Open Access Article
Open Access ArticleUser-friendly 3D bioassays with cell-containing hydrogel modules: narrowing the gap between microfluidic bioassays and clinical end-users' needs
Do-Hyun
Lee
a,
Chae Yun
Bae
a,
Seyong
Kwon
a and
Je-Kyun
Park
 *ab
*ab
aDepartment of Bio and Brain Engineering, Korea Advanced Institute of Science and Technology (KAIST), 291 Daehak-ro, Yuseong-gu, Daejeon 305–701, Republic of Korea. E-mail: jekyun@kaist.ac.kr; Fax: +82 42 350 4310; Tel: +82 42 350 4315
bKAIST Institute for the NanoCentury, 291 Daehak-ro, Yuseong-gu, Daejeon 305–701, Republic of Korea
First published on 30th March 2015
Abstract
Cell-containing hydrogel modules as cell–hydrogel microunits for creating a physiologically relevant 3D in vivo-like microenvironment with multiple cell types and unique extracellular matrix (ECM) compositions facilitate long-term cell maintenance and bioassays. To date, there have been many important advances in microfluidic bioassays, which incorporate hydrogel scaffolds into surface-accessible microchambers, driven by the strong demand for the application of spatiotemporally defined biochemical stimuli to construct in vivo-like conditions and perform real-time imaging of cell–matrix interactions. In keeping with the trend of fostering collaborations among biologists, clinicians, and microfluidic engineers, it is essential to create a simpler approach for coupling cell-containing hydrogel modules and an automated bioassay platform in a user-friendly format. In this article, we review recent progress in hydrogel-incorporated microfluidics for long-term cell maintenance and discuss some of the simpler and user-friendly 3D bioassay techniques combined with cell-containing hydrogel modules that can be applied to mutually beneficial collaborations with non-engineers. We anticipate that this modular and user-friendly format interfaced with existing laboratory infrastructure will help address several clinical questions in ways that extend well beyond the current 2D cell-culture systems.
Introduction
Because the cellular function in the body is essentially the basic criterion for discriminating patient status and sub-classifications of disease states, the ability to monitor cellular functions via cell-based assays can facilitate disease screening and personalized/tailored therapies. Currently, the field of cell-based assays has three major motivations in both academic research and the pharmaceutical industry: (i) more accurate assessment, based on improved cellular function and morphology,1 (ii) cell-based toxicity screening for in vitro studies to replace complex in vivo models,2 and (iii) cost reduction in late-stage drug failures to commercialize drugs efficiently.3 To date, cell-based assays have been generally carried out using ‘traditional’ well-plate-based 2D monolayer cultures; however, only different cellular functions are shown distinctively compared with their native environment in vivo due to the morphologically disparate cell phenotypes. In contrast to 2D cell cultures, 3D cell cultures have shown great importance in terms of culture conditions, such as the diffusion-limited transport of nutrients and oxygen, regulation of molecular gradients in concentration of metabolites, and maintenance of microenvironments for co-culture and long-term maintenance of cells.4,5 In particular, the mass transport of nutrients and metabolites in 3D cell cultures plays a critical role in cell proliferation and is allowed to occur in the long-term maintenance of isolated cell lines over weeks-to-months rather than around 24 h in 2D culture.6,7 Additionally, cell cultures in tissue-like 3D conditions better mimic in vivo-like culturing conditions, incorporating native extracellular matrix (ECM) structures, to better replicate the drug sensitivity trends of cancer cells in vivo.8To close this technological gap, the evolution of comprehensive microfluidic solutions offers the promise of systematic establishment of a 3D microenvironment in high-throughput systems, based on several advantages, including precise fluid handling, low reagent consumption, and potentially massive parallelization of experiments.9 For example, the design of cellular microenvironments by continuously controlling both nutrients and metabolites has been implemented using microfluidic components (e.g., mixers, valves, and gradient generators) to accelerate the realization of microfluidic perfusion cultures.10 However, these state-of-the-art microfluidic perfusion culture systems are not appropriate for long-term maintenance of cells due to the material inconsistency of poly(dimethylsiloxane) (PDMS), medium evaporation, cell loss due to high shear stress, and lack of ECM proteins.11
To overcome these challenges, various strategies have been developed with the aim of building cell-containing hydrogel “modules” as cell–hydrogel microunits encapsulating heterotypic cell types and unique ECM compositions to create a more physiologically relevant 3D microenvironment.12 In addition, much effort has been focused on the development of hydrogel-incorporating microfluidic cell culture assays, which allow integrative analyses of cellular interactions with ECM scaffolds in stable molecular concentration gradients,13in situ monitoring of cellular morphogenesis within a well-controlled 3D microenvironment,14 and further understanding of how co-cultured cells affect each other's function after long-term maintenance.15
However, despite these advantages, the major challenges related to 3D biofunctional assays based on the 3D microstructure of cell-containing hydrogel modules remain. Few of these microfluidic approaches have been adopted in 3D bioassays for the following reasons: (i) although various 3D cell culture techniques have been developed, conventional optical detection strategies still depend on 2D endpoint detection. That is, few analytical methods adequately capture the full complexity of, and measure cell activities within, the 3D microstructure of cell–hydrogel units. (ii) Microfluidic devices require a continuous flow to generate precise shear profiles, and thus external pumps and sophisticated fluid handling systems are needed. (iii) It is difficult to recover the encapsulated cells, when necessary, from the microdevice for transfer to the macroworld for further post-assay processing.
