Open Access Article
Open Access ArticleBridging cells of three colors with two bio-orthogonal click reactions†
Yue
Yuan‡
a,
Di
Li‡
b,
Jia
Zhang
a,
Xianmin
Chen
a,
Chi
Zhang
c,
Zhanling
Ding
a,
Lin
Wang
d,
Xueqian
Zhang
a,
Junhua
Yuan
c,
Yinmei
Li
b,
Yanbiao
Kang
a and
Gaolin
Liang
*a
aCAS Key Laboratory of Soft Matter Chemistry, Department of Chemistry, University of Science and Technology of China, Hefei, Anhui 230026, China. E-mail: gliang@ustc.edu.cn
bHefei National Laboratory for Physical Sciences at the Microscale, Department of Optics and Optical Engineering, University of Science and Technology of China, Hefei, Anhui 230026, China
cHefei National Laboratory for Physical Sciences at the Microscale, Department of Physics, University of Science and Technology of China, Hefei, Anhui 230026, China
dSchool of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230027, China
First published on 28th July 2015
Abstract
Cell–cell interactions play a crucial role in the development and function of multicellular organisms. To study cell–cell interactions in vitro, it is a big challenge for researchers to artificially build up cell junctions to bridge different types of cells for this purpose. Herein, by employing two orthogonal click reactions, we rationally designed four click reagents Mal-CBT, Mal-Cys, Mal-Alkyne, and Mal-N3 and successfully applied them to bridge cells of three colors. Orthogonality between these two click reactions was validated in solution and characterized with HPLC and ESI-MS analyses. After modifications of fluorescent protein-expressing prokaryotic Escherichia coli (E. coli) cells (or eukaryotic HEK 293T cells) of three colors with the reagents Mal-Cys, Mal-CBT and Mal-Alkyne, or Mal-N3, the cells were sequentially bridged. The HEK 293T cells showed a higher efficiency of cell bridging than the E. coli cells. Finally, using optical tweezers, we quantitatively measured the bridging probability between Mal-Cys-modified and Mal-CBT-modified HEK 293 cells, as well as the rupture force between two bridged cells. We found that the CBT–Cys click reaction markedly improved the efficiency of cell bridging and the rupture force between two bridged cells was measured to be 153.8 pN at a force-loading rate of 49 pN s−1. Our results demonstrate that it is possible to use two (or n) orthogonal click reactions to bridge three (or n + 1) types of cells. Taking the biological importance of cell junctions into consideration, we anticipate that our method of bridging three types of cells with two bio-orthogonal click reactions will be a useful tool for biologists to study cell–cell interactions with more convenience and efficiency.
Introduction
Cell–cell interactions between cell surfaces play a crucial role in the development and function of multicellular organisms. Through these interactions, cells are able to communicate with each other in response to their microenvironmental change. But the loss of communication between cells can result in uncontrollable cell growth and cancer.1–3 Cell interactions can be stabilized through cell junctions. Cell junctions (or intercellular bridges) are one type of structure that exist within the tissue of some multicellular organisms (e.g., animals), and are especially abundant in epithelial tissues.4–6 Normally, cell junctions consist of multi-protein complexes.4 On the one hand, cell membranes in the tissues of junctional surfaces are normally so permeable that many cellular ions and molecules may diffuse freely from one cell interior to the next.5 On the other hand, cell junctions build up a paracellular barrier between cells and control paracellular transport.7 When considering the biological importance of cell interactions, it is a big challenge for people to artificially build up cell junctions to bridge different types of cells and study their behavior. For example, to bridge three types of cells with a chemical approach, two