A fluorescent light-up nanoparticle probe with aggregation-induced emission characteristics and tumor-acidity responsiveness for targeted imaging and selective suppression of cancer cells†
Dan
Ding‡
ab,
Ryan T. K.
Kwok‡
c,
Youyong
Yuan
a,
Guangxue
Feng
a,
Ben Zhong
Tang
*cd and
Bin
Liu
*ae
aDepartment of Chemical and Biomolecular Engineering, National University of Singapore, 117576, Singapore. E-mail: cheliub@nus.edu.sg; Fax: +65 67791936; Tel: +65 65168049
bState Key Laboratory of Medicinal Chemical Biology, Key Laboratory of Bioactive Materials, Ministry of Education, College of Life Sciences, Nankai University, Tianjin, 300071, China
cDepartment of Chemistry, Institute for Advanced Study, Division of Biomedical Engineering, Division of Life Science, State Key Laboratory of Molecular Neuroscience and Institute of Molecular Functional Materials, The Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong, China. E-mail: tangbenz@ust.hk
dSCUT–HKUST Joint Research Laboratory, Guangdong Innovative Research Team, State Key Laboratory of Luminescent Materials and Devices, South China University of Technology, Guangzhou, 510640, China
eInstitute of Materials Research and Engineering (A*STAR), 3 Research Link, Singapore 117602
First published on 7th October 2014
Abstract
A pH-responsive light-up nanoparticle probe with aggregation-induced emission (AIE) features was designed and synthesized. The probe carries negative charges and shows very weak fluorescence under physiological conditions. In a tumor acidic extracellular microenvironment, the nanoparticle probe can switch to positive surface charge and thus significantly light up cancer cells, allowing for targeted imaging and selective suppression of cancer cells. As AIE nanoparticles are known for high fluorescence in the aggregate state, this study represents the first example of light-up AIE nanoparticle probe design.
Conceptual insightsFluorogens with aggregation-induced emission (AIE) characteristics have recently emerged as a new class of fluorescent materials for biosensing and bioimaging applications. AIE fluorogens show weak or no fluorescence as molecular species, but intense fluorescence as aggregates. They have attracted considerable attention in the fabrication of molecular fluorescent light-up probes for specific biomarker detection or biological process imaging with high signal-to-background ratios. It is well-known that nanoparticle probes may have distinct advantages over molecular probes, e.g., they can be easily internalized into cells through endocytosis and passively targeted to tumor tissues via an enhanced permeability and retention (EPR) effect. However, the bright fluorescence of the AIE aggregates has hampered the development of nanoparticle-based fluorescent light-up probes. In this contribution, by using a simple molecular design strategy, we have developed the first AIE fluorescent light-up nanoparticle probe, which not only possesses tumor-acidity responsiveness with good performance in targeted cancer imaging, but also shows selective inhibition of cancer cells. The smart probe design together with the excellent performance will inspire more exciting research in tumor theranostic platform development, which will further expand the applications of AIE fluorogens in biomedical research. |
The development of fluorescent light-up bioprobes is of great importance in diagnosis and tracking of many diseases, especially cancers, as these probes allow sensitive, simple and specific detection of analytes in biological environments.1 Particularly, designing light-up probes that can respond to stimuli in tumor microenvironments would open new avenues to intelligent fluorescent probes for the advancement of cancer imaging. Among the stimuli in tumor microenvironments, pH-responsiveness is of particular interest and it has been extensively investigated in controlled drug delivery for better cancer targeting and treatments.2 This is because the pH value in a tumor extracellular environment (pHe) is slightly acidic (6.5–7.2) as compared to the blood and normal tissues (∼7.4).3 Enthusiastic research studies on pH-triggered drug delivery systems inspired us to explore new pH-responsive fluorescent light-up probes that are capable of imaging cancer cells more intelligently and more efficiently. To date, most of the pH-responsive fluorescent probes are designed for targeting intracellular pH at ∼5.5, and only a few studies have focused on those that respond to a tumor extracellular acidic microenvironment.4
Recently, we have developed a novel class of organic fluorogens with unique aggregation-induced emission (AIE) features.5 AIE fluorogens generally possess rotating units (e.g., phenyl rings). In solution, the low-frequency motions of the rotating units lead to fast non-radiative decay of the excited states, which make the AIE fluorogens non-emissive as molecular species. However, in the aggregate state, the intramolecular motion of the rotating units is restricted due to steric hindrance, which opens the radiative pathway, endowing the AIE fluorogens with bright fluorescence.6 On the basis of the restriction of the intramolecular rotation (RIR) mechanism, the AIE fluorogens provide a simple and effective strategy to develop molecular probes that are able to specifically light up upon interaction with biomolecules in solutions and in cells with high signal-to-background ratios.7 On the other hand, the RIR mechanism limits the applications of AIE fluorogens in the development of nanoparticle-based fluorescent light-up probes, as the confinement of AIE fluorogens in the nanoparticles leads to high background fluorescence. However, it is well known that nanoparticle probes can be easily internalized into cells through endocytosis.8 More importantly, as compared to molecular probes, nanoparticle probes can be passively targeted to tumor tissues via an enhanced permeability and retention (EPR) effect, which enables the probe to achieve longer blood circulation with lower toxicity to normal tissues.9 Despite the great advantages, it is technically challenging to develop AIE fluorescent light-up nanoparticle probes that can specifically light up in the presence of analytes.