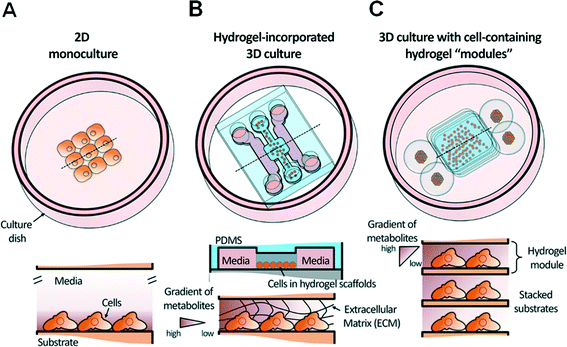
Thus, in order to lower the barrier to entry for biologists and clinicians and promote wider adoption of microfluidics in biological laboratories, it is crucial to establish a simpler approach for the coupling of cell-containing hydrogel modules and an automated bioassay platform in a user-friendly format. From this perspective, we provide an overview of progress over the past decade with a focus on recent progress in the development and application of hydrogel-incorporated 3D cell culture and microfluidic bioassays. Fig. 1 shows a schematic overview of a conventional 2D cell monoculture, a hydrogel-incorporated 3D cell culture, and a 3D culture with cell-containing hydrogel modules that can be enhanced using microfluidics. We discuss various case studies to focus on barriers to the adoption of microfluidic technologies in 3D bioassays that aim to replace traditional macroscale assays in biological and clinical research. Finally, we discuss positive future directions of simpler 3D bioassay techniques in a user-friendly format that can be applied to mutually beneficial collaborations with clinicians.
Cell-containing hydrogel modules for 3D cell culture
Hydrogels are a promising class of soft materials that exhibit intrinsic diffusion permeability to nutrients, metabolites, and oxygen, and can be tailored to resemble native ECMs mechanically.16 Ling et al. first fabricated cell–hydrogel scaffolds that facilitate the exchange of nutrients and waste products without concern for diffusion depth in the bulk hydrogel by using standard soft lithographic techniques.17 Since then, numerous specialized engineering tools and techniques have been introduced to fabricate hydrogel-based cellular modules, such as cellular microfibers, microcapsules, and sheets as culture units (Fig. 2). For example, several studies have reported the guidance of various types of cells in natural-ECM-encapsulated thin and long fibers. In a recent study, Kang et al. sought to create microfibers with tunable, morphological, structural, and chemical features using a programmable flow control system, allowing the generation of 3D structures with controlled cellular organization.18 Onoe et al. reported the fabrication of meter-long microfibers encapsulating primary pancreatic islet cells that were transplantable to diabetic mice for the treatment of diabetes mellitus.19 Cell-laden hydrogel microcapsules have been used as a monodisperse culture unit for 3D cell culture via cell microencapsulation technology. Specifically, microfluidic methods have been developed to create spheroidal aggregates involving multiple types of cells with controllable size and shape, due to the controllability of the diameter, inner structure (e.g., core–shell structure), and spherical morphology of the microcapsules.15,20,21 The hydrogel-based spherical scaffolds containing 3D cultured cells could also be administered into a target tissue as implantable and injectable forms, increasing the usability in modern cell-based therapeutics. In addition, several schemes of 2D, freestanding, and microarchitectured hydrogel sheets have been developed over time to take advantage of long-term freestanding cell culture. Leng et al. reported a one-step tessellation of planar hydrogel sheets including two-directional patterned primary cells with precise spatiotemporal control.22 Recently, the form of “freestanding biopaper” was expanded towards various 3D cell culture applications through layer-by-layer assembly and was demonstrated as a toolkit for biofunctional assays and cell proliferation assays.23,24 An artificial 3D hepatic tissue reconstruction was demonstrated by assembling cellular hydrogel biopaper modules.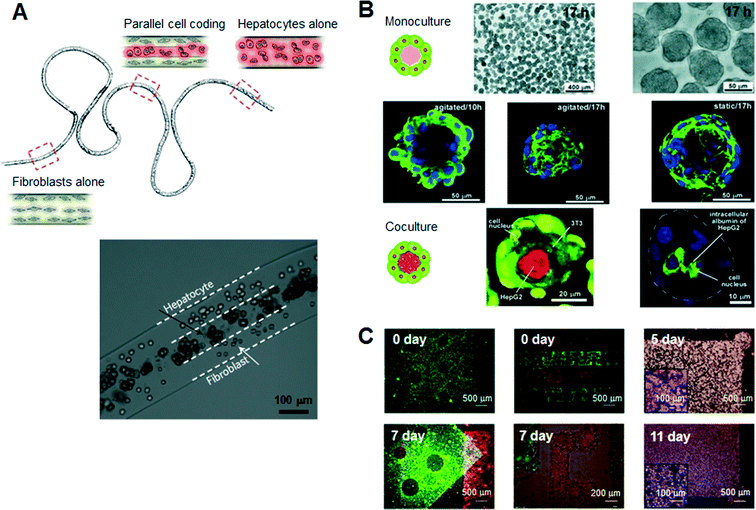 | ||
