Combining surface topography with polymer chemistry: exploring new interfacial biological phenomena
Dan
Li
,
Qing
Zheng
,
Yanwei
Wang
* and
Hong
Chen
*
Jiangsu Key Laboratory of Advanced Functional Polymer Design and Application, Department of Polymer Science and Engineering, College of Chemistry, Chemical Engineering and Materials Science, Soochow University, 199 Ren'ai Road, Suzhou 215123, P. R. China. E-mail: ywwang@suda.edu.cn; chenh@suda.edu.cn; Fax: +86-512-65880583; Tel: +86-512-6588082
First published on 29th July 2013
Abstract
The review focuses on the combination of surface topography and surface chemical modification with the grafting of polymer chains to develop optimal material interfaces for biological and biomedical applications. Understanding how surface chemistry and topography correlate with the interfacial properties and biological functions of a material is important for the development of biomaterials. Synergies between these two properties are known to exist, but have not been exploited extensively for biomaterial design. Preliminary studies suggest that the combination of surface topography and chemistry may not only enhance surface properties, but may also give biological properties that are opposite to those of the corresponding smooth surface, and even other unexpected biological properties. This review summarizes some recent studies in this area, mostly carried out in our own laboratory, as examples to illustrate how synergistic properties and functions may be obtained by combining surface topography with polymer chemistry. It is hoped that this review will stimulate a more thorough exploration of the topography–chemistry synergy as a means of injecting “new life” into efforts to develop novel bio-functional surfaces.
1 Introduction
Surface and interfacial properties are vital to the design and development of materials for biological and biomedical applications.1–3 This is because biomaterials first interact with the biological environment at the interface between the two, i.e. at the biointerface. Numerous examples have shown that the surface properties of a material determine the interactions such as proteinadsorption, cell adhesion and proliferation.4–11 Generally, the surface and interfacial properties of biomaterials depend both on surface chemistry and surface topography.12–19 Surface chemistry may be the main factor that influences interactions with biomolecules and cells, and surface bio-functionality has been achieved mainly by surface chemical modification. Classic examples include incorporating bio-inert polymers to weaken interactions with proteins and cells,20–24 grafting stimuli-sensitive polymers to regulate proteinadsorption and cell adhesion in response to environmental factors,8,9,25,26 and immobilizing polycations to kill bacteria or load drugs for intracellular delivery, etc.22,27–29In addition to surface chemistry, topography is also known to substantially affect the bulk properties of a material.6,7,30,31 For example, the introduction of nanostructure makes a hydrophobic surface more hydrophobic, and makes a hydrophilic surface more hydrophilic, as first explained by the Wenzel model.32–35 For biological applications, the introduction of surface topography usually increases the area of contact, which is essential for drug release, bio-separation and bio-detection applications. Indeed, surface topography may enhance biological properties that originate from surface chemistry. For instance, the surface of Pluronic®-blended polyurethane is protein-resistant, and introducing lotus leaf-like topography can further reduce proteinadsorption even though the effective surface area is increased.36 There are also examples of the opposite behaviour. A nano-structured surface coated with fluorinated polyurethane showed low platelet adhesion and activation, while the corresponding smooth surface showed very high platelet adhesion and activation, and such an improvement in biocompatibility was attributed to the special nanostructure and the superhydrophobicity derived from it.37 Therefore, the combined effects of surface chemistry and surface topography can be complicated and can lead to effects other than the simple enhancement of the individual effects.
Understanding how surface chemistry and topography correlate with the interfacial properties and biological functions of a material is critical for the design and development of novel biomaterials. It requires research and exploration involving different areas: creating surfaces with well-defined micro-, nano- and hierarchical structures,38–43 controlled surface chemical modification,44–50 characterization of surface topography and surface chemistry,13,51–53 and quantification of the materials’ biological performance at the biointerface.54,55 All these are active research areas. Among them, polymers are playing an increasingly important part both in the preparation of well-defined surface structures and in controlled surface chemical modification.21,24,44,45,56 In particular, immobilizing polymer chains on a solid surface can not only bring rich chemical properties to the surface, but can also alter mechanical properties and environmental responses of the surface.57–60 Moreover, polymer chains can often be easily functionalized, which result in the possibility of binding specific ligands to induce desired biological responses for a variety of biomedical applications.
Recent technologies for creating surfaces with well-defined topography and chemistry combined with sensitive surface characterization techniques have unquestionably deepened our understanding of surface chemical and topographical effects on cell behaviour. Synergistic properties and functions between surface chemistry and topography are known to exist,61–64 but have not been exploited extensively for biomaterial design purposes. In recent years, our laboratory has investigated the synergistic effects of topography and surface chemistry on the interactions of surfaces with biological systems, especially with proteins and cells. Two kinds of topographical surfaces have been studied: porous gold and vertically-aligned nanowire arrays. In this review, these two kinds of topographical surface will be used as examples to illustrate the synergy of surface topography with polymer chemistry. Other types of topographical surface are under investigation in various laboratories including surfaces prepared by soft lithography, etching, self-assembly, etc.65,66 These will not be discussed here.
This review is organized as follows. First, an overview of polymer chemistry for surface modification is given (section 2), followed by a brief summary of methods for the preparation and analysis of topographical surfaces (section 3). We will not go into detail in these two sections since several excellent reviews are already available.41–50 Instead, the focus will be on recent efforts to explore the synergistic effects of surface topography and chemical modification by polymer grafting. This is done in section 4 using examples involving judicious combinations of surface topography and polymer chemistry to provide specific interfacial biological functions. The review concludes with a brief discussion of future perspectives (section 5).
2 Polymer chemistry for surface modification
Different methods can be employed for surface modification, depending on the surface properties required. Coating and grafting with natural or synthetic polymers have proved to be effective methods in this regard and are being used increasingly.44–50 At the present time the development of polymer chemistry has made it possible to synthesize with relative ease homo-polymers, copolymers, polymers of branched and other non-linear architectures, conductive polymers and stimuli-responsive polymers in a more and more controlled fashion. Immobilization of these polymers on solid substrates can give surfaces of diverse and interesting functionalities.59,67As depicted in Fig. 1, both physical and chemical methods are available for the immobilization of thin layers of polymers on solid surfaces. Early work focused on physisorption.68–71 A classical example is the physisorption of polystyrene-b-poly(ethylene oxide) (PS-b-PEO) from a toluene solution to a mica surface. The PEO segments are preferentially adsorbed to the mica while the PS blocks interact preferentially with the solvent. Another example is the layer-by-layer assembly of polyelectrolyte multilayers by the alternative adsorption of polycations and polyanions via electrostatic interactions, which has been shown to be suitable for modifying the surface properties of biomaterials.72–74 Although the physisorbed blocks can be unstable under certain conditions of solvent and temperature and can be displaced by other adsorbents, this method remains attractive because of its simplicity.
