Integrating a thermoresponsive copolymer with host–guest interactions for fabricating molecular recognition surfaces†
Xiujuan
Shi
a,
Gaojian
Chen
*ab,
Lin
Yuan
a,
Zengchao
Tang
a,
Wei
Liu
a,
Qiang
Zhang
c,
David M.
Haddleton
c and
Hong
Chen
*a
aThe Key Lab of Health Chemistry and Molecular Diagnosis of Suzhou, College of Chemistry, Chemical Engineering and Materials Science, Soochow University, Suzhou 215123, P. R. China. E-mail: chenh@suda.edu.cn; Fax: +86 512 65880827; Tel: +86 512 65880827
bCenter for Soft Condensed Matter Physics and Interdisciplinary Research, Soochow University, Suzhou, 215006, P. R. China. E-mail: gchen@suda.edu.cn; Tel: +86 512 65884406
cDepartment of Chemistry, University of Warwick, Coventry CV4 7AL, UK
First published on 16th June 2014
Abstract
A platform capable of integrating variable molecular recognition moieties, having tunable function and regenerable/reusable ability has been developed to build bio-functional surfaces. More specifically, mannose and biotin-modified β-CD were incorporated into poly(N-isopropylacrylamide-co-1-adamantan-1-ylmethyl acrylate) [poly(NIPAAm-co-Ada)] surfaces by host–guest interactions to investigate their specific interaction with ConA and avidin, respectively. The surfaces have showed thermoresponsively tunable recognition for specific proteins, while showing resistance to nonspecific protein adsorption. By varying the Ada content, the regulation of specific protein adsorption in different temperature ranges could be achieved. The poly(NIPAAm-co-Ada) surfaces can be easily regenerated and reused for bio-functionalization.
Conceptual insightsThis work presented the design of a platform capable of integrating variable molecular recognition moieties, having tunable function and regenerable/reusable ability. These features are important in the applications of bio-related devices, considering their sophisticated fabrication and complex surface modification. The key to the design is PNIPAAm and host–guest interactions. The antifouling properties and smart switching functions of PNIPAAm were simultaneously introduced to improve the signal-to-noise ratio, and the host–guest interaction was adopted to reversibly integrate variable molecular-recognition moieties. Two model surfaces were fabricated utilizing the aforementioned approach and they displayed excellent reusability and thermoresponsively tunable recognition for specific proteins, while showing resistance to nonspecific proteins. The versatile approach emphasizes the possibilities of its introduction in biomedical and biotechnological applications. |
Introduction
Molecular recognition, which plays an important role in biological systems and is observed in between sugar–lectin, antigen–antibody, DNA–protein, RNA–ribosome, etc., is essential for life. Therefore, it has been widely used in biosensors, bioseparation, bioanalysis, microfluidic devices and biomaterials for human health.1–7 To improve the selectivity, accuracy, controllability, and reproducibility of devices, it is very important to understand, control and utilize the specific and nonspecific interactions between molecules in recognition.7–13 The decrease of nonspecific protein adsorption imparts biocompatibility to blood contacting materials, and reduces false signals in biosensors; the selective and tunable protein adsorption enhances the function of materials and the localized controllability. Poly(N-isopropylacrylamide) (PNIPAAm) is a classical thermoresponsive polymer for controlling protein adsorption and cell adhesion corresponding to its swelling/shrinking status at temperatures below and above the lower