Surface conformations of an anti-ricin aptamer and its affinity for ricin determined by atomic force microscopy and surface plasmon resonance†
B.
Wang
a,
Z.
Lou
b,
B.
Park
c,
Y.
Kwon
d,
H.
Zhang
b and
B.
Xu
*a
aSingle Molecule Study Laboratory, Faculty of Engineering and Nanoscale Science and Engineering Center, University of Georgia, Athens, GA 30602, USA. E-mail: bxu@engr.uga.edu
bCollege of Materials Science and Technology, Nanjing University of Aeronautics and Astronautics, Nanjing, 210016, P. R. China
cUSDA-ARS, Russell Research Center, Athens, GA 30605, USA
dAvian Disease Division, Animal and Plant Quarantine Agency, Anyang, The Republic of Korea
First published on 11th August 2014
Abstract
We used atomic force microscopy (AFM) and surface plasmon resonance (SPR) to study the surface conformations of an anti-ricin aptamer and its specific binding affinity for ricin molecules. The effect of surface modification of the Au(111) substrate on the aptamer affinity was also estimated. The AFM topography images had a resolution high enough to distinguish different aptamer conformations. The specific binding site on the aptamer molecule was clearly located by the AFM recognition images. The aptamer on a Au(111) surface modified with carboxymethylated-dextran (CD) showed both similarities to and differences from the one without CD modification. The influence of CD modification was evaluated using AFM images of various aptamer conformations on the Au(111) surface. The affinity between ricin and the anti-ricin aptamer was estimated using the off-rate values measured using AFM and SPR. The SPR measurements of the ricin sample were conducted in the range from 83.3 pM to 8.33 nM, and the limit of detection was estimated as 25 pM (1.5 ng mL−1). The off-rate values of the ricin–aptamer interactions were estimated using both single-molecule dynamic force spectroscopy (DFS) and SPR as (7.3 ± 0.4) × 10−4 s−1 and (1.82 ± 0.067) × 10−2 s−1, respectively. The results show that single-molecule measurements can obtain different reaction parameters from bulk solution measurements. In AFM single-molecule measurements, the various conformations of the aptamer immobilized on the gold surface determined the availability of each specific binding site to the ricin molecules. The SPR bulk solution measurements averaged the signals from specific and non-specific interactions. AFM images and DFS measurements provide more specific information on the interactions of individual aptamer and ricin molecules.
Introduction
The 3-dimensional conformations of proteins and nucleic acids are directly related to their functions and interactions.1,2 The surface immobilization of these biomolecules may lead to changes in their conformations and orientations on a substrate, and in turn cause different affinities to their target species in biosensor measurements.3,4 Therefore, morphology and interaction information are both important for the study of biomolecules on biosensor surfaces. Atomic force microscopy (AFM) has the ability to detect a single biomolecule on a substrate surface with a minute amount of sample and provide high resolution images and interaction information.5,6 SPR is another widely used surface detection technique with the advantages of ease of operation and rapid detection, so it has become widely used for biosensing.7,8 Surface modification is an important issue in SPR detection in order to reduce non-specific interactions between the probe molecule and substrate.9,10 It has been widely accepted that linker or spacer molecules on the substrate will improve the binding affinity of the probe molecule. However, the surface modification may also have a negative effect on the properties of the probe molecule, and SPR data cannot show these detailed changes because of the limitations of bulk measurements. AFM single-molecule measurements have the ability to reveal detailed properties of biomolecules with less external interference from the bulk solution environment.11 Therefore, the comparison of experimental results from SPR and AFM should be an important criterion for the development of future SPR biosensors using probes with complex properties such as aptamers.12,13Nucleic acid aptamers have been widely used as binding reagents for the label free detection of biomolecules.14,15 Compared to antibodies, aptamers have advantages such as easy synthesis, low cost, and better stability.4,8 Therefore, aptamers can be integrated into various detection platforms and provide fast detection and analysis in versatile applications related to biosafety and biosecurity, such as the detection of toxins in food products and the public environment.16,17 Based on the fast development of aptamer research and its integration into many nanotechnologies, aptamer-based biosensors will provide more versatile methods and devices for high throughput and label-free detection in biosafety applications.18,19
Ricin is a type-2 ribosome-inactivating protein (RIP) with two chains connected by a disulfide bond.20 The relatively easy access to ricin from caster-oil manufacture makes it a more dangerous and frequently used toxin species. Ricin has been used as a poisoning agent by the military, by criminals, and in recent terrorism cases, but it also has great potential in the development of immunotoxins to treat cancers.21,22 Here we used a certain anti-ricin aptamer to detect ricin in two platforms: AFM and SPR. AFM provided high resolution images of the aptamer on the Au(111) surface, with and without carboxymethylated-dextran (CD) modification. The possible negative influence of the surface modification method on the aptamer specificity and sensitivity to ricin can be directly studied from the AFM topography and recognition images. AFM dynamic force spectroscopy (DFS) can be used to estimate the off-rate of the aptamer–ricin interactions5 and be compared with the off-rate obtained from SPR measurements. Currently, only AFM can provide information about the morphologies and interactions of the aptamer and ricin at the single-molecule level in a single platform. The comparison of the measured off-rate values from AFM–DFS and SPR also provides evidence for the possible influence of surface modification on aptamers on the Au(111) surface (Fig. 1a and b). The results will be used as a reference for the further development of label-free biosensor devices on the micrometer and nanometer scales.