| Fig. 2 Typical hydrogel-based cellular modules, such as cellular microfibers, microcapsules, and sheets as culture units. (A) Schematic of a periodically coded fiber with primary rat hepatocytes, fibroblasts or a mixture of hepatocytes and fibroblasts. The bottom figure shows a magnified image of a co-culture region that consists of multiple parallel layers of hepatocytes or fibroblasts. Adapted with permission from Macmillan Publishers Ltd: Nat. Mater. Kang et al.18 copyright (2011). (B) Microscopic view of the monocultured cell-laden collagen microcapsules and fluorescence confocal microscopy of co-cultured microcapsules encapsulating NIH 3T3 and HepG2 cells. Adapted with permission from Matsunaga et al.15 copyright (2011) John Wiley and Sons. (C) Microscopic images of a freestanding cellular hydrogel biopaper of calcium alginate containing HepG2 cells, and demonstration of the assembly of cellular hydrogel biopapers. Five pieces of the microhole-perforated biopaper are stacked with guided alignment in the size-fitting square assembly well and can be destacked without structural destruction. Adapted with permission from Lee et al.23 copyright (2012) John Wiley and Sons. | ||
More in vivo-like bioassays through hydrogel-incorporated microfluidic platforms
Several microfluidic approaches have been regarded as better methods for the functional assessment of cells in vitro in terms of micro-patterning of cells, precise control of reagents, rapidity and accuracy of assays, and easy observation of physiological characteristics between cells and the surrounding environment. Early developments in 3D cell culture systems were focused on the microfluidic formation of 3D ECM scaffolds. Kim et al. used a sheath flow to form ‘Puramatrix’ (peptide hydrogel) scaffolds hydrodynamically, resulting in the 3D immobilization and encapsulation of human hepatocellular carcinoma (HepG2) cells with no additional surface treatment (Fig. 3A).25 They performed in situ cell-based dose-dependent cytotoxicity assays according to the concentration of the toxicant, Triton X-100, based on the evaluation of the linear concentration gradients across the peptide scaffold. Lii et al. reported a pneumatic valve combined with an individually addressable array of 3D ECMs containing undifferentiated mouse embryonic stem cells to deliver reagents and exchange diffusible factors between the chambers for studying chamber-to-chamber communication of diffusible factors.26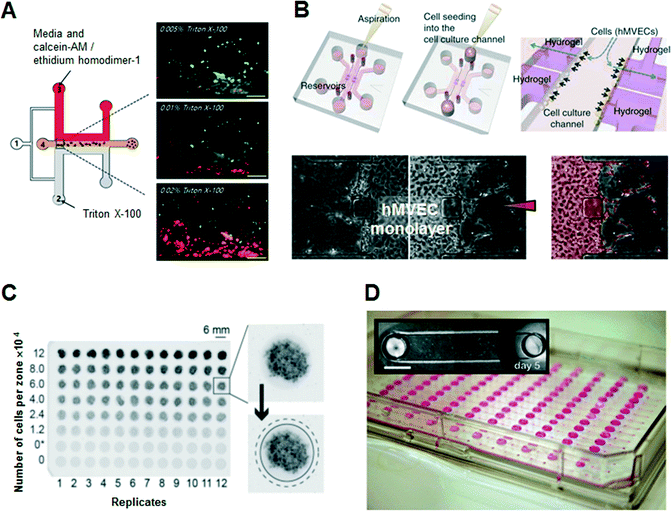 | ||
| Fig. 3 Examples of hydrogel-incorporated 3D microfluidic bioassay platforms and 3D bioassay platforms with a user-friendly microfluidics. (A) In situ dose-dependent cytotoxicity tests using human hepatocellular carcinoma cells (HepG2) according to the linear concentration gradient of Triton X-100 at the cross-sectional area of the peptide scaffolds. Adapted with permission from Kim et al.25 copyright (2007), with permission from Springer Science and Business Media. (B) Schematic of a hydrogel-incorporating microfluidic assay device. After aspiration and addition of a cell suspension of human microvascular endothelial cells (hMVECs), hMVECs become attached to the side of the collagen scaffold by interstitial flow due to the pressure difference between the middle cell channel and the side control channels. In 1-day culture, cells form an intact monolayer in the channel and on the collagen walls. Angiogenic response (segmented in pale red) from the monolayer was induced by the human vascular endothelial growth factor (VEGF) diffusion gradient from the right channel. Adapted with permission from Macmillan Publishers Ltd: Nat. Protoc. Shin et al.27 copyright (2012). (C) Photographs of stacked 96-zone paper plates that contain eight concentrations of MDA-MB-231 cells within Matrigel scaffolds. The average intensity of the black color in the image is proportional to the GFP fluorescence intensity in the sample. Adapted from Derda et al.32 with permission from PLoS One. (D) Compartmentalized microfluidic cell culture arrays based on surface tension driven passive pumping using a traditional pipette. Arrays are interfaced with a 96-tip liquid handling instrument. Photographs of an array of 192 microfluidic channels each with two access ports positioned according microtiter plate standards. Adapted from Meyvantsson et al.36 with permission from The Royal Society of Chemistry. | ||
