γ-Oxo-1-pyrenebutyric acid used for fluorescent detection of serum albumins and trypsin†
Jing
Wang‡
ad,
Hai-Bo
Liu‡
b,
Shinkyu
Park
a,
So Young
Kim
c,
Taiha
Joo
c and
Chang-Sik
Ha
*a
aDepartment of Polymer Science and Engineering, Pusan National University, Busan, 609-735, South Korea. E-mail: csha@pusan.ac.kr; Fax: +82-51-514-4331; Tel: +82-51-510-2407
bDepartment of Chemical and Biomolecular Engineering, Pusan National University, Busan 609-735, South Korea
cDepartment of Chemistry, Pohang University of Science and Technology (POSTECH), Pohang, 790-784, South Korea
dDepartment of Biology and Chemistry, City University of Hong Kong, 83 Tat Chee Avenue, Kowloon, Hong Kong, China
First published on 27th March 2012
Abstract
Fluorescence spectroscopy, one of the most informative analytical techniques, has played and continues to play a key role in modern research due to its high sensitivity, rapid response rate, and relatively low cost. Herein we report its application to the detection of the proteins bovine serum albumin (BSA) and human serum albumin (HSA) as well as a protease (trypsin). The detection is based on a fluorescent molecule: γ-oxo-1-pyrenebutyric acid (OPBA), which exhibits a quenched fluorescent change at 455 nm toward serum albumins (SAs). OPBA interacted with SAs with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry and a strong affinity due to π-stacking, hydrophobic interactions, and hydrogen bonding interactions. The microenvironment created by HSA and BSA played an important role in their respective interactions with OPBA. In addition, OPBA can be used for monitoring trypsin by the cleavage of HSA or BSA in the presence of copper ions, and the successful cleavage of SAs was demonstrated by the SDS polyacrylamide gel electrophoresis (PAGE) results. This indirect detection of trypsin might be developed as a strategy for sensors without substrate selection. All of these tests do not require sophisticated instrumentation and should be applicable to standard fluorescence assays.
1 stoichiometry and a strong affinity due to π-stacking, hydrophobic interactions, and hydrogen bonding interactions. The microenvironment created by HSA and BSA played an important role in their respective interactions with OPBA. In addition, OPBA can be used for monitoring trypsin by the cleavage of HSA or BSA in the presence of copper ions, and the successful cleavage of SAs was demonstrated by the SDS polyacrylamide gel electrophoresis (PAGE) results. This indirect detection of trypsin might be developed as a strategy for sensors without substrate selection. All of these tests do not require sophisticated instrumentation and should be applicable to standard fluorescence assays.
Introduction
Proteins play fundamental roles in sustaining life and are closely related to the origin, evolution and metabolism of life. Serum albumins (SAs) are the most abundant proteins in plasma. As the major soluble protein constituents of the circulatory system, SAs have many physiological functions.1 Proteases, also known as proteinases or proteolytic enzymes, are a large group of enzymes that occur naturally in all organisms. Trypsin is a serine protease found in the digestive system of many vertebrates and it plays a key role in controlling pancreatic exocrine function.2 Thus, the development of analytical tools to detect the biologically important analytes (serum albumins and trypsin) with easy handling, rapid monitoring, high sensitivity, and good binding linearity for both qualitative and quantitative analyses is of particular importance.3–5 Several methods, including atomic absorption spectroscopy, inductively coupled plasma atomic emission spectrometry, electrochemical sensing, and the use of piezoelectric quartz crystals, make it possible to detect low limits. However, these methods require expensive equipment and involve time-consuming and laborious procedures that can be carried out only by trained professionals.Alternatively, analytical techniques based on fluorescent detection are commonly appreciated for their versatility and sensitivity (up to 1000-fold higher than absorption spectrophotometry).6 Fluorescence probes based on pyrene chromophores have attracted considerable attention over the last decades because of their intense vibrational fluorescence,7 as well as their unique monomer and excimer emissions at considerably different wavelengths, which are highly attractive for probing chemical or biological analytes.8 However, less attention9 was paid to the special class of pyrene derivatives with broad structureless emission peaks: with a carbonyl group substituted on the α-position of the pyrene chromophore. Their photophysical properties are sensitive to medium polarity and hydrogen-bonding characteristics of microenvironments,10 especially in sensing systems for proteins. In addition, few reports have worked on developing probes with desirable multifunctional sensing properties.11 Thus, developing new and multifunctional applications of the α-carbonyl substituted pyrene derivatives for sensing systems is attractive.
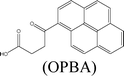
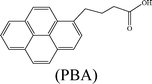
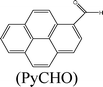
Herein, we describe the dual sensing properties of γ-oxo-1-pyrenebutyric acid (OPBA) (Scheme 1), with α-carbonyl substitution with respect to the proteins bovine (BSA) and human (HSA) serum albumin, and a protease (trypsin). HSA and BSA quenched the emission of OPBA at approximately 455 nm. HSA and BSA interacted with OPBA via π-stacking, hydrophobic, and hydrogen bonding interactions, as demonstrated from the fluorescence experimental results and the model between HSA and OPBA; in addition, the microenvironment created by the serum albumins (SAs) played an important role in the complex formation between OPBA and SAs. SDS polyacrylamide gel electrophoresis (PAGE) results illustrated that HSA and BSA were successfully cleaved by adding trypsin to a solution of OPBA/BSA/Cu2+ (1 μM/1 μM/100 μM). Correspondingly, the microenvironment of HSA and BSA that was favorable for the interaction between OPBA and SAs was interrupted. The quenched emission of OPBA, induced by the SAs, was recovered and the added trypsin was detected. The Cu2+ was used to remove any effect from the amino acids or peptide fragments created in the process of cleaving SAs. Similar experimental results were observed for HSA and BSA as they are similar in sequence and conformation, and only differ in the number of tryptophan residues.12 Pyrene, 1-pyrene-carboxaldehyde (PyCHO) and 1-pyrenebutyric acid (PBA) (Scheme 1) were also studied in this work in order to investigate the importance of the substitution groups of the carbonyl and carboxyl in the realization of probing versatile analytes.