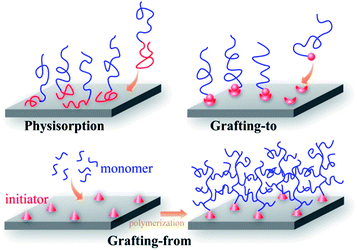 | ||
| Fig. 1 Schematic illustration of different methods for immobilizing polymer chains on solid surfaces: physisorption, “grafting-to” and “grafting-from” (surface initiated polymerization). | ||
The covalent bonding of polymer chains to surfaces has attracted interest because of the relative stability of the layers. In general, covalently attached polymer layers can be prepared by either the “grafting-to” or the “grafting-from” method (see Fig. 1).44,67 “Grafting-to” involves the chemical reaction of preformed, functionalized polymers with surfaces containing complementary functional groups. The main advantages of this method are simplicity and the possibility to characterize the preformed polymers in detail. Grafting can be performed using polymer solutions or melts and, in general, higher concentrations of polymer lead to higher grafting densities, but it has been argued that “grafting-to” cannot produce dense polymer layers (brushes) because the attachment of new chains may be impeded by the presence of previously attached chains. This problem is avoided in the “grafting-from” method, which involves the in situpolymerization of the monomer on an initiator functionalized surface. Many living and conventional vinylpolymerization techniques have been applied using this method.45,46,48 The graft density is determined by the initiator density which can be relatively high since the initiator is generally of a small size. The “grafting-from” technique may suffer complications because of low initiator efficiency, and the variable rate of diffusion of monomer to active polymerization sites. Side reactions may be more important than in bulk polymerization because of the high local concentration of polymer chains in the grafted layers; e.g., bimolecular termination in radical polymerization.75–78 Hence, “grafting-from” may lead to a broader chain length distribution.
While most of the syntheses of polymer brushes reported in the literature were conducted on flat surfaces, brushes can also be formed on surfaces with topographic features as demonstrated by work in our laboratory (see section 4). Surface topography may affect the polymerization kinetics, but this aspect has not been studied to any significant extent, either experimentally or theoretically.
A variety of methods have been developed to characterize the surface chemistry of polymer-modified flat surfaces. Chemical composition can be analyzed by X-ray photoelectron spectroscopy (XPS), infrared spectroscopy and time-of-flight secondary ion mass spectrometry (TOF-SIMS). A useful technique for qualitative and quantitative characterization of surface chemistry on a solid substrate with complex topography is transmission mode Fourier transform infrared (FTIR) spectroscopy. If the substrate does not appreciably absorb light near the characteristic frequency of the C–H stretching peak, which is the footprint of carbon–hydrogen bonds in polymer chains, then by proper calibration the measured FTIR absorbance peak can be used to calculate the amount of polymer on the substrate. For qualitative characterization of surface chemistry, contact angle measurements are extremely useful and can be conducted both on flat and rough surfaces. Ellipsometry is a convenient and accurate tool to determine the thickness of polymer thin films. However, the accuracy of the measurement of film thickness is limited when the substrate is rough. For polymer-modified topographical surfaces, atomic force spectroscopy (AFM),79 and in some cases, transmission electron microscopy (TEM), can give accurate measurements of film thickness.64
3 Surface topography
Very few materials exist that have atomically smooth surfaces. The majority of materials have a surface “landscape” made up of undulations and steeper gradients and pores.30 These features constitute the topography of the surface. The biological interactions of a surface have been found to depend on its topography. Many studies have been conducted to determine the effect of topography on proteinadsorption and cell behaviour, including protein amount, conformation and distribution on topographical surfaces,80 effects of contact guidance on cell orientation,81etc. We will not go into the details of those studies since there are already several excellent reviews on this topic.6,7,30,31,42Topography may be introduced at the surface of a material by different patterning strategies, such as photolithography, scanning beam lithography, molding, embossing, printing and self-assembly.41,43 It may also be introduced by the inherent relaxation of the material, or by mechanical roughening and wet chemical etchingetc.30 Recent studies in our laboratory have focused on two kinds of topographical surface (see Fig. 2), namely gold nanoparticle layers (GNPL) and vertically-aligned nanowire arrays, such as silicon nanowire arrays (SiNWAs). Details of the preparation, characterization and applications of those two types of surfaces are available in the literature (for GNPL79,82–84 and for SiNWAs62,64,85–91).
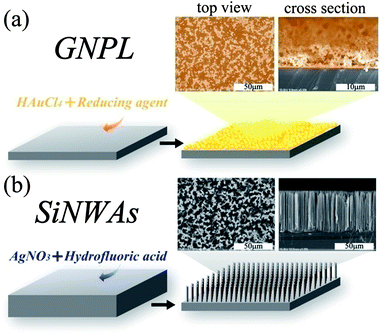 | ||
| Fig. 2 Schematic illustration and SEM images of two kinds of topographical surfaces studied in our laboratory: (a) gold nanoparticle layers (GNPL) and (b) silicon nanowire arrays (SiNWAs). | ||
The characterization of surface topography is of key importance when investigating the effects of various topographies on the interactions with biological objects. This includes the measurements of surface roughness, specific surface area, scale and shape of the feature, etc. For regular surface topographies, such properties can be obtained by microscopic techniques. However, the characterization becomes more difficult in the case of irregular ones. Surface roughness is an important parameter that can characterize an irregular topographical surface. There exist different types of roughness measurement apparatus such as the atomic force microscope (AFM), stylus profilometer, and optical apparatus.13 In particular, AFM offers an incomparable resolution and is the ideal tool to describe topography at the nanometre scale. The most often used roughness parameters in the biomaterials field are the arithmetic average roughness, Ra, and the root mean square roughness, Rq. However, it should be noted that the value of a roughness parameter depends on the scale of the measurement. An example is shown in Fig. 3, where roughness parameters Ra and Rq were obtained from AFM height images and then plotted as a function of the evaluation length. This problem was studied in depth by Bigerelle et al.,13,53 and a multi-scale roughness parameter evaluation method was developed in their studies to obtain more detailed descriptions of the surface topography.
