Development of nanoparticle probes for multiplex SERS imaging of cell surface proteins†
David C.
Kennedy
a,
Kelly A.
Hoop
b,
Li-Lin
Tay
c and
John Paul
Pezacki
*ab
aSteacie Institute for Molecular Sciences, National Research Council Canada, 100 Sussex Drive, Ottawa, K1A 0R6, Canada. E-mail: John.Pezacki@nrc-cnrc.gc.ca
bDepartment of Chemistry, University of Ottawa, 10 Marie Curie, Ottawa, K1N 6N5, Canada
cInstitute for Microstructural Sciences, National Research Council Canada, 1200 Montreal Road, M-50, Ottawa, K1A 0R6, Canada
First published on 18th May 2010
Abstract
Multiplexed SERS imaging of receptor proteins on the surface of mammalian cells has been carried out using functionalized silver nanoparticles. Deconvolution of four differently functionalized nanoparticles is readily achieved, and using this approach, receptor co-localization can be probed and protein–protein interactions can be elucidated at the surface of cells.
There are already numerous reports on detecting cancer cells using anti-EGFR targeted nanomaterials in a single detection approach.1–4 There are, however, few reports on the development of probes that can carry out in vitro and in vivo imaging of cellular targets in a multiplexed approach, despite considerable interest.5–7 To achieve this, surface-enhanced Raman scattering (SERS) nanotags have been developed and shown to be viable materials for making highly sensitive non-invasive SERS measurements in vivo.6 Nanomaterials designed for cellular imaging must meet a number of specific criteria.8 The materials must be water-soluble and stable, they must also be highly specific, biocompatible and stable towards substitution. For in vivo imaging, additional constraints apply as the materials must not only be highly target specific, but also non-toxic towards the host. Also, they must not alter the biochemistry of host cells as non-toxic metals are known to do.9
The ability to detect multiple markers simultaneously is heralded as an important achievement for making early diagnoses for many diseases. In order to expand the utility of such materials to probe not just the presence of cellular targets, but to understand their molecular biology on the surface of cells, the size of the nanoparticles (NPs) utilized must be reduced in order to achieve meaningful spatial resolution. The size of the particles should thus be comparable to the proteins and substrates they target on cell surfaces—typically less than 20 nm in diameter. Many of the core–shell type of SERS probes reported in the literatures are typically bigger than this ideal dimension which limits their applications in cellular imaging applications.10–14 Here we report on the design of SERS probes with colloidal Ag NPs in the 25 nm size range. Although SERS enhancement diminishes as particle size decreases, aggregation of NPs generates highly enhanced SERS active “hot-sites”.15,16 Additionally, while SERS provides a large enhancement to the Raman emission of the reporter molecule, efforts to develop reporter molecules that exhibit unique spectral signatures from each other can lead to the design of more efficient multiplexed probes for determination of the localization of the probe in or on cells. Multiplexed SERS imaging is advantageous over fluorescence imaging because it does not suffer from photo-bleaching and the broad emission profiles of many fluorescent labels limit their multiplexing capability. Furthermore, gold and silver NPs tend to be less toxic than fluorescent quantum dots making SERS probes ideal for multiplexed imaging. Here we describe the development of several new SERS nanoprobes that are compatible with the goal of monitoring protein–protein interactions on cell surfaces in a multiplexed fashion.
We have previously shown that mercaptomethylbenzyl nitrile functionalized NPs are an excellent marker for cell surface imaging.15 A mixed ligand coating that contains the reporter molecule for imaging, a short-chain polyethylene glycol molecule to enhance water solubility and a short-chain succinimide ester that is used to bind targeting antibodies to the NP has been used to effectively report on the aggregation state of β2-adrenergic receptors on cell surfaces.15 Additionally, we have adapted this same NP design to incorporate carborane molecules as potential therapeutics, transforming the NP contrast agent into a drug delivery vehicle as well.4 The carborane and nitrile reporter molecules each contain unique Raman stretches that arise in the spectroscopically silent region of the cell (1700–2800 cm−1). These nanoprobes have enormous potential as multimodal imaging agents and therapeutics. In addition to SERS imaging, NPs can be exploited for their luminescent properties and light scattering ability as well as in thermal ablation therapy.