 | ||
| Scheme 1 Chemical structures of pyrene derivatives. | ||
Results and discussion
Fluorescence sensing of SAs
Testing the photophysical properties (excitation and emission spectra) of OPBA was conducted in aqueous solutions. Excitation of OPBA with a carbonyl group on the α-position of the pyrene chromophore at 340 nm showed a broad, reasonably intense monomer emission at 455 nm which was attributed to the π–π* emitting state (Fig. 1).9–13 Upon titration with BSA and HSA, the emission of OPBA at 455 nm decreased significantly and almost disappeared in the presence of 1.0 equivalent of BSA or HSA. This result was attributed to the formation of the complex between OPBA and SAs (Fig. 1, Table 1).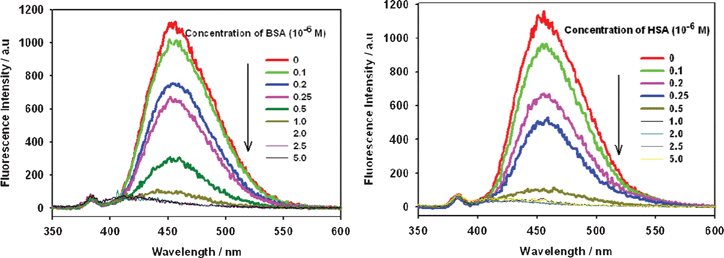 | ||
| Fig. 1 Fluorescence emission spectra of OPBA (1 × 10−6 M) in the presence of BSA (left) or HSA (right) at various concentrations in aqueous solutions. Excitation wavelength was 340 nm. | ||
| Samples | λ em/nma | I/I0d | Quenched (%)d | Enhanced (%)d | I/I0e | Quenched (%)e | Enhanced (%)e |
|---|---|---|---|---|---|---|---|
| a Excitation wavelength was 340 nm. b The emission of PBA was monitored at 375 nm. c The emission of pyrene was monitored at 372 nm. d I and I0 were the fluorescence of the samples in the presence and absence of 1.0 equivalents of BSA. e I and I0 were the fluorescence of the samples in the presence and absence of 1.0 equivalents of HSA. | |||||||
| OPBA | 455 | 0.08 | 92 | — | 0.04 | 96 | — |
| PBAb | 375, 396 | 0.38 | 62 | — | 0.35 | 65 | — |
| PyCHO | 470 | 0.43 | 57 | — | 0.47 | 53 | — |
| Pyrenec | 372, 392 | 2.32 | — | 132 | 1.42 | — | 42 |
In the range of 0–5 × 10−7 M, the I455nmversus the concentration of BSA or HSA exhibits a linear relationship (R = 0.998 or R = 0.990), thus, over this concentration range, OPBA can be used for the quantitative detection of BSA and HSA in aqueous solution (Fig. S1, ESI†). The detection limit of OPBA toward BSA and HSA was found to be less than 1.0 × 10−7 M (Fig. 1). Monitoring the fluorescence emission at 455 nm, OPBA exhibited the excitation peaks at 280 nm, 350 nm and a shoulder peak in the range of 380 nm–400 nm (Fig. S2, ESI†); all of these excitation peaks were quenched by increasing the concentration of SAs (Fig. S2, ESI†).
In addition, the relationship between the excitation wavelength and the quenching behavior of OPBA toward BSA was studied (Fig. 1, Fig. 2 and Fig. S3, ESI†). OPBA showed maximum fluorescence emission intensity for the excitation wavelengths of 280 nm, 340 nm, and 360 nm. As shown in Fig. 2, the fluorescence intensity of OPBA decreased quickly at the excitation wavelength of 340 nm. Thus, an excitation wavelength of 340 nm was used for all of the following experiments. Moreover, the fluorescence emission intensity of OPAB (1 × 10−6 M) at 455 nm remained stable over 72 h after excitation with 7 different excitation wavelengths: 280 nm, 295 nm, 320 nm, 340 nm, 360 nm, 380 nm, and 400 nm.
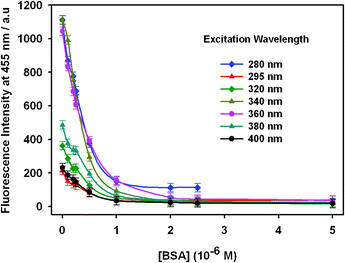 | ||
| Fig. 2 The fluorescence emission of OPBA (1 × 10−6 M) at 455 nm in the presence of BSA at various concentrations in aqueous solutions. The excitation wavelengths were 280 nm, 295 nm, 320 nm, 360 nm, 380 nm, and 400 nm. | ||
Emission quenching studies
Generally, fluorescent quenching can occur by two different mechanisms, static quenching and dynamic (collisional) quenching.14 Collisional quenching only affects the excited states of the fluorophores, and thus no changes in the absorption spectra are expected. In contrast, ground-state complex formation will frequently result in perturbation of the absorption spectra of the fluorophore especially if the interaction occurs by hydrogen bonding.The effect of BSA on the absorption spectra of OPBA in aqueous solution was explored. OPBA showed two intense peaks (centered at 280 nm and 354 nm) and a shoulder peak (Fig. S4, ESI†), the absorption spectra of OPBA changed with increasing BSA concentration. For instance, in the presence of 5.0 equivalents of BSA, the absorption peak at 354 nm increased and red shifted to 358 nm, and the absorbance of the shoulder peak increased. The absorbance of OPBA at 280 nm will not be discussed because it overlaps with the absorbance of BSA. As shown in Fig. S4, ESI†, there were changes in the absorption spectra of OPBA that supported the static quenching.15c,16
In addition, lifetime measurements of OPBA and OPBA with 0.25 equivalents of BSA were carried out in order to clarify the interaction mechanism. OPBA exhibited a double-exponential decay with lifetimes of τ1 = 0.113 ns and τ2 = 0.820 ns, and pre-exponential factors of A1 = 0.349 and A2 = 0.651. Whereas double-exponential decay with lifetimes of 0.095 ns (τ1) and 0.840 ns (τ2), and pre-exponential factors A1 = 0.427 and A2 = 0.573, were observed in the presence of 0.25 equivalents of BSA (Fig. S5, ESI†). At this stage, the above experimental results supported the presence of static quenching mechanism, although the existence of a dynamic quenching mechanism could not be excluded due to the upward curvature of Fig. 3 with an intercept value other than 1.
