Assembling Mn:ZnSe quantum dots-siRNA nanoplexes for gene silencing in tumor cells†
Yucheng
Wang
a,
Chengbin
Yang
a,
Rui
Hu
a,
Hui Ting
Toh
b,
Xin
Liu
c,
Guimiao
Lin
d,
Feng
Yin
a,
Ho Sup
Yoon
be and
Ken-Tye
Yong
*a
aSchool of Electrical and Electronic Engineering, Nanyang Technological University, Singapore 639798, Singapore. E-mail: ktyong@ntu.edu.sg; Tel: +65-6790-5444
bDivision of Structural Biology & Biochemistry, School of Biological Sciences, Nanyang Technological University, Singapore 639798, Singapore
cDepartment of Chemical and Biological Engineering, University at Buffalo (SUNY), Buffalo, New York 14260, USA
dThe Engineering Lab of Synthetic Biology and the Key Lab of Biomedical Engineering, School of Medicine, Shenzhen University, Shenzhen, 518060, China
eDepartment of Genetic Engineering, College of Life Sciences, Kyung Hee University, Yongin-si Gyeonggi-do, 446-701, Republic of Korea
First published on 24th September 2014
Abstract
In this work, we demonstrate the use of manganese doped zinc selenide QDs (Mn:ZnSe d-dots) for gene delivery in vitro. Specifically, the d-dots were prepared as nanoplexes for facilitating the intracellular delivery of small interfering RNA (siRNA) molecules to pancreatic cancer cells (Panc-1), thereby inducing sequence-specific silencing of oncogenic K-Ras mutations in pancreatic carcinoma. For nanoplex preparation, a layer-by-layer (LBL) assembling method was adopted to modify the d-dot surface with cationic polymer poly(allylamine hydrochloride) (PAH) or polyethylenimine (PEI) for generating positive surface potential for complexing with K-Ras siRNA molecules. Owing to the unique and stable PL properties of the d-dots, siRNA transfection and the subsequent intracellular release profile from the d-dot/polymer-siRNA nanoplexes were monitored by fluorescence imaging. Quantitative results from flow cytometry study suggested that a high gene transfection efficiency was achieved. The expression of the mutant K-Ras mRNA in Panc-1 cells was observed to be significantly suppressed upon transfecting them with the nanoplex formulation. More importantly, cell viability studies showed that the d-dot/PAH nanoplexes were biocompatible and non-toxic even at concentrations as high as 160 μg mL−1. Furthermore, the amine-terminated surface could be further modified to obtain multiple bio-functions. Based on these results, we envision that the designed d-dot nanoplexes can be developed as a flexible nanoplatform for both fundamental and practical clinical research applications.
Introduction
Pancreatic cancer is one of the most common cancers with a 5-year survival rate of less than 5%.1 As this disease is extremely difficult to treat with conventional therapies, development of novel therapeutic approaches is urgently needed. Due to the advancement of cancer biology in the past few decades, RNA interference (RNAi), which is a sequence specific post-transcriptional gene silencing mechanism induced by double-stranded RNA (dsRNA), has been developed as a novel platform for gene therapy of disease.2 Chemically synthesized small-interfering RNAs (siRNAs) can be delivered to tumor cells for initiating the specific degradation of target messenger RNA (mRNA) of complementary sequences, thereby inhibiting the expression of the corresponding cancer-promoting genes.2,3 However, the application of naked siRNA molecules is challenged by the limited uptake efficiency and the short half-life due to nuclease-mediated degradation.4 Recently, viral vectors and synthetic materials, such as liposomes, peptide dendrimers, polymers and inorganic nanoparticles (e.g. noble metals, metal oxides, nanocarbons and mesoporous silica), have been investigated as potential carriers for gene delivery.5–14 Among them, fluorescent QDs have been extensively investigated owing to their chemical stability and large surface area for the loading of gene materials. Most importantly, because of their exceptional optical properties, such as a broad excitation profile, tunable colour and high resistance to photobleaching,15–18 QD-based nanocarriers can serve as a multifunctional delivery system as well as as imaging probes for long-term tracking and monitoring of the siRNA transfection process in situ. This will allow one to investigate the gene silencing mechanism in detail.19–21 However, most of the reported QD-based gene vectors were developed using cadmium-based QDs, and their potential toxicity remains a major debated and unsettled issue for them to be translated for in vivo and clinical research. Some research groups have revealed the breakdown and degradation of QDs in biological systems.22–24 The release of the cadmium ions from the particle surface to the biological environment may induce severe acute toxic effects. Although several in vitro and in vivo studies have demonstrated that suitable protective inorganic and organic coatings on the QD surface will significantly reduce the nanoparticle toxicity, long-term studies are still needed to understand the ultimate fate of the QDs in vivo.25,26To overcome this challenge, in this study, we developed manganese doped zinc selenide QDs (Mn:ZnSe d-dots) as biocompatible nanocarriers for in vitro gene delivery. These Mn:ZnSe d-dots are more acceptable for real-life biomedical applications compared to traditional types of QDs that contain toxic heavy metals (e.g. CdSe, CdTe, PbS and PbSe).27 For genetic therapy of pancreatic cancers, we designed an siRNA sequence specifically targeting the mutant K-Ras gene with a point mutation at codon 12, which is present in approximately 90% of all types of pancreatic cancers and is associated with increased cell proliferation and resistance to apoptosis.28–30 Mn:ZnSe d-dots-siRNA nanoplexes were constructed for efficient loading and traceable delivery of the K-Ras siRNA into pancreatic cancer cells. Specifically, the d-dot surface was modified with cationic polymer poly(allylamine hydrochloride) (PAH) or polyethylenimine (PEI), and subsequently complexed with siRNA molecules for forming nanoplexes. The nanoplexes were colloidally stable for weeks and the photoluminescence from the d-dots was highly stable against photobleaching. The expression of the mutant K-Ras mRNA in Panc-1 cells was significantly suppressed upon transfecting them with the nanoplexes. More importantly, cell viability studies showed that the d-dot/PAH nanoplexes were biocompatible and non-toxic.
