Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Identification of a fluorometabolite from Streptomyces sp. MA37: (2R3S4S)-5-fluoro-2,3,4-trihydroxypentanoic acid†
Long
Ma‡
a,
Axel
Bartholome
a,
Ming Him
Tong
b,
Zhiwei
Qin
b,
Yi
Yu
c,
Thomas
Shepherd
d,
Kwaku
Kyeremeh
e,
Hai
Deng
*b and
David
O'Hagan
*a
aEaStChem School of Chemistry, University of St Andrews, North Haugh, St Andrews KY169ST, UK. E-mail: do1@st-and.ac.uk
bMarine Biodiscovery Centre, Department of Chemistry, University of Aberdeen, Meston Walk, Aberdeen AB24 3UE, UK. E-mail: h.deng@abdn.ac.uk
cKey Laboratory of Combinatory Biosynthesis and Drug Discovery (Ministry of Education), School of Pharmaceutical Sciences, Wuhan University, 185 East Lake Road, Wuhan 430071, P. R. China
dThe James Hutton Institute, Invergowrie, Dundee, DD2 5DA, UK
eDepartment of Chemistry, University of Ghana, FGO Torto Building, Legon, Ghana
First published on 3rd December 2014
Abstract
(2R3S4S)-5-Fluoro-2,3,4-trihydroxypentanoic acid (5-FHPA) has been discovered as a new fluorometabolite in the soil bacterium Streptomyces sp. MA37. Exogenous addition of 5-fluoro-5-deoxy-D-ribose (5-FDR) into the cell free extract of MA37 demonstrated that 5-FDR was an intermediate to a range of unidentified fluorometabolites, distinct from fluoroacetate (FAc) and 4-fluorothreonine (4-FT). Bioinformatics analysis allowed identification of a gene cluster (fdr), encoding a pathway to the biosynthesis of 5-FHPA. Over-expression and in vitro assay of FdrC indicated that FdrC is a NAD+ dependent dehydrogenase responsible for oxidation of 5-FDR into 5-fluoro-5-deoxy-lactone, followed by hydrolysis to 5-FHPA. The identity of 5-FHPA in the fermentation broth was confirmed by synthesis of a reference compound and then co-correlation by 19F-NMR and GC-MS analysis. The occurrence of 5-FHPA proves the existence of a new fluorometabolite pathway.
Introduction
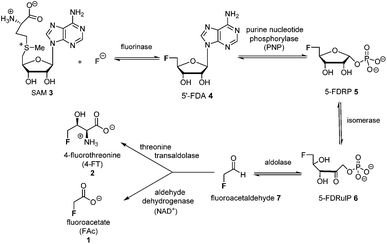
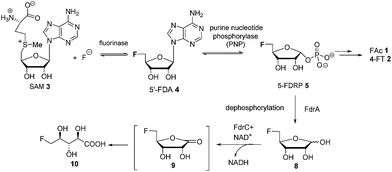
The introduction of fluorine into organic molecules can substantially modulate their physicochemical properties.1 About 30% of current drugs, including many top sellers, contain at least one fluorine atom.2 In contrast to man-made molecules, fluorinated natural products are extremely rare.3Fluoroacetate (FAc) 1 is the most ubiquitous fluorometabolite found as a toxic self-defence agent in many tropical and sub-tropical plants.4 In 1986, the actinomycete soil bacterium Streptomyces cattleya was found to secrete FAc 1 and 4-fluorothreonine (4-FT) 2 as part of its metabolic profile when grown in the presence of inorganic fluoride.5 Over the last decade, intermediates on the fluorometabolite pathway in S. cattleya have been uncovered and are shown in Scheme 1.6 The formation of the C–F bond is catalysed by the fluorinase, which mediates the biotransformation of S-adenesyl-L-methionine (SAM) 3 and inorganic fluoride into 5′-fluoro-5-deoxy-adenosine (5′-FDA) 4.7 5′-FDA is then phosphorylated generating 5-fluoro-5-deoxy-ribose phosphate (5-FDRP) 5,8 followed by ring opening to generate 5-fluoro-5-deoxy-ribulose-phosphate (5-FDRulP) 6.9 An aldolase catalyses a retro-aldol reaction to generate fluoroacetaldehyde 7, the last common intermediate on the pathway.10 Fluoroacetaldehyde 7 is either oxidised to FAc 1,11 or biotransformed into 4-FT 2 catalysed by a PLP-dependent transaldolase.12 The fluorinase gene remained a sole representative in the genome databases for a decade, however between 2012–2014, three fluorinase genes appeared in the