002/jia2.25185/full.
- C. Tumpach, A. Rhodes, Y. Kim, J. Ong, H. Liu and D. Chibo, et al., Adaptation of Droplet Digital PCR-Based HIV Transcription Profiling to Digital PCR and Association of HIV Transcription and Total or Intact HIV DNA, Viruses, 2023, 15(7), 1606 CrossRef CAS PubMed.
- S. A. Lee, S. Telwatte, H. Hatano, A. D. M. Kashuba, M. L. Cottrell and R. Hoh, et al., Antiretroviral Therapy Concentrations Differ in Gut vs. Lymph Node Tissues and Are Associated with HIV Viral Transcription by a Novel RT-ddPCR Assay, J. Acquired Immune Defic. Syndr., 2020, 83(5), 530–537 CrossRef CAS PubMed.
- R. K. Gupta, D. Peppa, A. L. Hill, C. Gálvez, M. Salgado and M. Pace, et al., Evidence for HIV-1 cure after CCR5Δ32/Δ32 allogeneic haemopoietic stem-cell transplantation 30 months post analytical treatment interruption: a case report, Lancet HIV, 2020, 7(5), e340–e347 CrossRef PubMed.
- C. Seeger and W. S. Mason, Hepatitis B Virus Biology, Microbiol. Mol. Biol. Rev., 2000, 64(1), 51–68 CrossRef CAS PubMed.
- G. P. Caviglia, M. L. Abate, A. Olivero, C. Rosso, A. Ciancio and E. Bugianesi, et al., Absolute quantification of intrahepatic HBV covalently-closed-circular DNA by droplet digital PCR, Dig. Liver Dis., 2017, 49(1), e36 CrossRef.
- J. T. Huang, Y. J. Liu, J. Wang, Z. G. Xu, Y. Yang and F. Shen, et al., Next generation digital PCR measurement of hepatitis B virus copy number in formalin-fixed paraffin-embedded hepatocellular carcinoma tissue, Clin. Chem., 2015, 61(1), 290–296 CrossRef CAS PubMed.
- U. Limothai, N. Chuaypen, K. Poovorawan, W. Chotiyaputta, T. Tanwandee and Y. Poovorawan, et al., Reverse transcriptase droplet digital PCR vs reverse transcriptase quantitative real-time PCR for serum HBV RNA quantification, J. Med. Virol., 2020, 92(12), 3365–3372 CrossRef CAS PubMed.
- R. Singh, G. Ramakrishna, M. K. Sharma, R. Kumar, E. Gupta and A. Rastogi, et al., Droplet digital PCR technique is ultrasensitive for the quantification of covalently closed circular DNA in the blood of chronic HBV-infected patients, Clin. Res. Hepatol. Gastroenterol., 2025, 49(3), 102531 CrossRef CAS PubMed.
- Y. Liu, A. L. Cathcart, W. E. Delaney and K. M. Kitrinos, Development of a digital droplet PCR assay to measure HBV DNA in patients receiving long-term TDF treatment, J. Virol. Methods, 2017, 249, 189–193 CrossRef CAS PubMed.
- S. Hayashi, M. Isogawa, K. Kawashima, K. Ito, N. Chuaypen and Y. Morine, et al., Droplet digital PCR assay provides intrahepatic HBV cccDNA quantification tool for clinical application, Sci. Rep., 2022, 12(1), 2133 CrossRef CAS PubMed.
- D. Li, V. Ho, C. F. Teng, H. W. Tsai, Y. Liu and S. Bae, et al., Novel digital droplet inverse PCR assay shows that natural clearance of hepatitis B infection is associated with fewer viral integrations, Emerging Microbes Infect., 2025, 14(1), 2450025 CrossRef PubMed.
- L. Piermatteo, R. Scutari, R. Chirichiello, M. Alkhatib, V. Malagnino and A. Bertoli, et al., Droplet digital PCR assay as an innovative and promising highly sensitive assay to unveil residual and cryptic HBV replication in peripheral compartment, Methods, 2022, 201, 74–81 CrossRef CAS PubMed.
- F. Villeret, F. Lebossé, S. Radenne, D. Samuel, B. Roche and J. Y. Mabrut, et al., Early intrahepatic recurrence of HBV infection in liver transplant recipients despite antiviral prophylaxis, JHEP Reports, 2023, 5(6), 100728 CrossRef PubMed.
- D. Mairiang, A. Songjaeng, P. Hansuealueang, Y. Malila, P. Lertsethtakarn and S. Silapong, et al., Application of one-step reverse transcription droplet digital pcr for dengue virus detection and quantification in clinical specimens, Diagnostics, 2021, 11(4), 639 CrossRef CAS PubMed.
- J. C. Rotondo, L. Oton-Gonzalez, C. Mazziotta, C. Lanzillotti, M. R. Iaquinta and M. Tognon, et al., Simultaneous detection and viral DNA load quantification of different human papillomavirus types in clinical specimens by the high analytical droplet digital PCR method, Front. Microbio., 2020, 11, 1–14 CrossRef PubMed.
- A. Villarmé, N. Ebran, T. Pace-Loscos, R. Schiappa, A. Mignot and A. Bozec, et al., Prognostic impact of intra tumoral HPV-16 viral load in oropharyngeal squamous cell carcinomas, Oral Oncol., 2024, 159, 107082 CrossRef PubMed.