Experimental section
Material
Mn-doped ZnSe quantum dots (Mn:ZnSe d-dots) were obtained from NN-labs and stored at 4 °C in the dark. The as-received QDs (1 mg mL−1 in water) were stabilized using mercaptopropionic acid (MPA) and dispersed in water. 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC), poly(allylamine hydrochloride) (PAH, Mw = 15 kDa), polyethylenimine (PEI, Mw = 1.8 kDa) and folic acid were purchased from Sigma-Aldrich. FAM-labeled small interference RNAs (K-Ras siRNAFAM, sense strand: 5′-FAM-GUUGGAGCUGAUGGCGUAGUU-3′; antisense: 5′ CUACGCCAUCAGCUCCAACUU-3′, italic bold indicates the K-Ras mutation site) were purchased from Shanghai GenePharma (China). 18.2 MΩ cm ultrapure water was used throughout the experiments.Preparation of QD-based siRNA vectors
The d-dots were developed as siRNA vectors by the layer-by-layer (LbL) assembly method. Cationic polyelectrolyte PAH was employed to modify the surface potential of the dots. 50 μL of d-dots stock solution (1 mg mL−1) was washed with ethanol and centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm. Next, 250 μL of PAH solution of different concentrations (0.05–1 mg mL−1, in DI water) was added to re-disperse the d-dot precipitate, followed by short sonication and vortexing for 20 min. After that, the d-dot/PAH0.05–1 particles were collected by centrifugation at 15
000 rpm. Next, 250 μL of PAH solution of different concentrations (0.05–1 mg mL−1, in DI water) was added to re-disperse the d-dot precipitate, followed by short sonication and vortexing for 20 min. After that, the d-dot/PAH0.05–1 particles were collected by centrifugation at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min to remove the non-adsorbed PAH. The resulting d-dot/PAH particles were dispersed in 200 μL of DEPC-treated water while aggregates were removed by centrifugation at 2000 rpm for 1 min. For siRNA loading, 100 μL of 10 μM K-Ras siRNAFAM solution was then introduced into the d-dot/PAH particle dispersion with gentle vortexing and left undisturbed for 40 min. Subsequently, the d-dot/PAH/siRNAFAM complex was further incubated with 8 μL of PAH solution (1 mg mL−1, in DI water) for 1 h to form d-dot/PAH/siRNAFAM/PAH nanoplexes. Centrifugation was used to further remove the excess PAH from the nanoplexe dispersion. The nanoplexes were redispersed in 100 μL of DEPC-treated water for cell transfection experiments. In a parallel experiment, PEI, a frequently reported polymeric gene transfecting material, was employed to form d-dot/PEI nanoplexes. To serve as a reference for d-dot/PAH, the d-dot/PEI nanoplexes were prepared following the same preparation method.
000 rpm for 10 min to remove the non-adsorbed PAH. The resulting d-dot/PAH particles were dispersed in 200 μL of DEPC-treated water while aggregates were removed by centrifugation at 2000 rpm for 1 min. For siRNA loading, 100 μL of 10 μM K-Ras siRNAFAM solution was then introduced into the d-dot/PAH particle dispersion with gentle vortexing and left undisturbed for 40 min. Subsequently, the d-dot/PAH/siRNAFAM complex was further incubated with 8 μL of PAH solution (1 mg mL−1, in DI water) for 1 h to form d-dot/PAH/siRNAFAM/PAH nanoplexes. Centrifugation was used to further remove the excess PAH from the nanoplexe dispersion. The nanoplexes were redispersed in 100 μL of DEPC-treated water for cell transfection experiments. In a parallel experiment, PEI, a frequently reported polymeric gene transfecting material, was employed to form d-dot/PEI nanoplexes. To serve as a reference for d-dot/PAH, the d-dot/PEI nanoplexes were prepared following the same preparation method.
Preparation of folic acid conjugated nanoplexes
Covalent conjugation of the d-dot/PAH nanoplexes with folic acid (FA) was conducted using the standard EDC/NHS condensation method. Firstly, 1.5 mg of FA was dissolved in 2 mL PBS (pH 7.4), followed by mixing with 200 μL EDC (0.1 M in PBS) and 200 μL of NHS (0.2 M in PBS) for 1 h, allowing the carboxyl groups of FA to be activated. Subsequently, 250 μL nanoplex solution was added and the mixture was gently stirred at room temperature for 2 hours. The FA-conjugated nanoplexes were obtained by centrifugation and washed with PBS 3 times to remove unreacted chemicals.Nanoparticle characterization
The UV-visible absorption spectra were obtained from a spectrophotometer (Shimadzu UV-2450). Photoluminescence (PL) spectra and lifetimes were collected using a Fluorolog-3 spectrofluorometer. Quantum yields (QYs) of the Mn:ZnSe QD were determined by comparing the integrated emission of diluted d-dots to CdSe QDs having matching absorbance. The QY of the CdSe reference sample was calibrated using rhodamine 6G. The hydrodynamic size distribution profile and the zeta potential of the nanoparticle formulation were measured using a particle size analyzer system (90Plus, Brookhaven Instruments). Fourier transform infrared (FT-IR) spectra were measured using a Shimadzu FT-IR spectrometer. All measurements were performed at room temperature.SiRNA transfection and gene expression analysis study