In this contribution, we report a rational design and synthesis of a surface charge-switchable light-up nanoparticle probe with AIE signatures for targeted imaging and selective suppression of cancer cells. The nanoparticle probe (Net-TPS-PEI-DMA, Scheme 1) is composed of an AIE fluorogen (TPS) and a pH-responsive charge-reversible polymer. Net-TPS-PEI-DMA is negatively charged and nearly non-emissive under physiological conditions (pH = 7.4). Upon exposure to a tumor acidic microenvironment (pHe = 6.5), the surface charge of the nanoparticle probe switches to positive. Electrostatic interactions between the positively charged nanoparticle residue and the negatively charged cell membranes or cell components not only promote its cellular uptake, but also activate the fluorescence of TPS, achieving targeted cancer cellular imaging in a high contrast and specific manner. The fluorescence of the nanoparticle probes is also observed in tumor tissues in vivo. Moreover, it is found that the nanoparticle probe could also suppress the cancer cells. To the best of our knowledge, this is the first report on synthesis and application of AIE fluorescent light-up nanoparticle probes, which will inspire more exciting research in the fields of AIE and bioimaging.
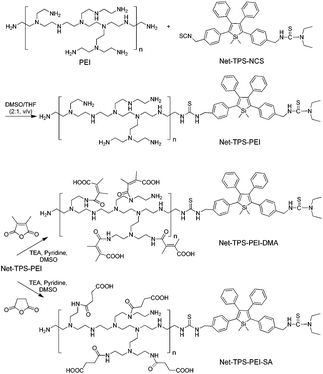
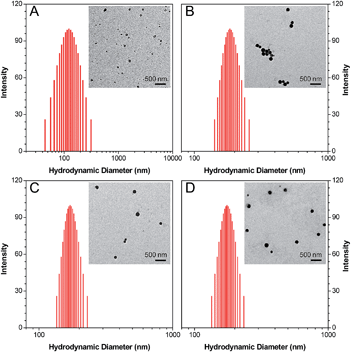
3-{4-[5-(4-(Isothiocyanatomethyl)phenyl)-1,1-dimethyl-3,4-diphenyl-silolyl]benzyl}-1,1-diethyl-thiourea (Net-TPS-NCS) was first synthesized in 63% yield. A detailed synthetic route and characterization (1H and 13C NMR and HRMS spectra) are shown in Scheme S1, Fig. S1 and S2 in the ESI.† Net-TPS-NCS was then conjugated to polyethyleneimine (PEI) to afford Net-TPS-PEI (Scheme 1). The product was characterized by 1H NMR (Fig. S3 in the ESI†). It is estimated from the 1H NMR spectrum that ∼4.5 of Net-TPS-NCS units were conjugated to one PEI chain on average. Net-TPS-PEI was further reacted with 2,3-dimethylmaleic anhydride (DMA) to convert the residual primary and secondary amines in PEI into amides, yielding Net-TPS-PEI-DMA (Scheme 1). The structure of Net-TPS-PEI-DMA was characterized by 1H NMR (Fig. S4 in the ESI†) and there were approximately 660 molecules of DMA on one PEI chain on average. As a control probe, Net-TPS-PEI-SA was also synthesized by replacing DMA with succinic anhydride (SA) to react with Net-TPS-PEI using the same reaction conditions as those used for Net-TPS-PEI-DMA. The 1H NMR spectrum of Net-TPS-PEI-SA is shown in Fig. S5 in the ESI.† It is noteworthy that the amide bonds formed between the primary or secondary amine and DMA are cleavable under a tumor acidic microenvironment at pHe ∼6.5 (Fig. S6 in the ESI†),10 while the amide bonds formed between amines and SA cannot be hydrolysed at this pH. Laser light scattering (LLS) results suggest that the average hydrodynamic diameters of Net-TPS-NCS, Net-TPS-PEI, Net-TPS-PEI-DMA and Net-TPS-PEI-SA are approximately 123, 184, 171 and 175 nm, respectively, in phosphate buffered saline (PBS) buffer at pH 7.4 (Fig. 1). Additionally, transmission electron microscopy (TEM) observations indicate that Net-TPS-PEI, Net-TPS-PEI-DMA and Net-TPS-PEI-SA are all well-dispersed and spherical in shape with uniform sizes (inset images in Fig. 1). In comparison, Net-TPS-NCS exhibits a