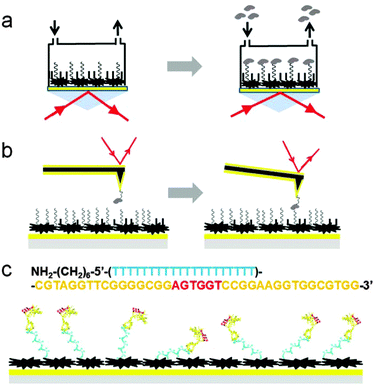 | ||
| Fig. 1 The experimental set-ups and aptamer immobilization for SPR and AFM. In both platforms in (a) and (b), the Au(111) surface was first blocked with carboxymethylated-dextran (large shapes in black). Next, the aptamer molecules (coils in black) were immobilized. Finally, the surface was blocked with ethanolamine (EA, short sticks in black). The ricin molecules (in grey) were detected in bulk solution SPR experiments (a) and in single-molecule AFM experiments (b). The aptamer sequence is shown in (c), with the predicted conformations of aptamer molecules on gold surface. The 20 (dT) spacer is in cyan, the simulated anti-ricin folded structure in yellow, and the predicted binding residues in red. The folded structure of the aptamer was predicted using AMBER 11.26 | ||
Experimental methods
Materials
The anti-ricin aptamer sequence was obtained from the literature.23 In both AFM and SPR experiments, the anti-ricin sequence was modified with twenty thymine bases as the spacer and an amine group as the linker at the 5′ end.24,25 The modified sequence was purchased from Integrated DNA Technologies (Coralville, IA, USA).The ricin sample was provided by Vector Laboratories (Burlingame, CA). In the AFM experiments, the ricin was attached by its amine group to the linker molecule and in turn to the gold coated tip. The polymer linker thiol-(polyethylene glycol)-acid (HS-PEG-COOH, M.W. 2000) was purchased from Creative PEGWorks (Winston Salem, NC, USA). The cystamine, carboxymethylated-dextran (CD), N-hydroxysuccinimide (NHS), and ethanolamine were all purchased from Sigma-Aldrich (St. Louis, MO, USA). The 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (EDC) was purchased from Fluka Chemicals (Sigma-Aldrich, St. Louis, MO, USA).
Phosphate buffered saline (PBS, pH 7.2) was purchased from Pierce (Thermo Scientific, Waltham, MA, USA). Triple deionized water was provided by a Barnstead Nanopure Diamond Laboratory Water System.