Panc-1 cells (ATCC® CRL-1469™) were cultured in Dulbecco's modified Eagle's medium (DMEM), supplemented with 10% fetal bovine serum (FBS, Hyclone), 100 μg mL−1 penicillin (Gibco) and 100 μg mL−1 streptomycin (Gibco). Cells were cultured at 37 °C under a humidified atmosphere with 5% CO2. Panc-1 cells were seeded in 6-well plates to approximately 30% cell confluence, and the culture medium was replaced with DMEM prior to treatment. D-dot/PAH (PEI)-siRNAFAM nanoplex dispersion was then added to the cell plates to give a final incubation concentration of the nanoplexes of around 10 μg mL−1. After 4 h of incubation at 37 °C under a humidified atmosphere with 5% CO2, the cells were washed with PBS three times and harvested for transfection efficiency determination. The cellular uptake efficiency was quantitatively evaluated using a FACS Calibur flow cytometer (Becton Dickinson, Mississauga, CA). For gene expression analysis at 48 h post-transfection, the treated cells were continuously cultured in DMEM with 10% FBS. The cells were harvested and washed with PBS. The total RNA was extracted using TRIzol reagent (Invitrogen) and quantitated using a spectrophotometer (Nano-Drop ND-1000). After that, RNA was reverse transcribed to cDNA using the reagent kit from Promega according to the vendor's instructions. Real time quantitative RT-PCR was then carried out for the analysis of K-Ras relative mRNA expression by normalizing against the expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which is one of the most commonly used housekeeping genes for gene expression comparisons. Forward and reverse primers used in real time RT-PCR were 5′-AGAGTGCCTTGACGATACAGC-3′, 5′-ACAAAGAAAGCCCTCCCCAGT-3′ for K-Ras mRNA, and 5′-ACCACAGTCCATGCCATCAC-3′, 5′-TCCACCACCCTGTTGCTGTA-3′ for GAPDH, respectively. In a parallel experiment, d-dot/PAH complex and free siRNAFAM with the same dosage level were introduced as negative controls while the commercial transfection reagent Oligofectamine™ (Invitrogen) coupled siRNAFAM was used as the positive control.Cell viability evaluation
Cell viability was measured by the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assays. Toxic effects of the d-dot nanocarriers were tested on different cell lines, including two human pancreatic cancer cell lines (Panc-1 and Miapaca-2), human breast cancer cells (MDA-MB-231) and mouse leukaemic monocyte macrophage cells (RAW 264.7). The cells were seeded in a 96-well plate at a density of 5000 cells per well and incubated with different concentrations of d-dot/PAH nanoplexes for 48 h. In a parallel experiment, different types of particles (i.e. d-dot, CdTe and CdTe/ZnS QDs, d-dot/PEI nanoplexes and d-dot/PAH-FA) were tested on the RAW 264.7 macrophage cell line and biocompatibility of these particles was compared at the same dosage addition. In each assay, 20 μL of 5 mg mL−1 MTT in PBS was added and the cells were incubated for 4 h. 150 μL of 100% dimethylsulfoxide (DMSO, Sigma) was then added to dissolve the precipitate with 5 min gentle shaking. Absorbance was then measured with a microplate reader (Bio-Rad) at a wavelength of 490 nm. The cell viability was calculated as the ratio of the absorbance of the sample well to that of the control well and was expressed as a percentage, assigning the viability of non-treated cells as 100%.Results and discussion
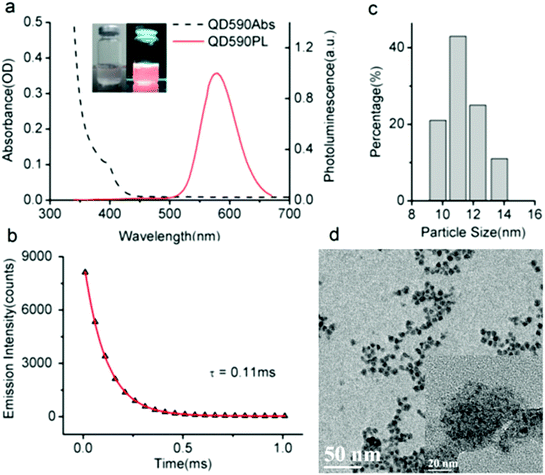
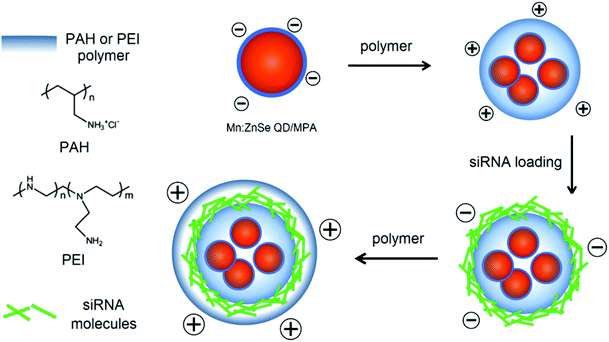
Fig. 1a shows the absorption and photoluminescent (PL) spectra of the MPA-stabilized Mn:ZnSe d-dots dispersion in water. The d-dots exhibit an absorption peak at 400 nm and a dopant emission peak at 590 nm with an FWHM (full width at half maximum) of 70 nm. Instead of band-edge transition in the host ZnSe matrix, the photogenerated exciton transfers energy into excited dopant states,31 which resulted in a large Stokes shift of ∼190 nm. This is desirable for biomedical imaging applications as it helps to minimize the self-absorption of the d-dots and allows one to easily differentiate them from the background autofluorescence of biological samples.32 The PL quantum yield (QY) of the d-dots was estimated to be around 20%. The PL decay curve could be fitted with a single exponential function with an estimated lifetime of τ = 0.11 ms (Fig. 1b). This long luminescence lifetime on the millisecond scale is consistent with literature reports,33 distinguishing the d-dots from common fluorescent QDs, as the optical transition in the Mn2+ centre (4T1–6A1) is spin-forbidden. Fig. 1c shows the hydrodynamic size distribution of the d-dots