critical solution temperature (LCST).14,15 The previous work of our group has found that PNIPAAm surfaces with high grafting density exhibit good protein-resistant properties in a certain narrow range (<15 nm) regardless of the temperature (above or below its LCST).16,17 So, we believe that the introduction of functional groups capable of molecular recognition into PNIPAAm chains can not only provide the ability to recognize specific proteins and repel nonspecific proteins, thereby improving the signal-to-noise ratio of bio-analytical devices, but also the ability to switch on/off specific protein adsorption, which can be potentially applied in smart switching.18–20There are generally two ways for incorporation of functional groups: covalent and non-covalent binding methods. The covalent method is relatively strong and enhances the stability of biomaterials, whilst non-covalent methods allow more flexibility, good reversibility, adaptivity, facile fabrication, etc.21–23 Host–guest interactions have been recently utilized as a new versatile and robust post-modification methodology for integrating functional moieties into biomaterials. The host–guest interactions based on β-cyclodextrin (β-CD) have the ability to tune the ligand valency, type, orientation and location, or even allow reversible binding.24–31 Amongst different host–guest pairs, β-CD and adamantane (Ada) are a strong pair and have been widely used as linkers for integration of biomolecules to construct functionalized bio-substrates.32–37 They can also be dissociated for the regeneration of biomaterials and sensor chips.37,38 Herein, poly(N-isopropylacrylamide-co-1-adamantan-1-ylmethyl acrylate) [poly(NIPAAm-co-Ada)] was used as a platform for attaching molecular recognition functionalities onto a silicon surface via host–guest interactions between adamantane and ligands bearing β-CD (CD(X)) (Scheme 1). A series of thermoresponsive and bio-functional surfaces can be built just by changing ligands on the β-CD ring. Therefore, the platform capable of integrating variable molecular recognition moieties, having tunable function and regenerable/reusable ability offers more potential for biomedical and biotechnological applications.
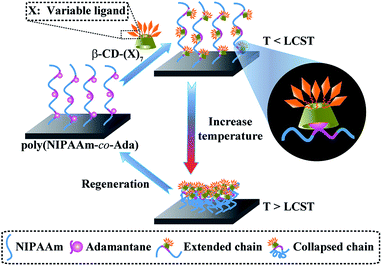 | ||
| Scheme 1 Schematic illustration of the preparation of bio-functional and thermoresponsive surfaces by the host–guest interaction between poly(NIPAAm-co-Ada) and β-CD-(X)7. | ||
Results and discussion
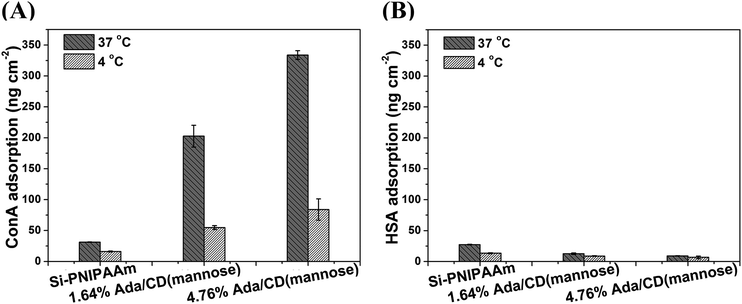
The surface-initiated process, grafting density and surface composition of poly(NIPAAm-co-Ada) on a silicon surface have been described in our previous work.39 Surfaces with the Ada content less than 5% were used in this work, as at higher Ada content, CD(mannose) is hard to complex with the poly(NIPAAm-co-Ada) layer due to steric hindrance.39 The thickness of all polymer layers was 10–12 nm, where the antifouling properties of PNIPAAm can be retained. In a-proof-of-concept study, mannose-modified β-CDs were incorporated onto poly(NIPAAm-co-Ada) layers to investigate their specific recognition with ConA. The discussion about the successful complexation of CD(mannose) and poly(NIPAAm-co-Ada) surfaces with different Ada feed ratios is described in the ESI.