However, these microfluidic bioassay platforms stated above are not capable of operating over physiological time frames or reconstituting the stabilized chemokine gradients needed to construct in vivo-like pathophysiological conditions. To overcome this limitation, Shin et al. developed a robust and versatile microfluidic bioassay platform consisting of hydrogel-incorporating chambers between two surface-accessible microchannels (Fig. 3B).27 Multiple cell types, including neuronal cells, hepatocytes, stem cells and floating cells, were isolated successfully to the hydrogel-incorporated microfluidic chamber with more in vivo-like appearances as well as high resolution and in situ imaging capabilities. Under spatiotemporally controlled biochemical and biophysical conditions, unexplored biological cellular interactions among cell populations were investigated, such as a 3D sprouting angiogenic response in the direction of increasing human vascular endothelial growth factor (VEGF) concentration. Bersini et al. developed a collagen gel-embedded 3D in vitro microfluidic model to analyze the extravasation of highly metastatic human breast cancer cells into an in vivo-like osteo-cell conditioned microenvironment.28 The tri-culture of bone marrow-derived mesenchymal stem cells, human breast cancer cells and endothelial cells provided quantitative results regarding the crosstalk between cancer and osteo-differentiated stem cells, such as the extravasation rate and the extravasated distance of breast cancer cells in the ECM. Cosson and Lutolf also described a hybrid system that combined stem cell culture in multiwell plates incorporating a microfluidic hydrogel chip.29 They tested and observed the spatiotemporally controlled induction of neurogenic differentiation of mouse embryonic stem cells by accurate delivery of a gradient of the morphogen retinoic acid from the gelatin-based hydrogel slab.
Meanwhile, PDMS, the most commonly used polymer for simple manufacture of microfluidic devices, is unfamiliar to biologists and clinicians, and somewhat inflexible for long-term cell maintenance due to medium evaporation and metabolite adsorption. Medium evaporation leads to osmolality shifts that prevent cell growth and development, and bubble propagation within the microchannel can causes cell lysis.30 Also, due to the hydrophobic nature of PDMS, non-specific protein adsorption can deplete protein levels within the culture medium significantly, leading to inhibition of cell signaling.31 To overcome these limited characteristics of PDMS as a substrate for cell culture and bioassays, various materials have recently been adopted for bioassay platforms, as destructible, cheap, and commercializable alternatives, such as thermoplastics, cyclo-olefin copolymers, and paper. Recently, thermoplastics such as polymethyl methacrylate and polystyrene have attracted attention as substitutes for PDMS in the development of more usable fabrication methods. Because polystyrene has long been used as a laboratory material for cell culture, biologists would prefer it for the thermoplastic microfluidic devices. Above all, patterned paper with well-defined channels, comprising hydrophilic paper bounded by a hydrophobic polymer, has attracted attention as a simple and inexpensive alternative. Derda et al. developed a 3D culture system, “cells-in-gels-in-paper”, that uses a wax-patterned paper as a scaffold to support cell-laden hydrogels enabling the stacking of multiple layers of paper that include hydrogel slabs containing cell suspensions (Fig. 3C).32 Furthermore, the stacking of multiple layers of paper was also demonstrated using co-cultured fibroblasts and cardiomyocytes that were suspended in hydrogels as a 3D in vitro model for cardiac ischemia.33 The patterned substrate with a standard 96-well format is an excellent example of a user-friendly solution for researchers in the biomedical and clinical community to design customized 3D culture and bioassay platforms.