genomes of sequenced microorganisms (Streptomyces sp. MA37, Actinoplanes sp. N902-109,13Norcardia brasiliensis13,14). PCR amplification of these genes from genomic DNA, or the expression of synthetic genes in E. coli demonstrated that these were all functional fluorinases. Most recently, the marine actinomycete Streptomyces xinghaiensis (NRRL B-24674) was also shown to have a functional fluorinase.15S. xinghaiensis was found to produce FAc 1 only (no 4-FT 2) and FAc 1 production is sea-salt dependent.15S. xinghaiensis is the first fluorometabolite producer from a marine microorganism. Most importantly, all fluorinase genes described after the disclosure of that obtained from S. cattleya have greater than 80% sequence identity to the original fluorinase. In culture, N. brasiliensis was unable to produce a trace of fluorometabolite under laboratory culture conditions.13,14
Streptomyces sp. MA37, an isolate from a Ghanaian soil sample, produces FAc 1 and 4-FT 2 in culture.13 Unlike S. cattleya and S. xinghaiensis, Streptomyces sp. MA37 also produced a range of unidentified fluorometabolites in addition to FAc 1 and 4-FT 2, as determined by 19F-NMR of a supernatant extract.13 The identity of these novel fluorometabolites is of interest given that this class of natural products is so rare.
Here we report that 5-FDRP 5 is a branch point metabolite between two pathways. Exogenous addition of 5-fluoro-5-deoxyribose (5-FDR) 8 to cell free extracts was found to support biosynthesis predominantly of the unidentified fluorometabolites and could not significantly support FAc 1 and 4-FT 2 biosynthesis. Genomic-driven analysis allowed identification of the fdr gene cluster, encoding elements of the new pathway. In vitro assay of over-expressed FdrC demonstrated that the protein is a NAD+ dependent dehydrogenase that oxidizes 5-FDR 8 to 5-fluoro-5-deoxy-D-ribono-γ-lactone (5-FRL) 9, followed by lactone hydrolysis to (2R3S4S)-5-fluoro-2,3,4-trihydroxypentanoic acid (5-FHPA) 10. This sequence from 5-FDRP forms the first steps of a new fluorometabolite pathway.
Results and discussion
5-FDRP 5 is known to be an intermediate in the biosynthesis of FAc 1 and 4-FT 2 in Streptomyces cattleya.8 In that study it was established that the de-phosphorylated free ribose 5-FDR 8 was unable to recover fluorometabolite biosynthesis in cell free extracts. It was unable to support fluorometabolite biosynthesis, and it was also unable to become phosphorylated to re-generate 5 and channel back into the fluorometabolite pathway to generate FAc 1 or 4-FT 2.8,16 Interestingly in the marine microorganism Salinispora tropica, a closely related pathway operates as illustrated in Scheme 2.17 This bacterium produces salinosporamide A 11 a chlorinated metabolite which has received attention as an anti-cancer therapeutic.18 Investigations into salinosporamide-A 11 biosynthesis have shown that the chlorine is introduced by a SAM-dependent chlorinase to generate 5′-ClDA 12, an enzyme closely analogous to the fluorinase of S. cattleya.19 The two pathways also share the second step. In S. cattleya, depurination of 5′-FDA generates 5-FDRP 5 and analogously in S. tropica 5′-ClRP 13 is generated. The two pathways appear to diverge at this point.17 For salinosporamide-A 11, SalN catalyses dephosphorylation of 5-ClRP 13 to generate the free sugar 5-chloro-5-deoxy-D-ribose (5-ClR) 14 as a key intermediate on the biosynthetic pathway to salinosporamide A 11. The free sugar does not appear to be relevant in S. cattleya. Interestingly, 5-FDR 8 was also found to support this biotransformation to form the analogous fluorinated derivative of salinosporamide A, demonstrating that the salinosporamide-A pathway can channel fluoromethyl as well as chloromethyl intermediates.20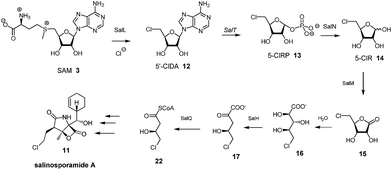 | ||