- A. Qvick, E. Andersson, A. Oldaeus Almerén, M. Waenerlund, B. Stenmark and C. Karlsson, et al., Sensitive and Specific Droplet Digital PCR Assays for Circulating Tumor HPV DNA: Development, Validation, and Clinical Application in HPV-Associated Cancers, Mol. Diagn. Ther., 2024, 28, 835–845 CrossRef CAS PubMed.
- T. Wu, R. Sun, J. Sun, X. Tao, J. Su and S. Kong, et al., Circulating tumor HPV DNA as a specific biomarker for cervical cancer, Int. J. Gynecol. Obstet., 2025, 169(1), 148–154 CrossRef CAS PubMed.
- V. K. Morris, W. Xiao, K. Lin, C. W. Wong, M. T. Wotman and E. B. Holliday, et al., Time Dependency for Human Papillomavirus Circulating Tumor DNA Detection after Chemoradiation as a Prognostic Biomarker for Localized Anal Cancer, Clin. Cancer Res., 2025, OF1–OF7 Search PubMed , Available from: https://aacrjournals.org/clincancerres/article/doi/10.1158/1078-0432.CCR-24-2575/751773/Time-Dependency-for-Human-Papillomavirus.
- A. Seo, W. Xiao, O. Gjyshi, K. Yoshida-Court, P. Wei and D. Swanson, et al., Human Papilloma Virus Circulating Cell-Free DNA Kinetics in Patients with Cervical Cancer Undergoing Definitive Chemoradiation, Clin. Cancer Res., 2025, 31(4), 697–706, DOI:10.1158/1078-0432.CCR-24-2343/751000/Human-Papilloma-Virus-Circulating-Cell-Free-DNA.
- F. Rosing, M. Plath, T. Proctor, D. Höfler, Y. Alt and C. Lucena-Porcel, et al., Post-treatment monitoring of surgically treated oropharyngeal squamous cell carcinoma patients using human papillomavirus cell-free DNA, Oral Oncol., 2025, 163, 107225 CrossRef CAS PubMed.
- B. Velapatiño, K. Locher, C. R. Belanger and M. K. Charles, Mycobacterium tuberculosis DNA detection in formalin-fixed paraffin-embedded tissue using Digital PCR: a proof-of-concept study, Diagn. Microbiol. Infect. Dis., 2025, 111(3), 116697 CrossRef PubMed.
- M. Lee, E. J. Lee, J. Kang, K. H. Lee, S. J. Lee and S. H. Hong, et al., Roles of Cancer Histology Type and HPV Genotype in HPV ctDNA Detection at Baseline in Cervical Cancer: Implications for Tumor Burden Assessment, Pathobiology, 2024, 123–132 CrossRef PubMed.
- F. T. Van Den Berg, N. A. Makoah, S. A. Ali, T. A. Scott, R. E. Mapengo and L. Z. Mutsvunguma, et al., AAV-Mediated Expression of Broadly Neutralizing and Vaccine-like Antibodies Targeting the HIV-1 Envelope V2 Region, Mol. Ther.--Methods Clin. Dev., 2019, 14, 100–112 CrossRef CAS PubMed.
- E. Abachin, S. Convers, S. Falque, R. Esson, L. Mallet and N. Nougarede, Comparison of reverse-transcriptase qPCR and droplet digital PCR for the quantification of dengue virus nucleic acid, Biologicals, 2018, 52, 49–54 CrossRef CAS PubMed.
- M. E. Navarro Sanchez, N. Devard, C. Houy, E. Abachin, S. Godard and R. Esson, et al., Multiplex reverse transcriptase droplet digital PCR for the simultaneous quantification of four dengue serotypes: Proof of concept study, Biologicals, 2020, 67, 62–68 CrossRef CAS PubMed.
- J. F. Gélinas, S. Kiesslich, R. Gilbert and A. A. Kamen, Titration methods for rVSV-based vaccine manufacturing, MethodsX, 2020, 7 Search PubMed.
- H. Pere, D. Veyer and V. Taly, et al., Clin. Chem., 2025, 71, 339–341 CrossRef CAS PubMed.
- World Health Organisation, Global Tuberculosis Report 2013, World Health Organization, 2013, p. 303.
- R. Ushio, M. Yamamoto, K. Nakashima, H. Watanabe, K. Nagai and Y. Shibata, et al., Digital PCR assay detection of circulating Mycobacterium tuberculosis DNA in pulmonary tuberculosis patient plasma, Tuberculosis, 2016, 99, 47–53 CrossRef CAS PubMed.
- L. Lyu, Z. Li, L. Pan, H. Jia, Q. Sun and Q. Liu, et al., Evaluation of digital PCR assay in detection of M.tuberculosis IS6110 and IS1081 in tuberculosis patients plasma, BMC Infect. Dis., 2020, 20(1), 657 CrossRef CAS PubMed.
- M. Yamamoto, R. Ushio, H. Watanabe, T. Tachibana, M. Tanaka and T. Yokose, et al., Detection of Mycobacterium tuberculosis-derived DNA in circulating cell-free DNA from a patient with disseminated infection using digital PCR, Int. J. Infect. Dis., 2018, 66, 80–82 CrossRef CAS PubMed.
- S. M. Cho, S. Shin, Y. Kim, W. Song, S. G. Hong and S. H. Jeong, et al., A novel approach for tuberculosis diagnosis using exosomal DNA and droplet digital PCR, Clin. Microbiol. Infect., 2020, 26(7), 942.e1–942.e5 CrossRef CAS PubMed.
