DOI:
10.1039/C3BM60123D
(Paper)
Biomater. Sci., 2013,
1, 1223-1234
Molecular weight and architectural dependence of well-defined star-shaped poly(lysine) as a gene delivery vector†
Received 10th May 2013, Accepted 18th July 2013
First published on 7th August 2013
Abstract
A series of well-defined star-shaped polypeptides were successfully synthesised by the ring opening polymerisation (ROP) of the N-carboxyanhydride (NCA) of ε-carbobenzyloxy-L-lysine (ZLL) using a range of generations of polypropylene imine (PPI) dendrimers as multifunctional initiators. The monomer feed ratio and dendrimer generation were varied to afford a series of polypeptide dendrimer hybrids with superior structural versatility and functionality. Subsequent protecting group removal yielded star-shaped poly(lysine) of controlled variation in polypeptide chain length and arm multiplicity. Star-shaped PLL polymers were used to prepare pDNA and siRNA to form “polyplexes” to determine their ability to complex different nucleic acid cargoes and were compared with linear PLL polyplex controls. Significant differences in size and surface charge were seen between star-shaped PLL polyplexes and linear PLL polyplexes for both cargoes. The star-shaped polypeptides were capable of more effective complexation of both nucleic acids at low N/P ratios compared to linear PLL as evidenced by zeta potential and electrophoretic data. This was particularly evident in siRNA polyplexes as linear PLL failed to completely complex siRNA into nanocomplexes of appropriate size for cell transfection i.e. <200 nm in size, while star poly(lysine) formed siRNA polyplexes <100 nm at certain N/P ratios, albeit strongly dependent on the particular molecular weight and architecture, as analysed by dynamic light scattering (DLS). Atomic force microscopy (AFM) identified discrete spherically shaped polyplexes for all star-shaped polypeptide-based polyplexes while linear PLL formed elongated irregular shaped complexes. This difference in morphology may go some way towards explaining the 300-fold increase in luciferase expression seen for star-shaped PLL polyplexes G5(64)-PLL40 compared to linear PLL pGLuc polyplexes in epithelial cells. Each of the PPI-PLL polymers appeared to be capable of protecting the nucleic acid cargoes from degradation by the relevant nuclease enzyme as effectively as the positive control polyethyleneimine (PEI) polyplexes. Overall the promising nucleic acid complexation, sizing, morphology and protection capacity of two different genetic “cargoes” highlight the potential of polypeptide dendrimer hybrids as gene delivery vectors.
Introduction
Gene delivery is an area which has generated immense interest in recent times.1,2 The ability to use genetic material such as plasmid DNA, siRNA and microRNA to better understand and potentially treat and prevent diseases that currently lack or have poor therapeutic or prophylactic modalities is very appealing.3 However the inability to deliver these nucleic acid-based “drugs” in a safe and efficient manner remains a significant challenge to their commercial and clinical translation. These biomolecules are large, polyanionic structures that can be easily degraded by enzymes and do not readily cross the target cell membrane to reach their intracellular sites of action. The use of viral and non-viral vectors has helped to overcome some of these hurdles but major obstacles remain.4 Although very efficient, viral delivery vectors possess the inherent immunogenic, safety and production concerns associated with them.5 Consequently the use of cationic non-viral delivery vectors has been extensively explored with natural and synthetic polymers including poly-L-lysine (PLL),6 chitosan,7 dimethylaminoethyl methacrylate (DMAEMA),8 poly(ethyleneimine) (PEI)9 and various dendrimers10,11 being harnessed for nucleic acid delivery.The concept of dendrimer facilitated gene therapy is of particular interest arising from the unique properties associated with these materials.12 Dendrimers are monodisperse and at higher generations are globular macromolecules that offer numerous functional groups within a highly defined compact structure. Some of the best-known cationic dendrimers include polypropylene imine (PPI) and poly(amido amine) (PAMAM) which contain peripheral ionisable primary amino groups, the number of which can be varied depending on the generation of dendrimers chosen. This cationic periphery permits the formation of electrostatically derived nanoparticle complexes with nucleic acids such as pDNA, whilst the inner tertiary amines enhance endosomal buffering capacity and therefore transfection.13 However some of the major hurdles, which have limited their use in biomedical applications to date is their inherent cytotoxicity, potential immunogenicity and size limitations.14
Star-shaped polymers provide an attractive alternative as unimolecular containers because the molecular weight and the number of polymeric arms can be tightly controlled by the choice of the polymerisation method and the multifunctional initiator.15–17 This facilitates manipulation of the shape and the cargo space available for drug loading. Particularly attractive are polypeptide based star polymers due to their improved biocompatibility, functionality and the ability to introduce stimuli responsiveness when required through inclusion of specific amino acid sequences, whilst maintaining a well-defined globular shape.18,19 A convenient method of polypeptide introduction is via N-carboxyanhydride (NCA) polymerisation which permits the versatile and well controlled growth of long chain polypeptides from a (multi) amino terminated initiator.20–30 NCA derived polypeptides have already demonstrated their potential biomedical applicability in areas such as tissue engineering,31 drug/gene delivery,32–35 medical adhesives,36 antimicrobial agents,37 and biorecognition applications.38–45
Cationic polypeptides such as linear poly(lysine) have also been investigated as a pDNA delivery vector owing to its cationic nature and potential biocompatibility.46,47 Such complexes, however, exhibited low transfection efficiencies and elevated cytotoxicity thus preventing their clinical application.48 The incorporation of poly(lysine) into a branched or star-shaped architecture may overcome these issues by creating a unimolecular nanoparticle with increased cargo capacity and protection. Pan et al. have reported the growth of short poly(lysine) chains from a fourth generation PAMAM dendrimer and their use as a pDNA transfection agent.49 These materials showed enhanced transfection efficiencies with reduced cytotoxicity thus highlighting the potential of such polypeptide–dendrimer hybrids. Dendritic poly(lysine) has also shown promise as a pDNA transfection agent but such materials are synthetically challenging and lack the structural versatility of a star-shaped polypeptide derived from the NCA/multifunctional initiator approach.50,51 We have recently reported the versatile synthesis of pH responsive star poly(glutamic acid) using various generations of PPI dendrimers as multifunctional initiators for NCA polymerisation.30 By this approach the number of arms and the total molecular weight of the star polypeptides were systematically varied. Herein, we report the synthesis and systematic study of a range of well-defined star-shaped lysine polymers of controlled molecular weights. We further investigate the potential of these materials as gene delivery vectors using pDNA and siRNA as suitable model cargoes. pDNA and siRNA are polyanionic biomolecules that present several different challenges in terms of their delivery to a cell.52 Structurally, the much larger pDNA readily collapses into compact nanoparticles with a polycationic vector whereas siRNA carrying much less negative charge tends to form weaker complexes. As a consequence it was interesting to determine if these star-shaped polypeptides were suitable as vectors for both cargoes in terms of their ability to efficiently package and protect these cargoes. The size, shape and morphology of these nanomaterials were determined by dynamic light scattering (DLS) and atomic force microscopy (AFM) in conjunction with ζ-potential and agarose/PAGE gel electrophoresis to measure complexation efficiency and stability. The star-shaped polypeptides were shown to effectively package and protect both pDNA and siRNA, and preliminary transfection studies compared their capacity to transfect pDNA into cells compared to linear PLL.