| Scheme 2 Early steps in salinosporamide A 11 biosynthesis.17 | ||
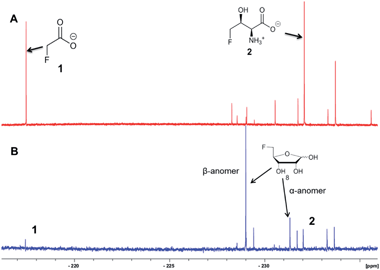
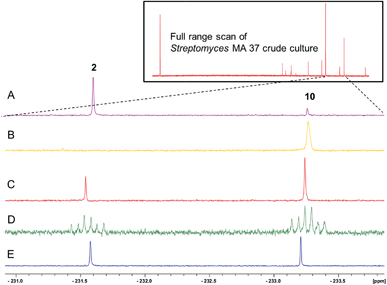
In this context it appeared appropriate to explore a role for 5-FDR 8 in Streptomyces sp. MA37 metabolism to establish if any of the additional unknown metabolites are derived from the free sugar. To this end, 5-FDR 8 was prepared by synthesis8 and was incubated with a cell-free extract (CFE) of Streptomyces sp. MA37. Products were monitored by 19F-NMR. The usual dominance of FAc 1 and 4-FT 2, found in full whole cell incubations with added fluoride ion was no longer the observed profile. Instead these signals were substantially diminished and some of the minor unknowns now dominated the 19F-NMR spectrum (Fig. 1). These unknown signals were not observed in control experiments using boiled CFE incubated with 5-FDR 8 or the CFE alone. Therefore 5-FDR 8 appears to be an intermediate to some of the fluorometabolites in Streptomyces sp. MA37, and does not appear to support FAc 1 or 4-FT 2 biosynthesis in a primary manner.
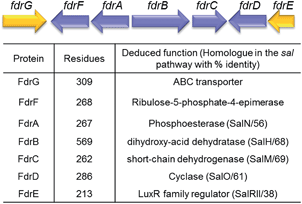
A homolog search of the Streptomyces sp. MA37 genome revealed an open reading frame (orf) fdrA that is predicted to encode a metal-dependent phosphoesterase, sharing high sequence identity (56% identity) with SalN of the salinosporamide biosynthetic pathway. Notably, immediately downstream of fdrA are two orfs, fdrB and fdrC, that are divergently transcribed. These are FdrB and FdrC which also share high sequence identities with SalH (68% identity) and SalM (69% identity), respectively (Fig. 2 and S1†) and are predicted to encode a dihydroxy-acid dehydratase and a short chain dehydrogenase (SDRs). SalM was found to catalyse the NAD+ dependant oxidation of 5-ClR 14 to 5-chlororibolactone 15, which is then hydrolysed to 5-chlororibonate 16 during studies exploring the biosynthesis of salinosporamide A 11.21
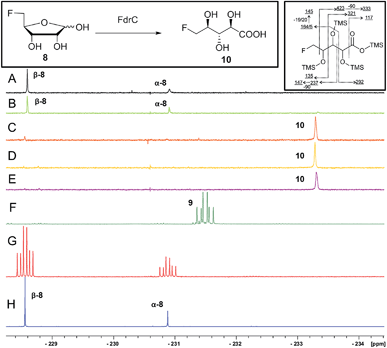
It was an objective in this study to explore the function of fdrC. Over-expression of a codon-optimized synthetic gene for fdrC in E. coli, with a His6 tag and a TEV protease cleavage site added, allowed isolation and purification of the coded protein. The resultant FdrC appeared on SDS-page with an estimated mol. wt ∼31 kDa (Fig. S2†). Incubations of the recombinant enzyme with 5-FDR 8 and NAD+ or NADP+, were followed by 19F NMR spectroscopy. When the assay was conducted in the absence of NAD+ or in the presence of NADP+, there was no turnover (Fig. 3A and B, respectively). Reactions with NAD+ however resulted in an efficient conversion of 5-FDR 8 to a new organofluorine compound (19F-NMR; −233.15 ppm, dt, 2JHF = 25 Hz, 3JHF = 47 Hz) demonstrating that FdrC is a NAD+ dependent enzyme (Fig. 3C). Given the similarity of this pathway to that in salinosporamide-A 11, it was anticipated that the enzymatic product might be carboxylic acid 10. This would arise by oxidation of 5-FDR 8 to lactone 5-FRL 9 and then hydrolysis to the ring opened carboxylic acid 5-FHPA 10. The identity of 5-FHPA 10 in the FdrC reaction mixture was confirmed by comparison with a synthetic sample. A synthesis of 5-FHPA 10 was carried out following the protocol22 illustrated in Scheme 3.