characterized by dynamic light scattering (DLS). The d-dots have a narrow size distribution with an average value of 11 nm. In addition, they have a negatively charged surface with a zeta potential value of −17 mV, owing to the carboxylic groups present on their surface. Fig. 1d shows the TEM image of the d-dots. The particles are crystalline and they have an average diameter of around 10 nm.The d-dots were developed as siRNA vectors through a Layer-by-Layer (LbL) assembly method. Scheme 1 illustrates the preparation steps, where cationic polymer poly(allylamine hydrochloride) (PAH) was employed for complexing with K-Ras siRNA molecules through electrostatic absorption. As compared to some other polymeric transfecting materials, such as PDDAC and PAMAM dendrimers, which have been reported with acute in vitro toxic effects, the PAH polymer could be more feasible for in vivo applications owing to its biocompatibility.7 It is worth noting that, by varying the PAH concentration (Fig. 2) the d-dot/PAH clusters can be prepared with varying hydrodynamic sizes (47–200 nm), and the surface zeta-potential was also tunable over a wide range (+20 to +48 mV). In addition, agarose gel electrophoresis was used to examine the loading of siRNA molecules onto d-dot/PAH particles (Fig. S1, ESI†). The results suggest that the d-dots without PAH coating cannot absorb negatively charged siRNA, while a higher loading efficiency was observed for d-dot/PAH clusters with higher zeta potential. In a parallel experiment, d-dot/PEI nanoplexes prepared under the same conditions exhibit higher zeta potential (+35 to +58 mV) than d-dot/PAH. The higher charge density of PEI can be more beneficial for loading of the oppositely charged gene material. After siRNA loading, a second PAH coating layer was applied, generating a surface with positive charges (∼+30 mV). Because of the primary amine groups present on PAH, the capping layer can promote cellular uptake or provide anchor moieties for further modification with functional molecules. In Fig. 3, we monitored the hydrodynamic size and zeta potential changes of the particles during LbL assembly. The surface charge reversals clearly indicated deposition of alternating layers of the positively charged PAH and negatively charged siRNAs. It is noticeable that the hydrodynamic size of the particles increases dramatically after coating of the first PAH layer. This observation, together with the TEM image (Fig. 1d, inset), indicate that multiple d-dots have been co-encapsulated to form a relatively large d-dot/PAH cluster. The hydrodynamic size change of the as-produced nanoplex formulations was monitored over a period of 2 weeks (Fig. S2†). Results suggest that the nanoplex possesses an excellent colloidal stability.
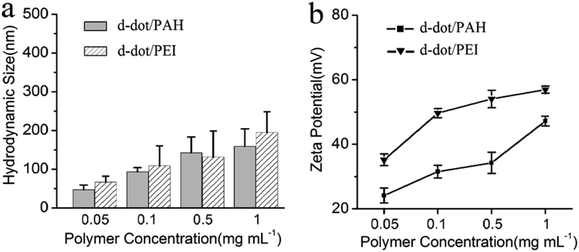 | ||
| Fig. 2 (a) Hydrodynamic size and (b) surface zeta potential of the d-dot/PAH and d-dot/PEI clusters prepared using polymer solutions of different concentrations. | ||
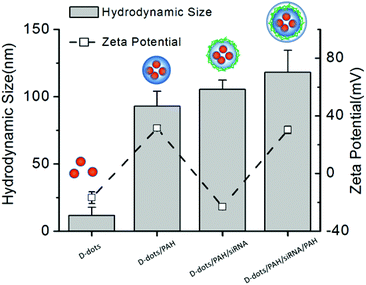 | ||
| Fig. 3 Monitoring the change in particle hydrodynamic size and zeta potential during the LBL construction steps to prepare d-dot/PAH0.1/siRNAFAM/PAH nanoplexes. | ||
As shown in Fig. S3,† we investigate the PL properties of the prepared d-dot/PAH-siRNAFAM nanoplexes, while PL spectra of d-dot/PAH and free siRNAFAM are also presented for comparison. The assembly of the nanoplex does not affect the characteristic peak wavelengths, and fluorescence resonance energy transfer (FRET) between the d-dots and FAM was not observed. This is quite different from the red emitting CdSe QD previously reported for gene release monitoring.19 For multichannel imaging purpose, narrow band-pass filter are normally employed to avoid crosstalk between adjacent channels, while the PL bandwidth of the fluorescent labels is required to be narrow. In our case, crosstalk between the FAM labels (520 nm) and d-dots (590 nm) signals was fundamentally avoided by employing different excitation wavelengths (i.e., UV for d-dot and blue light for FAM), because neither of the excitations can activate the two labels simultaneously. More importantly, owing to the large Stokes shift of the d-dots, the light source for FAM excitation can be selected over a wide wavelength range between 400 and 500 nm without activating the d-dots. In view of this, the d-dot shows an advantage over conventional band-edge emitting QD, as it may simplify the requirements for the optical system and provide a chance to include more optical channels. Additionally, the photostability of the nanoplex was also investigated. Fluorescence images shown in Fig. S4† suggest that, compared with FAM fluorophores, the inorganic d-dots are more suitable probes for long term optical tracing owing to their high resistance against photobleaching.
