Paired diagnostic and pharmacodynamic analysis of rare non-small cell lung cancer cells enabled by the VerIFAST platform
Benjamin P.
Casavant†
a,
Lindsay N.
Strotman†
a,
Jacob J.
Tokar
a,
Stephanie M.
Thiede
b,
Anne M.
Traynor
bc,
J. Scott
Ferguson
bc,
Joshua M.
Lang
bc and
David J.
Beebe
*ac
aDepartment of Biomedical Engineering, Wisconsin Institutes for Medical Research, University of Wisconsin-Madison, 1111 Highland Ave., Madison, WI, USA. E-mail: djbeebe@wisc.edu; Tel: +1 608 262 2260
bDepartment of Medicine, Wisconsin Institutes for Medical Research, University of Wisconsin-Madison, 1111 Highland Ave., Madison, WI, USA
cCarbone Cancer Center, Wisconsin Institutes for Medical Research, University of Wisconsin-Madison, 1111 Highland Ave., Madison, WI, USA
First published on 10th September 2013
Abstract
Lung cancer is the leading cause of cancer-related deaths in the United States and worldwide. This has led to major research initiatives focusing on improving early diagnosis rate, as well as the development of pharmacodynamic biomarkers. However, broad clinical integration of these approaches is limited due to the invasive nature of lung biopsies, needle aspirates and resections. Recently, an advance for sampling suspicious lung nodules to collect mini-bronchoalveolar lavage (mBAL) samples was shown to be diagnostically relevant but limited by standard cytology techniques leading to low sensitivity and specificity. In addition, a second non-invasive method that holds great promise is the collection of circulating tumor cells, a rare population of tumor cells that have shed into peripheral circulation from primary or metastatic tumor sites, from blood. Here, we utilize a recently published platform, VerIFAST, for the capture and proteomic analysis of rare cells, to isolate cells of interest from lung cancer patients using both mBAL and blood samples. The VerIFAST platform leverages surface tension at the microscale to pin aqueous and oil fluids in adjacent chambers to create a virtual filter between two aqueous fluids. In this manuscript, the VerIFAST was further enhanced to include oil pinning, which allowed on-device tumbling, further eliminating a laborious and time consuming step that could result in increased sample loss. Finally, we further developed the base assays used in standard histopathologic assays for diagnostic and pharmacodynamic analysis of these rare lung cancer cells. Specifically, we examined thyroid transcription factor-1 (TTF-1) signal intensity, in which loss is associated with more aggressive disease, and epidermal growth factor receptor (EGFR) signal intensity, which is a high value therapeutic target in lung cancer.
Introduction
Lung cancer is the leading cause of cancer-related death in the United States and worldwide.1 Non-small cell lung cancer (NSCLC) represents 88% of lung cancer diagnoses, with small cell lung cancer (SCLC) comprising the remaining 12%. The most frequently diagnosed subtype of NSCLC is adenocarcinoma. Before metastasis occurs and the tumor is <30 mm, the 5-year survival is near 77% (Stage 1A).2 When the primary tumor is >30 mm or metastasis occurs, that survival rate drops to between 58% and 9% (Stages IB–IV). These clinical observations have led to major research initiatives focused on improving the early diagnosis rate as well as the development of pharmacodynamic biomarkers that enable precision medical care for patients with advanced disease.3 Recent advances in these areas have involved high content molecular analyses from tumor cells isolated from lung biopsies. For example, Sequist et al. performed serial tumor biopsies from patients with advanced NSCLC for paired histologic and genomic analysis.4 These authors identified unexpected histologic subtypes of lung cancer in serial biopsies that altered therapeutic management and improved patient outcomes. However, broad clinical integration of these approaches is limited due to the nature of these invasive lung biopsies or resections including, but not limited to, hemorrhage, infection, and pneumothorax.5 These complications also occur with significantly higher frequency on lung lesions <4 cm in size, as is commonly found in early stage disease.6 Improving cancer care for patients across all stages of lung cancer will require the development of minimally invasive techniques for tumor sampling and rare cell analysis.One recent advance for sampling suspicious lung nodules is known as electromagnetic navigation bronchoscopy (ENB).7 ENB utilizes advanced hardware and software to guide bronchoscopic tools directly to suspicious lung nodules for the early diagnosis of malignancy. Following nodule visualization, a mini-bronchoalveolar lavage (mBAL) uses 20–50 mL of saline solution to wash cells from the area of the nodule. The collection of mBAL during ENB thus allows sampling of cells in proximity with very small lung nodules in a significantly less invasive manner than fine needle aspirates or core needle biopsies. This method has previously been shown to be diagnostically relevant, as the isolation of tumor cells has revealed insight into the genetics of malignant tumors.8,9 However, ENB and mBAL have been limited by standard cytology techniques to identify tumor cells in a complex mBAL specimen that includes leukocytes, stromal and non-malignant epithelial cells. Thus, the relatively low sensitivity and specificity of these assays for rare tumor cells in heterogeneous mBAL samples limits broad utility for diagnostic purposes.