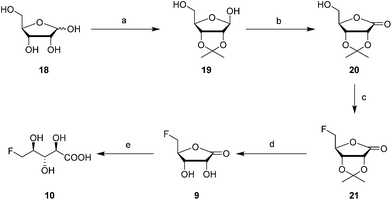
The 2′- and 3′-hydroxyl groups of D-ribose 18 were protected by preparation of acetonide 19.22 Oxidation of the anomeric alcohol to give lactone 20 was efficiently accomplished using iodine in DCM in the presence of potassium carbonate,23 and then fluorination with Deoxofluor® afforded fluorolactone 21 in good yield.24 Acetonide hydrolysis using TFA in water gave 5-FRL 9 and finally hydrolysis was accomplished with aqueous LiOH to give 10. This synthetic 5-FHPA 10 was found to be identical by 19F-NMR to the enzyme (FdrC) reaction product.
The 19F-NMR signals became coincident when a sample of the enzyme reaction mixture was spiked into the supernatant of the Streptomyces sp. MA37 fermentation (Fig. 4C). The synthetic reference of 5-FHPA 10 was also used to confirm this species as a component of the product mixture from Streptomyces sp. MA37. This was achieved by global persilylation of the components of the CFE using N-methyl-N-(trimethylsilyl)-trifluoroacetamide (MSTFA). The extract was then analysed by GC-MS (Fig. 3 and S3†). Comparison with the persilylated derivative of synthetic 5-FHPA 10 revealed a constituent of the CFE with an identical fragmentation pattern and retention time (Fig. S3†). Therefore these studies are consistent with 5-FHPA 10 as the product of FdrC enzyme, and as an end product in fluorometabolite biosynthesis in Streptomyces sp. MA37.
Incubation of synthetic 5-FRL 9 with FdrC (Fig. 3D) resulted in its complete conversion to 10, suggesting lactone hydrolysis is also catalysed by the enzyme. Two step enzymatic catalysis is probably also the case for SalM (conversion of 15 to 16) on the salinosporamide A 11 pathway.25
A study of the reaction kinetics for FdrC mediated NAD+ oxidation indicates that FdrC has a higher affinity for D-ribose over 5-FDR 8 as measured by Km, but overall both riboses oxidise with a similar efficiency (kcat/Km) of less than 0.4 μm−1 min−1 (Table 1 and Fig. S4†).
| V max (μM min−1) | K m (μM) | k cat (min−1) | Specificity constant (kcat/Km) (μM−1 min−1) | |
|---|---|---|---|---|
| 5-FDR 8 | 4.96 ± 0.25 | 2.73 ± 0.73 | 0.58 | 0.21 |
| D-ribose 18 | 2.73 ± 0.05 | 0.84 ± 0.14 | 0.32 | 0.38 |
This study identifies 10 as a novel fluorometabolite and extends the very small collection of this rare class of natural products. Organic chemists have only identified five unique fluorine containing natural products so far.3a The pathway to 10 branches from that already established for FAc 1 and 4-FT 2 biosynthesis. The branch point occurs at 5-FDRP 5, whereby phosphorolysis generates 5-FDR 8. This sugar is then oxidised by FdrC to 5-FRL 9, which undergoes hydrolysis to generate 5-FHPA 10 (Scheme 4).