Apparatus
In the SPR experiments, the sensor film was coated with Cys self-assembled monolayers (SAMs) via S–Au linkages by spreading a droplet (20 μL) consisting of cystamine hydrochloride (20 mM) onto the Au film and leaving overnight at room temperature. Then, a mixed solution of 15 mM EDC, 75 mM NHS, and 10 mg mL−1 CD was dropped onto the cystamine-modified Au surface and left for 2.5 h before cleaning the Au surface with deionized water. After the sensor chip was mounted on the SPR prism with the matching oil, 100 μL of a mixture of 15 mM NHS and 75 mM EDC in deionized water was injected into the cuvette and the mixture left for 10 minutes to activate the carboxyl group of CD. Afterwards, the aptamer solution (1 μM in PBS) was injected into the flow cell for its immobilization on the CD-modified Au surface. Finally, to deactivate the remaining active sites on the sensor chip surface, 100 μL of 1 M EA in deionized water was injected. With continuous flushing of the flow cell with deionized water, ricin solutions at different concentrations were injected into the BI-2000 SPR system (Biosensing Instrument, Tempe, AZ) for the SPR measurements (Fig. 1a, right part). The off-rate of the aptamer interaction with ricin in the SPR measurements was estimated using the data analysis and kinetics analysis software shipped with the BI-2000 system. The flow rate was controlled at 20 μL per minute and all the detection solutions were injected at the same rate of 20 μL per minute with a unified volume of 100 μL.For the AFM experiments, two surface modification methods were used and their AFM images were compared to investigate the possible influence of CD modification on the aptamer conformations on the Au surface. In the first method (M1), the thiol-modified aptamer was directly immobilized on the gold substrate. In the AFM imaging and DFS experiments, the TopMAC mode and PicoTREC module (Agilent Technologies, Santa Clara, CA, USA) were used to obtain the topography and recognition images, respectively. The ricin was attached to the gold coated AFM tip with a HS-PEG-COOH linker molecule using an EDC/NHS coupling reaction.5 The same tip was used to measure the DFS force–distance curves. Five loading rates were used to estimate the off-rate, and 300 curves were collected for each loading rate. In the second method (M2), the amine-modified aptamer was attached to the thin film of CD on the Au surface. Therefore, the aptamer molecules were immobilized on the Au(111) surface by the same method as in the SPR measurements.
The off-rate calculation for the SPR measurements was conducted using the commercial program “BI-KA” shipped with the SPR instrument. The off-rate calculation for the AFM–DFS was calculated using the same method mentioned in previous publications.5
Results and discussion
High resolution AFM images
The high resolution topography and recognition images showed the conformations of individual aptamer molecules on the Au(111) surface that was modified by M1 (Fig. 2a and b) and by M2 (Fig. 2c and d). The aptamer has a stable folding conformation, but the topography images revealed different conformations of the aptamer on the Au(111) surface. In the topography images, the length of a single aptamer molecule varies from 10 nm to 20 nm, and the width varies from 5 nm to 15 nm. Some aptamers maintained their folded structures and had shorter shapes, while others extended their single strands on the gold surface and had longer shapes. These various round-shaped and stripe-shaped images were caused by the flexibility of the aptamer. In particular, the non-binding parts of the aptamer, including the 20 (dT) spacer, have more degrees of freedom to change their conformations in the buffer and show various orientations or levels of extension. However, most of the binding parts of the aptamer were still active and showed corresponding recognition signals in the recognition image (Fig. 2b). Therefore, the binding conformations and specificity of the ricin–aptamer interactions have been directly shown in the topography and recognition images of AFM alone. | ||
| Fig. 2 The AFM topography and recognition images of the aptamer on the Au(111) surface with and without CD modification. (a) The representative topography image of the aptamer on the Au(111) surface. (b) The corresponding recognition image on the Au(111) surface. (c) The representative topography image of the aptamer on the CD-modified Au(111) surface. (d) The corresponding recognition image on the CD-modified Au(111) surface. The scan size of all the images is 500 × 500 nm2. Nine small areas were circled in (a) and (b) in the corresponding topography and recognition images. These highlighted areas showed the difference between the topography images and recognition images caused by different aptamer conformations. Aptamer conformations similar to numbers 1, 3, 4, 7, and 8 in (a) and (b) can also be found in (c) and (d). The detailed aptamer conformations and their binding sites for ricin are shown in the magnified topography and recognition images in Fig. 3. | ||
The AFM images on the CD-modified Au surface showed very similar gold terrace morphologies, although the thin film of CD made the edges of the gold terraces blur and added some film features on the gold surface (Fig. 2c). The aptamers still showed clear folding conformations and recognition signals (Fig. 2d). The numbered aptamers 1, 3, 4, 7, and 8 from Fig. 2a and b match similar aptamers in Fig. 2c and d. However, more aptamer molecules stayed very close to each other on the CD-modified Au surface than on the one without CD modification, so the surface distribution of the aptamers may be influenced by the CD modification. The additional CD thin film was certainly not as flat as the original Au(111) surface, so some aptamer molecules were buried inside the small holes generated by the CD modification, and some aptamers stretched their folded structures and changed their conformations. Therefore, a smaller number of aptamers on the CD-modified Au(111) surface maintained their activity towards ricin, and consequently showed fewer recognition images with their corresponding topography images. The AFM images proved that the CD modification, M2, did influence the surface morphology and aptamer activities towards ricin.
