Controllable cell adhesion, growth and orientation on layered silk protein films†
Felix Bauer‡,
Stefanie Wohlrab‡ and
Thomas Scheibel*
Lehrstuhl Biomaterialien, Universität Bayreuth, Universitätsstraße 30, 95440 Bayreuth, Germany. E-mail: Thomas.scheibel@uni-bayreuth.de; Fax: +49 921 557346; Tel: +49 921 557361
First published on 29th July 2013
Abstract
Due to their mechanical stability, biocompatibility and biodegradability, silks are promising materials for various biomedical applications including tissue engineering. Since the shape and the organisation of cells in and on scaffolds both affect their function, we tested patterned silk scaffolds made of three different silk proteins concerning their influence on cell adhesion, growth and orientation. Two different cell lines, BALB/3T3 fibroblasts and C2C12 myoblasts, showed controllable cell adhesion as well as orientation dependent on the silk proteins used and patterns made. Surprisingly, the presence of the integrin binding motif RGD did not influence cell adhesion and orientation on structured silk films, although it did so significantly on flat films.
Introduction
Silks are protein-based fibres produced by various arthropods including spiders and insects. They are well known for their mechanical stability, biocompatibility and biodegradability, which allow their use in biomedical applications.1 In contrast to obtaining silk proteins from natural sources, recombinantly produced silk proteins have the advantages of a defined composition, constantly high purity, and quality.2 Further, recombinant silk proteins can be processed into different morphologies such as fibres, nonwoven-meshes, capsules, particles, hydrogels, foams, and films.3,4Previously it has been shown that BALB/3T3 fibroblast adhesion is weak on flat eADF4(C16) (a recombinantly produced engineered spider silk protein) films,5 based on the lack of specific motifs for cell attachment in the protein's primary structure. Introduction of such motifs can improve cell adhesion on flat silk films.6–9 eADF4(C16) modified with the integrin binding motif RGD yielded significantly improved cell adhesion and proliferation.8
In tissue engineering it is further essential to restore the tissue structure and organisation. In most organs the cells and the extracellular matrix (ECM) are aligned, being essential for their cellular function and mechanical properties.10–12
The surface topography of a substrate is known to influence the adhesion and polarisation of cells. In order to align different cell types, various scaffolds are currently in use, such as contact guidance materials like micro-/nanogrooved scaffolds or fiber mesh matrices made of polymeric fibers.13–17 Most promising for tissue engineering applications are adhesive micro patterns produced by micro contact printing or other photolithographic methods. Parallel grooves for example promote topographical anchorage and orientation of the cells, which is important e.g. for bone implants, skin transplants, or artificial nerve tubes.13,14,16 Patterned structures allow to align different cell types such as osteoblasts, cardiac cells, muscle cells, corneal epithelial cells, and fibroblasts.10,18–20
Here, we used structured films made of two different silk proteins (Fig. 1) to test the influence of chosen proteins and morphology on BALB/3T3 fibroblasts. As a second model, mouse myoblast cells from muscle were tested, which fuse to myotubes at high cell densities and need alignment to generate strength and tension.20,21
![Schematic diagram of eADF4(C16), ntagCysC16-c(RGDfK), and N[AS]8C (A) and of films cast from proteins with identical and opposite charges (B).](/image/article/2013/BM/c3bm60114e/c3bm60114e-f1.gif) | ||
| Fig. 1 Schematic diagram of eADF4(C16), ntagCysC16-c(RGDfK), and N[AS]8C (A) and of films cast from proteins with identical and opposite charges (B). | ||
Materials and methods
Protein production
The spider silk protein eADF4(C16) was produced and purified as described by Huemmerich et al.26 Chemical coupling of synthesized cyclic RGD to ntagCysC16-c(RGDfK) was performed as described previously.8 The production of the recombinant lacewing egg stalk protein N[AS]8C was described by Bauer and Scheibel.27Coupling of NHS-fluorescein to N[AS]8C
1 mg of lyophilised N[AS]8C was dissolved in 6 M GdmSCN and dialysed against 20 mM HEPES (pH 7). For coupling, a 5-fold molar excess of NHS-fluorescein was added to the protein solution. After two hours of incubation at room temperature N[AS]8C was precipitated with ammonium sulphate, subsequently washed with distilled water, and lyophilised.Production of films
For production of unstructured films as well as for the ground layer films, lyophilized proteins were dissolved in formic acid and cast onto glass slides (1% (w/v), 0.15 mg cm−2) followed by evaporation of the solvent. Since spider silk films made from formic acid are water-insoluble, no post-treatment was necessary.24To prove the water stability of films made of the recombinant lacewing egg stalk protein, 1% (w/v) N[AS]8C was dissolved in formic acid and cast into a 48 well plate. After drying, the films were incubated in water for 24 hours. Afterwards, wells with films and blank wells were stained with Coomassie Brilliant Blue, and the presence of films was inspected visually.