A second method for minimally invasive sampling of tumor cells is through the use of standard blood draws for the capture and analysis of circulating tumor cells (CTCs).10 CTCs are a rare population of tumor cells shed into peripheral circulation from primary and metastatic tumor sites that may both contribute to the development of metastatic disease and reflect the heterogeneity that likely exists between various tumor deposits. This hematogenous dissemination of tumor cells has long been recognized as one route by which solid tumors can create secondary metastatic sites – the main cause of patient mortality.11 CTCs present an alternative method to collect lung cancer cells with a less invasive approach than the mBAL methods, but present a more difficult and heterogeneous population, as lung CTCs can exist at approximately 5 cells per 10 million peripheral cells.12 The frequency of CTCs in peripheral blood is also significantly lower in early versus late stage disease. Though the enumeration of CTCs has been linked to prognostic relevance in both NSCLC and SCLC13–15 their utility can be expanded towards pharmacodynamic and other biologically and clinically relevant endpoints.
Unfortunately, the clinical utility of these minimally invasive techniques is often limited by the small amount of tumor cells recovered for standard cytologic and histopathologic analysis.16 Thus advancing minimally invasive tumor sampling into clinical utility requires technologic advances that permit high content, cellular analyses on these rare cell populations. With physical characteristic scales enabling high precision relative to macroscale techniques, microfluidics is well positioned to capture and assess these rare cell populations with minimal sample loss. These microfluidic methods include the CTC Herringbone chip,17 the Micro-Hall Detector,18 DEP approaches,19 among others. Emerging studies continue to highlight the need for more sophisticated methods of CTC capture and analysis that go beyond enumeration.20 However, the ability to use these methods on patient samples other than blood (i.e. bone marrow analysis, urine samples, mBAL) for rare cell capture and analysis has not been fully explored.
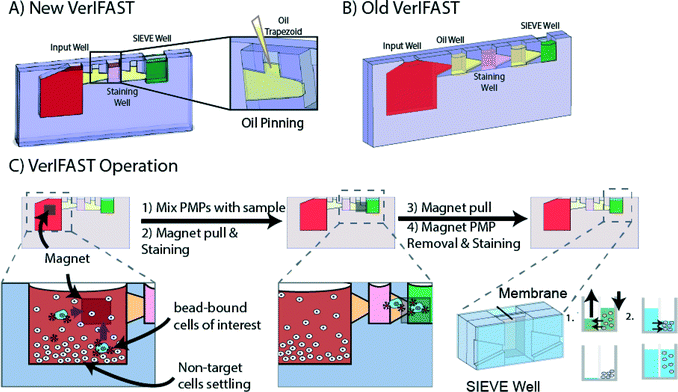
We have recently published a technique for the capture and proteomic analysis of rare cells.21 The VerIFAST platform leverages surface tension at the microscale to pin aqueous and oil fluids in adjacent chambers to create a virtual filter between two aqueous wells.22,23 Using paramagnetic particles (PMPs) with attached antibody, specific cell populations can be targeted and isolated from complex backgrounds through a simple traverse of the oil barrier. Further, this platform integrated a microporous membrane into an aqueous chamber, enabling multiple fluid transfers without the need for cell transfer or centrifugation. This integrated platform enabled removal of PMPs used for capture as well as multi-step molecular assays with essentially no cell loss during processing or analysis. We previously demonstrated the capacity of this platform to perform complex extracellular and intracellular staining for any protein target of interest.