† As Concanavalin A (ConA, 104–112 kDa, pI = 4.5–5.5) binds to α-D-mannosides specifically, while human serum albumin (HSA, 66.5 kDa, pI = 4.9) does not and has a comparable molecular weight and pI to ConA, it was chosen as a model to study the specific recognition of ConA and nonspecific protein adsorption on α-D-mannose-decorated thermoresponsive surfaces. The 125I-radiolabeling method was adopted in this work as it is very sensitive and accurate to quantify the adsorbed protein on surfaces, and has been successfully used for labeling ConA.40,41 We checked the dynamic adsorption of ConA on CD(mannose)-modified surfaces with incubation time, and its adsorption reached an equilibrium after about two hours (Fig. S5†); so in this research three-hour adsorption was adopted.In order to investigate the effect of the sugar content on protein adsorption at temperatures above and below LCST, 37 and 4 °C were chosen as the LCST of all mannose-decorated surfaces obtained within the temperature range (Fig. S2†). At 37 °C, above the LCST, although PNIPAAm is in a collapsed conformation, the adsorption of ConA and HSA for Si-PNIPAAm was 31.2 and 27.4 ng cm−2, respectively (Fig. 1A and B), which are still at a very low level, indicating the antifouling properties of PNIPAAm. With the increase of sugar content, the specific ConA adsorption increased to 334 ng cm−2, while the nonspecific HSA adsorption decreased to 9.0 ng cm−2, due to the nonspecific protein-repelling properties of glycopolymers.42–44 In addition, the sugar density regulated by the Ada content had great influence on protein adsorption. For sugar surfaces with 1.64% and 4.76% Ada feed ratios, ConA adsorption increased 5.5 and 9.7 times of that on the PNIPAAm surface (Fig. 1A); while HSA adsorption decreased 54% and 67% compared with that on the PNIPAAm surface (Fig. 1B). As cyclodextrin may attach to surfaces by interacting with alkyl side chains,45 we compared the ConA adsorption on the PNIPAAm surface without Ada moieties before and after immersion with CD(mannose) solution. The surfaces were found to exhibit almost the same low level of ConA adsorption. In addition, control experiments for ConA adsorption on copolymer surfaces of poly(NIPAAm-co-Ada) at 37 °C also showed a very low level of ConA adsorption, below 37 ng cm−2, which was comparable to that on the PNIPAAm surface (Fig. S4†). These results demonstrated that it was the host–guest interaction between cyclodextrin and Ada that integrated CD(mannose) onto the copolymer surface to form a glycopolymer surface, thereby increasing the specific protein adsorption and decreasing the nonspecific protein adsorption. Moreover, the mannose content on surfaces can be tuned by varying Ada feed ratios. At 4 °C, below the LCST, the adsorption of HSA for all the surfaces was very low, slightly lower than the protein adsorption above the LCST (Fig. 1B); for specific ConA adsorption, the biggest reduction was observed for the 4.76% Ada/CD(mannose) surface, which reduced ∼75% compared with that at 37 °C (Fig. 1A), indicating notable thermoresponsive protein adsorption. The significant difference in recognizing ConA at different temperatures will be beneficial for preparing responsive biomaterials/devices. The temperature-induced tunable adsorption could be explained by conformational change of polymer chains, indicated by the variation of surface wettability (Fig. S6†). Above the LCST, PNIPAAm chains collapsed, exposing the bulky and hydrophilic CD(mannose) and forming many glycoclusters, which can enhance the affinity with lectins as a result of the “cluster glycoside effect” (mainly the chelating effect and the statistical effect).46–49 Therefore, a large amount of ConA was adsorbed on surfaces above LCST. Whilst below the LCST, the hydrated and extended chains can not only prevent the approach of CD(mannose) to ConA, but also weaken the binding constant by decreasing the sugar density on the surface. So ConA was repelled by the hydrated PNIPAAm chains.
