Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Triptycene-based small molecules modulate (CAG)·(CTG) repeat junctions†
Stephanie A.
Barros
and
David M.
Chenoweth
*
Department of Chemistry, University of Pennsylvania, 231 South 34th Street, Philadelphia, PA 19104-6323, USA. E-mail: dcheno@sas.upenn.edu
First published on 10th June 2015
Abstract
Nucleic acid three-way junctions (3WJs) play key roles in biological processes such as nucleic acid replication in addition to being implicated as dynamic transient intermediates in trinucleotide repeat sequences. Structural modulation of specific nucleic acid junctions could allow for control of biological processes and disease states at the nucleic acid level. Trinucleotide repeat expansions are associated with several neurodegenerative diseases where dynamic slippage is thought to occur during replication, forming transient 3WJ intermediates with the complementary strand. Here, we report triptycene-based molecules that bind to a d(CAG)·(CTG) repeat using a gel shift assay, fluorescence-quenching and circular dichroism.
Introduction
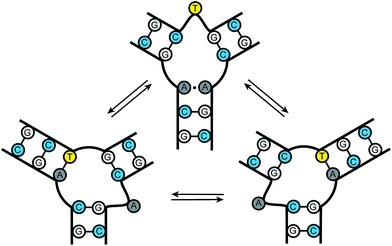
Nucleic acid junctions play important roles in biological processes and serve as key structural motifs in nanotechnology and aptamer-based sensing applications.1 In biology, three-way junctions (3WJs) are found as transient intermediates during replication, recombination, and DNA damage repair.2 Junctions are also present in several viral genomes, such as HIV-1, HCV, and adeno-associated virus in addition to playing key roles in viral assembly.3 Nucleic acid junctions are also prevalent in the emerging field of DNA and RNA nanotechnology where the bacteriophage phi29 pRNA containing RNA three-way junctions provide a particularly impressive example.3d–g Furthermore, they occur in trinucleotide repeat expansions found in unstable genomic DNA associated with neurodegenerative diseases.4 The development of structure and sequence-specific nucleic acid binding molecules remains an important challenge in chemical biology.5 The ability to target specific motifs using small molecules would allow for the precise control of biological processes and the possibility of modulating disease states.DNA trinucleotide repeats are present throughout the genome. Expansions of these repeats, however, are associated with a number of neurodegenerative diseases, including Huntington's disease, spinobulbar muscular atrophy, and myotonic dystrophy.6 Current models of triplet repeat expansion disease suggest slippage during DNA synthesis by the formation of dynamic DNA hairpin structures. As the length of the repeat increases, the growing hairpin structure gains thermodynamic stability, with repeat length providing an important positively correlated diagnostic for disease severity.6 Slipped-out (CAG)n·(CTG)n repeats have been implicated in the pathogenesis of triplet repeat expansion diseases such as Huntington's disease and several others.4,6 These “slipped-out” regions are dynamic and occur along the duplex, forming three-way junctions. Current models suggest that one arm of the junction contains the excess repeats while the other arms maximize complementary pairing between adjacent strands. A dynamic ensemble of conformations are possible at the slipped junction interface, where base pairing interactions differ with each state. The slipped-out (CAG)n repeat in Fig. 1 has been shown to contain one unpaired base at the center of the junction. Previous NMR studies have demonstrated that this sequence can adopt a multitude of conformations, where dynamic single nucleotide bulges at the junction interface interconvert between structures.4c Small molecule probes could provide important tools for gaining molecular level insight into the dynamics and repair processes associated with trinucleotide repeat junctions; however, probes of this kind are currently unknown.
Recently, we reported a new class of triptycene-based three-way junction (3WJ) binders.7 Here, we apply the triptycene scaffold as a first step toward developing new tools to recognize trinucleotide repeat junctions. We assessed the ability of our new triptycene based molecules to modulate the structure of d(CAG)n repeats using gel shift, fluorescence quenching, and circular dichroism (CD).