bio-orthogonal reactions are needed to fulfill this. Nevertheless, to the best of our knowledge, there has been no report on bridging three types of cells directly with two bio-orthogonal click reactions.Bio-orthogonal reactions are chemical reactions where the participating functional groups selectively react with each other under biocompatible conditions while neither interacting with nor interfering with the biological system.8 The use of bio-orthogonal chemistry to probe biomolecules in living systems typically follows a two-step process: first, a small chemical reporter (e.g., an aldehyde, azide, alkyne, or alkene) is site-selectively introduced into the biomolecule(s) of interest via an appropriate biosynthetic or biochemical pathway; then, a biophysical probe carrying the cognate reactive group bio-orthogonally reacts with the reporter in situ to selectively ligate the pre-tagged biomolecule(s) of interest.9 Using bio-orthogonal approaches, people have gained new insights into a wide range of biological processes, such as glycome imaging,10 protein lipidation and lipid trafficking,11 and activity-based protein profiling.12,13 In the early years of this century, Sharpless and co-workers defined a click reaction as one that is highly selective and efficient, wide in scope, easy to perform, uses readily available reagents, and is insensitive to oxygen and water. Work-up and isolation of the product of a click reaction must be simple, without requiring chromatographic purification.14–16 To date, click reactions have shown their specific advantages in the bio-orthogonal probing of biomarkers, cell labeling, and tumor-targeted imaging.17–22
Considering the biological importance of cell interactions, we proposed to choose two bio-orthogonal click reactions to bridge three types of cells. Firstly, a thiol-based click condensation reaction between 2-cyanobenzothiazole (CBT) and D-cysteine (D-Cys) developed by Rao and co-workers was chosen for this purpose.23–25 This click reaction takes place in the luciferin-regenerating pathway of a firefly body with high efficiency and biocompatibility. It has been successfully applied in the preparation of oligomeric nanostructures, molecular imaging (e.g., optical imaging, nuclear imaging, and magnetic resonance imaging), biomolecular detection, and has other potentialities.26–28 At pH 7.4 in water, this click reaction occurs spontaneously between the CBT–Cys pair to yield a thiazole ring which covalently bridges two types of cells. Then, a Cu(I)-catalyzed azide–alkyne click cycloaddition was chosen to bio-orthogonally bridge the third type of cell, because the azide–alkyne click cycloaddition has been extensively exploited and widely used in bioconjugate applications.29 After cell bridging, the bridging probability between two cells and the rupture force of bridged cells were quantitatively measured using optical tweezers. Optical tweezers have been widely used in single-molecule biology30 and cytobiology31,32 due to their advantage in non-contact manipulation and their ability to measure piconewton forces quantitatively.
Results and discussion
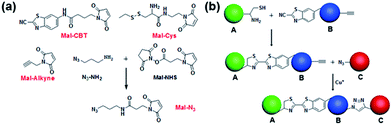
Despite the fact that the individual CBT–Cys condensation and azide–alkyne cycloaddition have been shown to be compatible with biological systems, their site-specific double incorporations have not yet been demonstrated. Therefore, we first validated the orthogonality between these two click reactions, and then used these two bio-orthogonal click reactions to bridge three types of cells. As shown in Fig. 1a, the compounds Mal-CBT, Mal-Cys, Mal-Alkyne, and Mal-N3 containing the bio-orthogonal functional groups for the click reactions and maleimide groups for conjugating the –SH groups on the cell walls (or membranes) were designed and synthesized. The synthetic routes and characterizations of these four compounds are shown in the ESI (Schemes S1–S4 and