Open Access Article
Open Access ArticleKurahyne, an acetylene-containing lipopeptide from a marine cyanobacterial assemblage of Lyngbya sp.†
Arihiro Iwasakia,
Osamu Ohnoa,
Shinpei Sumimotob,
Shoichiro Sudab and
Kiyotake Suenaga*a
aDepartment of Chemistry, Keio University, 3-14-1, Hiyoshi, Kohoku-ku, Yokohama, Kanagawa 223-8522, Japan. E-mail: suenaga@chem.keio.ac.jp
bFaculty of Science, University of Ryukyus, 1 Senbaru, Nishihara, Okinawa 903-0213, Japan
First published on 21st February 2014
Abstract
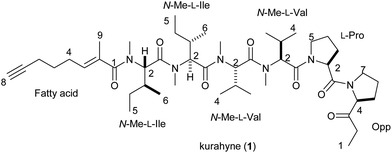
Kurahyne, a new acetylene-containing lipopeptide, was isolated from a cyanobacterial assemblage that mostly consisted of Lyngbya sp. Its structure was elucidated by spectroscopic analyses and chiral HPLC analyses of hydrolysis products. Kurahyne inhibited the growth of human cancer cells and induced apoptosis in HeLa cells, and it seemed to localize in mitochondria.
Some peptides derived from marine organisms have attracted attention due to their remarkable bioactivities.1 Marine cyanobacteria, in particular, produce a variety of novel lipopeptides.2 In our continuing search for new bioactive substances from marine cyanobacteria,3 we investigated the constituents of a cyanobacterial assemblage that mostly consisted of Lyngbya sp. collected at Okinawa, Japan, and isolated an acetylene-containing lipopeptide, kurahyne (1). Structurally, 1 contains a C8-alkynoate moiety and a C7-ketone moiety. The same C8-alkynoate unit has only been found in dragonamide E,4 and the same C7-ketone unit has only been found in bisebromoamides.5 Kurahyne (1) was found to inhibit the growth of human cancer cells and to induce apoptosis in HeLa cells.
The marine cyanobacterial samples6 (2.3 kg, wet weight) were collected at Kuraha, Okinawa, and were extracted with methanol. The extract was filtered, concentrated, and partitioned between EtOAc and H2O. The EtOAc-soluble material was further partitioned between 90% aqueous MeOH and hexane. The material obtained from the aqueous MeOH portion was subjected to fractionation with reversed-phase column chromatography (ODS silica gel, MeOH–H2O) and reversed-phase HPLC (Cosmosil Cholester, MeCN–H2O; Cosmosil 5C18-MS-II, MeOH–H2O) to give kurahyne (1) (29.9 mg) as a colorless oil. The molecular formula of 1 was found to be C47H78N6O7 by HRESIMS (m/z 839.5991, calcd for C47H79N6O7 [M + H]+ 839.6010). The NMR data for 1 are summarized in Table 1.
| Unit | Position | δCb | δHc (J in Hz) | COSY | HMBC | NOESY |
|---|---|---|---|---|---|---|
| a 1H-13C connectivities were determined by HMQC method.b Measured at 100 MHz.c Measured at 400 MHz.d These carbon signals are interchangeable, respectively.e These carbon signals are interchangeable, respectively.f These carbon signals are interchangeable, respectively.g These carbon signals are interchangeable, respectively.h These carbon signals are interchangeable, respectively. | ||||||
| Opp | 1 | 7.7 | 0.96, 3H, dd (7.8, 7.8) | 2a, 2b | 2, 3 | 2a, 2b |