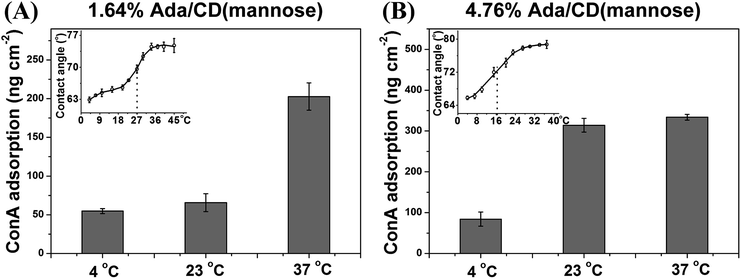
The thermoresponsive molecular recognition ability can be easily tuned at different temperature ranges by changing the Ada content. For example, the LCST of 1.64% Ada/CD(mannose) is ∼27 °C, (Fig. 2A, inset), therefore the ConA adsorption can be regulated between 37 and 23 °C. As shown in Fig. 2A, ConA adsorption on the 1.64% Ada/CD(mannose) surface reduced 68% from 37 °C to 23 °C. For comparison, ConA adsorption on 4.76% Ada/CD(mannose) surfaces showed almost no change at the two temperatures, as it has a lower LCST of ∼16.5 °C (Fig. 2B, inset). In fact, the 4.76% Ada/CD(mannose) surface showed a reduction of 73% for ConA adsorption from 23 °C to 4 °C (Fig. 2B). Generally speaking, the temperature rise is probable to increase protein adsorption itself, but unlikely to be in a significant way. This is because the observed changes in ConA adsorption were minor when 1.64% Ada/CD(mannose) and 4.76% Ada/CD(mannose) surfaces were changed from 4 °C to 23 °C and from 23 °C to 37 °C, respectively. Therefore, it is the conformational changes of thermoresponsive polymers triggered by the temperature that mainly control the arrangement of CD(mannose) and therefore regulate protein adsorption. And by simply adjusting the Ada content to acquire thermoresponsive glycopolymer surfaces with different LCSTs, the regulation of specific protein adsorption in different temperature ranges can be achieved.
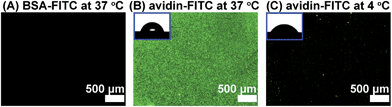
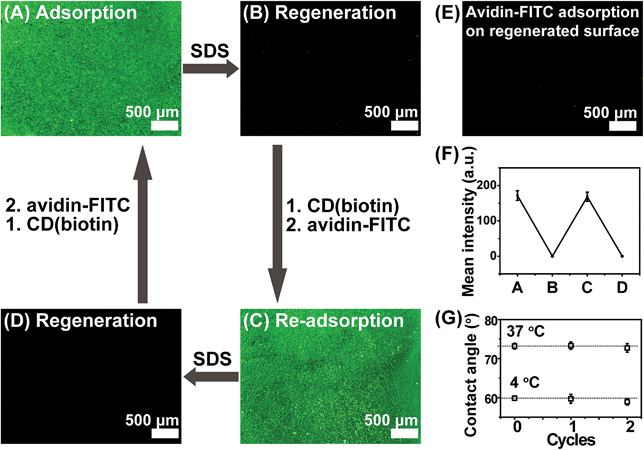
In order to prove the versatility of the poly(NIPAAm-co-Ada) platform, a biotin-modified β-CD ring was also introduced onto the surface, forming a biotin-functionalized thermoresponsive surface. The specific recognition ability of the biotin-modified surface was investigated using fluorescein-labeled avidin and BSA. β-CD-(biotin)7 (CD(biotin)) was synthesized and the conjugation ratio of biotin to β-CD was calculated to be 6.39 according to the integral values obtained from the H protons of β-CD at C1 and of the 1,2,3-triazole group in 1H NMR spectra (Fig. S7†). CD(biotin) was introduced onto 1.64% Ada surfaces via host–guest interactions to form CD(biotin)-complexed surfaces, which also showed a tunable avidin-recognition ability. The results of FITC-labeled avidin adsorption showed that the 1.64% Ada/CD(biotin) surface had much higher fluorescence intensity for avidin adsorption at 37 °C (Fig. 3B) than that at 4 °C (Fig. 3C), which was attributed to the temperature-induced chain conformation change revealed by the surface wettability change (Fig. 3B and C, insets, from 73.2 ± 1.1° to 58.4 ± 0.6°). Nevertheless, Si-PNIPAAm and Si-1.64% Ada surfaces (Fig. S8A and S8B†) exhibited negligible fluorescence intensity. These results demonstrated that avidin recognition to CD(biotin)-conjugated poly(NIPAAm-co-Ada) surfaces was switched on when the temperature was above the LCST and switched off when the temperature was below the LCST. With regard to the nonspecific BSA-FITC adsorption at 37 °C, there was almost no green fluorescence on the 1.64% Ada/CD(biotin) surface (Fig. 3A), indicating its ability of repelling nonspecific protein adsorption. The binding constant between biotin and avidin is ∼1015 M−1, which is one of the strongest known non-covalent bonds and does not break easily once formed.50 Conversely, the desorption of avidin can be realized by dissociating the complexion between β-CD and Ada using sodium dodecylsulfate solutions (SDS).38 As shown in Fig. 4, the exposure to 2% SDS resulted in almost complete desorption of avidin-FITC from poly(NIPAAm-co-Ada)/CD(biotin) surfaces (Fig. 