Antimicrobial properties of enzymatically triggered self-assembling aromatic peptide amphiphiles†
Meghan Hughes*a,
Sisir Debnatha,
Charles W. Knappb and
Rein V. Ulijn*a
aWestCHEM, Department of Pure and Applied Chemistry, University of Strathclyde, 295 Cathedral Street, Glasgow, G1 1XL, UK. E-mail: rein.ulijn@strath.ac.uk; meghan.hughes@strath.ac.uk
bCivil and Environmental Engineering, University of Strathclyde, 50 Richmond Street, Glasgow, G1 1XN, UK
First published on 26th July 2013
Abstract
The combination of catalysis and self-assembly influences many key processes in living systems. Synthetic analogues of such systems may provide opportunities to direct biological processes. Previously, it has been demonstrated that enzyme triggered assembly of peptide derivatives can influence bacterial cell death by intracellular fibre formation. In this article, we discuss the self-assembly of 9-fluorenylmethyloxycarbonyl (Fmoc) protected dipeptide amphiphiles, FY, YT, YS, YN and YQ, designing phosphorylated precursors to be alkaline phosphatase responsive. We use microscopy techniques, fluorescence and FTIR to demonstrate differences in molecular assembly and nanoscale architecture in vitro – indicating fibre formation of FY, YT, YS and YN, and spherical self-assembled structures of YQ. As the enzyme is naturally occurring in E. coli, we manipulate conditions to over-express the enzyme and demonstrate the conversion of precursors to self-assembling aromatic peptide amphiphiles in vivo. Furthermore, we test whether antimicrobial activity can be differentially controlled by the introduction of varying aromatic peptide amphiphiles, with the results indicating a similar antimicrobial response for each treatment.
Introduction
Molecular self-assembly and supramolecular fibres play key roles in biological systems. For example, the extracellular matrix (ECM) is a fibrous network composed of a range of proteins which is key in the structural support of tissues and may also regulate gene expression, differentiation and proliferation.1,2 The intracellular cytoskeleton is also composed of fibrous networks that regulate transport, cell movement and differentiation. These fibrous networks are highly dynamic and regulated by enzymatic catalysis. In recent years, enzyme catalysis combined with self-assembled nanomaterials based on peptides have become of interest for use intracellularly and extra-cellularly as a synthetic approach to mimic the functionality of naturally occurring supramolecular fibres.2Aromatic peptide amphiphiles have emerged as simple and versatile building blocks which can form self-assembled nanostructures. These are generally less than five amino acid residues in length and chemically modified with an aromatic group to aid assembly. These amphiphiles self-assemble in aqueous environments via a combination of hydrophobic, π-stacking, hydrogen bonding and electrostatic interactions,3–13 and typically form nano-scale fibrous network structures. They have been successfully interfaced with biological systems, with an additional desirability associated with the relatively simplistic modification of both chemical and mechanical properties necessary for function.14–17
The assembly/dis-assembly of such structures can be triggered by including an enzyme responsive functional group.18,19 Enzymes, such as phosphatases,20–27 esterases28–30 and proteases,31–36 have been utilised to trigger the self-assembly of aromatic peptide amphiphiles by converting non-assembling precursors into self-assembling components.
