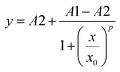Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Direct interfacial Y731 oxidation in α2 by a photoβ2 subunit of E. coli class Ia ribonucleotide reductase†
David Y.
Song
 a,
Arturo A.
Pizano
a,
Patrick G.
Holder
a,
Arturo A.
Pizano
a,
Patrick G.
Holder
 a,
JoAnne
Stubbe
*b and
Daniel G.
Nocera
*a
a,
JoAnne
Stubbe
*b and
Daniel G.
Nocera
*a
aDepartment of Chemistry and Chemical Biology, 12 Oxford Street, Cambridge, MA 02138-2902, USA. E-mail: dnocera@fas.harvard.edu
bDepartment of Chemistry, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139-4307, USA. E-mail: stubbe@mit.edu
First published on 8th June 2015
Abstract
Proton-coupled electron transfer (PCET) is a fundamental mechanism important in a wide range of biological processes including the universal reaction catalysed by ribonucleotide reductases (RNRs) in making de novo, the building blocks required for DNA replication and repair. These enzymes catalyse the conversion of nucleoside diphosphates (NDPs) to deoxynucleoside diphosphates (dNDPs). In the class Ia RNRs, NDP reduction involves a tyrosyl radical mediated oxidation occurring over 35 Å across the interface of the two required subunits (β2 and α2) involving multiple PCET steps and the conserved tyrosine triad [Y356(β2)–Y731(α2)–Y730(α2)]. We report the synthesis of an active photochemical RNR (photoRNR) complex in which a Re(I)-tricarbonyl phenanthroline ([Re]) photooxidant is attached site-specifically to the Cys in the Y356C-(β2) subunit and an ionizable, 2,3,5-trifluorotyrosine (2,3,5-F3Y) is incorporated in place of Y731 in α2. This intersubunit PCET pathway is investigated by ns laser spectroscopy on [Re356]-β2:2,3,5-F3Y731-α2 in the presence of substrate, CDP, and effector, ATP. This experiment has allowed analysis of the photoinjection of a radical into α2 from β2 in the absence of the interfacial Y356 residue. The system is competent for light-dependent substrate turnover. Time-resolved emission experiments reveal an intimate dependence of the rate of radical injection on the protonation state at position Y731(α2), which in turn highlights the importance of a well-coordinated proton exit channel involving the key residues, Y356 and Y731, at the subunit interface.
Introduction
Enzymatic control of coupled proton and electron transfer1–4 is critical in managing biological processes ranging from energy storage (photosystem II)5–9 and conversion (cytochrome c oxidase)10 to the synthesis of DNA precursors (ribonucleotide reductase).11–14 To better understand biological PCET, we have undertaken studies of this process in class Ia RNRs, which catalyse the conversion of nucleoside diphosphates (NDPs) to deoxynucleoside diphosphates (dNDPs)—a process required for synthesis and repair of DNA in all organisms.15,16 Catalysis by the class I RNRs proceeds by a radical mechanism requiring coupling of radical transport over 35 Å involving PCET across the two subunits to substrate turnover. The long distance, reversibility, and rate-limiting conformational gating of radical transport have made study of this process challenging. To overcome this challenge, we have developed two methodologies: photoRNRs17–21 and site-specific incorporation of unnatural amino acids in place of pathway residues.22–24E. coli class Ia RNR has served as the paradigm for this long distance radical transport. It is composed of two homodimeric subunits: α2 and β2. A docking model for this complex,25 substantiated by recent biochemical and biophysical studies,26,27 has provided the working model for the radical transport pathway shown in Fig. 1. The active site for NDP reduction resides in α2, where the cysteine radical (C439˙) must be transiently generated during each turnover by the essential diferric-tyrosyl radical (Y122˙) cofactor in β2. This long range oxidation requires a multi-step radical hopping mechanism that involves a specific pathway including four tyrosines (Y122 and Y356 in β2; Y731 and Y730 in α2)11,28 and potentially W48 in β2.11
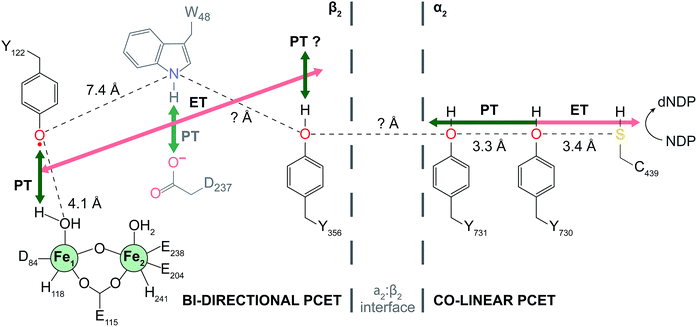 | ||