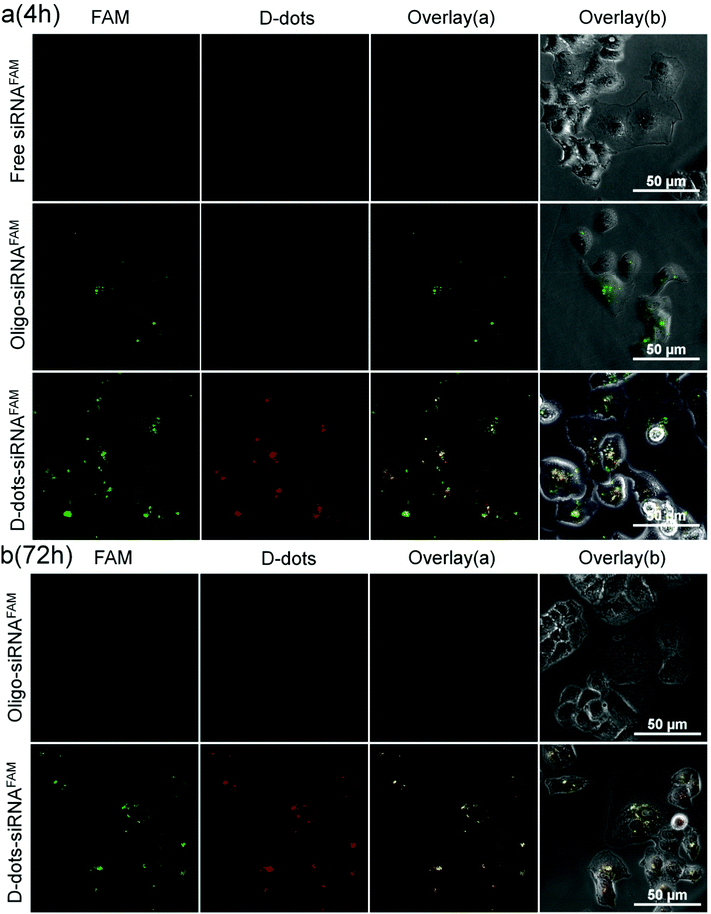
The d-dot/PAH nanoplex formulation was applied to deliver siRNAs that target the mutant oncogenic K-Ras gene in pancreatic cancer cells. In Fig. 4a, the successful delivery of the siRNAs can be easily identified through the green fluorescence from the FAM label at 4 h post transfection. Similar results were also observed in d-dot/PEI-siRNAFAM treated cells (Fig. S5†) and the positive control group using the commercially available transfection reagent Oligofectamine™. As a comparison, there is no fluorescence signal in the cells treated with free siRNAs, suggesting that the naked siRNAs are not able to penetrate the cell membrane without the assistance of transfection agents. More interestingly, the overlay of the FAM and the d-dot channels shows both co-localization (yellow, a merge of green and red) and delocalization within the d-dot/polymer-siRNAFAM treated cells. This indicates that the siRNAs were slowly released from the d-dots nanoplex surface. By further monitoring cells at 72 h post transfection, a variation in the intracellular distribution of d-dots can be observed (Fig. 4b). Furthermore, unlike the Oligofectamine™ formulation, where no FAM signals were detected at 72 h post transfection, the FAM fluorescence signals remained visible in the cells transfected with the d-dot/polymer nanoplexes (Fig. 4b and Fig. S4†). This suggests that a fraction of siRNAFAM molecules that are tightly bound to the d-dot/PAH (PEI) cluster surface have survived from the degradation process within cells, and they may need a longer time to be released to the cytoplasm. Cationic polymer coating plays an essential role in siRNA loading and release kinetics. Insufficient coating will result in a less positively charged nanoplex and relatively weak siRNA surface binding. In contrast, a higher siRNA loading efficiency can be achieved using a thicker polymer coating layer. However, this approach will cause a decrease in the siRNA release rate since the siRNA will be tightly bound by the strong electrostatic force. This situation will certainly influence the therapeutic activity of the nanoplex formulation. Our results suggest that a zeta potential between +20 mV and +40 mV is an optimum range for the nanoplex to successfully and effectively deliver siRNAs to the cells.
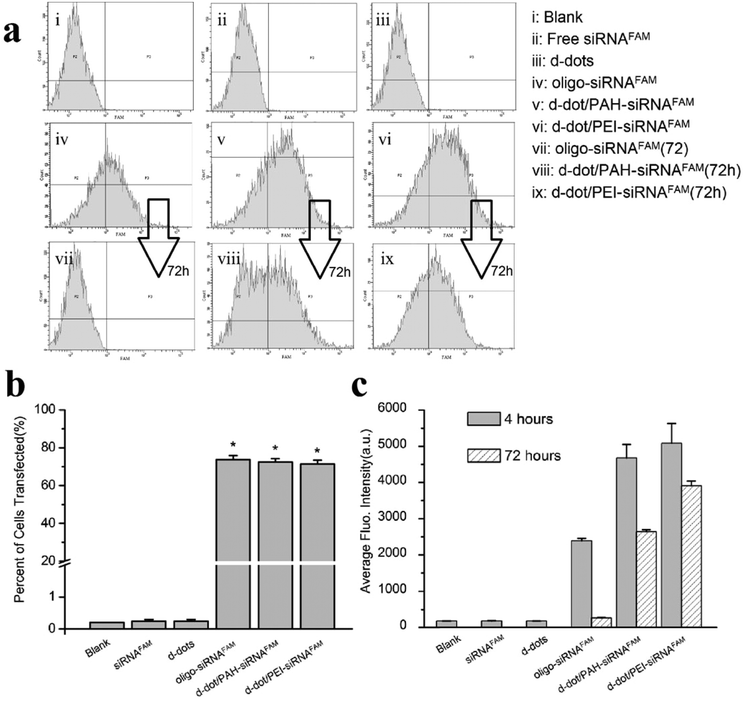
The transfection efficiency of the siRNAs into Panc-1 cells was quantitatively evaluated by flow cytometry analysis. Fig. 5a shows the representative plots of the FAM intensity in cells treated with different formulations. After 4 hours incubation, comparable transfection efficiencies of 73.7%, 72.3% and 71.3% were observed for cells transfected with the oligo agent, d-dot/PAH and d-dot/PEI nanoplexes, respectively (Fig. 5b). These results are consistent with the fluorescence imaging analysis, indicating considerable accumulation of siRNAs inside the cells. In contrast, no evident FAM signal was detected for the cases of free siRNAFAM, d-dot nanoplexes only and blank cell groups. Fig. 5c shows the average fluorescence intensity per cell count recorded in the flow cytometry evaluation. Because the average FAM intensity from the d-dot/PAH and d-dot/PEI treatment groups is two times higher than that of Oligo-siRNAFAM, we consider that siRNA transfection by the two d-dot formulations is more effective. It is also noticeable that, after 72 hours of incubation, fluorescence signals almost disappeared in oligo transfected cells (Fig. 5a and c). In contrast, around 56% and 77% of the intensity still remained for d-dot/PAH and d-dot/PEI transfected cells. These observations are in accordance with our imaging results (Fig. 4b and Fig. S4†), suggesting the protection and the partial release of siRNAs inside the cells.