In setting out to design new reporter molecules, several factors were considered. Among the many factors that effect the observed Raman enhancement, proximity of the marker in question to the particle surface is one of them.15 We chose a simple scaffold where functionalized mercaptomethylbenzene derivatives were used as chemically distinct Raman reporters. We have previously published the synthesis of (mercaptomethyl)benzonitrile and on its use as a Raman reporter on Ag NPs to probe cell surface receptors.15 Here we prepared three analogous compounds with unique Raman sensitive functional groups. The NPs require additional ligands in order to target and stabilize them in aqueous media; however, the reporter molecules synthesized from benzene derivatives (which have higher scattering cross-sections) generate strong SERS spectra. The four distinct molecules that were synthesized and probed were: 4-(mercaptomethyl)benzonitrile (MMBN), d7-mercaptomethyl benzene (DMMB), 4-(mercaptomethyl)ethynylbenzene (MMByne) and 4-(mercaptomethyl)nitrobenzene (MMBNO) (Fig. 1).
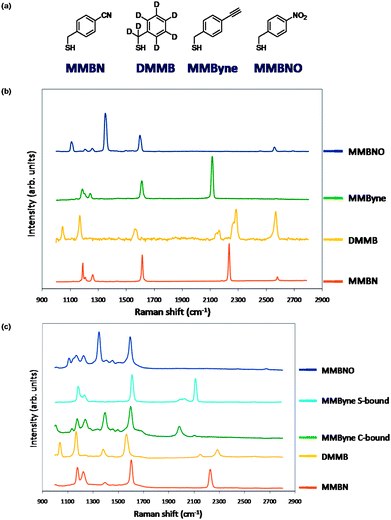 | ||
| Fig. 1 (a) Structures of 4-(mercaptomethyl)benzene-derived reporter molecules. (b) Raman spectra of the 4-(mercaptomethyl)benzene-derived reporter molecules. (c) SERS spectra of functionalized Ag NPs derived using the 4-(mercaptomethyl)benzene-derived reporter molecules. | ||
The spectra of the 4 ligands are shown in Fig. 1. The spectra for these molecules in the region from 1700–2800 cm−1 show chemically distinct reporter signals for MMBN, DMMB and MMByne. The peak at ∼2600 cm−1 is assigned to the S–H stretch. MMBNO lacks signals in this region; however, the signals that arise from the nitro group (1350 and 1600 cm−1) are very strong and the peak at 1350 cm−1 is unique to this molecule while the peak at 1600 cm−1 overlaps with the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C ring mode observed in all of the reporter molecules. Nitro compounds are often employed as SERS contrast agents, and in the absence of interference in the fingerprint region, NPs containing the nitro ligand could be used in multiplexed imaging experiments. All four ligands were then used to functionalize the surface of Ag NPs, and the SERS spectra of the resulting NPs are shown in Fig. 1. The proximity of the functional groups to the NP surface likely plays an important role in the strength of the observed signals. Of note, the spectrum for the MMByne functionalized NP appeared to initially be dominated by alkyne coordination rather than thiol coordination to the NP. The shift in the C
C ring mode observed in all of the reporter molecules. Nitro compounds are often employed as SERS contrast agents, and in the absence of interference in the fingerprint region, NPs containing the nitro ligand could be used in multiplexed imaging experiments. All four ligands were then used to functionalize the surface of Ag NPs, and the SERS spectra of the resulting NPs are shown in Fig. 1. The proximity of the functional groups to the NP surface likely plays an important role in the strength of the observed signals. Of note, the spectrum for the MMByne functionalized NP appeared to initially be dominated by alkyne coordination rather than thiol coordination to the NP. The shift in the C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C stretching mode to lower wavenumber is consistent with terminal alkyne coordination. Terminal alkynes can coordinate silver, and the resulting metal coordinated alkyne lowers the energy of the C
C stretching mode to lower wavenumber is consistent with terminal alkyne coordination. Terminal alkynes can coordinate silver, and the resulting metal coordinated alkyne lowers the energy of the C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C bond and thus the observed signal by about 100–200 cm−1.17 In order to prove this, we synthesized NPs, lacking the thiol, using phenylacetylene (see ESI†). The spectrum of these NPs was consistent with those observed for MMByne, indicating that the alkyne was binding. In order to synthesize thiol functionalized NPs, we repeated the NP functionalization process under reducing conditions and at elevated pH. It was found that raising the pH did not affect the coordination of the alkyne; however, adding NaBH4 to the buffer resulted in a species that was predominantly thiolate