Results and discussion
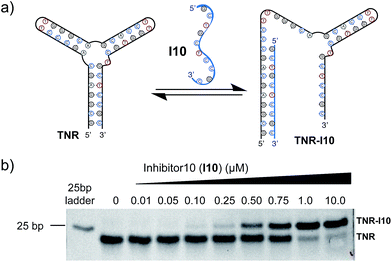
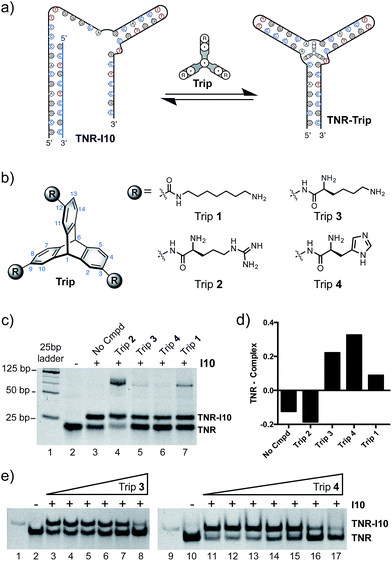
Utilizing a well-studied (CAG)·(CTG) repeat sequence known to form slipped-DNA three-way junctions, we developed a competitive inhibitor-based gel shift assay. This assay was inspired by previous work from our laboratory and seminal studies from the Plaxco and Ricci laboratories.1i,7 We utilized the junction forming (CAG)·(CTG) repeat sequence (TNR) and an inhibitor strand (I10) shown in Fig. 2a. The optimal inhibitor strand was 10 base pairs long and complementary to the 5′-end of the junction. Titration of inhibitor strand I10 into the folded junction resulted in a concentration dependent supershift of the band corresponding to hybridization of the TNR junction with I10, forming a larger molecular weight complex (TNR–I10) (Fig. 2b). Addition of small molecules capable of binding the TNR junction leads to I10 strand displacement from the TNR–I10 complex in a concentration dependent manner (Fig. 3a).Inspired by nucleic acid binding proteins, we synthesized a new class of triptycene molecules containing amino acids commonly found at the protein–nucleic acid interface. Analysis of these interfaces reveals an abundance of positively charged amino acid residues such as arginine, lysine, and histidine. We synthesized triptycene derivatives substituted with arginine, lysine, and histidine (Trip 2–4) as shown in Fig. 3b. Compounds 2–4 were assessed using a competitive displacement gel shift assay and compared to Trip 1, which was previously shown to significantly stabilize 3WJs (Fig. 3). Folded TNR 3WJ was incubated with I10, followed by addition of triptycenes 1–4 at the same concentration. Triptycenes 1–4 were able to reform the junction to varying degrees (Fig. 3c and d). Trip 3 and Trip 4 were the most effective, while Trip 1 was slightly less effective and Trip 2 did not show reformation of the junction. Interestingly, addition of Trip 1 and Trip 2 resulted in a slower moving band on the gel, indicating formation of a higher order structure. Trip 3 and Trip 4 resulted in the most significant inhibitor (I10) displacement, shifting TNR–I10 to the TNR–Trip complex and reforming the junction. A full titration of Trip 3 and Trip 4 with pre-incubated TNR–I10 was then performed (Fig. 3e). Concentration dependent displacement of the inhibitor strand (I10) resulted in reformation of the junction.
