DOI:
10.1039/C5RA23614B
(Paper)
RSC Adv., 2016,
6, 4302-4309
A highly selective marker reaction for measuring the activity of human carboxylesterase 1 in complex biological samples†
Received
11th November 2015
, Accepted 17th December 2015
First published on 21st December 2015
Abstract
Human carboxylesterase 1 (hCE1), plays pivotal roles in endobiotics homeostasis and xenobiotic metabolism. The plasma level of hCE1 could serve as a useful serologic biomarker for several hepatic diseases, such as hepatocellular carcinoma. However, no probe reaction has been fruitfully used to determine the activity of trace hCE1 in human plasma. This study aims to design and develop a highly selective marker reaction for measuring the enzymatic activities of hCE1 in complex biological samples including human plasma. N-(4-Methyl butyrate)-4-hydroxy-1,8-naphthalimide (NMHN), which contains a small alcohol group and a large acyl moiety, was intentionally designed based on the substrate preference of hCE1. NMHN could be easily hydrolyzed by hCE1 with very high catalytic efficacy under physiological conditions, while both reaction-phenotyping assays and chemical inhibition assays demonstrated that this reaction exhibited super selectivity towards hCE1 over other human hydrolases. Furthermore, the marker reaction possessed ideal kinetic behaviour (classic Michaelis–Menten) with high intrinsic clearance (5.8 mL min−1 mg−1). Based on this probe, a rapid hCE1 quantification method was developed and fully validated, which was successfully applied to determine the real activities of hCE1 in various biological samples including human plasma. Our findings afforded a promising tool for measuring the real activity of hCE1 and laid a solid foundation for further investigations on the biological functions of hCE1 in complex biological samples.
Introduction
Carboxylesterases (CEs, E.C.3.1.1.1), a well conserved multigene family of α/β-hydrolase fold enzymes, catalyze the hydrolysis of a vast number of structurally diverse endogenous and exogenous compounds including fatty acid esters, environmental toxins and ester-containing drugs.1 In humans, the majority of CEs belong to the CE1 and CE2 gene families, while human carboxylesterase 1 (hCE1) and carboxylesterase 2 (hCE2) have been identified as the two major CEs distributed in human tissues.2 hCE1 and hCE2 are distinct in substrate specificity, tissue distribution, and gene regulation. Generally, hCE1 preferentially hydrolyzes substrates with a small alcohol group and a large acyl group, while hCE2 preferentially catalyzes substrates with a large alcohol group and a small acyl group.3,4 hCE1 is abundantly expressed in the liver, and less so in the intestines, kidneys, lungs, testes, heart, monocytes, and macrophages, while hCE2 is mainly expressed in the gastrointestinal tract and at a relatively low level in the liver.5 Notably, a trace amount of hCE1 could also be detected in human plasma, and its level would be elevated in patients with hepatocellular carcinoma (HCC), thus plasma hCE1 could serve as a novel serologic biomarker candidate for HCC.6
As a top ten most abundant protein distributed in human liver, hCE1 is of particular importance for human health because it plays pivotal roles in endobiotics homeostasis and metabolic defence systems. Besides its essential role in hydrolysis of many structurally diverse endobiotics and xenobiotics, hCE1 also plays a non-negligible role in various physiological processes such as lipid homeostasis, testosterone synthesis, retinol biosynthesis, trafficking and retention of proteins in the endoplasmic reticulum.7–9 Dysfunction of hCE1 in human body has been proved to be closely related to many diseases, such as atherosclerosis, leydig cell tumour and hypercholesterolemia.10–12 Particularly, the expression of hCE1 at the mRNA and protein level and its hydrolytic activity could be easily modulated by many factors, e.g., the genetic factors, drug exposure, and other environmental factors, as well as pathological factors.13 The large inter-individual variability in both expression and function of hCE1 significantly affects the clinical outcome (including efficacy and toxicity) of the drugs mainly hydrolyzed by hCE-1, as well as the biological functions of hCE1 and relevant physiological processes.14 Hence, it is crucial to develop a practical method for precise measurement of hCE1 in complex biological systems such as human tissues, cancer cell lines, and plasma.