| Fig. 1 Current model of radical transport pathway in class Ia RNR leading to nucleotide reduction as determined by the docking model16 and diagonal distance measurements acquired by PELDOR spectroscopy.34 Key redox active amino acids and known distance measurements involved in PCET pathway are shown. Residue W48 is grayed indicating the absence of experimental evidence supporting its participation in the PCET radical mechanism. Residue Y356 is shown at the interface for illustrative purposes. Salmon arrows indicate electron transfer (ET) and green arrows represent proton transfer (PT). In α2, co-linear PCET is denoted by the dual-colored arrow, and the proposed bi-directional PCET in β2 is indicated by the orthogonality of the ET and PT pathways. | ||
Recent attention has focused on the detection of the proposed transient radical intermediates and identification of the operative PCET mechanism at each site. Mössbauer studies have established that Y122˙ reduction in β2 is triggered by binding of substrate and effector to α2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 29 and involves proton donation from the water at Fe1 (Fig. 1). This process involves orthogonal PCET wherein the proton and electron come from different residues. High-field electron paramagnetic resonance (Hf EPR) and deuterium electron nuclear double resonance (ENDOR) have provided atomic level resolution of local hydrogen bond interactions, specifically the co-linearity of the PCET within α2. Additionally, significant shifts in gx values together with the assignment of hyperfine coupling features from the ENDOR spectra of various amino-substituted RNR mutants propose an important role for electrostatics at the α2:β2 interface.30 However, the disordered C-terminal tail of β2 where Y356 resides has made interrogation of the chemistry at the subunit interface challenging (Fig. 1).
29 and involves proton donation from the water at Fe1 (Fig. 1). This process involves orthogonal PCET wherein the proton and electron come from different residues. High-field electron paramagnetic resonance (Hf EPR) and deuterium electron nuclear double resonance (ENDOR) have provided atomic level resolution of local hydrogen bond interactions, specifically the co-linearity of the PCET within α2. Additionally, significant shifts in gx values together with the assignment of hyperfine coupling features from the ENDOR spectra of various amino-substituted RNR mutants propose an important role for electrostatics at the α2:β2 interface.30 However, the disordered C-terminal tail of β2 where Y356 resides has made interrogation of the chemistry at the subunit interface challenging (Fig. 1).
Rate limiting conformational gating in RNR obscures radical transport across the subunit interface, prompting us to develop photoRNRs to trigger radical initiation with light to avoid this gating and to potentially enable the observation of Y˙ at the interface. Radical injection kinetics were initially made possible using a 19mer peptide photoRNR, which corresponded to the identical 19 residues of the C-terminal tail of β2 along with a modification that appended a photooxidant (rhenium phenanthroline [Re]) adjacent to Y356 or fluorinated derivatives. This peptide photoRNR enables nucleotide reduction in the presence of α2 and light and allows for observation of radical injection into α2.21 Radical injection was only realized in the presence of an intact Y731–Y730 dyad within α2, providing important support for co-linear PCET within this subunit.
More recently, photoRNRs have been generated in which the peptide with the photooxidant is replaced by the full-length β2 containing a site-specifically incorporated [Re] photooxidant at residue 355 using a S355C-β2 mutant.31 Transient absorption spectroscopy on the active [Re355]-β2:α2 complex in comparison to the control [Re355]-Y356F-β2:α2 complex allowed for the assignment of a photogenerated ˙Y356.32 In addition, further modification of [Re355]-β2 by installation of an unnatural 2,3,5-F3Y in place of Y356 in β2 yielded the first direct measurement of ˙Y propagation kinetics through the active RNR complex.33 Comparison of ˙Y decay transient kinetics in the [Re355]-2,3,5-F3Y356-β2:α2 complex in the presence of substrate, either [3′-1H]-CDP or [3′-2H]-CDP, and effector ATP, revealed an observed rate constant of 1.4 × 104 s−1 and unmasked for the first time an isotope effect on cleavage of the 3′ C–H of the substrate. The photoRNRs thus circumvent the masking of radicals by conformational gating and thus have provided insight regarding radical transport and nucleotide reduction chemistry not accessible by any other method.