The degree of this influence on the aptamer activity can be quantitatively studied using the AFM topography and recognition images. When one aptamer molecule showed its topography image but not its corresponding recognition image, that aptamer had lost its activity to ricin. Therefore, a large number of AFM images of individual aptamers were counted to obtain the recognition ratios of the aptamer on the Au(111) surface with and without CD modification. The statistical results comparing the aptamer morphologies and the availabilities of their binding sites are shown in S1 and Fig. S1 (ESI†). The recognition ratio of the aptamer on the CD-modified Au(111) surface is calculated as 75.8%, while the recognition ratio of the aptamer on the Au(111) surface is calculated as 93.6%. This significant decrease in the recognition ratio is mainly due to the changes in the aptamer conformations on the CD film and the reduced exposure of the aptamer active residues caused by CD modification. So far, only AFM has the capability to investigate these detailed changes in surface properties and reveal the different activities of individual aptamer molecules on modified substrates.
In order to further investigate the aptamer conformations on the Au(111) surface, nine small areas were selected from Fig. 2 as the representative images of single aptamer molecules. They were compared with the simulated aptamer structure in different orientations (Fig. S2 and S3 in ESI†). The details of the topography and recognition images in Fig. 3 revealed the significant influence of different conformations on their binding activities to ricin. For example, the aptamer 1 in Fig. 2a and b appeared rod-shaped in the topography images. However, the recognition images showed strong signals in the middle of each structure and weak signals on the two ends of each structure. These round-shaped recognition signals indicated that the ricin binding site was located in the middle of each aptamer conformation. The two ends of each aptamer conformation had only very weak interactions with ricin, but the topography image still showed the structure outline. The top-view and side-view (rotated 90° from the top-view) of aptamer 1 in Fig. 3 showed that the position of the ricin binding site is pointing towards the solution. Therefore, it can easily contact the ricin molecule on the AFM tip. In Fig. 2a, the recognition signal of aptamer 3 is on one end of the structure, while the other end showed a clear topography image but very weak recognition image. The other predicted binding sites for ricin were highlighted in the simulated aptamer structures in Fig. S2 (ESI†). An aggregate of two or three aptamer molecules may exist in the selected areas 8 and 9 (see Fig. 3 enlarged AFM image 8 and Fig. S3 in ESI†). The AFM topography images cannot provide resolutions high enough to distinguish the possible aggregates of molecules of such small size. However, the recognition images showed clear signals of the small binding sites and in turn proved the aggregation. In the comparison of the experimental images and simulated folded structures, the aggregation can be reconstructed with the simulated aptamer structures in different orientations on the gold surface. The non-binding parts of the two aptamers separated the smaller recognition signals, and provide enough resolution in the recognition images to determine the number of molecules in the aggregates, which corresponds to the number of recognition spots. This analysis also confirmed the specificity of the aptamer binding site for the ricin on the AFM tip.
 | ||
| Fig. 3 The magnified AFM topography and recognition images of the aptamer on the Au(111) surface (left column) and the CD-modified Au(111) surface (right column) from Fig. 2. For each numbered area, the topography image is on the left and the recognition image is on the right. The AFM images in each row show a similar alignment of the aptamer active residues (in red). The image size for each area from 1 to 7 is 25 × 25 nm2. The image size for area 8 is 30 × 30 nm2. The molecular simulations of the aptamer folding conformations are shown in the middle column, with the non-active aptamer sequence in yellow and spacer sequence in blue. The simulated structures in top-view are shown on the left and the side-views (rotated 90°) are on the right. | ||
The aptamer folding conformations simulated from molecular dynamics may not accurately represent all of the aptamers observed on the Au(111) surface due to the limits of the simulation methods. For example, some aptamers can be distorted and randomly oriented since they are inherently more flexible than proteins. However, the simulated molecular outlines and the orientations of the active sites are very helpful for the interpretation of the different topography and recognition images in Fig. 2 and 3. Here, AFM successfully distinguished different aptamer conformations on the Au(111) surfaces with and without CD modification. The detailed activity information from the recognition images revealed the influence of CD modification on the aptamer–ricin interactions.