relatively irregular spherical morphology with a smaller size and broader size distribution. These results reveal that all the AIE probes in this study are of nanosize in aqueous media, which are different from the well-established AIE-active molecular probes.7 The proposed mechanism of Net-TPS-PEI-DMA and Net-TPS-PEI-SA nanoparticle formation is as follows. As Net-TPS-PEI-DMA and Net-TPS-PEI-SA are amphiphilic molecules, the hydrophobic domains of the Net-TPS-PEI-DMA or Net-TPS-PEI-SA molecules tend to be entangled with each other, acting as the interior of the nanoparticles. The negatively charged carboxylic groups of the molecules are exposed to water due to their hydrophilic properties, which serve as the outer layers to stabilize the nanoparticles.
To verify that Net-TPS-PEI-DMA could switch its surface properties in response to pH through cleavage of the amide bonds, we investigated the surface zeta potential changes of the nanoparticle probe over time in buffers at pH 6.5 and 7.4. As shown in Fig. 2A, upon incubation of Net-TPS-PEI-DMA in buffer at pH 6.5, the zeta potential of the suspension is negative (−9.6 ± 2.8 mV). The zeta potential increases quickly as the time elapses, which reaches a plateau with a positive zeta potential within 30 min (5.9 ± 1.7 mV). In comparison, Net-TPS-PEI-DMA reveals a constant negative surface charge in buffer at pH 7.4 even after 2 h incubation (Fig. 2B). As control experiments, anionic Net-TPS-PEI-SA and cationic Net-TPS-PEI show a negligible change in surface charges in both pH 6.5 and 7.4 buffers over time. These results indicate that acid-responsive cleavage of the amide bonds could only occur in Net-TPS-PEI-DMA in a tumor acidic extracellular microenvironment.
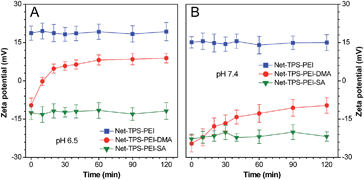 | ||
| Fig. 2 Zeta potential changes of the nanoparticle probes after incubation in buffers at (A) pH 6.5 and (B) pH 7.4 over time. | ||
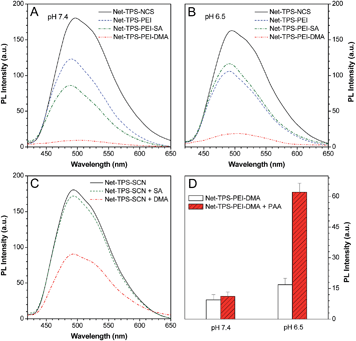
The optical properties of Net-TPS-NCS, Net-TPS-PEI, Net-TPS-PEI-DMA and Net-TPS-PEI-SA nanoprobes incubated in PBS buffer at pH 7.4 and 6.5 for 30 min were studied. All the nanoparticle probes were tested at the same concentration based on 5 μM TPS. As shown in Fig. S7A in the ESI,† in PBS buffer at pH 7.4, the Net-TPS-PEI, Net-TPS-PEI-DMA and Net-TPS-PEI-SA nanoparticle probes show similar absorption spectra with a maximum centered at ∼360 nm (similar to the absorption maximum of Net-TPS-NCS in THF), which is blue-shifted as compared to that of Net-TPS-NCS (∼375 nm). The difference is due to the Net-TPS-NCS aggregation formation in the aqueous media, while it is relatively well dispersed in the PEI matrices due to the small amount of the dye used for conjugation. Additionally, the absorption spectra of the four nanoparticle probes do not change upon incubation in the buffer at pH 6.5 (Fig. S7B in the ESI†). The photoluminescence (PL) spectra of the nanoparticle probes incubated in PBS buffers at pH 7.4 and 6.5 are depicted in Fig. 3A and B, respectively. Net-TPS-NCS emits the strongest fluorescence at both pH (with a quantum yield (Φ) of ∼18%), whereas the fluorescence intensity of Net-TPS-PEI is around 65% to that of Net-TPS-NCS at both pH. This fluorescence decrement should be due to the highly positive charges of PEI, which reduce the aggregation degree of TPS as compared to the neutral Net-TPS-NCS itself in water. Moreover, Net-TPS-PEI-SA shows slightly different fluorescence intensity as compared to that of Net-TPS-PEI at pH 7.4 and 6.5. Despite the high surface charges, both Net-TPS-PEI and Net-TPS-PEI-SA show relatively bright fluorescence, and they exist as nanoparticles in aqueous media, rather than as molecular species.