- H. D. Gliddon, M. Kaforou, M. Alikian, D. Habgood-Coote, C. Zhou and T. Oni, et al., Identification of Reduced Host Transcriptomic Signatures for Tuberculosis Disease and Digital PCR-Based Validation and Quantification, Front. Immunol., 2021, 12, 637164 CrossRef CAS PubMed.
- M. Belay, B. Tulu, S. Younis, D. A. Jolliffe, D. Tayachew and H. Manwandu, et al., Detection of Mycobacterium tuberculosis complex DNA in CD34-positive peripheral blood mononuclear cells of asymptomatic tuberculosis contacts: an observational study, Lancet Microbe, 2021, 2(6), e267–e275 CrossRef CAS PubMed.
- A. G. Adami, J. F. Gallo, J. M. W. Pinhata, M. C. Martins, C. M. S. Giampaglia and R. S. de Oliveira, Modified protocol for drug susceptibility testing of MGIT cultures of Mycobacterium tuberculosis by the MGIT 960, Diagn. Microbiol. Infect. Dis., 2017, 87(2), 108–111 CrossRef CAS PubMed.
- J. Luo, M. Luo, J. Li, J. Yu, H. Yang and X. Yi, et al., Rapid direct drug susceptibility testing of Mycobacterium tuberculosis based on culture droplet digital polymerase chain reaction, Int. J. Tuberc. Lung Dis., 2019, 23(2), 219–225 CrossRef CAS PubMed.
- S. Zhang, X. Chen, Z. Lin, Y. Tan, B. Liang and Y. Pan, et al., Quantification of Isoniazid-Heteroresistant Mycobacterium tuberculosis Using Droplet Digital PCR, J. Clin. Microbiol., 2023, 61(6), e0188422 CrossRef PubMed.
- C. Fleischmann, A. Scherag, N. K. J. Adhikari, C. S. Hartog, T. Tsaganos and P. Schlattmann, et al., Assessment of global incidence and mortality of hospital-treated sepsis current estimates and limitations, Am. J. Respir. Crit. Care Med., 2016, 193(3), 259–272 CrossRef CAS PubMed.
- M. Singer, C. S. Deutschman, C. Seymour, M. Shankar-Hari, D. Annane and M. Bauer, et al., The third international consensus definitions for sepsis and septic shock (sepsis-3), JAMA, J. Am. Med. Assoc., 2016, 315, 801–810 CrossRef CAS PubMed.
- L. E. Huerta and T. W. Rice, Pathologic Difference between Sepsis and Bloodstream Infections, J. Appl. Lab. Med., 2019, 3, 654–663 CrossRef CAS PubMed.
- W. V. Kern and S. Rieg, Burden of bacterial bloodstream infection—a brief update on epidemiology and significance of multidrug-resistant pathogens, Clin. Microbiol. Infect., 2020, 26, 151–157 CrossRef CAS PubMed.
- V. X. Liu, V. Fielding-Singh, J. D. Greene, J. M. Baker, T. J. Iwashyna and J. Bhattacharya, et al., The timing of early antibiotics and hospital mortality in sepsis, Am. J. Respir. Crit. Care Med., 2017, 196(7), 856–863 CrossRef PubMed.
- P. R. Murray and H. Masur, Current approaches to the diagnosis of bacterial and fungal bloodstream infections in the intensive care unit, Crit. Care Med., 2012, 40(12), 3277–3282 CrossRef CAS PubMed.
- Y. Li, J. Xiao, L. Xia, X. Sun, J. Li and H. Bai, Plasma cell-free DNA Droplet Digital PCR provides rapid and efficient infectious microbiology diagnosis for febrile haematological patients, Front. Cell. Infect. Microbiol., 2025, 15, 1522426 CrossRef PubMed.
- R. Almansa, S. Martín, M. Martin-Fernandez, M. Heredia-Rodríguez, E. Gómez-Sánchez and M. Aragón, et al., Combined quantification of procalcitonin and HLA-DR improves sepsis detection in surgical patients, Sci. Rep., 2018, 8(1), 11999 CrossRef PubMed.
- E. Tamayo, R. Almansa, E. Carrasco, A. Ávila-Alonso, A. Rodríguez-Fernández and J. Wain, et al., Quantification of IgM molecular response by droplet digital PCR as a potential tool for the early diagnosis of sepsis, Critical Care, 2014, 18, 433 CrossRef PubMed.
- A. P. Tedim, I. Merino, A. Ortega, M. Domínguez-Gil, J. M. Eiros and J. F. Bermejo-Martín, Quantification of bacterial DNA in blood using droplet digital PCR: a pilot study, Diagn. Microbiol. Infect. Dis., 2024, 108(1), 116075 CrossRef CAS PubMed.
- Y. Zheng, J. Jin, Z. Shao, J. Liu, R. Zhang and R. Sun, et al., Development and clinical validation of a droplet digital PCR assay for detecting Acinetobacter baumannii and Klebsiella pneumoniae in patients with suspected bloodstream infections, Microbiology, 2021, 10(6), e1247 CAS.
- S. Jiang, D. Zhao, C. Wang, X. Liu, Q. Yang and X. Bao, et al., Clinical evaluation of droplet digital PCR in the early identification of suspected sepsis patients in the emergency department: a prospective observational study, Front. Cell. Infect. Microbiol., 2024, 14, 1358801 CrossRef CAS PubMed.
