Specific detection of the cleavage activity of mycobacterial enzymes using a quantum dot based DNA nanosensor†
Morten Leth
Jepsen
ab,
Charlotte
Harmsen
b,
Adwait Anand
Godbole
c,
Valakunja
Nagaraja
c,
Birgitta R.
Knudsen
ab and
Yi-Ping
Ho
*ab
aInterdisciplinary Nanoscience Center (iNANO), Aarhus University, Aarhus 8000 C, Denmark. E-mail: megan.ypho@inano.au.dk
bDepartment of Molecular Biology and Genetics, Aarhus University, Aarhus 8000 C, Denmark
cDepartment of Microbiology and Cell Biology, Indian Institute of Science, Bangalore 560 012, India
First published on 13th November 2015
Abstract
We present a quantum dot based DNA nanosensor specifically targeting the cleavage step in the reaction cycle of the essential DNA-modifying enzyme, mycobacterial topoisomerase I. The design takes advantages of the unique photophysical properties of quantum dots to generate visible fluorescence recovery upon specific cleavage by mycobacterial topoisomerase I. This report, for the first time, demonstrates the possibility to quantify the cleavage activity of the mycobacterial enzyme without the pre-processing sample purification or post-processing signal amplification. The cleavage induced signal response has also proven reliable in biological matrices, such as whole cell extracts prepared from Escherichia coli and human Caco-2 cells. It is expected that the assay may contribute to the clinical diagnostics of bacterial diseases, as well as the evaluation of treatment outcomes.
Introduction
Rapid and quantitative detection of pathogenic bacteria, such as Mycobacterium tuberculosis, the etiological agent of tuberculosis (TB), is critical for effective antibiotic treatment and prevention of the disease spreading at an early infection stage. While an array of diagnostic approaches has been developed for this purpose, challenges remain particularly when accessibility and time-to-results are considered collectively. For instance, although polymerase chain reaction (PCR)1,2 and sequencing are promising for highly sensitive identification of bacteria, both techniques are expensive and require extensive resources. For diagnosis of bacterial diseases, including TB, culturing the bacteria from the specimen/samples followed by microscopic examinations of colonies is the most commonly adapted clinical gold standard.3,4 However, the long culture time of bacteria (up to 8 weeks in the case of mycobacteria5) often delays reporting. A handful of technological innovations have been recently introduced for identifying the presence of bacteria in general, including quartz crystal microbalance (QCM),6,7 surface plasmon resonance (SPR),8,9 electrochemical impedance spectroscopy (EIS),10 surface enhanced Raman scattering (SERS),11,12 and fluorescence spectroscopy.13 However, the performance of these techniques relies heavily on complex purification procedures to isolate bacteria from the biological matrices, hampering their applications in the field settings. Therefore, there is a critical need for a generic, accurate, and more importantly, a user-friendly assay that allows fast and quantitative detection of the bacterial pathogens in biological samples.To this end, we report the development of a novel use of quantum dot (QD) based DNA nanosensors for the rapid detection of bacterial enzymes, which are essential for bacterial cell survival. More specifically, the proposed assay directly measures the cleavage activity of mycobacterial topoisomerase I (TopoI) as a proof-of-concept quantifiable indication of viable mycobacterial cells. Topoisomerases are a ubiquitous class of enzymes, which maintain topological homeostasis during a variety of DNA transaction processes such as transcription, replication and chromosome segregation. The reaction mechanism is a multi-step process consisting of DNA binding, cleavage, strand passage and religation. The mycobacterial TopoI shows properties which are markedly distinct from its Escherichia coli (E. coli) counterpart.14–17 For instance, the mycobacterial enzyme can bind to and cleave double-, as well as single-stranded DNA with comparable affinity. The binding and cleavage are sequence specific and the hexameric sequence is referred to as the Strong Topoisomerase Site (STS).16,17 The site specific