Although Net-TPS-PEI-DMA has a size on the nanoscale (∼171 nm), it shows much weaker fluorescence than TPS-PEI-SA in PBS buffer at pH 7.4 with a quite low Φ of 0.85% (Fig. 3A). To understand this unique observation, we incubated DMA or SA in sodium hydroxide solution (pH 8.5) for 12 h to yield the corresponding acids, and the resulting solutions were subsequently added into the Net-TPS-NCS suspension in PBS buffer. As shown in Fig. 3C, the fluorescence intensity of Net-TPS-NCS does not change with addition of the hydrolyzed SA. On the other hand, the Net-TPS-NCS fluorescence is significantly weakened upon addition of the hydrolyzed DMA. As DMA has a similar chemical structure to SA, this result suggests that the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C in DMA can quench the TPS fluorescence, making Net-TPS-PEI-DMA a pH-responsive light-up nanoparticle probe. Therefore, DMA plays dual roles in the light-up nanoprobe design: firstly, it responds to an acidic environment; secondly, it serves as a quencher to quench the TPS fluorescence via an exciton annihilation process associated with the n–π electronic conjugation of the C
C in DMA can quench the TPS fluorescence, making Net-TPS-PEI-DMA a pH-responsive light-up nanoparticle probe. Therefore, DMA plays dual roles in the light-up nanoprobe design: firstly, it responds to an acidic environment; secondly, it serves as a quencher to quench the TPS fluorescence via an exciton annihilation process associated with the n–π electronic conjugation of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C group.11 Upon incubation of Net-TPS-PEI-DMA in PBS buffer at pH 6.5 for 30 min, the fluorescence of the probe increases by ∼1-fold as compared to that at pH 7.4 (Fig. 3B). This result reveals that only the cleavage of amide bonds cannot significantly switch on the nanoparticle fluorescence, as the released C
C group.11 Upon incubation of Net-TPS-PEI-DMA in PBS buffer at pH 6.5 for 30 min, the fluorescence of the probe increases by ∼1-fold as compared to that at pH 7.4 (Fig. 3B). This result reveals that only the cleavage of amide bonds cannot significantly switch on the nanoparticle fluorescence, as the released C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C groups can still act as a quencher in the system. When anionic molecules, such as poly(acrylic acid) (PAA) was added into Net-TPS-PEI-DMA in PBS buffer at pH 6.5, strong fluorescence was observed (Φ = 6.6%; Fig. 3D). The fluorescence enhancement is because the electrostatic interactions between the positively charged cleavage residues of Net-TPS-PEI-DMA and the negatively charged PAA lead to formation of large aggregates (∼360 nm), which restrict the rotations of the phenyl rings in TPS and activate the emission of the probe. In contrast, addition of PAA into the solution of Net-TPS-PEI-DMA in buffer at pH 7.4 shows almost no fluorescence change.