4B). This desorption was due to the dissociation between β-CD and Ada as proved by the negligible avidin adsorption on control surfaces (Fig. 4E), i.e. the regenerated surfaces without being immersed in CD(biotin) solution. In addition, the regenerated surfaces after conjugation with CD(biotin) could further adsorb avidin-FITC with almost the same fluorescence intensity as the first time (Fig. 4C). Moreover, the surfaces with re-adsorbed avidin-FITC could be regenerated again upon exposure to SDS (Fig. 4D). During cycles, the fluorescence intensity of avidin-adsorbed surfaces reduced by almost 99.9% after being washed with SDS each time (Fig. 4F).
We also measured the regeneration efficiency of ConA-adsorbed surfaces using the radio-labelling method, and almost no proteins remained on surfaces after being washed with SDS each time (Fig. S9†). Therefore, it is suggested that this system is capable of being regenerated and thus reused. Moreover, we studied the thermoresponsivity of poly(NIPAAm-co-Ada) surfaces following regeneration with SDS to check the stability of the surfaces. The regenerated surfaces remained almost the same WCAs at both 37 and 4 °C as the pristine surfaces, indicating the efficiency of regeneration and the stability of surfaces (Fig. 4G). Actually, other molecular recognition functionalities may be introduced onto the regenerated surfaces. Thus, the poly(NIPAAm-co-Ada) platform provides the feasibility of integrating functional groups and the ease of recovery via host–guest interactions. These are important and desirable features in the applications of bio-related devices, considering their sophisticated fabrication and complex surface modification.
Conclusions
In summary, we have developed a versatile platform of poly(NIPAAm-co-Ada) surfaces to integrate ligand-functionalized β-CDs, allowing for the preparation of various thermoresponsive and bio-functional surfaces. Different molecular recognition functionalities, i.e. mannose and biotin-modified β-CDs, were synthesized as models to investigate their specific recognition with ConA and avidin, respectively. The CD(mannose)-conjugated poly(NIPAAm-co-Ada) surface showed low nonspecific protein adsorption even less than that on the PNIPAAm surface, and presented thermoresponsively tunable ConA adsorption. By varying the Ada content, the regulation of ConA adsorption in different temperature ranges could be achieved. In addition, CD(biotin)-conjugated poly(NIPAAm-co-Ada) surfaces also showed a selective and thermoresponsively controlled avidin adsorption. So, there are four advantages of this system: (1) well-controlled ability to recognize specific biomolecules by changing the temperature; (2) easy and versatile functionalization by integrating a ligand-decorated β-CD ring; (3) prevention of nonspecific protein adsorption; (4) allowing surface regeneration by simply washing with SDS. The performance of this system indicates the potential to be used in biosensors, bioanalysis, bioseparation, microfluidic devices, etc.Acknowledgements
This work was supported by the National Science Fund for Distinguished Young Scholars (21125418), the National Natural Science Foundation of China (21374069 and 21334004), the Project of Scientific and Technologic Infrastructure of Suzhou (SZS201207), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). D.M.H. is a Wolfson Royal Society Fellow.Notes and references
- S. Choi, M. Goryll, L. Y. M. Sin, P. K. Wong and J. Chae, Microfluid. Nanofluid., 2011, 10, 231–247 CrossRef CAS.
- H. Xia, B. Mathew, T. John, H. Hegab and J. Feng, Biomed. Microdevices, 2013, 15, 519–530 CrossRef CAS PubMed.