coordinated to the NP, and the peak at 2125 cm−1 is the dominant peak in the region of interest in the SERS spectrum of the resulting species (see ESI†). The MMByne particle may also be a useful construct for carrying out bioorthogonal chemical labelling. Recently there have been several reports of using azide functionalized NPs to carry out click chemistry.18–20 Here we have an alkyne functionalized NP that should be able to carry out the same chemistry but allowing the azide tag to be part of the biomolecule of interest. This is important as it has been shown that azides can be readily incorporated into sugars and expressed in cellular systems.27 This alkyne probe is a potential tool for imaging such SERS probes that can be combined in a multiplexed manner to observe proteins associated with the azide functionalized tag. This is also a chemically sensitive method for performing click chemistry. Click chemistry on this alkyne will result in loss of the alkyne signal in the SERS spectrum. These probes have the potential to not only report on localization of targets, but also could be developed to report on processes resulting in changes in the functional groups near the NP surface.
C bond and thus the observed signal by about 100–200 cm−1.17 In order to prove this, we synthesized NPs, lacking the thiol, using phenylacetylene (see ESI†). The spectrum of these NPs was consistent with those observed for MMByne, indicating that the alkyne was binding. In order to synthesize thiol functionalized NPs, we repeated the NP functionalization process under reducing conditions and at elevated pH. It was found that raising the pH did not affect the coordination of the alkyne; however, adding NaBH4 to the buffer resulted in a species that was predominantly thiolate coordinated to the NP, and the peak at 2125 cm−1 is the dominant peak in the region of interest in the SERS spectrum of the resulting species (see ESI†). The MMByne particle may also be a useful construct for carrying out bioorthogonal chemical labelling. Recently there have been several reports of using azide functionalized NPs to carry out click chemistry.18–20 Here we have an alkyne functionalized NP that should be able to carry out the same chemistry but allowing the azide tag to be part of the biomolecule of interest. This is important as it has been shown that azides can be readily incorporated into sugars and expressed in cellular systems.27 This alkyne probe is a potential tool for imaging such SERS probes that can be combined in a multiplexed manner to observe proteins associated with the azide functionalized tag. This is also a chemically sensitive method for performing click chemistry. Click chemistry on this alkyne will result in loss of the alkyne signal in the SERS spectrum. These probes have the potential to not only report on localization of targets, but also could be developed to report on processes resulting in changes in the functional groups near the NP surface.
We attempted to simplify our previously designed system by incorporating our labels into amino acids that could be conjugated to dithiobis(succinimidyl propionate) (DTSP). Amino acid functionalized NPs tend to be water-soluble and stable,21–23 and short chain peptides can be used to target a wide range of biological markers.24 A single molecule monolayer approach has been reported recently25 and the use of DTSP functionalized NPs as biosensors has also been explored.26 This would allow a single molecule to be used to functionalize the NPs as opposed to three, and put more of the desired marker onto the NP surface further enhancing the observed signals. We selected, when available, phenylalanine derivatives for conjugation to form the DTSP-reporters because the resulting SERS signals were stronger than those of other amino acid derivatives. We again selected reporters that would give rise to signals in the spectroscopically silent region of the cell (1700–2800 cm−1) in order to ensure that detected signals could be unambiguously attributed to the functionalized NPs especially under conditions where confocal imaging is not possible. We selected the same reporter groups as in the previous system with the following Raman reporter functional groups: C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N, NO2, C
N, NO2, C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C, and C–D.
C, and C–D.