cleavage property is used in the proposed nanosensor to distinguish the activity of the mycobacterial TopoI, and hence the presence of mycobacteria, from the activity of TopoI from other species. The visual detection is made possible by the unique photophysical properties of the QDs, monodisperse semiconductor nanocrystals, that emit strong fluorescence under UV excitation.18,19 The designed DNA substrate consists of a single-stranded oligonucleotide, with the STS recognition sequence and is dual-labelled with biotin and Cy5 on the 5′ and 3′ end, respectively. Upon pre-assembly with the streptavidin functionalized QD, forming the DNA nanosensor, energy transfer due to the mechanism of Förster Resonance Energy Transfer (FRET)20,21 from the QD (emission peak at 605 nm) to Cy522–25 would lead to quenching of the QD fluorescence, shown as “the initial stage” in Fig. 1. Similar to the sensing principle of proteolytic activity using QDs,26,27 upon specific recognition of mycobacterial TopoI, the cleavage would then turn the FRET “off”, leading to a recovery of QD fluorescence. The increase of QD emission thus represents the presence of “active” mycobacterial TopoI, and may serve as a direct indication of viable mycobacterial cells. Purified Mycobacterium smegmatis topoisomerase I (MsTopoI), from non-pathogenic M. smegmatis, is selected as a model enzyme, for its similar biochemical characteristics with the TopoI from its pathogenic relative, M. tuberculosis.17 This report, for the first time, demonstrates the possibility to quantify the cleavage activities of mycobacterial enzyme via the response from the QD-based DNA nanosensors without the need of separation or amplification. The cleavage induced signal response has also proven reliable in biological crude samples containing human cell extracts of Caco-2 and E. coli. Besides the clear clinical value in diagnosing bacterial diseases, and evaluating the treatment outcome, the developed methodology may be promoted for monitoring different target enzymes present in various kinds of microorganisms. It is also therefore expected to generate benefits in other sectors, such as food industries, homeland security and agriculture.
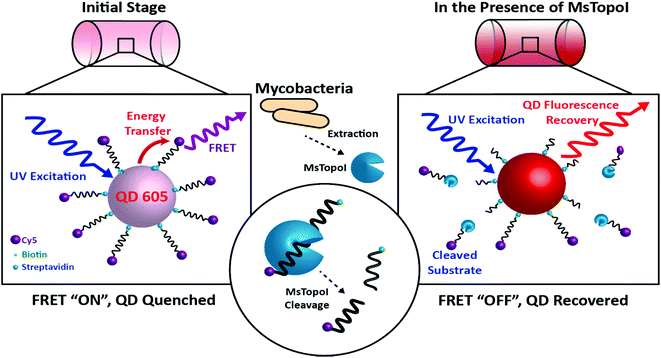 | ||
| Fig. 1 The principle of the proposed DNA nanosensor: Left: initial stage (FRET “ON”, QD quenched): the MsTopoI DNA substrates, consisting of the previously validated recognition sequence (ref. 31), are dual-functionalized with biotin and Cy5 on the 5′ and 3′ end, respectively. The DNA nanosensor is assembled by conjugating the MsTopoI DNA substrates onto the QD through biotin–streptavidin interactions in an optimized ratio. The QD fluorescence is quenched by the FRET-mediated energy transfer. Right: in the presence of MsTopoI (FRET “OFF”, QD recovered): MsTopoI binds and cleaves on the recognition site, which leads to the diffuse-away of Cy5 fluorophore. The cleavage activity of MsTopoI is quantitatively determined by the level of recovered QD fluorescence. | ||
Experimental
Preparation of DNA nanosensors
The MsTopoI specific DNA substrate (sequence, labelling and the cleavage site listed in Table 1) were synthesized, dual labeled, and HPLC purified by Integrated DNA Technologies, Coralville, Iowa, USA. This substrate included a previously validated cleavage site of MsTopoI (CGCT^TG, ^: cleavage site).31| MsTopoI substrate |
|---|
| 5′-biotin-CAG TGA GCG AGC TTC CGC T ^ TG ACA TCC CAA TA-Cy5-3′ (^: cleavage site) |
Streptavidin-functionalized CdSe–ZnS QD conjugates (Qdot® 605 ITK™ Streptavidin Conjugate) were purchased from Life Technologies. The DNA nanosensors were prepared by conjugating the MsTopoI DNA substrates onto QDs via biotin–streptavidin interactions in the conjugation buffer (40 mM Tris-HCl, 200 mM NaCl and 1 mM EDTA, pH = 8). The molar ratio of MsTopoI DNA substrates to QDs was maintained at a ratio of 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (1.25 pmol of MsTopoI DNA substrate and 50 fmol of QD). The ratio was empirically determined by where the optimal quenching of QD fluorescence occurred (ESI†).
1 (1.25 pmol of MsTopoI DNA substrate and 50 fmol of QD). The ratio was empirically determined by where the optimal quenching of QD fluorescence occurred (ESI†).