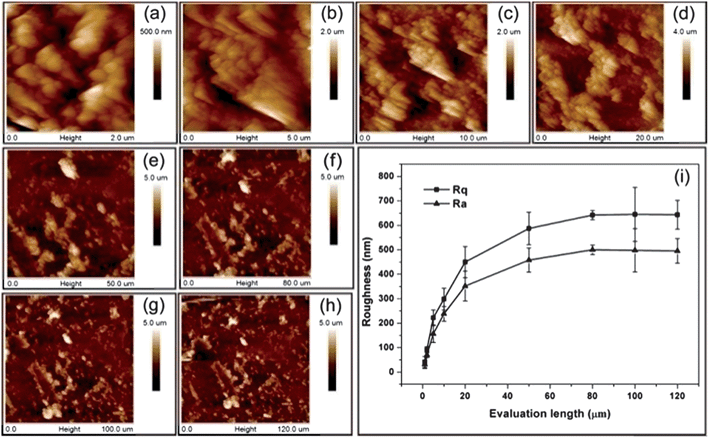 | ||
| Fig. 3 AFM height images of GNPL surfaces for different evaluation lengths: (a–h) 2, 5, 10, 20, 50, 80, 100, and 120 μm and (i) roughness parameters Rq and Ravs. evaluation length as a description of the surface topography. Adapted with permission from ref. 79. Copyright 2012 American Chemical Society. | ||
4 Combining surface topography with polymer chemistry
This section will focus on the combination of topographical surfaces and surface-grafted/coated polymers, which act together in modulating the interactions of the surface and biomolecules/cells. GNPL and vertically-aligned nanowire arrays are used as examples. The content is organized according to the function of the polymers, in the order of antifouling polymers, stimuli-responsive polymers and polymers as diffusion barriers for drug release. While the properties and functions are derived from polymer chemistry, synergistic effects are reflected in the change of the biological properties with the introduction of surface topography. Note that there are other methods to combine surface topography with polymers. The most direct approach may be to construct topography on polymer substrates viasoft lithography.92,93 This kind of topographical polymer surface is commonly used to investigate the effects of surface topography alone on the interactions between the surface and biological objects. Another method is the fabrication of patterned polymer brushes through top-down lithography and/or bottom-up polymer synthesis.94 These surfaces have also been used to build biomolecules or cell patterns.95–974.1 POEGMA-modified GNPL surfaces
In work with GNPL we applied the particles to the inner surfaces of an ELISA plate; the GNPL-modified surfaces were found to have high binding capacity for proteins and to maintain the biological activity of adsorbed proteins. When the GNPL modified-ELISA plates were used for the detection of carcinoembryonic antigen (CEA) in plasma, the ELISA signals were significantly amplified and the limit of detection was decreased compared to the unmodified plates.82,98 We then attempted to exploit this amplification effect for the isolation and enrichment of circulating tumor cells.83 A Ramos cell-specific aptamer was attached to the GNPL surface. The aptamer-modified surface showed high selectivity for Ramos cells in serum-free mixtures with CEM cells. This effect was attributed to the high aptamer density associated with the high specific area of the GNPL–gold surface. However, the Ramos cell selectivity was much lower under serum-containing conditions, presumably due to nonspecific adsorption of serum proteins shielding the aptamer and inhibiting the interactions between aptamer molecules and target cells. To avoid this problem, protein resistant elements were attached to the surface. The aptamer was immobilized on the GNPL surface via an antifouling spacer, poly(oligo(ethylene glycol) methacrylate) (POEGMA). Good selectivity for Ramos cells in serum-containing medium was observed on the POEGMA–aptamer surface, and the selectivity increased with increasing surface roughness.83 Therefore, both the antifouling properties of POEGMA and the high specific area of the GNPL surface contribute to the good selectivity for Ramos cells. This study suggests that surfaces having appropriate combinations of topography and chemistry can be useful for regulating cell–surface interactions.In a preliminary study to investigate the synergy of GNPL topography with bio-inert polymer modification, we carried out proteinadsorption and cell adhesion measurements on POEGMA-modified GNPL surfaces.79 POEGMA chains were immobilized on GNPL surfaces by surface-initiated atom transfer radical polymerization (SI-ATRP). It was found that similarly low amounts of cell-adhesive proteins were adsorbed on POEGMA-modified smooth and topographical surfaces, while the cell density on the GNPL–POEGMA surface was much higher than on the corresponding smooth surface (see Fig. 4). This result indicates that the GNPL topography can promote cell adhesion even though the surface chemistry does not favour the adsorption of cell-adhesive proteins. However, it was found that minimal amounts of adsorbed protein are necessary to support initial cell adhesion. Although cell spreading was constrained because of the lack of adsorbed proteins, the cells were more firmly attached and more stable than those on the POEGMA-modified smooth surface.79 From this study it was concluded that on the POEGMA-modified GNPL surface, POEGMA inhibited cell spreading by limiting proteinadsorption, while the GNPL topography promoted cell adhesion and stability on the surface. Surface topography and surface chemistry thus appear to affect different aspects of cell–surface interactions. In a follow-up study the cell adhesive peptide GRGDY was introduced to the surface using POEGMA as a bioinert spacer.99 This surface was shown to be strongly protein-resistant but also strongly cell-adhesive, presumably due to GRGDY–cell receptor interactions.
 | ||
| Fig. 4 Fluorescence and SEM images (scale bar = 50 μm) of L02 and BEL-7402 cells on surfaces. Cells that are spreading on the GNPL surface are marked by red ovals in B3 and D3. The shape and filopodia of the cells on POEGMA-modified surfaces were observed by confocal microscopy and are shown as insets in A2, A4, C2, and C4 (scale bar = 10 μm). The column headings (from left to right), smooth Au, sAu–POEGMA, GNPL and GNPL–POEGMA, refer to a smooth gold surface, a POEGMA-modified smooth gold surface, a GNPL surface and a POEGMA-modified GNPL surface, respectively. Reprinted with permission from ref. 79. Copyright 2012 American Chemical Society. | ||
4.2 Vertically aligned nanomaterial arrays modified with stimuli-responsive polymers
Recent efforts in this area in our laboratory involve combining 3D nanostructured SiNWAs with surface-grafting of stimuli-responsive polymers. Stimuli-responsive polymers are also known as smart polymers, and they can undergo conformation or phase changes in response to variations in environmental conditions (such as pH, temperature, electronic field and light).58 By immobilizing such polymers on a solid surface, stimuli-responsive changes in the surface and interfacial properties such as wettability, layer thickness and biomolecular adhesion can be achieved.56–59 We have been particularly interested in using stimuli-responsive polymers to control proteinadsorption and cell adhesion through the manipulation of environmental conditions.What is the effect of surface topography on the stimuli-responsiveness of such smart polymers when they are immobilized on a solid surface? Previous reports have shown that the stimuli-responsiveness of surface wettability can be greatly enhanced by introducing nanoscale roughness,31,33 but how about proteinadsorption and other bio-related properties? Can their stimuli-responsiveness be enhanced as well when nanoscale topography is introduced on substrate surfaces?
Surfaces modified with polyelectrolytes are believed to have great potential for the controlled binding and release of proteins because of their pH-responsive properties.100 As shown in Fig. 5, by combining nanostructured SiNWAs with the pH responsiveness of poly(methacrylic acid) (PMAA), which is a typical weak polyacid, we achieved not only an extremely high capacity for lysozyme binding at pH 4 (∼80-fold increase compared to PMAA-modified smooth silicon, presumably due to an enlarged surface area in the case of SiNWAs), but also an enhancement effect for the pH responsiveness.100 Notably, increasing the pH to 9 resulted in the release of ∼90% of the adsorbed lysozyme without loss of protein activity. The effect of pH change from 4 to 9 on lysozyme adsorption on the nanostructured surface was greater by a factor of ∼70 compared to the smooth Si–PMAA surface.
