Synthesis and hybridization properties of 2′-O-methylated oligoribonucleotides incorporating 2′-O-naphthyluridines†
Mitsuo
Sekine
*,
Yusuke
Oeda
,
Yoshihiro
Iijima
,
Haruhiko
Taguchi
,
Akihiro
Ohkubo
and
Kohji
Seio
Department of Life Science, Tokyo Institute of Technology, 4259, Nagatsuta, Midoriku, Yokohama 226-8501, Japan. E-mail: msekine@bio.titech.ac.jp; Fax: +81 45 924 5772; Tel: +81 45 924 5706
First published on 29th October 2010
Abstract
2′-O-(1-Naphthyl)uridine and 2′-O-(2-naphthyl)uridine were synthesized by a microwave-mediated reaction of 2,2′-anhydrouridine with naphthols. Using the 3′-phosphoramidite building blocks, these 2′-O-aryluridine derivatives were incorporated into 2′-O-methylated oligoribonucleotides. Incorporation of five 2′-O-(2-naphthyl)uridines into a 2′-O-methylated RNA sense strand significantly increased the thermostability of the duplex with a 2′-O-methylated RNA antisense strand. Circular dichroism spectroscopy and molecular dynamic simulation of the duplexes formed between the modified RNAs and 2′-O-methyl RNAs suggested that there are π–π interactions between two neighboring naphthyl groups in a sequence of the five consecutively modified nucleosides.
Introduction
2′-O-Modified ribonucleosides have been previously used as monomer components of antisense molecules for thermal stabilization of duplexes formed with complementary RNAs in antisense/antigene and RNAi strategies.1–3 Among them, the 2′-O-alkylated species, exemplified by 2′-O-methoxyethyl ribonucleosides, is one of the most frequently used.4 In contrast, only a few 2′-O-aryl substituted ribonucleoside derivatives have been used as monomer components for these approaches.5,6 It was reported that oligodeoxynucleotide derivatives incorporating the simplest 2′-O-phenyluridine lost a significant amount of the hybridization affinity for the complementary DNA strands.5 On the other hand, Wang and coworkers have extensively reported hybridization and the biological properties of oligoribonucleotides incorporating 2′-O-(2,4-dinitrophenyl)ribonucleosides.6a These 2′-O-(2,4-dinitrophenyl) (DNP)-modified RNA derivatives were obtained by reacting RNA oligomers with 1-fluoro-2,4-dinitrobenzene (F-DNP).6b They reported that DNP-RNA/RNA duplexes showed higher thermostability than unmodified RNA/RNA duplexes,6c and DNP-RNA acquired remarkable resistance to RNases and phosphodiesterases6e as well as an excellent membrane permeability even in the absence of transfection agents.6e–f They also showed strong antisense effects on the expression of mRNA of RIa/PKA in cancer cells6f–h and life-prolongation effects of SCID mice bearing human breast cancer6f or mice infected with Moloney murine leukemia virus.6i Since the synthesis of DNP-RNA was based on the controlled reaction of RNA with F-DNP to attain 70% modification, the real modified structure effective for this observation was unclear. Therefore, it is desirable to develop a general method for the synthesis of 2′-O-arylated RNA oligomers to clarify systematically the effect of aryl groups on the hybridization ability and enzyme resistance of such materials (Fig. 1). With this background, we studied the synthesis of oligoribonucleotides modified with three kinds of 2′-O-aryluridine derivatives.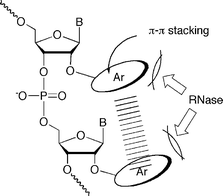 | ||
| Fig. 1 Illustration of 2′-O-arylated RNA oligomers capable of intramolecular stacking interaction. | ||
In this paper, we report the incorporation of these 2′-O-modified ribonucleosides into 2′-O-methylated oligoribonucleotides as well as their unique hybridization and enzymatic properties. In addition, the detailed structural analysis of these modified oligoribonucleotides is also reported based on circular dichroism (CD) spectroscopy and molecular dynamic (MD) simulations of model compounds.
Results and discussion
Synthesis of 2′-O-naphthyluridine 3′-phosphoramidite building blocks
To expand the use of aromatic substituents as 2′-O-modifiers, we focused on the use of 1- and 2-naphthyl groups as the 2′-O-substituents. It seems that these groups interact with themselves more effectively when 2′-O-naphthylribonucleosides are arranged in a consecutive base sequence. It is of great importance to examine whether the resulting stacking interaction enhances the hybridization affinity for complementary RNA strands as well as whether increased bulkiness of the aromatic ring increases the resistance to enzymatic digestion. Therefore, we synthesized the 2′-O-phenyluridine, 2′-O-(1-naphthyl)uridine and 2′-O-(2-naphthyl)uridine 3′-phosphoramidite building blocks 4a–c, as shown in Scheme 1.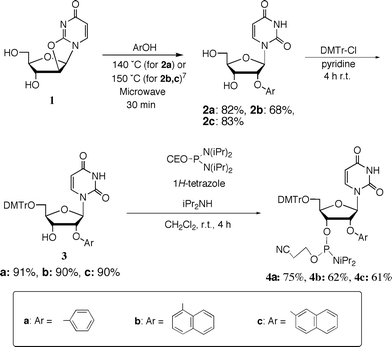 | ||
| Scheme 1 Synthesis of 2′-O-aryluridine 3′-phosphoramidite derivatives 4a–c. | ||
Quite recently, we reported a convenient method for the synthesis of 2′-O-aryluridines from 2,2′-anhydrouridine by microwave-mediated reactions.7 2′-O-Phenyluridine (2a: Ph), 2′-O-(1-naphthyl)uridine (2b: 1Np) and 2′-O-(2-naphthyl)uridine (2c: 2Np) were synthesized by this method. The usual tritylation of 2a–c gave 5′-O-dimethoxytritylated products (3a–c). The phosphoramidite derivatives 4a–c could be synthesized easily by the usual 3′-phosphitylation. Purification using silica gel column chromatography was very easy as the lipophilicity of these products is very high.
Synthesis of oligoribonucleotides incorporating 2′-O-naphthyluridine derivatives
By the general procedure of the phosphoramidite approach,8 we synthesized 2′-O-methylated oligoribonucleotides (RNAs5a–c, RNAs6a–c and RNAs7a–c) incorporating these modified nucleoside derivatives (Fig. 2 and Table 1) and then purified them by HPLC. In the nucleotide sequence, 2′-O-methylribonucleosides were used, except for the modified ribonucleosides.| RNA | sequence | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a Ph: 2′-O-phenyluridine, 1Np: 2′-O-(1-naphthyl)uridine, 2Np: 2′-O-(2-naphthyl)uridine, underlined letter: 2′-O-methylribonucleoside | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sense Strand | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| RNA 8 | 3′-(CUGAAAAACUGA)-5′ | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| RNA 9 | 3′-(![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) )-5′ )-5′ |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Antisense Strand | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| RNA 10 | 5′-(![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) )-3′ )-3′ |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| RNA 5a | 5′-(![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) Ph Ph ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) )-3′ )-3′ |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| RNA 5b | 5′-(![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) PhPhPh PhPhPh ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) )-3′ )-3′ |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| RNA 5c | 5′-(![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) PhPhPhPhPh PhPhPhPhPh ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) )-3′ )-3′ |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| RNA 6a | 5′-(![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) 1Np 1Np ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) )-3′ )-3′ |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| RNA 6b | 5′-(![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) 1Np1Np1Np 1Np1Np1Np ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) )-3′ )-3′ |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| RNA 6c | 5′-(![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) 1Np1Np1Np1Np1Np 1Np1Np1Np1Np1Np ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) )-3′ )-3′ |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| RNA 7a | 5′-(![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) U 2Np U 2Np ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) )-3′ )-3′ |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| RNA 7b | 5′-(![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) U 2Np2Np2Np U 2Np2Np2Np ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) )-3′ )-3′ |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| RNA 7c | 5′-(![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) 2Np2Np2Np2Np2Np 2Np2Np2Np2Np2Np ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) )-3′ )-3′ |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
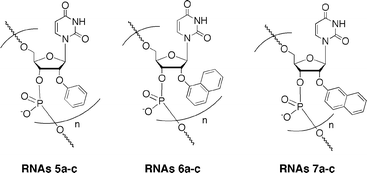 | ||
| Fig. 2 2′-O-Methylated RNA 12-mers (RNAs5a–c, RNAs6a–c, and RNAs7a–c) incorporating 2′-O-aryluridines; a: n = 1, b: n = 3, c: n = 5 (for the full structures see Table 1). | ||
Hybridization ability of 2′-O-methyl-RNA 12mers incorporating 2′-O-aryluridines
As mRNAs are targets in the antisense or RNAi strategies,1 we examined the hybridization ability of 2′-O-methylated oligoribonucleotides (RNAsXa–c: X = 5–7) incorporating 2′-O-aryluridines (2a–c) toward the complementary RNA 12mer [3′-(CUGAAAAACUGA)-5′] (RNA8). When RNAs5a, 5b and 5c were hybridized with RNA8, the resulting duplexes showed markedly lower Tm values with ΔTm per one modification of −2.3, −3.4 and −3.7 °C, respectively, than the unmodified RNA 12-mer (RNA10), as shown in Fig. 3A. A similar tendency was obtained in the case of the 2′-O-(1-naphthyl) modified RNAs6a–c that showed ΔTm values per one modification of −1.9, −4.4 and −4.4 °C for RNAs6a, 6b and 6c, respectively (Fig. 3B).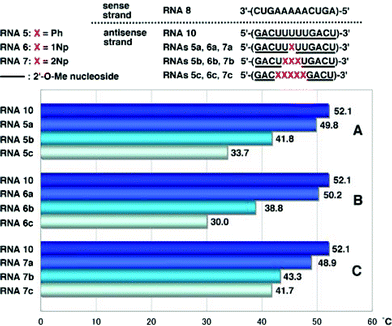 | ||
| Fig. 3 T m values of duplexes of RNA8 with RNA10, RNAs5a–c, RNAs6a–c and RNAs7a–c. | ||
Interestingly, it was found that the hybridization affinity of 2′-O-(2-naphthyl) modified RNAs7a–c for the complementary RNA8 did not decrease as significantly as it did for RNAs6a–c. Since partial incorporation of 2′-O-methylribonucleosides into the sense strands of siRNAs did not affect the RNAi activity, as reported by Tuschl,9 the sense strand was changed from RNA8 to 2′-O-methylated RNA strand (RNA9). It is also known that the 5′-terminal site of antisense strands of siRNAs should have weaker binding ability to the sense sequence.1,2 Therefore, it is of interest to see if oligoribonucleotides incorporating 2′-O-arylribonucleosides can have increased binding affinity for complementary 2′-O-methylated RNA strands. The results obtained by hybridization experiments using modified RNAs5a–c, RNAs6a–c and RNAs7a–c, are shown in Fig. 4.
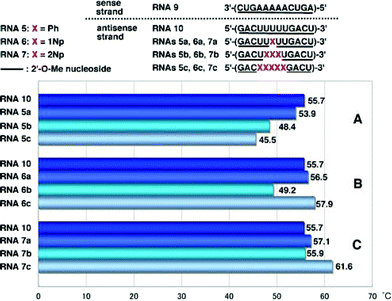 | ||
| Fig. 4 T m values of duplexes of 2′-O-methylated RNA9 with RNA10, RNAs5a–c, RNAs6a–c and RNAs7a–c. | ||
In the case of duplexes of RNAs5a–c, 6a–c and 7a–c with 2′-O-unmodified RNA8, Tm values decreased unexceptionally with an increase in the number of modified nucleosides, as shown in Fig. 3A–C. However, RNAs5–7 showed strange behavior when hybridized with RNA9. Tm values of RNA9/RNAs5a–c duplexes decreased with an increase in the number of Ph, as shown in Fig. 4A, but the degree of the drop of Tm was moderate compared with that of RNA8/RNAs5a–c (see Fig. 3A). RNAs5a, 5b and 5c showed ΔTm values per one modification of −1.8, −2.4 and −2.0 °C, respectively. In a series of RNAs6a–c, unexpected behavior was observed as shown in Fig. 4B: RNA6a incorporating one 1Np showed a slightly increased Tm compared with that of the unmodified RNA10, while incorporation of three 1Nps (when RNA6b was used) resulted in a sharp drop in Tm, but five-point incorporation of 1Nps (when RNA6c was used) resulted in a significant recovery of the Tm value, exceeding the original Tm of the unmodified RNA10, with an increase of 2.2 °C (ΔTm value per one modification = 0.4 °C).
On the other hand, RNA7a with one 2Np gave a slightly increased Tm value (Fig. 4A) and RNA7b with three 2Nps gave almost the same Tm value as that of RNA10 (Fig. 4B). In contrast to these results, RNA7c incorporating five 2NPs showed a more significant increase (ΔTm = 5.9 °C, ΔTm per one modification = 1.2 °C) in the binding affinity for fully 2′-O-methylated RNA9 (Fig. 4C).
Sugar puckering of modified nucleosides
Since sugar puckering is an essential factor in regulation of RNA duplex stability,10 we also measured the ratio of the C3′-endo (N) and C2′-endo (S) conformers in 2′-O-modified nucleosides 2a–c by NMR analysis. N (%) values were calculated by the following equation: N (%) = 100 × J3′,4′/(J1′,2′ + J3′,4′).11 These results are summarized in Table 2.To our surprise, these 2′-O-arylated uridine derivatives exist in an almost 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of the two conformers, showing conformational flexibility. Apparently, modified RNA oligomers containing these 2′-O-aryluridines require conformational change to A-type sugar puckering (C3′-endo) when hybridized with the target complementary RNA oligomer. This conformational change induces energy loss. Nonetheless, it is of great interest that the duplex formed between 2′-O-methylated RNA9 and 2′-O-methylated RNA7c containing five 2′-O-(2-naphthyl)uridines showed remarkably higher thermostability than that derived from RNA9 and RNA10.
1 ratio of the two conformers, showing conformational flexibility. Apparently, modified RNA oligomers containing these 2′-O-aryluridines require conformational change to A-type sugar puckering (C3′-endo) when hybridized with the target complementary RNA oligomer. This conformational change induces energy loss. Nonetheless, it is of great interest that the duplex formed between 2′-O-methylated RNA9 and 2′-O-methylated RNA7c containing five 2′-O-(2-naphthyl)uridines showed remarkably higher thermostability than that derived from RNA9 and RNA10.
CD spectra of single stranded modified RNAs and their double stranded duplexes with RNA 8 and 2′-O-methylated RNA 9
To understand why the 2′-O-methylated RNA9/RNA7c duplex where five 2′-O-(2-naphthyl)uridines were incorporated into the antisense strand showed unexpectedly high thermostability compared with the unmodified RNA/RNA duplexes, we conducted detailed experiments on modified RNA single strands by CD spectroscopy.12As shown in Fig. 5, when one or more 2′-O-phenyluridines were incorporated into the RNA antisense strand, the intensity of the positive and negative Cotton effects changed although the total shape remained almost unchanged (Fig. 5A). Therefore, there is no significant interaction of the phenyl group with itself or with other nucleobases in the single strand. In contrast to this result, RNAs6a–c containing 2′-O-(1-naphthyl)uridines exhibited characteristic strong negative Cotton effects at around 240 nm (Fig. 5B). These effects might be due to some interactions between the neighboring 1-naphthyl groups or between this group and the nearby base residues. In addition to this characteristic peak, RNA6c showed a clear positive Cotton effect at 215 nm. Since 2′-O-(1-naphthyl)uridine (2b) has a strong UV λmax at 215 nm, this eminent peak might be based on the geometrically regulated 1-naphthyl group in the single strand. In the case of RNAs7a–c containing 2′-O-(2-naphthyl)uridines (2Np), broad monotonous negative Cotton effects were observed at around 230–240 nm (Fig. 5C).
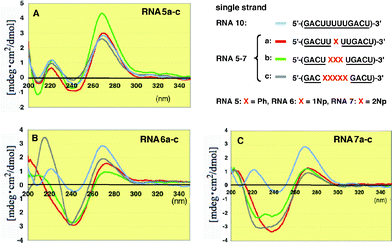 | ||
| Fig. 5 CD spectra of single stranded 2′-O-methylated RNAs5a–c, 6a–c and 7a–c incorporating Ph, 1Np and 2Np, respectively, where 2′-O-methyluridines were replaced by 2′-O-aryluridines (X). | ||
The duplexes of the complementary unmodified RNA8 with 2′-O-phenyl- and 2′-O-(1-naphthyl) modified RNAs5a–c and RNAs6a–c showed roughly similar whole CD patterns, although the peak intensities significantly changed at 270 nm and 220–240 nm, as shown in Fig. 6A and B, respectively.
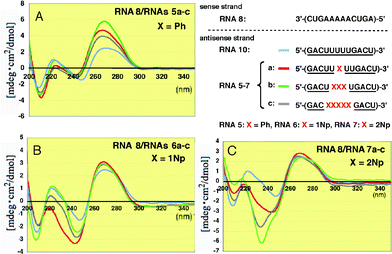 | ||
| Fig. 6 CD spectra of RNA–RNA duplexes formed between unmodified RNA8 and 2′-O-methylated RNAs5a–c, 6a–c and 7a–c incorporating Ph, 1Np and 2Np, respectively, where 2′-O-methyluridines were replaced by 2′-O-aryluridines (X). | ||
In sharp contrast, the CD spectra of the duplexes containing 2′-O-(2-naphthyl) modified RNAs7a–c showed dramatically different CD patterns at around 230 nm (Fig. 6C). In particular, the strong negative Cotton effect at 234 nm observed in RNAs7b and 7c was characteristic. This result suggests there is an interaction between two neighboring 2-naphthyl groups since 2Np has a strong λmax at 225 nm in its UV spectrum (Fig. S1†).
When the modified RNAs5a–c and RNAs6a–c were hybridized with the 2′-O-methylated RNA9, the overall CD patterns were basically similar to those observed in the unmodified RNA8, as shown in Fig. 7, but the peak intensities at 260 nm increased more markedly than those seen in Fig. 6. It seems that this result suggests the conformational change in the base residues becomes somewhat more restricted.
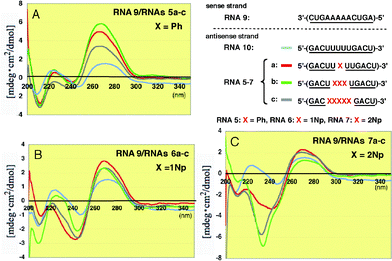 | ||
| Fig. 7 CD spectra of RNA–RNA duplexes formed between 2′-O-methylated RNA9 and 2′-O-methylated RNAs5a–c, 6a–c and 7a–c incorporating Ph, 1Np and 2Np, respectively, where 2′-O-methyluridines were replaced by 2′-O-aryluridines (X). | ||
MD simulation of duplexes incorporating modified nucleosides
To confirm whether the 2′-O-methyl group in the sense strand affects the thermostability of modified RNA–RNA duplexes, MD simulation of RNA duplexes containing modified nucleosides was carried out using the standard AMBER system13,14 (for the details of the procedure see Experimental Section) on a supercomputer (Tsubame, Tokyo Tech.).In the case of 5′-(![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif)
![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) 1Np
1Np![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif)
![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) )-3′/3′-(CUGAAAAACUGA)-5′ where the underline denotes 2′-O-methylribonucleosides, we selected the typical structures during a 5000 ps MD simulation after 50 ps equilibrium. These structures showed that the stabilized forms were of two main structures; 1-naphthyl group extruded to the outer space (Fig. 8B) in the first, and in the second one it covered the minor groove (Fig. 8C).
)-3′/3′-(CUGAAAAACUGA)-5′ where the underline denotes 2′-O-methylribonucleosides, we selected the typical structures during a 5000 ps MD simulation after 50 ps equilibrium. These structures showed that the stabilized forms were of two main structures; 1-naphthyl group extruded to the outer space (Fig. 8B) in the first, and in the second one it covered the minor groove (Fig. 8C).
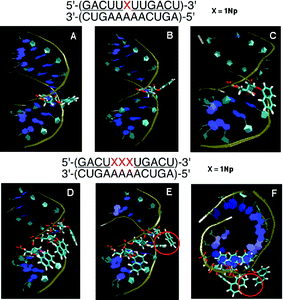 | ||
| Fig. 8 MD simulation of duplexes of an unmodified RNA 12-mer with a 2′-O-methyl-RNA 12-mer incorporating one and three 1Nps. Panel A: Initial structure before MD simulation. Panels B and C: Representative snapshots during MD simulation of the duplex incorporating one 1Np where the 1Np group was seen in two forms: protruded (B) and covered (C). Panel D: Initial structure before MD simulation. Panels E and F: Representative snapshots of MD simulation of the duplex incorporating three 1Nps where 1-naphthyl groups were orientated in different directions without stacking interactions. | ||
When MD simulation of the duplex of 5′-(![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) 1Np1Np 1Np
1Np1Np 1Np![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) )-3′/3′-(CUGAAAAACUGA)-5′ containing three 1Nps was performed, each 1-naphthyl group existed independently in conformations similar to those observed in the above simulation. No π–π stacking between two 1-naphthyl groups could be seen (Fig. 8F) but a CH–π-type interaction-like structure15 was often observed in the snapshots (Fig. 8E).
)-3′/3′-(CUGAAAAACUGA)-5′ containing three 1Nps was performed, each 1-naphthyl group existed independently in conformations similar to those observed in the above simulation. No π–π stacking between two 1-naphthyl groups could be seen (Fig. 8F) but a CH–π-type interaction-like structure15 was often observed in the snapshots (Fig. 8E).
In the duplex of 5′-(![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif)
![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) 2Np
2Np![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif)
![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) )-3′/3′-(CUGAAAAACUGA)-5′ containing a 2Np, two types of structures (Fig. 9B and C) were observed as described in the case of the 1-naphthyl group. When the three modified nucleosides were arranged in the RNA duplex, the apparent π–π interaction between two 2-naphthyl groups in the double strand was observed, as shown by the red circle in Fig. 9E and F, but the three 2-naphthyl groups never stack in a consecutive manner. Only neighboring two of the three 2-naphthyl groups allow π–π stacking interaction. This interaction might increase the duplex stability.
)-3′/3′-(CUGAAAAACUGA)-5′ containing a 2Np, two types of structures (Fig. 9B and C) were observed as described in the case of the 1-naphthyl group. When the three modified nucleosides were arranged in the RNA duplex, the apparent π–π interaction between two 2-naphthyl groups in the double strand was observed, as shown by the red circle in Fig. 9E and F, but the three 2-naphthyl groups never stack in a consecutive manner. Only neighboring two of the three 2-naphthyl groups allow π–π stacking interaction. This interaction might increase the duplex stability.
Next, MD simulation was performed after the unmodified RNA strand was replaced with fully 2′-O-methylated RNA. A similar MD simulation of 5′-(![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) 2Np2Np2Np
2Np2Np2Np ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) )-3′/3′-(
)-3′/3′-(![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) )-5′ suggested that the 2-naphthyl group interacts with the nearby 2′-O-methyl groups of the complementary strand using hydrophobic interaction, as shown in Fig. 10. This effect also contributes to further stabilization of the duplex. In conclusion, π–π interaction and the hydrophobic effect would be essential for stabilization of RNA–RNA duplexes incorporating 2′-O-(2-naphthyl)uridines.
)-5′ suggested that the 2-naphthyl group interacts with the nearby 2′-O-methyl groups of the complementary strand using hydrophobic interaction, as shown in Fig. 10. This effect also contributes to further stabilization of the duplex. In conclusion, π–π interaction and the hydrophobic effect would be essential for stabilization of RNA–RNA duplexes incorporating 2′-O-(2-naphthyl)uridines.
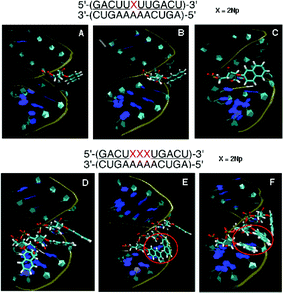 | ||
| Fig. 9 MD simulation of duplexes of an unmodified RNA 12-mer with a 2′-O-methyl-RNA 12-mer incorporating one and three 2′-O-(2-naphthyl)uridines. Panel A: Initial structure before MD simulation. Panels B and C: Representative snapshots during MD simulation of the duplex incorporating one 2Np where 2-naphthyl groups were seen in two forms: protruded (B) and covered (C). Panel D: Initial structure before MD simulation. Panels E and F: Representative snapshots during MD simulation of the duplex incorporating three 2Nps where two of three 2-naphthyl groups were stacked, as shown by the red circle. | ||
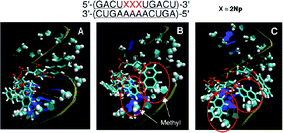 | ||
| Fig. 10 Snapshots of the typical orientation of 2-naphthyl groups in a fully 2′-O-methyl modified RNA duplex containing three consecutive 2Nps after MD simulation. | ||
Nuclease resistance of modified oligomers
We examined the enzymatic property of 2′-O-naphthyl modified oligoribonucleotide derivatives using snake venom phosphodiesterase (SVP) and spleen phosphodiesterase (SPD). As substrates, four dinucleoside monophosphate derivatives 11–14 were synthesized using a liquid-phase synthesis (Fig. 11).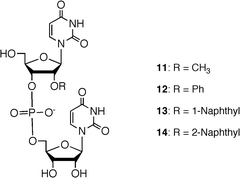 | ||
| Fig. 11 Structures of 2′-O-modified UpU derivatives as substrates for phosphodiesterases. | ||
Results of enzymatic reactions of these substrates are shown in Fig. 12. In the digestion of 11–14 with SVP, surprisingly, 2′-O-phenyl- and 2′-O-(2-naphthyl) modified dimers 12 and 14 were digested most rapidly, as shown in Fig. 12A. The digestion was ca. three time faster than that of the simplest 2′-O-methylated species 11. 2′-O-(1-naphthyl) modified species 13 exhibited a degradation rate similar to that of 11.
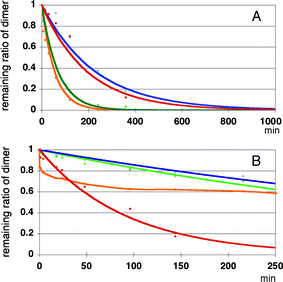 | ||
| Fig. 12 Enzyme resistance of modified UpU toward snake venom phosphodiesterase (Panel A) and calf-spleen phosphodiesterase (Panel B). Red: compound 11, green: compound 12, blue: compound 13 and orange: compound 14. | ||
On the other hand, enzymatic reactions of 11–14 with SPD gave entirely different results, as shown in Fig. 12B. At 140 min, ca. 80% of the 2′-O-methylated UpU 11 was digested while 2′-O-phenyl- and 2′-O-(1-naphthyl) modified UpU derivatives 11 and 13 proved to be the most resistant among all the derivatives to this enzyme showing ca. 20% digestion at 140 min, and ca. 40% of the 2′-O-(2-naphthyl) modified dimer 14 was digested at this moment.
From the above mentioned results, 2′-O-(1-naphthyl) modified UpU proved to be most resistant to the two phosphodiesterases.
Base discrimination ability of 2′-O-naphthylribonucleoside derivatives in RNA oligomers
To examine whether one-point modification of an RNA oligomer, 5′-(GACUUXUUGACU)-3′, affects the base recognition ability of A against G, C and U, we measured Tm values of the duplexes of modified RNA 12-mers (RNAs5a, 6a and 7a) with complementary RNA8 and mismatched RNA 12-mers (RNAs15–17) having G, C, and U, respectively, at the central position. These results are summarized in Fig. 13.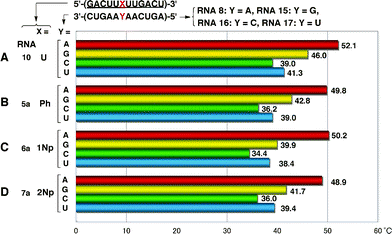 | ||
| Fig. 13 Base discrimination ability of 2′-O-aryl one-point modified RNA 12-mers. | ||
The base-discrimination ability of modified RNAs was evaluated using the difference between the Tm value of the duplex of fully 2′-O-methylated RNA10 with complementary RNA8 and the next largest Tm observed among the three mismatched duplexes. As shown in Fig. 13, the recognition patterns of all four RNAs10 and 5a–7a were basically similar to each other, but 2′-O-aryl modified RNA5a–7a exhibited better base recognition abilities than 2′-O-methyl modified RNA10, which showed the poorest discrimination ability with ΔTm = 6.1 °C. In particular, RNA6a showed the biggest difference (ΔTm = 10.3 °C) in discrimination between RNA8 (Y = A) and RNA15 (Y = G). These results suggested that replacement of ribonucleosides with 2′-O-aryluridines could enhance the base recognition ability of original RNA sequences. It is interesting if the formation of the mismatched base pair G–U could be suppressed by 2′-O-aryl modification. Further studies are needed to generalize this effect.
Conclusions
Aromatic groups have an inherent property capable of a potential π–π stacking interaction with themselves. For this interaction, 1-naphthyl and 2-naphthyl groups are more accessible than a phenyl group because the former have planar structures two times as wide as the latter. The results of MD simulation of modified RNA/RNA duplexes suggested that there might be a π–π interaction in the double-stranded duplex.The significant increase or recovery in Tm value in the modified RNA/2′-O-methylated RNA duplexes should lead us to reconsider the synthetic value of 2′-O-arylated RNAs as alternatives to widespread 2′-O-alkyl substituted siRNA or antisense molecules. Our present research also implies that, since incorporation of 1Np or 2Np into 2′-O-methylated RNA oligomers did not affect its base recognition ability, such a 2′-O-aryluridine block would be used as a new site for modification of oligonucleotides with a wide variety of functional groups that can be attached to the aromatic ring using halogenated or aminated aromatic substituents. We are now studying further applications using these new 2′-O-aryluridine derivatives.
Experimental
General remarks
1H, 13C and 31P NMR spectra were obtained at 500, 126 and 203 MHz, respectively. Chemical shifts were measured from tetramethylsilane (0.0 ppm) or DMSO-d6 (2.49 ppm) for 1H NMR, CDCl3 (77.0 ppm) or DMSO-d6 (39.7 ppm) for 13C NMR and 85% phosphoric acid (0.0 ppm) for 31P NMR. UV spectra were recorded on a U-2000 spectrometer. Column chromatography was performed with silica gel C-200 purchased from Wako Co. Ltd., and a minipump of a goldfish bowl was conveniently used to attain sufficient pressure for rapid chromatographic separation. HPLC was performed using the following systems. Reversed-exchange HPLC was done on a Waters Alliance system with a Waters 3D UV detector and a Waters XTerra MS C18 column (4.6 × 150 mm). A linear gradient (0–10%) of solvent I (0.03 M ammonium acetate buffer (pH 7.0)) in solvent II (CH3CN) was used at 50 °C at a rate of 1.0 mL min−1 for 30 min. Anion-exchange HPLC was done on a Shimadzu SLC-10A connected with LC-10 AD VP, CTO-10A, SPD-M10A, and a Gen-Pak™ FAX column (Waters, 4.6 × 100 mm). A linear gradient (0–30%) of solvent III (1 M NaCl in 25 mM phosphate buffer (pH 6.0) containing 10% CH3CN) in solvent IV (25 mM phosphate buffer (pH 6.0) containing 10% CH3CN) was used at 50 °C at a flow rate of 1.0 mL min−1 for 30 min. For a large-scale synthesis, preparative HPLC was performed with a JAIGEL GS-310 column using LC-9021R with a UV detector S-3110 at the flow rate of 5 ml min−1. ESI mass spectrometry was performed using MarinerTM (PerSeptive Biosystems Inc.). MALDI-TOF mass spectrometry was performed using a Bruker Daltonics [Matrix: 3-hydoroxypicolinic acid (100 mg ml−1) in H2O–diammonium hydrogen citrate (100 mg ml−1) in H2O (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v)]. MD simulation was carried out with Sun Dire X4600 using AMBER ver 7.0.16
1, v/v)]. MD simulation was carried out with Sun Dire X4600 using AMBER ver 7.0.16
T m analysis of duplexes
T m analysis was performed using a UV spectrometer. An appropriate oligomer (1 μM) and its complementary strand (1 μM) were dissolved in 10 mM phosphate buffer (pH 7.0) containing 0.1 M NaCl and 0.1 mM EDTA. The solution was kept at 85 °C or 70 °C for 5 min and the temperature was decreased to 10 °C at a rate of 0.5 °C min−1. After that, UV absorbance was measured at an interval of 1 °C by increasing the temperature to 85 °C or 70 °C. An UV melting curve was obtained by smoothing the original data 7 times using Dtavitzky–Golay method (25 points). Differentiation of this UV melting curve gave the Tm value. concentration of modified oligonucleotides was calculated using the ε values of the unmodified oligonucleotides. The average data of the Tm values thus obtained was calculated and used for discussion.2′-O-Phenyl-5′-O-(4,4′-dimethoxytrityl)uridine (3a)
Compound 2a (100 mg, 0.32 mmol) was coevaporated 3 times with anhydrous pyridine and dissolved in anhydrous pyridine (800 μl, 0.4 M) under argon atmosphere. 4,4′-Dimethoxytrityl chloride (126.9 mg, 0.37 mmol) was added to the above solution, and the mixture was stirred for 4 h. After the reaction, NaHCO3 (aq) was added to the mixture and the solution was extracted with ethyl acetate. The organic layer was washed thrice with brine and dried over anhydrous Na2SO4. After filtration, the filtrate was evaporated in vacuo. The residue was purified by N60 silica gel column chromatography (hexane/CHCl3 50–75%) to give compound 3a (91%).1H NMR (CDCl3, 500 MHz) δ 3.56–3.63 (2H, m), 3.80 (6H, s), 4.24–4.25 (1H, m), 4.68 (1H, q, J = 5.9 Hz), 4.88 (1H, t, J = 4.0 Hz), 5.30 (1H,d, J = 8.1 Hz), 6.19 (1H, d, J = 3.9 Hz), 6.85 (4H, d, J = 8.6 Hz), 7.08 (3H, d, J = 7.6 Hz), 7.26–7.35 (9H, m), 7.40 (2H, d, J = 7.6 Hz), 7.95 (1H, d, J = 8.3 Hz), 8.30 (1H, s); 13C NMR (CDCl3) δ 55.4, 62.3, 69.9, 81.3, 83.7, 87.3, 102.7, 113.5, 116.6, 123.3, 127.4, 128.2, 128.3, 130.1, 130.3, 130.4, 135.1, 135.3, 140.3, 144.4, 150.0, 157.0, 158.9, 159.0, 162.7. ESI-MS m/z calcd for C36H34N2NaO8 [M+Na] 645.2207, found 645.2041.
2′-O-(1-Naphthyl)-5′-O-(4,4′-dimethoxytrityl)uridine (3b)
Compound 3b was synthesized from 2b according to the same procedure as that described for compound 3a.1H NMR (CDCl3, 500 MHz) δ 3.60–3.68 (2H, m), 3.80 (6H, s), 4.39–4.40 (1H, m), 4.80 (1H, t, J = 4.9 Hz), 5.08 (1H, t, J = 4.4 Hz), 5.30 (1H, d, J = 8.1 Hz), 6.37 (1H, d, J = 3.9 Hz), 6.86 (4H, dd, J = 2.7 Hz, 9.2 Hz), 7.12 (1H, d, 7.8 Hz), 7.26–7.33 (7H, m), 7.38–7.42 (3H, m), 7.52–7.58 (3H, m), 7.84–7.86 (1H, m), 7.93–7.95 (1H, d, J = 8.3 Hz), 8.19–8.20 (1H, d, 9.3 Hz); 13C NMR (CDCl3) δ 55.4, 55.5, 62.6, 70.1, 81.5, 84.0, 87.1, 87.5, 102.8, 108.1, 113.6, 121.5, 122.9, 125.7, 125.8, 126.3, 127.0, 127.5, 128.0, 128.3, 130.3, 134.9, 135.1, 135.3, 140.2, 144.3, 150.0, 152.6, 159.0, 162.5. ESI-MS m/z calcd for C40H36N2NaO8 [M+Na] 695.2364, found 695.2382.
2′-O-(2-Naphthyl)-5′-O-(4,4′-dimethoxytrityl)uridine (3c)
Compound 3c was synthesized from 2c according to the same procedure as that described for compound 3a.1H NMR (CDCl3, 500 MHz) δ 3.57–3.66 (2H, m), 3.79 (6H, s), 4.29–4.30 (1H, m), 4.76 (1H, t, J = 5.0 Hz), 5.06 (1H, t, J = 4.2 Hz), 5.33 (1H, d, J = 8.1 Hz), 6.28 (1H, d, J = 3.7 Hz), 6.86 (4H, d, J = 7.8 Hz), 7.24–7.28 (2H, m), 7.31–7.34 (6H, m), 7.38–7.47 (5H, m), 7.66 (1H, d, J = 8.3 Hz), 7.79–7.81 (2H, m), 8.00 (1H, d, J = 8.3 Hz); 13C NMR (CDCl3) δ 55.4, 62.5, 69.9, 81.1, 83.7, 87.2, 87.6, 102.8, 110.1, 113.6, 118.7, 124.9, 127.1, 127.2, 127.5, 127.9, 128.3, 128.4, 130.0, 130.3, 130.5, 134.3, 135.1, 135.3, 140.3, 144.3, 150.0, 154.7, 159.0, 159.0, 162.5. ESI-MS m/z calcd for C40H36N2NaO8 [M+Na] 695.2364, found 695.2579.
2′-O-Phenyl-5′-O-(4,4′-dimethoxytrityl)uridine-3′-(2-cyanoethyl N,N-diisopropyl)phosphoramidite (4a)
Compound 3a (100 mg, 0.16 mmol) was coevaporated thrice with each of anhydrous pyridine and anhydrous toluene, and then dissolved in anhydrous CH2Cl2 (400 μl, 0.4 M) under argon atmosphere. Diisopropylamine (14 μl, 0.10 mmol), 1H-tetrazole (6.7 mg, 0.10 mmol) and 2-cyanoethyl N,N,N′,N′-tetraisopropylphosphorodiamidite (60.3 mg, 0.20 mmol) were added to this solution. After the reaction, the mixture was diluted with CH2Cl2 and quenched by NaHCO3 (aq). The solution was extracted with CH2Cl2 and washed with brine. The organic layer was dried over anhydrous Na2SO4. After filtration, the filtrate was evaporated in vacuo. The residue was purified by RP-HPLC using acetonitrile as an eluent to yield compound 4a (75%).1H NMR (CDCl3, 500 MHz) δ 0.99 (6H, d, J = 6.6 Hz), 1.09 (6H, t, J = 7.6 Hz), 1.27 (2H, J = 6.8 Hz), 2.45 (2H, m), 3.43–3.75 (6H, m), 3.80 (6H, d, J = 4.4 Hz), 4.35 (1H, t, J = 2.7 Hz), 4.74 (1H, m), 5.00 (1H, d, J = 4.2 Hz), 5.28 (1H, t, J = 9.3 Hz), 6.21 (1H, d, J = 3.7 Hz), 6.85 (4H, t, J = 8.1 Hz), 7.03 (3H, m), 7.24–7.33 (9H, m), 7.41 (2H, m), 7.95(1H, t, J = 8.3 Hz);13C NMR (CDCl3, 125 MHz) δ 20.3, 20.4, 24.6, 24.7, 43.2, 43.3, 55.4, 55.4, 55.4, 58.3, 58.5, 61.9, 80.2, 83.0, 87.4, 88.0, 102.6, 113.5, 116.1, 116.3, 117.8, 122.1, 122.2, 127.4, 128.2, 128.4, 128.4, 129.7, 130.4, 130.4, 135.1, 135.2, 140.2, 144.3, 150.2, 157.9, 158.9, 162.9; 31P NMR (CDCl3, 203 MHz) δ 151.9, 151.7. ESI-MS m/z calcd for C45H52N4O9P [M+H] 823.3466, found 823.3455.
2′-O-(1-Naphthyl)-5′-O-(4,4′-dimethoxytrityl)-3′-(2-cyanoethyl N,N-diisopropyl)phosphoramidite (4b)
Compound 4b was synthesized from 3b according to the same procedure as that described for compound 4a.1H NMR (CDCl3, 500 MHz) δ 0.72 (3H, d, J = 6.6 Hz), 0.93 (3H, d, J = 6.6 Hz), 0.99 (3H, d, J = 6.6 Hz), 1.28 (3H, t, J = 6.8 Hz), 2.15 (1H, m), 2.35 (1H, m), 3.38 (2H, m), 3.49–3.62 (3H, m), 3.69 (1H, t, J = 9.2 Hz), 3.80 (6H, d, J = 4.6 Hz), 4.53 (1H, m), 4.90 (1H, m), 5.15 (1H, m), 5.32 (1H, dd, J = 8.2, 16.5 Hz), 6.41 (1H, dd, J = 4.0, 8.2 Hz), 6.86 (4H, t, J = 8.2 Hz), 7.10 (1H, dd, J = 7.6, 12.5 Hz), 7.26–7.49 (14H, m), 7.78 (1H, dd, J = 7.6, 15.6 Hz), 7.99 (1H, dd, J = 8.2, 13.3 Hz), 8.26 (1H, dd, J = 3.8, 7.7 Hz); 13C NMR (CDCl3, 125 MHz) δ 20.0, 20.4, 24.1, 24.6, 43.2, 43.3, 43.4, 55.4, 57.7, 57.8, 58.1, 58.2, 62.2, 70.5, 70.6, 70.8, 70.9, 79.4, 80.0, 83.4, 83.7, 87.4, 87.5, 87.7, 102.8, 106.8, 113.3, 113.5, 117.6, 117.7, 121.5, 121.6, 122.3, 122.5, 125.5, 125.6, 125.8, 126.0, 126.6, 126.7, 127.4, 127.6, 128.2, 128.4, 129.3, 130.3, 134.7, 135.1, 135.3, 140.2, 144.3, 144.4, 150.5, 153.1, 153.2, 158.9, 163.2; 31P NMR (CDCl3, 203 MHz) δ 151.8. ESI-MS m/z calcd for C49H54N4O9P [M+H] 873.3623, found 873.3533.
2′-O-(2-Naphthyl)-5′-O-(4,4′-dimethoxytrityl)uridine-3′-(2-cyanoethyl N,N-diisopropyl)phosphoramidite (4c)
Compound 4c was synthesized from 3c using the same procedure as that described for compound 4a.1H NMR (CDCl3, 500 MHz) δ 0.90–1.19 (12H, m), 1.98 (4H, dd, J = 5.4, 10.5 Hz), 2.18 (1H, d, J = 2.9 Hz), 2.35-2.45 (1H, m), 3.36–3.41 (1H, m), 3.46–3.70 (5H, m), 3.77–3.82 (7H, m), 4.43 (1H, m), 4.86 (1H, m), 5.15 (1H, m), 5.33 (1H, dd, J = 8.1, 12.5 Hz), 6.30 (1H, m), 6.85 (4H, m), 7.21–7.47 (13H, m), 7.61 (1H, dd, J = 3.7, 8.1 Hz), 7.75 (1H, m), 8.02 (1H, d, J = 8.3 Hz);
13C NMR (CDCl3, 125 MHz) δ 20.4, 24.5, 24.7, 25.4, 43.2, 43.3, 43.3, 43.4, 55.4, 55.4, 57.9, 58.4, 62.0, 62.2, 70.5, 70.7, 76.9, 77.2, 77.4, 79.2, 80.0, 83.4, 87.5, 87.9, 102.7, 102.7, 109.3, 113.5, 117.6, 117.8, 119.0, 119.3, 124.3, 126.6, 126.7, 127.1, 127.1, 127.4, 127.7, 128.2, 128.3, 128.4, 129.6, 129.7, 130.4, 134.4, 135.1, 135.3, 140.2, 140.4, 144.3, 144.4, 150.3, 155.4, 155.6, 158.9; 31P NMR (CDCl3, 203 MHz) δ 152.0, 151.8. ESI-MS m/z calcd for C49H53N4NaO9P [M+Na] 895.3442, found 895.3304.
Synthesis of 2′-O-methylated oligoribonucleotides
2′-O-Methylated oligoribonucleotides were synthesized using an Applied Biosystems 392 oligonucleotide synthesizer, starting from CPG-supported 2′-OMe-U derivative (1 μmol). Chain extension was carried out with 2′-O-aryluridine phosphoramidite units (4a–c) or 2′-O-methylribonucleoside phosphoramidite units (PacA, isopropyl-Pac-G, and acetyl-C) purchased from Glen Research. Coupling time was set to 10 min. A 0.45 M solution of 1H-tetrazole in CH3CN was used as the reaction activator. After the extension, deprotection and excision from CPG were carried out with NH3 (aq) for 12 h, and the mixture was filtered. The excess ammonia was removed from the filtrate in vacuo, and the residue was purified by a C18 cartridge column and anion-exchange HPLC to yield 2′-O-methylated oligoribonucleotides 5a–c, 6a–c, and 7a–c.Nuclease resistance assay
The nuclease stability of 2′-O-arylated dinucleotide was evaluated by treatment with SVP or bovine SPD (purchased from Sigma). The SVP assay (5 × 6 U/ml) was performed in a 50 mM Tris-HCl buffer (pH 8.5, 72 mM NaCl, 14 mM MgCl2) at 37 °C. The bovine SPD assay (0.2 U/ml) was performed in a 30 mM NaOAc buffer (pH 6.0) at 37 °C. A 50 μmol sample of oligonucleotide was used in each assay. After digestion, the enzyme was deactivated by heating at 100 °C for 5 min. The solution was filtered and analyzed by reverse-phase HPLC.Circular dichroism (CD) analysis of single stranded oligonucleotides
Each oligonucleotide was dissolved in a 10 mM sodium phosphate buffer (pH 7.0, containing 0.1 M NaCl and 0.1 mM EDTA) and placed in the quartz cell (0.5 cm). Measurements were carried out by a spectropolarimeter at 10 °C. The absorbance spectra from 350 to 200 nm were collected 8 times at intervals of 1 nm at a rate of 100 nm min−1.Molecular dynamics simulations
The MD calculations were performed for the RNA duplexes, 2′-O-methyl-5′-(GACXXXXXGACU)-3′ (S1)/2′-OH-3′-(CUGAAAAACUGA)-5v (S2)/2′-O-methyl-3′-r(CUGAAAAACUGA)-5′ (S3) where X is U or 2′-O-Aryll-U, respectively, using the reported force field parameters for 2′-O-methyl-RNAs.17 The MD simulations were performed using the duplexes S1/S2, S1/S3. The MD simulations were performed using AMBER 7 program with the ff94 force field18 and the TIP3P water model.19 The partial atomic charges for 2′-O-Aryl-U were determined at the HF/6-31G level of calculations,20 and are described in the ESI.† The starting structures of A-type RNAs were generated using the NUCGEN module of AMBER software. The starting structure was contained in a solvent box consisting of water, sodium ions, and chloride ions. The solvent box was extended by 12 Å from the non-hydrogen atoms of the nucleic acids. To neutralize the negative charges of the duplexes, sodium cations were placed near each phosphate group using the additions command of the Leap module. In addition, another 8 sodium and 8 chloride ions were added at random positions to yield approximately 0.1 M NaCl concentrations. Molecular dynamics simulations were performed using the SANDER module of AMBER software. Minimization of the starting models was performed with the positional restraint of 500 kcal mol−1 on the non-hydrogen atoms of the oligonucleotides. After 200 steps of minimization, equilibrations were performed by eight 50 ps MD calculations during which the positional restraints were reduced to 20, 10, 5, 2, 1, 0.5, 0.1, and 0.01 kcal mol−1. The integration time step was 1 fs, and SHAKE21 was used to constrain all covalent bonds involving hydrogen atoms. In all calculations, electrostatic interactions were treated by the particle mesh Ewald method,22,23 and the cut-off of distance was set as 9.0 Å for Lennard-Jones interactions. After equilibrations, the final 5 ns calculation was performed for each of the duplexes without the positional restraints on the non-hydrogen atoms.Acknowledgements
This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan, and in part by Health Sciences Research Grants for Research on Psychiatric and Neurological Diseases and Mental Health from the Ministry of Health, Labor and Welfare of Japan and the global COE project.Notes and references
- (a) J. K. Watt, G. F. Deleavey and M. J. Damha, Drug Discovery Today, 2008, 13, 842–855 CrossRef CAS; (b) J. K. Watts, G. F. Deleavey and M. J. Damha, Drug Discovery Today, 2008, 13, 842–855 CrossRef CAS; (c) D. R. Corey, J. Clin. Invest., 2007, 117, 3615–3622 CrossRef CAS; (d) D. Bumcrot, M. Manoharan, V. Koleliansky and D. W. Y. Sah, Nat. Chem. Biol., 2006, 2, 711–719 CrossRef CAS; (e) R. I. Mahato, K. Cheng and R. V. Guntaka, Expert Opin. Drug Delivery, 2005, 2, 3–28 Search PubMed; (f) M. Manoharan, Curr. Opin. Chem. Biol., 2004, 8, 570–579 CrossRef CAS; (g) M. Manoharan, Antisense Nucleic Acid Drug Dev., 2002, 12, 103–128 CrossRef CAS; (h) M. Manoharan, Biochim. Biophys. Acta-Gene Struct. Express., 1999, 1489, 117–130 Search PubMed.
- (a) S. Obika, Chem. Pharm. Bull., 2004, 52, 1399–1404 CrossRef CAS; (b) T. Imanishi and S. Obiya, J. Synth. Org. Chem., 1999, 57, 969–980 CAS; (c) J. Wengel, Acc. Chem. Res., 1999, 32, 301–310 CrossRef CAS; (d) M. Petersen and J. Wengel, Trends Biotechnol., 2003, 21, 74–81 CrossRef CAS.
- (a) E. A. Lesnik, C. J. Guinosso, A. M. Kawasaki, H. Sasmor, M. Zounes, L. L. Cummins, D. J. Ecker and S. M. Freier, Biochemistry, 1993, 32, 7832–7838 CrossRef CAS; (b) M. Grotli, M. Douglas, R. Eritja and B. S. Sproat, Tetrahedron, 1998, 54, 5899–5914 CrossRef CAS; (c) M. Meldgaard and J. Wengel, J. Chem. Soc., Perkin Trans. 1, 2000, 3539–3554 RSC; (d) R. H. Griffey, B. P. Monia, L. L. Cummins, S. Freier, M. J. Greig, C. J. Guinosso, E. Lesnik, S. M. Manalili, V. Mohan, S. Owens, S., B. R. Ross, H. Sasmor, E. Wancewicz, K. Weiler, P. D. Wheeler and P. D. Cook, J. Med. Chem., 1996, 39, 5100–5109 CrossRef CAS; (e) T. P. Prakash, A. Puschl, E. Lesnik, V. Mohan, V. Tereshko, M. Egli and M. Manoharan, Org. Lett., 2004, 6, 1971–1974 CAS.