Experimental
Materials
All air and moisture sensitive compounds were handled under a nitrogen atmosphere using general Schlenk-line techniques. α-Pinene (98%) bis(trichloromethyl) carbonate (triphosgene) 99% and benzylamine were purchased from Sigma Aldrich. H-Lys(Z)-OH were supplied by Bachem. PPI (polypropylene imine) dendrimers generations 2–5 were purchased from SyMO-Chem BV (The Netherlands). Chloroform (anhydrous), ethyl acetate (anhydrous), dimethylformamide, tetrahydrofuran, n-heptane, and diethyl ether were supplied by Sigma Aldrich. All chemicals were used without any purification unless otherwise noted. Chloroform and ethyl acetate were used directly from the bottle and stored under an inert, dry atmosphere. ε-Carbobenzyloxy-L-lysine NCA was synthesised by following a literature procedure.53 siGENOME non-targeting siRNA #4 (5′ AUGAACGUGAAUUGCUCAA 3′) was obtained from Dharmacon and the pGL3-control vector plasmid was sourced from Promega.Star poly-ε-carbobenzyloxy-L-lysine ((G2-5) PPI-PZLL)
As a reference procedure, the NCA of ε-carbobenzyloxy-L-lysine (ZLL) (2.02 g, 6.6 mmol) was dissolved in 25 mL CHCl3 in a Schlenk tube under a nitrogen atmosphere. A solution of G2 PPI dendrimer (30 mg, 2.07 × 10−2 mmol) in 2 mL CHCl3 was quickly charged to the dissolved NCA solution via a syringe. The solution was stirred for 24 h at room temperature. Depending on the type of the initiator, the used molar amounts of the initiator and the monomer were adjusted to achieve the desired molecular weight of the star-shaped polypeptides. The polymer was precipitated into an excess of cold diethyl ether and dried under vacuum (yield: 95%).Star poly-L-lysine (PPI-PLL)
G2(8)-PZLL40 (1.2 g) was dissolved in 12 mL of trifluoroacetic acid (TFA). A 6-fold excess with respect to ε-carbobenzyloxy-L-lysine of a 33% solution of HBr in acetic acid (5 mL) was added slowly to the reaction mixture. After 16 h, the solution was precipitated into diethyl ether. The precipitate was redissolved in ethanol and precipitated thrice into diethyl ether. The polymer was dissolved in deionised water and dialyzed (MWCO 8000) for 3 days. The polymer was lyophilized. Yield: 68%. Deprotection was confirmed by 1H NMR due to the absence of signals at 7.2 ppm (benzyl group) and 5.0 ppm (CH2) (Fig. S-1 ESI†).Linear poly-ε-carbobenzyloxy-L-lysine (L-PLL)
In a Schlenk tube under a nitrogen atmosphere, a solution of the NCA of ε-carbobenzyloxy-L-lysine (ZLL (0.92 g, 3 mmol) in CHCl3 (30 mL) was prepared. A solution of benzylamine initiator (1 mg, 9.33 × 10−3 mmol) in 1 mL CHCl3 was also prepared and charged to the reaction solution via a syringe. The solution was stirred at room temperature until no further increase in molecular weight was achieved. The molecular weight was monitored by SEC by precipitating 1 mL (63 mg) of the reaction solution into an excess of cold diethyl ether. Upon no increase in molecular weight, the polymer was precipitated into an excess of cold diethyl ether and dried under vacuum (yield: 70%). For the deprotection the same procedure as for PPI-PZLL was applied (yield: 70%).Methods
Nuclear magnetic resonance (NMR) spectra were recorded using a Bruker Avance 400 (400 MHz) spectrometer at room temperature in CDCl3 and d-TFA as solvents. Attenuated total reflection (ATR) FTIR measurements were performed using a Perkin-Elmer Spectrum 100 in the spectral region of 650–4000 cm−1 and were obtained from 4 scans with a resolution of 2 cm−1. A background measurement was taken before the sample was loaded onto the ATR for measurement. Size exclusion chromatography (SEC) was performed using an Agilent 1200 system in conjunction with two PSS GRAM analytical (8 × 300 and 8 × 100, 10 μ) columns, a Wyatt Dawn Heleos 8 multi angle light scattering detector (MALS) and a Wyatt Optilab rEX differential refractive index detector (DRI) with a 658 nm light source. The eluent was DMF containing 0.1 M LiBr at a flow rate of 1 mL min−1. The column temperature was set to 40 °C with the MALS detector at 35 °C and the DRI detector at 40 °C. Molar masses and dispersities were calculated from the MALS signal by the Astra software (Wyatt) using the refractive index increment (dn/dc) of linear poly-(ε-carbobenzyloxy-L-lysine) (PZLL) of 0.101.54 All samples for GPC analysis were prepared with a concentration of 2 mg mL−1 and were filtered through a 0.45 μm PTFE filter (13 mm, PP housing, Whatman) prior to injection.Plasmid preparation
The pGL3-control vector plasmid (Promega) consists of 5256 bp and contains the firefly luciferase gene and an ampicillin resistance gene that are controlled by an SV40 promoter/enhancer. The plasmids were replicated in the high-copy DH5-α Escherichia coli strain grown in selective ampicillin (50 μg mL−1) supplemented Luria–Bertani medium, isolated by alkaline lysis followed by anion exchange chromatography using the Giga Qiagen kit (Qiagen) according to the manufacturer's protocol. The purity of the plasmid and the integrity of the cDNA insert were determined by agarose gel electrophoresis and UV spectroscopy (E 260/280 nm ratio). After isolation, the pDNA was dissolved to an end concentration of 1.2 μg mL−1 Tris HCl buffer (pH 8.0). The purity was assayed by 1% agarose gel electrophoresis. This was then used in the preparation of the pDNA polyplexes.Preparation of siRNA and pDNA polyplexes
siGENOME non-targeting siRNA #4 (5′ AUGAACGUGAAUUGCUCAA 3′) was obtained from Dharmacon and used in the preparation of the polyplexes. The siRNA/pDNA star-shaped PLL polyplexes were freshly prepared prior to use. The ratio of dendrimer to pDNA/siRNA in the polyplexes was calculated according to the molar ratio of the amino group in the dendrimer to the phosphate group in pDNA/siRNA, i.e., the N![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P ratio. To prepare the polyplexes, solutions of G2-PPI-PLL or G5-PPI-PLL prepared in buffer (Tris HCl buffer pH 7.4 and 10 mM NaCl) were mixed with pDNA/siRNA at various N
P ratio. To prepare the polyplexes, solutions of G2-PPI-PLL or G5-PPI-PLL prepared in buffer (Tris HCl buffer pH 7.4 and 10 mM NaCl) were mixed with pDNA/siRNA at various N![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P ratios (1, 5, 10, 20 and 100). These mixtures were incubated at room temperature for 30 min for polyplex formation, diluted and analysed.
P ratios (1, 5, 10, 20 and 100). These mixtures were incubated at room temperature for 30 min for polyplex formation, diluted and analysed.Particle size and zeta potential analysis
The polyplexes were prepared in triplicate at various N![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P ratios as described, and their particle size and surface charge (ζ-potential) were measured using a Malvern Zetasizer Nano ZS (Malvern Instruments, Ltd, United Kingdom).
P ratios as described, and their particle size and surface charge (ζ-potential) were measured using a Malvern Zetasizer Nano ZS (Malvern Instruments, Ltd, United Kingdom).Atomic force microscopy (AFM)
Particle size and morphology were also determined using atomic force microscopy (AFM, Asylum MFP-3D-BIO, DIT Focas Institute, Dublin, Ireland). For AFM, the polyplexes were prepared at the N![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P ratio of 20 by adding the polypeptide solution to pDNA/siRNA to give a final pDNA/siRNA concentration of 10 ng μL−1. After incubation at room temperature for 30 min, 2 μL of the complex solution was diluted into 198 μL H2O and deposited onto the surface of freshly cleaved mica sheets (Agar Scientific, G250-3, 11 × 11 mm mica). After 3 min, excess solution was removed by careful absorption onto filter paper (Whatman, Fisher Scientific, Ireland) and the mica was further dried at room temperature for 24 h before analysis by AFM. Samples were imaged using an MFP-3D BIO AFM (Asylum Research). Micromasch silicon NSC18 cantilevers were used. Tips were 225 nm long and had a typical resonant frequency of 75 kHz. The AFM operated in the AC mode (alternate contact) in order to minimise the tip sample interaction. Typical free air amplitudes were ∼800 mV and a high amplitude set-point relative to the set-point was maintained to minimise cell/tip damage. All samples were imaged in air at relative humidity. The images obtained contained 512 pixels per scan line.