C groups can still act as a quencher in the system. When anionic molecules, such as poly(acrylic acid) (PAA) was added into Net-TPS-PEI-DMA in PBS buffer at pH 6.5, strong fluorescence was observed (Φ = 6.6%; Fig. 3D). The fluorescence enhancement is because the electrostatic interactions between the positively charged cleavage residues of Net-TPS-PEI-DMA and the negatively charged PAA lead to formation of large aggregates (∼360 nm), which restrict the rotations of the phenyl rings in TPS and activate the emission of the probe. In contrast, addition of PAA into the solution of Net-TPS-PEI-DMA in buffer at pH 7.4 shows almost no fluorescence change.
As cell membranes are generally negatively charged, we hypothesized that the charge-reversal of Net-TPS-PEI-DMA in a tumor extracellular acidic microenvironment could not only enhance its internalization by cancer cells, but also turn-on its fluorescence due to the electrostatic interactions between the positively charged residues of Net-TPS-PEI-DMA and negatively charged cancer cell membranes or cell components (Scheme 2).
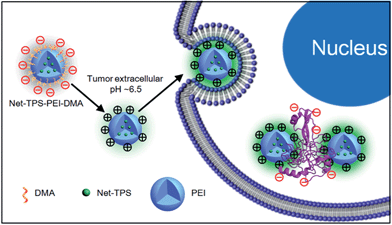 | ||
| Scheme 2 Schematic illustration of Net-TPS-PEI-DMA as a pH-responsive light-up nanoparticle probe for targeted cancer cell imaging. | ||
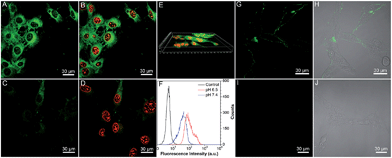
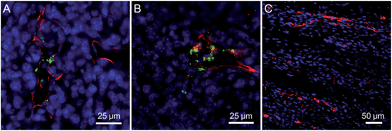
To verify our hypothesis, the application of Net-TPS-PEI-DMA for targeted cancer cell imaging was investigated using confocal laser scanning microscopy (CLSM). After incubation with Net-TPS-PEI-DMA (5 μM based on TPS) in the culture medium at pH 6.5 and 7.4 for 1 h at 37 °C, respectively, MCF-7 breast cancer cells were fixed and the cell nuclei were stained with propidium iodide (PI). As shown in Fig. 4A and B, intense green fluorescence is observed inside the MCF-7 cancer cells at pH 6.5, while much weaker fluorescence is observed at pH 7.4 (Fig. 4C and D). The 3D confocal image of MCF-7 cells incubated with Net-TPS-PEI-DMA at pH 6.5 reveals that the fluorescence is mainly from the cell cytoplasm (Fig. 4E). Additionally, the flow cytometry data indicate that the average fluorescence intensity of each cell incubated with Net-TPS-PEI-DMA at pH 6.5 is ∼3.0-fold higher as compared to that at pH 7.4 (Fig. 4F). These results reveal that Net-TPS-PEI-DMA can serve as a light-up nanoparticle probe for targeted cancer cell imaging.
We next treated the MCF-7 cancer cells with Net-TPS-PEI-DMA at 4 °C in culture medium at pH 7.4 and 6.5, respectively. There is obvious green fluorescence distributed on the MCF-7 cell membrane upon incubation at pH 6.5 owing to the electrostatic interaction between cleaved Net-TPS-PEI-DMA residues and the cell membrane (Fig. 4G and H), whereas almost no visible signal is observed at pH 7.4 (Fig. 4I and J). This result further verifies the surface charge switchable as well as light-up properties of Net-TPS-PEI-DMA nanoparticles. Moreover, as the low incubation temperature (4 °C) can suppress energy-dependent endocytosis,12 the results also suggest that the cleaved Net-TPS-PEI-DMA residues are internalized into cancer cells through the endocytosis mechanism.