The reduction of the S–S bond occurs spontaneously during nanoparticle functionalization and the succinimide esters survive the labelling process and can then be readily reacted with free amines for antibody functionalization. Here we selected four substrates: 4-cyano-phenylalanine, d5-phenylalanine, d2-glycine, and propargyl amine to conjugate to DTSP (see ESI† for details and spectra). DTSP was also conjugated to 4-nitro-phenylalanine; however, the resulting Raman spectrum of this sample showed evidence of photo-decomposition thus this molecule was not pursued as a potential marker. Difficulties with using alkyne-functionalized amino acids to derive DTSP-labelled NPs led us to use propargyl amine as a simpler approach to test the utility of this substrate for potential SERS activity. Both alkyl and aryl C–D bonds were explored using deuterated-glycine and -phenylalanine to probe the effect the aryl ring has on the observed SERS signal.
Of note, for the d2-glycine sample two sharp vibrational bands (2175 and 2257 cm−1) and a third broad and weak band (2100 cm−1) are observed for C–D stretch, while for the d5-phenylalanine sample only one broad resonance is observed for C–D (2298 cm−1). The 4-cyano-phenylalanine sample gives a sharp signal at 2236 cm−1 while the propargyl amine sample gives a sharp peak at 2125 cm−1. Next Ag NPs were functionalized with these ligands in order to determine if the resulting SERS spectra could be useful for the development of multiplexed biological SERS imaging. The spectra (see ESI†) of the 4-cyano-phenylalanine and d5-phenylalanine samples are generally the same in the region of interest between 1700 and 2800 cm−1 compared to the free ligands; however, the characteristic C–D peaks for the d2-glycine sample are broadened and their intensity ratio altered.
Additionally, the peak for the propargyl amine sample that was at 2125 cm−1 is now much weaker and a large broad peak centered at ∼2000 cm−1 is now present. This new broad peak is consistent with the formation of a Ag–alkyne bond. These DTSP-derived functionalized NPs may be useful as cellular imaging agents and such materials are already in use as biosensors,26 however, the SERS signals are currently too weak for probing cell surface proteins in a multiplexed approach.
To show that mercaptomethyl-derived NPs could be used to carry out multiple detection in a SERS experiment, we mixed equal amounts of the 4 NPs derived from MMBN, MMBNO, DMMB and MMByne. The resulting mixture was then spotted on a glass slide and probed under a Raman microscope. The SERS spectrum of the mixed sol was obtained from a large aggregate and shown as the black trace in Fig. 2. This spectrum contains all the major bands from the four components. We fitted the mixed spectrum to four of the individual components using the direct classic least square (DCLS) routine in Labspec 5. This deconvolution optimizes the linear combination of spectra from individual components that best represent the acquired mixed spectrum. The multiplication scalar factor is indicative of the percent composition of the four component spectra. From the DCLS deconvolution, the mixed spectrum (black trace) is determined to be composed of 47.3% of MMByne, 11.6% of MMBN, 5.6% of DMMB and 34.7% of MMBNO. Of course, it is expected that these ratio will change from spot to spot as the observed mixed spectrum of each probed area will depend heavily on the nanoscale environment of the probed area (1 µm in diameter). Multivariate deconvolution is a powerful technique but the simplicity and uniqueness in the proposed molecules do allow for very rapid identification of each Raman reporter molecule. For example, the 2230 cm−1 of MMBN, 965 cm−1 band of DMMB, 2020 cm−1 of MMByne and 1350 cm−1 of MMBNO are all unique and non-overlapping which serve as good candidate for univariate identification of specific reporter molecules. This experiment suggests that these functionalized molecules can be used as probes for multiplexed SERS imaging.