P ratio of 20 by adding the polypeptide solution to pDNA/siRNA to give a final pDNA/siRNA concentration of 10 ng μL−1. After incubation at room temperature for 30 min, 2 μL of the complex solution was diluted into 198 μL H2O and deposited onto the surface of freshly cleaved mica sheets (Agar Scientific, G250-3, 11 × 11 mm mica). After 3 min, excess solution was removed by careful absorption onto filter paper (Whatman, Fisher Scientific, Ireland) and the mica was further dried at room temperature for 24 h before analysis by AFM. Samples were imaged using an MFP-3D BIO AFM (Asylum Research). Micromasch silicon NSC18 cantilevers were used. Tips were 225 nm long and had a typical resonant frequency of 75 kHz. The AFM operated in the AC mode (alternate contact) in order to minimise the tip sample interaction. Typical free air amplitudes were ∼800 mV and a high amplitude set-point relative to the set-point was maintained to minimise cell/tip damage. All samples were imaged in air at relative humidity. The images obtained contained 512 pixels per scan line.Gel retardation assay
The pDNA polyplexes were prepared as outlined above, mixed with a 6× loading dye (Promega G190A-Blue/orange 6×) and analysed by running 1% (w/v) agarose gel electrophoresis at 100 V for 1 h in TBE buffer (Tris–borate–EDTA buffer, Gibco, Biosciences, Ireland) with 0.5 μg mL−1 ethidium bromide. Visualisation was obtained by UV transillumination (G. Box, Syngene, UK). The siRNA polyplexes were also prepared as before, mixed with loading dye (Promega G190A-Blue/orange 6×) and analysed by running 20% non-denaturing polyacrylamide gel in TBE buffer at 100 V for 1 h. The gel was then stained upon completion using Gelstar nucleic acid gel stain for 30 min. Visualisation was then obtained by UV transillumination (G. Box, Syngene, UK).DNase I protection assay
DNase I protection assay was performed to investigate the ability of star-shaped polypeptides to protect pDNA from endonuclease degradation. Samples of polyplexes were prepared as before and diluted to 10 μL, and 1.5 μL of 10× reaction buffer (as supplied) was then added. Samples were treated with 1 μg of DNase I (1 U μL−1, Fermentas) at 37 °C for 30 min. 2 μL of EDTA (50 mM) and 8 μL of sodium dodecyl sulfate (SDS, 1.25% w/v) were then added and the samples were incubated at 37 °C for a further 2 hours to allow complete DNA dissociation from complexes. Prior to running 1% agarose gel electrophoresis as before, 6× loading dye (Promega G190A-Blue/orange 6×) was added to these samples and the amount of pDNA released from the complexes was then visualized by UV transillumination (G. Box, Syngene, UK). Naked pDNA with or without DNase I treatment were used as positive and negative controls.RNase protection assay
RNase protection assay was performed to investigate the ability of star-shaped polypeptides to protect siRNA from endonuclease degradation. Samples of polyplexes were prepared as before and diluted to 10 μL. Samples were treated with 1 μg of RNase (1 U μL−1, Fermentas) and 1 μL of EDTA (50 mM) at 37 °C for 30 min. 8 μL of sodium dodecyl sulfate (SDS, 1.25% w/v) were then added and the samples were incubated at 37 °C for a further 2 hours to allow complete siRNA dissociation from complexes. The polyplexes were analysed using a 20% non-denaturing polyacrylamide gel in TBE buffer at 100 V for 1 h. The gel was then stained upon completion using Gelstar nucleic acid gel stain for 30 min. The amount of siRNA released from the complexes was then visualized by UV transillumination (G. Box, Syngene, UK). Naked siRNA with or without RNase treatment was used as positive and negative controls.pDNA transfections of Calu-3 cells using linear and star-shaped G5(64)-PLL40 polyplexes
Non differentiated airway epithelial cells (Calu-3 cells) were seeded at a density of 5 × 104 cells per well in 48-well plates 24 h prior to transfection. The reporter gene pGaussia luciferase was used for the subsequent transfections and the plasmid was isolated using a Qiagen maxi prep kit as per the manufacturer's instructions. Polyplexes of linear PLL at N/P 2 and G5(64)-PLL40 at various N/P ratios were prepared by mixing the star-shaped polypeptide G5(64)-PLL40 in 100 μL Opti-MEM and incubated at room temperature for 30 min. The cells were then transfected with these polyplexes for 4 hours. After 4 h, 150 μL of serum-containing media was added to each well and incubated for 24, 120, and 168 hours respectively at 37 °C and 5% CO2. Following this, luciferase activity of each well was analysed using a luciferase assay kit (Thermo Fischer Scientific) as per manufacturers instructions.Results and discussion
Synthesis of star poly(lysine)
The ROP of α-amino acid NCAs via primary amines is a well understood method for the synthesis of well defined synthetic linear polypeptides.55–57 Building on previous work conducted in our group, the use of multifunctional amino terminated dendrimers was employed to synthesise well defined star-shaped poly(lysine).30 By employing four different generations of PPI dendrimers and varying the monomer feed ratio of ε-carbobenzyloxy-L-lysine (ZLL) NCA it was possible to create a star-shaped poly(lysine) whose size, shape, arm length and molecular weight could be readily controlled (Fig. 1). To develop a systematic library of star polypeptides and demonstrate the versatility of the system, polymers with a similar polypeptide arm length (40 monomer units), but different degrees of branching (Table 1, entries 2–5) were synthesized from the different generations of PPI dendrimers (e.g. G5(64)-PZLL40). 1H NMR (S-1) confirmed the successful synthesis of PZLL and its conjugation to the dendrimer although PPI dendrimer peaks were difficult to observe for the higher molecular weight polymers due to overlapping with the broad polymer peaks and its low concentration in comparison to the high density of PZLL. Furthermore inaccessibility of the hydrophobic PPI core to D2O may account for the lack of visible peaks for PPI in 1H NMR. It was thus not possible to confirm NCA initiation from all of the primary amino sites although the agreement with the expected linear increase of molecular weights with increasing dendrimer generation suggests high initiation efficiency (Fig. 2). This library of materials was further completed by a 64 arm PZLL with only 5 repeating units per arm (Table 1, entry 6; G5(64)-PZLL5) and thus comparable to G2(8)-PZLL40 in total molecular weight yet with more and shorter arms. Finally, a linear PZLL was synthesised (Table 1, entry 1; L(1)-PZLL320). Deprotection of the described polymers afforded linear and star-shaped poly(lysine) possessing amino functionalities. Quantitative deprotection was confirmed by 1H NMR and FTIR (Fig. S3 and S4†). Polymers 1–6 were subsequently investigated as gene delivery vectors.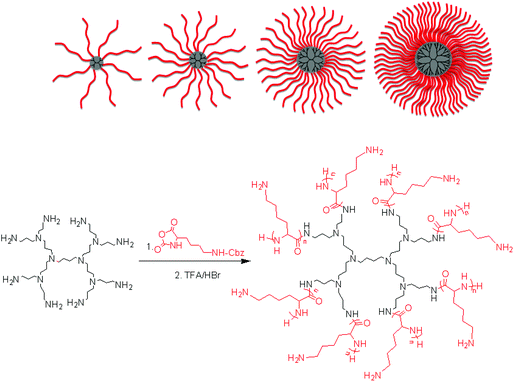 |
| | Fig. 1 Synthesis of poly(L-lysine) star polypeptides by NCA polymerisation using second to fifth generation PPI dendrimers as initiators. | |
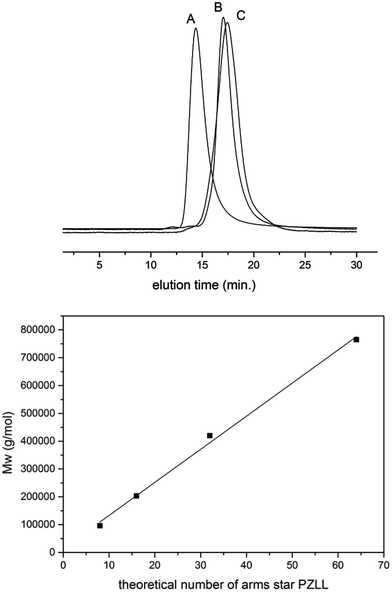 |
| | Fig. 2 SEC traces of selected star-shaped and linear poly-ε-carbobenzyloxy-L-lysine (A: G5(64)-PZLL40; B: G2(8)-PZLL40; C: G5(64)-PZLL5) and weight average molar mass (Mw) of star-shaped PZLL obtained by initiation of NCA from different generations of PPI dendrimers. The line represents a linear fit. | |
Table 1 Star-shaped PZLL by initiation from PPI dendrimers (L: linear initiator benzylamine; G5(64)-PZLL40 = initiator generation 5 dendrimer with maximum 64 arms and a theoretical arm length of 40 amino acids). Dispersities of all polymers < 1.2 (SEC MALS detection)
| Entry | Polymer | NCA/NH2 | NCA/dendrimer | Mw/g mol−1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a a | Mnth![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) b/g mol−1 b/g mol−1 |
|---|
| Determined by SEC (DMF) using MALS detection using the dn/dc of linear PZLL of 0.101. Calculated assuming initiation from all amino groups and quantitative conversion: c(monomer)/c(dendrimer) × M(monomer) + M(dendrimer). |
|---|
| 1 | L(1)-PZLL320 | 320 | 320 | 69![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 | 83![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 800 |
| 2 | G2(8)-PZLL40 | 40 | 320 | 96![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 | 83![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 800 |
| 3 | G3(16)-PZLL40 | 40 | 640 | 203![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 | 167![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 700 |
| 4 | G4(32)-PZLL40 | 40 | 1280 | 420![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 | 335![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 400 |
| 5 | G5(64)-PZLL40 | 40 | 2560 | 765![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 | 670![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 700 |
| 6 | G5(64)-PZLL5 | 5 | 320 | 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 | 83![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 800 |
Particle size, surface charge and morphology of pDNA-PLL polyplexes
