DOI:
10.1039/C4RA01936A
(Paper)
RSC Adv., 2014,
4, 20382-20392
Synthesis and biological evaluation of quinoline–imidazole hybrids as potent telomerase inhibitors: a promising class of antitumor agents
Received
6th March 2014
, Accepted 24th April 2014
First published on 24th April 2014
Abstract
A series of 1-isoquinoline-2-styryl-5-nitroimidazole derivatives has been synthesized and screened for telomerase inhibitory as well as antiproliferative properties on four variant cancer cell lines (U251, Hela, HepG-2 and A549). The bioassay results showed that most of the designed compounds exhibited moderate to potent in vitro inhibitory activity in the enzymatic and cellular assays, of which compounds 3u and 3v were revealed the most potent telomerase inhibitors, with IC50 values of 0.98 μM and 1.92 μM, respectively, compared to staurosporine. Docking simulation was performed to position compounds 3u and 3v into the telomerase active site to determine the probable binding pose. Twenty three compounds were scrutinized by the CoMFA and CoMSIA techniques of 3D Quantitative Structure–Activity Relationship (QSAR). These new findings along with molecular docking observations could provide important information that compounds 3u and 3v with potent inhibitory activity in tumor growth inhibition may be potential anticancer agents.
1. Introduction
Telomeres are dynamic DNA–protein complexes that cap the ends of linear chromosomes consisting of TTAGGG repeats and telomere-binding proteins. Telomeres protect the chromosome ends from degradation, recombination and DNA repair activities.1,2 Telomerase is the most important enzyme involved in telomere maintenance in tumor cells identified so far.3,4 Telomeres shorten after each round of replication and serve as a “mitotic clock”, which limits the number of replications in normal cells. This block can be bypassed in cancer cells by up regulation of telomerase, leading to unlimited proliferative capacity.5,6
Telomeres are differentially regulated in normal somatic cells compared to cancer cells, primarily through differing levels of active telomerase enzyme. The correlation between the detection of high levels of active telomerase in cancer cells and its absence or down-regulation in surrounding noncancerous cells has been well established7–9 and the activation of telomerase has been identified as one of the key stages in oncogenic transformation. Therefore, inhibition of telomerase has been approached through different strategies, ranging from antisense-based inhibitors to random screening of synthetic and natural products.10,11
Since the discovery of telomerase in cancer cells, although scientists have designed various telomerase inhibitors with certain effects, there has not been an efficient pilot structure. The main reason is that conclusive evidence of telomerase as a tumor site specific marker has not been proved, thus inhibitors can't be accurately designed. In 2008, Andrew J. G. revealed the key active site of telomerase [three dimensional structure of telomerase reverse transcriptase (TERT) protein catalytic subunit: 3DU6.pdb] and, the scientists can focus on the clear target to try a new molecular designing scheme.12,13 However, so far, few reports based on leading compounds targeting 3DU6 were published.
Quinolin derivatives have been known for a long time to exert multiple biological effects, in particular to act as anticancer agents14,15 and as inhibitors of DNA polymerases like telomerase inhibitors (Fig. 1).16–19 Moreover, nitroimidazole has been studied extensively for use as radio sensitisers due to its affinity for hypoxic tumors and molecular markers of hypoxic regions in solid tumours.20 2-Nitroimidazole derivatives, such as metronidazole and etanidazole, have been reported to have potent radiosensitising ability.21–24 In addition, to our knowledge, few reports available in literature which described the anticancer activity of novel compounds having modifications at C-2 of the nitromidazole. Now it is apparent that hydroxyl and nitro group of metronidazole play a key role in the metabolic activation. Nitromidazole derivatives substituted in the 2-methyl group may be promising for developing new, potent, and safe lead compounds.25,26
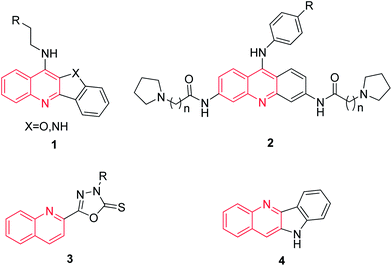 |
| | Fig. 1 Some reported chemical structures as telomerase inhibitors. | |
To explore novel and potent telomerase inhibitors for cancer chemotherapy, we hereby report a new series of isoquinoline derivatives bearing nitroimidazole with the in vitro antitumour studies. Docking simulations are performed using the X-ray crystallographic structure of the telomerase in complex with an inhibitor to explore the binding modes of these compounds at the active site.
2. Results and discussion
2.1 Chemistry
A series of 1-(isoquinoline-1-carboxylate)-2-styryl-5-nitroimidazole derivatives were synthesized by the routes outlined in Scheme 1. According to method A, compounds 1a–w were synthesized by the reaction of metronidazole with different substituted benzaldehyde using DMSO as solvent by adding rapidly a stirred solution of sodium methoxide in methanol at room temperature. Synthesized compounds 1a–w were treated with 4-methylbenzoyl chloride using triethylamine as catalyst in CH2Cl2 which resulted to compounds 2a–w which were further subjected to column chromatography for purification. Subsequently, isoquinoline-1-carboxylic acid, compounds 2a–w, and K2CO3 were taken in DMF and refluxed to provide the desired compounds 3a–w. In the interim, the desired products 3a–w could also be synthesized following another pathway depicted in method B. Compound 5 was prepared according the modified procedure of Kümmerle et al.27. Active isoquinoline-1-carboxylic acid and metronidazole were dissolved in CH2Cl2 followed by drop wise addition triethylamine and compound 6 was obtained with yield of 85%. Then, at room temperature, the target compounds 3a–w were synthesized by the reaction of compound 6 with substituted benzaldehydes in DMSO by adding rapidly a stirred solution of sodium methoxide in methanol. All of the synthetic compounds gave satisfactory analytical and spectroscopic data, which were in full accordance with their depicted structures.
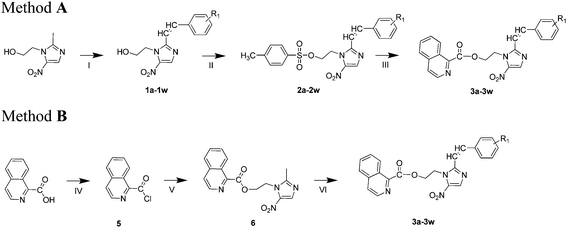 |
| | Scheme 1 General method of the preparation of compounds 3a–w. Reagents and conditions: method A: (I) sodium methoxide, different aldehydes, DMSO, methanol, room temperature, 4 h; (II) 4-toluene sulfonyl chloride; CH2Cl2, TEA, room temperature, 5 h. (III) Isoquinoline-1-carboxylic acid, DMF, K2CO3, reflux, overnight; method B: (IV) SOCl2, DMF, reflux, 4 h; (V) CH2Cl2, TEA, 8 h. (VI) NaOH, different aldehydes, DMSO, methanol, room temperature, overnight. | |
2.2 Biological activity
2.2.1 Antiproliferative activity. All the synthesized compounds 3a–w were screened for their antitumour activities against four cultured cell lines, compared to the potent positive control erlotinib and lapatinib under identical conditions for comparison in vitro, which were human pulmonary adenocarcinoma cells (A549), human breast cancer cells (MCF-7), human glioma cells (U251) and human cervical cancer cell line (Hela) by CCK-8 method. The outcomes were shown in Table 1. According to Table 1, most of the synthesized compounds exhibited a better anticancer activity against one or more of the cell lines used. Most significantly, compounds 3u and 3v displayed the most potent anticancer activities.