Over the past several years, methods, e.g., western blotting and mass spectrometry-based proteomic technique, have been developed for hCE1 detection and quantification.15,16 However, these techniques are usually time-consuming, labour-intensive, and costly, and the protocol is rather sophisticated and limited by some stringent requirements (such as expensive devices and skilful handling).17,18 Moreover, these methods can only evaluate the protein level rather than the activity of hCE1. Compared with antibody and proteomic techniques, activity-based non-fluorescent and fluorescent probes can selectively and directly measure the enzymatic activity of hCE1 under physiological conditions, and thus have attracted increasing attention in recent years.19 Although several therapeutic drugs such as oseltamivir and clopidogrel could be used as small molecule probes to measure the catalytic activity of hCE-1, poor sensitivity and low reaction rates of these probes strongly limited their applications in detecting trace amount of hCE-1 in real samples.20,21 Recently, a fluorescent probe for hCE1 (BMBT) has been developed, but it was unsuitable for measuring trace amount of hCE1 in human plasma due to its short emission wavelength and the involvement of albumin in BMBT hydrolysis.22,23 Thus, it is highly desirable to develop a practical, sensitive and selective marker reaction for the precise measurement of the real function of hCE1 in complex biological samples.
In this study, a new molecule probe for hCE1 was intentionally designed and developed on the basis of the substrate preference of hCE1. The probe N-(4-methyl butyrate)-4-hydroxy-1,8-naphthalimide (NMHN) contains a small alcohol group and a large acyl moiety. In order to explore whether NMHN hydrolysis could serve as a specific marker reaction for highly selective detection of hCE-1 in complex biological samples, the probe reaction and its quantification method were well characterized and fully validated in terms of specificity, kinetic behaviour, sensitivity, linearity, precision, and recovery. After validation, NMHN hydrolysis was successfully applied to measure hCE1 activity in complex biological systems such as human liver preparations, cancer cell homogenates and human plasma.
Results
Identification of NMHN hydrolytic product
The detailed synthesis route and structural characterization of NMHN are shown in the ESI data (Scheme S1, Fig. S1–S3†). NMHN could be easily synthesized with high yield by using 4-bromo-1,8-naphthalic anhydride as the starting material. Upon addition of hCE1 containing human liver microsomes (HLMs), the formyl ester bond is readily hydrolyzed by the enzyme after the specific substrate recognition, leading to the release of a single metabolite (see Scheme 1). The metabolite of NMHN was fully identified as N-(3-carboxy propyl)-4-hydroxy-1,8-naphthalimide (NCHN) based on the comparison of LC retention times (6.3 min), UV spectra (the maximum absorption wavelengths 244 nm) and MS/MS spectra (m/z 298 Da, aglycone's mass – 14) with the authentic standard (Fig. 1). In addition, both NMHN and its hydrolysis product NCHN exhibited good UV absorption and fluorescent emission properties, suggesting that both ultraviolet and fluorescence detector could be used to measure the enzymatic activity of hCE1.