Fig. S1–S7†). Since compound Mal-N3 is unstable, we used Mal-NHS and N3-NH2 to freshly prepare Mal-N3 for the cell-bridging experiments. The feasibility of this reaction was tested and is shown in the ESI (Scheme S4 and Fig. S6–S7†). Fig. 1b schematically shows the procedure of cell-bridging. In detail, a Mal-Cys-modified cell A was bridged with cell B that was modified with both Mal-CBT and Mal-Alkynevia CBT–Cys condensation under reducing conditions. Then the Mal-Alkyne modified cell B on the A–B cell pair was bridged with a Mal-N3-modified cell C under the catalysis of Cu+ to form the A–B–C cell complex.Validation of orthogonality between CBT–Cys condensation and azide–alkyne cycloaddition
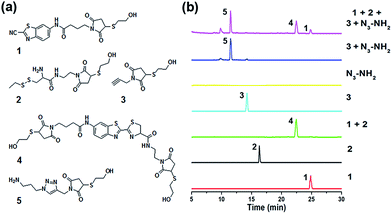
When the compound Mal-Cys was treated with reducing agents (e.g., tris(2-carboxyethyl)-phosphine, TCEP) to expose the cysteine motif for click condensation, the resulting –SH group would also react with its own maleimide motif. Therefore, to validate the orthogonality between the two click reactions, we firstly used mercaptoethanol (MCH) to react with the maleimide groups of compounds Mal-CBT, Mal-Cys, and Mal-Alkyne to prepare the respective compounds 1, 2, and 3, as shown in Fig. 2a. The synthetic routes and characterizations of 1, 2, and 3 are shown in the ESI (Schemes S5–S7 and Fig. S8–S10†). 100 μM 1, 100 μM 2, and 400 μM TCEP were together dissolved in 500 μL phosphate buffer (pH 7.4, 0.1 M), then the solution mixture was shaken for 1 hour at room temperature (RT) before being injected into a high-performance liquid chromatography (HPLC) system for analysis. As shown by the green HPLC trace in Fig. 2b, only the expected product 4 of the click reaction between 1 and 2 was cleanly obtained, which was confirmed by the electron-spray ionization mass spectrometry (ESI-MS) result in Fig. S11 (ESI†). Similarly, 100 μM 3, 100 μM N3-NH2, 1 mM CuSO4, and 7 mM NaVc were dissolved in 500 μL phosphate buffer (pH 7.4, 0.1 M) and then the solution mixture was shaken for 1 hour at RT. HPLC analysis indicated that only the expected product 5 of the click reaction between 3 and N3-NH2 was cleanly obtained (blue HPLC trace in Fig. 2b), proven by the ESI-MS result in Fig. S12 (ESI†). Then we investigated the orthogonality of these two click reactions. 50 μM 1, 50 μM 2, 50 μM 3, and 200 μM TCEP were dissolved in 500 μL phosphate buffer (pH 7.4, 0.1 M) and shaken for 30 min at RT. Then 100 μM N3-NH2, 0.5 mM CuSO4, and 3.5 mM NaVc were added to the above mixture and shaken for another 1 hour at RT. As shown by the magenta HPLC trace in Fig. 2b, except the expected compounds 4 and 5, no new compound appeared on the trace, suggesting that the CBT–Cys click condensation and the azide–alkyne click cycloaddition are orthogonal. To further confirm the completion of the reactions, we studied the kinetics of these CBT–Cys click condensation and azide–alkyne click cycloaddition reactions and their second-order rate constants were found to be 27.8 M−1 s−1 and 19.7 M−1 s−1 in these conditions, respectively (Fig. S13 and S14†).Bridging prokaryotic cells in three colors
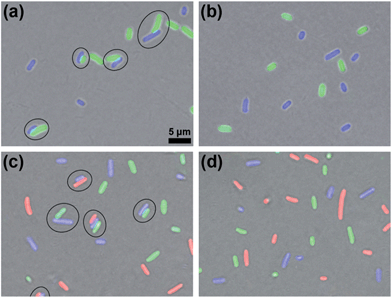
After validating the orthogonality between these two click reactions, we started to use them to bridge prokaryotic cells in three colors. Three colors of Escherichia coli (E. coli) cells that respectively express green (eGFP), yellow (eYFP), or red (mCherry, RFP) fluorescent proteins were used for the following experiments. Before being conjugated to click reagents, the cells were incubated with 1 mM TCEP at 37 °C for 30 min to expose the –SH groups on their surface proteins, then washed three times with phosphate-buffered saline (PBS, pH 7.4, 10 mM) by centrifugation at 8000 rpm and 4 °C. Then the GFP+ cells with 0.5 mM Mal-Cys, the YFP+ cells with 1 mM Mal-CBT and 1 mM Mal-Alkyne were respectively incubated at 37 °C for 1 h, and washed with PBS three times by centrifugation