Parallelized and automated 3D bioassays with user-friendly microfluidics
Researchers in biological research laboratories and the pharmaceutical industry have been using microfluidic devices in recent years, because they reduce costs and shorten process time in many steps of cell-based bioassays due to the miniaturization of fluidic systems. However, non-engineering researchers are currently confronted with incompatibility, because microfluidic engineers intended a multiplexed and sophisticated bioassay platform through microfluidic techniques, whereas end-users (biologists or clinicians) need a simple and convenient kit based on the conventional well-plate design and pipetting. Although conventional microfluidic devices have been applied to various types of cell-based assays via accurate control of fluids, they require complex external equipment, such as syringe pumps and pneumatic fluidic handling systems, which must be operated by highly trained personnel. Thus, these platforms have a limited capacity for widespread use outside the engineer's laboratory. To support methods compatible with existing liquid-dispensing equipment in a common biological laboratory, various ideas and concepts in the field of microfluidic technology have been suggested. Integrated multiple bioreactors in a multiwell plate format, known as well-plate microfluidic devices, have been introduced to examine the chemotactic responses of leukemia cells.34 Because the setup, operation, and detection of these well-plate microfluidic devices are compatible with conventional cell culture techniques, these modular approaches can provide simple methods for interfacing with cell cultures. Domansky et al. introduced a perfused multiwell plate containing an ECM-coated scaffold, enabling the circulation of culture medium for long-term maintenance of differentiated hepatocytes and liver sinusoidal endothelial cells according to oxygen consumption and transport.35 The open wells, built-in micropumps, and the perfused multiwell plate facilitated the operation and integration into the conventional incubator and bioassay kit. By constructing a model of oxygen consumption and transport, the relevant operating parameters for culturing primary liver cells were predicted.A small volume of liquid can be controlled in a simple microchannel design with passive pumping that requires only pipetting, instead of syringe pumps. Meyvantsson et al. suggested an automated cell culture microdevice based on surface tension-driven pumping with straightforward pipette operation, termed “tubeless microfluidics” (Fig. 3D).36 The device gives compartmentalized microfluidic cell culture arrays and thus microfluidic operations are possible through integration with existing laboratory infrastructures. This technique has been used to pattern endothelial cell-lined lumens through ECMs in various microchannel geometries for quantitative angiogenesis assays.37 Recently, they improved fluidic control in an open type of microchannel that uses surface tension to fill and maintain a fluid in microscale structures devoid of a ceiling and floor, known as suspended microfluidics.38 This approach was used to create arrays of collagen membranes as an ECM, establishing horizontal microtranswells for cellular invasion and metabolomics assays. Open microfluidics with high accessibility and robustness (tubeless or suspended microfluidics) ensures high-throughput multiplexed screening assays to evaluate cell growth within 3D ECMs.
According to the preferences of end-users, microfluidic researchers should improve and develop a more easily accessible and more universally applicable device. In particular, more microfluidic culture devices should be integrated with existing laboratory infrastructure, such as single or multichannel pipettes, off-the-shelf polystyrene substrates, and immunofluorescence reagents; this is desirable for a wider community of end-users. Modular microfluidics, as an approach for the construction of a microfluidic device to facilitate the customization and operation of microfluidic systems by non-experts, enables the interconnection of various microfluidic components in an easy and reliable manner. Several examples of pluggable modules include fit-to-flow world-to-chip interconnections,39 microfluidic D-subminiature connectors,40 and a microfluidic breadboard.41 The key advantages of this “add-on” modular architecture are (i) portability of the culture device that can be adapted to standard cell culture laboratory procedures for frequent transfer between workstations (e.g., cell culture benches, microscopes, and incubators) and (ii) ease of use by non-engineers in biology and clinical laboratories. Another intriguing technology, termed “3D printed microfluidics”, has been demonstrated in the stereolithography-based microfabrication of fully digital and intrinsically modular plastic microdevices with complex 3D microfluidic features.42 Non-engineers can easily operate the 3D-printed user-friendly fluid automation devices, which are capable of cell-based bioassays to replace the laborious manual handling processes in current use.