Production of patterned two protein films
The patterns were made using photolithographically produced templates to generate PDMS (polydimethylsiloxane) negatives. First, silicon wafers were spin coated with a photoresist resulting in a 25 μm thick layer. After curing, the wafer was exposed with the desired mask and subsequently treated with gamma-(4-fluorophenyl)-gamma-butyrolactone to remove the photoresist from the undesired areas. As a result, grooves with a width of 50 μm and ridges with a width of 20 μm with a height difference of 25 μm were retained (Fig. 2A).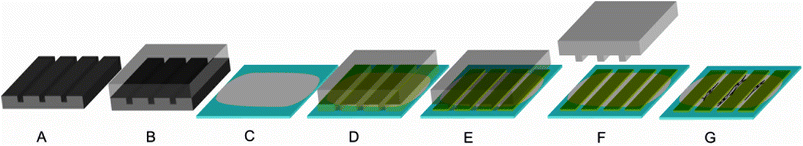 | ||
| Fig. 2 Production of patterned films (the ridges have a width of 50 μm and a height of less than 1 μm, whereas the grooves (i.e. spacing between the ridges) have a width of 20 μm): A: a silicon wafer was used as a template to process a PDMS stamp (B); C: a film was cast on a glass slide to form a ground layer; D: a PDMS stamp was placed on the ground layer protein film; E: a protein solution with a second protein was soaked into the channels of the PDMS stamp by capillary forces; F: after drying, the PDMS stamp was removed, leaving ridges of the second protein; G: cells preferentially adhere and align on the ground layer but not on the ridges. | ||
Then, polydimethylsiloxane (PDMS) moulds were created by using a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of PDMS prepolymer and curing agent. The mixture was degassed for 20 minutes and afterwards poured onto the wafer. The moulds were cured for 90 minutes at 80 °C, peeled from the wafer, and cut into pieces (Fig. 2B).
1 mixture of PDMS prepolymer and curing agent. The mixture was degassed for 20 minutes and afterwards poured onto the wafer. The moulds were cured for 90 minutes at 80 °C, peeled from the wafer, and cut into pieces (Fig. 2B).
A protein ground layer was cast on a glass slide (see Production of films). To deposit a protein stripe (ridge), the PDMS moulds were placed with their structured side downwards on the ground layer films (Fig. 2C and D). A droplet of a 1% (w/v) solution of the second protein in formic acid was deposited at the open side of the channels (the ground layer remains stable as seen when using the same protein as the ground layer and the ridges of the latter peel off (ESI Fig. 1†)). Based on capillary forces the channels were filled with the protein solution (Fig. 2E). After drying, the mould was gently removed (Fig. 2F) and the films were investigated microscopically.
Cell culture experiments
For cell culture experiments BALB/3T3 mouse fibroblasts and C2C12 mouse myoblasts (European Collection of Cell Cultures) were cultured in DMEM media (Biochrom, Berlin, Germany) supplemented with 10% foetal bovine serum (Biochrom, Berlin, Germany), 1% (v/v) GlutaMAX (Gibco, Grand Island, USA), and 0.1% (v/v) gentamicin sulphate (Sigma-Aldrich, Seelze, Germany). An incubator was used to control the atmosphere (5% CO2, 95% humidity) to maintain the cells.The glass slides containing the protein films were placed in six well plates, and 5000 cells per cm2 were seeded per film. The medium was changed once a day. Cells were cultured for up to 96 hours.