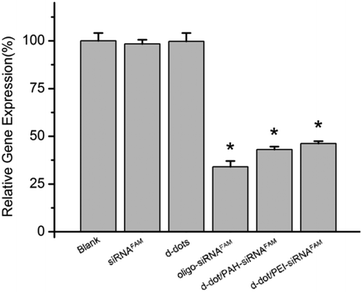
Regulated expression of the targeted mRNA, regarded as the direct proof of a successful RNAi process, was carried out to evaluate the therapeutic capability of the nanoplex formulation. Fig. 6 shows the relative expression levels of mutant K-Ras mRNA in Panc-1 cells treated with different formulations after 72 hours of transfection. No evident differences in the expression levels were observed in the cells treated with d-dot/polymer complexes only or free siRNAs. In contrast, the expression level of the mutant K-Ras mRNA in cells transfected with Oligo-siRNA and d-dot/PAH-siRNA was found to be significantly suppressed to 34% and 43%, respectively. In comparison with d-dot/PAH, the silencing effect was not further promoted using d-dot/PEI (46%), although it induced a higher transfection efficiency. These results, together with the residual FAM intensity detected by flow cytometry at 72 h post-transfection, imply that the d-dot/PAH complex with a lower zeta potential could be more favourable than d-dot/PEI for gene release. Considering that the zeta potential of the d-dot complex is essential for siRNA binding and the value of the d-dot/polymer is tunable over a wide range, the release kinetics of siRNA should be optimized in the future to obtain an improved knockdown efficiency or sustained release for long term gene treatment.
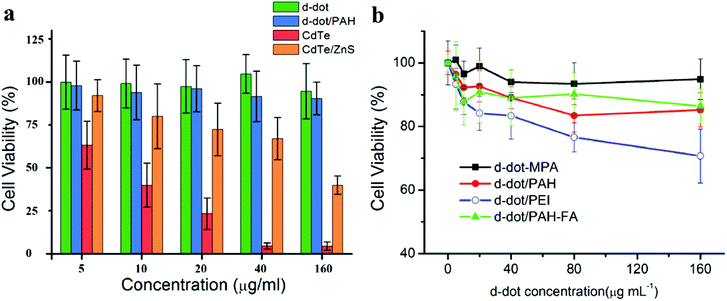
MTT assays were conducted to evaluate the cytotoxicity of d-dots and d-dot/polymer complexes. As shown in Fig. 7a, in addition to the d-dots, cytotoxicity evaluations for MPA stabilized CdTe and CdTe/ZnS core/shell QDs, which have similar physical and optical properties, were also performed in this study as positive controls. The test was firstly conducted using the macrophage cell line (RAW 264.7). Because macrophage cells internalized foreign particles nonspecifically through the phagocytosis process, the cytotoxicity evaluation could be less sensitive towards the discrepancy in the internalization/accumulation rate of the particles with different surface modifications. At 48 h post treatment, CdTe QDs without ZnS shell capping exhibit severe toxicity. The 50% cell viability (IC50) was determined to be less than 10 μg mL−1. Comparatively, ZnS capped CdTe QDs were found to be less toxic. The IC50 was greatly improved to above 40 μg mL−1 although the influence of the particles on cell viability was still pronounced, especially at high dosage concentrations. The ZnS capping layer may protect the Cd-core from fast degradation; however, it cannot prevent the release of heavy metal ions to the biological environment especially when the QDs are exposed to intracellular oxidative conditions. In contrast, after 48 h of incubation with d-dots, the viability of the treated cells was maintained over 90% for concentrations as high as 160 μg mL−1. These results indicate that the d-dots are highly biocompatible. Furthermore, the viabilities of the RAW 264.7 cells treated with d-dot/polymer nanoplexes are shown in Fig. 7b. It is observable that the toxicity of the d-dot nanoplexes was mainly determined by the polymer coating, rather than the inorganic d-dots. In comparison with the d-dot/PEI reference group, higher viabilities were evaluated for cells treated with equal amounts of d-dot/PAH nanoplex. To further confirm the biocompatibility of the d-dot/PAH, we investigated the toxic effects of the d-dot/PAH complexes on different cell lines, including human breast cancer cells (MDA-MB-231) and two human pancreatic cancer cell lines (Panc-1 and Mia PaCa-2). At 48 hours after exposure, all the four cell lines maintained over 80% viability across a wide range of dosages up to 160 μg mL−1 (Fig. S6†). These results suggested that the as-prepared d-dot/PAH complexes (∼+30 mV) are highly biocompatible and will be a promising optical contrast agent for biomedical applications ranging from imaging to drug delivery.