 |
| | Scheme 1 Schematic illustration of NMHN hydrolysis for hCE1 detection. | |
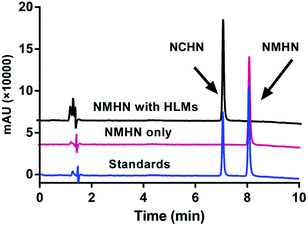 |
| | Fig. 1 LC-UV chromatograms of NMHN (50 μM) hydrolysis samples in the presence or absence of human liver microsomes (0.5 mg mL−1), with detection wavelength set at 247 nm. HLMs: pooled human liver microsome. | |
Assays with various enzymes with ester hydrolytic activity
To evaluate the selectivity of NMHN as the specific substrate towards hCE-1, reaction phenotyping assays were performed firstly. Various ester hydrolytic enzymes were used, including recombinant hCE1, hCE2, paraoxonases (PON1 and PON2), bovine serum albumin (BSA), human serum albumin (HSA), acetylcholinesterase (AChE), butyrylcholinesterase (BChE), chymotrypsin, lysozyme, pepsin, trypsin, carbonic anhydrase (CA), α1-acid glycoprotein (AGG), proteinase K, platelet-activating factor acetylhydrolase (PAF-AH), retinol binding protein 4 (RBP-4), and lipsase.24–37 Notably, to explore whether HSA in plasma could participate in NMHN hydrolysis, 0.5 mg mL−1 HSA was added in the incubation system, which is comparable to the HSA concentration of 1% human plasma. As shown in Fig. 2, only hCE1 displayed excellent efficacy towards NMHN hydrolysis, while NCHN could not be detected upon addition of HSA and other species. These results demonstrated that NMHN has high selectivity for hCE1 over other species, which is the foundation of subsequent applications.
 |
| | Fig. 2 Specificity of NMHN hydrolysis. | |
Chemical inhibition assays
To further confirm the selectivity of NMHN towards hCE1 in complex biological samples, the inhibitory effects of various selective esterase inhibitors on NMHN hydrolysis were investigated in pooled HLMs. As shown in Fig. 3a, the formation of NCHN in pooled HLMs could be strongly inhibited by BNPP (a potent inhibitor of hCEs), while LPA (a strong inhibitor of hCE2), HA (a powerful selective inhibitor of AChE) and EDTA (PONs inhibitor)38–41 displayed negligible inhibitory effects on the formation of NCHN. Furthermore, the dose-dependent inhibition of BNPPon NMHN hydrolysis in both HLMs and recombinant hCE1 was evaluated. As depicted in Fig. 3b, the inhibitory tendency of BNPP was similar between HLMs and hCE1, with the IC50 values of 28.15 and 21.24 nM, respectively, indicating that NMHN could be used to screen potential hCE1 inhibitors by using HLMs as the enzyme source instead of more expensive recombinant hCE1. These results indicated that hCE1 was the primary enzyme responsible for NMHN hydrolysis in HLMs.
 |
| | Fig. 3 (a) Inhibitory effects of selective esterase inhibitors on NMHN hydrolysis in HLMs. (b) Dose-dependent inhibition curves of BNPP on NMHN hydrolysis reaction in HLMs and hCE1, respectively. | |
Kinetic analysis
Besides excellent selectivity, other characteristics such as kinetic parameters and enzymatic kinetic behaviour can also affect the quantitative applications of the activity-based probes.42–44 To characterize the kinetic behaviour of NMHN towards hCE1, NMHN hydrolysis was performed in pooled HLMs and recombinant human hCE1. Preliminary experiments were carried out to ensure that the formation of NCHN was in the linear range of both protein concentrations (0–5 μg mL−1) and incubation time (0–60 min) (data not shown). NMHN hydrolysis in both HLMs and hCE1 followed the classic Michaelis–Menten kinetics, which are evidenced by the corresponding Eadie–Hofstee plots in the inset (Fig. 4). The kinetic parameters of NMHN hydrolysis in these enzyme sources were listed in Table 1, the Km value for NMHN hydrolysis in hCE1 and HLMs were 53.11 μM and 83.01 μM, respectively. The intrinsic clearance (CLint) values were also calculated to be as high as 5.8 and 12.9 mL min−1 mg−1 protein in hCE1 and HLMs, respectively, which were much greater than the reported values for other probes, such as oseltamivir (0.58 mL min−1 mg−1 protein for hCE1) and clopidogrel (0.06 mL min−1 mg−1 protein for hCE1).20,45 In addition, a comparison is presented in Table S1, ESI† between the specific probes used to determine hCE1. These findings indicated that hCE1-mediated NMHN hydrolysis possessed ideal kinetic behaviours with much higher intrinsic clearance, which inspired us to apply this marker reaction for quantitative measurement of hCE1 in complex biological samples.