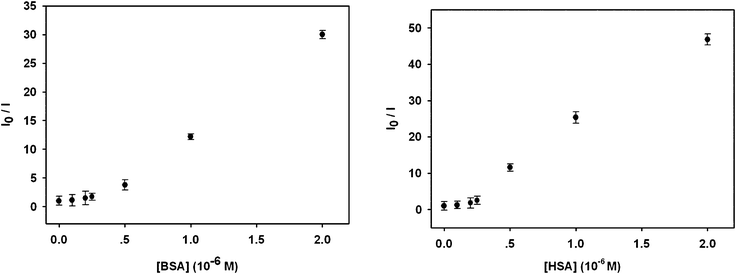 | ||
| Fig. 3 The fluorescent response of OPBA toward BSA (left) and HSA (right) at various concentrations in aqueous solutions. I0 and I are the fluorescence intensity values of OPBA (1.0 × 10−6 M) at 455 nm in the absence and presence of different amounts of BSA (left) and HSA (right), respectively. | ||
The influence of pH on the fluorescence of OPBA was shown in Fig. S6, ESI† and the pKa value of OPBA was calculated to be approximately 5.2. The isoelectric points (pI) of BSA and HSA are ∼4.817a and ∼4.9,17b respectively. OPBA and SAs were both negatively charged at pH 7.5, therefore an electrostatic interaction between OPBA and SAs might be excluded. Lysozyme,18 with a high pI value of ∼11, was chosen to further investigate the effect of electrostatic interactions. As shown in Fig. S7, ESI†, lysozyme (positively charged at pH 7.5) induced no changes in the fluorescence emission of OPBA. Thus, an electrostatic interaction was not the predominant driving force for the formation of the complex between OPBA and SAs.19
Pyrene, which is a typical hydrophobic fluorescent dye,20 was chosen for investigating the hydrophobic interaction, while PyCHO and PBA were selected in order to investigate substitution of the carbonyl and carboxyl groups on OPBA (Scheme 1).
With the addition of increasing concentrations of SAs, the typical monomer emissions at 372 nm and 392 nm were dramatically enhanced (Fig. S8, ESI†) and the excitation peaks of pyrene were red-shifted (Fig. S9, ESI†). These results suggest that a hydrophobic environment surrounded the pyrene chromophore, and that the protection of Py by SAs, π-stacking, and hydrophobic interactions were involved in the interaction between OPBA and SAs.21,22 The value of I3/I1 (Fig. S8, ESI†) also suggested that a hydrophobic microenvironment was around the pyrene chromophore.23 Quenched emission and excitation spectra were observed for both the alkyl-substituted PBA containing a carboxyl group and PyCHO upon addition of SAs to an aqueous solution of PBA or PyCHO (Table 1, Fig. S10 and S11, ESI†). However, 1.0 equivalents of BSA or 1.0 equivalents of HSA could not fully quench the emission of PBA and PyCHO. Thus, both the carbonyl and carboxyl groups on OPBA played an important role in its interaction with SAs and in the quenched fluorescence intensity after adding SAs. The interaction may be attributed to a hydrogen bonding effect between the SAs and both the carboxyl and carbonyl groups of OPBA.9c
When excited at 295 nm, BSA (or HSA) presents a strong emission at approximately 340 nm which was attributed to the tryptophan residues on BSA (or HSA).12,24 After adding OPBA to the solution of SAs, the emission at 340 nm was quenched dramatically (Fig. S12, ESI† and Table 2). Therefore, an interaction between OPBA and the tryptophan residues on SAs was confirmed. In addition, there was a possibility of an internal filter being involved in the decrease in the BSA fluorescence intensity by OPBA (Fig. S12, ESI†) because both the protein and OPBA absorb at 295 nm (Fig. S2, ESI†) and therefore the dramatic decrease in the intensity of fluorescence at 340 nm could partly be apparent.
| Added samples | I/I0a | Quenched (%)a | I/I0b | Quenched (%)b |
|---|---|---|---|---|
| a I and I0 were the fluorescence of BSA at 340 nm in the presence and absence of the added samples. b I and I0 were the fluorescence of HSA at 340 nm in the presence and absence of the added samples. | ||||
| OPBA | 0.1 | 90 | 0.2 | 80 |
| PBA | 0.14 | 86 | 0.26 | 74 |
| PyCHO | 0.75 | 25 | 0.8 | 20 |
| Pyrene | 0.75 | 25 | 0.83 | 17 |
The modified Stern–Volmer follows in eqn (1):19a,25
| log(I0 − I)/I = logK + nlog[Q] | (1) |
where I0 and I are the fluorescence intensity of SAs at 340 nm in the absence and presence of OPBA, respectively, K is the binding (or quenching) constant between SAs and OPBA, [Q] is the concentration of OPBA, and n is the number of binding sites per BSA or HSA.