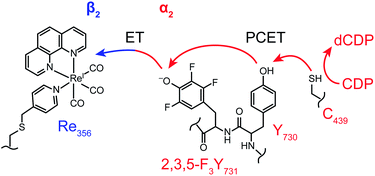
In this work, the [Re] photooxidant is attached to Cys in the Y356C-β2 mutant and this modified [Re356]-β2 subunit is associated to α2 in which Y731 is site-specifically replaced with 2,3,5-F3Y. This [Re356]-β2:2,3,5-F3Y731-α2 complex together with the [Re356]-β2:wt-α2 and [Re356]-β2:Y731F-α2 control complexes are studied in the presence of CDP and ATP. In contrast to previous photoRNR systems, installation of [Re] at position 356 enables direct interfacial generation of a tyrosyl radical at position 731. Additionally, by leveraging the greater acidity of 2,3,5-F3Y to enable deprotonation at neutral pH, this residue furnishes an ionizable reporter that varies with experimental pH. In turn, site-selective removal of a single proton at position Y731(α) provides the first protein:protein scaffold of RNR that permits the investigation of the effect of a modified proton microenvironment on radical transport on transient time scales (sub μs) at the interface. In the absence of Y356, radical injection is only achieved when position Y731 is deprotonated. In addition to confirming the complexity of RNR in maintaining a well-organized PCET pathway, this work introduces and highlights the importance of a well-defined proton exit channel out of α2 involving the key pathway residues, Y356 and Y731, at the subunit interface.
Experimental
Modified RNR subunits were constructed, expressed, purified, modified, and characterized as previously reported or with minor modification.19,23,24,31,35 Protein concentrations were measured by absorbance at 280 nm using: εα2 = 189![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1, εβ2-apo = 121
000 M−1 cm−1, εβ2-apo = 121![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1, εβ2-holo = 131
000 M−1 cm−1, εβ2-holo = 131![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1, and εβ2-[Re] = 189
000 M−1 cm−1, and εβ2-[Re] = 189![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1. Purity of protein constructs was assessed by SDS-PAGE (Fig. S1†). All measurements were conducted in assay buffer at pH 7.6 (50 mM HEPES, 15 MgSO4, 1 mM EDTA; unless otherwise specified). Measurement of the dissociation constant (KD) between [Re356]-β2 and wt-α2 was performed by a spectrophotometric competitive inhibition assay as previously reported.17,21 Measurement of the pKa of the phenolic proton of 2,3,5-F3Y731 within the assembled 2,3,5-F3Y731-α2:[Re356]-β2 complex was performed by fluorometric titration as previously reported.21 The details of methods that deviate from published procedures are provided in the ESI.† Similarly, photoinitiated nucleotide reduction activity assays were performed according to published methods.17–19,32 Error bars represent 2σ resulting from photolysis on ≥three independent samples.
000 M−1 cm−1. Purity of protein constructs was assessed by SDS-PAGE (Fig. S1†). All measurements were conducted in assay buffer at pH 7.6 (50 mM HEPES, 15 MgSO4, 1 mM EDTA; unless otherwise specified). Measurement of the dissociation constant (KD) between [Re356]-β2 and wt-α2 was performed by a spectrophotometric competitive inhibition assay as previously reported.17,21 Measurement of the pKa of the phenolic proton of 2,3,5-F3Y731 within the assembled 2,3,5-F3Y731-α2:[Re356]-β2 complex was performed by fluorometric titration as previously reported.21 The details of methods that deviate from published procedures are provided in the ESI.† Similarly, photoinitiated nucleotide reduction activity assays were performed according to published methods.17–19,32 Error bars represent 2σ resulting from photolysis on ≥three independent samples.
Time-resolved spectroscopic measurements were performed using a home-built nanosecond laser system previously described.21,31–33 Each sample was prepared prior to photolysis and measurements were performed in triplicate. The calculation of the uncertainty in experimental measurements to 95% confidence limits (2σ) is described in the ESI (eqn S1–S5†).