In this report, we further enhance the VerIFAST's capabilities by integrating unique oil-pinning interfaces to enable chip tumbling without disruption of the aqueous-oil interfaces between each well and reduces the footprint of this device. We demonstrate the ability of the enhanced VerIFAST microfluidic platform to integrate with minimally invasive techniques in use for patients with lung cancer: mBAL samples and peripheral blood (CTC) samples. We further develop the base assays used in standard histopathologic assays for diagnostic and pharmacodynamic analysis of these rare lung cancer cells. The thyroid transcription factor-1 (TTF-1) is a diagnostic marker identified in primary lung adenocarcinomas and loss of expression correlates with more aggressive disease.24 TTF-1 expression was evaluated in mBAL as a diagnostic assay and CTC samples as a confirmatory marker of a malignant cell. We pair TTF-1 analysis with protein analysis and quantification of staining intensity of the epidermal growth factor receptor (EGFR), which, when mutated by an activating deletion or point mutation in the kinase domain of exons 19 or 21, serves a high value therapeutic target in lung cancer.25 These two targets have been shown to have utility as diagnostic, predictive and pharmacodynamic biomarkers.26
Methods and materials
Device fabrication
The device was fabricated via CNC milling (PCNC770, Tormach, USA) from 2 mm thick polystyrene (PS, Goodfellow, UK). The height and widths varied for each well with differing volume capacities. The input well held 200 μL, whereas subsequent wells consisting of the staining well and sieve well held 30 μL and 50 μL respectively. The wells were connected via a trapezoid that had a 300 mm depth and height that tapered from 2 mm to 0.8 mm with a channel above for oil loading. A second PS layer containing an additional 50 μL well was solvent bonded using acetonitrile (Sigma-Aldrich, USA) to the first PS piece, with the membrane sandwiched between. The sieve well used for paramagnetic particle (PMP) removal and intracellular staining employed an 8 μm microporous membrane (Part PET8025100, Sterlitech, USA). A pressure sensitive adhesive was applied to each side of the device to contain the fluids (MicroAmp, Applied Biosystems, USA).Paramagnetic particle preparation
Streptavidin coupled PMPs from the Dynabeads® FlowComp™ Flexi (Life Technologies, USA) at a concentration of 250 μg per a reaction were used for all experiments. The PMPs were washed twice and resuspended in 0.01% Tween-20 in phosphate buffered solution (PBS). 0.4 μg mL−1 of epithelial cell adhesion molecule (EpCAM, R&D Systems, USA) biotinylated according to the Dynabeads® FlowComp™ Flexi manufacturer's directions were added to the solution. The PMPs and antibodies were mixed for 30 minutes at RT followed by three washes and resuspension in 0.1% BSA in PBS.Cell culture
The H358 bronchoalveolar NSCLC cell line and the A549 adenocarcinoma NSCLC cell line (ATCC, USA) were cultured in Cornig Cellgro® RPMI 1640 Medium (VWR, USA) containing 10% fetal bovine serum and 1% Pen-Strep. All cells were cultured at 37 °C and maintained under 5% CO2 in polystyrene flasks until confluent. Cells were released using a 0.05% trypsin/EDTA solution and collected via centrifugation.Immunohistochemistry
All cell lines were stained with epithelial cell adhesion molecule conjugated to PE (EpCAM, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 dilution, Santa Cruz Biotechnology, USA), Hoecsht stain (1
10 dilution, Santa Cruz Biotechnology, USA), Hoecsht stain (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250), epidermal growth factor receptor conjugated to FITC (EGFR, 1
250), epidermal growth factor receptor conjugated to FITC (EGFR, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, Abcam, USA) and transcription termination factor (TTF-1, 1
50, Abcam, USA) and transcription termination factor (TTF-1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, Santa Cruz Biotechnology, USA). Both the TTF-1 and EGFR primary antibody were located throughout the cell. A secondary antibody labeled with AlexaFlour-488 (Life Technology, USA) was used at a dilution of 1
50, Santa Cruz Biotechnology, USA). Both the TTF-1 and EGFR primary antibody were located throughout the cell. A secondary antibody labeled with AlexaFlour-488 (Life Technology, USA) was used at a dilution of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 for TTF-1. Samples were imaged using a Nikon Eclipse Ti (Nikon) and images were acquired with NIS Elements AR 4.10 software (Nikon). Image processing was completed using Image J, briefly a ROI was created for the EpCAM stain and used as a mask for TTF-1 and EGFR to measure signal intensity. The mean signal intensity was multiplied by the area. If cells clumped the total intensity was divided by the number of nuclei present before multiplication by area.
100 for TTF-1. Samples were imaged using a Nikon Eclipse Ti (Nikon) and images were acquired with NIS Elements AR 4.10 software (Nikon). Image processing was completed using Image J, briefly a ROI was created for the EpCAM stain and used as a mask for TTF-1 and EGFR to measure signal intensity. The mean signal intensity was multiplied by the area. If cells clumped the total intensity was divided by the number of nuclei present before multiplication by area.