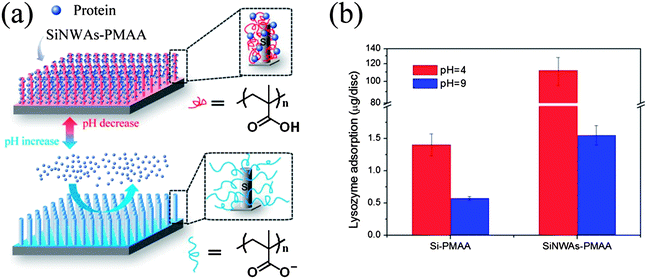 | ||
| Fig. 5 (a) Schematic illustration of the pH-responsive conformation change and proteinadsorption properties of PMAA grafted on SiNWAs surface. (b) Adsorption of lysozyme from phosphate-buffered saline (PBS) at pH = 4 and pH = 9. A comparison is made between Si–PMAA and SiNWAs–PMAA, which refer to PMAA-modified smooth silicon and PMAA-modified SiNWAs surfaces, respectively. | ||
In another study,101 step-wise control of proteinadsorption and bacterial attachment was achieved on poly[2-(N,N′-dimethylamino)ethyl methacrylate] (PDMAEMA)-grafted silicon nanowire arrays by varying the ionic strength. Furthermore, quaternization of the PDMAEMA gave a high concentration of positive charges on the surface, which was then found to be strongly anti-bacterial. Bacterial adhesion on the quaternized SiNWAs–PDMAEMA surface was much higher than on the analogous smooth surface ((34.6 ± 0.39) × 106vs. (5.0 ± 0.15) × 106 cells cm−2), and 95% of the adhered cells were killed compared to less than 45% for the smooth surface.87
Poly(N-isopropylacrylamide) (PNIPAAm) is a well-known thermo-responsive polymer with a lower critical solution temperature (LCST) of ∼32 °C. On surfaces modified with PNIPAAm, the surface wettability, proteinadsorption and cell adhesion properties can be controlled by varying the temperature.102,103 It has been demonstrated that the combination of nanostructure and surface grafting of PNIPAAm can enhance the thermal responsiveness of the surface wettability, and reversible switching between super-hydrophilicity and super-hydrophobicity can thus be achieved.34 It is thus expected that the effects of temperature on the biological properties of PNIPAAm-modified surfaces may also be enhanced by the introduction of surface topography. Recent data from our lab, however, do not support this expectation. Fibrinogen adsorption at 23 °C and 37 °C showed no clear difference, and compared with the unmodified arrays, the amount of proteinadsorption was reduced by more than 99% on the PNIPAAm-modified silicon nanowire arrays (SiNWAs–PNIPAAm) surface.62 Similar observations were also obtained for platelet adhesion. Chen et al. found that the SiNWAs–PNIPAAm surface was non-adhesive to platelets both below and above the LCST, while a smooth PNIPAAm surface was non-adhesive only below the LCST.104 This is presumably due to the trapping of water in the interstices of the nanowires resulting in the formation of a strong hydration layer which prevents the adhesion of platelets on the surface. Clearly, the vertically aligned nanoscale topography plays a key role in keeping water molecules on the SiNWAs–PNIPAAm surface.
Interestingly although the PNIPAAm-modified SiNWAs surface did not show thermal responsiveness for the adsorption of native proteins, it was thermally responsive to denatured proteins. We found that denatured proteins were adsorbed selectively to SiNWAs–PNIPAAm from native/denatured protein mixtures of either the same or different proteins.88 Even more interestingly, it was found that by temperature cycling above and below PNIPAAm's LCST (consisting of repeated cycles of 10 °C for 3 min and 40 °C for 3 min), the uptake of denatured protein was further increased, while the amount of native proteins and their enzymatic activity were maintained at nearly their original levels (see Fig. 6). These results may stimulate applications of the SiNWAs–PNIPAAm surface for native/denatured protein separation.
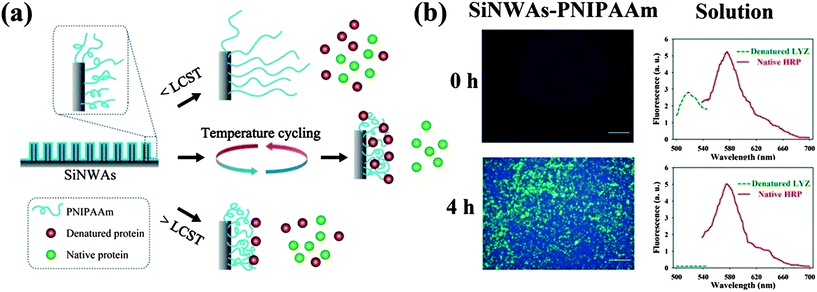 | ||
| Fig. 6 (a) Schematic illustration of the PNIPAAm-modified silicon nanowire arrays surface (SiNWAs–PNIPAAm) and its interaction with native and denatured proteins at temperatures below the LCST of PNIPAAm (<LCST), above the LCST of PNIPAAm (>LCST), and under temperature cycling conditions. (b) Fluorescence images of the adsorption of denatured lysozyme (denatured LYZ) and native horseradish peroxidase (native HRP) on PNIPAAm-modified SiNWAs and the corresponding fluorescence intensity in the solution phase for different incubation periods (0 h and 4 h). The protein mixture contains denatured LYZ labelled with fluorescein isothiocyanate (green colour) and native HRP labelled with rhodamine (red colour). Adapted with permission from ref. 88. © IOP Publishing. Reproduced by permission of IOP Publishing. All rights reserved. | ||
The SiNWAs–PNIPAAm surface was also found to have significant and reversible thermo-responsiveness to bacteria (E. coli) with high adhesion at 42 °C and low adhesion at 4 °C; also the attached cells could be almost completely released by decreasing the temperature.64 Taking advantage of this behavior, we used the SiNWAs–PNIPAAm surface as a nano-catalyst to promote DNA transformation; it was shown that the catalytic efficiency was increased by >400-fold compared to traditional methods.64 The achievement of this unique biological function is attributed to the combination of several features of the PNIPAAm-modified SiNWAs as illustrated in Fig. 7. First, the dramatic switch in surface wettability as a function of temperature plays a central role in the highly efficient DNA transformation process. At 4 °C, the surface is strongly hydrophilic and the PNIPAAm chains stretch into the water phase, similar to poly(ethylene glycol) (PEG) in the chemical transformation method. DNA precipitation on the E. coli cell surfaces is thus promoted. Heating to 42 °C (the optimal temperature for DNA transformation) makes the surface strongly hydrophobic, leading to a high cell density and an increasing the number of cells being available to be transformed. After transformation, decreasing the temperature to 4 °C makes the surface super-hydrophilic again and almost all of the cells are released, thus facilitating molecular screening of the transformants and recycling of the nano-catalyst. Second, the relatively high thermal conductivity of silicon is important for DNA transformation. During the transformation process at 42 °C, the plasmid DNA carried by the attached cells can be heat shocked quickly and uniformly, thereby avoiding the problem of non-uniform temperature distribution encountered in the chemical transformation method. In addition, the short transformation time (90 s) at 42 °C helps to preserve the viability of the attached cells for DNA transformation. Coincidentally, almost all of the steps in the transformation process (Fig. 7), including DNA acquisition, cell adhesion, DNA transfer, and transformant release, are facilitated by the SiNWAs–PNIPAAm platform. This is a clear example of achieving an important biological function by combining nano-topography and surface modification with a polymer.