In 2007, Bing Xu's group first demonstrated in vivo self-assembly of aromatic peptide amphiphiles by introducing precursors responsive to enzymes within biological systems. The addition of phosphatase responsive precursors of gelators to blood samples resulted in hydrogelation of the biological specimens.27 By exploiting the naturally occurring phosphatases37 and esterases30 in bacterial and mammalian cell systems respectively, the group demonstrated supramolecular assembly within cells. Consequently, the cultures displayed a depletion in cell numbers and cell inactivity (death) attributed to nanostructure formation within the cells, highlighting potential for selective cell reduction and anti-microbial materials.30,37 Further to this, Gao et al. successfully imaged the enzymatically triggered self-assembly of aromatic peptide amphiphiles nano fibres within HeLa cells using complementary microscopy techniques.38
In 2012, we demonstrated differential supramolecular structure formation of four self-assembling aromatic peptide amphiphiles, Fmoc-YT, Fmoc-YS, Fmoc-YN and Fmoc-YQ, triggered using an esterase enzyme, subtilisin.28 Fmoc-YT, Fmoc-YS and Fmoc-YN formed nanoscale fibres with different network properties, whereas Fmoc-YQ self-assembled to form spherical structures.28 This work describes a series of analogous self-assembling aromatic peptide amphiphile precursors designed to be responsive to alkaline phosphatase – an enzyme which has been thoroughly studied and known to be expressed in the periplasmic space of E. coli cells.39–42 Combining peptide design with in vivo enzymatic triggers for self-assembly, we investigated whether the treatment of E. coli with different aromatic peptide amphiphiles would result in differential anti-microbial properties.
Results and discussion
In vitro analysis
The alkaline phosphatase triggered self-assembly of Fmoc-FY-OH (1) in vitro has previously been demonstrated (see Fig. 1).43,44 Under these conditions, the phosphorylated precursor is unable to create supramolecular assemblies due to electrostatic repulsion between deprotonated phosphate groups. After treatment with alkaline phosphatase, a clear 10 mM solution transforms to a translucent hydrogel, with compound 1 yielding 96% from its precursor when monitoring the conversion using HPLC. Spectroscopic analysis confirmed that the molecular association is of π-stacked Fmoc groups using fluorescence emission spectroscopy (Fig. 2(a)). The emission spectra of the starting materials displayed a shoulder peak at 325 nm, representing the residual unassembled Fmoc group, and a prominent peak at 375 nm indicating the spherical aggregates as previously reported by Sadownik et al.43 After 24 hours, the Fmoc peak has disappeared, and a bathochromic shift of the aggregate peak from 375 nm to 400 nm is evident accompanied by quenching, indicating the formation of extended π-stacked aggregates. Additionally, the FTIR absorbance spectra indicate hydrogen bonding between the peptide units (Fig. 3). The amide I region of the spectra shows 2 prominent bands ∼1630 cm−1, indicative of β-sheet formation, and ∼1680 cm−1, arising from the carbamate moiety of the Fmoc hydrogen bonding into the β-sheet configuration.45 The self-assembly of molecular units of 1 results in the formation of nanoscale fibres observable via TEM (see Fig. 4(a)), similar to previously reported aromatic peptide amphiphiles.5,8,29,34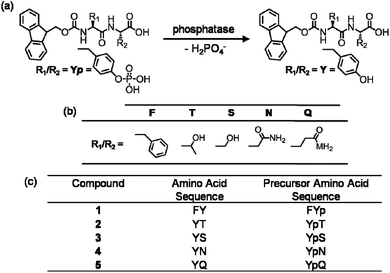 | ||