at 8000 rpm and 4 °C. Then the Mal-Cys-treated GFP+ cells (15.4% of Mal-Cys was loaded onto the GFP+ cells, as calculated by HPLC analysis), together with the Mal-CBT and Mal-Alkyne-treated YFP+ cells, were shaken with 0.5 mM TCEP in PBS at 37 °C for 2 h. The CBT–Cys click reaction efficiency under these conditions in the presence of the cells was calculated to be 76.5% by HPLC analysis. As shown in Fig. 3a, half of the GFP+ cells were bridged with YFP+ cells (8 of the total 16 cells in the view field). Note here that the blue fluorescence emission from the YFP+ cells results from the fluorescent compound Mal-CBT. Interestingly, in the absence of TCEP, the click reagent-treated GFP+ and YFP+ cells were not bridged but randomly scattered in the microscopic field, as shown Fig. 3b. This echoes that TCEP is needed to cleave the disulfide bond of Mal-Cys to initiate the click condensation. After 10 mM N3-NH2 was shaken with 10 mM Mal-NHS in PBS at RT for 1 h, the obtained Mal-N3 (diluted to 1 mM) was incubated with RFP+ cells at 37 °C for 1 h. Then the Mal-N3-treated RFP+ cells were shaken together with the above bridged GFP+ cells and YFP+ cells in PBS containing 1 mM CuSO4, and 7 mM NaVc at 37 °C for 2 h. As shown in Fig. 3c, some of the RFP+ cells were bridged with a single YFP+ cell or with YFP+ cells of a YFP+–GFP+ cell pair, suggesting that the azide–alkyne click cycloaddition really happened. A microscopy image of the bridged prokaryotic cells at lower magnification with more cells is shown in Fig. S15 (ESI†). Stability studies indicated that the conjunctions between the bridged prokaryotic cells were stable in PBS for 48 h at RT. Interestingly, we could not find RFP+ cells directly bridging with GFP+ cells, echoing that the CBT–Cys click condensation and the azide–alkyne click cycloaddition are orthogonal. If the RFP+ cells were not modified with Mal-N3, mixing the cells with the above control YFP+ cells and GFP+ cells shown in Fig. 3b did not result in any bridging between the cells, as shown in Fig. 3d.Bridging eukaryotic cells of three colors
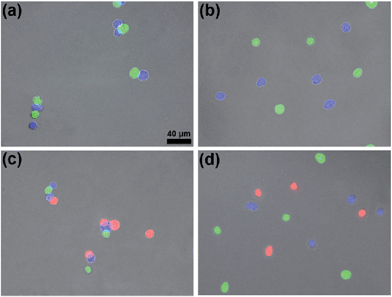
Since the content of proteins in the cytoderm of prokaryotic cells is usually much lower than that in the outer membrane of eukaryotic cells, we applied these two orthogonal click reactions to bridge eukaryotic cells with higher efficiency. HEK 293T cells respectively transfected with green, blue, or red (DsRed) fluorescent proteins were fixed with 4% paraformaldehyde and used for the following experiments. Before being conjugated with click reagents, HEK 293T cells were incubated with 100 μM TCEP at 37 °C for 30 min, then washed three times with PBS by centrifugation at 3000 rpm and 4 °C. After the incubation of GFP+ cells with 100 μM Mal-Cys, BFP+ cells with both 200 μM Mal-CBT and 200 μM Mal-Alkyne at 37 °C for 1 h, the cells were washed three times with PBS by centrifugation at 3000 rpm and 4 °C (22.6% of Mal-Cys was loaded onto the GFP+ cells, as calculated by HPLC analysis). Then the GFP+ cells and BFP+ cells were shaken together in PBS in the presence of 100 μM TCEP at 37 °C for 1.5 h. The CBT–Cys click reaction efficiency in these conditions in the presence of the cells was calculated to be 73.0% by HPLC analysis. As shown in Fig. 4a, most of the GFP+ cells were bridged with BFP+ cells (10 of the total 11 cells in the field). In the absence of TCEP, the click reagent-treated GFP+ and BFP+ cells were not bridged but randomly scattered in the microscopic field, as shown in Fig. 4b. Freshly prepared Mal-N3 at 200 μM was incubated with RFP+ cells at 37 °C for 1 h. Then the RFP+ cells were shaken with the above bridged GFP+ cells and BFP+ cells in PBS in the presence of 100 μM CuSO4 and 700 μM NaVc at 37 °C for 1.5 h. As shown in Fig. 4c, most of the RFP+ cells were bridged with either a single BFP+ cell or the BFP+ cells of a BFP+–GFP+ cell pair, suggesting that the azide–alkyne click cycloaddition really happened. A microscopy image of the bridged eukaryotic cells at higher density with more cells is shown in Fig. S16 (ESI†). Stability studies indicated that the conjunctions between the bridged eukaryotic cells were stable in PBS for 48 h at RT. Similarly, we could not find RFP+ cells directly bridging with GFP+ cells, additionally showing that the CBT–Cys click condensation and the azide–alkyne click cycloaddition are orthogonal. Without Mal-N3 modification, RFP+ cells incubated with the above control BFP+ cells and GFP+ cells shown in Fig. 4b did not result in any bridging between the cells in three colors, as shown in Fig. 4d. Interestingly, when the Mal-CBT-modified BFP+-HEK 293T cells were replaced with Mal-CBT-modified HepG2 cells, we found that three colors of two different cell lines (HEK 293T cells in green and red, HepG2 in blue) could also be bridged together (Fig. S17†).Distribution of the rupture force of bridged eukaryotic cells
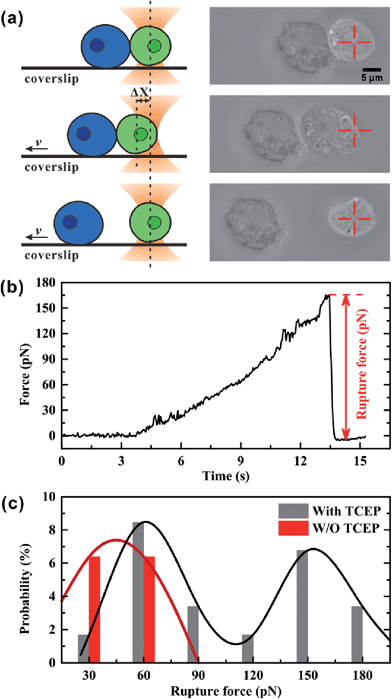
We used optical tweezers to measure the rupture force between Mal-Cys-treated GFP+ HEK 293T cells and Mal-CBT-treated BFP+ HEK 293T cells in the presence (experimental group) or absence (control group) of 100 μM TCEP. In the experiment, the GFP+ cells were distinguished from the BFP+ cells under a fluorescence microscope at first. Then, one GFP+ cell was trapped by optical tweezers and manipulated to contact a BFP+ cell, which was adhered to a coverslip, for 20 s. After that, the coverslip started to move at a velocity (v) of 1.4 μm s−1, but the movement of the trapped GFP+ cell along with the fixed BFP+ cell would be hindered by an increasing trapping force F, which was induced by the increasing displacement of the GFP+ cell departing from the trap center (Δx). From then on, there may be three situations. One situation was that there was no junction between the two cells and so the two cells were separated immediately, as shown in Fig. S18 and Video S1 (ESI†). For the control group, 80.9% of all contact-separation events fell into this situation, while 33.9% of experimental events did, as shown in Table S2 (ESI†). The great difference in this probability between these two groups of cells demonstrated that the click condensation reaction can bridge eukaryotic cells with a very high efficiency. The second situation was that there was a junction between the GFP+ and BFP+ cells but the bridged cells could be separated by the trapping force, as shown in Fig. 5a and Video S2 (ESI†). In this case, the optical tweezer-trapped GFP+ cell moved along with the BFP+ cell at the beginning and at a force-loading rate of 49 pN s−1. Once the trapping force was higher than the rupture force of the bridged GFP+–BFP+ cells, the two cells would be separated and the trapping force would drop to zero rapidly, as shown in Fig. 5b. The highest trapping force during stretching could be regarded as the rupture force. Fig. 5c illustrates the histogram of the rupture forces. There was only one histogram peak at a value of 45.0 pN for the control group. However, for the experimental group, besides a histogram peak at a value of 60.6 pN close to that of the control group, there was an extra histogram peak at 153.8 pN which was of the same order of magnitude as the force previously reported for a silicon–carbon bond.33 Therefore, this extra peak in the experimental group should be ascribed to the covalent bond formed in the CBT–Cys click reaction between the bridged GFP+–BFP+ cells. The third situation was that the bridged GFP+–BFP+ cells could not be ruptured by the trapping force before the GFP+ cell escaped from the optical trap, as shown in Fig. S19 and Video S3 (ESI†). In this case, the rupture force exceeded the measuring range of our optical tweezers (about 200 pN). For the control group, 6.4% of all contact-separation events fell into this situation. This might be ascribed to the non-specific