ζ-Potential analysis (Fig. 3 and S5†) demonstrated that the star-shaped PLL could successfully form cationic polyplexes with pDNA at N/P 2–5 whereas linear PLL required higher ratios of N/P 10–20 to neutralise the polyanionic charge of the pDNA. No noticeable effect on pDNA complexation was observed between the different star-shaped architectures. For successful pDNA transfection, nanoparticles <200 nm are required. Nanoparticles with sizes greater than 200 nm will be readily removed from the body by the reticuloendothelial system (RES)58 and will not be efficiently endocytosed by cell membranes. The particle size (Fig. 4 and S6†) of cationic pDNA polyplexes formed was investigated over a range of N/P ratios using dynamic light scattering (DLS). Around charge neutrality, i.e. complete pDNA neutralisation (as indicated by a ζ-potential around 0 mV in Fig. 3 and S5†), large sized pDNA polyplexes were observed which would be expected due to a lack of colloidal stability. For pDNA it was observed that at N/P 5–100, G2(8)-PLL40 and G5(64)-PLL5 could form nanoparticles <100 nm in size with the densely branched, long armed G3(16), G4(32) and G5(64)-PLL40 forming slightly larger nanoparticles over this N/P ratio range. Linear PLL could also compact pDNA into nanoparticles <200 nm but only at higher N/P ratios (N/P >10) than the star-shaped PLL. Atomic force microscopy was used to investigate the morphology of the pDNA polyplexes as the shape of the complexes can impact on gene delivery efficiency.59 It was observed that star-shaped PLL could efficiently complex pDNA into discrete, compact, spherically shaped nanoparticles with sizes in the range 50–150 nm (Fig. 5 and S7†). In comparison linear PLL/pDNA complexes were irregular and elongated in shape. pDNA-L(1)-PLL320 polyplexes displayed sizes >200 nm, supporting the DLS data. These irregular and large polyplexes would not be expected to facilitate efficient gene transfection.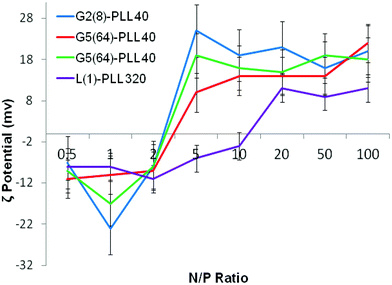 |
| | Fig. 3 ζ-Potential of G2(8)-PLL40, G5(64)-PLL40, G5(64)-PZLL5 and L(1)-PLL320 polypeptide/pDNA polyplexes over a range of N/P ratios. | |
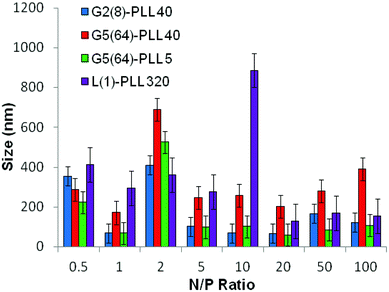 |
| | Fig. 4 Particle size of G2(8)-PLL40, G5(64)-PLL40, G5(64)-PZLL5 and L(1)-PLL320 polypeptide/pDNA polyplexes over a range of N/P ratios. | |
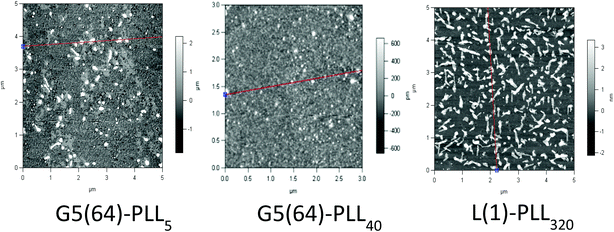 |
| | Fig. 5 Atomic force microscopy of G5(64)-PZLL5, G5(64)-PLL40, and L(1)-PLL320 polypeptide/pDNA polyplexes at N/P 20. | |
Particle size, surface charge and morphology of siRNA-PLL polyplexes
siRNA are much shorter chain oligonucleotides and therefore interact with and form very different polyplexes to those fabricated with pDNA. L(1)-PLL320 failed to effectively complex siRNA even at very high PLL![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) siRNA N/P ratios up to 300 (Fig. 6). The inability of linear PLL to neutralise siRNA and form a discrete compact cationic nanostructure clearly relates to its linear architecture as star-shaped polypeptides very effectively neutralised the polyanionic siRNA at low N/P ratios <5. The molecular weight and architecture had a significant impact on the overall surface charge of the siRNA polyplexes formed. For example G2(8)-PLL40 and G5(64)-PLL5, with similar molecular weights, both form cationic siRNA polyplexes at N/P 3 whereas the much larger G3(16), G4(32) and G5(64)-PLL40 required a higher polymer
siRNA N/P ratios up to 300 (Fig. 6). The inability of linear PLL to neutralise siRNA and form a discrete compact cationic nanostructure clearly relates to its linear architecture as star-shaped polypeptides very effectively neutralised the polyanionic siRNA at low N/P ratios <5. The molecular weight and architecture had a significant impact on the overall surface charge of the siRNA polyplexes formed. For example G2(8)-PLL40 and G5(64)-PLL5, with similar molecular weights, both form cationic siRNA polyplexes at N/P 3 whereas the much larger G3(16), G4(32) and G5(64)-PLL40 required a higher polymer![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) siRNA ratio of N/P 5 (Fig. 6 and S8†). For example, G5(64)-PLL40 with a large core and up to 64 long lysing arms overall possesses more lysine units than G2(8)-PLL40 and G5(64)-PLL5. G2(8)-PLL40 has a small core with only up to 8 long arms and G5(64)-PLL5 has a large core with up to 64 short arms, making their overall molecular weight and number of lysine residues smaller. Therefore, it can be determined from this and the L(1)-PLL320 data that it is not simply the number of cationic lysine residues presented that impacts on siRNA complexation but how they are presented. The large dendrimer core and dense polypeptide shell of G5(64)-PLL40 may limit siRNA interaction with PPI-PLL with the siRNA being retained upon the periphery of this star-shaped polypeptide. In contrast the long but relatively few arms of G2(8)-PLL40 and the many but short arms of G5(64)-PLL5 result in a decreased polypeptide density, enabling greater exposure of siRNA to lysine units and potentially the dendrimer core even though these star-shaped polypeptides have fewer overall cationic units than G5(64)-PLL40. This difference in ability to complex siRNA between the star-shaped PLL was also evident in the particle size analysis (Fig. 7 and S9†). siRNA polyplexes composed of G2(8)-PLL40 and G5(64)-PLL5 could form discrete nanoparticles <100 nm at N/P 5–100 whereas those composed of L(1)-PLL320 and G3(16), G4(32) and G5(64)-PLL40 could not form polyplexes <200 nm at N/P 1–100. AFM analysis (Fig. 8 and S10†) of star-shaped PLL siRNA polyplexes showed the efficient complexation and subsequent formation of spherically shaped compact nanoparticles (50–150 nm) at N/P 20. L(1)-PLL320, however, formed large elongated siRNA polyplexes (>200 nm) a feature which may inhibit efficient or complete siRNA complexation as observed in ζ-potential analysis of the described polyplex. A small amount of aggregation was observed for siRNA polyplexes of G2(8)-PLL40 (Fig. 8) which may be an artefact of sample preparation, i.e. as the mica sheet was dried, the gradual loss of solvent may have promoted minor aggregation. In spite of this, the star-shaped siRNA-polyplexes were predominantly finely dispersed.
siRNA ratio of N/P 5 (Fig. 6 and S8†). For example, G5(64)-PLL40 with a large core and up to 64 long lysing arms overall possesses more lysine units than G2(8)-PLL40 and G5(64)-PLL5. G2(8)-PLL40 has a small core with only up to 8 long arms and G5(64)-PLL5 has a large core with up to 64 short arms, making their overall molecular weight and number of lysine residues smaller. Therefore, it can be determined from this and the L(1)-PLL320 data that it is not simply the number of cationic lysine residues presented that impacts on siRNA complexation but how they are presented. The large dendrimer core and dense polypeptide shell of G5(64)-PLL40 may limit siRNA interaction with PPI-PLL with the siRNA being retained upon the periphery of this star-shaped polypeptide. In contrast the long but relatively few arms of G2(8)-PLL40 and the many but short arms of G5(64)-PLL5 result in a decreased polypeptide density, enabling greater exposure of siRNA to lysine units and potentially the dendrimer core even though these star-shaped polypeptides have fewer overall cationic units than G5(64)-PLL40. This difference in ability to complex siRNA between the star-shaped PLL was also evident in the particle size analysis (Fig. 7 and S9†). siRNA polyplexes composed of G2(8)-PLL40 and G5(64)-PLL5 could form discrete nanoparticles <100 nm at N/P 5–100 whereas those composed of L(1)-PLL320 and G3(16), G4(32) and G5(64)-PLL40 could not form polyplexes <200 nm at N/P 1–100. AFM analysis (Fig. 8 and S10†) of star-shaped PLL siRNA polyplexes showed the efficient complexation and subsequent formation of spherically shaped compact nanoparticles (50–150 nm) at N/P 20. L(1)-PLL320, however, formed large elongated siRNA polyplexes (>200 nm) a feature which may inhibit efficient or complete siRNA complexation as observed in ζ-potential analysis of the described polyplex. A small amount of aggregation was observed for siRNA polyplexes of G2(8)-PLL40 (Fig. 8) which may be an artefact of sample preparation, i.e. as the mica sheet was dried, the gradual loss of solvent may have promoted minor aggregation. In spite of this, the star-shaped siRNA-polyplexes were predominantly finely dispersed.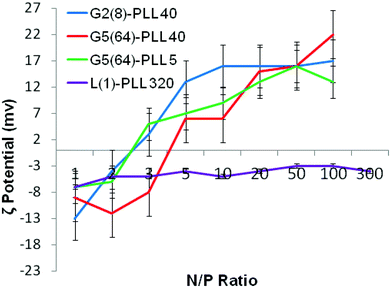 |
| | Fig. 6 ζ-Potential of G2(8)-PLL40, G5(64)-PLL40, G5(64)-PZLL5 and L(1)-PLL320 polypeptide/siRNA polyplexes over a range of N/P ratios. | |
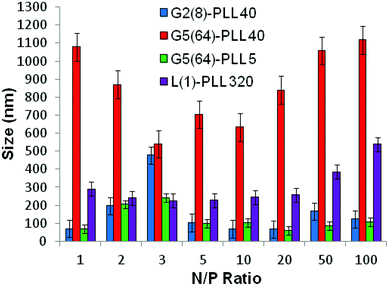 |
| | Fig. 7 Particle size of G2(8)-PLL40, G5(64)-PLL40, G5(64)-PZLL5 and L(1)-PLL320 polypeptide/siRNA polyplexes over a range of N/P ratios. | |
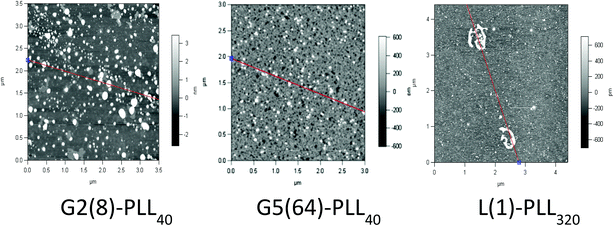 |
| | Fig. 8 Atomic force microscopy of G2(8)-PLL40, G5(64)-PLL40 and L(1)-PLL320 polypeptide/siRNA polyplexes at N/P 20. | |
Gel retardation assays of siRNA/pDNA polyplexes
The cationic nature of the star-shaped polypeptides provides a useful platform for the delivery of nucleic acids into cells. Gel retardation assays were used to evaluate and further confirm (Fig. 3–8) the ability of both linear and three of the star-shaped polypeptides G2(8)-PLL40, G5(64)-PLL5 and G5(64)-PLL40 to form complexes with the two different nucleic acid cargoes under investigation, i.e. plasmid DNA and siRNA. The assays were carried out at different N/P ratios for pDNA (N/P: 0.5, 1, 5, 10, 20) and siRNA (N/P: 1, 2, 3, 5, 10, 20, 50) based on the literature and the previous particle size and zeta potential studies.From Fig. 9 and S11† it was noted that all polymers, both linear and star-shaped PLL, successfully complexed pDNA at N/P 5. This is evidenced by the lack of a pDNA band seen on the gels (Fig. 9) in lanes 6, 7 and 8 which represent N/P ratios of 5, 10 and 20 respectively. This observation corroborates the particle size and zeta potential data (Fig. 3 and 4). The effective interaction seen for all the polylysines most likely arises from the large number of nucleic acids presented by pDNA that enables effective electrostatic complexation with all of the described polymers. The nature of the nanoparticle produced by this complexation, however, is structure dependent as evidenced in Fig. 4, 5, S6 and S7.† The PEI/pDNA complex and uncomplexed pDNA were used as a reference and a control respectively.