The application of the Net-TPS-PEI-DMA nanoparticle probe in in vivo tumor imaging was also studied. In this experiment, 4T1 breast cancer cells were subcutaneously inoculated into the right flank of the mice, establishing the tumor-bearing model animals. After intravenous administration of the Net-TPS-PEI-DMA nanoparticle probe for 2 h and 6 h, respectively, the mice were sacrificed and the tumor as well as the muscle in the mouse leg were sliced for vasculature staining and CLSM imaging. As depicted in Fig. 5A and B, the CLSM images from the tumor slices indicate that bright green fluorescent dots from Net-TPS-PEI-DMA nanoparticle aggregation are located close to the tumor blood vessels (red fluorescence) and many are distributed in the tumor cells (blue fluorescence refers to cell nuclei). These results demonstrate that Net-TPS-PEI-DMA nanoparticles can be accumulated in the tumor tissues from blood circulation and turn on their fluorescence in the tumor acidic extracellular microenvironment. In contrast, no fluorescence is detected in the muscle tissues with normal physiological conditions (Fig. 5C), demonstrating the capability of the Net-TPS-PEI-DMA nanoparticle probe in in vivo tumor imaging.
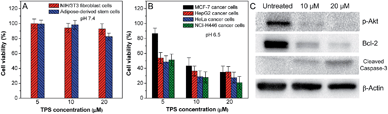
The cytotoxicities of the nanoparticle probes against normal as well as cancer cells were investigated after 24 h incubation using a MTT cell-viability assay. As shown in Fig. S8 in the ESI,† Net-TPS-PEI shows relatively high cytotoxicity to both normal NIH/3T3 fibroblast cells at pH 7.4 and MCF-7 cancer cells at pH 6.5 at concentrations of 5, 10 and 20 μM based on TPS. This result is in accordance with the reported cytotoxicity of PEI-based materials, which is ascribed to the highly positive charges of PEI.13 After conjugation with DMA, the anionic Net-TPS-PEI-DMA shows no obvious cytotoxicity to normal cell lines such as NIH/3T3 fibroblast cells and adipose-derived stem cells at pH 7.4 at concentrations of 5, 10 and 20 μM based on TPS (Fig. 6A). However, when Net-TPS-PEI-DMA was incubated at pH 6.5, the probe switches to positive charge, and the resulting probe shows significant cytotoxicity to four different cancer cell lines including MCF-7 breast cancer cells, human hepatocellular carcinoma cells (HepG2), human cervical cancer cells (HeLa) and human lung cancer cells (NCI-H446) at the tested concentrations (Fig. 6B). These results reveal that Net-TPS-PEI-DMA could be used as an anticancer drug to selectively kill the cancer cells in the tumor acidic extracellular microenvironment.
In order to understand the possible mechanism of the anticancer effect of the nanoparticle probes, western blot analysis was performed upon treatment of NCI-H446 cancer cells with Net-TPS-PEI-DMA at pH 6.5. As shown in Fig. 6C, it is found that the cleaved Net-TPS-PEI-DMA residues suppress the expression of the activated form of Akt proteins (phosphate-Akt), down-regulate the expression of Bcl-2 (a protective protein against apoptosis), and induce activation of caspase-3 proteins (a key mediator of cell apoptosis). This result elucidates that the cleaved Net-TPS-PEI-DMA residues result in the cytotoxicity of cancer cells by inhibition of the Akt pathway, which triggers the apoptotic cascade.14
In summary, we report a light-up nanoparticle probe of Net-TPS-PEI-DMA with AIE features that responds to a tumor acidic extracellular microenvironment. The nanoparticle probe is almost non-fluorescent and carries negative charges under physiological conditions. Under acidic conditions (i.e., pH = 6.5), the surface charges of Net-TPS-PEI-DMA switch to positive, which endow the nanoparticle probe with the abilities to be internalized into the cancer cells as well as significantly turn on its fluorescence. The in vitro and in vivo experiments demonstrate that Net-TPS-PEI-DMA can significantly light up the cancer cells, which is able to serve as an efficient pH-responsive light-up nanoparticle probe for targeted cancer cell imaging and in vivo tumor imaging. The cytotoxicity results also reveal that Net-TPS-PEI-DMA shows low cytotoxicity to normal cells and relatively high cytotoxicity to cancer cells. The high cytotoxicity against cancer cells is also demonstrated to be through the suppression of the Akt pathway and activation of the apoptotic pathway.