Towards simple hydrogel-incorporated 3D bioassays
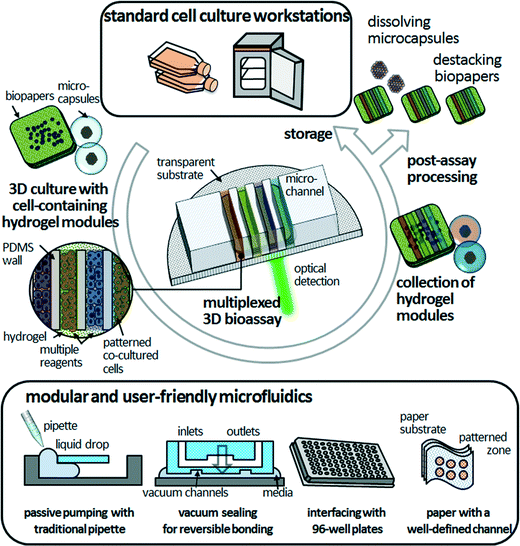
3D bioassay platforms with cell-containing hydrogel modules offer the promise of significant advantages over existing hydrogel-incorporated microfluidic devices, particularly long-term cell maintenance, co-culture of multiple cell types, and organization of cellular arrangements that can duplicate those in vivo. For simplicity and versatility of fabrication, culturing, manipulation, and assembly, freestanding cell-containing hydrogel modules could provide unprecedented tools for 3D bioassays. We expect that the modular and user-friendly microfluidics will facilitate the robust assembly and simple disassembly of the PDMS microfluidic devices and cell-containing hydrogel modules in a reversible manner (Fig. 4). Modular microfluidics, as described in the previous section, are already making an impact in terms of their technological capabilities for reversible sealing (e.g., adhesive tape-based bonding, vacuum sealing, and bonding with threaded screws).43–46 Such “detachable” microfluidic devices might be compatible with researchers who are not specialists in microfluidics. The next critical step is to increase the compatibility of 3D bioassay platforms that enable post-assay processing—such as enzyme-linked immunosorbent assay (ELISA), immunohistochemistry and Western blotting—to investigate cellular functions. Because the cell-containing hydrogel modules are mechanically stable without morphological distortion during long-term freestanding cell culture and microfluidic assays, end-users can culture heterotypic cell types to various culture stages, assemble them in microfluidic devices on demand for multiplexed assays in an in vivo-like 3D microenvironment, retrieve them from the hydrogel modules, and determine the functionality of target cells. “Simplifying” the processing in time and space is an opportunity to develop functional bioassays of 3D cultured cells that may be useful for biomedical and clinical researchers.Although hydrogel-incorporated 3D bioassay platforms open up the opportunities to address unanswered biological and clinical questions, development of analytical methods and tools remains a challenge. In microfluidic 3D cell culture systems, cells are located precisely within ECM scaffolds, thus confocal laser microscopy can be integrated with these systems straightforwardly to conduct live-cell assays. However, most of the bioassay kits are designed for 2D cell cultures, which cause difficulties with optical detection in the z-direction. Also, they depend on antibody-based biomarkers and are designed as endpoint tests for drug sensitivity and cellular functions,6 leading to cell death due to cell fixation. Thus, it becomes more difficult to accomplish post-assay processing to explore cellular functions, as mentioned above. Imaging technologies that can be applied for 3D cell samples include not only light scattering or confocal fluorescence detection, but also ionizing radiation, magnetic fields, and ultrasound.47 Live-cell monitoring based on non-invasive and label-free techniques such as Raman spectroscopy is a non-destructive analytical method with increased penetration depth.48 However, even with these constraints, hydrogel-incorporated 3D bioassay platforms would benefit cell-based drug screening in terms of mimicking more closely the in vivo microenvironment and contributing multiple factors to the processes of cellular morphogenesis.27 Hydrogel networks allow precise biomolecule delivery through the hydrogel layer, leading to a spatiotemporally controlled cellular response in stable long-term biochemical gradients.29 This should be valuable for the study of drug interactions, determining drug candidates, and biomarker identification. Furthermore, high-throughput and fully automated 3D assays would enable multiple cellular assays and multiplexed detection, leading to more rapid evaluation of drug candidate toxicity and human metabolism and cost reductions for late-stage drug failures.
Future outlook
In the future, hydrogel-incorporated 3D bioassay platforms may be used for clinical applications in ways that extend well beyond conventional 2D cell-culture systems. The advantage of rapid and accurate 3D functional cellular phenotyping with physiological relevance by recreating biological interfaces seen in vivo can determine the functional state of diverse subpopulations of target cells and can provide meaningful information for fundamental science and diagnostics. The proposed platforms may also be used for drug safety, drug discovery and toxicity testing with advances in high-throughput and multiplexed microfluidic assays. Eventually, these may decrease the research and development costs for new pharmaceuticals and increase the predictability of new drugs prior to their undergoing animal testing and clinical trials.The discussed hydrogel-incorporated 3D bioassay platforms are relatively new, and much work remains in terms of constructing physiologically relevant 3D in vivo-like microenvironments. Hydrogel-incorporated 3D microfluidic bioassays are also a promising technology for long-term cell maintenance in a 3D microenvironment and analysis of cellular function and morphology. The combination of hydrogel-incorporated 3D bioassay platforms and cell-containing hydrogel modules—including microfibers, microcapsules, sheet modules and microfluidic platforms—provides unique tools to assess 3D cell maintenance and has the potential to change the paradigm for in vitro assessments of cell biofunctionality.