Based on eqn (1), the binding (or quenching) constant between BSA (or HSA) and OPBA was established to be KOPBA–BSA of 1.26 × 106 M−1 (KOPBA–HSA of 1.52 × 106 M−1) and a n of 1.135 (1.323) suggested that one OPBA chromophore interacts with one BSA or HSA molecule (Fig. 4). However, free tryptophan amino acids did not induce any changes in the emission of OPBA (Fig. S13, ESI†), this result indicated that the microenvironment created by the macromolecules (BSA and HSA) was important for their respective interactions with OPBA.19 Pyrene (Fig. S14, ESI†) and PyCHO (Fig. S15, ESI†) only partially quenched the emission of SAs at 340 nm, while the emission of SAs at 340 nm was significantly quenched after adding PBA (Fig. S16 , ESI†and Table 2).
![Plot of log [(I0 − I)/I] versus log [OPBA]. I0 and I are the fluorescence intensity of BSA (left, 1.0 × 10−6 M) or HSA (right, 1.0 × 10−6 M) in the absence and presence of different amounts of OPBA, respectively.](/image/article/2012/RA/c2ra01011a/c2ra01011a-f4.gif) | ||
| Fig. 4 Plot of log [(I0 − I)/I] versus log [OPBA]. I0 and I are the fluorescence intensity of BSA (left, 1.0 × 10−6 M) or HSA (right, 1.0 × 10−6 M) in the absence and presence of different amounts of OPBA, respectively. | ||
The emission of OPBA/BSA (1 μM/1 μM) was almost unaffected by various non-protein substances, such as inorganic salts (sodium chloride, sodium acetate, sodium carbonate, sodium phosphate, zinc chloride, HEPES), detergents (sodium dodecyl sulfate (SDS)), the chelating compound EDTA, reductant glucose and organic solvents (acetone, chloroform, ethanol and methanol). In the presence of the other investigated proteins, such as lysozyme, β-gal, GOx, FalDH and MAPH, minimal or no changes of the OPBA emission at 455 nm was observed (Fig. S7 and S17, ESI†). All of the above results revealed that OPBA can be used as a probe for serum albumins (BSA and HSA).
Molecular modeling study
Molecular docking is one of the essential tools used for explaining correlations between a molecule’s structure and its biological activity. Docking can be performed routinely if both the structures of a protein and a molecule are known. No crystals suitable for X-ray diffraction studies have yet been obtained for BSA. Therefore, molecular docking study for HSA26 and pyrene derivatives were performed to understand the structure–activity relationship. According to the molecule binding sites, it is confirmed that there is a positive correlation between the docking results and the fluorescence data. The identified hydrogen bonds or polar interactions with the binding pocket in HSA and binding energy (in kcal mol−1) of the tested molecules are summarized in Table 3. These results indicate that several important amino acids residues, in particular, Lys195, Lys199, Trp214, Arg218, Arg222, and Lys436 in HSA, could specifically develop hydrogen bonds with the tested molecules. Apart from those amino acid residues in HSA, pyrene derivatives were completely buried into pockets formed by hydrophobic amino acids (Fig. 5). The emission of pyrene was enhanced after adding SAs because of the protection of the chromophore by HSA through π-stacking and hydrophobic interactions (Table 1). In the presence of 1.0 equivalents of SAs, the emissions of OPBA, PBA and PyCHO were quenched (Table 1) due to the hydrogen bonding effect (Table 3) between these molecules with SAs. The emission of OPBA were quenched to a greater degree than PyCHO or PBA because of the coexistence of the carbonyl and the carboxyl groups on OPBA. The emission of SAs at 340 nm, was decreased more by OPBA and PBA than by pyrene and PyCHO (Table 2). This may be due to the smaller molecular volumes of PyCHO and pyrene, which resulted in less steric hindrance for SAs (Table 3). These results indicate that the interactions between these compounds and SAs may be attributed to hydrogen bond formation and the steric hindrance of these aromatic groups in the hydrophobic binding pocket of SAs (Tables 1 and 2, Fig. 5).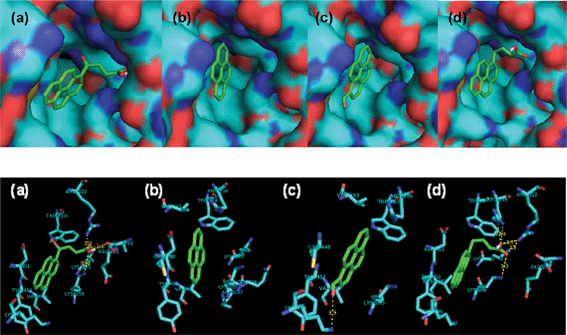 | ||
| Fig. 5 Models of complexes generated using the Ligand Fit docking program. (a) HSA–OPBA; (b) HSA–pyrene; (c) HSA–PyCHO; (d) HSA–PBA. Green dashed lines indicate hydrogen bonds. | ||
| Tested molecules | Volumea/Å3 | H-bondb | Involved amino acidc | Binding energyd/kcal mol−1 |
|---|---|---|---|---|
| a Volume of the tested molecules. b Amino acid residues involved binding to the tested molecule via hydrogen bonds. c Amino acid residues in the binding pocket. d Estimated free energy of binding; an index of docking score. | ||||
|
268 | Lys195;Lys199; Trp214;Arg222; Ala291 | Lys195; Lys199; Trp214;Arg222; Ala291;Glu292; Lys436;Tyr452; Val455 | −8.82 |
|
266 | Lys195;Arg218; Arg222 | Lys195;Lys199; Trp214;Arg218; Arg222;Ala291; Glu292;Lys436; Asp451;Tyr452; Val455 | −9.30 |
|
207 | Lys436 | Lys195;Trp214; Arg218;Ala343; Lys436;Cys448; Asp451;Tyr452; Val455 | −10.5 |
|
189 | None | Lys195;Trp214; Arg218;Val343; Cys448;Asp451; Tyr452; Val455 | −10.7 |
Cu2+-supported fluorescence detection of protease: trypsin
