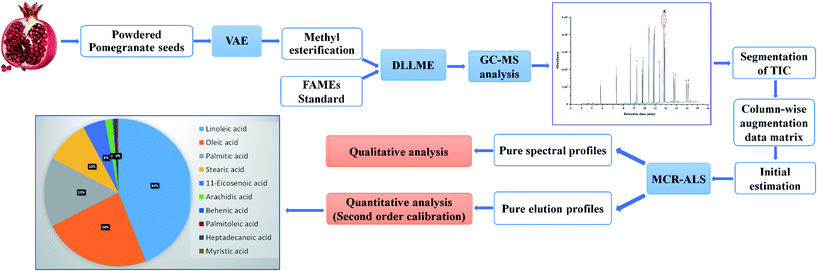
Second-order calibration for the determination of fatty acids in pomegranate seeds by vortex-assisted extraction-dispersive liquid–liquid micro-extraction and gas chromatography-mass spectrometry
Mohammad Ahmadvanda,
Hassan Sereshtia and
Hadi Parastar*b
aDepartment of Chemistry, Faculty of Science, University of Tehran, Tehran, Iran
bDepartment of Chemistry, Sharif University of Technology, P.O. Box 11155-3516, Tehran, Iran. E-mail: h.parastar@sharif.edu; h.parastar@gmail.com; Fax: +98-21-66005718; Tel: +98-21-66165306
First published on 22nd December 2014
Abstract
Multivariate curve resolution-alternating least squares (MCR-ALS) as a second-order calibration algorithm was proposed for the simultaneous analysis of eighteen fatty acid methyl esters (FAMEs) in a standard mixture and pomegranate seed sample using vortex-assisted extraction-dispersive liquid–liquid microextraction (VAE-DLLME), followed by gas chromatography-mass spectrometry (GC-MS). The chemometric resolution, identification and quantification of the target FAMEs in the standard mixture and real sample (i.e., the pomegranate seed) were carried out successfully in the presence of some uncalibrated interferences. The lack of fit (LOF) and reverse match factor (RMF) were used for the evaluation of the MCR-ALS results of the calibration samples. The LOF (%) and RMF values were in the ranges of 7.24–24.36 and 708–977, respectively. In addition, the regression coefficients (R2) and relative errors (REs, %) of the calibration curves of different FAs were in the satisfactory range of 0.9934–0.9989 and 3.70–7.45, respectively. The application of the proposed strategy to the pomegranate seed extract showed that linoleic acid, oleic acid, palmitic acid, stearic acid and cis-11-eicosenoic acid were the main fatty acids in the pomegranate seed with concentrations of 9098.0, 4873.0, 3147.0, 1960.0 and 1019.0 mg kg−1, respectively, and relative standard deviations (RSD, %) between 0.15 and 11.57. It is concluded that MCR-ALS combined with VAE-DLLME-GC-MS is a fast and simple strategy for the qualitative and quantitative analysis of complex samples such as natural products.
1 Introduction
The determination and quantification of fatty acids (FAs) in different sample matrices, including blue crab,1 fish oil,2 beer,3 alcoholic beverages and tobaccos,4 raw spirits5 and wild mushroom,6 have attracted the attention of health and nutrition researchers owing to their significant role in biological tissues. Pomegranate is one of the native fruits of Iran with an annual production of about 700![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 tonnes. Regarding the high content of FAs and other bioactive components, pomegranate seeds can be used as a rich source of FAs. Hence, the extraction and characterization of pomegranate seed compounds have attracted considerable attention in recent years.7,8 The amount of some FAs in pomegranate seeds has already been obtained by different extraction methods, such as Soxhlet extraction, supercritical fluid extraction (SFE) and ultrasonic-assisted solvent extraction (UASE).9–11 However, quantification of the FAs has always been a challenging task, due to insufficient sensitivity, the presence of a variety of polyunsaturated fatty acids (PUFA) isomers, and due to the co-eluted components.12,13
000 tonnes. Regarding the high content of FAs and other bioactive components, pomegranate seeds can be used as a rich source of FAs. Hence, the extraction and characterization of pomegranate seed compounds have attracted considerable attention in recent years.7,8 The amount of some FAs in pomegranate seeds has already been obtained by different extraction methods, such as Soxhlet extraction, supercritical fluid extraction (SFE) and ultrasonic-assisted solvent extraction (UASE).9–11 However, quantification of the FAs has always been a challenging task, due to insufficient sensitivity, the presence of a variety of polyunsaturated fatty acids (PUFA) isomers, and due to the co-eluted components.12,13
Among different extraction techniques, dispersive liquid–liquid microextraction (DLLME) was introduced by Assadi and co-workers in 2006 as a simple and efficient extraction/preconcentration technique.14 In general, the DLLME technique consists of two simple steps: (1) rapid injection of an appropriate mixture of the extractor and disperser solvent in an aqueous sample containing the sought analyte(s); and (2) centrifugation of the cloudy solution to separate the phases. In the first step, a stable cloudy solution (containing very fine droplets of extraction solvent dispersed in the aqueous phase) is formed. Owing to the large surface area between the extractor solvent and the aqueous sample, an equilibrium state is quickly achieved. In the second step, after centrifugation, the organic phase is separated and is analyzed by an appropriate instrumental technique.15 Although, less than one decade has elapsed since the introduction of DLLME, it has been frequently used for the extraction and preconcentration of a broad range of organic and inorganic compounds from different sample matrices, such as from foods, and environmental and biological samples.16–18
Gas chromatography coupled to mass spectrometry (GC-MS) is the most frequently used instrument for the identification and quantification of FAs in animal and plant samples.19 The derivatization of the FAs to their methyl esters (FAMEs) is a primary step in GC separation, in order to increase the resolution and sensitivity of the FAs' analysis. Very often, overlapping regions in the GC profiles of the FAMEs (especially for unsaturated FAMEs) can be observed due to the complexity of FAMEs matrices. Therefore, finding proper conditions for the comprehensive separation, identification and quantification of saturated and unsaturated FAMEs, particularly in the presence of other interferences, is a very difficult task.