Device operation
5 μL silicon oil (Fisher Scientific, USA) was added to the first trapezoid were it pinned via interfacial tension. PMPs and sample supplemented with 2 mM EDTA, 0.001% Tween-20, and 0.1% BSA in PBS was added to the input well. The device containing the sample was mixed on a tumbler for 30 min at 4 °C, in order for the cells to bind to the PMPs. Next, the second trapezoid was filled with silicon oil before the addition of a staining cocktail. The staining cocktail consisted of EpCAM conjugated to PE (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 dilution, Santa Cruz Biotechnology, USA), a second EpCAM conjugated to PE (1
10 dilution, Santa Cruz Biotechnology, USA), a second EpCAM conjugated to PE (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution, Abcam, USA) to further increase EpCAM signal, Hoescht (1
100 dilution, Abcam, USA) to further increase EpCAM signal, Hoescht (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250) and CD45 conjugated to Alexa Fluor 647 (1
250) and CD45 conjugated to Alexa Fluor 647 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 dilution, Biolegend, USA). A handheld magnet (K&J Magnetics, USA) transferred the PMPs into the staining well were it was incubated for 30 minutes at 4 °C. Finally, 100 μL PMP release buffer from the Dynabeads® FlowComp™ Flexi kit was added to the sieve well and the same magnet used to transfer the PMPs into this well. The magnet was then used to pull the excess unbound PMPs through the membrane to the back of the sieve well. After 30 minutes at RT in the release buffer to further remove excess PMPs from cells, they were removed. Cells were permeabilized for 30 minutes by the addition of 1% Tween 20 in PBS to the sieve well. Following permeabilization, a primary antibody, either TTF-1 or EGFR-FITC, was added and incubated overnight at 4 °C in a humidified environment. After 24 hours, the samples were washed 3 times with 0.1% BSA in PBS. For the TTF-1 samples, a secondary antibody labeled with AlexaFlour-488 was added for 1 hour at RT followed by 5 washes with 0.1% BSA in PBS. Samples were imaged within device.
10 dilution, Biolegend, USA). A handheld magnet (K&J Magnetics, USA) transferred the PMPs into the staining well were it was incubated for 30 minutes at 4 °C. Finally, 100 μL PMP release buffer from the Dynabeads® FlowComp™ Flexi kit was added to the sieve well and the same magnet used to transfer the PMPs into this well. The magnet was then used to pull the excess unbound PMPs through the membrane to the back of the sieve well. After 30 minutes at RT in the release buffer to further remove excess PMPs from cells, they were removed. Cells were permeabilized for 30 minutes by the addition of 1% Tween 20 in PBS to the sieve well. Following permeabilization, a primary antibody, either TTF-1 or EGFR-FITC, was added and incubated overnight at 4 °C in a humidified environment. After 24 hours, the samples were washed 3 times with 0.1% BSA in PBS. For the TTF-1 samples, a secondary antibody labeled with AlexaFlour-488 was added for 1 hour at RT followed by 5 washes with 0.1% BSA in PBS. Samples were imaged within device.
Cell capture efficiency
For quantification experiments H358 cells were incubated for ten minutes with 2 mM calcein AM (Life Technologies, USA) in serum free RPMI media. The cells were centrifuged and washed once in PBS, then counted with a hemocytometer and re-suspended in PBS. H358 cells were spiked in PBS with 0.1% BSA and 2 mM EDTA at 10, 100 and 1000 cells per a device. Cells were captured and processed as described above without additional staining.Patient samples
Lung lavage and blood samples were collected from NSCLC patients who had signed their respective Informed Consent documents under two separate University of Wisconsin IRB approved protocols. For sample processing the sample was split into two for either EGFR or TFF-1 staining. Lung lavage samples were centrifuged at 1200 × g for 5 minutes. The resulting cell pellets were fixed (BD Cytofix™, USA) for 30 minutes on ice before being washed once in PBS. Blood samples underwent a standard density centrifugation (Ficoll-Paque, GE, USA) to isolate peripheral mononuclear blood cells (PBMCs), which were then fixed and washed once in PBS. Samples were then processed using the device as described above. Circulating tumor cells (CTCs) and cells of interest in the lung lavage samples were defined as having an intact nucleic, EpCAM positive and CD45 negative. Image processing was completed as described above.Results and discussion
VerIFAST innovation