| Fig. 1 (a) Reaction schematic of enzymatic dephosphorylation by alkaline phosphatase. (b) Table of amino acid side chains. (c) The amino acid sequences of compounds 1–5 and their precursors: Fmoc-FY-OH, Fmoc-YT-OH, Fmoc-YS-OH, Fmoc-YN-OH and Fmoc-YQ-OH respectively. | ||
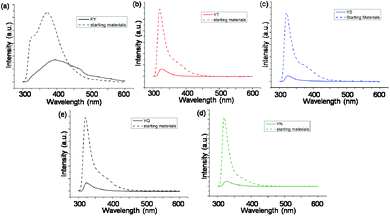 | ||
| Fig. 2 (a)–(e) Fluorescence emission spectra of 10 mM samples of 1, 2, 3, 4 and 5 respectively at 24 hours overlaid with emission of the precursor solution. | ||
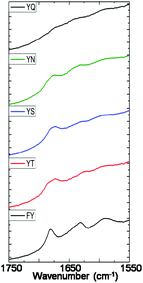 | ||
| Fig. 3 Amide I region of FTIR absorbance spectra of 10 mM samples of 1, 2, 3, 4 and 5 measured after 24 hours of enzyme addition. | ||
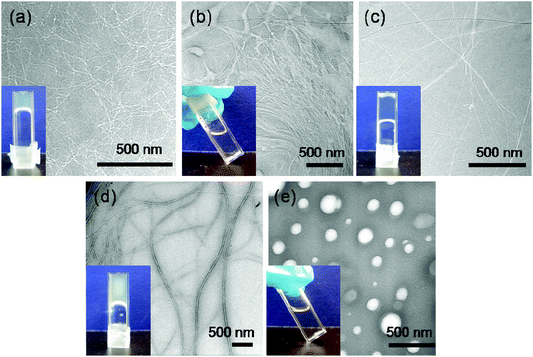 | ||
| Fig. 4 (a)–(e) TEM images of self-assembled structures of 10 mM samples of 1, 2, 3, 4 and 5 respectively after 24 hours of enzyme addition. Inset: hydrogelation of respective samples tested by vial inversion at 24 hours. | ||
Recently, we described the molecular self-assembly of subtilisin triggered assembly of Fmoc-YT-OH (2), Fmoc-YS-OH (3), Fmoc-YN-OH (4) and Fmoc-YQ-OH (5) from methyl ester precursors.28 Analogous to these systems, four alkaline phosphatase responsive aromatic peptide amphiphiles were designed and synthesised, Fmoc-YpT-OH, Fmoc-YpS-OH, Fmoc-YpN-OH and Fmoc-YpQ-OH (see Fig. 1). The self-assembly of the aromatic peptide amphiphiles Fmoc-YX-OH were biocatalytically triggered by enzymatic dephosphorylation of Fmoc-YpX-OH precursor using alkaline phosphatase at 10 mM concentration in 0.6 M sodium phosphate buffer pH 8, giving rise to four self-assembling aromatic amphiphiles, 2, 3, 4 and 5. The conversion of precursors to the desired Fmoc-YX-OH dipeptide was monitored using HPLC, with 2, 3, 4 and 5 yielding 97.1%, 95.0%, 99.9% and 99.9% respectively, after 24 hours of enzyme addition. At this time, each of the four suspensions had been transformed to: 2, a clear viscous liquid; 3, a clear self-supporting gel; 4, a translucent self-supporting gel; and 5, a clear solution, indicating that molecular self-assembly had occurred (see Fig. 4(b)–(e), inset). In vitro spectroscopic and microscopic analysis of 2, 3, 4 and 5 revealed identical molecular association and nanoscale structure as described for their subtilisin triggered counterpart.28 2, 3 and 4 assemble via extended π-stacked aggregates of the Fmoc-groups observable by monitoring the fluorescence emission spectra (Fig. 2(b)–(d)). Each of the starting materials displayed a prominent peak at 325 nm representing the Fmoc functionality and a shoulder peak at ∼375 nm indicating spherical aggregate formation of the starting materials.43 After 24 hours, 2, 3 and 4 displayed considerable quenching, which is consistent with extended π-stacked aggregate formation of Fmoc-groups. Furthermore, the combination of absorbance bands present for 2, 3 and 4 at ∼1675 cm−1 and 1625 cm−1 in the FTIR absorbance spectra indicates the carbamate and amide carbonyls of the peptide component respectively stabilised by β-sheet formation (Fig. 3). The absorbance bands associated with β-sheet formation are considerably less pronounced compared to other peptide amphiphiles studied,8,34 and are consistent with previous studies of self-assembly of identical Fmoc protected amino acid sequences.28 The supramolecular association of 2, 3 and 4 results in the formation of nanoscale fibres observable via TEM (Fig. 4(b)–(d)). In contrast, 5 self-assembles to form spherical structures established by TEM imaging (see Fig. 4(e)). Fluorescence emission spectroscopy indicates a blue-shift of the Fmoc peak from 325 nm in the starting materials spectra to a shorter wavelength after 24 hours (Fig. 2(e)). This, in combination with the absence of observable bands in the amide I region of the FTIR absorbance spectra (Fig. 3), indicates that the aggregate formation is due to the hydrophobic collapse rather than π–β assemblies, with the β-sheet formation most likely restricted due to the extended length of the glutamine side chain in 5 compared with 2, 3 and 4 – which is consistent with previous studies.28