| 2a | 33.44c | 2.29, 1H, dq (18.7, 7.8) | 1, 2b | 1, 3 | 1, 2b, 4 | |
| 2b | 2.07, 1H, dq (18.7, 7.8) | 1, 2a | 1, 3 | 1, 2a, 4 | ||
| 3 | 208.5 | |||||
| 4 | 64.1 | 4.41, 1H, dd (8.6, 4.7) | 5a, 5b | 5, 6 | 2a, 2b, 5a, 5b | |
| 5a | 27.77d | 1.35, 1H, m | 4, 5b, 6a, 6b | 3, 4, 6, 7 | 4 | |
| 5b | 1.18, 1H, m | 4, 5a, 6a, 6b | 3, 6, 7 | 4 | ||
| 6a | 25.01e | 1.43, 1H, m | 5a, 5b, 6b, 7a, 7b | 4, 5, 7 | 7b | |
| 6b | 1.31, 1H, m | 5a, 5b, 6a, 7a, 7b | 4, 5, 7 | 7a | ||
| 7a | 46.7 | 3.45, 1H, m | 6a, 6b, 7b | 4, 5, 6 | 6b, 7b, 2 (Pro) | |
| 7b | 3.05, 1H, m | 6a, 6b, 7a | 5, 6 | 6a, 7a, 2 (Pro) | ||
| Pro | 1 | 170.3 | ||||
| 2 | 58.1 | 4.50, 1H, dd (8.5, 4.0) | 3a, 3b | 3, 4 | 3a, 3b, 7a (Opp), 7b (Opp) | |
| 3a | 28.6 | 1.82, 1H, m | 2, 3b, 4a, 4b | 1, 2, 4, 5 | 2, 4b, 5b | |
| 3b | 1.64, 1H, m | 2, 3a, 4a, 4b | 1, 2, 4, 5 | 2, 4a | ||
| 4a | 24.96e | 1.81, 1H, m | 3a, 3b, 4b, 5a, 5b | 2, 3, 5 | 3b | |
| 4b | 1.43, 1H, m | 3a, 3b, 4a, 5a, 5b | 2, 5 | 3a, 5a | ||
| 5a | 47.9 | 4.03, 1H, m | 4a, 4b, 5b | 2, 3, 4 | 4b, 5b, 2 (N-Me-Val-1) | |
| 5b | 3.72, 1H, m | 4a, 4b, 5a | 3, 4 | 3a, 5a, 2 (N-Me-Val-1), 4 (N-Me-Val-1) | ||
| N-Me-Val-1 | 1 | 169.2 | ||||
| 2 | 59.7 | 5.40, 1H, d (11.2) | 3 | 1, 3, 4, 5, N-Me | 3, 4, 5, N-Me, 5a (Pro), 5b (Pro) | |
| 3 | 28.1 | 2.47, 1H, m | 2, 4, 5 | 1, 2, 4 | 2, 4, 5, N-Me | |
| 4 | 19.7 | 1.22, 3H, d (6.5) | 3 | 2, 3, 5 | 2, 3, 5b (Pro) | |
| 5 | 18.6 | 0.77, 3H, d (7.0) | 3 | 2, 3, 4 | 2, 3, N-Me | |
| N-Me | 30.9 | 3.38, 3H, s | 2, 1 (N-Me-Val-2) | 2, 3, 5, 2 (N-Me-Val-2) | ||
| N-Me-Val-2 | 1 | 170.88f | ||||
| 2 | 58.6 | 5.49, 1H, d (10.7) | 3 | 1, 3, 4, 5, N-Me (N-Me-Val-1) | 3, 4, 5, N-Me, N-Me (N-Me-Val-1) | |
| 3 | 27.71d | 2.55, 1H, m | 2, 4, 5 | 2, 4, 5 | 2, 4, 5, N-Me | |
| 4 | 19.9 | 0.94, 3H, d (6.4) | 3 | 2, 3, 5 | 2, 3 | |
| 5 | 18.0 | 0.79, 3H, d (7.0) | 3 | 2, 3, 4 | 2, 3, N-Me | |
| N-Me | 30.70g | 3.27, 3H, s | 2, 1 (N-Me-Ile-1) | 2, 3, 5, 2 (N-Me-Ile-1) | ||
| N-Me-Ile-1 | 1 | 170.6 | ||||
| 2 | 57.0 | 5.53, 1H, d (11.2) | 3 | 1, 3, 4, 6, N-Me (N-Me-Val-2) | 3, 4a, 5, 6, N-Me (N-Me-Val-2) | |
| 3 | 33.42h | 2.37, 1H, m | 4a, 4b, 6 | 2 | 2, 5, 6, N-Me | |
| 4a | 24.1 | 1.35, 1H, m | 3, 4b, 5 | 3, 5, 6 | 2 | |
| 4b | 1.03, 1H, m | 3, 4a, 5 | 3, 5, 6 | |||
| 5 | 10.9 | 0.86, 3H, m | 4a, 4b | 3, 4 | 2, 3, N-Me | |
| 6 | 15.77 | 0.86, 3H, m | 3 | 2, 3, 4 | 2, 3 | |
| N-Me | 30.63g | 3.26, 3H, s | 2, 1 (N-Me-Ile-2) | 3, 5, 2 (N-Me-Ile-2) | ||
| N-Me-Ile-2 | 1 | 170.91f | ||||
| 2 | 56.6 | 5.54, 1H, d (11.2) | 3 | 1, 3, 4, 6, N-Me (N-Me-Ile-1), 1 (fatty acid) | 3, 4a, 4b, 5, 6, N-Me (N-Me-Ile-1) | |
| 3 | 33.1h | 2.37, 1H, m | 4a, 4b, 6 | 2 | 2, 4a, 5, 6, N-Me | |
| 4a | 24.6 | 1.39, 1H, m | 3, 4b, 5 | 2, 3, 5, 6 | 2, 3, 4b, N-Me | |
| 4b | 1.19, 1H, m | 3, 4a, 5 | 3, 5, 6 | 2, 4a | ||
| 5 | 11.0 | 0.90, 3H, m | 4a, 4b | 3, 4 | 2, 3 | |
| 6 | 15.83 | 0.92, 3H, d (8.5) | 3 | 2, 3, 4 | 2, 3 | |
| N-Me | 31.9 | 2.86, 3H, s | 2, 1 (fatty acid) | 3, 4a, 3 (fatty acid), 9 (fatty acid) | ||