Calcein-AM staining
BALB/3T3 fibroblasts were stained with 2 mM calcein acetoxymethyl ester (calcein A/M) (Invitrogen, Eugene, Oregon, USA). Live cells were analyzed with a LeicaDMI3000 B fluorescence microscope after 15 min of incubation at 37 °C (Leica, Wetzlar, Germany).Microscopy
To confirm the existence of the protein ridges and to measure the height of the ridges, fluorescein was coupled to N[AS]8C prior to inflating the mould. Subsequently, the films were observed in a confocal laser scanning microscope (DMI 6000 CS Leica).Cell growth and alignment were observed with a Leica DMI 3000B microscope equipped with a camera. Images were made with and without cells at 48 h and at 96 h after fibroblast seeding.
Picture analysis
To analyse the preferences of the cells to bind to different regions of the protein films, all adhered cells were marked in Powerpoint software by a line across the longest axis of the spread cells symbolising their orientation (e.g. 267 fibroblasts were present on the surface, 218 were spread and marked with a line, ESI Fig. 3†). Afterwards, spread cells were counted on each protein type and the percentage of cells on the bottom layer was calculated.The orientation of the marked cells was measured using ImageJ software. The angle of the cells was measured relative to the groove direction of the film and plotted in 5° steps. The distribution and orientation of the cells were calculated from a minimum of three pictures.
Results and discussion
Employed proteins
To produce stable silk films with distinct topographies, proteins were chosen with different net charges (Fig. 1). N[AS]8C, an engineered protein based on the lacewing egg stalk protein MalXB2, was used as a polycationic silk protein and the spider silk protein eADF4(C16) with or without an RGD-modification as a polyanionic silk protein.5,8Production of flat silk surfaces
The preparation of silk films with a flat surface is straightforward and has been shown previously.22 Here, formic acid was chosen as a solvent for the proteins.23–25 Previously, it was shown that eADF4(C16) films made from formic acid solutions are water-insoluble due to a high β-sheet content enabling direct use in cell culture without further post treatment.24 The water stability of N[AS]8C films made from formic acid solution was confirmed by Coomassie staining of the films after intensive washing with water (data not shown).Fibroblast and myoblast adhesion on flat silk surfaces
BALB/3T3 mouse fibroblasts were seeded on eADF4(C16), ntagCysC16-c(RGDfK), a variant of eADF4(C16) with a chemically coupled, cyclic RGD-peptide,8 and N[AS]8C films respectively.As shown previously, BALB/3T3 fibroblasts adhered weakly on flat eADF4(C16) films, resulting in clustering of the cells (Fig. 3A),5,8 based on the low surface roughness and the lack of adhesion motifs in the primary structure of eADF4(C16).
![A–C: BALB/3T3 mouse fibroblasts cultured on films made of eADF4(C16) (A), ntagCysC16-c(RGDfK) (B) or N[AS]8C (C) with a cell seeding density of 5000 cells per cm2 after 24 hours of incubation. D–F: C2C12 myoblasts cultured on films made of eADF4(C16) (D), ntagCysC16-c(RGDfK) (E) or N[AS]8C (F) with a cell seeding density of 5000 cells per cm2 after 24 hours of incubation. Scale bars: 100 μm.](/image/article/2013/BM/c3bm60114e/c3bm60114e-f3.gif) | ||
| Fig. 3 A–C: BALB/3T3 mouse fibroblasts cultured on films made of eADF4(C16) (A), ntagCysC16-c(RGDfK) (B) or N[AS]8C (C) with a cell seeding density of 5000 cells per cm2 after 24 hours of incubation. D–F: C2C12 myoblasts cultured on films made of eADF4(C16) (D), ntagCysC16-c(RGDfK) (E) or N[AS]8C (F) with a cell seeding density of 5000 cells per cm2 after 24 hours of incubation. Scale bars: 100 μm. | ||
Good adhesion of BALB/3T3 fibroblasts was achieved on flat films of RGD-modified eADF4(C16). On such ntagCysC16-c(RGDfK) films, BALB/3T3 fibroblasts showed a spread morphology (Fig. 3B), and the proliferation rate and doubling time were similar to those of the positive control (treated cell culture plates).8
Films made of the recombinant lacewing egg stalk protein N[AS]8C showed a low surface roughness similar to films made of eADF4(C16), and fibroblast adhesion was very low on such films (Fig. 3C).