Results and discussion
PhotoRNRs: [Re356]-β2:α2 and [Re356]-β2:2,3,5-F3Y731-α2
To probe radical initiation across the α2:β2 interface, specific variants of each subunit were required. To directly target the intersubunit radical transport step of Y731 oxidation and subsequent radical injection into α2, we chose to circumvent Y356 oxidation entirely. In contrast to previous systems where photooxidants were placed adjacent to Y356 at position 355, [Re] replaces Y356 in this study. The new construct maintains the mutations C268S and C305S, and preserves catalytic activity. Additionally, the mutation, Y356C, thus enables alkylation with [Re]-Br to yield [Re356]-β2. To examine the effect of a proton at position Y731, this residue was replaced with 2,3,5-F3Y, solution pKa = 6.4, compared with Y (pKa = ∼10). The photoRNR construct is illustrated in Fig. 2. These two subunit modifications allow for direct oxidization of Y731 by a photoβ2.Construction, expression, isolation, and labelling to generate [Re356]-β2 were performed as previously reported with minor modifications.31 Unlabelled and reconstituted Y356C-β2 (holo) is inactive (0.16(5) U) towards nucleotide reduction (wt-β2 activity = 6000–8000 U), as expected given the absence of Y356. We note that labelling does not preclude binding (KD, = 0.43(11) μM), as measured in the competitive inhibition assay shown in Fig. S2.† This value is in agreement with previously reported values for active photoRNR β2 mutants,32 and not significantly altered from wt-RNR.12,36
[Re356]-β2 displays similar spectroscopic and photophysical properties, as reported previously for [Re355]-β2.31 Fig. S3† shows typical absorption and emission spectra for [Re356]-β2; the absorption spectrum is dominated by the characteristic features of rhenium(I) tricarbonyl bisimine compounds and the emission originates from a triplet metal-to-ligand charge transfer state. The absorption spectrum of [Re356]-β2 is accurately re-constructed as the sum of Y356C-β2 and twice the [Re]-Br absorption spectrum, as expected for a construct with two [Re] molecules per β2 dimer (Fig. S3†).
Construction of 2,3,5-F3Y731-α2 was achieved using in vivo nonsense suppression methodology previously developed to install fluorotyrosine reporters in class Ia RNR.24 The specific activity of 2,3,5-F3Y731-α2 under catalytic conditions (375(5) U) is diminished relative to wt-α2 (1800–2500 U), while under single-turnover activity (2.75(4) equiv. dCDP/α2) is comparable to wt-α2 (∼3 dCDP equiv./α2). This decrease in catalytic activity, though comparable to that of wild-type (wt), is consistent with previous reports of this mutant.37
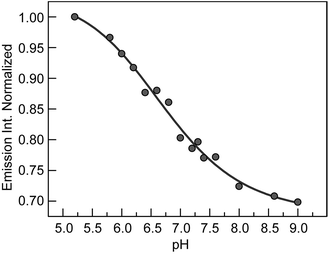
The deprotonation of 2,3,5-F3Y731 is expected to perturb the rate of radical generation at position Y731. The kinetic penalty associated with proton transfer is alleviated by removal of the proton when experimental pH > pKa, thus requiring measurement of the precise pKa of 2,3,5-F3Y731 in the assembled construct. As previously reported,19–21,32,33,38 the increase in the rate of photooxidation for tyrosinate relative to tyrosine,39 makes [Re] emission a reporter of the protonation state of nearby tyrosine residues. Fluorometric titration of the [Re356]-β2:2,3,5-F3Y731-α2 complex reveals the pKa of 2,3,5-F3Y731 to be 6.7(1), Fig. 3. Accordingly, ∼90% of 2,3,5-F3Y731 residues are deprotonated under experimental conditions at the optimal operating pH for RNR (pH 7.6). Given the thermodynamically unfavourable acidity of the tyrosyl radical cation (pKa = −2) a PCET process managing both the electron and proton transfers is mandated.40,41
Photoinitiated substrate turnover
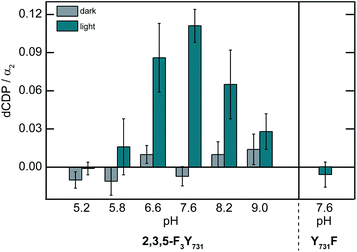
To establish that the photoRNR construct is competent to generate dCDP, the [Re356]-β2:2,3,5-F3Y731-α2 complex in the presence of [3H]-CDP and ATP was photolyzed for 10 min (λ > 313 nm) and dCDP was measured by scintillation counting. The results of this single turnover experiment are shown in Fig. 4. Perturbation of the enzyme by the introduction of [Re] results in a reduced level of turnover that is 5–10% relative to wt-RNR under the same pH conditions. Notwithstanding, the presence of photogenerated products establish the relevance of [Re356]-β2:2,3,5-F3Y731-α2 complex to the natural enzyme. Attenuated enzymatic activity is also detected at pH > 7.6 which is consistent with the observed pH rate profiles for the wt enzyme and fluorotyrosine derivatized RNR constructs.42Radical injection kinetics