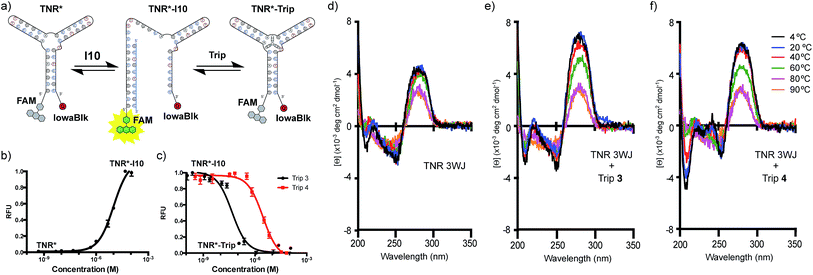
Next, we tested the ability of Trip 3 and 4 to induce fluorescence-quenching upon junction formation using a double labelled (CAG)·(CTG) repeat oligonucleotide. The TNR oligonucleotide was labelled with a FAM fluorophore on the 5′-end and an IowaBlack quencher on the 3′-end (TNR*) (Fig. 4a). Formation of the junction results in little to no fluorescence due to the proximity of the quencher and fluorophore, resulting in efficient contact quenching. The junction was incubated with the 10 bp inhibitor strand (I10). As expected, titration of I10 resulted in an increase in fluorescence, consistent with disruption of the folded junction, in which the fluorophore and quencher are separated in space (TNR*–I10) (Fig. 4b). Pre-formation of the open inhibited state of the junction (TNR*–I10) followed by titration of Trip 3 or Trip 4, resulted in a concentration dependent decrease in fluorescence (Fig. 4c). The decrease in signal indicates that the fluorophore and quencher are in close proximity due to refolding of the junction. Due to the competitive nature of this assay, only apparent Kd values are reported. The apparent Kd values of Trip 3 and Trip 4 were determined to be 52.8 nM and 2.36 μM, respectively.
Temperature-dependent circular dichroism (CD) was used to further characterize the interaction of Trip 3 and Trip 4 with the TNR junction. The CD spectra are indicative of B-DNA, showing positive signals at 280 nm due to base stacking and negative signals at 250 nm due to the right-handed helicity (Fig. 4d–f).8 As the temperature increased, the positive band at 280 nm decreased and the negative band at 250 nm increased, consistent with melting of the DNA structure (Fig. 4d). Upon addition of Trip 3 and Trip 4, distinct spectral changes are observed, resulting in an increased signal at 240 nm as well as a more positive signal at 280 nm and more negative signal at 210 nm (Fig. 4e and f). The increase at 280 nm is consistent with enhanced base stacking and increased helicity. Studies have shown that CAG slip-outs in a 3WJ are less paired and adopt more of an open loop structure.9 The increased helicity observed in the CD spectrum may be due to increased base pairing interactions in the slip-out region upon addition of Trip 3 and Trip 4.
Conclusions
In summary, we have described triptycene-based molecules that bind to d(CAG)·(CTG) trinucleotide repeats. Trip 3 and Trip 4 bind to a model (CAG)·(CTG) repeat as determined by gel shift and fluorescence-quenching experiments. The CD spectra are also consistent with enhanced helicity of the slipped out junction upon addition of Trip 3 and 4. This new class of nucleic acid binding small molecule may serve as inspiration for creating valuable probes for diseases associated with trinucleotide repeat expansions. Trinucleotide repeat nucleic acid sequences are associated with a large number (>30) of inherited human muscular and neurological diseases. The trinucleotide repeat tract length is dynamic and often correlates with disease severity, where short stable tracts are commonplace in the non-affected population. Longer unstable triplet repeat tracts are more prone to expansion as opposed to contraction, in addition to being predisposed to generational transmission.6c Trinucleotide repeat