interactions between the GFP+ and BFP+ cells. For the experimental group, the probability of this case was 40.7%, which was much higher than that of the control group, as shown in Table S2 (ESI†). We presume that this might be caused by multiple click reactions in one pair of bridged GFP+–BFP+ cells and also by non-specific interactions, and the former should be dominant. In this work, 71 contact-separation events covering the above three situations were measured by optical tweezers for the experimental group, and 63 contact-separation events were measured for the control group.Conclusions
In summary, employing two click reactions (click condensation of CBT–Cys and click cycloaddition of azide–alkyne), we rationally designed four click reagents Mal-CBT, Mal-Cys, Mal-Alkyne, and Mal-N3 for bridging cells of three colors. Orthogonality between these two click reactions was validated in solution, and characterized with HPLC and ESI-MS analyses. After modifications of different colors of fluorescent protein prokaryotic cells of E. coli with the reagents Mal-Cys, Mal-CBT and Mal-Alkyne, or Mal-N3, the cells were sequentially bridged. We also modified GFP+-, BFP+-, or RFP+-eukaryotic cells of HEK 293T cells with these four click reagents and sequentially bridged them with higher efficiency. Using optical tweezers, we quantitatively measured the bridging efficiency of the eukaryotic cells and the rupture force of bridged cells was found to be 153.8 pN. In consideration of the biological importance of cell junctions, we believe that our method of bridging three (or n + 1) types of cells with two (or n) bio-orthogonal click reactions might aid biologists to study cell–cell interactions in vitro with more convenience and efficiency.Experimental
Materials and methods
All the starting materials were obtained from Sigma-Aldrich, Adamas, or Sangon Biotech. Commercially available reagents were used without further purification, unless noted otherwise. All chemicals were of reagent grade or better. pBAD-GFP expresses eGFP from an arabinose-inducible promotor on a pBAD vector. pVS132 expresses eYFP from a IPTG-inducible promotor on a pTrc99a vector. pAmCherry1 was expressed from the plasmid pAmCherry1-N1. The fluorescent plasmids transfected to HEK 293T cells were generously provided by Prof. Bing Hu and Prof. Guoqiang Bi of University of Science and Technology of China (USTC). PBS used for cell experiments was prepared with PBS pills from Sangon Biotech. Milli-Q water (18.2 MΩ cm) was used throughout the experiment. The spectra of electrospray ionization-mass spectrometry (ESI-MS) were recorded on a LTQ Orbitrap mass spectrometer (Thermo Fisher). 1H NMR spectra were obtained on a 400 MHz Bruker AV 400 or a 300 MHz Bruker AV 300. High resolution ESI/MS spectra were obtained on a GCT premier mass spectrometer (Waters). HPLC analyses were performed on an Agilent 1200 HPLC system equipped with a G1322A pump and an in-line diode array UV detector using an Agilent Zorbax 300SB-C18 RP column with CH3CN (0.1% of trifluoroacetic acid (TFA)) and water (0.1% of TFA) as the eluent. Fluorescence microscopy images were taken under a fluorescence microscope OLMPUS IX71.Synthetic procedures
A mixture of 4-maleimidobutyric acid (18.1 mg, 0.1 mmol), HBTU (41.7 mg, 0.1 mmol), HOBT (13.5 mg, 0.1 mmol) in DMF (2 mL) was stirred for 30 min in the presence of DIPEA (11.1 mg, 0.1 mmol), then CBT (26.3 mg, 0.15 mmol), dissolved in 1 mL of DMF, was added into the mixture dropwise. After overnight stirring, the compound Mal-CBT (10.5 mg, yield: 31%) was obtained following HPLC purification. 1H NMR of compound Mal-CBT (d3-CD3CN, 300 MHz, Fig. S1†): 8.69 (d, J = 2.00 Hz, 1H), 8.11 (d, J = 9.01 Hz, 1H), 7.63 (dd, J1 = 2.05 Hz, J2 = 9.01 Hz, 1H), 6.75 (s, 2H), 3.54 (t, J = 6.88 Hz, 2H), 2.39 (t, J = 7.60 Hz, 2H), 1.92 (m, 2H). MS: calculated for C16H12N4O3S [(M + H)+]: 341.07; obsvd. ESI-MS: m/z 341.02 (Fig. S2†).