We finish with a commentary on the genes that surround fdrA–C in the gene cluster (Fig. 2). The fdr cluster was found located in a different scaffold of the draft genome assembly of the MA37 strain, indicating that it is physically remote to the fl gene cluster in the previous report.13 The orf fdrD is located immediately downstream of fdrC, encoding a putative cyclase that shares high sequence identity (61% identity) with SalO. Knockout of salO had no obvious effect on salinosporamide A 11 production,25 suggesting that salO is not involved directly in salinosporamide A synthesis, so a role for FdrD is not obvious. The gene fdrE is a LuxR family regulator gene and it shares moderate sequence identity (38% identity) with SalRII, a pathway-specific regulator in the biosynthesis of salinosporamide A. Over-expression of SalRII led to a significant increase in the production of salinosporamide A 11 thus there are perhaps prospects for up-regulation of fluorometabolism by over-expression of this gene.25 Immediately upstream of fdrA lie two orfs, fdrF and fdrG. FdrG belongs to the ABC transporter family and an obvious role is not clear. The gene fdrF is predicted to encode a ribulose-5-phosphate-4-epimerase, which may be relevant to the downstream metabolism of 5-FDR 8. Interestingly, the main difference between the newly-identified fdr cluster in the MA37 and the sal cluster in S. tropica is that there is no homolog of salQ in the proximity of the fdr cluster or anywhere in the draft genome of MA37. SalQ, a putative α-oxoacid ferredoxin oxidoreductase, was proposed to be the key enzyme catalysing oxidative decarboxylation from a 5-carbon intermediate 5-chloro-hydroxy-2-oxopentanoate 17 to a 4-carbon intermediate 4-chloro-3-hydroxybutyryl-CoA 21. This observation suggested that 5-FHPA 10 cannot be metabolised further and accumulated extracellularly, consistent with our chemical identification of 5-FHPA 10 as one of the most abundant fluorometabolite in the supernatant of the MA37 culture. We are currently focussing on establishing the biochemical steps and structure of the other fluorometabolites on this branch pathway.
Conclusions
In conclusion, 5-FHPA 10 is identified as a new fluorometabolite branching from the established fluorinase pathway to FAc 1 and 4-FT 2. In silico analysis enabled identification of a biosynthetically relevant gene cluster. In vitro assay of over-expressed FdrC demonstrated that it can oxidise (NAD+ dependant) 5-FDR 8 to its corresponding 5-FRL 9 followed by hydrolysis to generate 5-FHPA 10. FdrC was similarly active with D-ribose. GC-MS analysis and correlation of synthetic or enzymatically prepared 5-FHPA 10 with the product of the supernatant of Streptomyces sp. MA37 indicated identical products, demonstrating that 5-FHPA 10 is a new natural product of Streptomyces sp. MA37. This is the first secure identification of a new fluorinated natural product since 1998.26–29Experimental section
19F-NMR analysis
The samples from enzyme reaction and the supernatants of Streptomyces sp. MA-37 fermentation were subjected to 19F-NMR analysis. The 19F-NMR spectra were recorded with and without proton decoupling on a Bruker AV-500 MHz instrument (19F at 470.3 MHz). The chemical shifts of 19F-NMR were calculated with respect to CFCl3.Cell-free extraction (CFE) reaction
The cells of Streptomyces sp. MA 37 was harvested by centrifugation (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm × 20 min) after 8 day fermentation. The cell pellets were washed three times using Tris–HCl buffer (20 mM, pH 7.5) supplemented with 10 mM MgCl2 to remove remnant culture media. The cells were re-suspended in the same buffer (0.1 g wet-cell weight per mL). The cells were disrupted by ultra-sonication (60% duty cycle for 30–60 s). Cell debris was removed by centrifugation (13
000 rpm × 20 min) after 8 day fermentation. The cell pellets were washed three times using Tris–HCl buffer (20 mM, pH 7.5) supplemented with 10 mM MgCl2 to remove remnant culture media. The cells were re-suspended in the same buffer (0.1 g wet-cell weight per mL). The cells were disrupted by ultra-sonication (60% duty cycle for 30–60 s). Cell debris was removed by centrifugation (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 30 min) and the resultant clear supernatant was used as the cell free extract for incubation experiments. The cell free extracts (1 mL) were supplemented with or without 5-FDR at 37 °C for 6 hours. At the end of the incubation period, protein was precipitated by heating the vial to 90 °C for 3 min and the protein was then removed by centrifugation. The supernatant was collected for 19F-NMR analysis.