The VerIFAST (Fig. 1A) utilizes the relative dominance of surface tension over gravity in the microscale to load immiscible phases side by side without density driven stratification, in order to eliminate laborious and time consuming wash and centrifugation steps required by most sample preparation protocols. VerIFAST builds upon a previous platform that was designed to take advantage of settling of non-target cells to help decrease non-specific carryover.21 This platform was further modified to incorporate a membrane (<8 μm) in the sieve well, which was used to remove unbound PMPs that might interfere with imagining and enable fluid exchanges for intracellular staining with no direct contact (Fig. 1B). Sample transfer to VerIFAST increases the workflow and represents a potential step where sample could be lost. To enable constant mixing and eliminate a transfer step we utilized oil pinning within the VerIFAST (Fig. 1A). In the original VerIFAST design, mixing within the device was hindered because the oil would creep into adjacent compartments during device tumbling. Therefore, we took advantage of the low aspect ratio of the trapezoid for device redesign to enable oil pinning. While most pinning or virtual wall formations have been described using aqueous fluids, little focus has centered on the use of oil.21,27,28 Here, we are able to exploit oil pinning due to the dominance of surface tension in the microscale. Finally, the inputs of each well were also geometrically constrained such that aqueous fluids remained pinned while mixing.To operate, oil is first pinned within the confines of the trapezoidal regions using a pipette through the respective input channels. Next, the input well, staining well and sieve well were loaded with the assay-specific buffers. Once the sample was in the input well, it was placed on a tumbler and incubated with PMPs to bind cells (EpCAM+) of interest. Following binding, a simple handheld magnet transferred the PMPs along the front side of the devices through the oil-filled trapezoidal region into the staining well that contained a cocktail of antibodies to enable determination of cells of interest in the sieve well. After the second incubation, the cell bound PMPs were transferred again along the front side of the VerIFAST to the sieve well containing release buffer. Once in the well, a brief incubation was needed to help remove cell-bound PMPs that could interfere with imaging. A magnet held on the backside of the device removed the unbound PMPs from the front of the sieve well to the back, while retaining cells in the front side of the sieve well. Using the backside of the sieve well, fluids could be easily added or removed without manipulating the cells, further reducing potential sample loss. This enabled permeabilization and intercellular staining for cell targets including EGFR and TTF-1. Overall, the operational simplicity and minimization of transfer and manipulation steps reduce the chance of loss of the rare sample and decrease laboriousness of the procedure.
Integrated capture and molecular analysis lung cancer cells
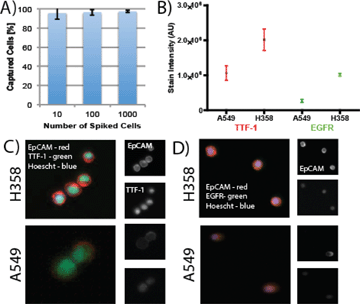
The VerIFAST utilizes a flexible, antibody-based capture methodology to isolate cells of interest from a heterogeneous population. Given the strength of EpCAM-based capture of lung cancer cells demonstrated by the Veridex platform and others, we evaluated the ability of VerIFAST to capture lung cells targeting this protein. As shown in Fig. 2A, the VerIFAST exhibits a capture efficiency of greater than 90% across three varying concentrations of H358 cells (Fig. 2A). In order to assess the levels of expression for TTF-1, a molecular marker with diagnostic relevance, and EGFR, contrived cell lines with varying expression of both were used to validate the assay (Fig. 2B). ImageJ was used for quantification, using EpCAM staining to define cellular boundaries, and measuring the fluorescent intensity of the stain interior to EpCAM. For TTF-1, the H358 NSCLC cell line was used for positive expression of TTF-1, with A549 cells demonstrating decreased expression of its protein (Fig. 2C). Using this analysis, the two cell populations were statistically different (n = 5, p < 0.05). The same analysis was applied to EGFR samples (Fig. 2D), of which there is variable expression of EGFR between the two cell lines.29 H358 cells were observed to express significantly higher EGFR than A549 (n = 5, p < 0.005), allowing the assessment of heterogeneous expression in patient samples.TTF-1 and EGFR expression in patients samples
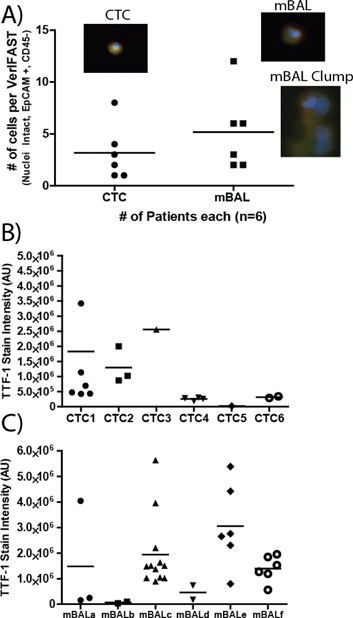
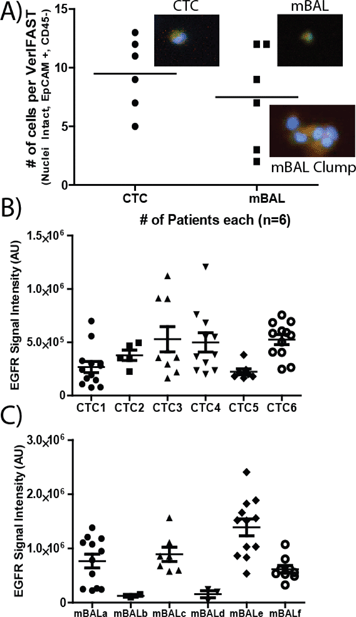