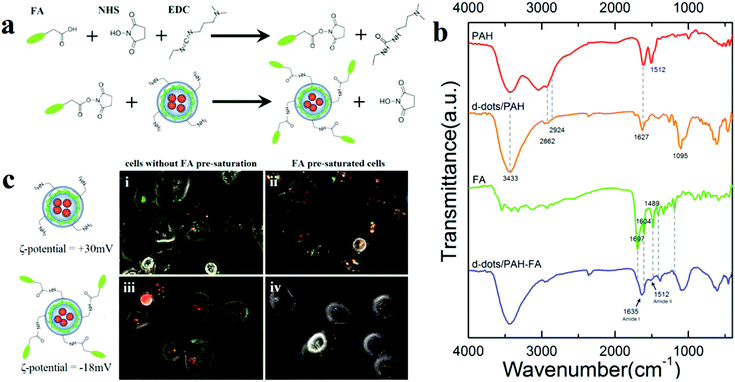
Because the PAH polymer exhibits plenty of primary amino groups, which can serve as anchor moieties to conjugate with different functional molecules, the d-dot complexes capped with a second PAH layer could be further modified to receive multiple bio-functions. For example, we conjugated folic acid (FA) on the d-dot/PAH to obtain an ability for cancer cell targeted gene delivery. Fig. 8a schematically illustrates the preparation of folic acid conjugated nanoplexes, where the EDC/NHS condensation method was applied for crosslinking. According to our results, after conjugation with FA, zeta potential of the nanoplex changed from +30 mV to a negative value (−18 mV). Compared with positive charges, a negatively charged surface helps to reduce non-specific internalization of the particle by cells.34 MTT results shown in Fig. 7b confirm the biocompatibility of these d-dot/PAH-FA formulations. Surface characterization of the FA conjugated d-dot/PAH was studied by FT-IR spectroscopy. For comparison, spectra of PAH, d-dot/PAH, FA and d-dot/PAH-FA are shown in Fig. 8b. The d-dot/PAH complexes exhibit characteristic peaks of amine groups in the PAH polymer including the N–H stretching mode at 3433 cm−1 and NH2 deformation vibration at 1627 cm−1. Appearance of the two bands at 2924 and 2862 cm−1 corresponded to asymmetric and symmetric stretching vibrations of –CH2, respectively. Compared with the PAH polymer in hydrochloride form, the NH3+ deformation vibration at 1512 cm−1 disappeared in d-dot/PAH after forming the complexes, accompanied by the presence of a new band at 1095 cm−1 which is assigned to the C–N stretching mode. Conjugation of folic acid onto d-dot/PAH was substantiated by the emergence of the amide I band at 1635 cm−1 and the amide II band at 1512 cm−1, which were attributed to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching and NH deformation in the secondary amides, respectively. Meanwhile, a series of characteristic IR absorption peaks of FA located at 1697 cm−1 (carboxyl), 1605 cm−1 (benzene), and 1481 cm−1 (hetero-ring) were also visible in the spectra of d-dot/PAH-FA. All these results substantiated that the folic acid molecules were successfully conjugated on the d-dot/PAH surface.
O stretching and NH deformation in the secondary amides, respectively. Meanwhile, a series of characteristic IR absorption peaks of FA located at 1697 cm−1 (carboxyl), 1605 cm−1 (benzene), and 1481 cm−1 (hetero-ring) were also visible in the spectra of d-dot/PAH-FA. All these results substantiated that the folic acid molecules were successfully conjugated on the d-dot/PAH surface.
To demonstrate the ability of the d-dot/PAH-FA for cancer cell targeted siRNA delivery, fluorescence images of Panc-1 cells treated with non-conjugated and FA-conjugated complexes are compared in Fig. 8c. Prior to transfections, Panc-1 cells in (ii) and (iv) were pre-saturated with free folic acid to block FA receptors (FAR) available on the cell surface. As shown in the fluorescence images, Panc-1 cells were stained with FA conjugated nanoplexes after 2 hours of incubation (iii), whereas minimal signal was observed from cells pre-saturated with FA (iv). These results suggested that uptake of the particles involves the interaction between FA and FAR. In contrast, for unconjugated nanoplexes, accumulations of the particles were observed in both unsaturated and FAR-blocked cells suggesting the non-specific nature of the uptake due to the positive surface charges. Considering that the FAR is overexpressed in many human cancer cell lines, these results indicated that the particles can be functionalized with folic acid for cancer cell targeted gene delivery. More importantly, these results exemplified that the amine groups present on the second PAH layer could be employed to link with molecules to obtain multiple bio-functions. For example, to explore the applications for in vivo gene delivery and tumor therapy, PEG should be conjugated for improved stability in serum and prolonged circulation time of the particles after intravascular administration.35
Conclusions
In summary, we have developed the Mn:ZnSe d-dot as a biocompatible nanocarrier for gene delivery in vitro. Using a d-dot/polymer nanoplex as a transfection agent, siRNAs targeting the mutant oncogenic K-Ras gene were delivered into pancreatic cancer cells for sequence specific gene therapy. Owing to the unique and stable PL properties of the d-dots, the delivery of siRNA and the subsequent intracellular release from the d-dot/polymer-siRNA nanoplexes can be monitored by fluorescence imaging. Quantitative results from our flow cytometry study suggested that a high gene transfection efficiency was achieved with the use of the prepared nanoplex formulation. Therapeutic effect was confirmed by the suppressed expression of the mutant K-Ras gene at the mRNA level. Cell viability studies demonstrated that the d-dot/PAH nanoplex formulation is highly biocompatible even at a concentration as high as 160 μg mL−1. This indicates that the d-dots can serve as a promising candidate for biomedical applications, although long term in vitro tests and in vivo experiments are required for further confirmation. The large surface area of the amine-terminated nanoplex presents plenty of opportunities for further bio-functionalization while maintaining a high siRNA loading efficiency.36 As an example, we demonstrated that the nanoplexes can be functionalized with folic acid for receptor-mediated cancer cell targeting and gene delivery. We envision that the developed d-dot nanoplex formulation will serve as a good platform in developing and optimizing the next generation nanoplex for targeted gene therapy of pancreatic cancer in vivo.Acknowledgements
The study was supported by Tier 1 Academic Research Funds (M4010360.040 RG29/10) and Tier 2 Research Grant MOE2010-T2-2-010 (4020020.040 ARC2/11) from the Singapore Ministry of Education.References
- A. Vincent, J. Herman, R. Schulick, R. H. Hruban and M. Goggins, Lancet, 2011, 378, 607–620 CrossRef.
- S. M. Elbashir, J. Harborth, W. Lendeckel, A. Yalcin, K. Weber and T. Tuschl, Nature, 2001, 411, 494–498 CrossRef CAS PubMed.
- A. de Fougerolles, H. P. Vornlocher, J. Maraganore and J. Lieberman, Nat. Rev. Drug Discovery, 2007, 6, 443–453 CrossRef CAS PubMed.
- K. A. Whitehead, R. Langer and D. G. Anderson, Nat. Rev. Drug Discovery, 2009, 8, 129–138 CrossRef CAS PubMed.
- R. L. Juliano, Curr. Opin. Mol. Ther., 2005, 7, 132–136 CAS.
- W. J. Song, J. Z. Du, T. M. Sun, P. Z. Zhang and J. Wang, Small, 2010, 6, 239–246 CrossRef CAS PubMed.
