The nature of the DNA template (single- versus double-stranded) affects the rate of aquation of a dinuclear Pt anticancer drug†
Murray S.
Davies
ac,
Susan J.
Berners-Price
*a,
John W.
Cox
b and
Nicholas
Farrell
*b
aDepartment of Chemistry, University of Western Australia, Crawley, WA 6009, Australia. E-mail: sbp@chem.uwa.edu.au; Fax: 61-8 9380 1005; Tel: 61-8-9380-3258
bDepartment of Chemistry, Virginia Commonwealth University, Richmond, VA 23284-2006, USA. E-mail: nfarrell@saturn.vcu.edu; Fax: 1-804-828-8599; Tel: 1-804-828-6320
cSchool of Pharmacy and Molecular Sciences, James Cook University, Townsville, QLD 4811, Australia
First published on 29th November 2002
Abstract
The rate of aquation of a dinuclear platinum anticancer agent is altered in the presence of template DNA with enhancement of hydrolysis in the presence of single-stranded over double-stranded DNA, emphasising how the alteration of chemical properties of small molecules in the presence of large host interactions is also dependent on the conformation and nature of that host.
There is growing interest in how DNA as a template affects kinetics of substitution reactions occurring within its domain.1 DNA exerts significant steric and electronic effects on small molecules interacting with it. Most examples to date have used duplex DNA as a template. Here we show that the nature of template DNA (single-stranded vs double-stranded) may also uniquely affect simple coordination chemistry processes such as aquation kinetics leading to substrate specificity. Transition metal cations may ‘pre-associate’ to DNA by electrostatic and hydrogen-bonding interactions prior to possible covalent bond formation. Only recently has such an effect been observed with the aquated form of cisplatin and natural substrate deoxyoligonucleotides using a quartz crystal microbalance2 and also between DNA and [Pt(NH3)4]2+ by ESI-mass spectrometry.3 Such pre-association may be responsible for many effects including local structure and sequence specificity.4
Polynuclear platinum complexes such as [{trans-PtCl(NH3)2}2{μ-(NH2(CH2)6NH2)}]2+ (1,1/t,t; 1), belong to a structurally novel set of anticancer complexes, including [{trans-PtCl(NH3)2}2{μ-trans-Pt(NH3)2(NH2(CH2)6NH2)}]4+ which has undergone Phase II clinical trials for the treatment of ovarian, gastric and lung tumors.5 Their Pt–DNA adducts are characterised by long-range intra- and inter-strand crosslinks, structurally distinct from those formed by cisplatin. More recently, pre-association of the charged complexes and double-stranded DNA template was observed by [1H,15N] HSQC NMR spectroscopy.6 A further distinction between the polynuclear complexes and cisplatin-based mononuclear agents is that they bind preferentially to single stranded DNA and RNA rather than to duplex DNA.7 Cisplatin, on the other hand, has shown a kinetic preference towards duplex DNA in studies with the oligonucleotide 5′-d(ATACATG(7)G(8)TACATA)-3′ (I) and the duplex (III) formed with its complementary strand 5′-d(TATG(25)TACCATG(18)TAT)-3′ (II).8,9
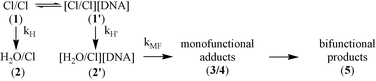
To examine the molecular basis for more rapid binding of di- and tri-nuclear Pt complexes to single stranded DNA we have used [1H,15N] HSQC NMR to compare the reaction of 15N-1 with I and III at 298 K, pH 5.4, under similar conditions to the cisplatin experiments (see ESI† ).‡ The reaction between 1 and III affords one major product, which was shown (via analysis of the NOESY NMR spectrum) to be the 5′→5′ 1,4-interstrand crosslink between G(8) and G(18) bases, existing in two conformational forms.10Fig. 1 shows the Pt–NH3 region of the [1H,15N] HSQC NMR spectra of the reactions of 15N-1 with the single strand I, and duplex III, recorded after 1.45 h. In both cases the 1H/15N peaks for 1 and the monoaqua monochloro complex 2 are deshielded (Δδ1H 0.02(4); 15N 0.21) with respect to the shifts in the absence of the DNA11 indicative of an electrostatic interaction. The peak for 1 is slightly less deshielded in the case of I, indicating a weaker association with the single strand than with the duplex.
![[1H,15N] HSQC NMR spectra of 1 after 1.45 h reaction with I (a) and III (b) at pH 5.4, 298 K. For assignments see Scheme 1. Peak 6 (δ 4.03/−63.9), seen only in the reaction with I, is assigned tentatively to an aquated monofunctional adduct. Peaks labeled ‘i’ are extraneous in the 15N-1 starting material.11 (The [1H,15N] NMR spectra of the final products of the reactions of 1 with both I and III are provided as ESI.)](/image/article/2003/CC/b209661g/b209661g-f1.gif) | ||
| Fig. 1 [1H,15N] HSQC NMR spectra of 1 after 1.45 h reaction with I (a) and III (b) at pH 5.4, 298 K. For assignments see Scheme 1. Peak 6 (δ 4.03/−63.9), seen only in the reaction with I, is assigned tentatively to an aquated monofunctional adduct. Peaks labeled ‘i’ are extraneous in the 15N-1 starting material.11 (The [1H,15N] NMR spectra of the final products of the reactions of 1 with both I and III are provided as ESI†.) | ||
 | ||
| Scheme 1 Reaction between 1 and I or III. | ||
For the duplex, analysis of the kinetic profiles allowed assignment of peaks (Fig. 1(b)) with time dependent profiles consistent with mono-functional adducts (DNA bound end) which can be assigned to the major 3′G (3a, δ 4.17/−59.9) and minor 5′G (4a, δ 4.30/−61.0) adducts.10
The final products of reaction of 1 with single-stranded DNA are multiple and a variety of cross-linked species are likely. Nevertheless, there are also two peaks assignable to monofunctional adducts. The minor peak (δ 4.31/−61.5, 4a) has similar 1H/15N shift to that of the 5′G monofunctional adduct of III, whereas the major peak (3a, δ 4.26/−60.7) has a very different shift to that of the 3′G adduct. The partner peaks (3b/4b, unbound end) for both monofunctional adducts of I are coincident (δ 3.92/−64.3) and are also slightly less strongly deshielded than the equivalent peaks in the reaction between 1 and III (δ 3.95/−64.4).
Identification of monofunctional adducts for both I and III allows comparison of kinetics of the initial covalent attachment. The volumes of the Pt–NH3 peaks in the [1H,15N] HSQC NMR spectra were measured at each time point, normalized and then used in a kinetic analysis of the reactions as reported previously.6 In the single-strand reaction the peak overlap is severe, but it is possible to obtain a reliable value for the contribution of the monofunctional adducts from the peak (3b/4b) for the unbound end, which is seen free from overlap throughout the time course. For the purposes of the kinetic fits, the concentrations of the combined monofunctional and final (bifunctional) adducts were calculated. A general reaction scheme is shown in Scheme 1 and the rate constants obtained by fitting the data are shown in Table 1. The time dependence plots for the two reactions are shown in Fig. 2.
| 1 a | Cisplatinb | |||
|---|---|---|---|---|
| Parameter | I | III | I | III |
| a pH 5.4 b pH 6, data from ref. 8 c The rate constants for monofunctional binding to the 3′ G and 5′ G are 1.5 ± 0.7 and 0.24 ± 0.11 M−1 s−1, respectively.10 d Binding to 3′G.8,9 e Binding to 5′ G.8,9 | ||||
| k H /10−5 s−1 | 5.64 ± 0.14 | 4.00 ± 0.03 | 1.73 ± 0.04 | 1.83 ± 0.03 |
| k MF/M−1 s−1 | 0.73 ± 0.25 | 1.49 ± 0.48c | 0.28 ± 0.05d | 0.47 ± 0.08d |
| 0.05 ± 0.025e | 0.14 ± 0.03e | |||
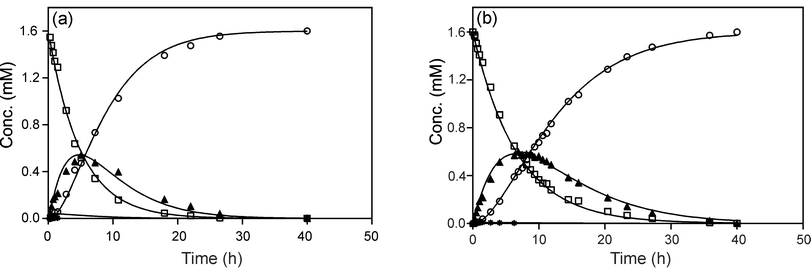 | ||
| Fig. 2 Plots of relative concentrations of species observed during reactions of 1 with I (a) and III (b) at 298 K, pH 5.4. Key: □ 1, * 2, ◆ 3/4, ○ 5. | ||
The rate constant for monofunctional binding of 1 to the DNA is in fact higher with duplex III than it is with I, explained by the enhanced nucleophilicity of –GG– base pairs in a double helix. However, the overall reaction is completed more rapidly in the case of the single strand I (∼30 h) than the duplex III (∼40 h). This observation is explained by the fact that the value of the pseudo first-order rate constant for the aquation of 1 (kH) is 1.4 times higher in the presence of I than it is with III and kHis rate-limiting. Thus, the retardation of aquation rate results in the observed preference for single-stranded binding. Little discrimination is observed for cisplatin8 with the same sequence.
These results show for the first time that the nature of DNA conformation may affect chemical properties such as aquation kinetics based on template modulation of the substrate. Interestingly, the rate of chloride aquation is significantly faster in poly(di/tri)nuclear complexes than for mononuclear cisplatin-based agents, but the equilibrium lies to the chloro form.11,12 At physiological conditions of chloride concentration and pH, significantly more polynuclear compound is in an ‘intact’ form. Single stranded DNA is present during transcription, replication, recombination and repair and is recognized by various single stranded DNA binding proteins. Further interesting biological applications resulting from attack on single stranded DNA include the modulation of antisense gene therapy whereby enhanced neutralization and stabilization of antisense oligonucleotides could positively affect cellular uptake and nuclease digestion.13
This work was supported by the Australian Research Council, US NIH (R01-CA78754), and CHE-9615727) and the American Cancer Society (RPG89-002-11-CDD.
Notes and references
- (a) M. Cusumano, M. L. Di Pietro, A. Giannetto, M. A. Messina and F. Romano, J. Am. Chem. Soc., 2001, 123, 1914 CrossRef CAS; (b) J. Kjellström and S. K. C. Elmroth, Inorg. Chem., 1999, 38, 6193 CrossRef.
- Y. Wang, N. Farrell and J. D. Burgess, J. Am. Chem. Soc., 2001, 123, 5576 CrossRef CAS.
- N. Carte, F. Legendre, E. Leize, N. Potier, F. Reeder, J.-C. Chottard and A. V. Dorsselaer, Anal. Biochem., 2000, 284, 77 CrossRef CAS.
- (a) D. Hamelberg, L. D. Williams and W. D. Wilson, J. Am. Chem. Soc., 2001, 123, 7745 CrossRef CAS; (b) L. D. Williams and L. J. Maher III, Ann. Rev. Biophy. Biomol. Struct., 2000, 29, 497 Search PubMed.