 |
| | Fig. 4 Enzyme kinetic profiles for NMHN hydrolysis in HLMs (a) and recombinant hCE1 (b). The corresponding Eadie–Hofstee plot (rate versus rate/NMHN concentration) is shown as an inset. | |
Table 1 Kinetic parameters derived for the formation of NCHN in HLM, and recombinant hCE1
| Enzyme |
Km μM |
Vmax nmol min−1 mg−1 |
CLint mL min−1 mg−1 |
| HLM |
53.11 |
684.3 |
12.9 |
| hCE1 |
83.01 |
484.9 |
5.8 |
Method validation
The excellent selectivity and ideal kinetic model of NMHN make it to be a promising marker reaction for measuring the real activity of hCE1 in biological samples. In these cases, the quantification methods, including a liquid chromatography with ultraviolet detection (LC-UV) and a liquid chromatography with fluorescent detection (LC-FD), were rigorously validated with respect to linearity, precision, and accuracy based on the relative peak area. The calibration curves constructed for NMHN and NCHN showed satisfactory linearity between the peak area and the concentration in the range of 0.5–50 μM, with the correlation coefficient (r2) greater than 0.999 (Fig. 5 and Table S2, ESI†). The lower limit of quantification (LOQ) of NCHN was 0.5 μM, suggesting that as low as 100 ng mL−1 hCE1 could be detected following 30 min incubation with NMHN (50 μM) under physiological conditions (Fig. S4, ESI†). For the intra- and inter-day precision and accuracy, the RSD and the deviation of accuracy were within the acceptable range of 4% and 5%, respectively (Table 2 and Table S3, ESI†). The recovery was complete with the value in the range of 99–110% (Table S4 and S5, ESI†).
 |
| | Fig. 5 (a) LC-UV standard curves of NCHN and NMHN. (b) LC-FD standard curves of NCHN and NMHN. The concentration ranges from 0.5 to 50 μM. | |
Table 2 Intra-and inter-day precision and accuracy of the LC-UV method for quantitative determination of NMHN and NCHN
| Analyte |
Added |
Intra-day |
Inter-day |
| Found |
RSD (%) |
(%) |
RSD (%) |
| NCHN |
1.0 |
1.03 ± 0.02 |
2.2 |
102.7 |
3.5 |
| 1.00 ± 0.02 |
1.6 |
100.1 |
|
| 1.00 ± 0.01 |
0.9 |
100.4 |
|
| 5.0 |
4.97 ± 0.06 |
1.3 |
99.4 |
1.2 |
| 4.99 ± 0.02 |
0.4 |
99.7 |
|
| 4.94 ± 0.04 |
0.9 |
98.9 |
|
| 20.0 |
20.49 ± 0.25 |
1.2 |
102.5 |
1.1 |
| 20.59 ± 0.11 |
0.6 |
103.0 |
|
| 20.68 ± 0.15 |
0.7 |
103.4 |
|
| NMHN |
1.0 |
1.05 ± 0.03 |
2.9 |
105.0 |
1.8 |
| 1.04 ± 0.01 |
0.6 |
103.5 |
|
| 1.04 ± 0.01 |
0.8 |
104.0 |
|
| 5.0 |
5.14 ± 0.05 |
1.1 |
102.8 |
2.1 |
| 5.10 ± 0.06 |
1.1 |
101.9 |
|
| 5.05 ± 0.04 |
0.8 |
101.1 |
|
| 20.0 |
20.33 ± 0.39 |
1.9 |
101.7 |
2.5 |
| 20.36 ± 0.10 |
0.5 |
101.8 |
|
| 20.71 ± 0.08 |
0.4 |
103.6 |
|
Biological applications
After method validation, NMHN hydrolysis reaction was used to determine the activity of hCE1 in the real biological samples including human liver preparations, cancer cell preparations and human plasma. As shown in Fig. 6a, NMHN hydrolysis reaction was applied to determine hCE1 activities in a panel of 12 HLMs from different individuals. There was about 4-fold differences between the highest and lowest activity values (963.7 to 3615.2 nmol min−1 mg−1 protein) in the 12 individual HLMs upon addition of 50 μM NMHN (close to the Km value for HLM mediated hydrolysis), which agreed well with activity variability in hCE1 levels obtained from HLMs.5,45 Furthermore, a strong correlation (R2 = 0.93, p < 0.0001) between the hydrolytic rate of NMHN and that of clopidogrel (a known hCE1 substrate) was presented in Fig. 6b. Three commonly used cancer cell lines, including A549, HepG2, and Caco-2, were cultured and assayed for determination of hCE1 protein expression and activity levels. As shown in Fig. 6c and d, these three cancer cell lines showed a general trend that both protein expression and activity were highest in HepG-2 cells and lowest in CaCo-2 cells, which agreed well with the previously reported expression level of hCE1 in human tissues (highest in the liver and to a lesser extent in lung and colon).46,47 In addition, we used this probe reaction for further investigation on hCE1 activity measurement in the plasma samples of 20 individuals. As shown in Fig. 7a, about 5-fold differences between the highest and lowest activity values (267 to 1281 nM min−1 mL−1 plasma) in the 20 individual plasma samples were detected. To the best of our knowledge, this is the first report that determines the real activity of hCE1 in human plasma. Furthermore, the formation of NCHN in cell preparations and human plasma could be selectively and strongly inhibited by BNPP, while the inhibitors of other human esterases displayed negligible inhibitory effects on this biotransformation (Fig. 7b and S5, ESI†). These findings strongly suggested that NMHN hydrolysis could be successfully used to measure the real activities of hCE1 even in a mixture of multiple human enzymes, and the quantification method is highly reliable.
 |
| | Fig. 6 (a) Hydrolysis activity of hCE1 in 12 individual HLMs using NMHN as the probe substrate. (b) Correlation analysis between the hydrolytic rates of clopidogrel and those of NMHN (n = 12). (c) Hydrolysis activities and protein expression level of hCE1 in A549, HepG2, and Caco-2 cell lines. (d) Protein expression level of hCE1 in A549, HepG2, and Caco-2 cell lines in western blot analysis. | |
 |
| | Fig. 7 (a) Activity of hCE1 in 20 individual plasma samples based on NMHN hydrolysis reaction. (b) Inhibitory effects of selective chemical inhibitors on NMHN hydrolysis in human plasma. | |
Conclusion
In summary, a specific marker reaction for highly selective sensing the activity of hCE1, one of the most important esterase in human, was developed and well characterized for the first time. NMHN could be highly selectively hydrolyzed by hCE1 over other hydrolases. NMHN hydrolysis followed ideal Michaelis–Menten kinetics and displayed much higher intrinsic clearance in comparison with other probes of hCE1, which are very helpful for the reliable quantification of hCE1 in biological samples. In addition, with the excellent selectivity and ideal kinetic behaviour, NMHN could be successfully applied to measure the real activity of hCE1 in biological samples containing multiple enzymes, such as tissue preparations, cell preparations, and human plasma. Furthermore, NMHN and its hydrolytic product NCHN could be easily and accurately detected by using both LC-UV and LC-FD methods. Our findings suggest that this marker reaction could be used as a sensible tool to determine the real activity of hCE1 in complex biological samples, which could be very useful in drug discovery and in exploring the biological functions of hCE1 in human diseases.