The affinity of the aptamer for ricin measured by AFM–DFS
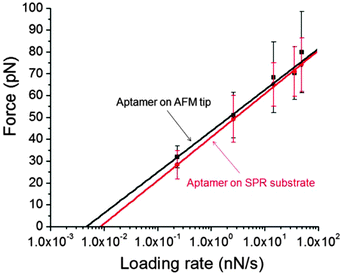
The affinity of the aptamer for ricin can be evaluated by the off-rate of the binding reaction (koff). At the single-molecule level, DFS was used to measure the unbinding force between the aptamer on the Au(111) surface and the ricin on the AFM tip. In previous DFS studies, we immobilized ricin on the Au(111) surface and attached the same anti-ricin sequence to the AFM tip.5 The same five loading rates were used in this DFS experiment and 300 force–distance curves were measured under each loading rate. The force histograms obtained at these five loading rates were compared with the previous ones using the aptamer-modified AFM tip (Fig. 4). The most probable unbinding force (F*) under each loading rate (R) was obtained using Gaussian fitting. No significant difference was observed from the histograms obtained using the aptamer and ricin-modified tips.The Bell–Evans single-barrier model was used to estimate the koff value from the F* vs. ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) R plot. To test the possible difference between the koff when using the ricin-modified tip and the koff when using the aptamer-modified tip, two F* vs. ln
R plot. To test the possible difference between the koff when using the ricin-modified tip and the koff when using the aptamer-modified tip, two F* vs. ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) R plots were constructed as shown in Fig. 5, with their corresponding error bars. The error bars for the data points under same loading rate showed large portions of overlay, which indicates that the deviations between the data from the two types of tip modification were not significant. The off-rate value obtained using the aptamer-modified tip (koff1) is (6.8 ± 0.9) × 10−4 s−1. The off-rate value obtained using the ricin-modified tip (koff2) is (7.3 ± 0.4) × 10−4 s−1.
R plots were constructed as shown in Fig. 5, with their corresponding error bars. The error bars for the data points under same loading rate showed large portions of overlay, which indicates that the deviations between the data from the two types of tip modification were not significant. The off-rate value obtained using the aptamer-modified tip (koff1) is (6.8 ± 0.9) × 10−4 s−1. The off-rate value obtained using the ricin-modified tip (koff2) is (7.3 ± 0.4) × 10−4 s−1.
Therefore, the affinities estimated by these two single-molecule experiments are considered to be in good agreement. These two off-rate values are also close to the one obtained using an antibody-modified AFM tip.27 The low off-rate values indicate a high affinity of the aptamer for ricin in the AFM single-molecule measurements. The CD modification should not significantly influence the DFS measurements, because the force–distance curves were obtained from aptamers with specific activity towards ricin. The aptamers that lost their activity did not show specific force–distance curves.