TTF-1 is utilized in standard pathologic analysis to confirm the diagnosis of primary adenocarcinoma of the lung from standard tissue biopsies. Because mBAL samples can be collected in an earlier disease state then CTCs and could also contain benign lung epithelial cells, molecular assays for TTF-1 on rare cells will be helpful in using these samples for the early diagnosis of primary adenocarcinoma of the lung. Fig. 3A shows the average number of cells collected from patients as well as examples of stained cells and clumps of interest that are EpCAM positive, CD45 negative and contain an intact nuclei. In five collected mBAL samples (Fig. 3C), the heterogeneity of TTF-1 samples was clear, as the fluorescent intensity of samples could range to nearly 5-fold differences in cellular expression of TTF-1. Interestingly, using H358 and A549 staining as positive and negative controls, respectively, only two mBAL samples displayed full loss of TTF-1 expression (below negative control A549 cells). In one of the samples, however, the expression varied and there was visibly a subset of cells that were below, but not the entire cell population (mBALa). Identical analysis of TTF-1 in CTC samples demonstrated significant heterogeneity across CTCs (Fig. 3B) that may reflect the different extent of tumor burden among the patients sampled, as well as possible epithelial-mesenchymal transitions30 that may characterize components of the metastatic process.Protein expression of EGFR is detected in up to 80% of NSLCC clinical specimens31 Cetuximab, a chimeric IgG1 monoclonal antibody that inhibits EGFR function, demonstrated clinical efficacy when NSCLC patients were selected by EGFR protein expression.32 The value of EGFR as a therapeutic target is heightened when activating mutations are present in the kinase domains of exons 19 and 21.33,34 Therefore, the ability to perform molecular assays on EGFR in lung cancer cells from patients in the least invasive manner, repeatedly in a longitudinal manner over the course of their disease, would permit optimal timing and selection of the most individualized EGFR-directed therapies. In Fig. 4, we demonstrate the ability of VerIFAST to detect EGFR protein expression in NSCLC patients by obtaining and analyzing clinical specimens without the invasive and potentially risky need for needle aspirate or core biopsies or by obtaining specimens from surgical resections. Signal intensity of EGFR was more consistent among the CTC samples (Fig. 4B), compared to more variable EGFR expression in the mBAL samples (Fig. 4C). This may be related to the fact that patients who underwent mBAL sampling had a lower disease stage, and therefore a lower tumor burden, compared to patients who underwent CTC sampling.
Conclusions
Here, we have presented a comprehensive platform that is able to collect and analyze lung cancer samples (both mBAL and blood) using collection techniques that are significantly less invasive than lung biopsies or resections. Leveraging the flexibility of the VerIFAST to use a variety of samples beyond blood, mBAL samples were collected and analyzed as an early collection and diagnostic technique. Blood samples were analyzed for CTCs as a second method towards the analysis of lung cancer. The ability of the VerIFAST to collect and analyze these patient samples is particularly useful for lung cancer, as minimally invasive sample collection that can be repeated serially allows for safer and longitudinal studies that will obviate the need for repeated biopsies and their attendant complications. TTF-1 was used as a diagnostic target, and revealed the heterogeneity of the lung lavage samples. The variability of EGFR expression in mBAL samples relative to CTC samples is an interesting observation, as patients that have CTCs are more likely to have advanced disease. This study was a proof-of-concept study for the use of mBAL and CTCs for the evaluation of EGFR and TTF-1, rather than only enumeration of discovered cells. The success of this study allows these samples to be used moving forward in a clinical trial to assess the validity of these endpoints. Further, the VerIFAST was enhanced with new features that facilitated, for the first time, the analysis of both mBAL samples and CTCs from blood samples from lung cancer patients.Acknowledgements
This work was supported by a grant from the Bill & Melinda Gates Foundation through the Grand Challenges in Global Health initiative, NIH Grant #5R33CA137673, NSF GRFP DGE-0718123, a UW Institute for Clinical and Translational Research-Type 1 Research Pilot Award, UW Carbone Cancer Center Investigator Initiated Pilot grant and a Prostate Cancer Foundation Young Investigator award to Dr. Lang. David J. Beebe holds equity in Salus Discovery, LLC, Bellbrook Labs, LLC and Ratio, Inc.References
- R. Siegel, D. Naishadham and A. Jemal, Ca-Cancer J. Clin., 2013, 63, 11–30 CrossRef PubMed.