Concomitantly, C2C12 myoblasts showed a similar behaviour on all three film types (Fig. 3D–F). On eADF4(C16) films as well as on N[AS]8C films, myoblasts revealed a spherical morphology and adhered weakly to the surface (Fig. 3D and F). The modification of eADF4(C16) with an RGD peptide resulted in spread morphology with improved cell adhesion (Fig. 3E).
Production of patterned films
To promote cell alignment on our silk film surfaces, patterned two-layer films made of various combinations of eADF4(C16), N[AS]8C, and ntagCysC16-c(RGDfK) were processed.The width between the ridges was adjusted to 20 μm, since widths between 10 μm and 20 μm were previously shown to be suitable for cell alignment, and the ridges had a width of 50 μm.15
In order to visualise the film patterns, N[AS]8C was chemically labelled with fluorescein before film formation. Confocal laser scanning microscopy (CLSM) of structured films with the labelled protein revealed a precise alignment of the two protein layers (Fig. 4A) and a thickness of the N[AS]8C layer (i.e. the height of the ridges) of less than one micron (Fig. 4B).
![Fluorescence microscopic images of a patterned film with fluorescein-coupled N[AS]8C protein as ridges (A: top view; B: side view). The gaps between ridges are clearly visible in B.](/image/article/2013/BM/c3bm60114e/c3bm60114e-f4.gif) | ||
| Fig. 4 Fluorescence microscopic images of a patterned film with fluorescein-coupled N[AS]8C protein as ridges (A: top view; B: side view). The gaps between ridges are clearly visible in B. | ||
First a ground layer was made of ntagCysC16-c(RGDfK) with good cell adhesion properties, and ridges were processed thereon using N[AS]8C with low adhesion properties. Next, films with eADF4(C16) as the ground layer and N[AS]8C as ridges were produced (Fig. 1B).
Fibroblast growth on patterned films
Cell adhesion was improved compared to unstructured films due to the rough edges of the structures. The orientation of the cells was measured after 48 hours and revealed a good alignment with the film structures (Fig. 5A). Cells adhered everywhere but with 63.2% of the cells in the grooves (Table 1).
![BALB/3T3 fibroblasts grown on structured films. A: orientation of fibroblasts grown on patterned films made of different protein combinations (ground layer protein/ridge protein) as depicted by the colour code after 48 hours of incubation; B: fluorescence microscopy of calcein AM stained cells, grown on a film with ntagCysC16-c(RGDfK) as the ground layer and N[AS]8C as ridges; C and D: light microscopic image after 48 hours of incubation using ntagCysC16-c(RGDfK) as the ground layer with N[AS]8C as ridges (C) and eADF4(C16) as the ground layer with N[AS]8C as ridges (D).](/image/article/2013/BM/c3bm60114e/c3bm60114e-f5.gif) | ||
| Fig. 5 BALB/3T3 fibroblasts grown on structured films. A: orientation of fibroblasts grown on patterned films made of different protein combinations (ground layer protein/ridge protein) as depicted by the colour code after 48 hours of incubation; B: fluorescence microscopy of calcein AM stained cells, grown on a film with ntagCysC16-c(RGDfK) as the ground layer and N[AS]8C as ridges; C and D: light microscopic image after 48 hours of incubation using ntagCysC16-c(RGDfK) as the ground layer with N[AS]8C as ridges (C) and eADF4(C16) as the ground layer with N[AS]8C as ridges (D). | ||
| ntagCysC16-c(RGDfK)/N[AS]8C | eADF4(C16)/N[AS]8C | eADF4(C16)/eADF4(C16) | |
|---|---|---|---|
| Cells in the grooves | 85.5% | 78.8% | 63.2% |
| Cells standardised per area in the grooves | 94.2% | 91.5% | 85.3% |
Cell adhesion on N[AS]8C films could not be quantified, since only a few cells adhered and were oriented with the axis of the ridges.
ntagCysC16-c(RGDfK) structured films showed no improved adhesion in comparison to unstructured ones due to the already high level of adhesion on the flat films.
Unfortunately, structured films made of one protein by this technique are highly unstable due to the identical charge of the proteins in both layers, and, therefore, the ridges peel off easily (ESI Fig. 1†).
On such films, 85.5% of the fibroblasts adhered on the ground layer (94.2% when standardised per area due to the 2.5 times larger area of the top layer), and most of the cells aligned along the axis of the ridges (Table 1, Fig. 5A, 5C and ESI Fig. 2†). Even the few cells adhering to the N[AS]8C ridges were mostly aligned.