- D. Dechtrirat, N. Gajovic-Eichelmann, F. F. Bier and F. W. Scheller, Adv. Funct. Mater., 2014, 24, 2233–2239 CrossRef CAS.
- J. Kirsch, C. Siltanen, Q. Zhou, A. Revzin and A. Simonian, Chem. Soc. Rev., 2013, 42, 8733–8768 RSC.
- C. Schou and N. H. Heegaard, Electrophoresis, 2006, 27, 44–59 CrossRef CAS PubMed.
- W. Qiang, W. Li, X. Li, X. Chen and D. Xu, Chem. Sci., 2014, 5, 3018–3024 RSC.
- F. Rusmini, Z. Zhong and J. Feijen, Biomacromolecules, 2007, 8, 1775–1789 CrossRef CAS PubMed.
- Q. Yu, Y. Zhang, H. Wang, J. Brash and H. Chen, Acta Biomater., 2011, 7, 1550–1557 CrossRef CAS PubMed.
- S. Jiang and Z. Cao, Adv. Mater., 2010, 22, 920–932 CrossRef CAS PubMed.
- H. Chen, L. Yuan, W. Song, Z. K. Wu and D. Li, Prog. Polym. Sci., 2008, 33, 1059–1087 CrossRef CAS PubMed.
- W. I. Wu, K. N. Sask, J. L. Brash and P. R. Selvaganapathy, Lab Chip, 2012, 12, 960–970 RSC.
- Y. Zhang, N. Islam, R. G. Carbonell and O. J. Rojas, Anal. Chem., 2012, 85, 1106–1113 CrossRef PubMed.
- N. J. Shirtcliffe, R. Toon and P. Roach, Methods Mol. Biol., 2013, 949, 241–268 CAS.
- K. Nagase, J. Kobayashi and T. Okano, J. R. Soc., Interface, 2009, 6, S293–S309 CrossRef CAS PubMed.
- D. L. Huber, R. P. Manginell, M. A. Samara, B. I. Kim and B. C. Bunker, Science, 2003, 301, 352–354 CrossRef CAS PubMed.
- Q. Yu, Y. X. Zhang, H. Chen, Z. Q. Wu, H. Huang and C. Cheng, Colloids Surf., B, 2010, 76, 468–474 CrossRef CAS PubMed.
- T. Zhao, H. Chen, J. Zheng, Q. Yu, Z. Wu and L. Yuan, Colloids Surf., B, 2011, 85, 26–31 CrossRef CAS PubMed.
- T. Sun and G. Qing, Adv. Mater., 2011, 23, H57–H77 CrossRef CAS PubMed.
- B. Nagel, A. Warsinke and M. Katterle, Langmuir, 2007, 23, 6807–6811 CrossRef CAS PubMed.
- E. H. Min, S. R. S. Ting, L. Billon and M. H. Stenzel, J. Polym. Sci., Part A: Polym. Chem., 2010, 48, 3440–3455 CrossRef CAS.
- K. Liu, Y. Kang, Z. Wang and X. Zhang, Adv. Mater., 2013, 25, 5530–5548 CrossRef CAS PubMed.
- X. J. Loh, Mater. Horiz., 2014, 1, 185–195 RSC.
- X. Liao, G. Chen and M. Jiang, Polym. Chem., 2013, 4, 1733–1745 RSC.
- D. Grünstein, M. Maglinao, R. Kikkeri, M. Collot, K. Barylyuk, B. Lepenies, F. Kamena, R. Zenobi and P. H. Seeberger, J. Am. Chem. Soc., 2011, 133, 13957–13966 CrossRef PubMed.
- K. Wei, J. Li, G. Chen and M. Jiang, ACS Macro Lett., 2013, 2, 278–283 CrossRef CAS.
- M. J. W. Ludden, X. Li, J. Greve, A. van Amerongen, M. Escalante, V. Subramaniam, D. N. Reinhoudt and J. Huskens, J. Am. Chem. Soc., 2008, 130, 6964–6973 CrossRef CAS PubMed.