Table 1 In vitro anticancer activities (IC50, μM) of compound 3a–w against human tumor cell lines
| Compounds |
R |
IC50 (μM) |
| A549 |
Hela |
U251 |
HepG-2 |
| 3a |
o-F |
0.41 |
0.35 |
0.47 |
0.46 |
| 3b |
m-F |
0.52 |
0.37 |
0.52 |
0.30 |
| 3c |
p-F |
0.44 |
0.20 |
0.39 |
0.31 |
| 3d |
o-Cl |
0.71 |
0.46 |
0.56 |
0.64 |
| 3e |
m-Cl |
0.58 |
0.59 |
0.56 |
0.57 |
| 3f |
p-Cl |
0.61 |
0.55 |
0.49 |
0.51 |
| 3g |
o-Br |
0.64 |
0.54 |
0.60 |
0.63 |
| 3h |
m-Br |
0.69 |
0.42 |
0.43 |
0.41 |
| 3i |
p-Br |
0.59 |
0.57 |
0.54 |
0.59 |
| 3j |
o-NO2 |
0.51 |
0.34 |
0.56 |
0.40 |
| 3k |
m-NO2 |
0.44 |
0.31 |
0.41 |
0.33 |
| 3l |
p-NO2 |
0.50 |
0.26 |
0.37 |
0.29 |
| 3m |
o-CH3 |
0.76 |
0.70 |
0.71 |
0.68 |
| 3n |
m-CH3 |
0.62 |
0.61 |
0.70 |
0.68 |
| 3o |
p-CH3 |
0.60 |
0.58 |
0.61 |
0.63 |
| 3p |
o-OCH3 |
0.81 |
0.74 |
0.68 |
0.78 |
| 3q |
m-OCH3 |
0.58 |
0.64 |
0.67 |
0.72 |
| 3r |
p-OCH3 |
0.60 |
0.56 |
0.47 |
0.59 |
| 3s |
2,4-2Cl |
0.60 |
0.43 |
0.51 |
0.40 |
| 3t |
2-F-6-Cl |
0.59 |
0.55 |
0.43 |
0.52 |
| 3u |
Fused Ph |
0.32 |
0.05 |
0.10 |
0.11 |
| 3v |
p-N(CH3)2 |
0.43 |
0.12 |
0.14 |
0.17 |
| 3w |
H |
0.55 |
0.38 |
0.53 |
0.38 |
| Tofacitinib |
— |
0.33 |
0.04 |
0.11 |
0.10 |
| Citrate |
— |
0.28 |
0.10 |
0.14 |
0.12 |
Structure–activity relationships in these studied compounds demonstrated that compound 3w, with a bare phenyl ring, showed more potent anticancer activity than compounds 3m–r containing methyl and methoxy groups. However, compared with compounds 3a–l with electron-withdrawing groups, compound 3w exhibited superior activity than compounds 3d–i whose substituted group were F and Cl, and inferior activity than compounds 3a–c and 3j–l with F and NO2 moiety, respectively. Interestingly, compound 3s, two chlorine groups on the phenyl ring, showed more potent activity than compounds 3d–f with only one chlorine moiety. Meanwhile, comparing compound 3t having two different halogen atoms (F and Cl) substituents and compounds 3a–f with only F or Cl atoms, we found that the potency order was compounds 3d–f < compound 3t < compounds 3a–c. And more, when the compounds were methoxyl/methyl-substituted derivatives, the activity gradient was ortho > meta > para. Compounds 3u and 3v, whose IC50 value were 0.32, 0.05, 0.10, 0.11 and 0.43, 0.12, 0.14, 0.17 μM against A549, Hela, U251 and HepG-2 respectively, with bulky substituted naphthyl and dimethylamiophenyl on the benzene ring, displayed higher activities than other substituents.
2.2.2 Analysis of apoptosis by Annexin V-PE fluorescence-activated cell sorting (FACS). To test whether the inhibition of cell growth of Hela was related to cell apoptosis, Hela cell apoptosis induced by compound 3u was determined using flow cytometry. The uptake of Annexin V-PE was significantly increased, and the uptake of normal cells was significantly decreased in a time-dependent manner. Finally the percentage of early apoptotic cells was markedly elevated in a density-dependent manner from 7.50% to 13.7% at 48 h (Fig. 2).
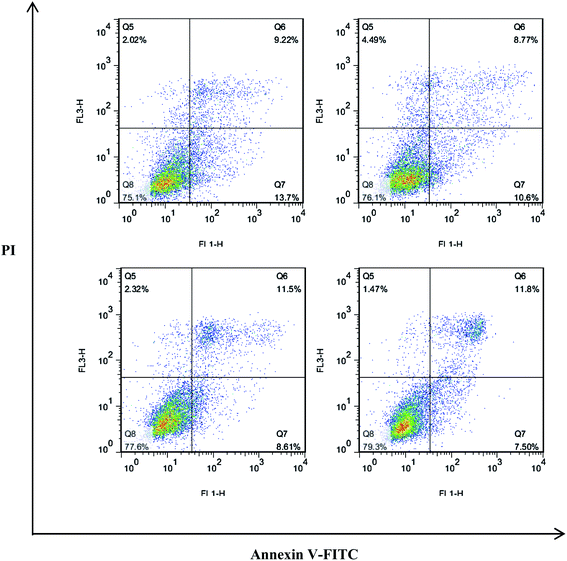 |
| | Fig. 2 Compound 3u induced apoptosis in Hela cells with the density of 1.6, 8.0, 40.0, 200.0 μg mL−1. Hela cells were treated with for 48 h. | |
2.2.3 In vitro telomerase inhibition activity. The telomerase inhibitory potency of compounds 3a–w was examined and the results were summarized in Table 2. Most of the tested compounds displayed good telomerase inhibitory activities and among them compounds 3u and 3v displayed the best inhibitory activity with an IC50 of 0.98 and 1.92 μM for telomerase which was comparable to the reference compound staurosporine. The results of telomerase inhibitory activity of the tested compounds correlated with the Structure–Activity Relationships (SAR) of their inhibitory effects on the cell proliferation assay. This suggested that the potent inhibitory effects of the synthetic compounds on the cell proliferation assay were causally related to their kinase inhibitory activities.
Table 2 Telomerase inhibitory activity and docking calculation of compounds 3a–w
| Compounds |
ΔGb (−kcal mol−1) |
IC50 (μM) |
SIa |
Compounds |
ΔGb (−kcal mol−1) |
IC50 (μM) |
SIa |
| CDocker energy |
Telomerase inhibition |
CDocker energy |
Telomerase inhibition |
| SI: selectivity index SI = log10(Cctrl/Ctest). Used as a positive control. |
| 3a |
45.65 |
7.42 |
−0.35 |
3m |
35.84 |
9.73 |
−0.47 |
| 3b |
44.24 |
7.51 |
−0.36 |
3n |
35.15 |
9.95 |
−0.8 |
| 3c |
49.17 |
4.53 |
−0.14 |
3o |
36.62 |
9.02 |
−0.44 |
| 3d |
36.21 |
9.21 |
−0.44 |
3p |
29.73 |
11.23 |
−0.53 |
| 3e |
37.10 |
8.56 |
−0.41 |
3q |
34.23 |
10.34 |
−0.49 |
| 3f |
39.05 |
8.38 |
−0.40 |
3r |
39.25 |
8.12 |
−0.39 |
| 3g |
36.22 |
8.84 |
−0.42 |
3s |
43.38 |
7.73 |
−0.37 |
| 3h |
42.11 |
7.96 |
−0.38 |
3t |
40.09 |
8.05 |
−0.39 |
| 3i |
37.03 |
8.67 |
−0.42 |
3u |
69.14 |
0.98 |
0.53 |
| 3j |
46.85 |
7.23 |
−0.34 |
3v |
53.23 |
1.92 |
0.24 |
| 3k |
48.23 |
6.03 |
−0.26 |
3w |
38.74 |
8.43 |
−0.41 |
| 3l |
51.02 |
3.75 |
−0.05 |
Staurosporineb |
— |
3.31 |
— |
2.3 Computational
2.3.1 Molecular docking. In order to explore probable interaction model of inhibitors and enzyme active site, molecular docking of the potent inhibitor compounds 3u and 3v into active binding site of telomerase was performed to simulate a binding model derived from TERT protein catalytic subunit (3DU6.pdb). All docking runs were applied CDocker Dock protocol of Discovery Studio 3.5. Binding models of compounds 3u and 3v and telomerase were presented in Fig. 3 and 4. The amino acid residues which had interaction with TERT protein catalytic subunit as well as bond length were labeled. In the binding mode, compound 3u was nicely bound to TERT protein catalytic subunit via four hydrogen bonds (angle O⋯H–N = 141.6°, distance = 5.3 Å with oxygen group of LYS189, angle O⋯H–O = 88.7°, distance = 5.9 Å with amino hydrogen of LYS189, angle N⋯H–O = 136.4°, distance = 3.6 Å with amino hydrogen of ALA255, angle N⋯H–O = 155.6°, distance = 3.5 Å with amino hydrogen of ALA255), four charge interactions and three π–cation interactions. The end group of ASP343, ASP254 and ASP251 were respectively formed four charge interactions with two nitrogen atoms. Meanwhile, the imidazole moiety, a five-membered heterocycle, formed a π–cation interaction with LYS189. Also the end amino cation of ARG194 formed two π–cation interactions with naphthyl ring. As for compound 3v, it also performed a nice bonding situation via one hydrogen bond (angle O⋯H–O = 161.6°, distance = 6.1 Å with oxygen group of LYS189), three charge interactions and three π–cation interactions. LYS189 formed three remarkable π–cation interactions with benzene ring, imidazole ring and isoquinoline ring respectively. Besides, simple to compound 3u, ASP343 and ASP251 were respectively formed three charge interactions with two nitrogen atoms. The docking calculation of all the compounds was also depicted in Table 2. The CDocker energy (energy of the ligand–receptor complexes) agreed with the telomerase inhibitory trend for all the synthesized compounds.