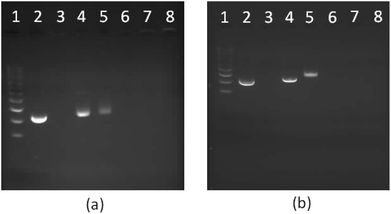 |
| | Fig. 9 1% Agarose gel electrophoresis of star-shaped PLL and linear PLL complexed with plasmid DNA to form polyplexes as a function of the N/P ratio. (a) G5(64)-PLL5; (b) L(1)-PLL320; lane 1: pDNA ladder, lane 2: pDNA, lane 3: PEI/pDNA (N/P 10), lanes 4–8: pDNA complexes at the N/P ratios of 0.5, 1, 5, 10, and 20. | |
Polypeptide architecture had a very significant effect on siRNA complexation (Fig. 10, S13†). L(1)-PLL320 was unable to completely complex siRNA even at N/P 50 as evidenced by the presence of an siRNA band in lane 9 of the gel in Fig. 10b. However, star-shaped PLL could successfully form polyplexes at ratios as low as N/P 1 as evidenced by the lack of an siRNA band seen in lanes 4–9 on the gel in Fig. 10a. Architectural variations within the star-shaped polypeptide family also displayed notable differences in siRNA complexation with G5(64)-PLL5 demonstrating gel retardation, i.e. complexation at the lowest N/P ratio of N/P 2 (Fig. 10a) while G5(64)-PLL40 and G2(8)-PLL40 fully retarded the siRNA at N/P20 and N/P 10 respectively (Fig. S-13†). Zeta potential analysis showed a similar trend with G5(64)-PLL40 neutralising siRNA at a slightly higher N/P ratio than G5(64)-PLL5 or G2(8)-PLL40 (Fig. 6) and producing the largest polyplexes over a range of N/P ratios (Fig. 7).
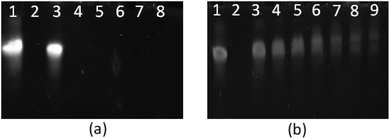 |
| | Fig. 10 PAGE gel electrophoresis of star-shaped PLL and linear PLL complexed with siRNA to form polyplexes as a function of the N/P ratio. (a) G5(64)-PLL5; (b) L(1)-PLL320; lane 1: siRNA, lane 2: PEI/siRNA (N/P 10), lanes 3–9: siRNA complexes at the N/P ratios of 1, 2, 3, 5, 10, 20, and 50. | |
Stability of polyplexes: DNase I and RNase I protection assays
Protection of the nucleic acid cargo from nuclease degradation is a vital prerequisite for any potential gene delivery vector. Molecular therapeutics such as pDNA and siRNA in particular are susceptible to hydrolysis and nuclease enzymes in vivo. Without sufficient molecular “cargo” protection gene therapy is inefficient or can be rendered completely ineffective. DNase and RNase degrade and nick pDNA and siRNA respectively resulting in extra banding or no bands at all (complete degradation) in gel electrophoresis. The ability of the star-shaped polypeptide architectures employed here to protect nucleic acids from this form of enzymatic breakdown was investigated using PEI and linear PLL as controls (Fig. 11, 12, S15 and S16†). The polyplexes were exposed to the relevant nuclease and then disrupted to enable the nucleic acids to travel down the gel. The same generations of star shaped were selected as used in the gel retardation study.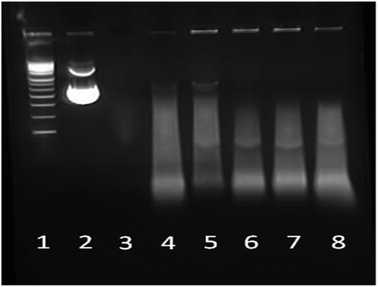 |
| | Fig. 11 1% Agarose gel electrophoresis of star-shaped PLL and linear PLL pDNA polyplexes at N/P 10 and their stability towards the enzyme DNase. Lane 1: pDNA ladder, lane 2: pDNA, lane 3: pDNA/DNase, lane 4: PEI/pDNA/DNase, lane 5: L(1)-PLL320/pDNA/DNase, lane 6: G2(8)-PLL40/pDNA/DNase, lane 7: G5(64)-PLL40/pDNA/DNase and lane 8: G5(64)-PLL5/pDNA/DNase. | |
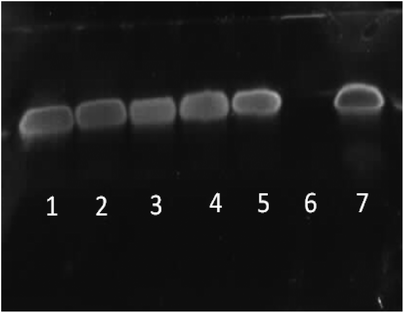 |
| | Fig. 12 PAGE gel electrophoresis of star-shaped PLL and linear PLL siRNA polyplexes at N/P 10 and their stability towards the enzyme RNase. Lane 1: G5(64)-PLL5/siRNA/RNase, lane 2: G5(64)-PLL40/siRNA/RNase, lane 3: G2(8)-PLL40/siRNA/RNase, lane 4: L(1)-PLL320/siRNA/RNase, lane 5: PEI/siRNA/RNase, lane 6: siRNA/RNase and lane 7: siRNA. | |
Naked pDNA is completely degraded in the presence of DNase emphasising the requirement and advantage of using a gene delivery vector (Fig. 11, lane 3). All of the star-shaped poly(lysine) formed tight complexes with pDNA and provided some degree of protection to the pDNA cargo from nuclease degradation similar to that seen for PEI, though smearing is seen in the pDNA that did not remain encapsulated (Fig. 11, lanes 6–8 and Fig. S15†). Their protective ability was similar to that of PEI at an N/P ratio of 10 has been shown to offer sufficient protection in vitro and in vivo to effect transfection. Interestingly from a stability perspective the star-shaped PLL was more stable to disruption by SDS than PEI as evidenced by the presence of DNA fluorescence in the wells of these samples (Fig. 11, lanes 5–8) while very little remains in the PEI sample (Fig. 11, lane 4).60 Chain length and degree of branching appeared to have minimal effect on the protective capacity of each polypeptide for pDNA with little difference observed amongst the star-shaped PLL (Fig. 11, lanes 6–8).
RNase stability assays indicated that each of the star-shaped polypeptides was capable of protecting siRNA from nuclease degradation via RNase (Fig. 12, lanes 1–3). Without complexation with a vector, naked siRNA was completely degraded by RNase (Fig. 12, lane 6). L(1)-PLL320 (Fig. 12, lane 4) also showed similar protective ability to that of the star-shaped polypeptides and PEI (Fig. 12, lane 5) despite its inability to completely complex siRNA at N/P 10 (Fig. 6). From observations of AFM images (Fig. 8) the L(1)-PLL320/siRNA complex adopts a very large coiled morphology, which may promote protection of the cargo by sterically hindering degradation by RNase. It was evident that disruption of these polyplexes was readily achieved owing to the sharp bands observed in Fig. 12 after siRNA decomplexation from the vectors. Such a feature possibly arises from the inherently small size of siRNA, allowing easier disruption of the polyplexes formed. The much larger pDNA, however, containing more nucleic acids, produced broad smeared bands as a consequence of its ability to form very strong complexes with the described polypeptides resulting in a complex more difficult to disrupt (Fig. 11).
pDNA transfections of Calu-3 cells using linear and star-shaped G5(64)-PLL40 polyplexes
Based on the physicochemical data outlined above, one of the star-shaped PLL generations, G5(64)-PLL40, was optimised for transfection using a reporter plasmid expressing luciferase, pGLuc, and compared to linear PLL at its previously determined N/P optimum of 2 for pDNA transfection.61Fig. 13 illustrates the results of this study with G5(64)-PLL40 successfully transfecting Calu-3 cells with the levels of expression dependent on the N/P ratio of the polyplexes used. An optimum N/P ratio of 5 was determined for this star-shaped polypeptide, similar to that found for other dendrimer-based gene vectors such as Superfect™. From the particle size analysis (Fig. 4), N/P ratios <5 for G5(64)-PLL40 with pDNA formed large nanoparticles >300 nm. Optical images taken of cells post treatment (S-13) indicated that at high N/P ratios, e.g. N/P50 (S-13D), there were signs of significant cellular toxicity, while at the optimum transfection an N/P ratio of 5 cells appears to be healthy and viable (S-13C). The highest levels of expression were seen at day 5 post transfection and these were significantly greater than for linear PLL at each of the N/P ratios 5, 10 and 20 (p < 0.05). These preliminary transfection studies indicate that the star-shaped PLL is capable of protecting pDNA, enhancing its cellular uptake and ultimately facilitating pDNA expression over prolonged periods, up to 5 days, which could enable applications in both conventional and regenerative medicine.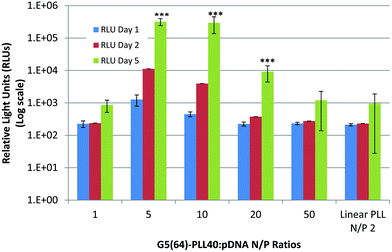 |
| | Fig. 13 Transfection of Calu-3 cells with pLuc/PLL polyplexes composed of linear PLL![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) pDNA (N/P 2) and star-shaped G5(64)-PLL40 over a range of N/P ratios (N/P 1–50). The expression of luciferase expressed as RLUs (log scale) was assessed 1, 2 or 5 days post transfection with the polyplexes (n = 3 ± S.D.). Data are represented as mean ± S.D. and were compared by a t-test (non-parametric, one-tailed) against linear PLL at day 5. Differences were considered to be highly significant at ***p < 0.001. pDNA (N/P 2) and star-shaped G5(64)-PLL40 over a range of N/P ratios (N/P 1–50). The expression of luciferase expressed as RLUs (log scale) was assessed 1, 2 or 5 days post transfection with the polyplexes (n = 3 ± S.D.). Data are represented as mean ± S.D. and were compared by a t-test (non-parametric, one-tailed) against linear PLL at day 5. Differences were considered to be highly significant at ***p < 0.001. | |
Comparison with hyperbranched polylysine (HBPL)
Linear polylysine and dendritic polylysine are lysine variants which have previously been investigated as gene delivery vectors.62,63 More recently, however, Klok et al. have utilised hyperbranched polylysine (HBPL) derivatives for gene transfection purposes.64 Although conceptually similar to the star-shaped polylysine materials reported here, these materials differ significantly in terms of structure and synthetic preparation. HBPL variants are irregular in shape and possess a very high degree of branching. This irregular or imperfect shape may promote better transfection efficiencies as has previously been seen for the now commercial Superfect™ or more simply “fractured” PAMAM dendrimers.65 Condensation of genes is enabled by the densely branched network of lysine units bearing accessible primary amino functionality. A major advantage of these materials is their relative synthetic ease in preparation via a one-pot polycondensation. However, purification and isolation of polymers of different molecular weights require column fractionation. Biodegradability of HBPL is readily achieved via enzymatic degradation affording a completely degradable system. The star-shaped polylysine materials presented here, however, consist of linear polylysine chains grown from a range of dendritic or branched cores enabling superior synthetic control. Controlled variation can be readily achieved in terms of branching, lysine density and molecular weight. The employment of suitable deprotection chemistry is a major drawback however. Cationic functionality is achieved via the lysine primary amino side chain whilst the PPI dendrimer core offers tertiary amino functionality which may enhance the buffering capacity of such materials, a step which is vital for the endosomal escape and overall success of transfection agents.66,67 The presence of this dendrimer core does, however, render these materials only partially degradable. Owing to the nature of this core shell type system, inaccessibility of the inner lysine units at the core may prove difficult. Such a feature may result in binding and structural differences between star-shaped polylysine and HBPL gene complexes.Conclusion
Star-shaped polypeptides are an attractive class of molecules for use as gene delivery vectors owing to their potential improved biocompatibility, structural characteristics and therapeutic “cargo” capacity. In this paper we describe the facile and systematic synthesis of a range of well-defined, monodisperse star-shaped poly(lysine) polymers boasting molecular weight, branching and polypeptide chain length versatility. The applicability of the described polypeptides as gene delivery vectors was subsequently investigated. From our study, it was apparent that polypeptide architecture had a significant impact on the nature of the polyplexes formed. In terms of pDNA, all star-shaped polypeptides efficiently packaged pDNA into discrete spherically shaped and positively charged nanocomplexes at low N/P ratios with sizes as low as <100 nm whereas the equivalent linear polypeptide required a higher complexation ratio. The complexation of siRNA with star-shaped PLL was greatly superior to linear PLL. In particular the small armed, less densely branched star polypeptides were most promising as both pDNA and siRNA delivery vectors owing to their optimum sizing and complexation data. All star polypeptides offered similar protection of pDNA and siRNA from nuclease degradation once complexed. However upon decomplexation it was noted that siRNA complexes were more readily disrupted in comparison to pDNA complexes, highlighting the differences in stability between polyplexes composed of the different nucleic acids. Overall the promising nucleic acid complexation, sizing, morphology and protection capacity of two different genetic “cargos” highlight the potential of polypeptide dendrimer hybrids as gene delivery vectors (GDVs). Their potential as GDVs was confirmed with preliminary pDNA transfection studies in which one of the G5(64)-PLL40 was found to be capable of successfully transfecting epithelial cells with pDNA, and importantly these levels were significantly greater compared to linear PLL with a 300-fold increase in expression seen over the 5 day study. The systematic study reported here may potentially lead to the identification and development of an ideal polypeptide dendrimer hybrid material for use as a versatile platform in gene delivery. Versatility may be readily enhanced through the facile control of polypeptide dimensions such as branching and molecular weight, end group and side chain modification using pegylation, glycosylation and attachment of ligands to further improve biocompatibility, cell targeting and overall efficacy of the delivery platform. As a consequence these materials may offer some advantages over previously reported lysine variants for gene delivery.Acknowledgements
This work was conducted under an IRCSET Embark Scholarship Award (M.B.). A.H. is a SFI Stokes Senior Lecturer (07/SK/B1241). S.A.C. is a PI in SFI Strategic Research Cluster (07/SRC/B1154).References
- E. W. Alton, U. Griesenbach and D. M. Geddes, Gene Ther., 1999, 6, 155–156 CrossRef CAS.
- D. N. Nguyen, J. J. Green, J. M. Chan, R. Langer and D. G. Anderson, Adv. Mater., 2009, 21, 847–867 CrossRef CAS.
- E. L. Tatum, Perspect. Biol. Med., 1966, 10, 19–32 CAS.
- S. Y. Wonga, J. M. Pelet and D. Putnam, Prog. Polym. Sci., 2007, 32, 799–837 CrossRef.
- E. Marshall, Science, 2000, 288, 951–957 CrossRef CAS.
- G. Y. Wu and C. H. Wu, J. Biol. Chem., 1987, 262, 4429–4432 CAS.
- M. Koping-Hoggard, Y. S. Melnikova, K. M. Varum, B. Lindman and P. Artursson, J. Gene Med., 2003, 5, 130–141 CrossRef CAS.
- T. K. Georgiou, M. Vamvakaki, L. A. Phylactou and C. S. Patrickios, Biomacromolecules, 2005, 6, 2990–2997 CrossRef CAS.
- E. G. Tierney, G. P. Duffy, A. J. Hibbitts, S. Cryan and F. J. O'Brien, J. Controlled Release, 2012, 158, 304–311 CrossRef CAS.
- O. Taratula, O. B. Garbuzenko, P. Kirkpatrick, I. Pandya, R. Savla, V. P. Pozharov, H. He and T. Minko, J. Controlled Release, 2009, 140, 284–293 CrossRef CAS.
- M. L. Patil, M. Zhang, O. Taratula, O. B. Garbuzenko, H. He and T. Minko, Biomacromolecules, 2009, 10, 258–266 CrossRef CAS.
- C. Dufes, I. F. Uchegbu and A. G. Schatzlein, Adv. Drug Delivery Rev., 2005, 57, 2177–2202 CrossRef CAS.
- Y. N. Xue, M. Liu, L. Peng, S. W. Huang and R. X. Zhuo, Macromol. Biosci., 2006, 10, 404–414 CrossRef.
- M. Mintzer and M. W. Grinstaff, Chem. Soc. Rev., 2011, 40, 173–190 RSC.
- H. Gao, S. Ohno and K. Matyaszewski, J. Am. Chem. Soc., 2006, 128, 15111–15113 CrossRef CAS.
- A. W. Bosman, R. Vestberg, A. Heumann, J. M. J. Frechet and C. Hawker, J. Am. Chem. Soc., 2003, 125, 715–728 CrossRef CAS.
- V. Y. Lee, K. Havenstrite, M. Tijo, M. McNeil, H. M. Blau, R. D. Miller and J. Sly, Adv. Mater., 2011, 23, 4509–4515 CrossRef CAS.