We checked the fluorescence responses of the OPBA probe versus 20 kinds of amino acids. Minimal or no emission changes of OPBA were observed upon adding the 20 kinds of amino acids to the aqueous solution of OPBA in the absence and presence of SAs (Fig. S18 and S19, ESI†). Trypsin can cleave serum albumins (SAs) into amino acids or peptide fragments.27 Thus, the interaction between OPBA and SAs might be interrupted; also, the OPBA chromophore could be released from SAs and the quenched emission of OPBA induced by SAs might be recovered by adding trypsin to the solution of OPBA/SAs. Correspondingly, the added trypsin can be detected.Trypsin (1.0 μg mL−1) was added to a solution of OPBA/BSA (1 μM/1 μM), however, minimal recovery of the OPBA fluorescence was observed within 25 min. The existence of the amino acids or peptide fragments produced from the cleaving of BSA by trypsin might hinder the recovery of OPBA fluorescence, even though the individual amino acids have minimal effect on the fluorescence of OPBA (Fig. S18 and S19, ESI†). Cu2+ ions (100 μM) were then added in order to chelate the amino acid or peptide fragments with a strong binding constant. The emission of OPBA at 455 nm increased immediately, while Cu2+ did not influence the emission of OPBA at 455 nm in the presence of BSA due to the stronger interaction between BSA and OPBA than between Cu2+ and BSA or OPBA (Fig. S20, ESI†).2
Based on the above preliminary experimental results, trypsin (2.5 μg mL−1, 5.0 μg mL−1, 10 μg mL−1, or 12.5 μg mL−1) was added to the complex of OPBA/SAs/Cu2+ (1 μM/1 μM/100 μM) in separate aqueous solutions. Lanes 2 and 4 in Fig. 6 showed BSA and HSA on an SDS-PAGE after staining with OPBA and pyrene; the addition of trypsin resulted in several new fragments (lanes 3 and 5 of Fig. 6). This demonstrated that HSA and BSA were successfully cleaved. The fluorescence spectra of OPBA was measured within 30 min and the emission of OPBA at 455 nm was monitored as a function of time. Upon addition of 10 μg mL−1 of trypsin, the emission at 455 nm gradually increased over 20 min, and cleaving equilibrium of BSA was achieved within 20 min (Fig. 7). The fluorescence intensity recovery of OPBA at 455 nm was 40%. The fluorescence of OPBA could not be fully recovered, which may due to some probes that could not be fully released from the protein and exposed to the lights, the presence of free Cu2+ ions, the cleaving rate of SAs by trypsin, etc. The concentration of the added trypsin also influenced the fluorescence recovery of OPBA at 455 nm (Fig. 7). As shown in Fig. 7, the fluorescence recovery of OPBA at 455 nm was higher at 10 μg mL−1 than 12.5 μg mL−1 trypsin. This difference, however, could be due to some deviations within the acceptable error range and thus does not affect the trend significantly if a proper amount of trypsin was used for cleavage (<10 μg mL−1). These results suggested that the OPBA/SAs/Cu2+ complex system can be used for the detection of trypsin. In addition, changing the OPBA to pyrene, PyCHO and PBA didn't influence the cleaving of SAs by trypsin. Thus, all of the pyrene derivatives studied in this work, after interacting with SAs, can be further applied for the detection of protease, while substrate selection can be avoided.27
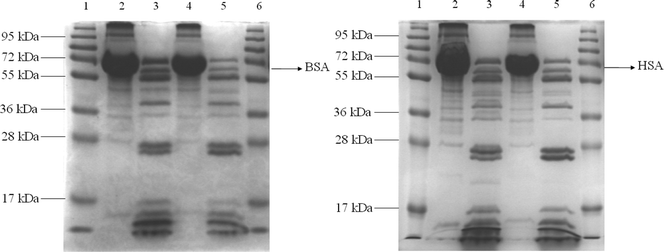 | ||
| Fig. 6 SDS-PAGE analysis. (Left) Lane 1: Protein marker; Lane 2: BSA+OPBA+Cu2+; Lane 3: BSA+OPBA+Cu2++trypsin; Lane 4: BSA+Pyrene+Cu2+; Lane 5: BSA+Pyrene+Cu2++trypsin; Lane 6: Protein marker. (Right) Lane 1: Protein marker; Lane 2: HSA+OPBA+Cu2+; Lane 3: HSA+OPBA+Cu2++trypsin; Lane 4: HSA+Pyrene+Cu2+; Lane 5: HSA+Pyrene+Cu2++trypsin; Lane 6: Protein marker. | ||
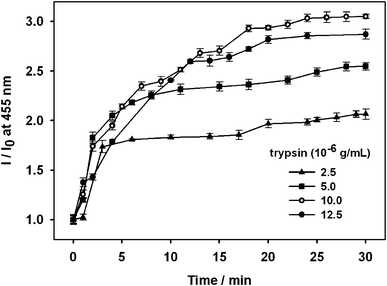 | ||
| Fig. 7 Dependence of the intensity ratio (I/I0) of OPBA emission at 455 nm on the concentration of trypsin with fixed OPBA (1.0 × 10−6 M), Cu2+ ions (1.0 × 10−4 M), and BSA (1.0 × 10−6 M) concentrations in aqueous solutions. I0 is the fluorescence intensity of the complex OPBA/BSA/Cu2+. I is the fluorescence intensity of the complex OPBA/BSA/Cu2+ in the presence of trypsin as a function of time. | ||
Experimental
General
Bovine serum albumin (BSA, lyophilized powder, essentially fatty acid free and essentially globulin free), human serum albumin (HSA, lyophilized powder, fatty acid free, globulin free), lysozyme, β-galactosidase (β-Gal), gluocose oxidase (GOx), monoclonal antipolyhistidine (MAPH), formaldehyde dehydrogenase (FalDH), γ-oxo-1-pyrenebutyric acid (OPBA), pyrene, 1-pyrene-carboxaldehyde (PyCHO), 1-pyrenebutyric acid (PBA), trypsin and 20 kinds of amino acids and all the other materials used in this work were purchased from Sigma-Aldrich. The UV–Vis spectra were measured using a Hitachi U-2010 spectrometer. The fluorescence emission spectra were measured using a Hitachi F-4500 spectrometer. Time-resolved fluorescence (TRF) was measured by the time-correlated single photon counting (TCSPC) method. The light source was a home-built cavity-dumped Ti![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) sapphire oscillator pumped by a frequency-doubled Nd
sapphire oscillator pumped by a frequency-doubled Nd![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) YVO4 laser (Verdi, Coherent, Inc.). The center wavelength was 760 nm, and the energy of the output pulses was 40 nJ at the repetition rate of 760 kHz. Pump pulses at 380 nm were generated by the second harmonic generation in a 300-μ β-barium borate (BBO) crystal. The fluorescence at 440 nm was collected by a parabolic mirror, dispersed by a monochromator, and detected with an avalanche photodiode. Magic angle detection was used to avoid the effect of polarization. The width of the instrument response function was 50 ps. HEPES (4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid, pH 7.0–8.0) buffer solutions were used to give the required pH 7.5 in this work. A pH 64 Radiometer (Copenhagen, Denmark) combined with a GK 2401 B electrode was used for the pH measurements.
YVO4 laser (Verdi, Coherent, Inc.). The center wavelength was 760 nm, and the energy of the output pulses was 40 nJ at the repetition rate of 760 kHz. Pump pulses at 380 nm were generated by the second harmonic generation in a 300-μ β-barium borate (BBO) crystal. The fluorescence at 440 nm was collected by a parabolic mirror, dispersed by a monochromator, and detected with an avalanche photodiode. Magic angle detection was used to avoid the effect of polarization. The width of the instrument response function was 50 ps. HEPES (4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid, pH 7.0–8.0) buffer solutions were used to give the required pH 7.5 in this work. A pH 64 Radiometer (Copenhagen, Denmark) combined with a GK 2401 B electrode was used for the pH measurements.
Preparation of stock solutions for absorbance and fluorescence studies
0.48 mM solutions of OPBA (0.0073 g, 0.024 mmol), pyrene (0.0049 g, 0.024 mmol), PBA (0.007 g, 0.024 mmol), and PyCHO (0.0055 g, 0.024 mmol) were prepared in DMSO (50 mL), then diluted 240 times with H2O to get the 2 μM stock solutions. The total volume of DMSO contained in the 2 μM stock solutions was below 0.5%. 10 μM stock solutions of BSA (0.01 g) and HSA (0.01 g) were prepared in H2O (15 mL). Lysozyme (0.0143 g) was dissolved in HEPES (4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid, pH 7.0–8.0) buffer solutions to obtain the 100 μM stock solution. 2000 μg mL−1 stock solutions of β-Gal (0.03 g), GOx (0.03 g), MAPH (0.03 g) and FalDH (0.03 g) were prepared in H2O (15 mL). All of the metal ion stock solutions were prepared at 1.0 mM in H2O. 10 mM stock solutions of 20 kinds of amino acids were prepared in H2O. 100 μg trypsin was dissolved in H2O to obtain the 50 μg mL−1 stock solution.Molecular docking study
The X-ray crystallographic structure of HSA (Protein Data Bank ID: 1bm0) was used as target protein in the molecular docking study. The version 4.0 of AutoDock was used for molecular docking study. Water was excluded from the workspace, and standard preparation of the molecules was used in the importation of the database. The addsol module of AutoDock was then used to generate the protein's PDBQS files, which included the solvation parameters. Affinity and electrostatic grids were generated by the autogrid module. The centers of the grid maps were assigned to the geometric center of the enzyme. The rest of the parameters were set to the default values. Docking of each compound was repeated at least five times to ensure conformation to the lowest energy state due to the iterative nature of the process.SDS-polyacrylamide gel electrophoresis (SDS-PAGE)
To study the binding mechanism of small molecules in BSA/HSA, the mixture of tested molecule and protein was incubated at R. T. (room temperature) for 30 min before the addition of trypsin. Soon afterwards, the mixture was treated with trypsin for cleavage. The protein integration information was obtained from SDS-PAGE by using 12% polyacrylamide gel (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 bis-acrylamide to acrylamide ratio) at 120 V. Coomassie Brilliant Blue R-250 was used to stain proteins.
30 bis-acrylamide to acrylamide ratio) at 120 V. Coomassie Brilliant Blue R-250 was used to stain proteins.
Conclusions
In summary, we have successfully developed new applications of γ-oxo-1-pyrenebutyric acid (OPBA) molecules in the sensing of dual analytes: proteins (BSA and HSA) and a protease (trypsin). The quenched emission of OPBA toward BSA and HSA was observed. The complex formed between OPBA and BSA (or HSA) involved π-stacking, hydrogen-bonding, and hydrophobic interactions, and it had 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry. The microenvironment created by BSA and HSA played an important role in their respective interactions with OPBA, as no influence on the fluorescent changes of OPBA by the free amino acid tryptophan was observed. Other proteins (lysozyme, β-gal, GOx, MAPH, FalDH) and non-protein substances induced minimal or no changes in the emission of OPBA at 455 nm, thus OPBA can be applied as a serum albumin (HSA and BSA) probe. In addition, the detection of HSA and BSA can be quantitatively realized by the probe OPBA. The model designs facilitated understanding the OPBA sensing system toward proteins.
1 stoichiometry. The microenvironment created by BSA and HSA played an important role in their respective interactions with OPBA, as no influence on the fluorescent changes of OPBA by the free amino acid tryptophan was observed. Other proteins (lysozyme, β-gal, GOx, MAPH, FalDH) and non-protein substances induced minimal or no changes in the emission of OPBA at 455 nm, thus OPBA can be applied as a serum albumin (HSA and BSA) probe. In addition, the detection of HSA and BSA can be quantitatively realized by the probe OPBA. The model designs facilitated understanding the OPBA sensing system toward proteins.
After adding trypsin to a mixture of OPBA/SAs/Cu2+, the SDS-PAGE results demonstrated that BSA and HSA were cleaved into amino acid or peptide fragments, which are strong Cu2+ chelators and form stable complexes with Cu2+ ions. Thus, the quenched fluorescence of OPBA recovered gradually within 20 min. This strategy suggested that all of the SA probes might be further applied for detecting trypsin, and it may also be extended to more sensing systems. Most importantly, substrate selection can be avoided using the present method. The fluorescence recovery of OPBA at 455 nm was only 40%, notwithstanding its limitation, this study does suggest that further studies for developing new and more effective sensors with this strategy are still necessary. Additional studies are now under way, which will be presented in a forthcoming work.
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) Grant funded by the Ministry of Education, Science and Technology, Korea (MEST) (Acceleration Research Program (No. 2012-0000108); the Pioneer Research Center Program (No. 2012-0000421/2012-0000422), a grant from the Fundamental R&D program for Core Technology of Materials funded by the Ministry of Knowledge Economy, Korea, and the MEST for the Brain Korea 21 Project and the scholarship for foreign students (J. W.). We sincerely thank Prof. Jeffrey I. Zink of the University of California Los Angeles for his valuable suggestions and input during the preparation of this manuscript.References
- N. Chadborn, J. Bryant, A. J. Bain and P. O'Shea, Biophys. J., 1999, 76, 2198 CrossRef CAS
.
- X. Lou, L. Zhang, J. Qin and Z. Li, Langmuir, 2010, 26, 1566 CrossRef CAS
.
-
(a) D. L. Ma, W. L. Wong, W. H. Chung, F. Y. Chan, P. K. So, T. S. Lai, Z. Y. Zhou, Y. C. Leung and K. Y. Wong, Angew. Chem., Int. Ed., 2008, 47, 3735 CrossRef CAS
; (b) P. Wu, E. L. M. Wong, D. L. Ma, G. S. M. Tong, K. M. Ng and C. M. Che, Chem.–Eur. J., 2009, 15, 3652 CrossRef CAS
.
-
(a) Y. Suzuki and K. Yokoyama, Angew. Chem., Int. Ed., 2007, 46, 4097 CrossRef CAS
; (b) J. J. Strunk, I. Gregor, Y. Becker, P. Lamken, S. Lata, A. Reichel, J. Enderlein and J. Piehler, Bioconjugate Chem., 2009, 20, 41 CrossRef CAS
.
-
(a) Y. Suzuki and K. Yokoyama, J. Am. Chem. Soc., 2005, 127, 17799 CrossRef CAS
; (b) J. X. Yan, A. T. Devenish, R. Wait, T. Stone, S. Lewis and S. Fowler, Proteomics, 2002, 2, 1682 CrossRef CAS
; (c) Q. Zeng, L. Y. Zhang, L. Zhen, J. G. Qin and B. Z. Tang, Polymer, 2009, 50, 434 CrossRef CAS
.
-
(a) R. W. Sinkeldam, N. J. Greco and Y. Tor, Chem. Rev., 2010, 110, 2579 CrossRef CAS
; (b) M. M. Adams and E. V. Anslyn, J. Am. Chem. Soc., 2009, 131, 17068 CrossRef CAS
; (c) A. Granzhan, H. Ihmels and G. Viola, J. Am. Chem. Soc., 2007, 129, 1254 CrossRef CAS
; (d) W. M. Nau, G. Ghale, A. Hennig, H. Bakirci and D. M. Bailey, J. Am. Chem. Soc., 2009, 131, 11558 CrossRef CAS
; (e) J. Gu, J. Chen and R. H. Schmehl, J. Am. Chem. Soc., 2010, 132, 7338 CrossRef CAS
; (f) Y. Xie, T. Maxson and Y. Tor, J. Am. Chem. Soc., 2010, 132, 11896 CrossRef CAS
; (g) K. Krishnamoorthy and T. P. Begley, J. Am. Chem. Soc., 2010, 132, 11608 CrossRef CAS
.
-
J. B. Birks, Photophysics of Aromatic Molecules, Wiley-Interscience, London, 1970 Search PubMed
.
-
(a) F. M. Winnik, Chem. Rev., 1993, 93, 587 CrossRef CAS
; (b) R. H. Yang, W. H. Chan, A. W. M. Lee, P. F. Xia, H. K. Zhang and K. A. Li, J. Am. Chem. Soc., 2003, 125, 2884 CrossRef CAS
; (c) S. Nishizawa, Y. Kato and N. Teramae, J. Am. Chem. Soc., 1999, 121, 9463 CrossRef CAS
; (d) B. Bodenant, F. Fages and M. H. Delville, J. Am. Chem. Soc., 1998, 120, 7511 CrossRef CAS
; (e) S. K. Kim, S. H. Lee, J. Y. Lee, R. A. Bartsch and J. S. Kim, J. Am. Chem. Soc., 2004, 126, 16499 CrossRef CAS
; (f) K. Fujimoto, Y. Muto and M. Inouye, Chem. Commun., 2005, 4780 RSC
; (g) A. Okamoto, T. Ichiba and I. Saito, J. Am. Chem. Soc., 2004, 126, 8364 CrossRef CAS
; (h) C. W. Rogers and M. O. Wolf, Angew. Chem., Int. Ed., 2002, 41, 1898 CrossRef CAS
; (i) K. M. Matkovich, L. M. Thorne, M. O. Wolf, T. C. S. Pace, C. Bohne and B. O. Patrick, Inorg. Chem., 2006, 45, 4610 CrossRef CAS
.
-
(a) A. Okamoto, K. Kanatani and I. Saito, J. Am. Chem. Soc., 2004, 126, 4820 CrossRef CAS
; (b) G. Jones and V. I. Vullev, Org. Lett., 2002, 4, 4001 CrossRef CAS
; (c) C. Armbruster, M. Knapp, K. Rechthaler, R. Schamschule, A. B. J. Parusel, G. Köhler and W. Wehrmann, J. Photochem. Photobiol., A, 1999, 125, 29 CrossRef CAS
; (d) S. S. Bag, R. Kundu, K. Matsumoto, Y. Saito and I. Saito, Bioorg. Med. Chem. Lett., 2010, 20, 3227 CrossRef CAS
.