SPR measurements
In the real-time SPR detection, six ricin concentrations of 0.083 nM (5 ng mL−1), 0.167 nM (10 ng mL−1), 0.83 nM (50 ng mL−1), 1.67 nM (100 ng mL−1), 4.17 nM (250 ng mL−1), and 8.33 nM (500 ng mL−1) were used to test the limit of detection and estimate the koff value. The angle change values increased with increasing ricin concentration. The representative SPR sensorgrams from these six ricin concentrations are shown in Fig. 6.The off-rate of the aptamer from ricin in the SPR measurements was estimated using an aptamer–ricin 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding mode in the kinetics analysis software. Fig. 7a shows the plot of ricin concentration vs. SPR signal obtained from the averaged angle shift values. The error bars represent the standard errors of three repeat injections for each concentration. High ricin concentrations showed large errors, which are the major sources of the error in the estimate of the koff value. Fig. 7b was used to estimate the limit of detection for the SPR measurement of ricin. The extrapolation of the fitted straight line at the x-axis showed that the limit of detection for ricin should be around 1.5 ng mL−1 (25 pM). The final calculated koff value in the SPR bulk solution measurement (koff3) is (1.82 ± 0.067) × 10−2 s−1. It is more than one order of magnitude larger than those obtained from the AFM single-molecule measurements. The deviation is also much larger than the those in the AFM experiments. Therefore, the difference in the off-rate values between the bulk solution and single-molecule measurements has been clearly revealed from the experimental data from AFM and SPR. Moreover, the AFM experimental results at the single-molecule level can be used to connect the difference in the off-rate values to the aptamer conformations on the Au(111) surface. The details of this are discussed in next section.
1 binding mode in the kinetics analysis software. Fig. 7a shows the plot of ricin concentration vs. SPR signal obtained from the averaged angle shift values. The error bars represent the standard errors of three repeat injections for each concentration. High ricin concentrations showed large errors, which are the major sources of the error in the estimate of the koff value. Fig. 7b was used to estimate the limit of detection for the SPR measurement of ricin. The extrapolation of the fitted straight line at the x-axis showed that the limit of detection for ricin should be around 1.5 ng mL−1 (25 pM). The final calculated koff value in the SPR bulk solution measurement (koff3) is (1.82 ± 0.067) × 10−2 s−1. It is more than one order of magnitude larger than those obtained from the AFM single-molecule measurements. The deviation is also much larger than the those in the AFM experiments. Therefore, the difference in the off-rate values between the bulk solution and single-molecule measurements has been clearly revealed from the experimental data from AFM and SPR. Moreover, the AFM experimental results at the single-molecule level can be used to connect the difference in the off-rate values to the aptamer conformations on the Au(111) surface. The details of this are discussed in next section.
Comparison of AFM and SPR measurements
Fig. 8 compares the koff values and their standard errors on the same scale. The koff values (koff1 and koff2) obtained from the two AFM single-molecule measurements don’t show a significant difference in statistics, but they are much smaller than that obtained using SPR bulk solution measurements (koff3). Moreover, the error for koff3 is larger than the values of koff1 and koff2. Based on this comparison, the SPR measurements showed relatively high sensitivity and specificity for aptamer–ricin detection. However, the off-rate estimate based on these bulk solution measurements was different from the AFM single-molecule measurements. This difference will have a greater impact on the measured parameters when the detection devices reach the nanometer scale, such as detection in nanoarrays or microfluidic channels. On the nanometer scale, the major factors that can cause the deviation include nonspecific interactions, influence from solvent, and interface properties.28,29 When the development of biosensors moves onto the nanometer scale, thorough study of the difference between bulk solution measurements and single-molecule measurements will provide important information for the further development of surface modifications and signal analysis.For this study of ricin–aptamer interactions in SPR, the large error value of koff is actually comparable to the koff values themselves in the single-molecule measurements. This large error is a common problem for bulk solution measurements, although SPR can detect ricin at the pM level. One source of this large error came from different aptamer conformations on the Au surface, partly caused by the CD modification. Fig. 2 clearly showed that some aptamer conformations were changed by the CD modification, which caused a different recognition ratio for the aptamer–ricin interactions. Fig. S2 and S3 (ESI†) show more AFM images that revealed various aptamer molecules with different orientations and aggregation conformations. The recognition signals in these images indicated different binding–unbinding interactions during the tip scanning. In the bulk solution measurements, all of these conformations have the opportunity to interact with ricin molecules with specific or non-specific forces, but the apparent values of the off-rate (koff3) and affinity are averaged based on large amounts of various aptamers. During the surface modification and aptamer immobilization, different chemical and physical environments on the substrate and in the solution could influence the flexible aptamer conformations on the gold surface and cause the change in conformations and orientations. Therefore, the averaged off-rate value (koff3) has a much larger experimental error than those from the single-molecule measurements.