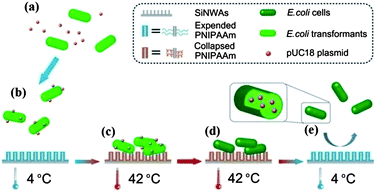 | ||
| Fig. 7 Schematic illustration of the application of PNIPAAm-modified silicon nanowire arrays (SiNWAs) as an efficient “nano-catalyst” platform for the DNA transformation of E. coli cells. (a) Mixing of DNA (pUC18 plasmid) with E. coli cells just before loading onto the PNIPAAm-modified SiNWAs. (b) Incubation of DNA and E. coli cells at 4 °C with the PNIPAAm-modified SiNWAs and the distribution of DNA on the cell surfaces. (c) Cell adhesion onto the PNIPAAm-modified silicon nanowire arrays at 42 °C. (d) Internalization of DNA into E. coli cells during heat shock. (e) Release of E. coli cells, including transformants, from the material's surface. Reprinted with permission from ref. 64. Copyright 2011 The Royal Society of Chemistry. | ||
4.3 Polymer-modified vertically aligned nanomaterial arrays for drug delivery
Vertically-aligned nano-structured arrays have also been explored for drug delivery applications. In general the drug is loaded in the spaces between or in the pores within the vertical elements, thus allowing higher loading compared to simple surface adsorption. Drug release from the pores is based on molecular diffusion and the release rate is regulated mainly by the pore size. Introducing release barriers after drug loading is an alternative way to regulate the release rate without reducing the loading, and polymer modification is effective in this regard. As a reservoir for loading the hydrophilic drugvancomycin, Simovic et al. prepared a porous anodic aluminum oxide (AAO) surface with a pore diameter of 80–90 nm and a pore depth 20 μm (Fig. 8a).105 After drug loading, a thin film of poly(allylamine) was formed on the top of the porous layer by plasma polymerization. The release rate could be regulated by adjusting the polymerization time, i.e. the chain length of the polymer. For the longest polymerization time (200 s), only half of the drug was released after 500 h, while from the uncoated surface the drug was completely released within 45 min. Recently, Gulati et al. developed this drug release strategy further, using vertically aligned titania nanotube arrays (TNTAs) but with a much simpler method of solution casting (dip coating) for polymer deposition.106 After loading the anti-inflammatory drugsindomethacin, chitosan or poly(lactic-co-glycolic acid) (PLGA), as biodegradable and biocompatible polymers, were dip-coated onto the TNTAs to create a barrier for drug release (Fig. 8b). Compared to the arrays without the coating, the polymer coated materials showed reduced burst release (77% vs. 20%) and extended overall release (4 days vs. >30 days).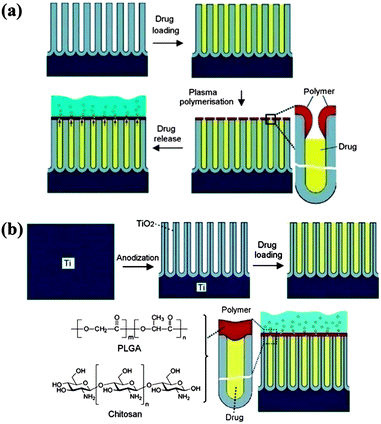 | ||
| Fig. 8 Two examples of vertically-aligned nano-structured arrays modified with polymers for drug delivery applications. (a) Plasma modification of a porous platform for controlled drug release. The four stages marked with arrows between them show (i) the anodic aluminium oxide (AAO) porous layer fabricated by electrochemical anodization, (ii) drug loading (vancomycin) inside the pores, (iii) the deposition of the plasma polymer layer (allylamine) on the top of the pores, and finally (iv) the release of the drug from the pores into solution. Reprinted with permission from ref. 105. Copyright 2010 The Royal Society of Chemistry. (b) Titania nanotube (TNT/Ti) implants modified with polymer film infrastructure. The four stages marked with arrows between them show (i) the Ti substrate, (ii) the bare TNT layer formed on the Ti substrate by electrochemical anodization, (iii) loading of the drug inside the TNT structures, and (iv) chitosan or PLGA polymer film coated on the TNT by dip coating (thin and thick) with the aim of controlling the drug release and improving the antibacterial properties and bone integration. The scheme shows the diffusion of drug molecules through the polymer matrix. Reprinted with permission from ref. 106. Copyright 2012 Elsevier. | ||
Vertically-aligned arrays of high aspect ratio are particularly useful for intracellular delivery because the shape of the array elements enables direct physical access to the interior of the cell. The delivery of various “bio-cargos”, including DNAs, RNAs, peptides, proteins and small molecules to a number of cell types has been demonstrated using nanofibre arrays or nanowire arrays.107–111 However, in these experiments the “bio-cargos” were loaded by adsorption to the surface, and hence the total releasable quantity was limited. Park et al. reported a simple intracellular delivery system based on carbon nanosyringe arrays (CNSAs).112 An amphiphilic copolymer was applied to the surface of the nanosyringes so that drug “cargos” could be readily loaded in the hollow cores by capillary action. Using this technology, efficient delivery of DNA and quantum dots to the cytoplasm of cancer cells and human mesenchymal stem cells was demonstrated.
5 Conclusion and future perspectives
Modification with polymers has been used to improve the biological properties of biomaterials for many years. The influence of the composition and structure of the surface grafted or coated polymers on interactions with biological systems has been studied extensively and has been exploited to regulate the biological properties of biomaterials. However, less attention has been paid to the effect of combining surface topography with polymer modification on the biointeractions of surfaces. Synergy between the two may be expected as suggested by the examples discussed in this article. While these investigations may be seen as preliminary, we believe that more emphasis should be placed on the topography–chemistry synergy as a means of injecting “new life” into efforts to develop novel bio-functional surfaces. Based on the examples discussed above, further efforts may be guided by the following possibilities.• The scale, shape and arrangement of surface topographical features may give a variety of synergistic effects when modified with polymers.
• The favourable biological properties of polymer modified surfaces may be further amplified by the optimized design of surface topography.
• The combination of surface topography and polymer grafting/coating may amplify the respective functions of the two factors, or suppress the effects of one of them.
• The excellent optical and electrical properties of surfaces resulting from the combination of surface topography and polymer coatings113 may be useful for bio-detection applications.