000 rpm, 30 min) and the resultant clear supernatant was used as the cell free extract for incubation experiments. The cell free extracts (1 mL) were supplemented with or without 5-FDR at 37 °C for 6 hours. At the end of the incubation period, protein was precipitated by heating the vial to 90 °C for 3 min and the protein was then removed by centrifugation. The supernatant was collected for 19F-NMR analysis.
Plasmid construction, over-expression and purification of Streptomyces sp. MA 37 FdrC enzyme
The plasmid encoding the chemically synthesised and codon-optimised fdrC gene was provided by the commercial supplier DNA 2.0 (CA, USA). AGGAGGTAAAACAT was incorporated as the ribosome binding site (RBS). T7 promoter and kanamycin-resistance gene were incorporated. A His-tag containing peptide (Met-Ser-Tyr2-His6-Asp-Tyr-Asp-IIe-Pro-Thr2) was fused to the N-terminus of the enzyme. A TEV proteinase cleavage site (Pro-Val-Phe-Ser-Gly) was engineered into the synthetic gene to enable cleavage of the His-tag. Two restriction sites of NcoI and EcoRI were designed to locate upstream and downstream of the fdrC gene, respectively. The plasmid was transformed using the heat shock method. A mixture of chemically competent bacteria and plasmid DNA was placed at 42 °C for 90 s and then placed back on ice. E. coli BL21(DE3) Gold cells, transformed with the synthetic fdrC plasmids were grown in Lysogeny broth containing 50 mg mL−1 kanamycin at 37 °C until cell density reached an absorbance at ∼0.6 at 600 nm. The culture was then cooled on ice for 30 min. A final concentration of 0.3 mM isopropylthiogalactoside (IPTG) was added as induction procedure for FdrC over-expression. The incubation was continued at 25 °C for ∼24 h. The resultant E. coli cells were collected and lysed. After centrifugation, the cell lysate was applied onto a bench-top column packed with Ni2+-charged His-Bind resin (Qiagen) for protein purification. Recombinant protein bound on the resin was firstly washed with a solution of Tris–HCl (20 mM, pH 8.0), imidazole (20 mM) and NaCl (0.5 M) buffer, followed by a further wash with a solution of Tris–HCl (20 mM, pH 8.0), imidazole (50 mM) and NaCl (0.5 M). The protein was eventually eluted with a solution of Tris–HCl (20 mM, pH 8.0), imidazole (400 mM) and NaCl (0.5 M). The protein concentration was measured by OD280 nm (Nanodrop). The extinction coefficient was determined using the ExPAsy ProtParam tool. The identity of the protein was confirmed by both polyacrylamide gel electrophoresis and ESI-MS analysis (Fig. S2†).ESIMS analysis of FdrC enzyme reaction
ESI-FT-MS was used to determine the identity of enzymatic products in the FdrC-mediated reactions. Reaction mixtures consisted of FdrC (0.625 mg mL−1), NAD+ (5 mM) and MgCl2 (10 mM) in the presence of either 5-FDR (10 mM) or D-ribose (10 mM) in the Tris–HCl buffer (1 mL, 25 mM, pH 7.8). The reactions were conducted overnight at 37 °C. Protein was removed by heating the vial to 90 °C for 3 min, followed by centrifugation (133![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 3 min). The supernatant was subjected for ESI-FT-MS analysis.