Optical tweezer measurements
Acknowledgements
This work was supported by Collaborative Innovation Center of Suzhou Nano Science and Technology, the Major program of Development Foundation of Hefei Center for Physical Science and Technology, the National Natural Science Foundation of China (Grants 21175122, 11374292, and 21375121), and Anhui Provincial Natural Science Foundation (Grant 1508085JGD06).Notes and references
- U. Rutishauser, A. Acheson, A. Hall, D. Mann and J. Sunshine, Science, 1988, 240, 53–57 CAS.
- T. G. Wolfsberg, P. Primakoff, D. G. Myles and J. M. White, J. Cell Biol., 1995, 131, 275–278 CrossRef CAS.
- G. A. Zimmerman, S. M. Prescott and T. M. McIntyre, Immunol. Today, 1992, 13, 93–100 CrossRef CAS.
- V. Kumar, R. S. Cotran and S. L. Robbins, Robbins Basic Pathology, Saunders, 2003 Search PubMed.
- W. R. Loewenstein and Y. Kanno, Nature, 1966, 209, 1248–1249 CrossRef CAS PubMed.
- J. Wiener, D. Spiro and W. R. Loewenstein, J. Cell Biol., 1964, 22, 587–598 CrossRef CAS.
- C. M. van Itallie and J. M. Anderson, Annu. Rev. Physiol., 2006, 68, 403–429 CrossRef CAS PubMed.
- E. M. Sletten and C. R. Bertozzi, Acc. Chem. Res., 2011, 44, 666–676 CrossRef CAS PubMed.
- W. Song, Y. Wang, J. Qu, M. M. Madden and Q. Lin, Angew. Chem., Int. Ed., 2008, 47, 2832–2835 CrossRef CAS PubMed.
- S. T. Laughlin and C. R. Bertozzi, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 12–17 CrossRef CAS PubMed.
- H. C. Hang, J. P. Wilson and G. Charron, Acc. Chem. Res., 2011, 44, 699–708 CrossRef CAS PubMed.
- B. F. Cravatt, A. T. Wright and J. W. Kozarich, Annu. Rev. Biochem., 2008, 77, 383–414 CrossRef CAS PubMed.
- D. Ye, A. J. Shuhendler, L. Cui, L. Tong, S. S. Tee, G. Tikhomirov, D. W. Felsher and J. Rao, Nat. Chem., 2014, 6, 519–526 CrossRef CAS PubMed.
- H. C. Kolb, M. G. Finn and K. B. Sharpless, Angew. Chem., Int. Ed., 2001, 40, 2004–2021 CrossRef CAS.
- H. C. Kolb and K. B. Sharpless, Drug Discovery Today, 2003, 8, 1128–1137 CrossRef CAS.
- Y. Yuan and G. L. Liang, Org. Biomol. Chem., 2014, 12, 865–871 CAS.
- M. D. Best, Biochemistry, 2009, 48, 6571–6584 CrossRef CAS PubMed.
- A. E. Speers, G. C. Adam and B. F. Cravatt, J. Am. Chem. Soc., 2003, 125, 4686–4687 CrossRef CAS PubMed.