Reaction of MsTopoI and QD DNA nanosensors
Recombinant MsTopoI was purified by transforming pPVN123 encoding MsTopoI in the E. coli BL21 expression strain. The protein was purified by two column chromatographic steps using heparin Sepharose followed by SP Sepharose as previously described.28 The validation of cleavage with the QD DNA nanosensors was conducted by mixing the DNA nanosensors (prepared as above described) with a titrated amount of purified MsTopoI. Titration of MsTopoI was prepared by diluting the purified MsTopoI in a dilution buffer (50 mM KCl, 1 mM EDTA, and 20 mM Tris-HCl, pH = 8). After the dilution, MsTopoI was then mixed with the DNA nanosensors and incubated at 37 °C for 60 minutes. EDTA was present in the reaction to deplete the divalent cations and thus inhibit DNA nucleases from non-specific cleavage.Measurement of relaxation activity of MsTopoI
The relaxation assay was performed with a 200 fmol PAD negatively supercoiled plasmid in the relaxation buffer (150 mM NaCl, 1 mM EDTA, 5 mM MgCl2, 5 mM CaCl2, and 10 mM Tris-HCl, pH = 7.5). Subsequently after MsTopoI was added to the mixture, the reaction was incubated at 37 °C for 60 minutes. After the incubation, the relaxation reaction was stopped by adding SDS (0.25% final) and proteinase K (20 μg) to digest MsTopoI at 37 °C for 60 minutes. DNA relaxation was assessed by gel electrophoresis in 1% agarose in TBE with ethidium bromide. Gels were subjected to electrophoresis at 10 V overnight, stained with ethidium bromide and visualized under UV light (Versadoc 5000, Bio-Rad, Hercules, CA).Preparation of human cell nuclear extract
Caco-2 cells (heterogeneous human epithelial colorectal adenocarcinoma cells) were cultured in complete medium (Minimum Essential Media, MEM-GlutaMAX, supplemented with non-essential amino acid, 20% fetal bovine serum (FBS), 1% penicillin, and 1% streptomycin). Cells were harvested with 0.5% Trypsin-EDTA (Gibco) and media was then discarded. Cells were subsequently washed with phosphate-buffered saline (PBS) prior to nuclear extraction. Extraction was executed following the previously established protocol.29 Briefly, 5 × 106 cells per mL of cells were incubated in 1 mL of lysis buffer (0.1% Igepal CA-630, 10 mM Tris-HCl (pH = 7.9), 10 mM MaCl2, 15 mM NaCl, and 1% PMSF (phenylmethyl sulfonyl fluoride)) on ice for 10 minutes. The lysate was then spun at 400g for 5 min and the supernatant containing cell debris was subsequently removed. The nuclear extraction was prepared by adding 50 μL of extraction buffer (0.5 M NaCl, 20 mM HEPES (pH = 7.9), 20% glycerol, and 1% PMSF) into the precipitate and incubated at 4 °C for 1 h. Followed by a final spin at 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200g for 10 min, the supernatant was collected as the nuclear cell extract and used for subsequent tests.
200g for 10 min, the supernatant was collected as the nuclear cell extract and used for subsequent tests.
E. coli expression of MsTopoI and preparation of enzyme extract from E. coli
The expression plasmid pPVN123 was transformed into E. coli BL21, and induced as previously described.28 Briefly, the expression plasmid pPVN123 was transformed into E. coli cells and induced at 37 °C for 3 h with 0.3 mM IPTG. The cells were harvested by centrifugation. To prepare the extract, the pellet was resuspended in ice cold PBS (1 g pellet in 2 mL PBS). The cell suspension was then mixed with equal volume of glass beads (212–300 μm, acid-washed, Sigma-Aldrich) and 2 μL mL−1 of PMSF. The E. coli were lysed by repeated vortexing of the cells vigorously with glass beads. In detail, the mixture was vortexed for 1 min and left on ice. This process was repeated 3 times, with 1 min incubation on ice in between each repetition. After extraction, the mixture was centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g (4 °C) for 5 min. The supernatant was then collected and used immediately. Untransformed E. coli was processed the same way and used as a negative control. To simulate the crude biological sample, extract from the E. coli expressing MsTopoI was titrated with untransformed E. coli in a final concentration of 25 pM in the reaction buffer. The mixed extract was tested for the cleavage activity as described in the previous section without further purification.