- H. Kitagawa, M. Kojima, K. Tadera, S. Kogasaki, K. Omori and T. Nomura, et al., Clinical diagnostic performance of droplet digital PCR for pathogen detection in patients with Escherichia coli bloodstream infection: a prospective observational study, BMC Infect. Dis., 2025, 25(1), 22 CrossRef CAS PubMed.
- I. Ziegler, S. Lindström, M. Källgren, K. Strålin and P. Mölling, 16S rDNA droplet digital PCR for monitoring bacterial DNAemia in bloodstream infections, PLoS One, 2019, 14(11), e0224656 CrossRef CAS PubMed.
- J. Shin, S. Shin, S. H. Jung, C. Park, S. Y. Cho and D. G. Lee, et al., Duplex dPCR system for rapid identification of gram-negative pathogens in the blood of patients with bloodstream infection: A culture-independent approach, J. Microbiol. Biotechnol., 2021, 31(11), 1481–1489 CrossRef CAS PubMed.
- Y. Zhao, K. Lin, H. Zhang, G. Yuan, Y. Zhang and J. Pan, et al., Evaluation of droplet digital PCR rapid detection method and precise diagnosis and treatment for suspected sepsis (PROGRESS): a study protocol for a multi-center pragmatic randomized controlled trial, BMC Infect. Dis., 2022, 22(1), 630 CrossRef CAS PubMed.
- Y. Peng, R. Xie, Y. Luo, P. Guo, Z. Wu and Y. Chen, et al., Clinical evaluation of a multiplex droplet digital PCR for diagnosing suspected bloodstream infections: a prospective study, Front. Cell. Infect. Microbiol., 2024, 14, 1489792 CrossRef PubMed.
- B. Chen, Y. Xie, N. Zhang, W. Li, C. Liu and D. Li, et al., Evaluation of Droplet Digital PCR Assay for the Diagnosis of Candidemia in Blood Samples, Front. Microbio., 2021, 12, 700008 CrossRef PubMed.
- J. Wu, B. Tang, Y. Qiu, R. Tan, J. Liu and J. Xia, et al., Clinical validation of a multiplex droplet digital PCR for diagnosing suspected bloodstream infections in ICU practice: a promising diagnostic tool, Crit. Care, 2022, 26(1), 243 CrossRef PubMed.
- K. Lin, Y. Zhao, B. Xu, S. Yu, Z. Fu and Y. Zhang, et al., Clinical Diagnostic Performance of Droplet Digital PCR for Suspected Bloodstream Infections, Microbiol. Spectrum, 2023, 11(1), e01378-22 CrossRef PubMed.
- J. Feng, X. Cui, B. Du, H. Zhao, Y. Feng and J. Cui, et al., Detection and Quantification of Klebsiella pneumoniae in Fecal Samples Using Digital Droplet PCR in Comparison with Real-Time PCR, Microbiol. Spectrum, 2023, 11(4), e04249-22 CrossRef PubMed.
- F. Zhou, S. Sun, X. Sun, Y. Chen and X. Yang, Rapid and sensitive identification of pleural and peritoneal infections by droplet digital PCR, Folia Microbiologica, 2020, 66, 213–219 CrossRef PubMed , Available from: https://www.medcalc.org/calc/.
- Y. Wouters, D. Dalloyaux, A. Christenhusz, H. M. J. Roelofs, H. F. Wertheim and C. P. Bleeker-Rovers, et al., Droplet digital polymerase chain reaction for rapid broad-spectrum detection of bloodstream infections, Microb. Biotechnol., 2020, 13(3), 657–668 CrossRef CAS PubMed.
- F. Song, J. V. Kuehl, A. Chandran and A. P. Arkin, A Simple, Cost-Effective, and Automation-Friendly Direct PCR Approach for Bacterial Community Analysis, mSystems, 2021, 6(5), e00224-21 CrossRef PubMed.
- H. Yuan, Y. Chao and H. C. Shum, Droplet and Microchamber-Based Digital Loop-Mediated Isothermal Amplification (dLAMP), Small, 2020, 16, e1904469 CrossRef PubMed.
- W. Yin, J. Zhuang, J. Li, L. Xia, K. Hu and J. Yin, et al., Digital Recombinase Polymerase Amplification, Digital Loop-Mediated Isothermal Amplification, and Digital CRISPR-Cas Assisted Assay: Current Status, Challenges, and Perspectives, Small, 2023, 19, e2303398 CrossRef PubMed.
- T. C. McMahon, B. W. Blais, A. Wong and C. D. Carrillo, Multiplexed single intact cell droplet digital PCR (MuSIC ddPCR) method for specific detection of enterohemorrhagic E. coli (EHEC) in food enrichment cultures, Front. Microbio., 2017, 8, 332 Search PubMed.
- H. Pan, K. Dong, L. Rao, L. Zhao, X. Wu and Y. Wang, et al., Quantitative detection of viable but nonculturable state Escherichia coli O157:H7 by ddPCR combined with propidium monoazide, Food Control, 2020, 112, 107140 CrossRef CAS.
- Y. Du, Z. Yan, K. Song, J. Jin, L. Xiao and Z. Sun, et al., Development and evaluation of a multiplex droplet digital polymerase chain reaction method for simultaneous detection of five biothreat pathogens, Front. Microbio., 2022, 13, 970973 CrossRef PubMed.
- D. C. Duffy, Digital detection of proteins, Lab Chip, 2023, 23, 818–847 RSC.
- H. Schröder, M. Grösche, M. Adler, M. Spengler and C. M. Niemeyer, Immuno-PCR with digital readout, Biochem. Biophys. Res. Commun., 2017, 488(2), 311–315 CrossRef PubMed.