| Fatty acid | 1 | 173.4 | ||||
| 2 | 133.6 | |||||
| 3 | 129.1 | 5.38, 1H, dt (7.5, 1.6) | 4, 9 | 1, 4, 9 | 4, 5, N-Me (N-Me-Ile-2) | |
| 4 | 26.6 | 1.96, 2H, m | 3, 5 | 2, 3, 5, 6 | 3, 9, 5 | |
| 5 | 28.0 | 1.34, 2H, m | 4, 6 | 3, 4, 6, 7 | 4, 6 | |
| 6 | 18.2 | 1.93, 2H, dt (7.3, 2.9) | 5, 8 | 4, 5, 7, 8 | 5 | |
| 7 | 83.8 | |||||
| 8 | 69.4 | 1.80, 1H, t (2.9) | 6 | |||
| 9 | 14.4 | 1.75, 3H, br s | 3 | 1, 2, 3 | 4, N-Me (N-Me-Ile-2) | |
The 1H NMR spectrum revealed the presence of four singlets corresponding to N-methyl amide substituents (δ 3.38, 3.27, 3.26, 2.86), one vinyl methyl group (δ 1.75), five methine groups corresponding to the α position of the amino acid residue (δ 5.54, 5.53, 5.49, 5.40, 4.50) and nine high-field methyl groups (δ 1.22, 0.96, 0.94, 0.92, 0.90, 0.86, 0.86, 0.79, 0.77). Additionally, the 1H NMR spectrum indicated an olefinic double triplet (δ 5.38) and a triplet characteristic of a terminal acetylene proton [δ 1.80 (J = 2.9 Hz)]. 1 possessed two 13C NMR absorptions consistent with a terminal acetylene (δ 83.8 and 69.4), and seven carbonyl signals (δ 208.5, 173.4, 170.91, 170.88, 170.6, 170.3, 169.2). Further analysis of the 1H NMR, 13C NMR, COSY, HMQC and HMBC spectra revealed the presence of proline, two N-methylvalines and two N-methylisoleucines. Additionally, the presence of residues derived from 2-(1-oxo-propyl)-pyrrolidine (Opp) and 2-methyloct-2-en-7-ynoic acid (fatty acid) was also established. Despite the presence of a terminal acetylene, an IR band of triple bond stretching was not observed (see ESI, S10†). The existence of an acetylene group was confirmed based on the magnitude of the coupling constant of C-8 of fatty acid (1JC–H = 253 Hz) on the INEPT spectrum (see ESI, S8 and 9†) and comparison of the 13C chemical shifts between 1 and dragonamide E4 possessing the same fatty acid moiety (see ESI, S20†).
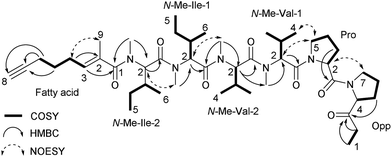
The sequence of these partial structures was determined based on HMBC and NOESY data (Table 1 and Fig. 1). A NOESY correlation between H-7 of Opp and H-2 of Pro connected these two residues. Two NOESY correlations observed at H-5 of Pro/H-2 of N-Me-Val-1 and H-5b of Pro/H-4 of N-Me-Val-1 revealed the connectivity between these two residues. Additional HMBC correlations, H-2 of N-Me-Val-2/N-Me of N-Me-Val-1 and N-Me of N-Me-Val-2/C-1 of N-Me-Ile-1, and NOESY correlations, N-Me of N-Me-Val-2/H-2 of N-Me-Ile-1 and N-Me of N-Me-Ile-1/H-2 of N-Me-Ile-2, expanded the sequence to Opp-Pro-N-Me-Val-N-Me-Val-N-Me-Ile-N-Me-Ile. Furthermore, an HMBC correlation at N-Me of N-Me-Ile-2/C-1 of fatty acid allowed us to determine the location of the fatty acid moiety. Finally, NOESY correlations, H-4 of fatty acid/H-9 of fatty acid, and the chemical shift of the vinyl methyl carbon (δ 14.4, C-9 of fatty acid) supported an E geometry for the C2–C3 olefinic bond in the fatty acid moiety, thereby completing the gross structure of kurahyne, as shown in Fig. 1.