- A. Samanta, M. C. A. Stuart and B. J. Ravoo, J. Am. Chem. Soc., 2012, 134, 19909–19914 CrossRef CAS PubMed.
- J. Voskuhl, J. Brinkmann and P. Jonkheijm, Curr. Opin. Chem. Biol., 2014, 18, 1–7 CrossRef CAS PubMed.
- Á. Martínez, C. Ortiz Mellet and J. M. García Fernández, Chem. Soc. Rev., 2013, 42, 4746–4773 RSC.
- P. Wan, Y. Wang, Y. Jiang, H. Xu and X. Zhang, Adv. Mater., 2009, 21, 4362–4365 CrossRef CAS.
- L. Yang, A. Gomez-Casado, J. F. Young, H. D. Nguyen, J. Cabanas-Danés, J. Huskens, L. Brunsveld and P. Jonkheijm, J. Am. Chem. Soc., 2012, 134, 19199–19206 CrossRef CAS PubMed.
- G. Chen and M. Jiang, Chem. Soc. Rev., 2011, 40, 2254–2266 RSC.
- M. Ortiz, A. Fragoso and C. K. O'Sullivan, Anal. Chem., 2011, 83, 2931–2938 CrossRef CAS PubMed.
- L. Wang, J. Lei, R. Ma and H. Ju, Anal. Chem., 2013, 85, 6505–6510 CrossRef CAS PubMed.
- H. Li, J. Frith and J. J. Cooper-White, Biomacromolecules, 2013, 15, 43–52 CrossRef PubMed.
- M. Ortiz, M. Torréns, A. Fragoso and C. K. O'Sullivan, Anal. Bioanal. Chem., 2012, 403, 195–202 CrossRef CAS PubMed.
- Y. Zhang, Q. Tu, D. E. Wang, Y. Chen, B. Lu, M. S. Yuan and J. Wang, New J. Chem., 2013, 37, 2358–2368 RSC.
- C. David, M. C. Millot, B. Sébille and Y. Lévy, Sens. Actuators, B, 2003, 90, 286–295 CrossRef CAS.
- X. J. Shi, G. J. Chen, Y. W. Wang, L. Yuan, Q. Zhang, D. M. Haddleton and H. Chen, Langmuir, 2013, 29, 14188–14195 CrossRef CAS PubMed.
- P. G. Phillips, P. Furmanski and M. Lubin, Exp. Cell Res., 1974, 86, 301–308 CrossRef CAS.
- Y. Okada, Biochim. Biophys. Acta, Biomembr., 1981, 648, 120–128 CrossRef CAS.
- M. X. Hu, L. S. Wan, Z. M. Liu, Z. W. Dai and Z. K. Xu, J. Mater. Chem., 2008, 18, 4663–4669 RSC.
- K. Yu, B. F. L. Lai and J. N. Kizhakkedathu, Adv. Healthcare Mater., 2012, 1, 199–213 CrossRef CAS PubMed.
- J. Yuan, J. Meng, Y. Kang, Q. Du and Y. Zhang, Appl. Surf. Sci., 2012, 258, 2856–2863 CrossRef CAS PubMed.
- A. Hashidzume and A. Harada, Polym. Chem., 2011, 2, 2146–2154 RSC.
- V. Wittmann and R. J. Pieters, Chem. Soc. Rev., 2013, 42, 4492–4503 RSC.
- T. Okada, C. Isobe, T. Wada, S. Ezaki and N. Minoura, Bioconjugate Chem., 2013, 24, 841–845 CrossRef CAS PubMed.
- X. L. Meng, Y. Fang, L. S. Wan, X. J. Huang and Z. K. Xu, Langmuir, 2012, 28, 13616–13623 CrossRef CAS PubMed.
- K. Yu, A. L. Creagh, C. A. Haynes and J. N. Kizhakkedathu, Anal. Chem., 2013, 85, 7786–7793 CrossRef CAS PubMed.
- N. M. Green, Biochem. J., 1963, 89, 585–591 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4mh00081a |
| This journal is © The Royal Society of Chemistry 2014 |