- (a) M. Teplova, G. Minasov, V. Tereshko, G. B. Inamati, P. D. Cook, Manoharan and M. Egli, Nat. Struct. Biol., 1999, 6, 535–539 CrossRef CAS; (b) R. S. Geary, T. A. Watanabe, L. Truong, S. Freier, E. A. Lesnik, N. B. Sioufi, H. Sasmor and M. A. A. Manoharan Levin, J. Pharm. Exp. Ther., 2001, 296, 890–897 CAS; (c) A. Sabahi, J. Guidry, G. B. Inamati and M. Manoharan, Nucleic Acids Res., 2001, 29, 2163–2170 CrossRef CAS.
- K. H. Altmann, N. M. Dean, D. Fabbro, S. M. Freier, T. Geiger, R. Haner, D. Husken, P. Martin, B. P. Monia, M. Muller, F. Natt, P. Nicklin, J. Phillips, U. Pieles, H. Sasmor and H. E. Moser, Chim, 1996, 50, 168–176 Search PubMed.
- (a) X. Chen, N. Dudgeon, L. Shen and J. H. Wang, Drug Discovery Today, 2005, 10, 587–593 CrossRef CAS; (b) I. Kang and J. H. Wang, J. Biol. Chem., 1994, 269, 12024–12031 CAS; (c) X. L. Chen, L. Shen and J. H. Wang, Oligonucleotides, 2004, 14, 90–99 CrossRef CAS; (d) M. H. Rahman, I. Kang, R. G. Waterbury, U. N. Narang, F. V. Bright and J. H. Wang, Anal. Chem., 1996, 68, 134–138 CrossRef CAS; (e) M. A. Ashun, Y. Hu, I. S. Kang and J. H. Wang, Antimicrob. Agents Chemother., 1996, 40, 2311–2317 CAS; (f) K. Ru, S. Schmitt, W. I. James and J. H. Wang, Oncol. Res., 1999, 11, 505–512 CAS; (g) K. Ru, M. L. Taub and J. H. Wang, Onco. Res., 1998, 10, 389–397 Search PubMed; (h) L. Shen, X. Chen and J. H. Wang, Antisense Nucleic Acid Drug Dev., 2003, 13, 67–74 CrossRef CAS; (i) A. Wang and J. Wang, Antisense Nucleic Acid Drug Dev, 1999, 9, 43–51 CAS.
- Y. Oeda, Y. Iijima, H. Taguchi, A. Ohkubo, K. Seio and M. Sekine, Org. Lett., 2009, 11, 5582–5585 CrossRef CAS.
- (a) S. L. Beaucage and M. H. Caruthers, in Current Protocols in Nucleic Acid Chemistry, ed. S. L. Beaucage, D. E. Bergstrom, G. D. Glick and R. A. Jones, John Wiley & Sons, New York, 2000, vol. I, pp. 3.3.1–3.3.20 Search PubMed; (b) S. L. Beaucage and R. P. Iyer, Tetrahedron, 1993, 49, 6123–6194 CrossRef CAS and references therein.
- S. M. Elbashir, J. Martinez, A. Patkaniowska, W. Lendeckel and T. Tuschl, EMBO J., 2001, 20, 6877–6888 CrossRef CAS.
- (a) W. Saenger, Principles of Nucleic Acid Structure, Springer, New York, 1984 Search PubMed; (b) M. Sekine, O. Kurasawa, K. Shohda, K. Seio and T. Wada, Eur. J. Org. Chem., 2001, 1989–1999 CrossRef CAS; (c) M. Sekine, O. Kurasawa, K. Shohda, K. Seio and T. Wada, J. Org. Chem., 2000, 65, 6515–6524 CrossRef CAS; (d) M. Sekine, O. Kurasawa, K. Shohda, K. Seio and T. Wada, J. Org. Chem., 2000, 65, 3571–3578 CrossRef CAS; (e) K. Shohda, I. Okamoto, T. Wada and M. Sekine, Bioorg. Med. Chem. Lett., 2000, 10, 1795–1798 CrossRef CAS.
- C. Altona and M. Sundaralingham, J. Am. Chem. Soc., 1973, 95, 2333–2344 CrossRef CAS.
- (a) K. Nakanishi, Circular Dichroism Principles and Applications, ed. N. Berova, R. W. Woody, VCH, New York, 1994 Search PubMed; (b) Analytical Applications of Circular Dichroism. Techniques and Instrumentation in Analytical Chemistry, N. Purdie, H. G. Brittain, Elsevier, Amsterdam, 1993 Search PubMed; (c) Circular Dichroism and the Conformational Analysis of Biomolecules, ed. G. D. Fasman, Plenum Press, New York, 1996 Search PubMed.
- T. E. Cheatham III and D. A. Case, in Computational Studies of RNA and DNA, ed. J. Sponer and F. Lankas, Springer, New York, 2006, pp. 45–71 Search PubMed.
- T. Sasami, R. Tawarada, A. Ohkubo, M. Sekine and K. Seio, THEOCHEM, 2009, 899, 54–60 CrossRef CAS.
- (a) M. Nishio, M. Hirota and Y. Umezawa, in The CH/p Interaction, Evidence, Nature, and Consequences, Methods in Stereochemical Analysis, ed. A. P. Marchand, Wiley-VCH, New York, 1998 Search PubMed; (b) J.-M. Lehn, Supramolecular Chemistry Concepts and Perspectives, Wiley-VCH, Weinheim, 1995 Search PubMed.
- D. A. Case, et al., AMBER 7, University of California, San Francisco, 2002 Search PubMed.
- P. Auffinger and E. Westhof, Angew. Chem., Int. Ed., 2001, 40, 4648–4650 CrossRef CAS.
- W. D. Cornell, P. Cieplak, C. I. Bayly, I. R. Gould, K. M. Merz Jr., D. M. Ferguson, D. C. Spellmeyer, T. Fox, J. W. Caldwell and P. A. Kollman, J. Am. Chem. Soc., 1995, 117, 5179–5197 CrossRef CAS.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, J. Chem. Phys., 1983, 79, 926–935 CrossRef CAS.
- C. L. Bayly, P. Cieplak, W. D. Cornell and P. A. Kollman, J. Phys. Chem., 1993, 97, 10269–10280 CrossRef CAS.
- J. Ryckaert, G. Ciccotti and H. J. C. Berendsen, J. Comput. Chem., 1977, 23, 327–341 CAS.
- T. Darden, D. York and L. Pedersen, J. Chem. Phys., 1993, 98, 10089–10092 CrossRef CAS.
- U. Essmann, L. Perera, M. L. Berkowitz, T. Darden, H. Lee and L. G. Pedersen, J. Chem. Phys., 1995, 103, 8577–8593 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: The detailed experimental section and the 1H and 13C data of all new products. See DOI: 10.1039/c0ob00248h |
| This journal is © The Royal Society of Chemistry 2011 |