000g (4 °C) for 5 min. The supernatant was then collected and used immediately. Untransformed E. coli was processed the same way and used as a negative control. To simulate the crude biological sample, extract from the E. coli expressing MsTopoI was titrated with untransformed E. coli in a final concentration of 25 pM in the reaction buffer. The mixed extract was tested for the cleavage activity as described in the previous section without further purification.
Measuring cleavage activity of MsTopoI via recovery of QD fluorescence
Fluorescence emission spectra and end-point measurements were achieved by a commercially available plate reader (FlexStation 3, multi-mode microplate reader, Molecular Devices, Sunnyvale, USA). The emissions for QD605 (bandpass 570–590 nm) and FRET-mediated Cy5 (660–680 nm) were measured upon excitation at 360 nm in 384-well plates.Calculation of recovery efficiency
Empirically, the recovery was characterized using the ratiometric FRET.25,30 The recovery efficiency (recovery %) was determined according to the following equation. Briefly, the obtained fluorescence was background subtracted and normalized by the difference of positive control (QD conjugated with unlabelled DNA substrates) and negative control (QD nanosensors in the absence of enzymes).At least three individual experiments, unless otherwise noted, were conducted for each condition. Error bars were calculated as the standard error of the mean from each independent experiment. The statistical significance was determined by using Prism 5.0 (GraphPad Software, La Jolla, CA).
Results and discussion
The configuration of the proposed DNA nanosensor and the model system: Mycobacterium smegmatis topoisomerase I (MsTopoI)
The site specific nature of mycobacterial TopoI is exploited for the proposed DNA sensors in this study. The working principle of the nanosensor is shown in Fig. 1. The MsTopoI DNA substrate, consisting of the previously validated recognition sequence,31 was dual-functionalized with biotin and Cy5 on the 5′ and 3′ end, respectively. The DNA nanosensor is assembled by conjugating the MsTopoI substrates onto the QD through biotin–streptavidin interactions in an optimized ratio. The QD fluorescence is initially quenched (Fig. 1, left), due to the FRET-mediated energy transferred to Cy5. Upon specific recognition of MsTopoI, the cleavage would then lead to a recovery of QD fluorescence (Fig. 1, right).To ease the sample handling at the development stage of the nanosensor, purified MsTopoI, from non-pathogenic M. smegmatis, was used as a model since it shares similar biochemical characteristics with the TopoI from its pathogenic relative, M. tuberculosis.17 Therefore, the optimization and development of the sensor for M. smegmatis can be easily transformed for the detection of other pathogenic mycobacteria in the future.
Characterization of the interaction between the dual-functionalized DNA substrates and MsTopoI
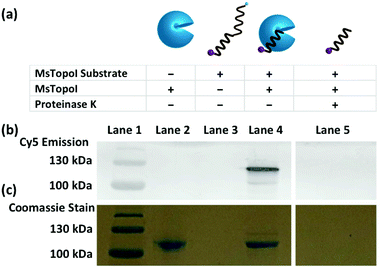
To validate whether the dual-functionalized substrate can be cleaved by MsTopoI, a cleavage assay was performed and the reaction products were analyzed using standard SDS polyacrylamide gel electrophoresis. To enhance the concentration of cleavage complexes and to avoid non-specific cleavage by e.g. nucleases, the cleavage reaction was performed in an EDTA containing buffer. This prevents religation by MsTopoI, due to depletion of divalent cations from the reaction mixture. Samples containing different combinations of entities were prepared (Fig. 2a), and retarded in a SDS polyacrylamide gel. The Cy5 emission and Coomassie stain were scanned for visualization of the retardations of the Cy5-labeled DNA substrates and MsTopoI, respectively (Fig. 2b and c). As evident from the gel scanning, interaction of MsTopoI with the Cy5-labeled DNA substrate under these conditions resulted in a Cy5 band (compare lanes 4 and 3, Fig. 2b) at a position corresponding to the approximate mobility of MsTopoI (both lanes 2 and 4 showed a mobility of around 110 kDa, the Coomassie stain is shown in Fig. 2c). This band disappeared upon proteinase K digestion of the reaction products (lane 5) clearly confirming its identity as a protein coupled product, which is consistent with the MsTopoI induced cleavage complex. Taken together, these results confirm that the enzyme can bind and cleave the designed cleavage substrate, forming a 5′ covalent intermediate as expected.28Detection limit determined with titrated purified enzymes