In vivo analysis
The conditions to over-express alkaline phosphatase in MC-1000 E. coli bacterial cells were firstly optimised by the inclusion of inosine within the medium (see ESI†). Previous studies by Csopak et al. demonstrating the enzyme over-expression were carried out using different media formulations.39 To allow for a direct comparison, we used identical media formulations, either with/without inosine, to ensure that the over-expression of the enzyme was due to the inclusion of the compound within the growth medium. The population growth rate appeared to be unaffected by the presence of the compound; however, the activity of the alkaline phosphatase increased 2-fold, from 0.128 a.u. min−1 mL−1 to 0.245 a.u. min−1 mL−1 using the enzymatic assay method modified from the Worthington Enzyme Manuel for in vivo purposes.46 Bacterial cell cultures which were grown in media (both with and without inosine) for 24 hours were then treated with 13.0 mg of Fmoc-FYp-OH and incubated at 37 °C for 24 hours. The cells were then separated from the media by centrifugation. The cells were lysed using acetonitrile; both the media and lysed cells were syringe filtered to make them suitable for HPLC analysis. A successful conversion from phosphorylated precursor to 1 at yields of 84.2% in a standard media formulation, and 99.5% for bacterial cells grown in the presence of inosine. 1 was predominantly present in bacterial cells rather than the media, similar to the observations made by Yang et al. in 2007.37 This was as expected given the increased hydrophobic character of 1 compared with its precursor which would more favourably be located with the more hydrophobic environment of the cells compared with the media (see ESI Fig. S2.2 and Table S2.1†). These initial results indicate that it is possible to enzymatically produce aromatic peptide amphiphiles in vivo, and that the media containing inosine was beneficial to these studies as it increased the enzymatic activity.Similarly, E. coli cultures grown in the presence of inosine were also incubated with the phosphorylated precursors of 2, 3, 4 and 5 for 24 hours at 37 °C. Each of the four systems was successfully converted by the bacterial alkaline phosphatase with 2, 3, 4 and 5 yielding 31.4%, 49.5%, 60.7% and 87.9% respectively. However, the products are predominantly found within the media with only a small concentration found within the cell – 2.3%, 5.3%, 0.6% and 2.1% of 2, 3, 4 and 5 respectively (see ESI Fig. S2.3 and Table S2.1†). This is in contrast to the Fmoc-FY-OH system, where the product was found to remain predominantly within the cells. This observation can be linked to the hydrophilicity of the generated aromatic peptide amphiphiles – 2, 3, 4 and 5 have terminating hydrophilic amino acid residues; they are much less hydrophobic than Fmoc-FY-OH. Therefore, they have a much higher affinity to partition in the aqueous media, rather than be retained within the more hydrophobic cell membranes.