- E. Y. Zhao, Z. X. Zhao, J. C. Wang, C. H. Yang, C. J. Chen, L. Y. Gao, Q. Feng, W. J. Hou, M. Y. Gao and Q. Zhang, Nanoscale, 2012, 4, 5102–5109 RSC.
- W. J. Li and F. C. Szoka, Pharm. Res., 2007, 24, 438–449 CrossRef CAS PubMed.
- V. P. Torchilin, Annu. Rev. Biomed. Eng., 2006, 8, 343–375 CrossRef CAS PubMed.
- J. M. Lee, T. J. Yoon and Y. S. Cho, Biomed. Res. Int., 2013 DOI:10.1155/2013/782041.
- A. Kwok, G. A. Eggimann, J. L. Reymond, T. Darbre and F. Hollfelder, ACS Nano, 2013, 7, 4668–4682 CrossRef CAS PubMed.
- B. Pan, D. Cui, P. Xu, C. Ozkan, G. Feng, M. Ozkan, T. Huang, B. Chu, Q. Li, R. He and G. Hu, Nanotechnology, 2009, 20 DOI:10.1088/0957-4484/20/12/125101.
- T. Kim and T. Hyeon, Nanotechnology, 2014, 25 DOI:10.1088/0957-4484/25/1/012001.
- S. B. Hartono, M. Yu, W. Gu, J. Yang, E. Strounina, X. Wang, S. Qiao and C. Yu, Nanotechnology, 2014, 25 DOI:10.1088/0957-4484/25/5/055701.
- B. Dubertret, P. Skourides, D. J. Norris, V. Noireaux, A. H. Brivanlou and A. Libchaber, Science, 2002, 298, 1759–1762 CrossRef CAS PubMed.
- X. Michalet, F. F. Pinaud, L. A. Bentolila, J. M. Tsay, S. Doose, J. J. Li, G. Sundaresan, A. M. Wu, S. S. Gambhir and S. Weiss, Science, 2005, 307, 538–544 CrossRef CAS PubMed.
- I. L. Medintz, H. T. Uyeda, E. R. Goldman and H. Mattoussi, Nat. Mater., 2005, 4, 435–446 CrossRef CAS PubMed.
- D. R. Larson, W. R. Zipfel, R. M. Williams, S. W. Clark, M. P. Bruchez, F. W. Wise and W. W. Webb, Science, 2003, 300, 1434–1436 CrossRef CAS PubMed.
- J. M. Li, M. X. Zhao, H. Su, Y. Y. Wang, C. P. Tan, L. N. Ji and Z. W. Mao, Biomaterials, 2011, 32, 7978–7987 CrossRef CAS PubMed.
- P. Subramaniam, S. J. Lee, S. Shah, S. Patel, V. Starovoytov and K. B. Lee, Adv. Mater., 2012, 24, 4014–4019 CrossRef CAS PubMed.
- Y. P. Ho and K. W. Leong, Nanoscale, 2010, 2, 60–68 RSC.
- A. M. Derfus, W. C. W. Chan and S. N. Bhatia, Nano Lett., 2004, 4, 11–18 CrossRef CAS.
- R. Hardman, Environ. Health Perspect., 2006, 114, 165–172 CrossRef.
- H. S. Choi, W. Liu, P. Misra, E. Tanaka, J. P. Zimmer, B. I. Ipe, M. G. Bawendi and J. V. Frangioni, Nat. Biotechnol., 2007, 25, 1165–1170 CrossRef CAS PubMed.
- R. Hu, W. C. Law, G. Lin, L. Ye, J. Liu, J. Liu, J. L. Reynolds and K. T. Yong, Theranostics, 2012, 2, 723 CrossRef CAS PubMed.
- C. Kirchner, T. Liedl, S. Kudera, T. Pellegrino, A. M. Javier, H. E. Gaub, S. Stolzle, N. Fertig and W. J. Parak, Nano Lett., 2005, 5, 331–338 CrossRef CAS PubMed.
- D. Zhu, X. Jiang, C. Zhao, X. Sun, J. Zhang and J.-J. Zhu, Chem. Commun., 2010, 46, 5226–5228 RSC.
- S. Guerrero, I. Casanova, L. Farre, A. Mazo, G. Capella and R. Mangues, Cancer Res., 2000, 60, 6750–6756 CAS.
- J. B. Fleming, G. L. Shen, S. E. Holloway, M. Davis and R. A. Brekken, Mol. Cancer Res., 2005, 3, 413–423 CrossRef CAS PubMed.
- T. R. Brummelkamp, R. Bernards and R. Agami, Cancer Cell, 2002, 2, 243–247 CrossRef CAS.
- N. Pradhan, D. M. Battaglia, Y. C. Liu and X. G. Peng, Nano Lett., 2007, 7, 312–317 CrossRef CAS PubMed.
- L. Jing, K. Ding, S. Kalytchuk, Y. Wang, R. Qiao, S. V. Kershaw, A. L. Rogach and M. Gao, J. Phys. Chem. C, 2013, 117, 18752–18761 CAS.
- C. L. Gan, Y. P. Zhang, D. Battaglia, X. G. Peng and M. Xiao, Appl. Phys. Lett., 2008, 92 DOI:10.1063/1.2945274.
- Z.-G. Yue, W. Wei, P.-P. Lv, H. Yue, L.-Y. Wang, Z.-G. Su and G.-H. Ma, Biomacromolecules, 2011, 12, 2440–2446 CrossRef CAS PubMed.
- H. Yin, R. L. Kanasty, A. A. Eltoukhy, A. J. Vegas, J. R. Dorkin and D. G. Anderson, Nat. Rev. Genet., 2014, 15, 541–555 CrossRef CAS PubMed.
- P. Zrazhevskiy, M. Sena and X. H. Gao, Chem. Soc. Rev., 2010, 39, 4326–4354 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4bm00306c |
| This journal is © The Royal Society of Chemistry 2015 |