The standard errors of the two single-molecule measurements are also different. As shown in Fig. 8, the aptamer-modified AFM tip gave a relatively large error bar for koff1, and the ricin-modified tip gave a relatively small error bar for koff2. When the ricin-modified tip interacted with the aptamers on the Au(111) surface, the PEG linker provided enough degrees of freedom for the ricin to form interactions, and the flexible aptamer structure also increased the probability of the aptamer binding site interacting with the ricin binding site. On the contrary, the ricin molecules immobilized on the Au(111) surface had fewer degrees of freedom because of the short linker molecule used in that study.30 Although the aptamer-modified tip had enough degrees of freedom, the more rigid ricin conformations on the substrate suffered from more interference in aptamer–ricin binding events. Therefore, the error bar for koff1 is larger than that for koff2.
Conclusions
We investigated the specific conformations and interactions of an anti-ricin aptamer in two different platforms, i.e. AFM and SPR instruments. The topography and recognition images obtained using AFM illustrated the various binding conformations and the specific binding site of the aptamer molecules on the Au(111) surface with and without CD modification. The off-rate of ricin–aptamer single-molecule interactions was found from AFM–DFS to be (7.3 ± 0.4) × 10−4 s−1. This off-rate value obtained using a ricin-modified AFM tip is statistically consistent with the one obtained using an aptamer-modified tip.5 This comparison shows that the off-rate value in DFS measurements is not affected by which molecule is attached to the tip and which one is immobilized on the substrate for the ricin–aptamer interactions. In the SPR experiments, the ricin sample solutions were detected by the aptamer immobilized on the CD-modified gold substrate at concentrations as low as 83.3 pM (5 ng mL−1). The off-rate of (1.82 ± 0.067) × 10−2 s−1 estimated from SPR sensorgrams is much larger than those from the DFS measurements. The very large standard error in the SPR result indicates that the bulk solution measurements were influenced by the various aptamer conformations at the single-molecule level. Additionally, the bulk solution measurements included more interference from non-specific interactions from modified environment conditions. The differences between single-molecule measurements and bulk solution measurements are significant, and the single-molecule AFM images provide more detailed information for individual aptamer binding conformations and activities on the CD-modified Au(111) surface. The AFM force measurements also provide more precise techniques to control the ricin–aptamer binding events. This work provides fundamental information on the aptamer as a probe molecule on a biosensor surface. The results will facilitate the understanding of the aptamer binding mechanism and the development of aptamer-based nanoarrays and other label-free detection devices using nanotechnologies.Acknowledgements
The authors would like to thank the partial support of this work by the US National Science Foundation (ECCS 1231967, CBET 1139057), the Animal and Plant Quarantine Agency, Republic of Korea (I-AD15-2010-13-01), and the Nature Science Foundation of Jiangsu Province, China (BK20131355).Notes and references
- Y.-H. Cai and H. Huang, Amino Acids, 2012, 43, 1141–1146 CrossRef CAS PubMed.
- E. A. Dethoff, J. Chugh, A. M. Mustoe and H. M. Al-Hashimi, Nature, 2012, 482, 322–330 CrossRef CAS PubMed.
- Y. Chen and R. M. Corn, J. Am. Chem. Soc., 2013, 135, 2072–2075 CrossRef CAS PubMed.
- A. Hayat, S. Andreescu and J.-L. Marty, Biosens. Bioelectron., 2013, 45, 168–173 CrossRef CAS PubMed.
- B. Wang, C. Guo, G. Chen, B. Park and B. Xu, Chem. Commun., 2012, 48, 1644–1646 RSC.
- Y. Lyubchenko, L. Shlyakhtenko, R. Harrington, P. Oden and S. Lindsay, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 2137–2140 CrossRef CAS.
- B. Liedberg, C. Nylander and I. Lundstrom, Sens. Actuators, 1983, 4, 299–304 CrossRef CAS.
- J. Homola, Chem. Rev., 2008, 108, 462–493 CrossRef CAS PubMed.
- M. J. Linman, A. Abbas and Q. Cheng, Analyst, 2010, 135, 2759–2767 RSC.
- J.-F. Masson, T. M. Battaglia, J. Cramer, S. Beaudoin, M. Sierks and K. S. Booksh, Anal. Bioanal. Chem., 2006, 386, 1951–1959 CrossRef CAS.
- A. R. Bizzarri and S. Cannistraro, Chem. Soc. Rev., 2010, 39, 734–749 RSC.