- F. C. Detterbeck, P. J. Mazzone, D. P. Naidich and P. B. Bach, Chest, 2013, 143, e78S–92S Search PubMed.
- D. A. Eberhard, G. Giaccone and B. E. Johnson, J. Clin. Oncol., 2008, 26, 983–994 CrossRef PubMed.
- L. V. Sequist, B. A. Waltman, D. Dias-Santagata, S. Digumarthy, A. B. Turke, P. Fidias, K. Bergethon, A. T. Shaw, S. Gettinger, A. K. Cosper, S. Akhavanfard, R. S. Heist, J. Temel, J. G. Christensen, J. C. Wain, T. J. Lynch, K. Vernovsky, E. J. Mark, M. Lanuti, A. J. Iafrate, M. Mino-Kenudson and J. A. Engelman, Sci. Transl. Med., 2011, 3, 75ra26 CrossRef PubMed.
- D. W. Hsia, K. W. Jensen, D. Curran-Everett and A. I. Musani, J. Bronchology Interv. Pulmonol., 2012, 19, 5–11 CrossRef PubMed.
- L. S. Poulou, P. Tsagouli, P. D. Ziakas, D. Politi, R. Trigidou and L. Thanos, Acta Radiol., 2013 Search PubMed.
- S. S. Dhillon and E. U. Dexter, J. Carcinog., 2012, 11, 19 CrossRef PubMed.
- S. A. Ahrendt, J. T. Chow, L. H. Xu, S. C. Yang, C. F. Eisenberger, M. Esteller, J. G. Herman, L. Wu, P. A. Decker, J. Jen and D. Sidransky, J. Natl. Cancer Inst., 1999, 91, 332–339 CrossRef CAS PubMed.
- D. Ha, H. Choi, F. A. Almeida, A. Arrossi, J. Brainard, J. Cicenia, C. Farver, T. Gildea, M. S. Machuzak and P. Mazzone, J. Bronchology Interv. Pulmonol., 2013, 20, 10–15 CrossRef PubMed.
- G. Attard and J. S. de Bono, Curr. Opin. Genet. Dev., 2011, 21, 50–58 CrossRef CAS PubMed.
- G. R. Mundy, Nat. Rev. Cancer, 2002, 2, 584–593 CrossRef CAS PubMed.
- V. S. Nair, K. V. Keu, M. S. Luttgen, A. Kolatkar, M. Vasanawala, W. Kuschner, K. Bethel, A. H. Iagaru, C. Hoh, J. B. Shrager, B. W. Loo Jr., L. Bazhenova, J. Nieva, S. S. Gambhir and P. Kuhn, PLoS One, 2013, 8, e67733 CAS.
- M. G. Krebs, J. M. Hou, R. Sloane, L. Lancashire, L. Priest, D. Nonaka, T. H. Ward, A. Backen, G. Clack, A. Hughes, M. Ranson, F. H. Blackhall and C. Dive, J. Thorac. Oncol., 2012, 7, 306–315 CrossRef PubMed.
- J. M. Hou, M. G. Krebs, L. Lancashire, R. Sloane, A. Backen, R. K. Swain, L. J. Priest, A. Greystoke, C. Zhou, K. Morris, T. Ward, F. H. Blackhall and C. Dive, J. Clin. Oncol., 2012, 30, 525–532 CrossRef PubMed.
- M. G. Krebs, R. Sloane, L. Priest, L. Lancashire, J. M. Hou, A. Greystoke, T. H. Ward, R. Ferraldeschi, A. Hughes, G. Clack, M. Ranson, C. Dive and F. H. Blackhall, J. Clin. Oncol., 2011, 29, 1556–1563 CrossRef PubMed.
- D. C. Danila, K. Pantel, M. Fleisher and H. I. Scher, Cancer J., 17, 438–450 CrossRef CAS PubMed.
- S. Nagrath, L. V. Sequist, S. Maheswaran, D. W. Bell, D. Irimia, L. Ulkus, M. R. Smith, E. L. Kwak, S. Digumarthy, A. Muzikansky, P. Ryan, U. J. Balis, R. G. Tompkins, D. A. Haber and M. Toner, Nature, 2007, 450, 1235–1239 CrossRef CAS PubMed.
- D. Issadore, J. Chung, H. Shao, M. Liong, A. A. Ghazani, C. M. Castro, R. Weissleder and H. Lee, Sci. Transl. Med., 2012, 4, 141ra192 CrossRef PubMed.