In summary, the rapid development of 3D microfluidic bioassay platforms and cell-containing hydrogel modules has delivered a paradigm shift in 3D cell culture and assay platforms over the past decade. Simplified and highly integrated microdevices coupled with cell-containing hydrogel modules and an automated bioassay platform in a user-friendly format would revolutionize fundamental and applied research in biological and clinical fields with interdisciplinary collaborations.
Acknowledgements
This research was supported by the National Leading Research Laboratory Program (grant NRF-2013R1A2A1A05006378) and the Converging Research Center Program (grant 2011K000864) through the National Research Foundation of Korea funded by the Ministry of Science, ICT and Future Planning. The authors also acknowledge the KAIST Systems Healthcare Program.Notes and references
- E. F. A. Brandon, C. D. Raap, I. Meijerman, J. H. Beijnen and J. H. M. Schellens, Toxicol. Appl. Pharmacol., 2003, 189, 233–246 CrossRef CAS.
- M. Y. Lee and J. S. Dordick, Curr. Opin. Biotechnol., 2006, 17, 619–627 CrossRef CAS PubMed.
- J. F. Pritchard, M. Jurima-Romet, M. L. J. Reimer, E. Mortimer, B. Rolfe and M. N. Cayen, Nat. Rev. Drug Discovery, 2003, 2, 542–553 CrossRef CAS PubMed.
- S. Zhang, Nat. Biotechnol., 2004, 22, 151–152 CrossRef CAS PubMed.
- K. L. Schmeichel and M. J. Bissell, J. Cell Sci., 2003, 116, 2377–2388 CrossRef CAS PubMed.
- Y.-C. Tung, A. Y. Hsiao, S. G. Allen, Y.-s. Torisawa, M. Ho and S. Takayama, Analyst, 2011, 136, 473–478 RSC.
- S. T. Koshy, T. C. Ferrante, S. A. Lewin and D. J. Mooney, Biomaterials, 2014, 35, 2477–2487 CrossRef CAS PubMed.
- T. Ohmori, J.-L. Yang, J. O. Price and C. L. Arteaga, Exp. Cell Res., 1998, 245, 350–359 CrossRef CAS PubMed.
- J. Voldman, M. L. Gray and M. A. Schmidt, Annu. Rev. Biomed. Eng., 1999, 1, 401–425 CrossRef CAS PubMed.
- J. El-Ali, P. K. Sorger and K. F. Jensen, Nature, 2006, 442, 403–411 CrossRef CAS PubMed.
- L. Kim, Y.-C. Toh, J. Voldman and H. Yu, Lab Chip, 2007, 7, 681–694 RSC.
- J. W. Nichol and A. Khademhosseini, Soft Matter, 2009, 5, 1312–1319 RSC.
- D. R. Albrecht, G. H. Underhill, T. B. Wassermann, R. L. Sah and S. N. Bhatia, Nat. Methods, 2006, 3, 369–375 CrossRef CAS PubMed.
- C. M. Nelson, M. M. VanDuijn, J. L. Inman, D. A. Fletcher and M. J. Bissell, Science, 2006, 314, 298–300 CrossRef CAS PubMed.
- Y. T. Matsunaga, Y. Morimoto and S. Takeuchi, Adv. Mater., 2011, 23, H90–H94 CrossRef CAS PubMed.
- H. Geckil, F. Xu, X. Zhang, S. Moon and U. Demirci, Nanomedicine, 2010, 5, 469–484 CrossRef CAS PubMed.
- Y. Ling, J. Rubin, Y. Deng, C. Huang, U. Demirci, J. M. Karp and A. Khademhosseini, Lab Chip, 2007, 7, 756–762 RSC.
- E. Kang, G. S. Jeong, Y. Y. Choi, K. H. Lee, A. Khademhosseini and S.-H. Lee, Nat. Mater., 2011, 10, 877–883 CrossRef CAS PubMed.
- H. Onoe, T. Okitsu, A. Itou, M. Kato-Negishi, R. Gojo, D. Kiriya, K. Sato, S. Miura, S. Iwanaga, K. Kuribayashi-Shigetomi, Y. T. Matsunaga, Y. Shimoyama and S. Takeuchi, Nat. Mater., 2013, 12, 584–590 CrossRef CAS PubMed.
- Y. Du, E. Lo, S. Ali and A. Khademhosseini, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 9522–9527 CrossRef CAS PubMed.
- P. Panda, S. Ali, E. Lo, B. G. Chung, T. A. Hatton, A. Khademhosseini and P. S. Doyle, Lab Chip, 2008, 8, 1056–1061 RSC.
- L. Leng, A. McAllister, B. Zhang, M. Radisic and A. Günther, Adv. Mater., 2012, 24, 3650–3658 CrossRef CAS PubMed.
- W. Lee, C. Y. Bae, S. Kwon, J. Son, J. Kim, Y. Jeong, S. S. Yoo and J. K. Park, Adv. Healthcare Mater., 2012, 1, 635–639 CrossRef CAS PubMed.