- S. Ray, G. Mehta and S. Srivastava, Proteomics, 2010, 10, 731–748 CrossRef CAS PubMed.
- Y.-H. Lao, K. Peck and L.-C. Chen, Anal. Chem., 2009, 81, 1741–1754 CrossRef PubMed.
- A. B. Iliuk, L. Hu and W. A. Tao, Anal. Chem., 2011, 83, 4440–4452 CrossRef CAS PubMed.
- B. Basnar, R. Elnathan and I. Willner, Anal. Chem., 2006, 78, 3638–3642 CrossRef CAS PubMed.
- A. Sassolas, L. J. Blum and B. D. Leca-Bouvier, Biosens. Bioelectron., 2011, 26, 3725–3736 CrossRef CAS PubMed.
- N. de-los-Santos-Alvarez, M. J. Lobo-Castanon, A. J. Miranda-Ordieres and P. Tunon-Blanco, TrAC, Trends Anal. Chem., 2008, 27, 437–446 CrossRef CAS PubMed.
- M. Piliarik, L. Parova and J. Homola, Biosens. Bioelectron., 2008, 24, 1399–1404 CrossRef PubMed.
- G. Lautner, Z. Balogh, V. Bardóczy, T. Mészáros and R. E. Gyurcsányi, Analyst, 2010, 135, 918–926 RSC.
- E. Rutenber, B. J. Katzin, S. Ernst, E. J. Collins, D. Mesna, M. P. Ready and J. D. Robertus, Proteins, 1991, 10, 240–250 CrossRef CAS PubMed.
- S. Worbs, K. Kohler, D. Pauly, M.-A. Avondet, M. Schaer, M. B. Dorner and B. G. Dorner, Toxins, 2011, 3, 1332–1372 CrossRef CAS PubMed.
- M. Friedman and R. Rasooly, Toxins, 2013, 5, 743–775 CrossRef CAS PubMed.
- J. Tang, J. Xie, N. Shao and Y. Yan, Electrophoresis, 2006, 27, 1303–1311 CrossRef CAS PubMed.
- S. Centi, S. Tombelli, M. Minunni and M. Mascini, Anal. Chem., 2007, 79, 1466–1473 CrossRef CAS PubMed.
- C. Polonschii, S. David, S. Tombelli, M. Mascini and M. Gheorghiu, Talanta, 2010, 80, 2157–2164 CrossRef CAS PubMed.
- D. A. Case, T. A. Darden, T. E. Cheatham III, C. L. Simmerling, J. Wang, R. E. Duke, R. Luo, M. Crowley, R. C. Walker, W. Zhang, K. M. Merz, B. Wang, S. Hayik, A. Roitberg, G. Seabra, I. Kolossvary, K. F. Wong, F. Paesani, J. Vanicek, J. Liu, X. Wu, S. R. Brozell, T. Steinbrecher, H. Gohlke, Q. Cai, X. Ye, J. Wang, M.-J. Hsieh, G. Cui, D. R. Roe, D. H. Mathews, M. G. Seetin, C. Sagui, V. Babin, T. Luchko, S. Gusarov, A. Kovalenko and P. A. Kollman, AMBER II, University of California, San Francisco, 2010 Search PubMed.
- G. Chen, J. Zhou, B. Park and B. Xu, Appl. Phys. Lett., 2009, 95, 043103 CrossRef PubMed.
- J. Liu, M. A. Eddings, A. R. Miles, R. Bukasov, B. K. Gale and J. S. Shumaker-Parry, Anal. Chem., 2009, 81, 4296–4301 CrossRef CAS PubMed.
- L. R. Brewer and P. R. Bianco, Nat. Methods, 2008, 5, 517–525 CrossRef CAS PubMed.
- B. Wang, C. Guo, M. Zhang, B. Park and B. Xu, J. Phys. Chem. B, 2012, 116, 5316–5322 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Statistical results of the activities of the aptamer molecules on the Au(111) surface with and without CD modification. Enlarged AFM images of selected aptamer conformations from Fig. 2a and b compared with the simulated aptamer folded structures. See DOI: 10.1039/c4cp03190c |
| This journal is © the Owner Societies 2015 |