- C. Zhang, K. Zheng, C. Li, R. Zhang, Y. Zhu and L. Xia, et al., Single-Molecule Protein Analysis by Centrifugal Droplet Immuno-PCR with Magnetic Nanoparticles, Anal. Chem., 2024, 96(5), 1872–1879 CrossRef CAS PubMed.
- B. C. Vanness and T. H. Linz, Multiplexed miRNA and Protein Analysis Using Digital Quantitative PCR in Microwell Arrays, Anal. Chem., 2024, 96(3), 1371–1379 CrossRef CAS PubMed.
- H. Li, J. Li, Z. Zhang, Q. Yang, H. Du and Q. Dong, et al., Digital Quantitative Detection for Heterogeneous Protein and mRNA Expression Patterns in Circulating Tumor Cells, Adv. Sci., 2025, 12(2), e2410120 CrossRef PubMed.
- J. Ko, Y. Wang, J. C. T. Carlson, A. Marquard, J. Gungabeesoon and A. Charest, et al., Single Extracellular Vesicle Protein Analysis Using Immuno-Droplet Digital Polymerase Chain Reaction Amplification, Adv. Biosyst., 2020, 4(12), e1900307 CrossRef PubMed.
- M. F. Abasıyanık, K. Wolfe, H. Van Phan, J. Lin, B. Laxman and S. R. White, et al., Ultrasensitive digital quantification of cytokines and bacteria predicts septic shock outcomes, Nat. Commun., 2020, 11(1), 2607 CrossRef PubMed.
- S. A. Byrnes, T. Huynh, T. C. Chang, C. E. Anderson, J. J. McDermott and C. I. Oncina, et al., Wash-Free, Digital Immunoassay in Polydisperse Droplets, Anal. Chem., 2020, 92(5), 3535–3543 CrossRef CAS PubMed.
- E. Navarro, G. Serrano-Heras, M. J. Castaño and J. Solera, Real-time PCR detection chemistry, Clin. Chim. Acta, 2015, 439, 231–250 CrossRef CAS PubMed.
- B. Leatham, K. McNall, H. K. K. Subramanian, L. Jacky, J. Alvarado and D. Yurk, et al., A rapid, multiplex digital PCR assay to detect gene variants and fusions in non-small cell lung cancer, Mol. Oncol., 2023, 17(11), 2221–2234 CrossRef CAS PubMed.
- T. Okada, Y. Mizukami, Y. Ono, H. Sato, A. Hayashi and H. Kawabata, et al., Digital PCR-based plasma cell-free DNA mutation analysis for early-stage pancreatic tumor diagnosis and surveillance, J. Gastroenterol., 2020, 55(12), 1183–1193 CrossRef CAS PubMed.
- S. Hussung, M. E. Hess, E. B. Haghighi, U. A. Wittel, M. Boerries and R. M. Fritsch, Integrated Analysis of Cell-Free DNA and Novel Protein Biomarkers for Stratification and Therapy Monitoring in Stage IV Pancreatic Cancer: A Preliminary Study, Diagnostics, 2024, 15(1), 49 CrossRef PubMed , Available from: http://www.ncbi.nlm.nih.gov/pubmed/39795577.
- C. Tan, X. Chen, F. Wang, D. Wang, Z. Cao and X. Zhu, et al., A multiplex droplet digital PCR assay for non-invasive prenatal testing of fetal aneuploidies, Analyst, 2019, 144(7), 2239–2247 RSC.
- S. Laššáková, P. Šenkyřík, E. Pazourková, A. Hořínek, P. Calda and M. Břešťák, et al., Rapid non-invasive prenatal screening test for trisomy 21 based on digital droplet PCR, Sci. Rep., 2023, 13(1), 22948 CrossRef PubMed.
- Y. Hashimoto, N. Masunaga, N. Kagara, K. Abe, T. Yoshinami and M. Tsukabe, et al., Detection of Ultra-Rare ESR1 Mutations in Primary Breast Cancer Using LNA-Clamp ddPCR, Cancers, 2023, 15(9), 2632 CrossRef CAS PubMed.
- Q. Yu, H. Jiang, X. Su, Z. Jiang, X. Liang and C. Zhang, et al., Development of Multiplex Drop-Off Digital PCR Assays for Hotspot Mutation Detection of KRAS, NRAS, BRAF, and PIK3CA in the Plasma of Colorectal Cancer Patients, J. Mol. Diagn., 2023, 25(6), 388–402 CrossRef CAS PubMed.
- J. Tanaka, T. Nakagawa, A. Shiratori, Y. Shimazaki, C. Uematsu and M. Kamahori, et al., KRAS genotyping by digital PCR combined with melting curve analysis, Sci. Rep., 2019, 9(1), 2626 CrossRef PubMed.
- J. Tanaka, T. Nakagawa, K. Harada, C. Morizane, H. Tanaka and S. Shiba, et al., Efficient and accurate KRAS genotyping using digital PCR combined with melting curve analysis for ctDNA from pancreatic cancer patients, Sci. Rep., 2023, 13(1), 3039 CrossRef CAS PubMed.
- X. Dai, M. Cao and Z. Wang, Digital Melting Curve Analysis for Multiplex Quantification of Nucleic Acids on Droplet Digital PCR, Biosensors, 2025, 15(1), 36 CrossRef CAS PubMed.