- C. Y. Bae, M.-k. Min, H. Kim and J.-K. Park, Lab Chip, 2014, 14, 2183–2190 RSC.
- M. S. Kim, J. H. Yeon and J.-K. Park, Biomed. Microdevices, 2007, 9, 25–34 CrossRef CAS PubMed.
- J. Lii, W.-J. Hsu, H. Parsa, A. Das, R. Rouse and S. K. Sia, Anal. Chem., 2008, 80, 3640–3647 CrossRef CAS PubMed.
- Y. Shin, S. Han, J. S. Jeon, K. Yamamoto, I. K. Zervantonakis, R. Sudo, R. D. Kamm and S. Chung, Nat. Protoc., 2012, 7, 1247–1259 CrossRef CAS PubMed.
- S. Bersini, J. S. Jeon, G. Dubini, C. Arrigoni, S. Chung, J. L. Charest, M. Moretti and R. D. Kamm, Biomaterials, 2014, 35, 2454–2461 CrossRef CAS PubMed.
- S. Cosson and M. P. Lutolf, Sci. Rep., 2014, 4, 4462 CAS.
- Y. S. Heo, L. M. Cabrera, J. W. Song, N. Futai, Y. C. Tung, G. D. Smith and S. Takayama, Anal. Chem., 2007, 79, 1126–1134 CrossRef CAS PubMed.
- G. M. Walker, H. C. Zeringue and D. J. Beebe, Lab Chip, 2004, 4, 91–97 RSC.
- R. Derda, S. K. Y. Tang, A. Laromaine, B. Mosadegh, E. Hong, M. Mwangi, A. Mammoto, D. E. Ingber and G. M. Whitesides, PLoS One, 2011, 6, e18940 CAS.
- B. Mosadegh, B. E. Dabiri, M. R. Lockett, R. Derda, P. Campbell, K. K. Parker and G. M. Whitesides, Adv. Healthcare Mater., 2014, 3, 1036–1043 CrossRef CAS PubMed.
- C. G. Sip, N. Bhattacharjee and A. Folch, Lab Chip, 2014, 14, 302–314 RSC.
- K. Domansky, W. Inman, J. Serdy, A. Dash, M. H. M. Lim and L. G. Griffith, Lab Chip, 2010, 10, 51–58 RSC.
- I. Meyvantsson, J. W. Warrick, S. Hayes, A. Skoien and D. J. Beebe, Lab Chip, 2008, 8, 717–724 RSC.
- L. L. Bischel, E. W. K. Young, B. R. Mader and D. J. Beebe, Biomaterials, 2013, 34, 1471–1477 CrossRef CAS PubMed.
- B. P. Casavant, E. Berthier, A. B. Theberge, J. Berthier, S. I. Montanez-Sauri, L. L. Bischel, K. Brakke, C. J. Hedman, W. Bushman, N. P. Keller and D. J. Beebe, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 10111–10116 CrossRef CAS PubMed.
- A. Chen and T. Pan, Lab Chip, 2011, 11, 727–732 RSC.
- A. Scott, A. K. Au, E. Vinckenbosch and A. Folch, Lab Chip, 2013, 13, 2036–2039 RSC.
- K. A. Shaikh, K. S. Ryu, E. D. Goluch, J. M. Nam, J. Liu, C. S. Thaxton, T. N. Chiesl, A. E. Barron, Y. Lu, C. A. Mirkin and C. Liu, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 9745–9750 CrossRef CAS PubMed.
- K. C. Bhargava, B. Thompson and N. Malmstadt, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 15013–15018 CrossRef CAS PubMed.
- D. Pakto, Z. Martonfalvi, B. Kovacs, F. Vonderviszt, M. Kellermayer and R. Horvath, Sens. Actuators, B, 2014, 196, 352–356 CrossRef PubMed.
- C. G. Sip, N. Bhattacharjee and A. Folch, Biomicrofluidics, 2011, 5, 022210 Search PubMed.
- P. Skafte-Pedersen, C. G. Sip, A. Folch and M. Dufva, J. Micromech. Microeng., 2013, 23, 055011 CrossRef.
- E. Wilhelm, C. Neumann, T. Duteenhofer, L. Pires and B. E. Rapp, Lab Chip, 2013, 13, 4343–4351 RSC.
- S. Y. Nam, L. M. Ricles, L. J. Suggs and S. Y. Emelianov, Tissue Eng., Part B, 2015, 21, 88–102 CrossRef PubMed.
- V. Charwat, K. Schütze, W. Holnthoner, A. Laverntieva, R. Gangnus, P. Hofbauer, C. Hoffmann, B. Angres and C. Kasper, J. Biotechnol., 2015 DOI:10.1016/j.jbiotec.2015.02.007.
| This journal is © The Royal Society of Chemistry 2015 |