It is expected that further development in technologies for creating surfaces with well-defined topography and chemistry will continue to promote studies combining surface topography with polymer chemistry to explore new interfacial biological phenomena and functions. For a clear understanding of the combined effects of surface topography and chemistry, the development of sensitive surface characterization techniques is also crucial. Compared to the characterization of polymer chains immobilized on flat surfaces, the characterization of chain length, layer thickness and grafting density of those on topographical surfaces, especially those of random roughness, is inherently more challenging. However, even though we are still far from a clear understanding of the combined effects of surface topography and chemistry, this remains an exciting task for the future, considering the importance of controlling surface interactions with biological substances in the development of biomaterials.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (21204055, 21204061), the National Science Fund for Distinguished Young Scholars (21125418), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and the Project of Scientific and Technological Infrastructure of Suzhou (SZS201207).Notes and references
- D. G. Castner and B. D. Ratner, Surf. Sci., 2002, 500, 28–60 CrossRef CAS
.
- M. Tirrell, E. Kokkoli and M. Biesalski, Surf. Sci., 2002, 500, 61–83 CrossRef CAS
.
- P. Roach, D. Eglin, K. Rohde and C. Perry, J. Mater. Sci.: Mater. Med., 2007, 18, 1263–1277 CrossRef CAS PubMed
.
- E. Ostuni, R. G. Chapman, R. E. Holmlin, S. Takayama and G. M. Whitesides, Langmuir, 2001, 17, 5605–5620 CrossRef CAS
.
- N. Nath, J. Hyun, H. Ma and A. Chilkoti, Surf. Sci., 2004, 570, 98–110 CrossRef CAS PubMed
.
- A. Curtis and C. Wilkinson, Biomaterials, 1997, 18, 1573–1583 CrossRef CAS
.
- A. S. G. Curtis and C. D. W. Wilkinson, J. Biomater. Sci., Polym. Ed., 1998, 9, 1313–1329 CrossRef CAS PubMed
.
- M. Yamato and T. Okano, Mater. Today, 2004, 7, 42–47 CrossRef CAS
.
- M. Yamato, Y. Akiyama, J. Kobayashi, J. Yang, A. Kikuchi and T. Okano, Prog. Polym. Sci., 2007, 32, 1123–1133 CrossRef CAS PubMed
.
- H. G. Craighead, C. D. James and A. M. P. Turner, Curr. Opin. Solid State Mater. Sci., 2001, 5, 177–184 CrossRef CAS
.
- K. Kulangara and K. W. Leong, Soft Matter, 2009, 5, 4072–4076 RSC
.
- K. Anselme, A. Ponche and M. Bigerelle, Proc. Inst. Mech. Eng., Part H, 2010, 224, 1487–1507 CrossRef CAS
.
- A. Ponche, M. Bigerelle and K. Anselme, Proc. Inst. Mech. Eng., Part H, 2010, 224, 1471–1486 CrossRef CAS
.
- H. Chen, L. Yuan, W. Song, Z. Wu and D. Li, Prog. Polym. Sci., 2008, 33, 1059–1087 CrossRef CAS PubMed
.
- L. Yuan, Q. Yu, D. Li and H. Chen, Macromol. Biosci., 2011, 11, 1031–1040 CrossRef CAS PubMed
.
-
L. Dan and C. Hong, in Proteins at Interfaces III State of the Art, American Chemical Society, 2012, ch. 13, vol. 1120, pp. 301–319 Search PubMed
.
- J. Y. Wong, J. B. Leach and X. Q. Brown, Surf. Sci., 2004, 570, 119–133 CrossRef CAS PubMed
.
- L. Tang, P. Thevenot and W. Hu, Curr. Top. Med. Chem., 2008, 8, 270–280 CrossRef
.
- J. Wu, Z. Mao, H. Tan, L. Han, T. Ren and C. Gao, Interface Focus, 2012, 2, 337–355 CrossRef PubMed
.
- C. J. Fristrup, K. Jankova, R. Eskimergen, J. T. Bukrinsky and S. Hvilsted, Polym. Chem., 2012, 3, 198–203 RSC
.
- M. Amiji and K. Park, J. Biomater. Sci., Polym. Ed., 1993, 4, 217–234 CrossRef CAS PubMed
.
- I. Banerjee, R. C. Pangule and R. S. Kane, Adv. Mater., 2011, 23, 690–718 CrossRef CAS PubMed
.
- Q. Yu, Y. Zhang, H. Wang, J. Brash and H. Chen, Acta Biomater., 2011, 7, 1550–1557 CrossRef CAS PubMed
.
- X. Liu, Y. Xu, Z. Wu and H. Chen, Macromol. Biosci., 2013, 13, 147–154 CrossRef CAS PubMed
.
- H. Kanazawa and T. Okano, J. Chromatogr., A, 2011, 1218, 8738–8747 CrossRef CAS PubMed
.
- D. L. Huber, R. P. Manginell, M. A. Samara, B.-I. Kim and B. C. Bunker, Science, 2003, 301, 352–354 CrossRef CAS PubMed
.
- S. J. Yuan, S. O. Pehkonen, Y. P. Ting, K. G. Neoh and E. T. Kang, ACS Appl. Mater. Interfaces, 2009, 1, 640–652 CAS
.
- P. Jain, J. Dai, S. Grajales, S. Saha, G. L. Baker and M. L. Bruening, Langmuir, 2007, 23, 11360–11365 CrossRef CAS PubMed
.
- G. Zhai, Z. L. Shi, E. T. Kang and K. G. Neoh, Macromol. Biosci., 2005, 5, 974–982 CrossRef CAS PubMed
.
- H. Assender, V. Bliznyuk and K. Porfyrakis, Science, 2002, 297, 973–976 CrossRef CAS PubMed
.
- P. Koegler, A. Clayton, H. Thissen, G. N. C. Santos and P. Kingshott, Adv. Drug Delivery Rev., 2012, 64, 1820–1839 CrossRef CAS PubMed
.
- D. Quéré, Annu. Rev. Mater. Res., 2008, 38, 71–99 CrossRef
.
- R. N. Wenzel, J. Phys. Colloid Chem., 1949, 53, 1466–1467 CrossRef CAS
.
- T. L. Sun, G. J. Wang, L. Feng, B. Q. Liu, Y. M. Ma, L. Jiang and D. B. Zhu, Angew. Chem., Int. Ed., 2004, 43, 357–360 CrossRef CAS PubMed
.
- T. Sun, L. Feng, X. Gao and L. Jiang, Acc. Chem. Res., 2006, 39, 487 CrossRef CAS
.
- J. Zheng, W. Song, H. Huang and H. Chen, Colloids Surf., B, 2010, 77, 234–239 CrossRef CAS PubMed
.
- T. L. Sun, H. Tan, D. Han, Q. Fu and L. Jiang, Small, 2005, 1, 959–963 CrossRef CAS PubMed
.
- D. Chen, G. Wang and J. Li, J. Phys. Chem. C, 2007, 111, 2351–2367 CAS
.