The photogeneration of product for the [Re356]-β2:2,3,5-F3Y731-α2 complex prompted us to undertake radical injection kinetics studies. Using the [Re]* emission lifetime as a reporter for radical injection, ns TA laser spectroscopy on the [Re356]-β2:α2 complexes in the presence of CDP and ATP was conducted. The emission decay lifetimes for each construct were measured at pH = 7.6, where maximum turnover was observed, and are summarized in Table 1; representative traces are included in Fig. S4.† As previously observed for [Re355]-β2,32 the [Re]* lifetime in [Re356]-β2 (τ = 507(3) ns) increases upon binding to Y731F-α2 (τo = 652(4) ns), consistent with the more hydrophobic environment engendered by the protein environment. The lifetime τo of the Y731F-α2 variant provides a reference for excited-state decay of [Re]* when it is located at the interface, but in the absence of quenching by the tyrosine located at position 731. Upon introduction of Y731, the [Re]* emission (τ = 629(4) ns) is quenched relative to Y731F-α2 according to the following equation: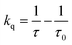 | (1) |
| [Re356]-β2: X731-α2 | τ 600 nm ns−1 | k q/104 s−1b |
|---|---|---|
| a Lifetime of emission decay measured on 10 μM [Re356]-β2, 25 μM α2 (as indicated), 1 mM CDP, 3 mM ATP in assay buffer (pH 7.6), λexc = 355 nm, λobs = 600 nm. Errors shown in parentheses represent 2σ resulting from measurement on ≥3 independent samples. b Emission quenching rate constant, kq, determined from eqn (1). Error in quenching rate constants calculated as shown in ESI. | ||
| F | 652(4) | — |
| Y | 629(4) | 5.6 (1.4) |
| 2,3,5-F 3 Y | 593(6) | 15.4 (2.2) |
Accordingly, this quenching rate constant, kq, is equivalent to the radical generation rate, and from eqn (1) it is calculated to be 5.6(1.4) × 104 s−1. In comparison to previously observed photooxidation of Y356 in [Re355]-β2 (kq = 4.1(1) × 105 s−1), the direct oxidation of Y731 by [Re356]*-β2 is slower possibly owing to an increased charge transfer distance, which is occurring across the subunit interface vs. at adjacent positions in previous investigations. Further analysis of the emission decay kinetics of [Re]* reveal a dependency on the rate of radical injection and the protonation state of tyrosine at position Y731(α). Replacement of Y731 with 2,3,5-F3Y731 enhances the quenching rate constant by a factor of three (τ = 593(7) ns (kq = 15.4(2.2) × 104 s−1). This acceleration of radical injection into α2 is in accordance with the differing protonation states of tyrosine in the two constructs. Under the experimental conditions of pH = 7.6, 2,3,5-F3Y731 is deprotonated and hence quenching occurs by ET rather than PCET, which results in faster tyrosine oxidation, despite being ∼50–100 mV more difficult to oxidize than native tyrosine at this pH (as determined from solution peak potentials (Ep) of N-acetyl and C-amide protected fluorotyrosines measured by differential pulse voltammetry).22 For clarity, obtaining precise single reside midpoint potentials in protein constructs is extremely challenging, and thus thermodynamic considerations must be guided from these measured Ep values,11 despite recent findings that suggest significant deviations of the formal reduction potential and solution Ep values.43
To investigate how the proton at position Y731 affects interfacial PCET and subsequent interfacial radical injection kinetics, emission quenching of the [Re]* within the [Re356]-β2:2,3,5-F3Y731-α2 complex was monitored over the activity accessible pH region of RNR. A plot of the [Re]* decay lifetimes for [Re356]-β2 alone and in the three [Re356]-β2:α2 variant complexes monitored as a function of pH are shown in Fig. S5;† representative emission decay traces are provided in Fig. S4.† The quenching rate constants, kq, as a function of pH may be determined from eqn (1); these rate constants are plotted in Fig. 5. Quenching by Y731(wt)-α2 (red circles,  ) and 2,3,5-F3Y731-α2 (light blue squares,
) and 2,3,5-F3Y731-α2 (light blue squares,  ) are referenced to the control, Y731F-α2. Within our error limits, little to no dependence of kq is observed for the [Re356]-β2:Y731(wt)-α2 complex (
) are referenced to the control, Y731F-α2. Within our error limits, little to no dependence of kq is observed for the [Re356]-β2:Y731(wt)-α2 complex ( ), as native tyrosine is protonated throughout the pH window. Conversely, a large pH dependence is observed for kq when [Re356]-β2:2,3,5-F3Y731-α2 is compared to Y731F (
), as native tyrosine is protonated throughout the pH window. Conversely, a large pH dependence is observed for kq when [Re356]-β2:2,3,5-F3Y731-α2 is compared to Y731F ( ). Guided by our measurement of the pKa of 2,3,5-F3Y731-α2, shown in Fig. 3, we ascribe the observed differences in quenching to the relative ratio of deprotonated:protonated forms of 2,3,5-F3Y731 as pH is varied. The large kqs at high pH is expected owing to deprotonation of F3Y731 whereas quenching at low pH approaches that of Y731 where both of the tyrosines are protonated.