- L. Miglietta, A. Moniri, I. Pennisi, K. Malpartida-Cardenas, H. Abbas and K. Hill-Cawthorne, et al., Coupling Machine Learning and High Throughput Multiplex Digital PCR Enables Accurate Detection of Carbapenem-Resistant Genes in Clinical Isolates, Front. Mol. Biosci., 2021, 8, 775299 CrossRef CAS PubMed.
- C. Li, N. Kang, S. Ye, W. Huang, X. Wang and C. Wang, et al., All-In-One OsciDrop Digital PCR System for Automated and Highly Multiplexed Molecular Diagnostics, Adv. Sci., 2024, 11(21), 2309557 CrossRef CAS PubMed.
- F. Schlenker, E. Kipf, N. Borst, T. Hutzenlaub, R. Zengerle and F. Von Stetten, et al., Virtual Fluorescence Color Channels by Selective Photobleaching in Digital PCR Applied to the Quantification of KRAS Point Mutations, Anal. Chem., 2021, 93(30), 10538–10545 CrossRef CAS PubMed.
- F. Schlenker, E. Kipf, M. Deuter, I. Höffkes, M. Lehnert and R. Zengerle, et al., Stringent base specific and optimization-free multiplex mediator probe DDPCR for the quantification of point mutations in circulating tumor DNA, Cancers, 2021, 13(22), 5742 CrossRef CAS PubMed.
- M. Neugebauer, S. Calabrese, S. Müller, T. T. Truong, P. Juelg and N. Borst, et al., Generic Reporter Sets for Colorimetric Multiplex dPCR Demonstrated with 6-Plex SNP Quantification Panels, Int. J. Mol. Sci., 2024, 25(16), 8968 CrossRef CAS PubMed.
- E. Y. Shum, J. H. Lai, S. Li, H. G. Lee, J. Soliman and V. K. Raol, et al., Next-Generation Digital Polymerase Chain Reaction: High-Dynamic-Range Single-Molecule DNA Counting via Ultrapartitioning, Anal. Chem., 2022, 94(51), 17868–17876 CrossRef CAS PubMed.
- N. Majumdar, T. Wessel and J. Marks, Digital PCR modeling for maximal sensitivity, dynamic range and measurement precision, PLoS One, 2015, 10(3), 1–17 CrossRef PubMed.
- W. W. Liu, Y. Zhu, Y. M. Feng, J. Fang and Q. Fang, Droplet-Based Multivolume Digital Polymerase Chain Reaction by a Surface-Assisted Multifactor Fluid Segmentation Approach, Anal. Chem., 2017, 89(1), 822–829 CrossRef CAS PubMed.
- M. Schulz, S. Probst, S. Calabrese, A. R. Homann, N. Borst and M. Weiss, et al., Versatile tool for droplet generation in standard reaction tubes by centrifugal step emulsification, Molecules, 2020, 25(8), 1914 CrossRef CAS PubMed.
- H. Si, G. Xu, F. Jing, P. Sun, D. Zhao and D. Wu, A multi-volume microfluidic device with no reagent loss for low-cost digital PCR application, Sens. Actuators, B, 2020, 318, 128197 CrossRef CAS.
- J. E. Kreutz, T. Munson, T. Huynh, F. Shen, W. Du and R. F. Ismagilov, Theoretical design and analysis of multivolume digital assays with wide dynamic range validated experimentally with microfluidic digital PCR, Anal. Chem., 2011, 83(21), 8158–8168 CrossRef CAS PubMed.
- Y. Luo, Q. Hu, Y. Yu, W. Lyu and F. Shen, Experimental investigation of confinement effect in single molecule amplification via real-time digital PCR on a multivolume droplet array SlipChip, Anal. Chim. Acta, 2024, 1304, 342541 CrossRef CAS PubMed.
- B. W. Buchan, D. A. Jobe, M. Mashock, D. Gerstbrein, M. L. Faron and N. A. Ledeboer, et al., Evaluation of a Novel Multiplex High-Definition PCR Assay for Detection of Tick-Borne Pathogens in Whole-Blood Specimens, J. Clin. Microbiol., 2019, e00513–e00519 CAS , Available from: https://journals.asm.org/journal/jcm.
- L. Jacky, D. Yurk, J. Alvarado, B. Leatham, J. Schwartz and J. Annaloro, et al., Virtual-Partition Digital PCR for High-Precision Chromosomal Counting Applications, Anal. Chem., 2021, 93(51), 17020–17029 CrossRef CAS PubMed.
- W. Trypsteen, M. Vynck, J. de Neve, P. Bonczkowski, M. Kiselinova and E. Malatinkova, et al., Ddpcrquant: Threshold determination for single channel droplet digital PCR experiments, Anal. Bioanal. Chem., 2015, 407(19), 5827–5834 CrossRef CAS PubMed.
- A. De Falco, C. M. Olinger, B. Klink, M. Mittelbronn and D. Stieber, Digital PCR cluster predictor: a universal R-package and shiny app for the automated analysis of multiplex digital PCR data, Bioinformatics, 2023, 39(5), btad282 CrossRef CAS PubMed.
- Y. Chen, W. De Spiegelaere, W. Trypsteen, J. Vandesompele, G. Wils and D. Gleerup, et al., Polytect: an automatic clustering and labeling method for multicolor digital PCR data, NAR: Genomics Bioinf., 2025, 7(1), lqaf015, DOI:10.1093/nargab/lqaf015/8063808.