- F. Xia and L. Jiang, Adv. Mater., 2008, 20, 2842–2858 CrossRef CAS
.
- K. Liu and L. Jiang, Nano Today, 2011, 6, 155–175 CrossRef CAS PubMed
.
- J. V. Barth, G. Costantini and K. Kern, Nature, 2005, 437, 671–679 CrossRef CAS PubMed
.
- M. M. Stevens and J. H. George, Science, 2005, 310, 1135–1138 CrossRef CAS PubMed
.
- B. D. Gates, Q. Xu, M. Stewart, D. Ryan, C. G. Willson and G. M. Whitesides, Chem. Rev., 2005, 105, 1171–1196 CrossRef CAS PubMed
.
- B. Zhao and W. J. Brittain, Prog. Polym. Sci., 2000, 25, 677–710 CrossRef CAS
.
- S. Edmondson, V. L. Osborne and W. T. Huck, Chem. Soc. Rev., 2004, 33, 14–22 RSC
.
- C. J. Fristrup, K. Jankova and S. Hvilsted, Soft Matter, 2009, 5, 4623–4634 RSC
.
- S. T. Milner, Science, 1991, 251, 905–914 CrossRef CAS PubMed
.
- R. Barbey, L. Lavanant, D. Paripovic, N. Schüwer, C. Sugnaux, S. Tugulu and H.-A. Klok, Chem. Rev., 2009, 109, 5437–5527 CrossRef CAS PubMed
.
- N. Ayres, Polym. Chem., 2010, 1, 769–777 RSC
.
- B. Yameen and A. Farrukh, Chem.–Asian J., 2013, 8, 1736–1753 CrossRef CAS PubMed
.
- J. M. Bennett, Meas. Sci. Technol., 1992, 3, 1119 CrossRef
.
-
T. V. Vorburger, J. A. Dagata, G. Wilkening and K. Iizuka, in Beam Effects, Surface Topography, and Depth Profiling in Surface Analysis, ed. A. Czanderna, T. Madey and C. Powell, Springer, US, 2002, ch. 4, vol. 5, pp. 275–354 Search PubMed
.
- M. Bigerelle, A. Van Gorp and A. Iost, Polym. Eng. Sci., 2008, 48, 1725–1736 CAS
.
- M. Rabe, D. Verdes and S. Seeger, Adv. Colloid Interface Sci., 2011, 162, 87–106 CrossRef CAS PubMed
.
- P. Roy, Z. Rajfur, P. Pomorski and K. Jacobson, Nat. Cell Biol., 2002, 4, E91–E96 CrossRef CAS PubMed
.
- S. V. Orski, K. H. Fries, S. K. Sontag and J. Locklin, J. Mater. Chem., 2011, 21, 14135–14149 RSC
.
- M. A. Stuart, W. T. Huck, J. Genzer, M. Muller, C. Ober, M. Stamm, G. B. Sukhorukov, I. Szleifer, V. V. Tsukruk, M. Urban, F. Winnik, S. Zauscher, I. Luzinov and S. Minko, Nat. Mater., 2010, 9, 101–113 CrossRef PubMed
.
- N. Nath and A. Chilkoti, Adv. Mater., 2002, 14, 1243–1247 CrossRef CAS
.
- S. Minko, J. Macromol. Sci., Polym. Rev., 2006, 46, 397–420 CrossRef CAS
.
- P. M. Mendes, Chem. Soc. Rev., 2008, 37, 2512–2529 RSC
.
- A. Calvo, B. Yameen, F. J. Williams, G. J. Soler-Illia and O. Azzaroni, J. Am. Chem. Soc., 2009, 131, 10866–10868 CrossRef CAS PubMed
.
- Q. Yu, X. Li, Y. Zhang, L. Yuan, T. Zhao and H. Chen, RSC Adv., 2011, 1, 262–269 RSC
.
- A. R. Ferhan, L. Guo, X. Zhou, P. Chen, S. Hong and D. H. Kim, Anal. Chem., 2013, 85, 4094–4099 CrossRef CAS PubMed
.
- L. Yuan, H. Wang, Q. Yu, Z. Wu, J. L. Brash and H. Chen, J. Mater. Chem., 2011, 21, 6148–6151 RSC
.
- X. Zhou, F. Boey, F. Huo, L. Huang and H. Zhang, Small, 2011, 7, 2273–2289 CrossRef CAS PubMed
.
- S. Tawfick, M. De Volder, D. Copic, S. J. Park, C. R. Oliver, E. S. Polsen, M. J. Roberts and A. J. Hart, Adv. Mater., 2012, 24, 1628–1674 CrossRef CAS PubMed
.
- W. J. Brittain and S. Minko, J. Polym. Sci., Part A: Polym. Chem., 2007, 45, 3505–3512 CrossRef CAS
.
- T. Cosgrove, T. G. Heath, K. Ryan and T. L. Crowley, Macromolecules, 1987, 20, 2879–2882 CrossRef CAS
.
- T. Cosgrove, T. G. Heath and K. Ryan, Langmuir, 1994, 10, 3500–3506 CrossRef CAS
.
- E. Parsonage, M. Tirrell, H. Watanabe and R. G. Nuzzo, Macromolecules, 1991, 24, 1987–1995 CrossRef CAS
.
- M. L. Hair, D. Guzonas and D. Boils, Macromolecules, 1991, 24, 341–342 CrossRef CAS
.
- L. Han, Z. Mao, J. Wu, Y. Guo, T. Ren and C. Gao, Colloids Surf., B, 2013, 111, 1–6 CrossRef CAS PubMed
.
- L. Han, Z. Mao, J. Wu, Y. Guo, T. Ren and C. Gao, Biomaterials, 2013, 34, 975–984 CrossRef CAS PubMed
.
- L. Han, Z. Mao, H. Wuliyasu, J. Wu, X. Gong, Y. Yang and C. Gao, Langmuir, 2011, 28, 193–199 CrossRef PubMed
.
- S. Turgman-Cohen and J. Genzer, J. Am. Chem. Soc., 2011, 133, 17567–17569 CrossRef CAS PubMed
.
- S. Turgman-Cohen and J. Genzer, Macromolecules, 2012, 45, 2128–2137 CrossRef CAS
.
- E. D. Bain, S. Turgman-Cohen and J. Genzer, Macromol. Theory Simul., 2013, 22, 8–30 CrossRef CAS
.
- X. Gao, W. Feng, S. Zhu, H. Sheardown and J. L. Brash, Macromol. React. Eng., 2010, 4, 235–250 CrossRef CAS
.
- X. Shi, Y. Wang, D. Li, L. Yuan, F. Zhou, B. Song, Z. Wu, H. Chen and J. L. Brash, Langmuir, 2012, 28, 17011–17018 CrossRef CAS PubMed
.
- W. Song and H. Chen, Chin. Sci. Bull., 2007, 52, 3169–3173 CrossRef CAS
.