000 rpm, 3 min). The supernatant was subjected for ESI-FT-MS analysis.
Enzyme kinetics measurements for FdrC
A UV-Vis spectroscopy-based assay was employed to monitor the enzyme activity of FdrC. The enzyme reactions were initiated by mixing FdrC (final concentration was 0.26 mg mL−1) with either 5-FDR or D-ribose, supplemented with 1 mM magnesium chloride and 2.5 mM nicotinamide adenine dinucleotide (NAD+) in the Tris–HCl buffer (25 mM, pH 7.8). The concentrations of the substrates used in the assay ranged from 0.5 μM to 200 μM. After mixing with the substrate, the UV spectra were recorded immediately, scanning from 300 nm to 450 nm every 0.2 minutes for 3 min at ambient temperature. An increase in absorbance at 340 nm was attributed to the production of NADH. Initial FdrC reaction velocities were plotted against substrate concentration and a Michaelis–Menten plot was established (see Fig. S4†). Enzyme kinetics parameters Vmax, Km, kcat, and specificity constant were all calculated accordingly and are listed in Table 1.Acknowledgements
DOH thanks EPSRC and the ERC for financial support. He also acknowledges the Royal Society for a Wolfson Merit Award. KK and HD thank the Leverhulme Trust; Royal Society Africa Award (AA090088).Notes and references
- D. O'Hagan, J. Fluorine Chem., 2010, 131, 1071–1081 CrossRef PubMed.
- V. A. Brunet and D. O'Hagan, Angew. Chem., Int. Ed., 2008, 47, 1179–1182 CrossRef CAS PubMed.
- (a) H. Deng and D. O'Hagan, Chem. Rev., 2014 DOI:10.1021/cr500209t; (b) H. Deng, D. O'Hagan and C. Schaffrath, Nat. Prod. Rep., 2004, 21, 773–784 RSC; (c) C. D. Murphy, Nat. Prod. Rep., 2006, 23, 147–152 RSC.
- D. O'Hagan, R. Perry, J. M. Lock, J. J. M. Meyer, L. Dasaradhi, J. T. G. Hamilton and D. B. Harper, Phytochemistry, 1993, 33, 1043–1046 CrossRef.
- M. Sanada, T. Miyano, S. Iwadare, J. M. Williamson, B. H. J. Arison, L. Smith, A. W. Douglas, J. M. Liesch and E. Inamine, J. Antibiot., 1986, 39, 259–265 CrossRef CAS.
- H. Deng and D. O'Hagan, Curr. Opin. Chem. Biol., 2008, 12, 582–592 CrossRef CAS PubMed.
- (a) D. O'Hagan, C. Schaffrath, S. L. Cobb, J. T. G. Hamilton and C. D. Murphy, Nature, 2002, 416, 279 CrossRef PubMed; (b) C. Schaffrath, H. Deng and D. O'Hagan, FEBS Lett., 2003, 547, 111–114 CrossRef CAS.
- S. L. Cobb, H. Deng, J. T. G. Hamilton, R. P. McGlinchey and D. O'Hagan, Chem. Commun., 2004, 592–593 RSC.
- M. Onega, R. P. McGlinchey, H. Deng, J. T. G. Hamilton and D. O'Hagan, Bioorg. Chem., 2007, 35, 375–385 CrossRef CAS PubMed.
- S. J. Moss, C. D. Murphy, D. O'Hagan, C. Schaffrath, J. T. G. Hamilton, W. C. McRoberts and D. B. Harper, Chem. Commun., 2000, 2281–2282 RSC.
- C. D. Murphy, S. J. Moss and D. O'Hagan, Appl. Environ. Microbiol., 2001, 67, 4919–4921 CrossRef CAS PubMed.
- (a) C. Murphy, D. O'Hagan and C. Schaffrath, Angew. Chem., Int. Ed., 2001, 4479–4481 CrossRef CAS; (b) H. Deng, S. M. Cross, R. P. McGlinchey, J. T. G. Hamilton and D. O'Hagan, Chem. Biol., 2008, 15, 1268–1276 CrossRef CAS PubMed.