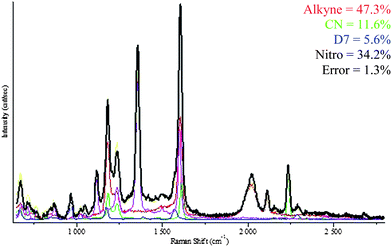 | ||
| Fig. 2 Multi-component SERS spectrum (black) and the deconvolution of this spectrum into its component spectra with percentages assigned to each component (listed in text). | ||
The MMBN probe has already been applied to NP imaging of cell receptors in vitro and so we chose DMMB to carry out a double surface receptor imaging experiment using these two SERS labels to detect β2-adrenergic receptors (β2-AR) and caveolin-3 (cav3) on the surface of rat cardiomyocytes (Fig. 3). We have previously shown that the MMBN functionalized NPs can successfully bind to β2-AR and report on its location and aggregation at the cell surface. Here we add to the mix DMMB functionalized NPs that target cav3 to address whether or not these two proteins co-localize at the cell surface. The results show that 17% of receptors are colocalized. This agrees generally with what has been previously reported27 and may provide a means by which to study changes in the degree of colocalization in response to receptor ligand binding and thus add to the discussion on the biological significance of these proteins in cell signalling.
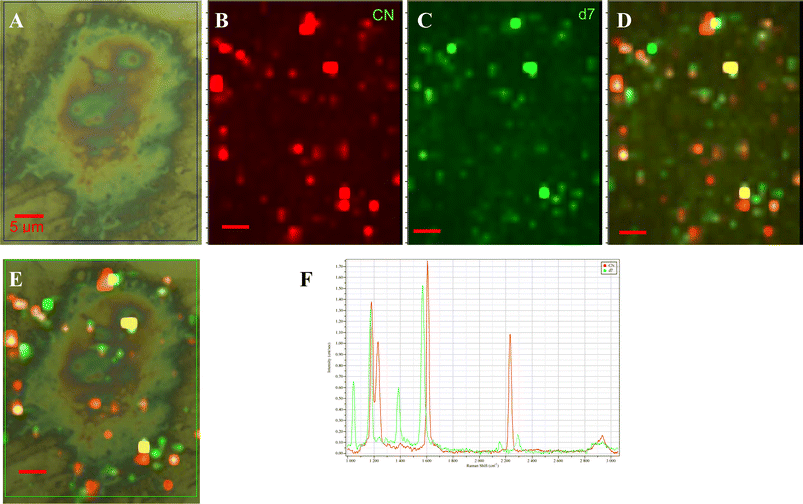 | ||
| Fig. 3 Multiplexed imaging of β2-adrenergic receptor and caveolin-3 on the surface of a rat cardiomyocyte using MMBN and DMMB functionalized silver NPs. (A) A bright field optical image of a cell. Deconvoluted images of the β2-AR receptor distribution labeled with MMBN-NPs (B) and the cav3 receptor distributions labeled with DMMB-NPs (C). (D) An overlay of (B) and (C) which shows all the labeled β2-AR and cav3 receptors. (E) Overlay of the (A) and (D) showing the location of labeled receptors on the cell surface. (F) The model spectra of MMBN and DMMB used in the image deconvolution. All scale bars are 5 µm. | ||
The DTSP-derived SERS probe molecules exhibit unique SERS spectra when bound to NPs, however, the broadened vibrational modes render them unsuitable for multiplexed experiments at this time. The smaller benzylthiol-derived molecules can be applied to imaging cell surface proteins in a multiplexed approach to probe multiple surface receptors simultaneously using SERS microscopy. We have shown that using this approach, four distinct reporters can be deconvoluted, and in cell imaging, the spatial relationship between proteins can be investigated using SERS microscopy to map the localization of the receptors.
References
- N. J. Durr, T. Larson, D. K. Smith, B. A. Korgel, K. Sokolov and A. Ben-Yakar, Nano Lett., 2007, 7, 941–945 CrossRef CAS.
- I. H. El-Sayed, X. Huang and M. A. El-Sayed, Nano Lett., 2005, 5, 829–834 CrossRef CAS.
- I. H. El-Sayed, X. Huang and M. A. El-Sayed, Cancer Lett., 2006, 239, 129–135 CrossRef CAS.
- D. C. Kennedy, D. R. Duguay, L. L. Tay, D. S. Richeson and J. P. Pezacki, Chem. Commun., 2009, 6750–6752 RSC.
- J. H. Kim, J. S. Kim, H. Choi, S. M. Lee, B. H. Jun, K. N. Yu, E. Kuk, Y. K. Kim, D. H. Jeong, M. H. Cho and Y. S. Lee, Anal. Chem., 2006, 78, 6967–6973 CrossRef CAS.
- C. L. Zavaleta, B. R. Smith, I. Walton, W. Doering, G. Davis, B. Shojaei, M. J. Natan and S. S. Gambhir, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 13511–13516 CrossRef CAS.
- G. B. Braun, S. J. Lee, T. Laurence, N. Fera, L. Fabris, G. C. Bazan, M. Moskovits and N. O. Reich, J. Phys. Chem. C, 2009, 113, 13622–13629 CrossRef CAS.
- E. Boisselier and D. Astruc, Chem. Soc. Rev., 2009, 38, 1759–1782 RSC.
- D. C. Kennedy, R. K. Lyn and J. P. Pezacki, J. Am. Chem. Soc., 2009, 131, 2444–2445 CrossRef CAS.
- R. A. Alvarez-Puebla, R. Contreras-Caceres, I. Pastoriza-Santos, J. Perez-Juste and L. M. Liz-Marzan, Angew. Chem., Int. Ed., 2009, 48, 138–143 CrossRef CAS.
- P. J. Huang, L. L. Tay, J. Tanha, S. Ryan and L. K. Chau, Chem.–Eur. J., 2009, 15, 9330–9334 CrossRef CAS.
- S. P. Mulvaney, M. D. Musick, C. D. Keating and M. J. Natan, Langmuir, 2003, 19, 4784–4790 CrossRef.
- S. J. Oldenburg, S. L. Westcott, R. D. Averitt and N. J. Halas, J. Chem. Phys., 1999, 111, 4729–4735 CrossRef CAS.
- B. Kustner, M. Gellner, M. Schutz, F. Schoppler, A. Marx, P. Strobel, P. Adam, C. Schmuck and S. Schlucker, Angew. Chem., Int. Ed., 2009, 48, 1950–1953 CrossRef.
- D. C. Kennedy, L.-L. Tay, R. K. Lyn, Y. Rouleau, J. Hulse and J. P. Pezacki, ACS Nano, 2009, 3, 2329–2339 CrossRef CAS.
- M. Moskovits, Rev. Mod. Phys., 1985, 57, 783–826 CrossRef CAS.
- S. S. Chui, M. F. Ng and C. M. Che, Chem.–Eur. J., 2005, 11, 1739–1749 CrossRef CAS.
- A. Gole and C. J. Murphy, Langmuir, 2008, 24, 266–272 CrossRef CAS.
- E. Boisselier, L. Salmon, J. Ruiz and D. Astruc, Chem. Commun., 2008, 5788–5790 RSC.
- H. Li, Y. Yao, C. Han and J. Zhan, Chem. Commun., 2009, 4812–4814 RSC.
- Q. Hu, L. L. Tay, M. Noestheden and J. P. Pezacki, J. Am. Chem. Soc., 2007, 129, 14–15 CrossRef CAS.
- C. J. Ackerson, P. D. Jadzinsky, G. J. Jensen and R. D. Kornberg, J. Am. Chem. Soc., 2006, 128, 2635–2640 CrossRef CAS.
- M. Zheng and X. Huang, J. Am. Chem. Soc., 2004, 126, 12047–12054 CrossRef CAS.
- L. Sun, D. Liu and Z. Wang, Langmuir, 2008, 24, 10293–10297 CrossRef CAS.
- C. Jehn, B. Kustner, P. Adam, A. Marx, P. Strobel, C. Schmuck and S. Schlucker, Phys. Chem. Chem. Phys., 2009, 11, 7499–7504 RSC.
- J.-H. Park, E. O. Ganbold, D. Uuriintuya, K. Lee and S.-W. Joo, Chem. Commun., 2009, 7354–7356 RSC.
- A. Ianoul, D. D. Grant, Y. Rouleau, M. Bani-Yaghoub, L. J. Johnston and J. P. Pezacki, Nat. Chem. Biol., 2005, 1, 196–202 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Detailed experimental procedures and characterization data are outlined. See DOI: 10.1039/c0nr00122h |
| This journal is © The Royal Society of Chemistry 2010 |