- M. N. Hatori, S. C. Kim and A. R. Abate, Particle-Templated Emulsification for Microfluidics-Free Digital Biology, Anal. Chem., 2018, 90(16), 9813–9820 CrossRef CAS PubMed.
- T. Heinrich, S. Toepfer, K. Steinmetzer, M. Ruettger, I. Walz and L. Kanitz, et al., DNA-Binding Magnetic Nanoreactor Beads for Digital PCR Analysis, Anal. Chem., 2023, 95(38), 14175–14183 CrossRef CAS PubMed.
- C. Sun, L. Liu, H. N. Vasudevan, K. C. Chang and A. R. Abate, Accurate Bulk Quantitation of Droplet Digital Polymerase Chain Reaction, Anal. Chem., 2021, 93(29), 9974–9979 CrossRef CAS PubMed.
- W. Zhang, L. Cui, Y. Wang, Z. Xie, Y. Wei and S. Zhu, et al., An Integrated ddPCR Lab-on-a-Disc Device for Rapid Screening of Infectious Diseases, Biosensors, 2024, 14(1), 2 CrossRef CAS PubMed.
- X. Zhang, S. Wang, J. Wang, X. Sun, J. Xue and Z. Wang, et al., A ddPCR platform based on a microfluidic chip with a dual-function flow-focusing structure for sample-to-result DNA quantification analysis, Lab Chip, 2023, 24(4), 738–750 RSC.
- L. Cao, X. Guo, P. Mao, Y. Ren, Z. Li and M. You, et al., A Portable Digital Loop-Mediated Isothermal Amplification Platform Based on Microgel Array and Hand-Held Reader, ACS Sens., 2021, 6(10), 3564–3574 CrossRef CAS PubMed.
- C. D. Ahrberg, J. W. Choi, J. M. Lee, K. G. Lee, S. J. Lee and A. Manz, et al., Plasmonic heating-based portable digital PCR system, Lab Chip, 2020, 20(19), 3560–3568 RSC.
- T. Gou, J. Hu, W. Wu, X. Ding, S. Zhou and W. Fang, et al., Smartphone-based mobile digital PCR device for DNA quantitative analysis with high accuracy, Biosens. Bioelectron., 2018, 120, 144–152 CrossRef CAS PubMed.
- X. Wu, J. Pan, X. Zhu, C. Hong, A. Hu and C. Zhu, et al., MS2device: Smartphone-facilitated mobile nucleic acid analysis on microfluidic device, Analyst, 2021, 146(12), 3823–3833 RSC.
- X. Liu, X. Wang, H. Zhang, Z. Yan, M. Gaňová and T. Lednický, et al., Smartphone integrated handheld (SPEED) digital polymerase chain reaction device, Biosens. Bioelectron., 2023, 232, 115319 CrossRef CAS PubMed.
- H. Zhang, X. Liu, X. Wang, Z. Yan, Y. Xu and M. Gaňová, et al., SPEED: an integrated, smartphone-operated, handheld digital PCR Device for point-of-care testing, Microsyst. Nanoeng., 2024, 10(1), 62 CrossRef PubMed.
- M. E. Dueck, R. Lin, A. Zayac, S. Gallagher, A. K. Chao and L. Jiang, et al., Precision cancer monitoring using a novel, fully integrated, microfluidic array partitioning digital PCR platform, Sci. Rep., 2019, 9(1), 19606 CrossRef CAS PubMed.
- J. Lamanna, E. Y. Scott, H. S. Edwards, M. D. Chamberlain, M. D. M. Dryden and J. Peng, et al., Digital microfluidic isolation of single cells for -Omics, Nat. Commun., 2020, 11(1), 5632 CrossRef CAS PubMed.
- B. B. Li, E. Y. Scott, M. Dean Chamberlain, B. T. V. Duong, S. Zhang and S. J. Done, et al., Cell invasion in digital microfluidic microgel systems, Sci. Adv., 2020, eaba9589 CrossRef CAS PubMed , Available from: https://www.science.org.
- Q. Ruan, W. Ruan, X. Lin, Y. Wang, F. Zou and L. Zhou, et al., Digital-WGS: Automated, highly efficient whole-genome sequencing of single cells by digital microfluidics, Sci. Adv., 2020, 6, eabd6454 CrossRef CAS PubMed , Available from: https://www.science.org.
- X. Xu, L. Cai, S. Liang, Q. Zhang, S. Lin and M. Li, et al., Digital microfluidics for biological analysis and applications, Lab Chip, 2023, 23, 1169–1191 RSC.
- C. Yang, X. Gan, Y. Zeng, Z. Xu, L. Xu and C. Hu, et al., Advanced design and applications of digital microfluidics in biomedical fields: An update of recent progress, Biosens. Bioelectron., 2023, 242, 115723 CrossRef CAS PubMed.
- FDA, 510(k) Substantial Equivalence Determination Decision Summary.
- S. Liang, C. Li, Y. Ning, R. Su, M. Li and Y. Huang, et al., DMF-Bimol: Counting mRNA and Protein Molecules in Single Cells with Digital Microfluidics, Anal. Chem., 2024, 96, 17253–17261 CrossRef CAS PubMed.
- H. Bai, J. Hu, T. Liu, L. Wan, C. Dong and D. Luo, et al., A sample-to-answer digital microfluidic multiplexed PCR system for syndromic pathogen detection in respiratory tract infection, Lab Chip, 2025, 25, 1552–1564 RSC.