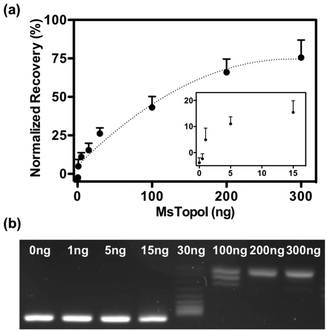
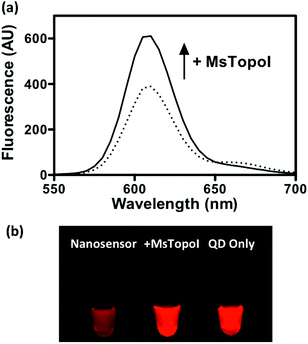
The performance of the sensor was evaluated by a titration experiment with purified MsTopoI in a buffer containing EDTA (as described in the Experimental section). As shown in Fig. 3a, the measured and recovered QD fluorescence was in positive correlation to the amount of purified MsTopoI. The detection limit was determined to be around 1–5 ng (Fig. 3a, inset). Relaxation of a negatively supercoiled substrate is typically used to determine the complete catalytic activity of type IA topoisomerases. This technique, however, may not detect activity at low amounts of enzyme. Indeed, as shown in Fig. 3b, the purified MsTopoI was able to relax supercoiled DNA in a concentration dependent manner. However, the detection limit of the relaxation assay of ∼15–30 ng is poorer than that of the proposed DNA nanosensor. Hence, in contrast to the relaxation assay which relies on specialized equipment for readout and has a higher limit for detection of activity, the fluorescence based assay may present the advantage of enabling the possibility of visualized measurement and detecting low level of enzymatic activity.The spectra shown in Fig. 4a further confirmed that the QD fluorescence (peak at 605 nm) was recovered by the addition of MsTopoI. As evidenced by the spectra, the ratiometric change depends largely on the QD recovery as compared to the disappearance of FRET-mediated Cy5 emission (see also ESI Fig. 2†). Owing to the unique photophysical properties of QD, the recovered fluorescence was clearly distinguishable under UV (360 nm) excitation in the presence of MsTopoI (Fig. 4b). The possibility to visualize the recovered fluorescence upon cleavage of MsTopoI may open up future possibilities in promoting this assay for tests under the point-of-care setting.
Performance evaluated in crude biological samples
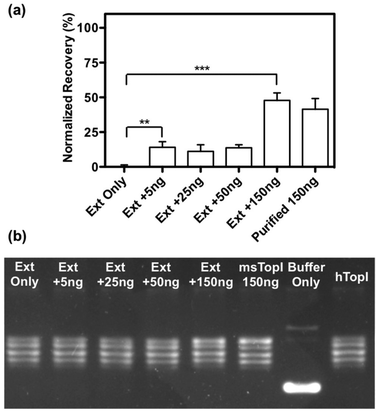
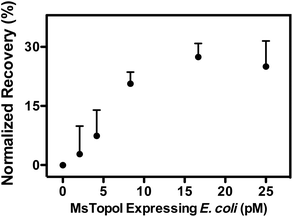
The subsequent effort investigates whether the nanosensor remains functional in crude biological samples by enabling specific detection of MsTopoI even in the context of human cell content. The nanosensor was tested using the purified MsTopoI spiked-in human nuclear extract from Caco-2 (derived from colorectal adenocarcinoma). As shown in Fig. 5a, the detection limit (∼5 ng) was minimally affected in the presence of human cell extracts, containing human topoisomerases (hTopI) along with other eukaryotic proteins. Also the sensor was highly specific towards MsTopoI and did not detect any of proteins/enzymes present in the human cell extract. In contrast with the proven specificity of the proposed QD nanosensor, it was not possible to distinguish MsTopoI relaxation activity from other relaxation activities present in the human cell extract from Caco-2. As shown in Fig. 5b, almost no difference was observed in the relaxation capability shown in the lanes containing either cell extracts mixed with different amounts of purified MsTopoI (5–150 ng), purified MsTopoI only (150 ng) or purified hTopI only. This is not surprising since it is well established that at least hTopI exhibit relaxation activity in the MsTopoI relaxation buffer.Similar cleavage experiments were also carried out using extracts from E. coli cells over-expressing MsTopoI while the untransformed E. coli was used as a negative control. The samples were prepared by mixing a titrated amount of extract from MsTopoI-expressing E. coli with extract from the untransformed E. coli in a total E. coli concentration of 25 pM. The mixed extract was tested for the cleavage activity without further purification. The results shown in Fig. 6 provide a validation that the proposed QD nanosensor remained quantifiable and functional with the biological crude sample in the current setup.