- V. I. Vullev and G. Jones II, Tetrahedron Lett., 2002, 43, 8611 CrossRef CAS
.
- Z. Li, Y. Q. Dong, J. W. Y. Lam, J. Sun, A. Qin, M. Häußler, Y. P. Dong, H. H. Y. Sung, I. D. Williams, H. S. Kwok and B. Z. Tang, Adv. Funct. Mater., 2009, 19, 905 CrossRef CAS
.
- G. Zolese, G. Falcioni, E. Bertoli, R. Galeazzi, M. Wozniak, Z. Wypych, E. Gratton and A. Ambrosini, Proteins: Struct., Funct., Genet., 2000, 40, 39 CrossRef CAS
.
-
(a) K. A. Fletcher, S. N. Baker, G. A. Baker and S. Pandey, New J. Chem., 2003, 27, 1706 RSC
; (b) P. Banerjee, S. Chatterjee, S. Pramanik and S. C. Bhattacharya, Colloids Surf., A, 2007, 302, 44 CrossRef CAS
; (c) Y. Saito, Y. Miyauchi, A. Okamoto and I. Saito, Tetrahedron Lett., 2004, 45, 7827 CrossRef CAS
; (d) L. Búcsiová, P. Hrdloviĉ and Š. Chmela, J. Photochem. Photobiol., A, 2001, 143, 59 CrossRef
; (e) A. P. de Silva, H. Q. N. Gunaratne, T. Gunnlaugsson, A. J. M. Huxley, C. P. McCoy, J. T. Rademacher and T. E. Rice, Chem. Rev., 1997, 97, 1515 CrossRef CAS
.
- Q. Zhou and T. M. Swager, J. Am. Chem. Soc., 1995, 117, 7017 CrossRef CAS
.
-
(a) L. Lu, R. Helgeson, R. M. Jones, D. McBranch and D. Whitten, J. Am. Chem. Soc., 2002, 124, 483 CrossRef CAS
; (b) K. Haskins-Glusac, M. R. Pinto, C. Tan and K. S. Schanze, J. Am. Chem. Soc., 2004, 126, 14964 CrossRef CAS
; (c) M. Rahman and H. J. Harmon, Spectrochim. Acta, Part A, 2006, 65, 901 CrossRef
; (d) K. Wang, W. Wang, J. H. No, Y. Zhang, Y. Zhang and E. Oldfield, J. Am. Chem. Soc., 2010, 132, 6719 CrossRef CAS
; (e) A. Divsalar, A. A. Saboury, H. Mansoori-Torshizi and B. Hemmatinejad, Bull. Korean Chem. Soc., 2006, 27, 1801 CrossRef CAS
.
-
(a) J. P. Rostron, G. Ulrich, P. Retailleau, A. Harriman and R. Ziessel, New J. Chem., 2005, 29, 1241 RSC
; (b) H. W. Li, B. Wang, Y. Q. Dang, L. Li and Y. Wu, Sens. Actuators, B, 2010, 148, 49 CrossRef
.
-
(a) K. H. Kim and M. H. Moon, Anal. Chem., 2009, 81, 1715 CrossRef CAS
; (b) M. JoséMartínez-Tomé, R. Esquembre, R. Mallavia and C. Reyes Mateo, Biomacromolecules, 2010, 11, 1494 CrossRef
.
-
(a) M. Park, S. S. Park, M. Selvaraj, D. Zhao and C. S. Ha, Microporous Mesoporous Mater., 2009, 124, 76 CrossRef CAS
; (b) B. Wang and C. Yu, Angew. Chem., Int. Ed., 2010, 49, 1485 CrossRef CAS
.
-
(a) Y. Li, H. W. Li, L. J. Ma, Y. Q. Dang and Y. Wu, Chem. Commun., 2010, 46, 3768 RSC
; (b) B. Ojha and G. Das, Chem. Commun., 2010, 46, 2079 RSC
.
- H. Zhang, Z. Li, P. Xu, R. Wu and Z. Jiao, Chem. Commun., 2010, 46, 6783 RSC
.
- C. V. Kumar, A. Buranaprapuk, H. C. Sze, S. Jockusch and N. J. Turro, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 5810 CrossRef CAS
.
-
(a) B. Haldar, A. Mallick and N. Chattopadhyay, J. Photochem. Photobiol., B, 2005, 80, 217 CrossRef CAS
; (b) T. Tsukamoto and T. Hikida, J. Photochem. Photobiol., A, 1996, 95, 271 CrossRef CAS
; (c) Y. Wei, S. Wang, S. Shuang and C. Dong, Talanta, 2010, 81, 1800 CrossRef CAS
.
- S. Zhang and Y. Zhao, J. Am. Chem. Soc., 2010, 132, 10642 CrossRef CAS
.
- T. Q. Wu, Q. Wu, S. Y. Guan, H. X. Su and Z. J. Cai, Biomacromolecules, 2007, 8, 1899 CrossRef CAS
.
-
(a) L. Liang, H. A. Tajmir-Tiahi and M. Subirade, Biomacromolecules, 2008, 9, 50 CrossRef CAS
; (b) Z. Liu, X. Zheng, X. Yang, E. Wang and J. Wang, Biophys. J., 2009, 96, 3917 CrossRef CAS
.
- H. X. Min and D. C. Carter, Nature, 1992, 358, 209 CrossRef
.
- L. An, L. Liu and S. Wang, Biomacromolecules, 2009, 10, 454 CrossRef CAS
.
Footnotes |
| † Electronic Supplementary Information (ESI) available. See DOI: 10.1039/c2ra01011a/ |
| ‡ Jing Wang and Hai-Bo Liu contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2012 |