- L. Malic, L. Clime, B. U. Moon, C. Nassif, D. Da Fonte and D. Brassard, et al., Sample-to-answer centrifugal microfluidic droplet PCR platform for quantitation of viral load, Lab Chip, 2024, 24, 4755 RSC.
- A. J. Politza, R. Nouri and W. Guan, Digital CRISPR systems for the next generation of nucleic acid quantification, TrAC, Trends Anal. Chem., 2023, 159, 116917 CrossRef CAS PubMed.
- J. F. Huggett, C. A. Foy, V. Benes, K. Emslie, J. A. Garson and R. Haynes, et al., The digital MIQE guidelines: Minimum information for publication of quantitative digital PCR experiments, Clin. Chem., 2013, 59(6), 892–902 CrossRef CAS PubMed.
- S. Ye, C. Li, X. Zheng, W. Huang, Y. Tao and Y. Yu, et al., OsciDrop: A Versatile Deterministic Droplet Generator, Anal. Chem., 2022, 94(6), 2918–2925 CrossRef CAS PubMed.
- S. Min, S. Shin and Y. J. Chung, Detection of KRAS mutations in plasma cell-free DNA of colorectal cancer patients and comparison with cancer panel data for tissue samples of the same cancers, Genomics Inform., 2019, 17(4), e42 CrossRef PubMed.
- D. Sefrioui, L. Beaussire, A. Perdrix, F. Clatot, P. Michel and T. Frebourg, et al., Direct circulating tumor DNA detection from unpurified plasma using a digital PCR platform, Clin. Biochem., 2017, 50(16–17), 963–966 CrossRef CAS PubMed.
- W. Y. Chan, J. H. Lee, A. Stewart, R. J. Diefenbach, M. Gonzalez and A. M. Menzies, et al., Circulating tumour DNA dynamics predict recurrence in stage III melanoma patients receiving neoadjuvant immunotherapy, J. Exp. Clin. Cancer Res., 2024, 43(1), 238 CrossRef CAS PubMed.
- Y. Wang, W. Gao, M. Wu, X. Zhang, W. Liu and Y. Zhou, et al., EGFR mutation detection of lung circulating tumor cells using a multifunctional microfluidic chip, Talanta, 2021, 225, 122057 CrossRef CAS PubMed.
- X. Wang, Y. Zhang, C. Niu, S. Wang, L. Li and Y. Guo, et al., Establishment of primary reference measurement procedures and reference materials for: EGFR variant detection in non-small cell lung cancer, Anal. Methods, 2021, 13(18), 2114–2123 RSC.
- H. Blons, K. Pallier, D. Le Corre, C. Danel, M. Tremblay-Gravel and C. Houdayer, et al., Genome wide SNP comparative analysis between EGFR and KRAS mutated NSCLC and characterization of two models of oncogenic cooperation in non-small cell lung carcinoma, BMC Med. Genomics, 2008, 1(1), 25 CrossRef PubMed.
- M. Tsuruoka, M. Ninomiya, J. Inoue, T. Iwata, A. Sano and K. Sato, et al., Changes in Mutations of Cell-Free DNA and Liver Tumor Tissue in Patients with Advanced Hepatocellular Carcinoma before and after Introduction of Lenvatinib, Oncology, 2024, 102(12), 1072–1083 CrossRef CAS PubMed.
- S. Giannoni-Luza, O. Acosta, A. G. Murillo Carrasco, P. Danos, J. M. Cotrina Concha and H. Guerra Miller, et al., Chip-based digital Polymerase Chain Reaction as quantitative technique for the detection of PIK3CA mutations in breast cancer patients, Heliyon, 2022, 8(11), e11396 CrossRef CAS PubMed.
- I. Keraite, V. Alvarez-Garcia, I. Garcia-Murillas, M. Beaney, N. C. Turner and C. Bartos, et al., PIK3CA mutation enrichment and quantitation from blood and tissue, Sci. Rep., 2020, 10(1), 17082 CrossRef CAS PubMed.
- M. Abe, H. Hiraki, T. Tsuyukubo, S. Ono, S. Maekawa and D. Tamura, et al., The Clinical Validity of Urinary Pellet DNA Monitoring for the Diagnosis of Recurrent Bladder Cancer, J. Mol. Diagn., 2024, 26(4), 278–291 CrossRef CAS PubMed.
- A. Brik, D. G. Weber, S. Casjens, P. Rozynek, S. Meier and T. Behrens, et al., Digital PCR for the Analysis of MYC Copy Number Variation in Lung Cancer, Dis. Markers, 2020, 2020, 4176376 Search PubMed.
- A. C. Hatch, J. S. Fisher, S. L. Pentoney, D. L. Yang and A. P. Lee, Tunable 3D droplet self-assembly for ultra-high-density digital micro-reactor arrays, Lab Chip, 2011, 11(15), 2509–2517 RSC.
- M. Schulz, J. Ruediger, E. Landmann, M. Bakheit, S. Frischmann and D. Rassler, et al., High Dynamic Range Digital Assay Enabled by Dual-Volume Centrifugal Step Emulsification, Anal. Chem., 2021, 93(5), 2854–2860 CrossRef CAS PubMed.
- B. Lin, T. Tian, Y. Lu, D. Liu, M. Huang and L. Zhu, et al., Tracing Tumor-Derived Exosomal PD-L1 by Dual-Aptamer Activated Proximity-Induced Droplet Digital PCR, Angew. Chem., Int. Ed., 2021, 60(14), 7582–7586 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |