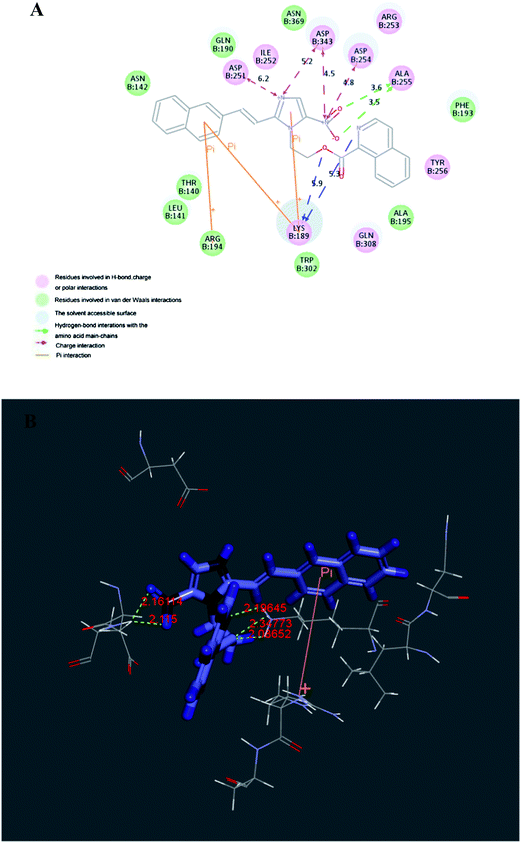 |
| | Fig. 3 Molecular docking modeling of compound 3u with telomerase: (A) 2D model of the interaction between compound 3u and the telomerase binding site. (B) 3D model of the interaction between compound 3u and the telomerase binding site. | |
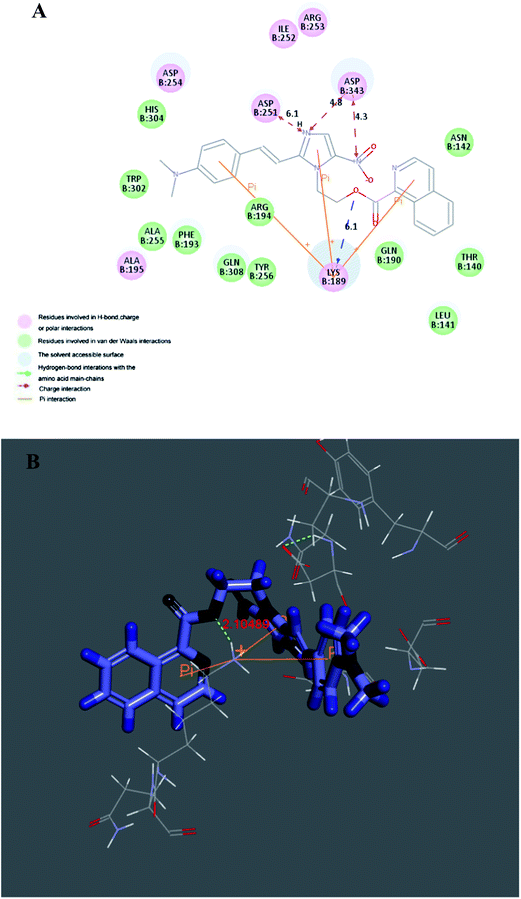 |
| | Fig. 4 Molecular docking modeling of compound 3v with telomerase: (A) 2D model of the interaction between compound 3v and the telomerase binding site. (B) 3D model of the interaction between compound 3v and the telomerase binding site. | |
2.3.2 3D-QSAR model. In order to acquire a systematic SAR profile on 23 compounds as antitumor agents and to explore the more powerful telomerase inhibitors, 3D-QSAR model was built to choose activity conformation of the designed molecular and reasonably evaluated the designed molecules by using the corresponding pIC50 values which were converted from the obtained IC50 (μM) values of telomerase inhibition and performed by built-in QSAR software of DS 3.5 (Discovery Studio 3.5, Accelrys, Co. Ltd). The way of this transformation was derived from an online calculator developed from an Indian medicinal chemistry lab (http://www.sanjeevslab.org/tools-IC50.html).22 The training and test set was divided by the random diverse molecules method of DS 3.5, in which the test set accounted for 83% while the training set was set to 17%. The training set composed 5 agents and 18 agents were consisted of the relative test set. The success of this model depended on docking study and the reliability of previous study about activities data.In default situation, the alignment conformation of each molecule that possessed the lowest CDocker_Interaction_Energy among the ten docked poses. The 3D-QSAR model generated from DS 3.5, defined the critical regions (steric or electrostatic) affecting the binding affinity. It was a PLS model that set up 230 independent variables (conventional R2 = 0.87). The graphical relationship of observed and predicted values had been illustrated in Fig. 5a, in which the plot of the observed IC50 versus the predicted values showed that this model could be used in prediction of activity.
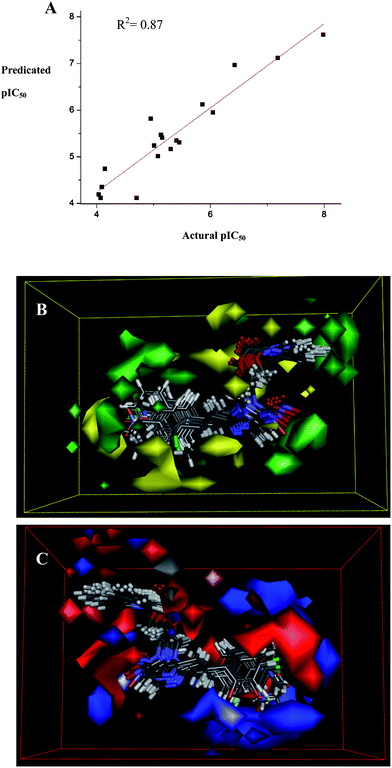 |
| | Fig. 5 (A) Using linear fitting curve to compare the predicted pIC50 value with that of experiment. (B) Isosurface of the 3D-QSAR model coefficients on electrostatic potential grids. The blue triangle mesh represents positive electrostatic potential and the red area represents negative electrostatic potential. (C) Isosurface of the 3D-QSAR model coefficients on van der Waals grids. The green triangle mesh representation indicates positive coefficients; the yellow triangle mesh indicates negative coefficients. | |
A contour plot of the electrostatic field region favorable (in blue) or unfavorable (red) for anticancer activity based on telomerase target was shown in Fig. 5b while the energy grids corresponding to the favorable (in green) or unfavorable (yellow) steric effects for the telomerase affinity were shown in Fig. 5c. It was widely acceptable that a better inhibitor based on the 3D-QSAR model should have strong van der Waals attraction in the green areas and a polar group in the blue electrostatic potential areas (which were dominant close to the skeleton). As shown in these two pictures, this promising model would provide a guideline to design and optimize more effective telomerase inhibitors based on the 1-(isoquinoline-1-carboxylate)-2-styryl-5-nitroimidazole derivatives and pave the way for us to further study in future.
3. Conclusion
In summary, a series of novel 1-(isoquinoline-1-carboxylate)-2-styryl-5-nitroimidazole derivatives 3a–w were synthesized and tested for their anticancer activities against A549, U251, Hela, HepG-2 and telomerase inhibitory activities. Preliminary results showed that most of the compounds displayed enhanced anticancer and inhibitory telomerase activities, with IC50 ranging from 0.05–0.81 μM and 0.98–9.95 μM respectively. Of all the studied compounds, 3u and 3v displayed the most potent anticancer activities which accorded exactly with telomerase kinase inhibitory activity. The docking simulation was performed to get the probable binding models and poses. The results indicated that compounds 3u and 3v which acted as potential telomerase inhibitors, can both bind well into the active site of telomerase. Finally, QSAR models were built with previous activity data and binding conformations to motivate current work as well as to provide a reliable tool for reasonable design of telomerase inhibitors in the future.
4. Experiments
4.1 Chemistry
All chemicals and reagents used in the current study were of analytical grade. The reactions were monitored by thin layer chromatography (TLC) on Merck precoated silica GF254 plates. Melting points (uncorrected) were determined on a XT4 MP apparatus (Taike Corp, Beijing, China). All the 1H NMR spectra were recorded on a Bruker DPX 300 model Spectrometer in DMSO-d6 and chemical shifts were reported in ppm (δ). Elemental analyses were performed on a CHN-O-Rapid instrument and were within ±0.4% of the theoretical values. ESI-MS spectra were recorded on a Mariner System 5304 Mass spectrometer.
4.2 General method of synthesis of compounds 3a–w
4.2.1 Method A. Reaction of metronidazole (12 mmol) with different substituted benzaldehyde (16 mmol) in 6 mL DMSO by adding rapidly a stirred solution of sodium methoxide (12.8 mmol) in methanol at room temperature resulted in the formation of target compounds 1a–w. To a stirred solution of compounds 1a–w (7 mmol) in CH2Cl2 (15 mL), 4-methylbenzoyl chloride (7 mmol) and triethylamine (10 mL) at room temperature were added, stirring for 5 hours. The precipitate was purified by column chromatography using EtOAc–hexane (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6) as eluent to give the purified compounds 2a–w. Equimolar proportions of the synthesized compounds 2a–w (5 mmol), isoquinoline-1-carboxylic acid (5 mmol) and K2CO3 (5 mmol) were dissolved in approximately 15 mL of DMF. The reaction solution was allowed to stir overnight at room temperature resulted in the precipitation of crude product was purified by column chromatography using EtOAc–hexane (1
6) as eluent to give the purified compounds 2a–w. Equimolar proportions of the synthesized compounds 2a–w (5 mmol), isoquinoline-1-carboxylic acid (5 mmol) and K2CO3 (5 mmol) were dissolved in approximately 15 mL of DMF. The reaction solution was allowed to stir overnight at room temperature resulted in the precipitation of crude product was purified by column chromatography using EtOAc–hexane (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9) as eluent to give the pure compounds 3a–w.
9) as eluent to give the pure compounds 3a–w.
4.2.2 Method B. SOCl2 (10 mL) was added to a stirred solution of isoquinoline-1-carboxylic acid (20 mmol) in anhydrous DMF (50 mL). The reaction solution was allowed to stir at room temperature for approximately 4 h. Then, active isoquinoline-1-carboxylic acid (15 mmol) and metronidazole (15 mmol) were dissolved in CH2Cl2 followed by drop wise addition triethylamine and compound 6 was obtained with yield of 85%. Then, at room temperature, compound 6 (10 mmol), different substituted benzaldehyde (12 mmol) and NaOH (15 mmol) were dissolved in DMSO (30 mL). The appropriate amount of water was added in the residue, and then filtered. The resulting solid was collected and washed with cold water, dried and crystallized from anhydrous ethanol to get the desired compounds. All of the synthetic compounds gave satisfactory analytical and spectroscopic data, which were in full accordance with their depicted structures.The spectroscopic data of all the compounds 3a–w are in good agreement with their proposed structures. The 1H NMR of compound 3a showed two triplet at δ 4.84 (J = 4.6 Hz) and δ 5.03 (J = 4.6 Hz) which is attributed to N–CH2 and O–CH2 proton respectively. A singlet appeared around at δ 8.29 was consistent with C–H proton in imidazole ring. 13C-NMR data also confirmed the assigned structure.
4.2.3 (E)-2-(2-(2-Fluorostyryl)-5-nitro-1H-imidazol-1-yl)ethyl isoquinoline-1-carboxylate (3a). Yield 69.2%. m.p. 187–189 °C; 1H NMR (DMSO-d6, 300 MHz) δ: 8.32 (d, J = 7.8 Hz, 1H), 8.25 (d, J = 5.7 Hz, 2H), 7.95–7.57 (m, 6H), 7.42–7.31 (m, 2H), 7.17–7.06 (m, 2H), 5.03 (t, J = 4.6 Hz, 2H), 4.84 (t, J = 4.6 Hz, 2H), ESI-MS: 433.12 [M + H]+. 13C NMR (DMSO-d6, 75 MHz) δ: 42.23, 65.31, 112.50, 124.62, 125.28, 125.97, 126.18, 127.21, 127.68, 128.20, 129.35, 129.48, 130.19, 131.21, 134.42, 135.40, 136.39, 138.37, 140.48, 141.92, 147.53, 150.72, 164.68. Anal. calcd for C23H17FN4O4: C, 63.89; H, 3.96; N, 12.96 (%); found: C, 63.71; H, 4.10; N, 11.83 (%).
4.2.4 (E)-2-(2-(3-Fluorostyryl)-5-nitro-1H-imidazol-1-yl)ethyl isoquinoline-1-carboxylate (3b). Yield 64.5%. m.p. 186–187 °C; 1H NMR (DMSO-d6, 300 MHz) δ: 8.37 (d, J = 4.5 Hz, 1H), 8.28 (t, J = 7.1 Hz, 2H), 7.97–7.89 (m, 2H), 7.78 (t, J = 7.5 Hz, 1H), 7.67 (t, J = 7.6 Hz, 1H), 7.53 (d, J = 7.8 Hz, 1H), 7.42 (d, J = 7.9 Hz, 2H), 7.37–7.27 (m, 2H), 7.13 (t, J = 7.9 Hz, 1H), 5.06 (t, J = 4.4 Hz, 2H), 4.85 (t, J = 4.2 Hz, 2H), ESI-MS: 433.12 [M + H]+. 13C NMR (DMSO-d6, 75 MHz) δ: 46.47, 66.51, 114.32, 123.64, 124.71, 125.28, 126.41, 127.56, 127.86, 128.19, 129.49, 129.74, 130.11, 131.80, 134.72, 135.18, 136.68, 137.30, 140.69, 141.78, 146.81, 150.90, 165.17. Anal. calcd for C23H17FN4O4: C, 63.89; H, 3.96; N, 12.96 (%); found: C, 63.93; H, 3.71; N, 13.02 (%).
4.2.5 (E)-2-(2-(4-Fluorostyryl)-5-nitro-1H-imidazol-1-yl)ethyl isoquinoline-1-carboxylate (3c). Yield 70.3%. m.p. 187–188 °C; 1H NMR (DMSO-d6, 300 MHz) δ: 8.35 (d, J = 6.2 Hz, 1H), 8.26 (t, J = 5.4 Hz, 2H), 7.95–7.62 (m, 4H), 7.45 (t, J = 5.3 Hz, 2H), 7.34–7.25 (m, 2H), 7.16 (t, J = 5.3 Hz, 2H), 5.04 (t, J = 6.3 Hz, 2H), 4.86 (t, J = 4.2 Hz, 2H), ESI-MS: 433.12 [M + H]+. 13C NMR (DMSO-d6, 75 MHz) δ: 42.70, 65.85, 115.45, 124.93, 125.55, 126.14, 127.34, 127.63, 128.73, 129.06, 129.75, 130.96, 135.71, 135.86, 136.90, 137.04, 140.52, 141.53, 147.54, 149.59, 164.03. Anal. calcd for C23H17FN4O4: C, 63.89; H, 3.96; N, 12.96 (%); found: C, 63.02; H, 4.13; N, 13.09 (%).
4.2.6 (E)-2-(2-(2-Chlorostyryl)-5-nitro-1H-imidazol-1-yl)ethyl isoquinoline-1-carboxylate (3d). Yield 68.1%. m.p. 187–189 °C; 1H NMR (DMSO-d6, 300 MHz) δ: 8.29 (d, J = 4.5 Hz, 1H), 8.22 (t, J = 6.2 Hz, 2H), 7.89–7.55 (m, 6H), 7.45 (d, J = 4.5 Hz, 2H), 7.21–7.05 (m, 2H), 5.06 (t, J = 6.4 Hz, 2H), 4.82 (t, J = 3.2 Hz, 2H), ESI-MS: 449.09 [M + H]+. 13C NMR (DMSO-d6, 75 MHz) δ: 46.54, 66.65, 113.87, 124.12, 125.95, 125.99, 126.44, 127.40, 127.84, 128.10, 129.61, 129.88, 130.16, 131.50, 134.66, 135.23, 136.58, 137.70, 140.82, 141.48, 147.26, 150.76, 164.08. Anal. calcd for C23H17ClN4O4: C, 61.54; H, 3.82; N, 12.48 (%); found: C, 61.63; H, 3.98; N, 12.34 (%).
4.2.7 (E)-2-(2-(3-Chlorostyryl)-5-nitro-1H-imidazol-1-yl)ethyl isoquinoline-1-carboxylate (3e). Yield 67.5%. m.p. 187–188 °C; 1H NMR (DMSO-d6, 300 MHz) δ: 8.32 (d, J = 3.3 Hz, 1H), 8.21 (t, J = 4.2 Hz, 2H), 7.95–7.86 (m, 2H), 7.75 (t, J = 6.3 Hz, 1H), 7.63 (t, J = 5.3 Hz, 1H), 7.49 (t, J = 3.3 Hz, 1H), 7.41–7.31 (m, 2H), 7.23 (d, J = 6.3 Hz, 2H), 7.14 (d, J = 5.2 Hz, 1H), 5.09 (t, J = 4.2 Hz, 2H), 4.83 (t, J = 5.1 Hz, 2H), ESI-MS: 449.09 [M + H]+. 13C NMR (DMSO-d6, 75 MHz) δ: 46.04, 66.85, 115.39, 123.94, 124.55, 125.20, 126.97, 127.60, 127.83, 128.57, 129.42, 129.89, 130.03, 131.37, 134.77, 135.62, 136.59, 138.96, 140.39, 141.75, 146.06, 151.98, 165.27. Anal. calcd for C23H17ClN4O4: C, 61.54; H, 3.82; N, 12.48 (%); found: C, 61.72; H, 4.04; N, 12.57 (%).
4.2.8 (E)-2-(2-(4-Chlorostyryl)-5-nitro-1H-imidazol-1-yl)ethyl isoquinoline-1-carboxylate (3f). Yield 65.2%. m.p. 187–189 °C; 1H NMR (DMSO-d6, 300 MHz) δ: 8.31 (d, J = 4.2 Hz, 1H), 8.24 (d, J = 4.5 Hz, 2H), 7.96–7.67 (m, 4H), 7.55 (t, J = 5.1 Hz, 2H), 7.35–7.27 (m, 2H), 7.12 (d, J = 7.2 Hz, 2H), 5.03 (t, J = 3.6 Hz, 2H), 4.82 (t, J = 3.3 Hz, 2H), ESI-MS: 449.09 [M + H]+. 13C NMR (DMSO-d6, 75 MHz) δ: 43.66, 66.44, 117.22, 123.29, 125.92, 126.25, 127.69, 127.87, 128.32, 129.16, 129.60, 131.02, 133.84, 134.46, 136.72, 137.26, 141.13, 142.68, 146.94, 150.05, 166.78. Anal. calcd for C23H17ClN4O4: C, 61.54; H, 3.82; N, 12.48 (%); found: C, 61.42; H, 3.75; N, 12.55 (%).
4.2.9 (E)-2-(2-(2-Bromostyryl)-5-nitro-1H-imidazol-1-yl)ethyl isoquinoline-1-carboxylate (3g). Yield 62.5%. m.p. 188–189 °C; 1H NMR (DMSO-d6, 300 MHz) δ: 8.34 (d, J = 3.3 Hz, 1H), 8.25 (d, J = 6.3 Hz, 2H), 7.95–7.62 (m, 4H), 7.58 (d, J = 4.6 Hz, 2H), 7.41 (t, J = 3.3 Hz, 2H), 7.24–7.06 (m, 2H), 5.09 (t, J = 5.4 Hz, 2H), 4.82 (t, J = 3.3 Hz, 2H), ESI-MS: 493.04 [M + H]+. 13C NMR (DMSO-d6, 75 MHz) δ: 44.32, 63.92, 113.90, 124.54, 125.78, 125.93, 126.03, 127.43, 127.80, 128.01, 129.06, 129.37, 130.77, 131.28, 135.53, 135.81, 136.51, 138.62, 139.12, 141.63, 148.17, 151.04, 165.43. Anal. calcd for C23H17BrN4O4: C, 56.00; H, 3.47; N, 11.36 (%); found: C, 56.23; H, 3.31; N, 11.47 (%).
4.2.10 (E)-2-(2-(3-Bromostyryl)-5-nitro-1H-imidazol-1-yl)ethyl isoquinoline-1-carboxylate (3h). Yield 65.3%. m.p. 186–188 °C; 1H NMR (DMSO-d6, 300 MHz) δ: 8.22 (d, J = 3.3 Hz, 1H), 8.15 (t, J = 5.1 Hz, 2H), 7.91–7.79 (m, 4H), 7.71 (t, J = 6.9 Hz, 1H), 7.57 (t, J = 5.1 Hz, 1H), 7.41 (t, J = 3.3 Hz, 1H), 7.24 (d, J = 5.4 Hz, 2H), 7.11 (d, J = 5.1 Hz, 1H), 5.09 (t, J = 4.5 Hz, 2H), 4.86 (t, J = 6.3 Hz, 2H), ESI-MS: 493.04 [M + H]+. 13C NMR (DMSO-d6, 75 MHz) δ: 43.73, 63.71, 114.11, 124.17, 125.49, 125.67, 126.11, 127.24, 127.89, 128.57, 129.27, 129.76, 130.43, 132.91, 135.64, 135.98, 136.09, 137.33, 139.17, 140.95, 147.81, 150.79, 167.07. Anal. calcd for C23H17BrN4O4: C, 56.00; H, 3.47; N, 11.36 (%); found: C, 56.14; H, 3.62; N, 11.21 (%).
4.2.11 (E)-2-(2-(4-Bromostyryl)-5-nitro-1H-imidazol-1-yl)ethyl isoquinoline-1-carboxylate (3i). Yield 67.3%. m.p. 1H NMR (DMSO-d6, 300 MHz) δ: 8.31 (d, J = 3.3 Hz, 1H), 8.23 (t, J = 4.2 Hz, 1H), 7.91–7.62 (m, 5H), 7.52 (t, J = 5.1 Hz, 2H), 7.31–7.23 (m, 2H), 7.13 (d, J = 7.2 Hz, 2H), 5.04 (t, J = 3.3 Hz, 2H), 4.84 (t, J = 3.6 Hz, 2H), ESI-MS: 449.09 [M + H]+. 13C NMR (DMSO-d6, 75 MHz) δ: 44.71, 64.51, 113.99, 124.48, 125.87, 125.90, 127.72, 127.97, 129.03, 129.30, 129.72, 131.21, 135.32, 135.74, 136.61, 138.55, 139.27, 141.71, 148.48, 150.99, 165.66. Anal. calcd for C23H17BrN4O4: C, 56.00; H, 3.47; N, 11.36 (%); found: C, 56.30; H, 3.72; N, 11.25 (%).
4.2.12 (E)-2-(5-Nitro-2-(2-nitrostyryl)-1H-imidazol-1-yl)ethyl isoquinoline-1-carboxylate (3j). Yield 64.7%. m.p. 186–188 °C; 1H NMR (DMSO-d6, 300 MHz) δ: 8.37 (d, J = 3.3 Hz, 1H), 8.31 (d, J = 6.3 Hz, 2H), 7.97–7.71 (m, 6H), 7.52 (d, J = 4.2 Hz, 2H), 7.34–7.12 (m, 2H), 5.06 (t, J = 5.7 Hz, 2H), 4.85 (t, J = 3.3 Hz, 2H), ESI-MS: 460.12 [M + H]+. 13C NMR (DMSO-d6, 75 MHz) δ: 47.31, 67.43, 114.36, 124.92, 125.25, 125.53, 126.73, 127.79, 127.93, 128.21, 129.38, 129.46, 130.33, 131.14, 134.52, 135.35, 136.63, 137.34, 140.91, 141.45, 146.29, 150.24, 165.15. Anal. calcd for C23H17N5O6: C, 60.13; H, 3.73; N, 15.24 (%); found: C, 60.29; H, 3.49; N, 15.07 (%).
4.2.13 (E)-2-(5-Nitro-2-(3-nitrostyryl)-1H-imidazol-1-yl)ethyl isoquinoline-1-carboxylate (3k). Yield 65.7%. m.p. 186–188 °C; 1H NMR (DMSO-d6, 300 MHz) δ: 8.25 (d, J = 3.6 Hz, 1H), 8.19 (t, J = 5.1 Hz, 2H), 7.96–7.82 (m, 2H), 7.75 (d, J = 5.1 Hz, 1H) 7.68 (t, J = 6.9 Hz, 1H), 7.55 (t, J = 5.1 Hz, 2H), 7.43 (t, J = 3.3 Hz, 1H), 7.26 (d, J = 5.4 Hz, 2H), 7.13 (d, J = 5.1 Hz, 1H), 5.06 (t, J = 4.2 Hz, 2H), 4.83 (t, J = 6.3 Hz, 2H), ESI-MS: 460.12 [M + H]+. 13C NMR (DMSO-d6, 75 MHz) δ: 45.07, 66.22, 114.13, 123.05, 124.01, 125.09, 126.02, 127.00, 127.35, 128.63, 129.02, 129.93, 130.29, 131.68, 134.51, 135.65, 136.40, 137.73, 140.21, 141.32, 146.69, 150.66, 165.98. Anal. calcd for C23H17N5O6: C, 60.13; H, 3.73; N, 15.24 (%); found: C, 60.01; H, 3.84; N, 15.30 (%).
4.2.14 (E)-2-(5-Nitro-2-(4-nitrostyryl)-1H-imidazol-1-yl)ethyl isoquinoline-1-carboxylate (3l). Yield 62.3%. m.p. 187–188 °C; 1H NMR (DMSO-d6, 300 MHz) δ: 8.26 (d, J = 3.9 Hz, 1H), 8.17 (t, J = 4.2 Hz, 2H), 7.95–7.68 (m, 4H), 7.55 (t, J = 5.4 Hz, 2H), 7.36–7.25 (m, 2H), 7.17 (d, J = 7.4 Hz, 2H), 5.06 (t, J = 3.3 Hz, 2H), 4.86 (t, J = 3.6 Hz, 2H), ESI-MS: 460.12 [M + H]+. 13C NMR (DMSO-d6, 75 MHz) δ: 45.41, 65.61, 116.36, 124.78, 125.10, 126.65, 127.08, 127.51, 128.83, 129.12, 129.38, 130.80, 132.99, 134.47, 136.01, 137.15, 140.56, 142.77, 146.82, 151.30, 167.74. Anal. calcd for C23H17N5O6: C, 60.13; H, 3.73; N, 15.24 (%); found: C, 60.20; H, 3.65; N, 15.39 (%).
4.2.15 (E)-2-(2-(2-Methylstyryl)-5-nitro-1H-imidazol-1-yl)ethyl isoquinoline-1-carboxylate (3m). Yield 65.4%. m.p. 186–188 °C. 1H NMR (DMSO-d6, 300 MHz) δ: 8.35 (d, J = 4.5 Hz, 1H), 8.27 (d, J = 5.4 Hz, 1H), 8.07–7.94 (m, 4H), 7.75 (t, J = 7.5 Hz, 1H), 7.62 (d, J = 14.5 Hz, 1H), 7.45–7.27 (m, 4H), 7.14 (d, J = 7.5 Hz, 1H), 5.09 (t, J = 4.5 Hz, 2H), 4.87 (t, J = 6.3 Hz, 2H), 2.45 (d, J = 4.2 Hz, 3H), ESI-MS: 429.15 [M + H]+. 13C NMR (DMSO-d6, 75 MHz) δ: 19.55, 44.72, 64.47, 114.76, 124.48, 125.87, 125.89, 126.29, 126.48, 127.69, 129.25, 129.55, 130.86, 131.20, 134.37, 135.32, 135.64, 136.61, 136.79, 139.26, 141.69, 148.49, 151.08, 165.66. Anal. calcd for C24H20N4O4: C, 67.28; H, 4.71; N, 13.08 (%); found: C, 67.03; H, 4.53; N, 13.20 (%).
4.2.16 (E)-2-(2-(3-Methylstyryl)-5-nitro-1H-imidazol-1-yl)ethyl isoquinoline-1-carboxylate (3n). Yield 65.9%. m.p. 187–189 °C. 1H NMR (DMSO-d6, 300 MHz) δ: 8.32 (d, J = 4.2 Hz, 1H), 8.26 (d, J = 5.4 Hz, 1H), 8.12–7.95 (m, 2H), 7.75 (t, J = 7.5 Hz, 1H), 7.69–7.60 (m, 2H), 7.51 (t, J = 4.5 Hz, 2H), 7.44–7.26 (m, 3H), 7.14 (d, J = 7.5 Hz, 1H), 5.05 (s, 2H), 4.85 (s, 2H), 2.36 (d, J = 4.2 Hz, 3H), ESI-MS: 429.15 [M + H]+. 13C NMR (DMSO-d6, 75 MHz) δ: 20.94, 46.19, 64.45, 115.72, 123.23, 124.58, 125.62, 126.12, 126.26, 127.54, 128.68, 129.68, 130.07, 130.98, 132.63, 134.37, 135.85, 136.32, 137.65, 139.02, 140.76, 145.93, 154.09, 167.00. Anal. calcd for C24H20N4O4: C, 67.28; H, 4.71; N, 13.08 (%); found: C, 67.35; H, 4.65; N, 12.91 (%).
4.2.17 (E)-2-(2-(4-Methylstyryl)-5-nitro-1H-imidazol-1-yl)ethyl isoquinoline-1-carboxylate (3o). Yield 67.9%. m.p. 187–189 °C. 1H NMR (DMSO-d6, 300 MHz) δ: 8.39–8.32 (m, 1H), 8.22 (d, J = 8.4 Hz, 1H), 8.00–7.92 (m, 2H), 7.79 (t, J = 7.5 Hz, 1H), 7.71–7.64 (m, 2H), 7.54 (d, J = 14.5 Hz, 1H), 7.42–7.24 (m, 4H), 7.11 (d, J = 7.5 Hz, 1H), 5.02 (s, 2H), 4.84 (s, 2H), 2.32 (d, J = 7.2 Hz, 3H), ESI-MS: 429.15 [M + H]+. 13C NMR (DMSO-d6, 75 MHz) δ: 22.34, 43.56, 66.24, 114.85, 124.08, 125.43, 126.96, 127.17, 127.26, 128.54, 129.64, 129.77, 130.82, 135.97, 136.05, 136.47, 137.12, 141.16, 141.88, 147.90, 148.99, 165.49. Anal. calcd for C24H20N4O4: C, 67.28; H, 4.71; N, 13.08 (%); found: C, 67.42; H, 4.83; N, 13.27 (%).
4.2.18 (E)-2-(2-(2-Methoxystyryl)-5-nitro-1H-imidazol-1-yl)ethyl isoquinoline-1-carboxylate (3p). Yield 62.4%. m.p. 187–188 °C. 1H NMR (DMSO-d6, 300 MHz) δ: 8.45 (d, J = 4.5 Hz, 1H), 8.32 (d, J = 5.4 Hz, 1H), 8.17–7.98 (m, 4H), 7.82 (t, J = 7.5 Hz, 2H), 7.62 (d, J = 4.5 Hz, 1H), 7.49–7.26 (m, 3H), 7.17 (d, J = 7.5 Hz, 1H), 5.05 (t, J = 6.0 Hz, 2H), 4.83 (t, J = 3.3 Hz, 2H), 3.55 (d, J = 4.2 Hz, 3H), ESI-MS: 445.14 [M + H]+. 13C NMR (DMSO-d6, 75 MHz) δ: 45.30, 56.01, 67.39, 113.62, 123.92, 124.53, 125.14, 126.09, 126.55, 127.36, 128.18, 129.19, 130.00, 130.44, 132.11, 134.89, 135.46, 136.15, 138.34, 140.48, 141.74, 146.79, 151.22, 165.01. Anal. calcd for C24H20N4O5: C, 64.86; H, 4.54; N, 12.61 (%); found: C, 64.61; H, 4.31; N, 12.55 (%).
4.2.19 (E)-2-(2-(3-Methoxystyryl)-5-nitro-1H-imidazol-1-yl)ethyl isoquinoline-1-carboxylate (3q). Yield 64.4%. m.p. 187–188 °C. 1H NMR (DMSO-d6, 300 MHz) δ: 8.35 (d, J = 4.5 Hz, 1H), 8.22 (d, J = 5.1 Hz, 1H), 8.11–7.93 (m, 2H), 7.81 (t, J = 7.5 Hz, 2H), 7.57 (d, J = 4.2 Hz, 2H), 7.46–7.24 (m, 2H), 7.12 (d, J = 7.5 Hz, 1H), 5.02 (t, J = 6.3 Hz, 2H), 4.81 (t, J = 3.3 Hz, 2H), 3.51 (d, J = 4.2 Hz, 3H), ESI-MS: 445.14 [M + H]+. 13C NMR (DMSO-d6, 75 MHz) δ: 46.60, 54.82, 66.35, 116.66, 122.81, 123.05, 124.95, 126.75, 127.16, 128.20, 129.10, 129.73, 130.15, 131.89, 132.52, 133.30, 135.40, 136.41, 139.70, 140.56, 142.92, 145.44, 153.78, 166.47. Anal. calcd for C24H20N4O5: C, 64.86; H, 4.54; N, 12.61 (%); found: C, 64.93; H, 4.67; N, 12.43 (%).
4.2.20 (E)-2-(2-(4-Methoxystyryl)-5-nitro-1H-imidazol-1-yl)ethyl isoquinoline-1-carboxylate (3r). Yield 62.4%. m.p. 186–188 °C. 1H NMR (DMSO-d6, 300 MHz) δ: 8.38 (d, J = 4.2 Hz, 1H), 8.25 (t, J = 5.1 Hz, 1H), 8.13–7.95 (m, 4H), 7.82 (t, J = 7.5 Hz, 1H), 7.54–7.43 (m, 2H), 7.46–7.24 (m, 3H), 7.12 (d, J = 7.5 Hz, 1H), 5.02 (t, J = 6.3 Hz, 2H), 4.81 (t, J = 3.3 Hz, 2H), 3.45 (d, J = 4.2 Hz, 3H), ESI-MS: 445.14 [M + H]+. 13C NMR (DMSO-d6, 75 MHz) δ: 46.33, 55.59, 64.86, 115.58, 123.95, 125.76, 126.71, 127.45, 127.52, 128.37, 128.94, 129.80, 130.41, 135.72, 136.07, 136.27, 138.31, 142.59, 143.75, 148.91, 151.03, 167.20. Anal. calcd for C24H20N4O5: C, 64.86; H, 4.54; N, 12.61 (%); found: C, 64.72; H, 4.49; N, 12.78 (%).
4.2.21 (E)-2-(2-(2,4-Dichlorostyryl)-5-nitro-1H-imidazol-1-yl)ethyl-7-1-carboxylate (3s). Yield 65.1%. m.p. 187–189 °C; 1H NMR (DMSO-d6, 300 MHz) δ: 8.27 (d, J = 4.2 Hz, 1H), 8.18 (t, J = 6.2 Hz, 1H), 7.84–7.56 (m, 5H), 7.42 (d, J = 4.5 Hz, 2H), 7.22–7.07 (m, 3H), 5.07 (t, J = 6.3 Hz, 2H), 4.84 (t, J = 3.3 Hz, 2H), ESI-MS: 485.07 [M + H]+. 13C NMR (DMSO-d6, 75 MHz) δ: 45.91, 66.48, 116.11, 121.17, 122.57, 124.61, 125.33, 126.71, 127.83, 128.18, 129.55, 129.69, 131.13, 131.50, 134.97, 135.03, 136.39, 137.80, 139.06, 140.42, 145.79, 153.14, 166.46. Anal. calcd for C23H16Cl2N4O4: C, 59.17; H, 3.35; N, 12.00 (%); found: C, 59.31; H, 3.23; N, 11.93 (%).
4.2.22 (E)-2-(2-(4-Chloro-2-fluorostyryl)-5-nitro-1H-imidazol-1-yl)ethyl isoquinoline-1-carboxylate (3t). Yield 67.1%. m.p. 187–189 °C; 1H NMR (DMSO-d6, 300 MHz) δ: 8.29 (d, J = 4.2 Hz, 1H), 8.17 (d, J = 6.3 Hz, 2H), 7.85–7.58 (m, 4H), 7.42 (d, J = 4.2 Hz, 2H), 7.24–7.09 (m, 3H), 5.05 (t, J = 6.3 Hz, 2H), 4.81 (t, J = 3.3 Hz, 2H), ESI-MS: 467.08 [M + H]+. 13C NMR (DMSO-d6, 75 MHz) δ: 43.64, 66.36, 115.67, 125.08, 125.86, 126.53, 126.87, 127.38, 128.90, 128.99, 129.04, 129.51, 130.74, 131.31, 135.43, 136.21, 136.96, 138.59, 139.88, 142.28, 147.77, 152.27, 165.22. Anal. calcd for C23H16ClFN4O4: C, 59.17; H, 3.45; N, 12.00 (%); found: C, 59.31; H, 3.23; N, 11.83 (%).
4.2.23 (E)-2-(2-(2-(Naphthalen-2-yl)vinyl)-5-nitro-1H-imidazol-1-yl)ethyl isoquinoline-1-carboxylate (3u). Yield 64.5%. m.p. 187–188 °C; 1H NMR (DMSO-d6, 300 MHz) δ: 8.29 (d, J = 4.8 Hz, 1H), 8.19 (t, J = 6.6 Hz, 2H), 7.88–7.55 (m, 6H), 7.43 (d, J = 4.2 Hz, 2H), 7.24–7.09 (m, 5H), 5.07 (t, J = 6.3 Hz, 2H), 4.85 (t, J = 3.3 Hz, 2H), ESI-MS: 465.15 [M + H]+. 13C NMR (DMSO-d6, 75 MHz) δ: 44.25, 64.84, 114.24, 122.41, 123.00, 123.15, 124.53, 125.87, 125.90, 126.07, 127.38, 127.46, 128.76, 129.01, 129.44, 130.95, 131.57, 132.10, 133.49, 134.67, 135.29, 136.70, 139.82, 141.34, 145.60, 152.90, 168.04. Anal. calcd for C27H20N4O4: C, 69.82; H, 4.34; N, 12.06 (%); found: C, 69.73; H, 4.59; N, 11.86 (%).
4.2.24 (E)-2-(2-(4-(Dimethylamino)styryl)-5-nitro-1H-imidazol-1-yl)ethyl isoquinoline-1-carboxylate (3v). Yield 66.3%. m.p. 186–188 °C. 1H NMR (DMSO-d6, MHz) δ: 8.29 (d, J = 3.6 Hz, 1H), 8.19 (d, J = 4.5 Hz, 1H), 7.88–7.67 (m, 4H), 7.52 (d, J = 5.4 Hz, 1H), 7.34–7.21 (m, 2H), 7.13 (d, J = 6.6 Hz, 2H), 5.05 (t, J = 3.6 Hz, 2H), 4.82 (t, J = 3.9 Hz, 2H), 1.54–1.22 (m, 6H) ESI-MS: 458.18 [M + H]+. 13C NMR (DMSO-d6, 75 MHz) δ: 41.53, 45.12, 65.71, 116.93, 123.32, 125.62, 126.24, 126.79, 127.36, 128.73, 129.26, 130.51, 130.65, 131.22, 135.54, 137.74, 138.27, 140.18, 142.84, 145.77, 154.02, 166.52. Anal. calcd for C25H23N5O4: C, 65.63; H, 5.07; N, 15.31 (%); found: C, 65.44; H, 4.86; N, 15.52 (%).
4.2.25 (E)-2-(5-Nitro-2-styryl-1H-imidazol-1-yl)ethyl isoquinoline-1-carboxylate (3w). Yield 63.3%. m.p. 186–188 °C. 1H NMR (DMSO-d6, MHz) δ: 8.26 (d, J = 6.6 Hz, 1H), 8.16 (d, J = 4.5 Hz, 1H), 7.85–7.63 (m, 4H), 7.48 (t, J = 5.4 Hz, 2H), 7.36–7.23 (m, 4H), 7.13 (d, J = 4.2 Hz, 2H), 5.08 (t, J = 3.6 Hz, 2H), 4.85 (t, J = 3.9 Hz, 2H), ESI-MS: 458.18 [M + H]+. 13C NMR (DMSO-d6, 75 MHz) δ: 42.83, 67.48, 115.86, 124.05, 125.08, 126.56, 126.92, 127.06, 128.64, 129.37, 130.20, 130.61, 134.97, 135.42, 136.99, 138.35, 140.21, 142.14, 146.66, 151.75, 165.89. Anal. calcd for C23H18N4O4: C, 66.66; H, 4.38; N, 13.52 (%); found: C, 66.83; H, 4.23; N, 13.69 (%).
5. Bioassay conditions
5.1 Telomerase inhibitory assay
Compounds 3a–w was assayed for telomerase inhibition by a modified TRAP assay, using a SGC-7901 cell extract. The telomerase assay methods used are the same as previously described.9
5.2 Cell proliferation assay
CCK-8 is much more convenient and helpful than MTT for analyzing cell proliferation, because it can be reduced to soluble formazan by dehydrogenase in mitochondria and has little toxicity to cells. Cell proliferation was determined using CCK-8 dye (Beyotime Inst Biotech, China) according to manufacture's instructions. Briefly, 1–5 × 103 cells per well were seeded in a 96-well plate, grown at 37 °C for 12 h. Subsequently, cells were treated with the target compounds at increasing concentrations in the presence of 10% FBS for 24 or 48 h. After 10 μL CCK-8 dye was added to each well, cells were incubated at 37 °C for 1–2 h and plates were read in a Victor-V multilabel counter (Perkin-Elmer) using the default europium detection protocol. Percent inhibition or IC50 values of compounds were calculated by comparison with DMSO-treated control wells.