- F. Zhou, L. Yuan, Y. Mei and H. Chen, Chin. Sci. Bull., 2011, 56, 977–981 CrossRef
.
- F. Zhou, L. Yuan, H. Wang, D. Li and H. Chen, Langmuir, 2011, 27, 2155–2158 CrossRef CAS PubMed
.
- Y. Wang, F. Zhou, X. Liu, L. Yuan, D. Li, Y. Wang and H. Chen, ACS Appl. Mater. Interfaces, 2013, 5, 3816–3823 CAS
.
- H. J. Bai, M. L. Shao, H. L. Gou, J. J. Xu and H. Y. Chen, Langmuir, 2009, 25, 10402–10407 CrossRef CAS PubMed
.
- H. Wang, W. Jiang, Y. Wang, X. Liu, J. Yao, L. Yuan, Z. Wu, D. Li, B. Song and H. Chen, Langmuir, 2013, 29, 3–7 CrossRef CAS PubMed
.
- H. Wang, W. Jiang, L. Yuan, L. Wang and H. Chen, ACS Appl. Mater. Interfaces, 2013, 5, 1800–1805 CAS
.
- H. Wang, L. Wang, P. Zhang, L. Yuan, Q. Yu and H. Chen, Colloids Surf., B, 2011, 83, 355–359 CrossRef CAS PubMed
.
- H. Wang, Y. Wang, L. Yuan, L. Wang, W. Yang, Z. Wu, D. Li and H. Chen, Nanotechnology, 2013, 24, 105101 CrossRef PubMed
.
- L. Chen, X. Liu, B. Su, J. Li, L. Jiang, D. Han and S. Wang, Adv. Mater., 2011, 23, 4376–4380 CrossRef CAS PubMed
.
- S. Wang, H. Wang, J. Jiao, K.-J. Chen, G. E. Owens, K.-i. Kamei, J. Sun, D. J. Sherman, C. P. Behrenbruch, H. Wu and H.-R. Tseng, Angew. Chem., Int. Ed., 2009, 48, 8970–8973 CrossRef CAS PubMed
.
- S. Wang, K. Liu, J. Liu, Z. T. F. Yu, X. Xu, L. Zhao, T. Lee, E. K. Lee, J. Reiss, Y.-K. Lee, L. W. K. Chung, J. Huang, M. Rettig, D. Seligson, K. N. Duraiswamy, C. K. F. Shen and H.-R. Tseng, Angew. Chem., Int. Ed., 2011, 50, 3084–3088 CrossRef CAS PubMed
.
- L. Han, Z. Mao, J. Wu, Y. Guo, T. Ren and C. Gao, Colloids Surf., B, 2013, 111, 1–6 CrossRef CAS PubMed
.
- H. Chen, W. Song, F. Zhou, Z. Wu, H. Huang, J. Zhang, Q. Lin and B. Yang, Colloids Surf., B, 2009, 71, 275–281 CrossRef CAS PubMed
.
- T. Chen, I. Amin and R. Jordan, Chem. Soc. Rev., 2012, 41, 3280–3296 RSC
.
- C. Li, A. Glidle, X. Yuan, Z. Hu, E. Pulleine, J. Cooper, W. Yang and H. Yin, Biomacromolecules, 2013, 14, 1278–1286 CrossRef CAS PubMed
.
- A. M. Telford, L. Meagher, V. Glattauer, T. R. Gengenbach, C. D. Easton and C. Neto, Biomacromolecules, 2012, 13, 2989–2996 CrossRef CAS PubMed
.
- J. Gan, H. Chen, F. Zhou, H. Huang, J. Zheng, W. Song, L. Yuan and Z. Wu, Colloids Surf., B, 2010, 76, 381–385 CrossRef CAS PubMed
.
- F. Zhou, M. Wang, L. Yuan, Z. Cheng, Z. Wu and H. Chen, Analyst, 2012, 137, 1779–1784 RSC
.
- F. Zhou, D. Li, Z. Wu, B. Song, L. Yuan and H. Chen, Macromol. Biosci., 2012, 12, 1391–1400 CrossRef CAS PubMed
.
- Q. Yu, H. Chen, Y. Zhang, L. Yuan, T. Zhao, X. Li and H. Wang, Langmuir, 2010, 26, 17812–17815 CrossRef CAS PubMed
.
- L. Wang, H. Wang, L. Yuan, W. Yang, Z. Wu and H. Chen, J. Mater. Chem., 2011, 21, 13920–13925 RSC
.
- N. Matsuda, T. Shimizu, M. Yamato and T. Okano, Adv. Mater., 2007, 19, 3089–3099 CrossRef CAS
.
- Q. Yu, Y. Zhang, H. Chen, Z. Wu, H. Huang and C. Cheng, Colloids Surf., B, 2010, 76, 468–474 CrossRef CAS PubMed
.
- L. Chen, M. Liu, H. Bai, P. Chen, F. Xia, D. Han and L. Jiang, J. Am. Chem. Soc., 2009, 131, 10467–10472 CrossRef CAS PubMed
.
- S. Simovic, D. Losic and K. Vasilev, Chem. Commun., 2010, 46, 1317–1319 RSC
.
- K. Gulati, S. Ramakrishnan, M. S. Aw, G. J. Atkins, D. M. Findlay and D. Losic, Acta Biomater., 2012, 8, 449–456 CrossRef CAS PubMed
.
- D. G. J. Mann, T. E. McKnight, J. T. McPherson, P. R. Hoyt, A. V. Melechko, M. L. Simpson and G. S. Sayler, ACS Nano, 2008, 2, 69–76 CrossRef CAS PubMed
.
- T. E. McKnight, A. V. Melechko, G. D. Griffin, M. A. Guillorn, V. I. Merkulov, F. Serna, D. K. Hensley, M. J. Doktycz, D. H. Lowndes and M. L. Simpson, Nanotechnology, 2003, 14, 551–556 CAS
.
- T. E. McKnight, A. V. Melechko, D. K. Hensley, D. G. J. Mann, G. D. Griffin and M. L. Simpson, Nano Lett., 2004, 4, 1213–1219 CrossRef CAS
.
- A. K. Shalek, J. T. Robinson, E. S. Karp, J. S. Lee, D. R. Ahn, M. H. Yoon, A. Sutton, M. Jorgolli, R. S. Gertner and T. S. Gujral, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 1870–1875 CrossRef CAS PubMed
.
- W. Kim, J. K. Ng, M. E. Kunitake, B. R. Conklin and P. Yang, J. Am. Chem. Soc., 2007, 129, 7228–7229 CrossRef CAS PubMed
.
- S. Park, Y.-S. Kim, W. B. Kim and S. Jon, Nano Lett., 2009, 9, 1325–1329 CrossRef CAS PubMed
.
- M. Lin, X. Hu, Z. Ma and L. Chen, Anal. Chim. Acta, 2012, 746, 63–69 CrossRef CAS PubMed
.
| This journal is © The Royal Society of Chemistry 2014 |