- J. M. Baskin, J. A. Prescher, S. T. Laughlin, N. J. Agard, P. V. Chang, I. A. Miller, A. Lo, J. A. Codelli and C. R. Bertozzi, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 16793–16797 CrossRef CAS PubMed.
- E. Saxon and C. R. Bertozzi, Science, 2000, 287, 2007–2010 CrossRef CAS.
- M. Mammen, S. K. Choi and G. M. Whitesides, Angew. Chem., Int. Ed., 1998, 37, 2755–2794 CrossRef CAS.
- Y. Xianyu, Z. Wang and X. Jiang, ACS Nano, 2014, 8, 12741–12747 CrossRef CAS PubMed.
- G. Liang, H. Ren and J. Rao, Nat. Chem., 2010, 2, 54–60 CrossRef CAS PubMed.
- G. Liang, J. Ronald, Y. Chen, D. Ye, P. Pandit, M. L. Ma, B. Rutt and J. Rao, Angew. Chem., Int. Ed., 2011, 50, 6283–6286 CrossRef CAS PubMed.
- D. Ye, A. J. Shuhendler, P. Pandit, K. D. Brewer, S. S. Tee, L. Cui, G. Tikhomirov, B. Rutt and J. Rao, Chem. Sci., 2014, 5, 3845–3852 RSC.
- Y. Yuan, H. Sun, S. Ge, M. Wang, H. Zhao, L. Wang, L. An, J. Zhang, H. Zhang, B. Hu, J. Wang and G. Liang, ACS Nano, 2015, 9, 761–768 CrossRef CAS PubMed.
- S. Liu, A. Tang, M. Xie, Y. Zhao, J. Jiang and G. Liang, Angew. Chem., Int. Ed., 2015, 54, 3639–3642 CrossRef CAS PubMed.
- Y. Yuan, J. Zhang, L. An, Q. Cao, Y. Deng and G. Liang, Biomaterials, 2014, 35, 7881–7886 CrossRef CAS PubMed.
- Q. Wang, T. R. Chan, R. Hilgraf, V. V. Fokin, K. B. Sharpless and M. G. Finn, J. Am. Chem. Soc., 2003, 125, 3192–3193 CrossRef CAS PubMed.
- J. R. Moffitt, Y. R. Chemla, S. B. Smith and C. Bustamante, Annu. Rev. Biochem., 2008, 77, 205–228 CrossRef CAS PubMed.
- M. Zhong, X. Wei, J. Zhou, Z. Wang and Y. Li, Nat. Commun., 2013, 4, 1768 CrossRef PubMed.
- P. Xia, J. Zhou, X. Song, B. Wu, X. Liu, D. Li, S. Zhang, Z. Wang, H. Yu, T. Ward, J. Zhang, Y. Li, X. Wang, Y. Chen, Z. Guo and X. Yao, J. Mol. Cell Biol., 2014, 6, 240–254 CrossRef CAS PubMed.
- S. W. Schmidt, M. K. Beyer and H. Clausen-Schaumann, J. Am. Chem. Soc., 2008, 130, 3664–3668 CrossRef CAS PubMed.
- E. H. White, H. Worther, H. H. Seliger and W. D. McElroy, J. Am. Chem. Soc., 1966, 88, 2015–2019 CrossRef CAS.
- B. Lin, J. Yu and S. A. Rice, Phys. Rev. E: Stat. Phys., Plasmas, Fluids, Relat. Interdiscip. Top., 2000, 62, 3909 CrossRef CAS.
- K. Visscher, S. P. Gross and S. M. Block, IEEE J. Sel. Top. Quantum Electron., 1996, 2, 1066–1076 CrossRef CAS.
- K. C. Vermeulen, G. J. Wuite, G. J. Stienen and C. F. Schmidt, Appl. Opt., 2006, 45, 1812–1819 CrossRef.
- R. C. Gonzalez, R. E. Woods and S. L. Eddins, Digital image processing using MATLAB, Pearson Education India, Delhi, India, 2004 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: ESI Schemes S1–S7; ESI Fig. S1–S19; ESI Tables S1 and S2; ESI Videos S1–S3. See DOI: 10.1039/c5sc01315a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2015 |