). Guided by our measurement of the pKa of 2,3,5-F3Y731-α2, shown in Fig. 3, we ascribe the observed differences in quenching to the relative ratio of deprotonated:protonated forms of 2,3,5-F3Y731 as pH is varied. The large kqs at high pH is expected owing to deprotonation of F3Y731 whereas quenching at low pH approaches that of Y731 where both of the tyrosines are protonated.
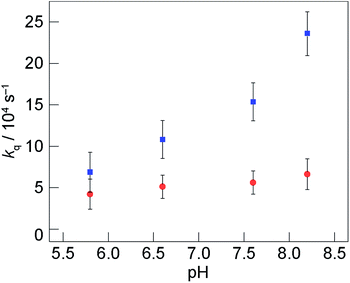 | ||
Fig. 5 Quenching rate constant of [Re]* by tyrosine, kq, monitored as function of pH: Y731(wt) relative to Y731F ( ) and 2,3,5-F3Y731 relative to Y731F ( ) and 2,3,5-F3Y731 relative to Y731F ( ). τo corresponds to the lifetime of [Re356]-β2:Y731F-α2. Error bars in kq represent 2σ resulting from error propagation on lifetime measurements as described in ESI.† ). τo corresponds to the lifetime of [Re356]-β2:Y731F-α2. Error bars in kq represent 2σ resulting from error propagation on lifetime measurements as described in ESI.† | ||
Scheme 1 summarizes the decay pathways for [Re]* in the different variants. The presence of tyrosine introduces an excited-state decay pathway via radical generation. For the case of Y731(wt)-α2, oxidation occurs by PCET whereas for 2,3,5-F3Y731-α2 occurs by ET. These differing mechanisms of tyrosine oxidation for the two variants in the absence of Y356 introduce the possibility that Y356 may be involved with facilitating proton removal at the interface, though future experiments are needed to establish its specific role. While a co-linear Y356–Y731 π-stacked mode where Y356 acts directly as the proton acceptor for Y731 is unlikely in light of recent Hf EPR/ENDOR data on amino-substituted tyrosine derivatives at various pathway positions, the strongly perturbed gx values of NH2Y356˙ indicate that Y356 may communicate with Y731 through a network of water molecules.30 Additional evidence supporting the involvement of Y356 in modulating Y731 oxidation was also observed in [Re355]-β2 construct.32 The photooxidation kinetics of Y731, which are summarized in Table S1,† indicate that Y731 radical generation is enhanced by the presence of Y356 as [Re355]-β2 oxidation of Y731 is 2.1(1.2) times faster for Y356 than for F356. The presence of Y356 may facilitate proton removal from α2via the interface, thus assisting in PCET.
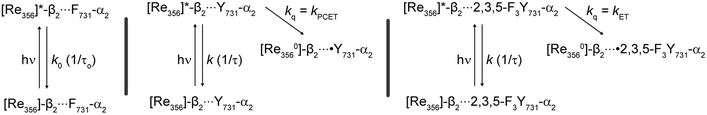 | ||
| Scheme 1 Excited-state deactivation pathways for [Re356]-β2 in the presence of indicated α2 subunit. | ||
In this directed study, whereby Y356 is absent by virtue of its replacement with [Re356], efficient injection of a radical into α2 is realized only when a proton is removed from the pathway by the introduction of 2,3,5-F3Y731. While this result does not implicate Y356 directly as the proton acceptor for Y731, it supports the contention that Y356 is in communication with Y731 at the α2:β2 subunit interface and that Y356 enables the PCET required for efficient radical transport. Further investigations of this contention are underway along with studies to assess the role of possible contributions from other residues, or perhaps metal ions that may also be involved in managing protons at the interface.
Conclusions
Replacement of Y356 by a [Re] photooxidant and installation of 2,3,5-F3Y at position 731 in α2 furnishes a photoRNR that specifically targets intersubunit radical transport. This construct supports photoinitiated substrate turnover, confirming its fidelity to the natural system. Time-resolved emission studies reveal that 2,3,5-F3Y731 is oxidized at a rate 3 times faster than native Y731 even though the non-natural amino acid is thermodynamically more difficult to oxidize at pH 7.6 (ΔEp ∼ 50–100 mV). These results emphasize the enzymatic imperative for coupling the proton and electron to allow for efficient radical transport. In conjunction with the parallel studies of [Re355]-β2, these results suggest the importance of a well-coordinated proton exit channel involving Y356 and Y731 as key interfacial residues for radical transport across the α2:β2 interface.Acknowledgements
DYS acknowledges the National Science Foundation's Division of Graduate Education Grant DGE-1144152. PGH thanks the National Institute of Health for a Post-Doctoral Fellowship (GM087034). This research was supported by the U.S. National Institute of Health Grants GM047274 (DGN) and GM029595 (JS).Notes and references
- A. Migliore, N. F. Polizzi, M. J. Therien and D. N. Beratan, Chem. Rev., 2014, 114, 3381–3645 CrossRef CAS PubMed.
- D. R. Weinberg, C. J. Gagliardi, J. F. Hull, C. F. Murphy, C. A. Kent, B. C. Westlake, A. Paul, D. H. Ess, D. G. McCafferty and T. J. Meyer, Chem. Rev., 2012, 112, 4016–4093 CrossRef CAS PubMed.
- S. Hammes-Schiffer and A. A. Stuchebrukhov, Chem. Rev., 2010, 110, 6939–6960 CrossRef CAS PubMed.
- S. Y. Reece and D. G. Nocera, Annu. Rev. Biochem., 2009, 78, 673–699 CrossRef CAS PubMed.
- J. Barber, Biochem. Soc. Trans., 2006, 34, 619–631 CrossRef CAS PubMed.
- C. C. Moser, C. C. Page, R. J. Cogdell, J. Barber, C. A. Wraight and P. L. Dutton, Adv. Protein Chem., 2003, 63, 71–109 CrossRef CAS.
- J. D. Megiatto Jr, D. D. Mendez-Hernandex, M. E. Tejeda-Ferrari, A. L. Teillout, M. J. Llansola-Portoles, G. Kodis, O. G. Poluektov, T. Rajh, V. Mujica, T. L Groy, D. Gust, T. A. Moore and A. L. Moore, Nat. Chem., 2014, 6, 423–428 CrossRef PubMed.
- B. A. Barry, J. Photochem. Photobiol., B, 2011, 104, 60–71 CrossRef CAS PubMed.
- J. L. Dempsey, J. R. Winkler and H. B. Gray, Chem. Rev., 2010, 110, 7024–7039 CrossRef CAS PubMed.
- V. R. I. Kaila, M. I. Verkhovsky and M. Wikstrom, Chem. Rev., 2010, 110, 7062–7081 CrossRef CAS PubMed.
- E. C. Minnihan, D. G. Nocera and J. Stubbe, Acc. Chem. Res., 2013, 46, 2524–2535 CrossRef CAS PubMed.
- J. Stubbe, D. G. Nocera, C. S. Yee and M. C. Y. Chang, Chem. Rev., 2003, 103, 2167–2202 CrossRef CAS PubMed.
- J. Stubbe and W. A. van der Donk, Chem. Rev., 1998, 98, 705–762 CrossRef CAS PubMed.
- P. E. M. Siegbahn, L. Eriksson, F. Himo and M. Pavlov, J. Phys. Chem. B, 1998, 102, 10622–10629 CrossRef CAS.
- P. Nordlund and P. Reichard, Annu. Rev. Biochem., 2006, 75, 681–706 CrossRef CAS PubMed.
- M. Kolberg, K. R. Strand, P. Graff and K. K. Andersson, Biochim. Biophys. Acta, 2004, 1699, 1–34 CrossRef CAS.
- M. C. Y. Chang, C. S. Yee, J. Stubbe and D. G. Nocera, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 6882–6887 CrossRef CAS PubMed.
- S. Y. Reece, M. R. Seyedsayamdost, J. Stubbe and D. G. Nocera, J. Am. Chem. Soc., 2007, 129, 8500–8509 CrossRef CAS PubMed.
- S. Y. Reece, M. R. Seyedsayamdost, J. Stubbe and D. G. Nocera, J. Am. Chem. Soc., 2007, 129, 13828–13830 CrossRef CAS PubMed.
- S. Y. Reece, D. A. Lutterman, M. R. Seyedsayamdost, J. Stubbe and D. G. Nocera, Biochemistry, 2009, 48, 5832–5838 CrossRef CAS PubMed.
- P. G. Holder, A. A. Pizano, B. L. Anderson, J. Stubbe and D. G. Nocera, J. Am. Chem. Soc., 2012, 134, 1172–1180 CrossRef CAS PubMed.