After confirming that the conversion to aromatic peptide amphiphiles had indeed taken place, and a fraction of the product is located within the cells, the bacterial cell response and viability was monitored using Live/Dead® cell staining. Each of the bacterial samples was grown in medium B (containing inosine) and treated with the self-assembling aromatic peptide amphiphile precursors of 1, 2, 3, 4 and 5. After 24 hours, the cells were treated with a solution of 1.67 μM SYTO 9 dye and 9.99 μM propidium iodide in DMSO to monitor cell viability: SYTO 9 staining live cells and propidium iodide staining inactive cells. The samples were viewed and counted using a fluorescent microscope (see Fig. 5). Untreated cultures demonstrated that only 6.1% of cells were inactive. However, cultures treated with aromatic peptide amphiphiles demonstrated a degree of increased cell inactivity, which we presume is linked to supramolecular structure formation or a temporary structure formation within the cells. The presence of 1 increased cell death to 34.9%; 2, 28.5%; 3, 33.1%; 4, 46.7%, and; 5, 24.9%, with each sample found to be statistically different from the untreated control illustrating the anti-microbial effects of treating E. coli bacterial cells with alkaline phosphatase responsive precursors of aromatic peptide amphiphiles. However, the resulting inactivity of treated cultures with precursors of 1, 2, 3, 4 or 5 were found not to be statistically different from each other, implicating that there is not a differential anti-microbial response between aromatic peptide amphiphiles tested on E. coli (see ESI†). It is interesting to note that the anti-microbial activity of 1 is similar to that of the more hydrophilic amphiphiles 2–5, despite it being retained by the cells to a much greater extent. Given that the enzymatic conversion occurs exclusively within the cells,47 this observation suggests that only a temporary formation of nanostructures, followed by dissolution and expulsion from the cell, is sufficient to trigger cell death. Alternatively, it may suggest that the role of the supramolecular structure formation is less important than anticipated. Alternatively, as only a small percentage of the self-assembling aromatic peptide amphiphiles 2, 3, 4 or 5 was found to be retained within the cells, one might speculate that the anti-microbial effect is due to the localised concentration of self-assembling aromatic peptide amphiphiles being sufficient for the (temporary) supramolecular structure formation, and in turn, cell inactivation.
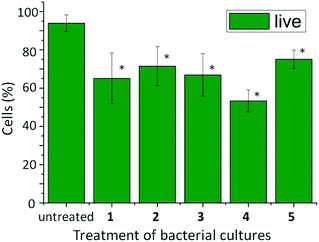 | ||
| Fig. 5 Percentage of active (live) cells in 20 mL E. coli bacterial cultures after treatment with 13.0 mg of the phosphorylated precursors of self-assembling aromatic peptide amphiphiles 1, 2, 3, 4 and 5. Data displayed are the average of three determinations ± std dev. One-way ANOVA was used to compare the treated cultures to the untreated control, with a P value of <0.05 considered to be significant (*). | ||
Conclusions
The production of peptide nanostructures has been demonstrated using alkaline phosphatase as an enzymatic trigger for the self-assembly of Fmoc-FY-OH (1), Fmoc-YT-OH (2), Fmoc-YS-OH (3), Fmoc-YN-OH (4), and Fmoc-YQ-OH (5). In vitro studies indicate molecular association and supramolecular structure formation which are consistent with previous studies,28,44 with the route to obtaining the self-assembling aromatic peptide amphiphile (esterase vs. phosphatase) having no pronounced effect on the resulting structures. It has been demonstrated that the inclusion of inosine within the culture media results in an over-expression of alkaline phosphatase when culturing E. coli. The non-assembling precursors can be converted to self-assembling aromatic peptide amphiphiles in vivo, with the location of the products being directly linked to the hydrophobicity of the products – the more hydrophobic Fmoc-FY-OH products accumulating within the cells and the hydrophilic Fmoc-YX-OH peptides being found in the media. Furthermore, the presence of the aromatic peptide amphiphiles within the cultured E. coli demonstrates an anti-microbial response. However, this response was insignificantly different regardless of the chemical structure of aromatic peptide amphiphiles.Notes and references
- C. Frantz, K. M. Stewart and V. M. Weaver, J. Cell Sci., 2010, 123, 4195 CrossRef CAS.
- M. M. Stevens and J. H. George, Science, 2005, 310, 1135 CrossRef CAS.
- V. Jayawarna, M. Ali, T. A. Jowitt, A. E. Miller, A. Saiani, J. E. Gough and R. V. Ulijn, Adv. Mater., 2006, 18, 611 CrossRef CAS.