Experimental
Reagents and materials
NMHN and NCHN were chemically synthesized, and the structure was fully characterized by 1H-NMR, 13C-NMR and high resolution mass spectrometry. Bis (4-nitrophenyl) phosphate (BNPP) and loperamide hydrochloride (LPA) were purchased from TCI (Tokyo, Japan). Huperzine A (HA) was purchased from Sichuan Weikeqi Biological technology Co., Ltd. (Chengdu, Sichuan, China). Ethylene diamine tetraacetic acid (EDTA) and clopidogrel were obtained from J&K Chemical Ltd. (Beijing, China). All other materials were of analytical grade. Human PON1 and PON2 were obtained from Bioworld (MN, USA). AChE, BChE, HSA, PAF-AH, RBP-4, pepsin, trypsin, proteinase K, AGG, lipase, and CA, were obtained from Sigma-Aldrich (St. Louis, MO, USA). Lysozyme, chymotrypsin, and BSA were purchased from Solarbio Science & Technology Co., Ltd. (Beijing, China). Two recombinant hCEs (hCE-1 and hCE-2) were purchased from BD Biosciences (Woburn, MA, USA). Pooled HLMs (n = 50) and a panel of individual HLMs (n = 12) were used. Horseradish peroxidase-conjugated goat anti-rabbit secondary antibody and polyclonal anti-hCE1 antibody were purchased from Abcam (Cambridge, UK).
Analytical instruments and conditions
NMHN and its hydrolysis product were analyzed by a UFLC system (Shimadzu, Kyoto, Japan), equipped with a CBM-20A communications bus module, a DGU-20 A5R vacuum degasser, two LC-30AD pumps, a SIL-30AC autosampler, a CTO-30A column oven, and a SPD-20A UV/vis detector. A Shim-pack XR-ODS (2.2 μm, I.D. 2.0 mm × 75.0 mm; Shimadzu, Japan) analytical column with an ODS guard column (2.2 μm, I.D. 2.0 mm × 5 mm; Shimadzu) was used to analyze the parent compound and its metabolite. Column temperature was kept at 40 °C and UV detector was set at 317 nm. The mobile phase consisted of acetonitrile (A) and 0.2% formic acid water (B) with the following gradient: 0 to 2 min, 90% B; 2 to 9 min, 90% B to 25% B; 9 to 10 min, 25% B; 10 to 13 min, 25% B to 5% B; and 13 to 16 min, balanced to 90% B.
Assays with various enzymes with ester hydrolytic activity
A panel of 18 commercial hydrolytic enzymes was used to screen whether they have NMHN hydrolytic activity. In brief, a mixture (total volume 0.2 mL) consisting of PBS buffer (pH 7.4, 50 mM), NMHN (50 μM, concentration corresponding to the apparent Km), and the hydrolytic enzyme was incubated for 15 min at 37 °C. The protein concentrations were as follows: carboxylesterase (hCE1 and hCE2, 5 μg mL−1), BSA (5 μg mL−1), AChE (5 μg mL−1), pepsin (5 μg mL−1), trypsin (5 μg mL−1), proteinase K (5 μg mL−1), AGG (5 μg mL−1), lysozyme (5 μg mL−1), chymotrypsin (5 μg mL−1), lipase (5 μg mL−1), CA (5 μg mL−1), PAF-AH (5 μg mL−1), BChE (10 U L−1), HSA (500 μg mL−1), RBP-4 (0.05 μg mL−1), PON1 and PON2 (0.05 μg mL−1) and human plasma (1%, v/v). The reaction was terminated by the addition of acetonitrile (200 μL) and the resulting mixture was centrifuged for 20 min at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g. The supernatant (10 μL) was taken and subjected to UFLC measurements.
000 × g. The supernatant (10 μL) was taken and subjected to UFLC measurements.
Chemical inhibition assays
In order to reveal the role of the involved hydrolytic enzyme in NMHN hydrolysis via HLMs, plasma, and cancer cell lines, chemical inhibition assays were performed. BNPP (a potent inhibitor of hCEs), LPA (a strong inhibitor of hCE2), HA (a powerful selective inhibitor of AChE) and EDTA (PONs inhibitor) were selected to investigate their potential inhibitory effects on NMHN hydrolysis in these biological samples containing hCE1. NMHN (50 μM, near Km value) was incubated with HLMs (4 μg mL−1), cell S9 (0.5 mg mL−1), or plasma (1%, v/v) in the absence (control) or presence of each inhibitor (100 μM). The IC50 values for the inhibition of BNPP on NMHN hydrolysis in HLMs and hCE1 were estimated using GraphPad Prism V6 (GraphPad Software, San Diego, CA).