- H. Deng, L. Ma, N. Bandaranayaka, Z. Qin, G. Mann, K. Kyeremeh, Y. Yu, T. Shepherd, J. H. Naismith and D. O'Hagan, ChemBioChem, 2014, 15, 364–368 CrossRef CAS PubMed.
- Y. Wang, Z. Deng and X. Qu, F1000Research, 2014, 61, 1–14 CAS.
- S. Huang, L. Ma, M. H. Tong, Y. Yu, D. O'Hagan and H. Deng, Org. Biomol. Chem., 2014, 12, 4828–4831 CAS.
- S. L. Cobb, H. Deng, J. T. G. Hamilton, R. P. McGlinchey, D. O'Hagan and C. Schaffrath, Bioorg. Chem., 2005, 33, 393–401 CrossRef CAS PubMed.
- A. S. Eustáquio, R. P. McGlinchey, Y. Liu, C. Hazzard, L. L. Beer, G. Florova, M. M. Alhamadsheh, A. Lechner, A. J. Kale, Y. Kobayashi, K. Reynolds and B. S. Moore, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 12295–12300 CrossRef PubMed.
- W. Fenical, P. R. Jensen, M. Palladino, K. S. Lam, G. K. Lloyd and B. C. Potts, Bioorg. Med. Chem., 2009, 17, 2175–2180 CrossRef CAS PubMed.
- A. S. Eustáquio, F. Pojer, J. P. Noel and B. S. Moore, Nat. Chem. Biol., 2008, 4, 69–74 CrossRef PubMed.
- A. S. Eustáquio, D. O'Hagan and B. S. Moore, J. Nat. Prod., 2010, 73, 378–382 CrossRef PubMed.
- A. J. Kale, R. P. McGlinchey and B. S. Moore, J. Biol. Chem., 2010, 285, 33710–33717 CrossRef CAS PubMed.
- Y. H. Jin and C. K. Chu, Tetrahedron Lett., 2002, 43, 4141–4143 CrossRef CAS.
- M. B. Fusaro, V. Chagnault, S. Josse and D. Postel, Tetrahedron, 2013, 69, 5880–5883 CrossRef CAS PubMed.
- P. Nasomjai, D. O'Hagan and A. M. Z. Slawin, Beilstein J. Org. Chem., 2009, 5, A37 Search PubMed.
- A. Lechner, A. S. Eustáquio, T. A. M. Gulder, M. Hafner and B. S. Moore, Chem. Biol., 2011, 18, 1527–1536 CrossRef CAS PubMed.
- Recently a fluoroaromatic was claimed as a new metabolite from a Streptomyces isolate,27 however it was subsequently shown to be an insecure identification.28 Also some derivatives of 5-fluorouracil were claimed29 as isolates from a sponge in the South China Sea, however these most likely arise from industrial contamination and then bio-accumulation. The last bona fide fluorometabolites to be identified are derivatives of ω-fluoro-oleate from seeds of the West African plant Dichapetalum toxicarium.30.
- N. Jaivel, C. Uvarani, R. Rajesh, D. Velmurugan and P. Marimuthu, J. Nat. Prod., 2014, 77, 2–8 CrossRef CAS PubMed.
- M. S. Ayoup, D. B. Cordes, A. M. Z. Slawin and D. O’Hagan, J. Nat. Prod., 2014, 77, 1249–1251 CrossRef CAS PubMed.
- X. H. Xu, G. M. Yao, Y. M. Li, C. J. Lin, X. Wang and C. H. Kong, J. Nat. Prod., 2003, 66, 285–288 CrossRef CAS PubMed.
- W. W. Christie, J. T. G. Hamilton and D. B. Harper, Chem. Phys. Lipids, 1998, 97, 41–47 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: The fdr gene cluster was deposited in EMBL Nucleotide Sequence Database under the accession no: LN612605. See DOI: 10.1039/c4sc03540b |
| ‡ Current address: School of Biotechnology, Tianjin University of Science & Technology, Tianjin 300457, P. R. China. |
| This journal is © The Royal Society of Chemistry 2015 |