repair outcomes are also affected by structural features present in slipped sequences, where the structure may determine which proteins are recruited for repair.10 Stabilization of a particular structure could lead to altered repair of slipped-out junctions. Addition of ligands that bind to these junctions may affect repair outcomes as well as recruitment of proteins. Small molecule probes will provide valuable tools to study these processes. Small molecule binding and stabilization or modulation of these dynamic structures could lead to the development of therapeutic agents for their associated diseases.Acknowledgements
This work was supported by funding from the University of Pennsylvania. Instruments supported by the National Science Foundation and the National Institutes of Health including HRMS (NIH RR-023444) and MALDI-MS (NSF MRI-0820996). S. A. B. wishes to thank the NIH for funding through the Chemistry–Biology Interface Training Program (T32 GM07133).References
- (a) L. A. Howell and M. Searcey, Chembiochem, 2009, 10, 2139–2143 CrossRef CAS PubMed; (b) A. K. Sharma and J. M. Heemstra, J. Am. Chem. Soc., 2011, 133, 12426–12429 CrossRef CAS PubMed; (c) H. Zhang, J. A. Endrizzi, Y. Shu, F. Haque, C. Sauter, L. S. Shlyakhtenko, Y. Lyubchenko, P. Guo and Y. Chi, RNAi, 2013, 19, 1226–1237 CrossRef CAS PubMed; (d) Y. N. Teo and E. T. Kool, Chem. Rev., 2012, 112, 4221–4245 CrossRef CAS PubMed; (e) F. Wang, C. H. Lu and I. Willner, Chem. Rev., 2014, 114, 2881–2941 CrossRef CAS PubMed; (f) M. N. Stojanovic, P. de Prada and D. W. Landry, J. Am. Chem. Soc., 2000, 122, 11547–11548 CrossRef CAS; (g) A. L. Benvin, Y. Creeger, G. W. Fisher, B. Ballou, A. S. Waggoner and B. A. Armitage, J. Am. Chem. Soc., 2007, 129, 2025–2034 CrossRef CAS PubMed; (h) C. A. Kellenberger, S. C. Wilson, J. Sales-Lee and M. C. Hammond, J. Am. Chem. Soc., 2013, 135, 4906–4909 CrossRef CAS PubMed; (i) A. Porchetta, A. Vallee-Belisle, K. W. Plaxco and F. Ricci, J. Am. Chem. Soc., 2012, 134, 20601–20604 CrossRef CAS PubMed.
- (a) M. R. Singleton, S. Scaife and D. B. Wigley, Cell, 2001, 107, 79–89 CrossRef CAS; (b) K. C. Woods, S. S. Martin, V. C. Chu and E. P. Baldwin, J. Mol. Biol., 2001, 313, 49–69 CrossRef CAS PubMed.
- (a) C. J. Leonard and K. I. Berns, Prog. Nucleic Acid Res. Mol. Biol., 1994, 48, 29–52 CAS; (b) J. M. Watts, K. K. Dang, R. J. Gorelick, C. W. Leonard, J. W. Bess Jr, R. Swanstrom, C. L. Burch and K. M. Weeks, Nature, 2009, 460, 711–716 CrossRef CAS PubMed; (c) Y. Bai, A. Tambe, K. Zhou and J. A. Doudna, eLife, 2014, 3, e03656 Search PubMed; (d) P. Guo, Nat. Nanotechnol., 2010, 5, 833–842 CrossRef CAS PubMed; (e) D. Shu, Y. Shu, F. Haque, S. Abdelmawla and P. Guo, Nat. Nanotechnol., 2011, 6, 658–667 CrossRef CAS PubMed; (f) L. Tian and Y. Weizmann, J. Am. Chem. Soc., 2013, 135, 1661–1664 CrossRef CAS PubMed; (g) M. Endo and H. Sugiyama, ChemBioChem, 2009, 10, 2420–2443 CrossRef CAS PubMed.
- (a) S. M. Mirkin, Nature, 2007, 447, 932–940 CrossRef CAS PubMed; (b) G. Liu, X. Chen, J. J. Bissler, R. R. Sinden and M. Leffak, Nat. Chem. Biol., 2010, 6, 652–659 CrossRef CAS PubMed; (c) M. M. Slean, K. Reddy, B. Wu, K. Nichol Edamura, M. Kekis, F. H. Nelissen, R. L. Aspers, M. Tessari, O. D. Scharer, S. S. Wijmenga and C. E. Pearson, Biochemistry, 2013, 52, 773–785 CrossRef CAS PubMed.