- A. Sulistio, P. A. Gurr, A. Blencowe and G. Qiao, Aust. J. Chem., 2012, 65, 978–984 CrossRef CAS.
- J. Huang and A. Heise, Chem. Soc. Rev., 2013, 42, 7373–7390 RSC.
- Y. Haba, A. Harada, T. Takagishi and K. Kono, Polymer, 2005, 46, 1813–1820 CrossRef CAS.
- C. Kojima, C. Regino, Y. Umeda, H. Kobayashi and K. Kono, Int. J. Pharm., 2010, 383, 293–296 CrossRef CAS.
- C. S. Cho, J. B. Cheon, Y. I. Jeong, I. S. Kim, S. H. Kim and T. Akaike, Macromol. Rapid Commun., 1997, 18, 361–369 CrossRef CAS.
- T. Borase, M. Iacono, S. I. Ali, P. D. Thornton and A. Heise, Polym. Chem., 2012, 3, 1267–1275 RSC.
- T. Borase, T. Ninjbadgar, A. Kapetanakis, S. Roche, R. O'Connor, C. Kerskens, A. Heise and D. F. Brougham, Angew. Chem., Int. Ed., 2013, 52, 3164–3167 CrossRef CAS.
- K. Inoue, H. Sakai, S. Ochi, T. Itaya and T. Tanigaki, J. Am. Chem. Soc., 1994, 116, 10783–10784 CrossRef CAS.
- G. Mihov, D. Grebel-Koehler, A. Lübbert, G. W. M. Vandermeulen, A. Herrmann, H. A. Klok and K. Müllen, Bioconjugate Chem., 2005, 16, 283–293 CrossRef CAS.
- H. Huang, J. Li, L. Liao, J. Li, L. Wu, C. Dong, P. Lai and D. Liu, Eur. Polym. J., 2012, 48, 696–704 CrossRef CAS.
- Q. Wang, J. Yu, Y. Yan, S. Xu, F. Wang, Q. Li, J. Wang, X. Zhang and D. Liu, Polym. Chem., 2012, 3, 1284–1290 RSC.
- D. Appelhans, H. Komber, R. Kirchner, J. Seidel, C. F. Huang, D. Voigt, D. Kuckling, F. C. Chang and B. Voit, Macromol. Rapid Commun., 2005, 26, 586–591 CrossRef CAS.
- M. Byrne, P. D. Thornton, S. A. Cryan and A. Heise, Polym. Chem., 2012, 3, 2825–2831 RSC.
- A. P. Nowak, V. Breedveld, L. Pakstis, B. Ozbas, D. J. Pine, D. Pochan and T. J. Deming, Nature, 2002, 417, 424–428 CrossRef CAS.
- E. P. Holowka, V. Z. Sun, D. T. Kamei and T. J. Deming, Nat. Mater., 2007, 6, 52–57 CrossRef CAS.
- A. Constancis, R. Meyrueix, N. Bryson, S. Huille, J. M. Grosselin and T. Gulik-Krzywicki, J. Colloid Interface Sci., 1999, 217, 357–368 CrossRef CAS.
- N. Gabrielson, H. Lu, L. Yin, D. Li, F. Wang and J. Cheng, Angew. Chem., Int. Ed., 2012, 51, 1143–1147 CrossRef CAS.
- J. Yen, Y. Zhang, N. P. Gabrielson, L. Yin, L. Guan, I. Chaudhury, H. Lu, F. Wang and J. Cheng, Biomater. Sci., 2013, 1, 719–727 RSC.
- M. Yu, J. Hwang and T. J. Deming, J. Am. Chem. Soc., 1999, 121, 5825–5826 CrossRef CAS.
- P. D. Thornton, R. Brannigan, J. Podporska, B. Quilty and A. Heise, J. Mater. Sci. Mater. Med., 2012, 23, 37–45 CrossRef CAS.
- C. Xiao, C. Zhao, P. He, Z. Tang, X. Chen and X. Jing, Macromol. Rapid Commun., 2010, 31, 991–997 CrossRef CAS.
- J. R. Kramer and T. J. Deming, J. Am. Chem. Soc., 2010, 132, 15068–15071 CrossRef CAS.
- J. Sun and H. Schlaad, Macromolecules, 2010, 43, 4445–4448 CrossRef CAS.
- H. Tang and D. Zhang, Biomacromolecules, 2010, 11, 1585–1592 CrossRef CAS.
- J. Huang, G. Habraken, F. Audouin and A. Heise, Macromolecules, 2010, 43, 6050–6057 CrossRef CAS.
- J. Huang, C. Bonduelle, J. Thévenot, S. Lecommandoux and A. Heise, J. Am. Chem. Soc., 2012, 134, 119–122 CrossRef CAS.
- D. Pati, N. Kalva, S. Das, G. Kumaraswamy, S. S. Gupta and A. V. Ambade, J. Am. Chem. Soc., 2012, 134, 7796–7802 CrossRef CAS.
- J. R. Kramer, A. R. Rodriguez, U. J. Choe, D. T. Kamei and T. J. Deming, Soft Matter, 2013, 9, 3389–3395 RSC.
- C. M. Ward, M. Pechar, D. Oupicky, K. Ulbrich and L. W. Seymour, J. Gene Med., 2002, 4, 536–547 CrossRef CAS.
- M. Mannisto, S. Vanderkerken, V. Toncheva, M. Elomaa, M. Ruponen, E. Schacht and A. Urtti, J. Controlled Release, 2002, 83, 169–182 CrossRef CAS.
- H. Lv, S. Zhang, B. Wang, S. Cui and J. Yan, J. Controlled Release, 2006, 114, 100–109 CrossRef CAS.
- S. Pan, C. Wang, X. Zeng, Y. Wen, H. Wu and M. Feng, Int. J. Pharm., 2011, 420, 206–215 CrossRef CAS.
- M. Yamagata, T. Kawano, K. Shiba, T. Mori, Y. Katayama and T. Niidome, Bioorg. Med. Chem., 2007, 15, 526–532 CrossRef CAS.
- K. Luo, C. Li, G. Wang, Y. Nie, B. He, Y. Wu and Z. Gu, J. Controlled Release, 2011, 155, 77–87 CrossRef CAS.
- C. Scholz and E. Wagner, J. Controlled Release, 2012, 161, 554–565 CrossRef CAS.
- G. J. M. Habraken, C. E. Koning and A. Heise, J. Polym. Sci., Part A: Polym. Chem., 2009, 47, 6883–6893 CrossRef CAS.
- A. Sulistio, A. Blencowe, A. Widjaya, X. Zhang and G. Qiao, Polym. Chem., 2012, 3, 224–234 RSC.
- H. R. Kricheldorf, Angew. Chem., Int. Ed., 2006, 45, 5752–5784 CrossRef CAS.
- N. Hadjichristidis, H. Iatrou, M. Pitsikalis and G. Sakellariou, Chem. Rev., 2009, 109, 5528–5578 CrossRef CAS.
- G. J. M. Habraken, A. Heise and P. D. Thornton, Macromol. Rapid Commun., 2012, 33, 272–286 CrossRef CAS.
- G. M. Soliman, A. Sharma, D. Maysinger and A. Kakkar, Chem. Commun., 2011, 47, 9572–9587 RSC.
- H. L. Fu, S. X. Cheng, X. Z. Zhang and R. X. Zhuo, J. Controlled Release, 2007, 124, 181–188 CrossRef CAS.
- J. L. Lee, C. W. Lo, S. M. Ka, A. Chen and W. S. Chen, J. Controlled Release, 2012, 160, 64–71 CrossRef CAS.
- S. A. Cryan and C. M. O'Driscoll, Pharm. Res., 2003, 20, 569–575 CrossRef CAS.
- D. W. Pack, A. S. Hoffman, S. Pun and P. S. Stayton, Nat. Rev. Drug Discovery, 2005, 4, 581–593 CrossRef CAS.
- M. Yamagata, T. Kawano, K. Shiba, T. Mori, Y. Katayama and T. Niidome, Bioorg. Med. Chem., 2007, 15, 526–532 CrossRef CAS.
- Z. Kadlecova, Y. Rajendra, M. Matasci, D. Hacker, L. Baldi, F. M. Wurm and H. A. Klok, Macromol. Biosci., 2012, 12, 794–804 CrossRef CAS.
- M. X. Tang, C. T. Redemann and F. C. Szoka, Bioconjugate Chem., 1996, 7, 703–714 CrossRef CAS.
- M. J. Shieh, C. L. Peng, P. J. Lou, C. H. Chiu, T. Y. Tsai, C. Y. Hsu, C. Y. Yeh and P. S. Lai, J. Controlled Release, 2008, 129, 200–206 CrossRef CAS.
- L. Tian, P. Nguyen and P. T. Hammond, Chem. Commun., 2006, 3489–3491 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c3bm60123d |
|
| This journal is © The Royal Society of Chemistry 2013 |
Click here to see how this site uses Cookies. View our privacy policy here. ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P ratio. To prepare the polyplexes, solutions of G2-PPI-PLL or G5-PPI-PLL prepared in buffer (Tris HCl buffer pH 7.4 and 10 mM NaCl) were mixed with pDNA/siRNA at various N
P ratio. To prepare the polyplexes, solutions of G2-PPI-PLL or G5-PPI-PLL prepared in buffer (Tris HCl buffer pH 7.4 and 10 mM NaCl) were mixed with pDNA/siRNA at various N![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P ratios (1, 5, 10, 20 and 100). These mixtures were incubated at room temperature for 30 min for polyplex formation, diluted and analysed.