Conclusions
DNA interacting enzymes, such as topoisomerases, are essential for all living organisms, due to their function of maintaining topological homeostasis during vital cellular functions.32 Measurement of the activity of topoisomerases has been reported as a reliable indication of infectious diseases.19,33 For example, malaria infection has been determined by the level of enzymatic activities based on isothermal conversion of single DNA cleavage–ligation events catalyzed specifically by the topoisomerase I of malaria causing plasmodium species.19 The presented QD-based nanosensor joins the league by providing a facile and rapid measurement through fluorescence generated by the cleavage activity of mycobacterial topoisomerase I, which eliminates washing steps and post-amplification as required in the previously developed platform.19,33 Significant clinical value is anticipated by extending this assay for the detection of mycobacteria-associated diseases, such as TB. In particular, an improved species specificity is anticipated when combining the effort with antibodies or peptide based recognition.34 Further, the proposed nanosensor is highly transformable for the detection of contaminating mycobacteria in water or food, which may have a broad appeal to the food industries and agricultural sectors.Acknowledgements
The authors would like to acknowledge the support from the Danish Research Councils (116325/FTP), Lundbeck Foundation (R95-A10275), Karen Elise Jensen Foundation, the Dagmar Marshalls Fond, the Aase and Ejnar Danielsens Foundation, the Arvid Nilssons Foundation, Marie & M. B. Richters Fond, Augustinus fonden, Aage og Johanne Louis-Hansens fond, and Familiens Erichsens mindefond. The research in Prof. V. Nagaraja's laboratory is supported by Department of Biotechnology, Government of India. We thank Dr Magnus Stougaard at the Aarhus University Hospital for kindly granting the use of the lab facilities.References
- G. Jenikova, J. Pazlarova and K. Demnerova, Int. Microbiol., 2000, 3, 225–229 CAS.
- D. Rodriguez-Lazaro, M. D'Agostino, A. Herrewegh, M. Pla, N. Cook and J. Ikonomopoulos, Int. J. Food Microbiol., 2005, 101, 93–104 CrossRef CAS PubMed.
- P. L. McDonough, S. J. Shin and D. H. Lein, J. Clin. Microbiol., 2000, 38, 1221–1226 CAS.
- R. Kumar, P. K. Surendran and N. Thampuran, Lett. Appl. Microbiol., 2008, 46, 221–226 CrossRef CAS PubMed.
- R. Ghodbane, D. Raoult and M. Drancourt, Sci. Rep., 2014, 4, 4236 Search PubMed.
- I. S. Park and N. Kim, Biosens. Bioelectron., 1998, 13, 1091–1097 CrossRef CAS PubMed.
- Z. Q. Shen, J. F. Wang, Z. G. Qiu, M. Jin, X. W. Wang, Z. L. Chen, J. W. Li and F. H. Cao, Biosens. Bioelectron., 2011, 26, 3376–3381 CrossRef CAS PubMed.
- A. D. Taylor, J. Ladd, Q. Yu, S. Chen, J. Homola and S. Jiang, Biosens. Bioelectron., 2006, 22, 752–758 CrossRef CAS PubMed.
- O. Torun, I. Hakki Boyaci, E. Temur and U. Tamer, Biosens. Bioelectron., 2012, 37, 53–60 CrossRef CAS PubMed.
- C. Ruan, L. Yang and Y. Li, Anal. Chem., 2002, 74, 4814–4820 CrossRef CAS PubMed.
- R. M. Jarvis and R. Goodacre, Chem. Soc. Rev., 2008, 37, 931–936 RSC.
- W. R. Premasiri, D. T. Moir, M. S. Klempner, N. Krieger, G. Jones and L. D. Ziegler, J. Phys. Chem. B, 2005, 109, 312–320 CrossRef CAS PubMed.
- M. D. Disney, J. Zheng, T. M. Swager and P. H. Seeberger, J. Am. Chem. Soc., 2004, 126, 13343–13346 CrossRef CAS PubMed.