Fortunately, over the past few decades, second-order calibration methods have attracted great attention from analytical chemists, due to their great achievements in improving the sensitivity, increasing the selectivity, and in the modelling of the analyte contribution in the presence of uncalibrated interferences.20 Various second-order multivariate calibration algorithms have been developed in the recent decade, and have been reviewed in the literature.21–23
Among different second-order calibration methods, multivariate curve resolution-alternating least squares (MCR-ALS) and parallel factor analysis 2 (PARAFAC2)24 have been proposed and extensively used to resolve multiple pure responses and concentrations of components present in unknown mixtures. However, different studies have been done to show the superiority of MCR-ALS over PARAFAC2 when fundamental chromatographic problems exist.25 PARAFAC2 is a variant of the PARAFAC technique, and has been applied to the chromatographic data. Strict trilinearity in the PARAFAC2 model is not necessary, and it does not allow for significant shape changes in the elution peak across different runs (the extent of the allowed peak deformation in practice is still unknown). PARAFAC2 allows for analysis of data when there are moderate elution time shifts in the chromatographic dimensions due to, for example, temperature programming or misalignment across samples. However, PARAFAC2 is computationally more complex and expensive, and it does not allow for constraints (e.g. non-negativity and/or unimodality) in the chromatographic direction, and therefore negative values and multimodal peaks may appear in the results. Also, applying constraints selectively only to some selected components is not possible.
The MCR-ALS has been applied to data collected from multi-component liquid chromatography-mass spectrometry (LC-MS),26 gas chromatography-mass spectrometry (GC-MS),2 and high performance liquid chromatography-diode array detection (HPLC-DAD)27 and fluorescence excitation-emission matrices (EEM).28 In MCR-ALS, the measured analytical signals are assumed to follow a generalized bilinear additive model, and the contribution of each component to the measured signal depends on its concentration and on its spectral profile. MCR-ALS can also be used to obtain quantitative analytical information, because any type of constraints can be easily applied to the solutions.
In the present contribution, MCR-ALS was used as a second-order calibration algorithm for the identification and quantification of FAMEs in the standard mixture and in the pomegranate seed samples. For this purpose, the simple microextraction method of VAE-DLLME, followed by GC-MS, was carried out on a standard mixture of FAMEs. Then, the data were analyzed by the MCR-ALS algorithm to overcome the chromatographic problems, to build calibration curves and to obtain the analytical figures of merit. Finally, the pomegranate seed FAMEs were investigated as the test sample for evaluation of the proposed calibration method.
2. Experimental
2.1 Chemicals and reagents
The fatty acid methyl esters (FAMEs) mixture (Grain FAME mix. 10 mg mL−1 in CH2Cl2) was purchased from Sigma Aldrich (St. Louis, MO, USA). Pentadecanoic acid, chloroform, methanol and sodium chloride with purity higher than 99.0% were purchased from Merck (Darmstadt, Germany). Pure helium (99.999%) was obtained from the Roham gas company (Tehran, Iran, http://www.roham.com).2.2 Sample preparation: extraction, esterification and preconcentration of the FAs
Fresh pomegranates were obtained from a market in Karaj (Alborz province, Iran). The pomegranates were crushed and their seeds were separated from the other parts. The dewatered seeds were washed with tap water several times for the removal of residuals and were then dried at room temperature for 48 h. The dried seeds were powdered by a household grinder and sieved using a 60-mesh sieve (pore size 0.3 mm). The obtained powder was stored in a glass vessel at 4 °C for the subsequent analyses.A portion of the seed powder (0.3 g) was placed into a 5 mL screw cap glass test tube and then 2 mL methanol (extraction solvent) was added to it. Afterwards, the mixture was subjected to vortexing using a vortex-mixer (Velp Scientifica, Milan, Italy) for 3 min. After the extraction, the solid particles were separated from the mixture by centrifugation, and the supernatant was transferred to another test tube. Then, pentadecanoic acid was added to it as an internal standard (5 mg L−1). In the next step, a simple esterification procedure was performed by adding one drop of concentrated H2SO4 to 1 mL of the supernatant fixed in a water bath (at 70 °C for 60 min). Then, 0.5 mL of the esterified solution was placed into a test tube and 28 μL chloroform (preconcentration solvent) was added to it. Afterwards, the solution was injected rapidly into a conical bottom test tube containing 2 mL aqueous NaCl solution (5% w/v). Accordingly, a cloudy solution containing tiny droplets of chloroform dispersed into the aqueous phase was formed. In this step, the analytes were extracted from the aqueous sample into the chloroform droplets. Then, the organic extraction phase was separated by centrifugation at 4000 rpm for 3 min. Finally, 0.5 μL of the lower phase (chloroform phase) was injected into GC-MS using a 1.0 μL microsyringe.
2.3 GC-MS analysis
A 6890 GC system coupled with a 5973 network mass selective detector (Agilent Technologies, Santa Clara, CA, USA) and equipped with a HP5-MS capillary fused silica column (30 m length; 0.25 mm I.D.; 0.25 μm film thicknesses, methyl 5% phenyl polysiloxane) was used for analysis of the samples. The temperature program started at 50 °C for 1 min, and then increased at the rate of 20 °C min−1 to 270 °C, where it was then held for 3 min. The total GC run time was 15 min. In this regard, the chromatographic conditions were chosen to achieve a reasonable chromatographic resolution and a short analysis time. The carrier gas (helium, 99.999%) was maintained at a constant pressure of 36 psi with a flow rate of 1 mL min−1 (4 min solvent delay). The other operating conditions were as follows: injection volume, 0.5 μL; split ratio, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; interface temperature, 250 °C; and ion source temperature, 230 °C. Mass spectra were taken at 70 eV ionization energy and in the full scan mode. The scanned mass range was set at 50–450 m/z. An enhanced ChemStation software package (G1701 DA-MSD, Rev. D.00.01.27) was used for the data collection and analysis in the GC-MS.
20; interface temperature, 250 °C; and ion source temperature, 230 °C. Mass spectra were taken at 70 eV ionization energy and in the full scan mode. The scanned mass range was set at 50–450 m/z. An enhanced ChemStation software package (G1701 DA-MSD, Rev. D.00.01.27) was used for the data collection and analysis in the GC-MS.
2.4 Chemometric analysis
Hyphenated systems such as HPLC-DAD, LC-MS and GC-MS produce a large volume of data that can be stored in a rectangular data arrays called second-order data matrices. These mixed data matrices contain information from two different data directions (i.e. rows and columns) and can be decomposed into the contribution of the pure component profiles of the constituents, by means of a simple bilinear data decomposition, which is defined as follow:| X = CST + E | (1) |
Eqn (1) was solved for C and ST, using an iterative algorithm based on two constrained linear least squares steps. It required an initial estimation of the concentration, C, or of the spectra, ST, profiles, which could be easily obtained using different methods or from the ‘purest’ data samples or variables (e.g. simple-to-use interactive self-modelling mixture analysis (SIMPLISMA)29). Although, finding unique solutions of eqn (1) is impossible when only the information about the data matrix X is provided, the use of constraints (e.g. non-negativity or any other previously known property or constraint about the nature of the component profiles) can significantly decrease this indeterminacy, and eventually totally eliminate it. A more detailed discussion about the MCR-ALS method can be found in previous work in the literature.30,31
One of the most important features of MCR-ALS is its potential to be extended to the analysis of higher-order data. The extended MCR-ALS bilinear decomposition is as follow:
| Xaug = [X1;X2;…;Xk] = [C1;C2;…;Ck]ST + [E1;E2;…;Ek] = CaugST + Eaug | (2) |
The results from the MCR-ALS, when applied to the chromatographic data analysis, are the pure elution and spectral profiles of the constituents of the analyzed samples. The resolved spectral profiles can be used to identify these components by comparing the resolved spectra with those of the authentic standards or standard spectra in available libraries. On the other hand, to obtain quantitative chromatographic information, the areas of the resolved elution profiles can be exploited for the quantification, especially in the case of the simultaneous analysis of several chromatographic runs. One of the interesting aspects of the MCR-ALS model in this work is its potential to model the baseline/background contribution instead of its correction before the analysis. In addition, the elution time shift can be handled by MCR-ALS. In other words, the presence of elution time shifts cannot affect the MCR-ALS solutions, because of the bilinear model assumption.
2.5 Quantitative analyses
In order to prepare the calibration samples, the original standard of the FAMEs were diluted to 20, 50, 100, 250, 500 and 1000-fold of the original in methanol. Then, pentadecanoic acid methyl ester as an internal standard (with the final concentration of 5.0 mg L−1) was added to each solution, which were treated in accordance with the proposed procedure (see Section 2.2). The obtained total ion chromatogram (TIC) of the standards was divided into sixteen segments (based on the emerging chromatographic peaks of the standard FAMEs) for simplifying the calculations. The matrices for each analyte in different standards were arranged in a column-wise augmented data matrix. The augmented data matrix was analyzed using MCR-ALS under the application of non-negativity, unimodality and spectral normalization constraints. Also, SIMPLISMA was used to estimate the initial spectral profiles.Quantification was performed based on the summation of the peak areas of the resolved profiles by MCR-ALS. Thus, the relative quantitative information for one target compound can be directly derived from the comparison of the MCR-ALS-resolved elution profiles for different samples under the assumption of the linear relation between the relative peak areas of the resolved elution profiles and their relative concentrations. It should be pointed out that the overall volume integration (OVI) was considered as the analyte signal for the peak area calculation.32 The OVI is preferred to the total peak area, because all of the mass spectral intensities are taken into account in the calculation. The calculated OVI for each of the FAMEs was divided by the OVI of the internal standard (i.e. pentadecanoic acid methyl ester) and was used to build the calibration curves. Fig. 1 shows the general procedure used in this study.
Data analyses were performed on an Intel (R) Core (TM) 2 Duo-based DELL (Vostro) personal computer with a 2.50 GHz CPU and 4 GB RAM. All the calculations were carried out using MATLAB 7.10.0 (The Mathworks Inc., MA, USA). The MCR-ALS toolbox is freely available from the homepage of MCR at http://www.mcrals.info/. MCRC software33 was used for the data preprocessing and local rank analysis. The library searches and spectral matching of the resolved pure components were conducted on the NIST MS database.34
3. Results and discussion
3.1 Optimization of the extraction procedure
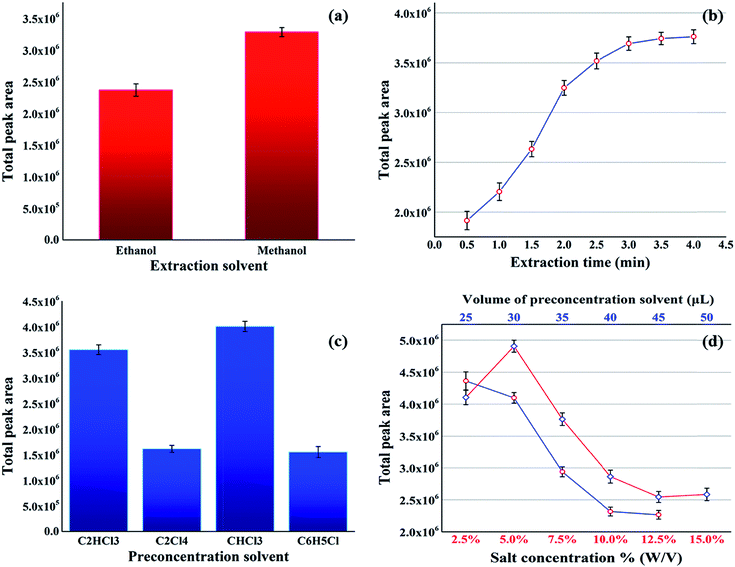
In order to achieve the maximum efficiency of the proposed method, the effective parameters, including type of extraction solvent, extraction time, type and volume of the preconcentration solvent, and salt concentration, were investigated and optimized. In addition, the total peak areas of the sought FAs were considered as the response for optimization.The extraction solvent was chosen based on its properties, such as ability to extract the sought analytes (FAs) and good miscibility with the organic preconcentration solvent and the aqueous sample solution (as it plays the role of disperser in DLLME step). Therefore, methanol and ethanol were tested as the potential extraction solvents in accordance with the proposed procedure in Section 2.2. The results showed that the maximum total peak area was produced using methanol (see Fig. 2a), thus this was used as the extraction solvent in the subsequent experiments.
The extraction time was the next parameter, and its influence was investigated in the range of 0.5–4 min. According to the results presented in Fig. 2b, 3 min was selected as the optimum extraction time for the subsequent experiments.
The appropriate preconcentration solvent was selected based on certain characteristics, such as extraction capability for the sought compounds, immiscibility with water, higher density than water, and suitability for gas chromatography analysis. Considering these properties, several solvents consisting of chloroform, chlorobenzene, trichloroethylene and tetrachloroethylene were evaluated for this purpose. As shown in Fig. 2c, among these solvents, the highest total peak area was generated using chloroform as the preconcentration solvent. Therefore, this was chosen as the most suitable preconcentration solvent in the proposed procedure. In the next step, the influence of the volume of the selected preconcentration solvent (chloroform) was studied in the range of 25–45 μL. An inspection of the results in Fig. 2d (blue line) shows that with the increasing volume of chloroform (extraction solvent), the response decreases due to the reduction in the concentration of the FAMEs. However, as the highest total peak areas were obtained in the region 25–30 μL, therefore, 28 μL was chosen as the optimized preconcentration solvent volume.
Finally, the effect of salt concentration on the preconcentration of the target analytes (FAs) was investigated in the range of 0–15% (w/v). Fig. 2d (red line) shows the increasing efficiency with the increasing salt concentration up to 5%. However, higher salt concentrations caused a decrease in the efficiency. The addition of salt in the first region (0–5%) decreased the solubility of analytes in the aqueous solution and enhanced the extraction efficiency by a salting-out effect. At concentrations above 5%, the increased viscosity of the aqueous solution overcame the salting-out effect, leading to a difficult mass transfer and low extraction efficiency.35
3.2 Resolution and quantification of FAMEs in the standard mixture solution
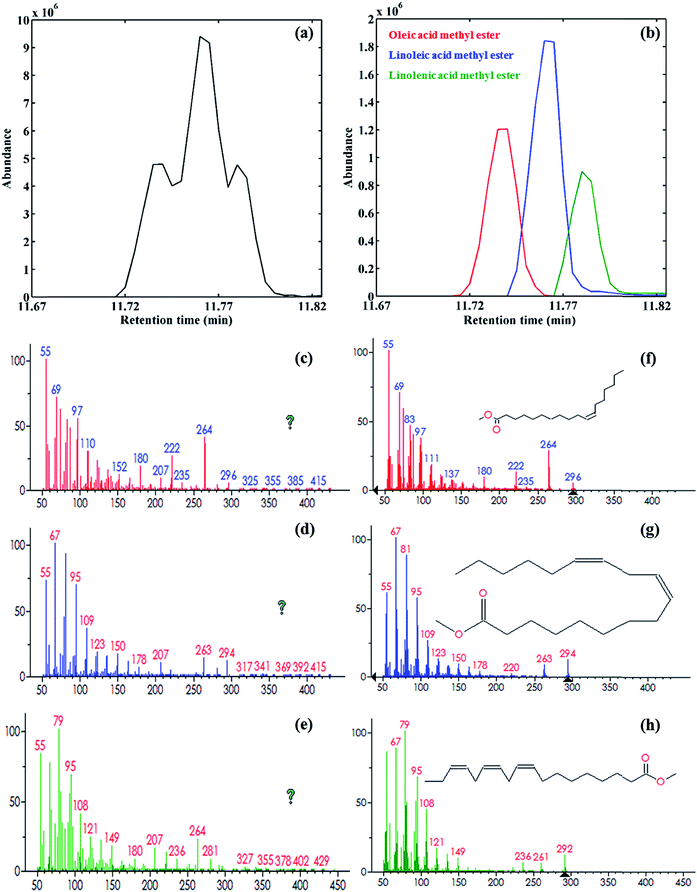
Under the optimized conditions, the standard FAMEs were extracted from the standard mixture, preconcentrated and then analyzed with GC-MS. The obtained chromatogram (see Fig. 3a) shows that most of the peaks are well separated, except for the region marked with ‘A’. In this region, three peaks related to the components 11, 12 and 13 overlap. The GC-MS TIC of the pomegranate seed extract depicted in Fig. 3b is very complex, consisting of a large number of peaks, together with different chromatographic problems, such as baseline/background contribution, low S/N signals and peak overlap. The black rectangles in Fig. 3b marked with ‘B’ (12.7–13.1 min) and ‘C’ (13.8–14.3 min) highlight two problematic regions in the TIC. These chromatographic regions will be analyzed to demonstrate the potential of the proposed strategy in this work.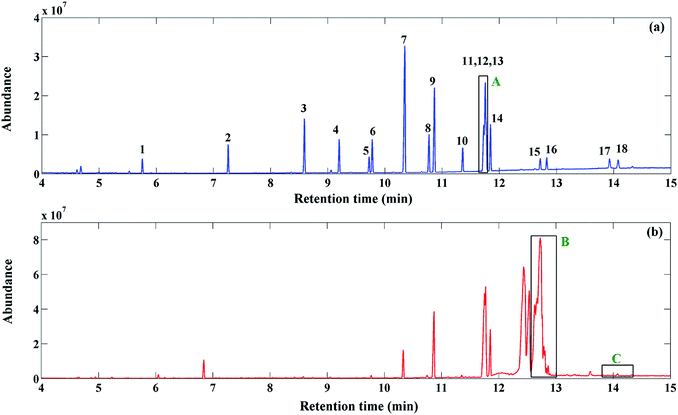 | ||
| Fig. 3 GC-MS TIC of FAMEs in (a) standard mixture and (b) pomegranate seed samples. The numbers are related to the components in Table 1. Regions A, B and C illustrate the three investigated problematic regions due to the co-eluted components, uncalibrated interferences and low S/N ratio, respectively. | ||
At first, the GC-MS data of the standard mixture samples of eighteen FAMEs were analyzed using the proposed strategy presented in Fig. 1. For this purpose, the second-order data obtained from the chromatographic regions containing the target analytes were exported to MATLAB, and then the GC-MS data for the different concentrations and replicates of the sought analytes were arranged in a column-wise augmented data matrix. The augmented data matrix was then analyzed using MCR-ALS under non-negativity, unimodality, spectral normalization and component correspondence constraints. In the analysis of the standard mixture samples, some of the components overlap. For example, region A in Fig. 3a, which is also shown in Fig. 4a, depicts a chromatographic region where some FAMEs are heavily overlapped in both the chromatographic and mass spectrometric dimensions. The singular values from the singular value decomposition (SVD)36 were used to find the number of chemical components in this region. As can be expected, three chemical components were recognized (the singular values for these three components were 1.53 × 107, 6.85 × 106 and 2.56 × 106, respectively). The SIMPLISMA was then used to obtain an initial estimate of the spectral profiles for these three components to start the ALS optimization. In the next step, MCR-ALS was applied under the application of non-negativity (to the chromatographic and spectral profiles), unimodality (to the chromatographic profiles), and normalization (to the spectral profiles) constraints. The MCR-ALS-resolved elution profiles for this chromatographic region are shown in Fig. 4b. The values for the lack of fit (LOF, %) and the explained variance (R2, %) were equal to 18.83% and 96.45%, respectively. The resolved mass spectral profiles for each component (see Fig. 4c–e) were matched with the stored mass spectra in the NIST MS library database, and from this, oleic acid methyl ester, linoleic acid methyl ester and linolenic acid methyl ester were respectively identified (see Fig. 4f–h).
The same procedure was used for the MCR-ALS resolution of all the other FAMEs, and their calibration curves were built up. Table 1 shows the retention times, the LOF, RMF, calibration equation, linear dynamic range, regression coefficient and relative errors (REs) of the calibration concentrations for the eighteen FAMEs. The two parameters of LOF and RMF were explored for evaluation of the MCR-ALS results, which were in the range of 7.24–24.36% and 708–977, respectively. It is important to note that in this study, MCR-ALS was performed by using raw data without any data preprocessing method, such as baseline correction or denoising. In addition, the R2 and REs were in the satisfactory ranges of 0.9934–0.9989 and 3.70–7.45%, respectively. All of these results confirm the validity of the proposed strategy.
| No. | tRa | Chemical name | Formula | LOFb (%) | RMFc | Calibration equation | Calibration range (mg L−1) | R2 (calibration) | REd (%) |
|---|---|---|---|---|---|---|---|---|---|
a Retention time (min).b 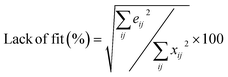 , where xij and eij are the elements of the matrices X and E, respectively.c Reverse match factor, the criterion of similarity between the resolved and library search mass spectra.d , where xij and eij are the elements of the matrices X and E, respectively.c Reverse match factor, the criterion of similarity between the resolved and library search mass spectra.d 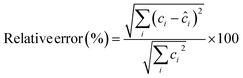 , where ci is the known concentration of standard i and ĉi is its calculated value using the calibration equation obtained from the overall volume integration (OVI) in TIC mode. , where ci is the known concentration of standard i and ĉi is its calculated value using the calibration equation obtained from the overall volume integration (OVI) in TIC mode. |
|||||||||
| 1 | 5.75 | Caprylic acid methyl ester | C9H18O2 | 7.25 | 969 | y = 0.0285x + 0.0139 | 0.19–9.5 | 0.9970 | 4.22 |
| 2 | 7.35 | Capric acid methyl ester | C11H22O2 | 8.87 | 971 | y = 0.0336x + 0.0239 | 0.32–16 | 0.9971 | 4.14 |
| 3 | 8.65 | Lauric acid methyl ester | C13H26O2 | 11.35 | 975 | y = 0.033x + 0.0577 | 0.64–32 | 0.9976 | 3.76 |
| 4 | 9.30 | Tridecanoic acid methyl ester | C14H28O2 | 8.99 | 964 | y = 0.0372x + 0.0358 | 0.32–16 | 0.9977 | 3.71 |
| 5 | 9.77 | Myristoleic acid methyl ester | C15H28O2 | 22.69 | 931 | y = 0.0401x + 0.0242 | 0.19–9.5 | 0.9955 | 5.10 |
| 6 | 9.84 | Myristic acid methyl ester | C15H30O2 | 10.93 | 955 | y = 0.0397x + 0.0339 | 0.32–16 | 0.9967 | 4.43 |
| 7 | 10.51 | Pentadecanoic acid methyl ester | C16H32O2 | 11.36 | 932 | Internal standard | — | — | — |
| 8 | 10.80 | Palmitoleic acid methyl ester | C17H32O2 | 14.39 | 977 | y = 0.0263x + 0.0795 | 0.64–32 | 0.9906 | 7.46 |
| 9 | 10.90 | Palmitic acid methyl ester | C17H34O2 | 8.43 | 959 | y = 0.0261x + 0.1301 | 1.3–65 | 0.9959 | 4.97 |
| 10 | 11.37 | Heptadecanoic acid methyl ester | C18H36O2 | 14.25 | 948 | y = 0.0345x + 0.0331 | 0.32–16 | 0.9934 | 6.22 |
| 11 | 11.75 | Oleic acid methyl ester | C19H36O2 | 18.83 | 708 | y = 0.0269x + 0.0637 | 1.3–65 | 0.9986 | 2.21 |
| 12 | 11.75 | Linoleic acid methyl ester | C19H34O2 | 18.83 | 708 | y = 0.0142x + 0.0363 | 2.22–111 | 0.9989 | 1.96 |
| 13 | 11.75 | Linolenic acid methyl ester | C19H32O2 | 18.83 | 718 | y = 0.0352x + 0.0442 | 0.64–32 | 0.9989 | 1.98 |
| 14 | 11.87 | Stearic acid methyl ester | C19H38O2 | 18.13 | 968 | y = 0.0264x + 0.0798 | 0.65–32.5 | 0.9953 | 5.29 |
| 15 | 12.73 | 11-Eicosenoic acid methyl ester | C21H40O2 | 24.36 | 944 | y = 0.0375x + 0.011 | 0.19–9.5 | 0.9964 | 4.61 |
| 16 | 12.84 | Arachidic acid methyl ester | C21H42O2 | 17.02 | 961 | y = 0.0375x + 0.018 | 0.19–9.5 | 0.9964 | 4.57 |
| 17 | 13.94 | Erucic acid methyl ester | C23H44O2 | 24.13 | 945 | y = 0.0395x + 0.0101 | 0.19–9.5 | 0.9956 | 5.07 |
| 18 | 14.10 | Behenic acid methyl ester | C23H46O2 | 17.70 | 959 | y = 0.0376x + 0.0205 | 0.19–9.5 | 0.9947 | 5.58 |
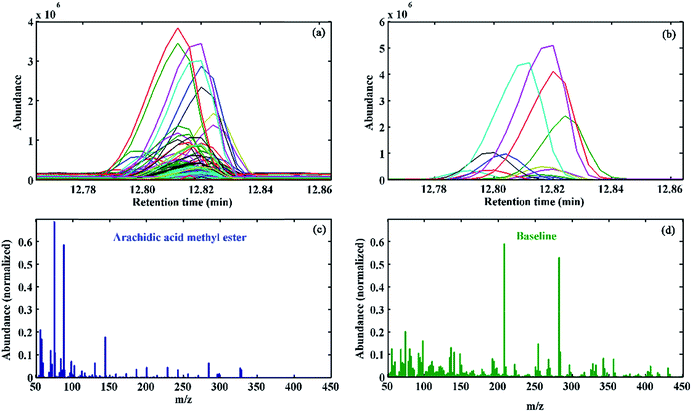
Another interesting aspect of MCR-ALS is its proper performance in the presence of elution time shifts. Elution time shifts are considered to be one of the most important problems in chromatography, which can be caused due to factors such as column ageing, minor changes in the mobile phase composition and instrumental drift. The common chemometric way to handle this problem is its correction by the most applicable alignment methods, such as dynamic time warping (DTW),37 correlation optimized warping (COW)38,39 and multivariate curve resolution-correlation optimized warping (MCR-COW).40,41 However, these methods are based on the selection of a reference chromatogram and the optimization of other parameters that are not trivial to choose in practice. Moreover, in the presence of interferences, alignment of the chromatograms becomes much more difficult. Therefore, elution time shifts modelling using MCR-ALS can be a more efficient and better strategy to correct them. As an example, Fig. 5a shows the overlaid TICs for a single-component chromatographic region in different calibration samples, where the presence of elution time shifts and a baseline/background contribution is evident. As demonstrated in Fig. 5b, MCR-ALS can properly recover the elution profiles for the target compounds in the presence of a significant elution time shift. In addition, baseline contributions in different standard samples were also properly modelled. The resolved mass spectral profile of the sought analyte and the baseline are respectively shown in Fig. 5c and d. The comparison of the resolved mass spectrum of target analyte with the standard one in the NIST MS database confirmed the presence of arachidic acid methyl ester with an acceptable RMF = 961.
3.3 Resolution and quantification of the FAMEs in the pomegranate seed
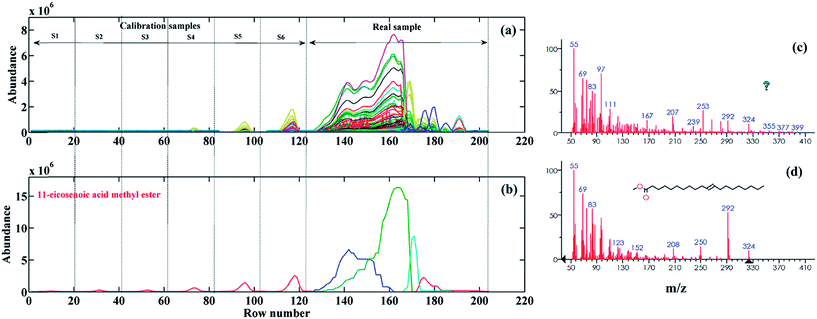
In order to evaluate the performance of the proposed second-order calibration method, the chemometric resolution, identification and quantification of FAMEs in pomegranate seeds were performed. The extraction and methyl esterification of FAs from pomegranate seeds were performed according to the procedure given in Section 2.2. The GC-MS analysis of the pomegranate seed sample was carried under the same conditions as the standard sample. As was shown earlier in Fig. 3b, the GC-MS TIC of the pomegranate seed seems to be very complex, with a large number of components and different chromatographic problems. Two highlighted chromatographic regions in Fig. 3b (B (12.7–13.1 min) and C (13.8–14.3 min)) were used to discuss how MCR-ALS can resolve these problematic regions. Region B shows a very complex chromatographic region with one of the calibrated FAMEs and some uncalibrated-unknown interferences that heavily overlap with the sought analyte. In addition, the peak cluster C illustrates the region of the pomegranate seed TIC where the S/N is too low due to the baseline drift.The most challenging region in the GC-MS TIC of the pomegranate seed is region B. To the best of our knowledge, this region belongs to the main component of the pomegranate seed, i.e. punicic acid ((Z,E,Z) 9,11,13-Octadecatrienoic acid), which constitutes about 70% of the total FAs of pomegranate seeds.8,10 The GC-MS data of the desired chromatographic region was column-wise augmented with the GC-MS data of the standard samples (S1–S6) of the target compound in this region (11-eicosenoic acid methyl ester) (see Fig. 6a). At first, the SVD was used for the determination of the number of chemical components involved in this region. The obtained singular values confirmed the presence of four chemical components in this region. To make sure about the number of chemical components in this data matrix, a local rank analysis method of an evolving factor analysis (EFA)42 was also used. The forward and backward EFA plots (for the sake of brevity, the results are not shown) confirmed the presence of the four chemical components previously estimated by SVD. SIMPLISMA was used to calculate the initial values of the mass spectral profiles to start the ALS optimization. Non-negativity (in both the chromatographic and mass spectroscopic dimensions), unimodality (in the chromatographic dimension), spectral normalization and component correspondence constraints were applied during the optimization to reduce the effects of rotational ambiguity. Since the external calibration strategy was used in this work, therefore, the contribution of the three unknown components in the real sample was set to zero in the calibration set using the component correspondence constraint, which significantly reduces the extent of the rotational ambiguity. Fig. 6b shows the resolved MCR-ALS elution profiles for standard and real samples. As can be seen, the pure contribution of 11-eicosenoic acid methyl ester in the pomegranate seed was successfully resolved by MCR-ALS (with an LOF = 18.72%), despite its heavy overlap with other components. The resolved mass spectral profile (see Fig. 6c) of the sought analyte was searched within the stored mass spectra of the NIST MS library and the presence of 11-eicosenoic acid methyl ester (see Fig. 6d) was confirmed with RMF 882. Afterwards, the resolved elution profile was used for the quantification of 11-eicosenoic acid methyl ester, and concentration (mg kg−1) and RSD (%) values of 1019.36 and 4.98 were, respectively, obtained (see Table 2).
| No. | Component | LOF (%) | RMF | FAs' amounta | RSDb% |
|---|---|---|---|---|---|
a Amount of fatty acids (mg kg−1) in pomegranate seeds.b Relative standard deviation based on OVIs. 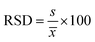 , where s is the standard deviation and , where s is the standard deviation and ![[x with combining macron]](https://www.rsc.org/images/entities/i_char_0078_0304.gif) is the mean calculated from the overall volume integration (OVI) in TIC mode.c Below the limit of quantification (LOQ).d Not detected.e Internal standard. is the mean calculated from the overall volume integration (OVI) in TIC mode.c Below the limit of quantification (LOQ).d Not detected.e Internal standard. |
|||||
| 1 | Caprylic acid | 7.28 | 969 | <LOQc | 0.22 |
| 2 | Capric acid | 8.88 | 971 | <LOQ | 5.11 |
| 3 | Lauric acid | 11.39 | 956 | <LOQ | 1.84 |
| 4 | Tridecanoic acid | 9.00 | 964 | <LOQ | 0.36 |
| 5 | Myristoleic acid | — | — | n.d.d | — |
| 6 | Myristic acid | 10.95 | 955 | 47.81 | 0.15 |
| 7 | Pentadecanoic acid | 11.24 | 943 | ISe | — |
| 8 | Palmitoleic acid | 24.61 | 969 | 84.20 | 11.57 |
| 9 | Palmitic acid | 12.22 | 957 | 3147.02 | 2.07 |
| 10 | Heptadecanoic acid | 14.35 | 948 | 51.92 | 3.33 |
| 11 | Oleic acid | 6.25 | 715 | 4873.71 | 5.78 |
| 12 | Linoleic acid | 6.25 | 717 | 9098.23 | 6.16 |
| 13 | Linolenic acid | — | — | n.d. | — |
| 14 | Stearic acid | 18.46 | 957 | 1960.16 | 1.97 |
| 15 | 11-Eicosenoic acid | 18.72 | 882 | 1019.36 | 4.98 |
| 16 | Arachidic acid | 21.62 | 961 | 319.80 | 6.09 |
| 17 | Erucic acid | 23.98 | 946 | 16.60 | 11.09 |
| 18 | Behenic acid | 17.77 | 959 | 93.81 | 0.61 |
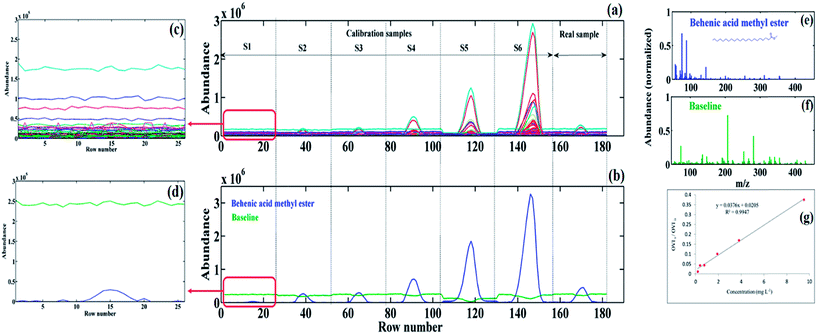
The last example shows the potential of MCR-ALS as a second-order calibration method to model the baseline drift, and therefore, to resolve the contribution of the minor components (low S/N signals) (i.e. the chromatographic region C in Fig. 3b). Fig. 7a shows the augmented data matrix for the target compound in the real sample, along with corresponding data matrices in the calibration samples. As can be seen, low S/N signals are observed due to the baseline drift for both standards (in low concentration levels, S1–S3) and for the pomegranate seed sample.
Fig. 7b illustrates the resolved MCR-ALS elution profiles in the standard and real samples. The green line in Fig. 7b shows the contribution of the baseline in different samples, which emphasizes one of the most important features of MCR-ALS for the modelling of baseline/background contribution instead of its correction before analysis. The GC-MS data (see Fig. 7a) and the resolved elution profiles (see Fig. 7b) for the S1 standard are enlarged to indicate the resolution of the baseline and analyte elution profiles (see Fig. 7c and d). As can be seen, the analyte signal in Fig. 7c is indistinguishable from the baseline, while after applying MCR-ALS, its contribution becomes recognizable (see Fig. 7d). The resolved spectral profile of the analyte and baseline are shown in Fig. 7e and f, respectively. The resolved spectral profile was matched with the NIST library search and the presence of behenic acid methyl ester with a RMF equal to 959 was confirmed. Fig. 7g shows the calibration curve of behenic acid methyl ester with R2 equals to 0.994, which is considered acceptable.
Table 2 represents the identified FAs in pomegranate seeds using MCR-ALS. The values of the LOF, RMF, concentration (mg kg−1) and RSD are also presented in this table. As can be seen, most of the FAs in the standard samples were also detected in the real sample, with a RMF higher than 900, which again confirms the reliability of the present method. However, the concentration of caprylic acid, capric acid, lauric acid and tridecanoic acid were below the limit of quantification (LOQ). In addition, myristoleic acid and linolenic acid were not detected in the pomegranate seed, whereas linoleic acid, oleic acid, palmitic acid, stearic acid and cis-11-eicosenoic acid were, respectively, the main detected fatty acids of pomegranate seed, with concentrations of 9098.0, 4873.0, 3147.0, 1960.0 and 1019.0 mg kg−1, and relative standard deviations (RSD%) between 0.15 and 11.57.
4. Conclusion
In the present study, the VAE-DLLME-GC-MS technique was used for the determination of the FAMEs in a standard mixture solution and in a pomegranate seed sample in less than 20 min. Then, the GC-MS data were column-wise augmented with the retention times as rows and the m/z values as the columns of this data matrix. Finally, MCR-ALS was performed with the application of proper constraints to obtain pure elution and mass spectral profiles, along with calibration curves of the target FAMEs. The typical problems associated with the GC-MS analysis of FAMEs, such as elution time shifts, baseline/background contribution, and peak overlaps were successfully solved using MCR-ALS bilinear modeling of the column-wise augmented data matrix. Furthermore, proper qualitative (RMF higher than 800) and quantitative (RE below 7.5% and RSD below 11.6%) results were obtained for both the standard mixture and pomegranate seed samples. Therefore, it can be concluded that the proposed combined strategy (VAE-DLLME-GC-MS coupled with MCR-ALS) is applicable for complex natural products containing different unknown interferences.Abbreviations
| COW | Correlation optimized warping |
| DLLME | Dispersive liquid–liquid micro-extraction |
| DTW | Dynamic time warping |
| EEM | Excitation-emission matrices |
| EFA | Evolving factor analysis |
| FA | Fatty acid |
| FAME | Fatty acid methyl ester |
| GC-MS | Gas chromatography-mass spectrometry |
| HPLC-DAD | High performance liquid chromatography-diode array detection |
| LC-MS | Liquid chromatography-mass spectrometry |
| LOF | Lack of fit |
| MCR-ALS | Multivariate curve resolution-alternating least squares |
| OVI | Overall volume integration |
| PARAFAC | Parallel factor analysis 2 |
| PUFA | Polyunsaturated fatty acid |
| RE | Relative error |
| RMF | Reverse match factor |
| RSD | Relative standard deviation |
| SFE | Supercritical fluid extraction |
| SIMPLISMA | Simple-to-use interactive self-modelling mixture analysis |
| SVD | Singular value decomposition |
| TIC | Total ion chromatogram |
| UASE | Ultrasonic-assisted solvent extraction |
| VAE | Vortex-assisted extraction |
Notes and references
- M. Çelik, C. Türeli, M. Çelik, Y. Yanar, Ü. Erdem and A. Küçükgülmez, Food Chem., 2004, 88, 271 CrossRef PubMed.
- M. Vosough and A. Salemi, Talanta, 2007, 73, 30 CrossRef CAS PubMed.
- T. Horák, J. Culík, P. Cejka, M. Jurková, V. Kellner, J. Dvorák and D. Hasková, J. Agric. Food Chem., 2009, 57, 11081 CrossRef PubMed.
- L.-K. Ng, Anal. Chim. Acta, 2002, 465, 309 CrossRef CAS.
- B. Plutowska and W. Wardencki, Anal. Chim. Acta, 2008, 613, 64 CrossRef CAS PubMed.
- I. Marekov, S. Momchilova, B. Grung and B. Nikolova-damyanova, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2012, 910, 54 CrossRef CAS PubMed.
- E. P. Lansky and R. A. Newman, J. Ethnopharmacol., 2007, 109, 177 CrossRef CAS PubMed.
- P. Jing, T. Ye, H. Shi, Y. Sheng, M. Slavin, B. Gao, L. Liu and L. (Lucy) Yu, Food Chem., 2012, 132, 1457 CrossRef CAS PubMed.
- A. Fadavi, M. Barzegar and M. H. Azizi, J. Food Compos. Anal., 2006, 19, 676 CrossRef CAS PubMed.
- H. Abbasi, K. Rezaei and L. Rashidi, J. Am. Oil Chem. Soc., 2008, 85, 83 CrossRef CAS PubMed.
- Y. Tian, Z. Xu, B. Zheng and Y. M. Lo, Ultrason. Sonochem., 2013, 20, 202 CrossRef CAS PubMed.
- G. Wei and E. Y. Zeng, Trends Anal. Chem., 2011, 30, 1429 CrossRef CAS PubMed.
- A. Saba, F. Mazzini, A. Raffaelli, A. Mattei and P. Salvadori, J. Agric. Food Chem., 2005, 53, 4867 CrossRef CAS PubMed.
- M. Rezaee, Y. Assadi, M. R. Milani Hosseini, E. Aghaee, F. Ahmadi and S. Berijani, J. Chromatogr. A, 2006, 1116, 1 CrossRef CAS PubMed.
- M. Rezaee, Y. Yamini and M. Faraji, J. Chromatogr. A, 2010, 1217, 2342 CrossRef CAS PubMed.
- P. Viñas, N. Campillo, I. López-García and M. Hernández-Córdoba, Anal. Bioanal. Chem., 2013, 406, 2067 CrossRef PubMed.
- S. Dadfarnia and A. M. H. Shabani, Anal. Chim. Acta, 2010, 658, 107 CrossRef CAS PubMed.
- A. V. Herrera-Herrera, M. Asensio-Ramos, J. Hernández-Borges and M. Á. Rodríguez-Delgado, Trends Anal. Chem., 2010, 29, 728 CrossRef CAS PubMed.
- T. Seppänen-Laakso, I. Laakso and R. Hiltunen, Anal. Chim. Acta, 2002, 465, 39 CrossRef.
- K. S. Booksh and B. R. Kowalski, Anal. Chem., 1994, 66, 782A CrossRef CAS.
- V. Gómez and M. P. Callao, Anal. Chim. Acta, 2008, 627, 169 CrossRef PubMed.
- A. C. Olivieri, G. M. Escandar and A. Peña, TrAC, Trends Anal. Chem., 2011, 30, 607 CrossRef CAS PubMed.
- A. C. Olivieri, Anal. Methods, 2012, 4, 1876 RSC.
- R. Bro, C. A. Andersson and H. A. L. Kiers, J. Chemom., 1999, 13, 295 CrossRef CAS.
- S. A. Bortolato and A. C. Olivieri, Anal. Chim. Acta, 2014, 842, 11 CrossRef CAS PubMed.
- E. Peré-Trepat, S. Lacorte and R. Tauler, Anal. Chim. Acta, 2007, 595, 228 CrossRef PubMed.
- M. Vosough and H. M. Esfahani, Talanta, 2013, 113, 68 CrossRef CAS PubMed.
- M. M. Bravo, L. F. Aguilar, W. V. Quiroz, A. C. Olivieri and G. M. Escandar, Microchem. J., 2013, 106, 95 CrossRef PubMed.
- W. Windig and J. Guilment, Anal. Chem., 1991, 63, 1425 CrossRef CAS.
- M. Jalali-Heravi and H. Parastar, Talanta, 2011, 85, 835 CrossRef CAS PubMed.
- H. Parastar and R. Tauler, Anal. Chem., 2014, 86, 286 CrossRef CAS PubMed.
- F. Gong, Y. Liang, H. Cui, F. T. Chau and B. T. P. Chan, J. Chromatogr. A, 2001, 909, 237 CrossRef CAS.
- M. Jalali-heravi, H. Parastar, M. Kamalzadeh, R. Tauler and J. Jaumot, Chemom. Intell. Lab. Syst., 2010, 104, 155 CrossRef CAS PubMed.
- NIST/EPA/NIH MS Library, v. 2.0d, Gaithersburg, MD, USA, 2005 Search PubMed.
- D. Ge and H. K. Lee, J. Chromatogr. A, 2012, 1263, 1 CrossRef CAS PubMed.
- D. L. Massart, B. G. M. Vandeginste, L. M. C. Buydens, S. De Jong, P. J. Lewi and J. Smeyers-Verbeke, Handbook of chemometrics and qualimetrics: Part A, Elsevier, Amsterdam, 1997 Search PubMed.
- A. Kassidas, J. F. MacGregor and P. A. Taylor, AIChE J., 1998, 44, 864 CrossRef CAS.
- N. P. V. Nielsen, J. M. Carstensen and J. Smedsgaard, J. Chromatogr. A, 1998, 805, 17 CrossRef CAS.
- G. Tomasi, F. van den Berg and C. Andersson, J. Chemom., 2004, 18, 231 CrossRef CAS.
- C. Tistaert and Y. Vander Heyden, Anal. Chem., 2012, 84, 5653 CrossRef CAS PubMed.
- H. Parastar and N. Akvan, Anal. Chim. Acta, 2014, 816, 18 CrossRef CAS PubMed.
- M. Maeder and A. D. Zuberbuehler, Anal. Chim. Acta, 1986, 181, 287 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2015 |