Acknowledgements
We thank the Singapore National Research Foundation (R-279-000-390-281), Singapore-MIT Alliance for Research and Technology (SMART) Innovation Grant (R279-000-378-592), the Institute of Materials Research and Engineering of A-Star, Singapore (IMRE/14-8P1110), the National University of Singapore (R279-000-415-112), the Research Grants Council of Hong Kong (HKUST2/CRF/10 and N_HKUST620/11), Guangdong Innovative Research Team Program of China (20110C0105067115) and the NSFC (81301311) for financial support.Notes and references
- (a) M. D. Shults and B. Imperiali, J. Am. Chem. Soc., 2003, 125, 14248 CrossRef CAS PubMed; (b) H. Kobayashi, M. Ogawa, R. Alford, P. L. Choyke and Y. Urano, Chem. Rev., 2010, 110, 2620 CrossRef CAS PubMed; (c) I. C. Sun, D. K. Eun, H. Koo, C. Y. Ko, H. S. Kim, D. K. Yi, K. Choi, I. C. Kwon, K. Kim and C. H. Ahn, Angew. Chem., Int. Ed., 2011, 50, 9348 CrossRef CAS PubMed; (d) Y. D. Zhuang, P. Y. Chiang, C. W. Wang and K. T. Tan, Angew. Chem., Int. Ed., 2013, 52, 8124 CrossRef CAS PubMed; (e) H. M. Wang, J. Liu, A. T. Han, N. N. Xiao, Z. S. Xue, G. Wang, J. F. Long, D. L. Kong, B. Liu, Z. M. Yang and D. Ding, ACS Nano, 2014, 8, 1475 CrossRef CAS PubMed.
- (a) D. Schmaljohann, Adv. Drug Delivery Rev., 2006, 58, 1655 CrossRef CAS PubMed; (b) Y. Y. Yuan, C. Q. Mao, X. J. Du, J. Z. Du, F. Wang and J. Wang, Adv. Mater., 2012, 24, 5476 CrossRef CAS PubMed; (c) M. Kamimura, J. O. Kim, A. V. Kabanov, T. K. Bronich and Y. Nagasaki, J. Controlled Release, 2012, 160, 486 CrossRef CAS PubMed; (d) W. She, N. Li, K. Luo, C. Guo, G. Wang, Y. Geng and Z. Gu, Biomaterials, 2013, 34, 2252 CrossRef CAS PubMed.
- (a) L. Gerweck and K. Seetharaman, Cancer Res., 1996, 56, 1194 CAS; (b) R. A. Cardone, V. Casavola and S. J. Reshkin, Nat. Rev. Cancer, 2005, 5, 786 CrossRef CAS PubMed.
- (a) Y. Wu, W. Zhang, J. Li and Y. Zhang, Am. J. Nucl. Med. Mol. Imaging, 2013, 3, 1 CAS; (b) Y. Zhao, T. Ji, H. Wang, S. Li, Y. Zhao and G. Nie, J. Controlled Release, 2014, 177, 11 CrossRef CAS PubMed; (c) Y. Y. Yuan, D. Ding, K. Li, J. Liu and B. Liu, Small, 2014, 10, 1967 CrossRef CAS PubMed.
- (a) J. D. Luo, Z. L. Xie, J. W. Y. Lam, L. Cheng, H. Y. Chen, C. F. Qiu, H. S. Kwok, X. W. Zhan, Y. Q. Liu, D. B. Zhu and B. Z. Tang, Chem. Commun., 2001, 1740 RSC; (b) Y. N. Hong, J. W. Y. Lam and B. Z. Tang, Chem. Commun., 2009, 4332 RSC; (c) Y. N. Hong, J. W. Y. Lam and B. Z. Tang, Chem. Soc. Rev., 2011, 40, 5361 RSC; (d) D. Ding, K. Li, B. Liu and B. Z. Tang, Acc. Chem. Res., 2013, 46, 2441 CrossRef CAS PubMed.
- (a) M. Wang, G. X. Zhang, D. Q. Zhang, D. B. Zhu and B. Z. Tang, J. Mater. Chem., 2010, 20, 1858 RSC; (b) D. Ding, C. C. Goh, G. X. Feng, Z. J. Zhao, J. Liu, R. R. Liu, N. Tomczak, J. L. Geng, B. Z. Tang, L. G. Ng and B. Liu, Adv. Mater., 2013, 25, 6083 CrossRef CAS PubMed; (c) K. Li and B. Liu, Chem. Soc. Rev., 2014, 43, 6570 RSC; (d) X. Q. Zhang, Z. G. Chi, Y. Zhang, S. W. Liu and J. R. Xu, J. Mater. Chem. C, 2013, 1, 3376 RSC; (e) Z. G. Chi, X. Q. Zhang, B. J. Xu, X. Zhou, C. P. Ma, Y. Zhang, S. W. Liu and J. R. Xu, Chem. Soc. Rev., 2012, 41, 3878 RSC.
- (a) H. B. Shi, J. Z. Liu, J. L. Geng, B. Z. Tang and B. Liu, J. Am. Chem. Soc., 2012, 134, 9569 CrossRef CAS PubMed; (b) H. B. Shi, R. Kowk, J. Z. Liu, B. G. Xing, B. Z. Tang and B. Liu, J. Am. Chem. Soc., 2012, 134, 17972 CrossRef CAS PubMed; (c) Y. Y. Yuan, R. Kwok, B. Z. Tang and B. Liu, J. Am. Chem. Soc., 2014, 136, 2546 CrossRef CAS PubMed; (d) Y. Y. Huang, F. Hu, R. Zhao, G. X. Zhang, H. Yang and D. Q. Zhang, Chem.–Eur. J., 2014, 20, 158 CrossRef CAS; (e) D. Ding, J. Liang, R. T. K. Kwok, M. Gao, G. X. Feng, Y. Y. Yuan, B. Z. Tang and B. Liu, J. Mater. Chem. B, 2014, 2, 231 RSC; (f) X. J. Wang, H. Liu, J. W. Li, K. G. Ding, Z. L. Lv, Y. G. Yang, H. Chen and X. M. Li, Chem.–Asian J., 2014, 9, 784 CrossRef CAS PubMed; (g) X. Q. Zhang, X. Y. Zhang, L. Tao, Z. G. Chi, J. R. Xu and Y. Wei, J. Mater. Chem. B, 2014, 2, 4398 RSC.
- W. Jiang, B. Y. S. Kim, J. T. Rutka and W. C. W. Chan, Nat. Nanotechnol., 2008, 3, 145 CrossRef CAS PubMed.
- O. C. Farokhzad and R. Langer, ACS Nano, 2009, 3, 16 CrossRef CAS PubMed.
- (a) P. S. Xu, E. A. Van Kirk, Y. H. Zhan, W. J. Murdoch, M. Radosz and Y. Q. Shen, Angew. Chem., Int. Ed., 2007, 46, 4999 CrossRef CAS PubMed; (b) Z. X. Zhou, Y. Q. Shen, J. B. Tang, M. H. Fan, E. A. Van Kirk, W. J. Murdoch and M. Radosz, Adv. Funct. Mater., 2009, 19, 3580 CrossRef CAS; (c) J. Z. Du, X. J. Du, C. Q. Mao and J. Wang, J. Am. Chem. Soc., 2011, 133, 17560 CrossRef CAS PubMed.
- Y. Liu, Y. Yu, J. W. Y. Lam, Y. Hong, M. Faisal, W. Z. Yuan and B. Z. Tang, Chem.–Eur. J., 2010, 16, 8433 CrossRef CAS PubMed.
- P. Watson, A. T. Jones and D. J. Stephens, Adv. Drug Delivery Rev., 2005, 57, 43 CrossRef CAS PubMed.
- (a) S. M. Moghimi, P. Symonds, J. C. Murray, A. C. Hunter, G. Debska and A. Szewczyk, Mol. Ther., 2005, 11, 990 CrossRef CAS PubMed; (b) S. Werth, B. Urban-Klein, L. Dai, S. Höbel, M. Grzelinski, U. Bakowsky, F. Czubayko and A. Aigner, J. Controlled Release, 2006, 112, 257 CrossRef CAS PubMed.
- (a) X. Li, X. Lu, H. Xu, Z. Zhu, H. Yin, X. Qian, R. Li, X. Jiang and B. Liu, Mol. Pharmaceutics, 2012, 9, 222 CrossRef CAS PubMed; (b) Y. F. Wang, C. Y. Chen, S. F. Chung, Y. H. Chiou and H. R. Lo, Cancer Chemother. Pharmacol., 2004, 54, 322 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) is available. See DOI: 10.1039/c4mh00164h |
| ‡ Both authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2015 |