- A. M. Smith, R. J. Williams, C. Tang, P. Coppo, R. F. Collins, M. L. Turner, A. Saiani and R. V. Ulijn, Adv. Mater., 2008, 20, 37 CrossRef CAS.
- C. Tang, A. M. Smith, R. F. Collins, R. V. Ulijn and A. Saiani, Langmuir, 2009, 25, 9447 CrossRef CAS.
- A. Mahler, M. Reches, M. Rechter, S. Cohen and E. Gazit, Adv. Mater., 2006, 18, 1365 CrossRef CAS.
- D. M. Ryan, T. M. Doran, S. B. Anderson and B. L. Nilsson, Langmuir, 2011, 27, 4029 CrossRef CAS.
- S. Debnath, A. Shome, D. Das and P. K. Das, J. Phys. Chem. B, 2010, 114, 4407 CrossRef CAS.
- L. Chen, K. Morris, A. Laybourn, D. Elias, M. R. Hicks, A. Rodger, L. Serpell and D. J. Adams, Langmuir, 2010, 26, 5232 CrossRef CAS.
- E. K. Johnson, D. J. Adams and P. J. Cameron, J. Mater. Chem., 2011, 21, 2024 RSC.
- G. Cheng, V. Castelletto, C. M. Moulton, G. E. Newby and I. W. Hamley, Langmuir, 2010, 26, 4990 CrossRef CAS.
- D. Bardelang, F. Camerel, J. C. Margeson, D. M. Leek, M. Schmutz, M. B. Zaman, K. Yu, D. V. Soldatov, R. Ziessel, C. I. Ratcliffe and J. A. Ripmeester, J. Am. Chem. Soc., 2008, 130, 3313 CrossRef CAS.
- C. Tang, R. V. Ulijn and A. Saiani, Langmuir, 2011, 27, 14438 CrossRef CAS.
- D. N. Woolfson and Z. N. Mahmoud, Chem. Soc. Rev., 2010, 39, 3464 RSC.
- J. H. van Esch, Langmuir, 2009, 25, 8392 CrossRef CAS.
- R. V. Ulijn and A. M. Smith, Chem. Soc. Rev., 2008, 37, 664 RSC.
- Z. Ge and S. Liu, Chem. Soc. Rev., 2013 10.1039/C3CS60048C.
- J. Hu, G. Zhang and S. Liu, Chem. Soc. Rev., 2012, 41, 5933 RSC.
- S. Roy and R. V. Ulijn, in Enzymatic Polymerisation, ed. A. R. A. H. A. Palmans, 2010, vol. 237, pp. 127–143 Search PubMed.
- Z. M. Yang, H. W. Gu, D. G. Fu, P. Gao, J. K. Lam and B. Xu, Adv. Mater., 2004, 16, 1440 CrossRef CAS.
- J. Gao, H. Wang, L. Wang, J. Wang, D. Kong and Z. Yang, J. Am. Chem. Soc., 2009, 131, 11286 CrossRef CAS.
- Z. M. Yang, G. L. Liang, L. Wang and B. Xu, J. Am. Chem. Soc., 2006, 128, 3038 CrossRef CAS.
- H. Wang, C. Ren, Z. Song, L. Wang, X. Chen and Z. Yang, Nanotechnology, 2010, 21, 225606 CrossRef.
- H. Wang, Z. Wang, D. Song, J. Wang, J. Gao, L. Wang, D. Kong and Z. Yang, Nanotechnology, 2010, 21, 155602 CrossRef.
- J. Gao, W. Zheng, D. Kong and Z. Yang, Soft Matter, 2011, 7, 10443 RSC.
- K. Thornton, A. M. Smith, C. L. R. Merry and R. V. Ulijn, Biochem. Soc. Trans., 2009, 37, 660 CrossRef CAS.