- T. Bhaduri, T. K. Bagui, D. Sikder and V. Nagaraja, J. Biol. Chem., 1998, 273, 13925–13932 CrossRef CAS PubMed.
- T. Bhaduri, S. Basak, D. Sikder and V. Nagaraja, FEBS Lett., 2000, 486, 126–130 CrossRef CAS PubMed.
- T. Bhaduri, D. Sikder and V. Nagaraja, Nucleic Acids Res., 1998, 26, 1668–1674 CrossRef CAS PubMed.
- A. A. Godbole, M. N. Leelaram, A. G. Bhat, P. Jain and V. Nagaraja, Arch. Biochem. Biophys., 2012, 528, 197–203 CrossRef CAS PubMed.
- W. C. W. Chan and S. M. Nie, Science, 1998, 281, 2016–2018 CrossRef CAS PubMed.
- S. Juul, C. J. Nielsen, R. Labouriau, A. Roy, C. Tesauro, P. W. Jensen, C. Harmsen, E. L. Kristoffersen, Y. L. Chiu, R. Frohlich, P. Fiorani, J. Cox-Singh, D. Tordrup, J. Koch, A. L. Bienvenu, A. Desideri, S. Picot, E. Petersen, K. W. Leong, Y. P. Ho, M. Stougaard and B. R. Knudsen, ACS Nano, 2012, 6, 10676–10683 CrossRef CAS PubMed.
- W. R. Algar and U. J. Krull, Anal. Chem., 2009, 81, 4113–4120 CrossRef CAS PubMed.
- I. L. Medintz, A. R. Clapp, H. Mattoussi, E. R. Goldman, B. Fisher and J. M. Mauro, Nat. Mater., 2003, 2, 630–638 CrossRef CAS PubMed.
- H. H. Chen, Y. P. Ho, X. Jiang, H. Q. Mao, T. H. Wang and K. W. Leong, Mol. Ther., 2008, 16, 324–332 CrossRef CAS PubMed.
- Y. P. Ho, H. H. Chen, K. W. Leong and T. H. Wang, J. Controlled Release, 2006, 116, 83–89 CrossRef CAS PubMed.
- M. L. Jepsen, A. Ottaviani, B. R. Knudsen and Y. P. Ho, RSC Adv., 2014, 4, 2491–2494 RSC.
- C. Y. Zhang, H. C. Yeh, M. T. Kuroki and T. H. Wang, Nat. Mater., 2005, 4, 826–831 CrossRef CAS PubMed.
- I. L. Medintz, A. R. Clapp, F. M. Brunel, T. Tiefenbrunn, H. Tetsuo Uyeda, E. L. Chang, J. R. Deschamps, P. E. Dawson and H. Mattoussi, Nat. Mater., 2006, 5, 581–589 CrossRef CAS PubMed.
- W. R. Algar, A. Malonoski, J. R. Deschamps, J. B. Blanco-Canosa, K. Susumu, M. H. Stewart, B. J. Johnson, P. E. Dawson and I. L. Medintz, Nano Lett., 2012, 12, 3793–3802 CrossRef CAS PubMed.
- P. Jain and V. Nagaraja, J. Mol. Biol., 2006, 357, 1409–1421 CrossRef CAS PubMed.
- M. Stougaard, J. S. Lohmann, A. Mancino, S. Celik, F. F. Andersen, J. Koch and B. R. Knudsen, ACS Nano, 2009, 3, 223–233 CrossRef CAS PubMed.
- E. Haustein, M. Jahnz and P. Schwille, ChemPhysChem, 2003, 4, 745–748 CrossRef CAS PubMed.
- D. Sikder and V. Nagaraja, Nucleic Acids Res., 2000, 28, 1830–1837 CrossRef CAS PubMed.
- J. C. Wang, Untangling the double helix, Cold Spring Harbor Laboratory Pr, 2009 Search PubMed.
- M. Stougaard and Y. P. Ho, Expert Rev. Mol. Diagn., 2014, 14, 1–3 CrossRef CAS PubMed.
- H. Yang, L. Qin, Y. Wang, B. Zhang, Z. Liu, H. Ma, J. Lu, X. Huang, D. Shi and Z. Hu, Int. J. Nanomedicine, 2015, 10, 77–88 CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Characterization of the QD-based DNA Nanosensor. See DOI: 10.1039/c5nr06326d |
| This journal is © The Royal Society of Chemistry 2016 |