- (a) D. M. Chenoweth, D. A. Harki and P. B. Dervan, Org. Lett., 2009, 11, 3590–3593 CrossRef CAS PubMed; (b) R. A. F. Rarig, M. N. Tran and D. M. Chenoweth, J. Am. Chem. Soc., 2013, 135, 9213–9219 CrossRef CAS PubMed; (c) P. B. Dervan, Bioorg. Med. Chem., 2001, 9, 2215–2235 CrossRef CAS; (d) D. M. Chenoweth, J. L. Meier and P. B. Dervan, Angew. Chem., Int. Ed., 2013, 52, 415–418 CrossRef CAS PubMed; (e) D. M. Chenoweth and P. B. Dervan, J. Am. Chem. Soc., 2010, 132, 14521–14529 CrossRef CAS PubMed; (f) D. M. Chenoweth and P. B. Dervan, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 13175–13179 CrossRef CAS PubMed; (g) A. Hirata, K. Nokihara, Y. Kawamoto, T. Bando, A. Sasaki, S. Ide, K. Maeshima, T. Kasama and H. Sugiyama, J. Am. Chem. Soc., 2014, 136, 11546–11554 CrossRef CAS PubMed; (h) S. Asamitsu, Y. Kawamoto, F. Hashiya, K. Hashiya, M. Yamamoto, S. Kizaki, T. Bando and H. Sugiyama, Bioorg. Med. Chem., 2014, 22, 4646–4657 CrossRef CAS PubMed; (i) D. M. Chenoweth, J. A. Poposki, M. A. Marques and P. B. Dervan, Bioorg. Med. Chem., 2007, 15, 759–770 CrossRef CAS PubMed.
- (a) C. E. Pearson and R. R. Sinden, Curr. Opin. Struct. Biol., 1998, 8, 321–330 CrossRef CAS; (b) C. E. Pearson, K. Nichol Edamura and J. D. Cleary, Nat. Rev. Genet., 2005, 6, 729–742 CrossRef CAS PubMed; (c) C. Lopez, J. D. Cleary and C. E. Pearson, Nat. Rev. Mol. Cell Biol., 2010, 11, 165–170 CrossRef PubMed.
- S. A. Barros and D. M. Chenoweth, Angew. Chem., Int. Ed., 2014, 53, 13746–13750 CrossRef CAS PubMed.
- G. D. Fasman, Circular Dichroism and then Conformational Analysis of Biomolecules, Plenum Press, New York, NY, 1996 Search PubMed.
- (a) N. N. Degtyareva, M. J. Reddish, B. Sengupta and J. T. Petty, Biochemistry, 2009, 48, 2340–2346 CrossRef CAS PubMed; (b) N. N. Degtyareva, C. A. Barber, B. Sengupta and J. T. Petty, Biochemistry, 2010, 49, 3024–3030 CrossRef CAS PubMed; (c) N. N. Degtyareva, C. A. Barber, M. J. Reddish and J. T. Petty, Biochemistry, 2011, 50, 458–465 CrossRef CAS PubMed.
- (a) M. Tam, S. E. Montgomery, M. Kekis, B. D. Stollar, G. B. Price and C. E. Pearson, J. Mol. Biol., 2003, 332, 585–600 CrossRef CAS; (b) C. E. Pearson, E. E. Eichler, D. Lorenzetti, S. F. Kramer, H. Y. Zoghbi, D. L. Nelson and R. R. Sinden, Biochemistry, 1998, 37, 2701–2708 CrossRef CAS PubMed; (c) C. E. Pearson, Y.-H. Wang, J. D. Friffith and R. R. Sinden, Nucleic Acids Res., 1998, 26, 816–823 CrossRef CAS PubMed; (d) C. E. Pearson, A. Ewel, S. Acharya, R. A. Fishel and R. R. Sinden, Hum. Mol. Genet., 1997, 6, 1117–1123 CrossRef CAS PubMed; (e) C. E. Pearson and R. R. Sinden, Biochemistry, 1996, 35, 5041–5053 CrossRef CAS PubMed; (f) C. E. Pearson, M. Tam, Y.-H. Wang, S. E. Montgomery, A. C. Dar, J. D. Cleary and K. Nichol, Nucleic Acids Res., 2002, 30, 4534–4547 CrossRef CAS PubMed; (g) G. B. Panigrahi, M. M. Slean, J. P. Simard, O. Gileadi and C. E. Pearson, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 12593–12598 CrossRef CAS PubMed; (h) G. B. Panigrahi, R. Lau, S. E. Montgomery, M. R. Leonard and C. E. Pearson, Nat. Struct. Mol. Biol., 2005, 12, 654–662 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5sc01595b |
| This journal is © The Royal Society of Chemistry 2015 |