To assign the absolute configurations of the eight chiral centers, we generated optically active fragments. Enantiomeric standards for Pro are commercially available, while those for other moieties must be synthesized in the laboratory by standard methods (N-Me-Val, N-Me-Ile, Opp).7 According to the previous paper,5a a direct acid hydrolysis of an Opp-containing compound resulted in the epimerization of C-4 of the Opp moiety. To prevent the racemization of Opp, reduction of the ketone with NaBH4 followed by acid hydrolysis afforded every amino acid component contained in 1 and 2-(1-hydroxypropyl)-pyrrolidine derived from an Opp moiety as a mixture of two diastereomers. Based on a comparison of the retention times of the obtained amino acids to those of authentic samples by chiral HPLC, all of the amino acid components in 1 were determined to be L-form. With regard to the Opp moiety, the absolute configuration was determined by reversed-phase HPLC analysis based on a comparison of the retention times of Marfey derivatives8 of the obtained diastereomeric alcohols to those of authentic samples. As a result of the analysis, the absolute configuration of the Opp moiety was elucidated to be 4S. The absolute stereochemistry of kurahyne was determined as shown in 1.
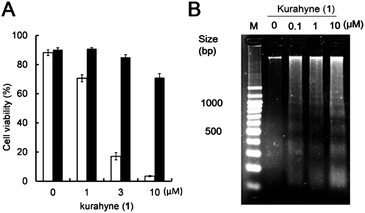
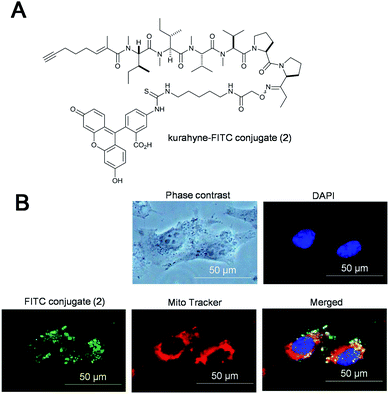
To evaluate the growth-inhibitory activities of kurahyne (1), an MTT assay with HeLa cells and HL60 cells was used. The cells were treated in 96-well plates with various concentrations of the compounds (0.01–10 μg mL−1 for HeLa cells, 0.001–10 μg mL−1 for HL60 cells) for 72 h. The data from these assays revealed that 1 inhibited the growth of both HeLa cells and HL60 cells, with IC50 values of 3.9 ± 1.1 μM and 1.5 ± 0.1 μM, respectively. Additionally, 1 showed cytotoxicity against HeLa cells, as determined using the trypan blue dye exclusion assay. Furthermore, cell death of these cells induced by 1 was suppressed in the presence of Z-VAD-FMK, an irreversible and cell-permeable inhibitor of caspases (Fig. 2A). Significant DNA laddering in these cells was observed in the presence of 1 (Fig. 2B). These results indicated that 1 induced apoptosis in HeLa cells. To examine the subcellular localization of 1, a kurahyne–fluorescein conjugate 2 was synthesized (Fig. 3A). Introduction of fluorescein to COOH-terminus of kurahyne retained its growth inhibitory activity (IC50: 85 ± 13 μM, HeLa cells). The staining sites of 2 seemed to be included in those of Mito Tracker Red in HeLa cells on the basis of fluorescence microscopic analysis (Fig. 3B). From this result, it is likely that the mechanism of action of 1 is mitochondrially-targeted.