Surprisingly, control experiments using eADF4(C16) instead of ntagCysC16-c(RGDfK) as the ground layer revealed similar results (Fig. 5D), with much higher cell adhesion numbers than for eADF4(C16)/eADF4(C16) films. The fibroblasts mostly grew on the eADF4(C16) area (78.8% of the fibroblasts (91.5% calculated per area)) and not on the N[AS]8C ridges (Table 1). Cell alignment was pronounced as well (Fig. 5A) accentuating the importance of substrate morphology for cell attachment. The adhesion of fibroblasts on eADF4(C16) films is much better on structured films than on flat ones (Fig. 3A), probably due to the microstructure at the interface of the two layers, which results from drying effects. In contrast, N[AS]8C seems to be a good ridge material as it is not a good substrate for cells, and, therefore, it directs cell adhesion to the desired ground layer areas. Images after 96 hours showed that cells proliferated well on the films and stayed on the ground protein layer as desired (Fig. 6A).
![A: BALB/3T3 fibroblasts grown on structured films using eADF4(C16) as the ground layer and N[AS]8C as ridges after 96 hours of incubation; B: C2C12 myoblasts grown on eADF4(C16)/N[AS]8C films after 48 hours of incubation.](/image/article/2013/BM/c3bm60114e/c3bm60114e-f6.gif) | ||
| Fig. 6 A: BALB/3T3 fibroblasts grown on structured films using eADF4(C16) as the ground layer and N[AS]8C as ridges after 96 hours of incubation; B: C2C12 myoblasts grown on eADF4(C16)/N[AS]8C films after 48 hours of incubation. | ||
In summary, structures are important for orientation of cells even if they are less than 1 μm in height (Fig. 4B).
Myoblast growth on patterned films
C2C12 myoblasts grown on patterned films made of ntagCysC16-c(RGDfK)/N[AS]8C and eADF4(C16)/N[AS]8C behaved similar to fibroblasts (Fig. 6B). C2C12 myoblasts mostly grew on the spider silk ground layers and not on the N[AS]8C ridges.It was impossible to calculate the cell angle and distribution due to the high cell density after 48 hours and the formation of myotubes.
The formation of myotubes in an aligned manner, however, is an important step towards skeletal muscle regeneration.
Conclusion
The beneficial influence of patterned scaffolds on fibroblast and myoblast adhesion and alignment was shown by our experiments with patterned films using one single protein. Fibroblasts as well as myoblasts adhere and proliferate much better on structured than on unstructured films made of the same protein. A width of 20 μm between ridges with a height of 1 μm is suitable to induce fibroblast as well as myoblast alignment in parallel to the grooved pattern (Fig. 5 and 6). On patterned eADF4(C16) films, fibroblasts grew on the ridges and in the grooves with slight preferences for the grooves. The deposition of N[AS]8C as stripe protein (ridges) with low fibroblast and myoblast adhesion properties increased cell binding in the grooves. Surprisingly, ntagCysC16-c(RGDfK) as the ground layer did not result in a significantly better cell binding compared to an eADF4(C16) ground layer. The unspecific cell adhesion, induced by surface roughness and structure, leads to strong cell adhesion in the case of the polyanionic spider silk proteins, but not in the case of the polycationic egg stalk silk protein. Surprisingly, in this context the specific interaction of the cells with RGD did not show significant benefits for patterned spider silk films.In the future, our results will have an impact on patterning of silk coatings e.g. for bone grafts, muscle cell cultures, or nerve guidance/regeneration.13,14
Acknowledgements
The authors thank Aniela Heidebrecht for the CLSM images, Martin Trebbin for the silicon wafer template and Martin Humenik for critically reading the manuscript. This work was supported by the DFG SCHE603/4 and the Bavarian State Ministry of the Environment and Public Health (U8793-2012/6-2).Notes and references
- M. Heim, L. Romer and T. Scheibel, Chem. Soc. Rev., 2010, 39, 156–164 RSC.
- A. Heidebrecht and T. Scheibel, Adv. Appl. Microbiol., 2013, 82, 115–153 CrossRef CAS.
- J. G. Hardy, L. M. Romer and T. R. Scheibel, Polymer, 2008, 49, 4309–4327 CrossRef CAS.