5.3 Flow cytometry
Cells (1.3 × 105 cells per mL) were cultured in the presence or not of novobiocin analogues at 200 μM. NVB at the same concentration served as reference inhibitor. After treatment for 48 and 72 h, cells were washed and fixed in PBS–ethanol (30/70). For cytofluorometric examination, cells (104 cells per mL) were incubated for 30 min in PBS/Triton X-100, 0.2%/EDTA 1 mM, and propidium iodide (PI) (50 μg mL−1) in PBS supplemented by RNase (0.5 mg mL−1). The number of cells in the different phases of the cell cycle was determined, and the percentage of apoptotic cells was quantified. Analyses were performed with a FACS Calibur (Becton Dickinson, Le Pont de Claix, France). Cell Quest software was used for data acquisition and analysis.
6. Virtual calculation
6.1 Experimental protocol of docking study
Molecular docking of compounds 3a–w into the three-dimensional TERT protein catalytic subunit (3DU6.pdb) was carried out using the Auto-Dock software package (version 3.5) as implemented through the graphical user interface Auto-DockTools (ADT 1.4.6).
6.2 3D-QSAR
Ligand-based 3D-QSAR approach was performed by QSAR software of DS 3.5 (Discovery Studio 3.5, Accelrys, Co. Ltd). The training sets were composed of inhibitors with the corresponding pIC50 values which were converted from the obtained IC50 (μM), and test sets comprised compounds of data sets. All the definition of the descriptors can be seen in the “Help” of DS 3.5 software and they were calculated by QSAR protocol of DS 3.5. The alignment conformation of each molecule was the one with lowest interaction energy in the docked results of CDocker. The predictive ability of 3D-QSAR modeling can be evaluated based on the cross-validated correlation coefficient, which qualifies the predictive ability of the models. Scrambled test (Y scrambling) was performed to investigate the risk of chance correlations. The inhibitory potencies of compounds were randomly reordered for 30 times and subject to leave-one-out validation test, respectively. The models were also validated by test sets, in which the compounds are not included in the training sets. Usually, one can believe that the modeling is reliable, when the R2 for test sets is larger than 0.6.
Acknowledgements
This work was supported by the project (no. BY2012136) from the Science & Technology Agency of Jiangsu Province and by the projects (no. CXY1213 & CXY1222) from the Science & Technology Bureau of Lianyuangang City of Jiangsu Province.
References
- C. Autexier and C. W. Greider, Trends Biochem. Sci., 1996, 21, 387 CrossRef CAS.
- E. H. Blackburn, Nature, 1991, 350, 569 CrossRef CAS PubMed.
- K. Masutomi, E. Y. Yu, S. Khurts, I. Ben-Porath, J. L. Currier, G. B. Metz, M. W. Brooks, S. Kaneko, S. Murakami and J. A. DeCaprio, Cell, 2003, 114, 241 CrossRef CAS.
- J. Wong and K. Collins, Lancet, 2003, 362, 983 CrossRef CAS.
- C. B. Harley, Mutat. Res., DNAging: Genet. Instab. Aging, 1991, 256, 271 CAS.
- J. W. Shay, W. E. Wright and H. Werbin, Biochim. Biophys. Acta, Rev. Cancer, 1991, 1072, 1 CrossRef CAS.
- C. M. Counter, H. W. Hirte, S. Bacchetti and C. B. Harley, Proc. Natl. Acad. Sci. U. S. A., 1994, 91, 2900 CrossRef CAS.
- W. C. Hahn, C. M. Counter, A. S. Lundberg, R. L. Beijersbergen, M. W. Brooks and R. A. Weinberg, Nature, 1999, 400, 464 CrossRef CAS PubMed.
- N. W. Kim, M. A. Piatyszek, K. R. Prowse, C. B. Harley, M. D. West, P. d. L. Ho, G. M. Coviello, W. E. Wright, S. L. Weinrich and J. W. Shay, Science, 1994, 266, 2011 CAS.
- S. Neidle and L. Kelland, Anti-Cancer Drug Des., 1999, 14, 341 CAS.
- P. J. Perry and T. C. Jenkins, Expert Opin. Invest. Drugs, 1999, 8, 1981 CrossRef CAS PubMed.
- M. Mason, A. Schuller and E. Skordalakes, Curr. Opin. Struct. Biol., 2011, 21, 92 CrossRef CAS PubMed.
- R. McLendon, A. Friedman, D. Bigner, E. G. Van Meir, D. J. Brat, G. M. Mastrogianakis, J. J. Olson, T. Mikkelsen, N. Lehman and K. Aldape, Nature, 2008, 455, 1061 CrossRef CAS PubMed.
- M. Hranjec, M. Kralj, I. Piantanida, M. Sedic, L. Šuman, K. Pavelic and G. Karminski-Zamola, J. Med. Chem., 2007, 50, 5696 CrossRef CAS PubMed.
- C. Pidathala, R. Amewu, B. n. d. Pacorel, G. L. Nixon, P. Gibbons, W. D. Hong, S. C. Leung, N. G. Berry, R. Sharma and P. A. Stocks, J. Med. Chem., 2012, 55, 1831 CrossRef CAS PubMed.
- M. J. Moore, C. M. Schultes, J. Cuesta, F. Cuenca, M. Gunaratnam, F. A. Tanious, W. D. Wilson and S. Neidle, J. Med. Chem., 2006, 49, 582 CrossRef CAS PubMed.
- J. Sun, A. J. Makawana and H.-L. Zhu, Mini-Rev. Med. Chem., 2013, 13, 1725 CrossRef CAS.
- J. Zhou, X. Zhu, Y. Lu, R. Deng, Z. Huang, Y. Mei, Y. Wang, W. Huang, Z. Liu and L. Gu, Oncogene, 2005, 25, 503 Search PubMed.
- J.-L. Zhou, Y.-J. Lu, T.-M. Ou, J.-M. Zhou, Z.-S. Huang, X.-F. Zhu, C.-J. Du, X.-Z. Bu, L. Ma and L.-Q. Gu, J. Med. Chem., 2005, 48, 7315 CrossRef CAS PubMed.
- J. Giglio, S. Fernández, A. Rey and H. Cerecetto, Bioorg. Med. Chem. Lett., 2011, 21, 394 CrossRef CAS PubMed.
- J. L. Born, B. R. Smith, N. Harper and C. J. Koch, Biochem. Pharmacol., 1992, 43, 1337 CrossRef CAS.
- Y. Duan, Z. Wang, Y. Sang, X. Tao and H. Zhu, Curr. Top. Med. Chem., 2013, 13, 3118 CrossRef CAS.
- Y. Luo, Y. Li, K.-M. Qiu, X. Lu, J. Fu and H.-L. Zhu, Bioorg. Med. Chem. Lett., 2011, 19, 6069 CrossRef CAS PubMed.
- Y. Qian, H.-J. Zhang, H. Zhang, C. Xu, J. Zhao and H.-L. Zhu, Bioorg. Med. Chem. Lett., 2010, 18, 4991 CrossRef CAS PubMed.
- R. J. Heath and C. O. Rock, Nat. Prod. Rep., 2002, 19, 581 RSC.
- J. A. Upcroft, L. A. Dunn, J. M. Wright, K. Benakli, P. Upcroft and P. Vanelle, Antimicrob. Agents Chemother., 2006, 50, 344 CrossRef CAS PubMed.
- A. E. Kϋx, M. Schmitt, S. V. Cardozo, C. Lugnier, P. Villa, A. B. Lopes, N. C. Romeiro, H. Justiniano, M. A. Martins and C. A. Fraga, J. Med. Chem., 2012, 55, 7525 CrossRef PubMed.
|
| This journal is © The Royal Society of Chemistry 2014 |
Click here to see how this site uses Cookies. View our privacy policy here. ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6) as eluent to give the purified compounds 2a–w. Equimolar proportions of the synthesized compounds 2a–w (5 mmol), isoquinoline-1-carboxylic acid (5 mmol) and K2CO3 (5 mmol) were dissolved in approximately 15 mL of DMF. The reaction solution was allowed to stir overnight at room temperature resulted in the precipitation of crude product was purified by column chromatography using EtOAc–hexane (1
6) as eluent to give the purified compounds 2a–w. Equimolar proportions of the synthesized compounds 2a–w (5 mmol), isoquinoline-1-carboxylic acid (5 mmol) and K2CO3 (5 mmol) were dissolved in approximately 15 mL of DMF. The reaction solution was allowed to stir overnight at room temperature resulted in the precipitation of crude product was purified by column chromatography using EtOAc–hexane (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9) as eluent to give the pure compounds 3a–w.
9) as eluent to give the pure compounds 3a–w.