- (a) G. Pratesi, P. Perego, D. Polizzi, S. C. Righetti, R. Supino, C. Caserini, C. Manzotti, F. C. Giuliani, G. Pezzoni, S. Tognella, S. Spinelli, N. Farrell and F. Zunino, Br. J. Cancer, 1999, 80, 1912 CrossRef CAS; (b) P. Perego, C. Caserini, L. Gatti, N. Carenini, S. Romanelli, R. Supino, D. Colangelo, I. Viano, R. Leone, S. Spinelli, G. Pezzoni, C. Manzotti, N. Farrell and F. Zunino, Mol. Pharmacol., 1999, 55, 528 Search PubMed; (c) P. M. Calvert, M. S. Highley, A. N. Hughes, E. R. Plummer, A. S. T. Azzabi, M. W. Verrill, M. G. Camboni, E. Verdi, A. Bernareggi, M. Zucchetti, A. M. Robinson, J. Carmichael and A. H. Calvert, Clin. Cancer Res., 1999, 5, 3796s Search PubMed; (d) C. Sessa, G. Capri, L. Gianni, F. Peccatori, G. Grasselli, J. Bauer, M. Zucchetti, L. Vigano, A. Gatti, C. Minoia, P. Liati, S. Van den Bosch, A. Bernareggi, G. Camboni and S. Marsoni, Ann Oncol., 2000, 11, 977 CrossRef CAS; (e) N. Farrell, Y. Qu, U. Bierbach, M. Valsecchi and E. Menta, in Cisplatin: Chemistry and Biochemistry of a Leading Anticancer Drug, ed. B. Lippert, Wiley-VCH, Zurich, 1999pp. 479–496 Search PubMed.
- J. W. Cox, S. J. Berners-Price, M. S. Davies, N. Farrell and Y. Qu, J. Am. Chem. Soc., 2001, 123, 1316 CrossRef CAS.
- M. B. G. Kloster, J. C. Hannis, D. C. Muddiman and N. Farrell, Biochemistry, 1999, 38, 14731 CrossRef CAS.
- S. J. Berners-Price, K. J. Barnham, U. Frey and P. J. Sadler, Chem.–Eur. J., 1996, 2, 1283 Search PubMed.
- F. Reeder, Z. Guo, P. d. S. Murdoch, A. Corazza, T. W. Hambley, S. J. Berners-Price, J. C. Chottard and P. J. Sadler, Eur. J. Biochem., 1997, 249, 370 CrossRef CAS.
- S. J. Berners-Price, M. S. Davies, J. W. Cox, D. S. Thomas and N. Farrell, Chem.—Eur. J., 2002, in press Search PubMed.
- M. S. Davies, J. W. Cox, S. J. Berners-Price, W. Barklage, Y. Qu and N. Farrell, Inorg. Chem., 2000, 39, 1710 CrossRef CAS.
- M. S. Davies, D. S. Thomas, A. Hegmans, S. J. Berners-Price and N. Farrell, Inorg. Chem., 2002, 41, 1101 CrossRef CAS.
- N. Farrell and M. B. G. Kloster, US Pat., 6,310,047, 2002.
Footnotes |
| † Electronic supplementary information (ESI) available: experimental conditions for the NMR reactions, the models used for the kinetic fits and [1H,15N] HSQC NMR spectra of the final products from reactions of 1 with the single strand (I) (before and after addition of the complementary strand (II)), and with the duplex (III). See http://www.rsc.org/suppdata/cc/b2/b209661g/ |
| ‡ A pH of 5.4 was chosen for the reaction of 1 with I and III, as it lies 0.2 pH units below the pKa value of the monoaquachloro form 2 (5.62),11 allowing a more meaningful comparison with the previous DNA platination reactions of I and III by cisplatin at pH 6 (pKa of cis-[PtCl(NH3)2(H2O)]+ is 6.41: S. J. Berners-Price, T. A. Frenkiel, U. Frey, J. D. Ranford, and P. J. Sadler, Chem. Commun. 1992, 789). |
| This journal is © The Royal Society of Chemistry 2003 |