- J. G. Hardy and T. R. Scheibel, J. Polym. Sci., Polym. Chem., 2009, 47, 3957–3963 CrossRef CAS.
- A. Leal-Egana, G. Lang, C. Mauerer, J. Wickinghoff, M. Weber, S. Geimer and T. Scheibel, Adv. Eng. Mater., 2012, 14, B67–B75 CrossRef.
- E. S. Gil, B. B. Mandal, S. H. Park, J. K. Marchant, F. G. Omenetto and D. L. Kaplan, Biomaterials, 2010, 31, 8953–8963 CrossRef CAS.
- E. Bini, C. W. Foo, J. Huang, V. Karageorgiou, B. Kitchel and D. L. Kaplan, Biomacromolecules, 2006, 7, 3139–3145 CrossRef CAS.
- S. Wohlrab, S. Muller, A. Schmidt, S. Neubauer, H. Kessler, A. Leal-Egana and T. Scheibel, Biomaterials, 2012, 33, 6650–6659 CrossRef CAS.
- A. Leal-Egana and T. Scheibel, J. Mater. Chem., 2012, 22, 14330–14336 RSC.
- E. S. Gil, S. H. Park, J. Marchant, F. Omenetto and D. L. Kaplan, Macromol. Biosci., 2010, 10, 664–673 CrossRef CAS.
- J. Torbet, M. Malbouyres, N. Builles, V. Justin, M. Roulet, O. Damour, A. Oldberg, F. Ruggiero and D. J. Hulmes, Biomaterials, 2007, 28, 4268–4276 CrossRef CAS.
- B. D. Lawrence, J. K. Marchant, M. A. Pindrus, F. G. Omenetto and D. L. Kaplan, Biomaterials, 2009, 30, 1299–1308 CrossRef CAS.
- D. Yucel, G. T. Kose and V. Hasirci, Biomacromolecules, 2010, 11, 3584–3591 CrossRef CAS.
- D. Yucel, G. T. Kose and V. Hasirci, Biomaterials, 2010, 31, 1596–1603 CrossRef CAS.
- J. L. Charest, A. J. Garcia and W. P. King, Biomaterials, 2007, 28, 2202–2210 CrossRef CAS.
- J. T. Zhang, J. Q. Nie, M. Muhlstadt, H. Gallagher, O. Pullig and K. D. Jandt, Adv. Funct. Mater., 2011, 21, 4079–4087 CrossRef CAS.
- S. Fujita, M. Ohshima and H. Iwata, J. R. Soc. Interface, 2009, 6((Suppl 3), S269–S277 CrossRef.
- B. Zhu, Q. Lu, J. Yin, J. Hu and Z. Wang, Tissue Eng., 2005, 11, 825–834 CrossRef CAS.
- T. C. McDevitt, K. A. Woodhouse, S. D. Hauschka, C. E. Murry and P. S. Stayton, J. Biomed. Mater. Res. A, 2003, 66, 586–595 Search PubMed.
- A. W. Feinberg, A. Feigel, S. S. Shevkoplyas, S. Sheehy, G. M. Whitesides and K. K. Parker, Science, 2007, 317, 1366–1370 CrossRef CAS.
- H. Fujita, K. Shimizu and E. Nagamori, Biotechnol. Bioeng., 2009, 103, 1034–1041 CrossRef CAS.
- D. Huemmerich, U. Slotta and T. Scheibel, Appl. Phys. A: Mater., 2006, 82, 219–222 CrossRef CAS.
- K. Spiess, S. Wohlrab and T. Scheibel, Soft Matter, 2010, 6, 4168–4174 RSC.
- K. Spiess, R. Ene, C. D. Keenan, J. Senker, F. Kremer and T. Scheibel, J. Mater. Chem., 2011, 21, 13594–13604 RSC.
- S. Wohlrab, K. Spiess and T. Scheibel, J. Mater. Chem., 2012, 22, 22050–22054 RSC.
- D. Huemmerich, C. W. Helsen, S. Quedzuweit, J. Oschmann, R. Rudolph and T. Scheibel, Biochemistry, 2004, 43, 13604–13612 CrossRef CAS.
- F. Bauer and T. Scheibel, Angew. Chem., Int. Ed., 2012, 51, 6521–6524 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c3bm60114e |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2013 |