- Z. M. Yang, G. L. Liang, M. L. Ma, Y. Gao and B. Xu, Small, 2007, 3, 558 CrossRef CAS.
- M. Hughes, L. S. Birchall, K. Zuberi, L. A. Aitken, S. Debnath, N. Javid and R. V. U. Ulijn, Soft Matter, 2012, 8, 11565 RSC.
- A. R. Hirst, S. Roy, M. Arora, A. K. Das, N. Hodson, P. Murray, S. Marshall, N. Javid, J. Sefcik, J. Boekhoven, J. H. van Esch, S. Santabarbara, N. T. Hunt and R. V. Ulijn, Nat. Chem., 2010, 2, 1089 CrossRef CAS.
- Z. Yang, K. Xu, Z. Guo, Z. Guo and B. Xu, Adv. Mater., 2007, 19, 3152 CrossRef CAS.
- M. Hughes, P. W. J. M. Frederix, J. Raeburn, L. S. Birchall, J. Sadownik, F. C. Coomer, I. H. Lin, E. J. Cussen, N. T. Hunt, T. Tuttle, S. J. Webb, D. J. Adams and R. V. Ulijn, Soft Matter, 2012, 8, 5595 RSC.
- M. Hughes, H. Xu, P. W. J. M. Frederix, A. M. Smith, N. T. Hunt, T. Tuttle, I. A. Kinloch and R. V. Ulijn, Soft Matter, 2011, 7, 10032 RSC.
- J. B. Guilbaud, E. Vey, S. Boothroyd, A. M. Smith, R. V. Ulijn, A. Saiani and A. F. Miller, Langmuir, 2010, 26, 11297 CrossRef CAS.
- A. K. Das, R. Collins and R. V. Ulijn, Small, 2008, 4, 279 CrossRef CAS.
- R. J. Williams, A. M. Smith, R. Collins, N. Hodson, A. K. Das and R. V. Ulijn, Nat. Nanotechnol., 2009, 4, 19 CrossRef CAS.
- S. Toledano, R. J. Williams, V. Jayawarna and R. V. Ulijn, J. Am. Chem. Soc., 2006, 128, 1070 CrossRef CAS.
- Z. Yang, G. Liang, Z. Guo, Z. Guo and B. Xu, Angew. Chem., Int. Ed., 2007, 46, 8216 CrossRef CAS.
- Y. Gao, J. Shi, D. Yuan and B. Xu, Nat. Commun., 2012, 3, 1033 CrossRef.
- H. Csopak, G. Garellick and B. Hallberg, Acta Chem. Scand., 1972, 26, 2401 CrossRef CAS.
- D. Chappele, M. Iwatsubo and M. Lazdunsk, Biochemistry, 1974, 13, 3754 CrossRef.
- H. N. Fernley, Nat. – New Biol., 1973, 241, 110 CAS.
- B. Stec, K. M. Holtz and E. R. Kantrowitz, J. Mol. Biol., 2000, 299, 1303 CrossRef CAS.
- J. W. Sadownik, J. Leckie and R. V. Ulijn, Chem. Commun., 2011, 47, 728 RSC.
- W. P. Wang, Z. M. Yang, S. Patanavanich, B. Xu and Y. Chau, Soft Matter, 2008, 4, 1617 RSC.
- S. Fleming, P. W. J. M. Frederix, I. R. Sasselli, N. T. Hunt, R. V. Ulijn and T. Tuttle, Langmuir, 2013 DOI:10.1021/1a400994v.
- V. Worthington, Worthington Enzyme Manual, Worthington Biochemical Corporation, Lakewood, 1993 Search PubMed.
- A. I. Derman and J. Beckwith, J. Bacteriol., 1995, 177, 3764 CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Methods and materials, NMR, enzymatic activity assays and HPLC. See DOI: 10.1039/c3bm60135h |
| This journal is © The Royal Society of Chemistry 2013 |