- M. R. Seyedsayamdost, S. Y. Reece, D. G. Nocera and J. Stubbe, J. Am. Chem. Soc., 2006, 128, 1569–1579 CrossRef CAS PubMed.
- M. R. Seyedsayamdost, C. S. Yee and J. Stubbe, Nat. Protoc., 2007, 2, 1225–1235 CrossRef CAS PubMed.
- E. C. Minnihan, D. D. Young, P. G. Schultz and J. Stubbe, J. Am. Chem. Soc., 2011, 133, 15942–15945 CrossRef CAS PubMed.
- M. Eriksson, U. Uhlin, S. Ramaswamy, M. Ekberg, K. Regnström, B. M. Sjöberg and H. Eklund, Structure, 1997, 5, 1077–1092 CrossRef CAS.
- N. Ando, E. J. Brignole, C. M. Zimanyi, M. A. Funk, K. Yokoyama, F. J. Asturias, J. Stubbe and C. L. Drennan, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 21046–21051 CrossRef CAS PubMed.
- E. C. Minnihan, N. Ando, E. J. Brignole, L. Olshansky, J. Chittuluru, F. J. Asurias, C. L. Drennan, D. G. Nocera and J. Stubbe, Proc. Natl. Acad. Sci. U. S. A., 2013, 109, 39–43 Search PubMed.
- T. U. Nick, W. Lee, S. Koßmann, F. Neese, J. Stubbe and M. Bennati, J. Am. Chem. Soc., 2015, 137, 289–298 CrossRef CAS PubMed.
- B. Wörsdörfer, D. A. Conner, K. Yokoyama, J. Livada, M. R. Seyedsayamdost, W. Jiang, A. Silakov, J. Stubbe, J. M. Bollinger Jr and C. Krebs, J. Am. Chem. Soc., 2013, 135, 8585–8593 CrossRef PubMed.
- T. Argirević, C. Riplinger, J. Stubbe, F. Neese and M. Bennati, J. Am. Chem. Soc., 2012, 134, 17661–17670 CrossRef PubMed.
- A. A. Pizano, D. A. Lutterman, P. G. Holder, T. S. Teets, J. Stubbe and D. G. Nocera, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 39–43 CrossRef CAS PubMed.
- A. A. Pizano, L. Olshansky, P. G. Holder, J. Stubbe and D. G. Nocera, J. Am. Chem. Soc., 2013, 135, 13250–13253 CrossRef CAS PubMed.
- L. Olshansky, A. A. Pizano, Y. Wei, J. Stubbe and D. G. Nocera, J. Am. Chem. Soc., 2014, 136, 16210–16216 CrossRef CAS PubMed.
- M. R. Seyedsayamdost, C. T. Y. Chan, V. Mugnaini, J. Stubbe and M. Bennati, J. Am. Chem. Soc., 2007, 129, 15478–15749 Search PubMed.
- J. Ge, G. Yu, M. A. Ator and J. Stubbe, Biochemistry, 2003, 42, 10071–10083 CrossRef CAS PubMed.
- M. C. Y. Chang, Ph.D. Thesis, Dept. of Chemistry, MIT, 2004.
- E. C. Minnihan, Ph.D. Thesis, Dept. of Chemistry, MIT, 2012.
- K. Yokoyama, U. Uhlin and J. Stubbe, J. Am. Chem. Soc., 2010, 132, 8385–8397 CrossRef CAS PubMed.
- T. Irebo, M. T. Zhang, T. F. Markle, A. M. Scott and L. Hammarström, J. Am. Chem. Soc., 2012, 134, 16247–16254 CrossRef CAS PubMed.
- M. Sjodin, S. Styring, B. Akermark, L. Sun and L. Hammarström, J. Am. Chem. Soc., 2000, 122, 3932–3936 CrossRef.
- C. Carra, N. Iordanova and S. Hammes-Schiffer, J. Am. Chem. Soc., 2003, 125, 10429–10436 CrossRef CAS PubMed.
- M. R. Seyedsayamdost, C. S. Yee, S. Y. Reece, D. G. Nocera and J. Stubbe, J. Am. Chem. Soc., 2009, 131, 1562–1568 CrossRef PubMed.
- K. R. Ravichandran, L. Liang, J. Stubbe and C. Tommos, Biochemistry, 2013, 52, 8907–8915 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures, calculation of uncertainty in data analysis, purity gels, determination of KD, spectroscopic characterization, time-resolved emission decay traces, and table from reference S8 are provided. See DOI: 10.1039/c5sc01125f |
| This journal is © The Royal Society of Chemistry 2015 |