- I. J. Vishal Gupta, M. Garza, V. O. Melnikova, D. K. Hasegawa, R. Pethig and D. W. Davis, Biomicrofluidics, 2012, 6, 24133 Search PubMed.
- U. Dharmasiri, M. A. Witek, A. A. Adams and S. A. Soper, Annu. Rev. Anal. Chem., 2010, 3, 409–431 CrossRef CAS PubMed.
- B. P. Casavant, D. J. Guckenberger, S. M. Berry, J. T. Tokar, J. M. Lang and D. J. Beebe, Lab Chip, 2013 Search PubMed.
- S. M. Berry, E. T. Alarid and D. J. Beebe, Lab Chip, 2011, 11, 1747–1753 RSC.
- S. M. Berry, L. N. Strotman, J. D. Kueck, E. T. Alarid and D. J. Beebe, Biomed. Microdevices, 2011, 13, 1033–1042 CrossRef PubMed.
- D. Tan, Q. Li, G. Deeb, N. Ramnath, H. K. Slocum, J. Brooks, R. Cheney, S. Wiseman, T. Anderson and G. Loewen, Hum. Pathol., 2003, 34, 597–604 CrossRef CAS.
- T. John, G. Liu and M. S. Tsao, Oncogene, 2009, S14–23 CrossRef CAS PubMed 28 Suppl 1.
- J. S. Lee, H. R. Kim, C. Y. Lee, M. Shin and H. S. Shim, Ann. Surg. Oncol., 2013 Search PubMed.
- J. Atencia and D. J. Beebe, Nature, 2005, 437, 648–655 CrossRef CAS PubMed.
- B. Zhao, N. O. Viernes, J. S. Moore and D. J. Beebe, J. Am. Chem. Soc., 2002, 124, 5284–5285 CrossRef CAS PubMed.
- S. Derer, P. Bauer, S. Lohse, A. H. Scheel, S. Berger, C. Kellner, M. Peipp and T. Valerius, J. Immunol., 2012, 189, 5230–5239 CrossRef CAS PubMed.
- R. L. Bitting, R. Boominathan, C. Rao, G. Kemeny, B. Foulk, M. A. Garcia-Blanco, M. Connelly and A. J. Armstrong, Methods, 2013 Search PubMed.
- T. Kanematsu, S. Yano, H. Uehara, Y. Bando and S. Sone, Oncol. Res., 2003, 13, 289–298 Search PubMed.
- R. Pirker, J. R. Pereira, A. Szczesna, J. von Pawel, M. Krzakowski, R. Ramlau, I. Vynnychenko, K. Park, C. T. Yu, V. Ganul, J. K. Roh, E. Bajetta, K. O'Byrne, F. de Marinis, W. Eberhardt, T. Goddemeier, M. Emig and U. Gatzemeier, Lancet, 2009, 373, 1525–1531 CrossRef CAS.
- F. A. Shepherd, J. Rodrigues Pereira, T. Ciuleanu, E. H. Tan, V. Hirsh, S. Thongprasert, D. Campos, S. Maoleekoonpiroj, M. Smylie, R. Martins, M. van Kooten, M. Dediu, B. Findlay, D. Tu, D. Johnston, A. Bezjak, G. Clark, P. Santabarbara and L. Seymour, N. Engl. J. Med., 2005, 353, 123–132 CrossRef CAS PubMed.
- R. Rosell, E. Carcereny, R. Gervais, A. Vergnenegre, B. Massuti, E. Felip, R. Palmero, R. Garcia-Gomez, C. Pallares, J. M. Sanchez, R. Porta, M. Cobo, P. Garrido, F. Longo, T. Moran, A. Insa, F. De Marinis, R. Corre, I. Bover, A. Illiano, E. Dansin, J. de Castro, M. Milella, N. Reguart, G. Altavilla, U. Jimenez, M. Provencio, M. A. Moreno, J. Terrasa, J. Munoz-Langa, J. Valdivia, D. Isla, M. Domine, O. Molinier, J. Mazieres, N. Baize, R. Garcia-Campelo, G. Robinet, D. Rodriguez-Abreu, G. Lopez-Vivanco, V. Gebbia, L. Ferrera-Delgado, P. Bombaron, R. Bernabe, A. Bearz, A. Artal, E. Cortesi, C. Rolfo, M. Sanchez-Ronco, A. Drozdowskyj, C. Queralt, I. de Aguirre, J. L. Ramirez, J. J. Sanchez, M. A. Molina, M. Taron and L. Paz-Ares, Lancet Oncol., 2012, 13, 239–246 CrossRef CAS.
Footnote |
| † These authors contributed equally to this work |
| This journal is © The Royal Society of Chemistry 2014 |