P ratios (1, 5, 10, 20 and 100). These mixtures were incubated at room temperature for 30 min for polyplex formation, diluted and analysed.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P ratios as described, and their particle size and surface charge (ζ-potential) were measured using a Malvern Zetasizer Nano ZS (Malvern Instruments, Ltd, United Kingdom).
P ratios as described, and their particle size and surface charge (ζ-potential) were measured using a Malvern Zetasizer Nano ZS (Malvern Instruments, Ltd, United Kingdom).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P ratio of 20 by adding the polypeptide solution to pDNA/siRNA to give a final pDNA/siRNA concentration of 10 ng μL−1. After incubation at room temperature for 30 min, 2 μL of the complex solution was diluted into 198 μL H2O and deposited onto the surface of freshly cleaved mica sheets (Agar Scientific, G250-3, 11 × 11 mm mica). After 3 min, excess solution was removed by careful absorption onto filter paper (Whatman, Fisher Scientific, Ireland) and the mica was further dried at room temperature for 24 h before analysis by AFM. Samples were imaged using an MFP-3D BIO AFM (Asylum Research). Micromasch silicon NSC18 cantilevers were used. Tips were 225 nm long and had a typical resonant frequency of 75 kHz. The AFM operated in the AC mode (alternate contact) in order to minimise the tip sample interaction. Typical free air amplitudes were ∼800 mV and a high amplitude set-point relative to the set-point was maintained to minimise cell/tip damage. All samples were imaged in air at relative humidity. The images obtained contained 512 pixels per scan line.
P ratio of 20 by adding the polypeptide solution to pDNA/siRNA to give a final pDNA/siRNA concentration of 10 ng μL−1. After incubation at room temperature for 30 min, 2 μL of the complex solution was diluted into 198 μL H2O and deposited onto the surface of freshly cleaved mica sheets (Agar Scientific, G250-3, 11 × 11 mm mica). After 3 min, excess solution was removed by careful absorption onto filter paper (Whatman, Fisher Scientific, Ireland) and the mica was further dried at room temperature for 24 h before analysis by AFM. Samples were imaged using an MFP-3D BIO AFM (Asylum Research). Micromasch silicon NSC18 cantilevers were used. Tips were 225 nm long and had a typical resonant frequency of 75 kHz. The AFM operated in the AC mode (alternate contact) in order to minimise the tip sample interaction. Typical free air amplitudes were ∼800 mV and a high amplitude set-point relative to the set-point was maintained to minimise cell/tip damage. All samples were imaged in air at relative humidity. The images obtained contained 512 pixels per scan line.

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a
a![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) b/g mol−1
b/g mol−1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800
800![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800
800![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700
700![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400
400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700
700![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800
800


![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) siRNA N/P ratios up to 300 (Fig. 6). The inability of linear PLL to neutralise siRNA and form a discrete compact cationic nanostructure clearly relates to its linear architecture as star-shaped polypeptides very effectively neutralised the polyanionic siRNA at low N/P ratios <5. The molecular weight and architecture had a significant impact on the overall surface charge of the siRNA polyplexes formed. For example G2(8)-PLL40 and G5(64)-PLL5, with similar molecular weights, both form cationic siRNA polyplexes at N/P 3 whereas the much larger G3(16), G4(32) and G5(64)-PLL40 required a higher polymer
siRNA N/P ratios up to 300 (Fig. 6). The inability of linear PLL to neutralise siRNA and form a discrete compact cationic nanostructure clearly relates to its linear architecture as star-shaped polypeptides very effectively neutralised the polyanionic siRNA at low N/P ratios <5. The molecular weight and architecture had a significant impact on the overall surface charge of the siRNA polyplexes formed. For example G2(8)-PLL40 and G5(64)-PLL5, with similar molecular weights, both form cationic siRNA polyplexes at N/P 3 whereas the much larger G3(16), G4(32) and G5(64)-PLL40 required a higher polymer![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) siRNA ratio of N/P 5 (Fig. 6 and S8†). For example, G5(64)-PLL40 with a large core and up to 64 long lysing arms overall possesses more lysine units than G2(8)-PLL40 and G5(64)-PLL5. G2(8)-PLL40 has a small core with only up to 8 long arms and G5(64)-PLL5 has a large core with up to 64 short arms, making their overall molecular weight and number of lysine residues smaller. Therefore, it can be determined from this and the L(1)-PLL320 data that it is not simply the number of cationic lysine residues presented that impacts on siRNA complexation but how they are presented. The large dendrimer core and dense polypeptide shell of G5(64)-PLL40 may limit siRNA interaction with PPI-PLL with the siRNA being retained upon the periphery of this star-shaped polypeptide. In contrast the long but relatively few arms of G2(8)-PLL40 and the many but short arms of G5(64)-PLL5 result in a decreased polypeptide density, enabling greater exposure of siRNA to lysine units and potentially the dendrimer core even though these star-shaped polypeptides have fewer overall cationic units than G5(64)-PLL40. This difference in ability to complex siRNA between the star-shaped PLL was also evident in the particle size analysis (Fig. 7 and S9†). siRNA polyplexes composed of G2(8)-PLL40 and G5(64)-PLL5 could form discrete nanoparticles <100 nm at N/P 5–100 whereas those composed of L(1)-PLL320 and G3(16), G4(32) and G5(64)-PLL40 could not form polyplexes <200 nm at N/P 1–100. AFM analysis (Fig. 8 and S10†) of star-shaped PLL siRNA polyplexes showed the efficient complexation and subsequent formation of spherically shaped compact nanoparticles (50–150 nm) at N/P 20. L(1)-PLL320, however, formed large elongated siRNA polyplexes (>200 nm) a feature which may inhibit efficient or complete siRNA complexation as observed in ζ-potential analysis of the described polyplex. A small amount of aggregation was observed for siRNA polyplexes of G2(8)-PLL40 (Fig. 8) which may be an artefact of sample preparation, i.e. as the mica sheet was dried, the gradual loss of solvent may have promoted minor aggregation. In spite of this, the star-shaped siRNA-polyplexes were predominantly finely dispersed.
siRNA ratio of N/P 5 (Fig. 6 and S8†). For example, G5(64)-PLL40 with a large core and up to 64 long lysing arms overall possesses more lysine units than G2(8)-PLL40 and G5(64)-PLL5. G2(8)-PLL40 has a small core with only up to 8 long arms and G5(64)-PLL5 has a large core with up to 64 short arms, making their overall molecular weight and number of lysine residues smaller. Therefore, it can be determined from this and the L(1)-PLL320 data that it is not simply the number of cationic lysine residues presented that impacts on siRNA complexation but how they are presented. The large dendrimer core and dense polypeptide shell of G5(64)-PLL40 may limit siRNA interaction with PPI-PLL with the siRNA being retained upon the periphery of this star-shaped polypeptide. In contrast the long but relatively few arms of G2(8)-PLL40 and the many but short arms of G5(64)-PLL5 result in a decreased polypeptide density, enabling greater exposure of siRNA to lysine units and potentially the dendrimer core even though these star-shaped polypeptides have fewer overall cationic units than G5(64)-PLL40. This difference in ability to complex siRNA between the star-shaped PLL was also evident in the particle size analysis (Fig. 7 and S9†). siRNA polyplexes composed of G2(8)-PLL40 and G5(64)-PLL5 could form discrete nanoparticles <100 nm at N/P 5–100 whereas those composed of L(1)-PLL320 and G3(16), G4(32) and G5(64)-PLL40 could not form polyplexes <200 nm at N/P 1–100. AFM analysis (Fig. 8 and S10†) of star-shaped PLL siRNA polyplexes showed the efficient complexation and subsequent formation of spherically shaped compact nanoparticles (50–150 nm) at N/P 20. L(1)-PLL320, however, formed large elongated siRNA polyplexes (>200 nm) a feature which may inhibit efficient or complete siRNA complexation as observed in ζ-potential analysis of the described polyplex. A small amount of aggregation was observed for siRNA polyplexes of G2(8)-PLL40 (Fig. 8) which may be an artefact of sample preparation, i.e. as the mica sheet was dried, the gradual loss of solvent may have promoted minor aggregation. In spite of this, the star-shaped siRNA-polyplexes were predominantly finely dispersed.







![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) pDNA (N/P 2) and star-shaped G5(64)-PLL40 over a range of N/P ratios (N/P 1–50). The expression of luciferase expressed as RLUs (log scale) was assessed 1, 2 or 5 days post transfection with the polyplexes (n = 3 ± S.D.). Data are represented as mean ± S.D. and were compared by a t-test (non-parametric, one-tailed) against linear PLL at day 5. Differences were considered to be highly significant at ***p < 0.001.
pDNA (N/P 2) and star-shaped G5(64)-PLL40 over a range of N/P ratios (N/P 1–50). The expression of luciferase expressed as RLUs (log scale) was assessed 1, 2 or 5 days post transfection with the polyplexes (n = 3 ± S.D.). Data are represented as mean ± S.D. and were compared by a t-test (non-parametric, one-tailed) against linear PLL at day 5. Differences were considered to be highly significant at ***p < 0.001.