In conclusion, kurahyne (1), a novel acetylene-containing lipopeptide, was isolated from a marine cyanobacterial assemblage that mostly consisted of Lyngbya sp. The structure of 1 was established by spectroscopic analysis and HPLC analysis of acid hydrolysates. The structural features of 1 are the presence of an Opp moiety and a 2-methyloct-2-en-7-ynoic acid moiety. To our knowledge, 1 is the first reported compound which possesses both of these moieties. 1 inhibited the growth of both HeLa cells and HL60 cells, with IC50 values of 3.9 ± 1.1 μM and 1.5 ± 0.1 μM, respectively. Furthermore, 1 was revealed to induce apoptosis in HeLa cells, and it seemed to localize in mitochondria. The detailed bioactive investigation of 1 is ongoing.
Acknowledgements
This work was supported in part by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (24310160 and 25750386) and the MEXT-Supported Program for the Strategic Research Foundation at Private Universities, 2009–2013 from the Ministry of Education, Culture, Sports, Science and Technology, Japan.Notes and references
- L. H. Zheng, Y. J. Wang, J. Sheng, F. Wang, Y. Zheng, X. K. Lin and M. Sun, Mar. Drugs, 2011, 9, 1840–1859 CrossRef CAS PubMed.
- (a) M. Tanighchi, J. K. Nunnery, N. Engene, E. Esquenazi, T. Byrum, P. C. Dorrestein and W. H. Gerwick, J. Nat. Prod., 2010, 73, 393–398 CrossRef PubMed; (b) H. Choi, A. R. Pereira, Z. Cao, C. F. Shuman, N. Engene, T. Byrum, T. Matainaho, T. T. Murray, A. Mangoni and W. H. Gerwick, J. Nat. Prod., 2010, 73, 1411–1421 CrossRef CAS PubMed; (c) E. Mevers, T. Byrum and W. H. Gerwick, J. Nat. Prod., 2013, 76, 1810–1814 CrossRef CAS PubMed.
- (a) A. Iwasaki, T. Teruya and K. Suenaga, Tetrahedron Lett., 2010, 51, 959–960 CrossRef CAS PubMed; (b) T. Teruya, H. Sasaki, K. Kitamura, T. Nakayama and K. Suenaga, Org. Lett., 2009, 11, 2421–2424 CrossRef CAS PubMed; (c) M. Morita, O. Ohno, T. Teruya, T. Yamori, T. Inuzuka and K. Suenaga, Tetrahedron, 2012, 30, 5984–5990 CrossRef PubMed.
- M. J. Balunas, R. G. Linington, K. Tidgewell, A. M. Fenner, L. D. Ureña, G. D. Togna, D. E. Kyle and W. H. Gerwick, J. Nat. Prod., 2010, 73, 60–66 CrossRef CAS PubMed.
- (a) T. Teruya, H. Sasaki, H. Fukazawa and K. Suenaga, Org. Lett., 2009, 11, 5062–5065 CrossRef CAS PubMed; (b) H. Sasaki, T. Teruya, H. Fukazawa and K. Suenaga, Tetrahedron, 2011, 67, 990–994 CrossRef CAS PubMed.
- The most cyanobacterium was morphologically classified into the genus Lyngbya because it was composed of short cells with a thick sheath. From the phylogenetic tree inferred from 959 bp of 16S rRNA gene sequences (see ESI, S16†) revealed that the present cyanobacterium (Maeda 130904A, accession no. AB857842) formed a clade with Oscillatoria miniata NAC8-50 (GU724208), and closely related with Trichodesmium spp.9 The identification described above was carried out using the same cyanobacterium collected at the same location in September 2013, and we confirmed that its extract contained 1. Despite our efforts, other minor cyanobacteria in the assemblage were not identified by 16S rRNA gene analysis. A detailed classification will be reported later.
- X. Gao, Y. Liu, S. Kwong, Z. Xu and T. Ye, Org. Lett., 2010, 12, 3018–3021 CrossRef CAS PubMed.
- P. Marfey, Carlsberg Res. Commun., 1984, 49, 591–596 CrossRef CAS.
- (a) S. Suda, R. Moriya, S. Sumimoto, O. Ohno and K. Suenaga, J. Mar. Sci. Tech., 2013, 21, 175–180 Search PubMed; (b) A. Iwasaki, S. Sumimoto, O. Ohno, S. Suda and K. Suenaga, Bull. Chem. Soc. Jpn. DOI:10.1246/bcsj.20140008.
Footnote |
| † Electronic supplementary information (ESI) available: 1H, 13C, COSY, NOESY, HMQC, HMBC and INEPT NMR spectra in C6D6 for kurahyne (1). IR spectrum. HPLC chromatograms for determination of absolute configurations. Detailed experimental procedures. See DOI: 10.1039/c4ra00132j |
| This journal is © The Royal Society of Chemistry 2014 |