Kinetic analysis
In order to fully characterize the kinetic behavior of NMHN hydrolysis in involved enzymes, kinetic analysis was performed in pooled HLMs and hCE1 enzyme. Preliminary experiments were carried out to ensure that the formation of hydrolysis product was in the linear range of both protein concentrations (0 to 2 μg mL−1) and incubation time (0 to 60 min). NMHN at a series of concentrations (0 to 1000 μM) was incubated with hCE1 (0.5 μg mL−1) and HLMs (1.0 μg mL−1) at 37 °C for 30 min. The Michaelis–Menten equation (eqn (1)) was employed to analyze the kinetic data. Vmax is the maximum reaction rate, V is the reaction rate, [S] is the substrate concentration, and Km is the substrate affinity constant. The model selection is based on Eadie–Hofstee plots. The apparent kinetic parameters were described as mean from individual duplicates. Data analysis was performed using GraphPad Prism V6.| |
 | (1) |
Method validation
To validate the assay method, limits of detection (LOD), LOQ, linearity, accuracy, precision, and matrix effects were determined according to the FDA guideline for method validation. LOD and LOQ were used to assess system sensitivity. Seven concentrations of each standard were used to construct the standard curves. The linearity of the method was assessed by linear regression analysis. To assess the intra- and inter-day precision and accuracy of the method, the relative standard deviation (RSD) for the peak area of six replicates of each Q.C. samples (1, 5, 20 μM) was calculated. The recovery was evaluated by comparing the analytical results of enzyme-containing samples at three concentrations with those of no enzyme standards that represent 100% recovery.
Inter-individual variability of NMHN hydrolysis in HLMs and correlation analysis
To investigate the inter-individual difference of NMHN hydrolysis in individual HLMs, the formation rates of NCHN were measured in a panel of HLMs prepared from 12 donors. The incubation conditions were the same as described above. The substrate concentration was 50 μM (near to Km value) and the final protein concentration was 1 μg mL−1. Clopidogrel (100 μM) was used as the probe substrate for hCE1, and the formation rates of the hydrolysis product in these individual HLMs (final protein concentration 5 μg mL−1) were also determined. The rates of NMHN hydrolysis in individual HLMs were correlated with that of clopidogrel hydrolysis. Correlation (Pearson) analysis was performed by GraphPad Prism V6. The linear regression coefficient (r) was calculated, and p < 0.01 was recognized as being statistically significant.
Inter-individual variability of NMHN hydrolysis in the individual plasma
The formation rates of NCHN were measured in plasma samples from 20 healthy donors. The incubation conditions were the same as described above. The substrate concentration was 50 μM (near to Km value) and the plasma volume was 10 μL.
Western blot analysis of hCE1 expression
Cells were cultured in 12-well plates and collected when they reached 90% confluence. The cell preparation samples were separated by sodium dodecyl sulphate (SDS)-polyacrylamide gel electrophoresis (10% acryl amide gels) and transferred onto polyvinylidene difluoride (PVDF) membranes using a wet blotting system. The membranes were blocked with TBST buffer (20 mM Tris, 150 mM NaCl, 0.05% Tween-20, 5% non-fat dry milk, pH 8.0) for 2 h, and incubated for 2 h with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution of a polyclonal anti-hCE1 antibody, followed by incubation with a 1
1000 dilution of a polyclonal anti-hCE1 antibody, followed by incubation with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3000 dilution of horseradish peroxidase-conjugated goat anti-rabbit secondary antibody for 2 h. The protein was detected by ECL chemiluminescence system.
3000 dilution of horseradish peroxidase-conjugated goat anti-rabbit secondary antibody for 2 h. The protein was detected by ECL chemiluminescence system.
Hydrolysis assays with cell homogenates
Cells were cultured in 175 cm2 flasks for 1–2 days, and collected when they were about 90% confluence. Then cells were harvested with a trypsin–EDTA solution, after washing with ice-cold PBS, they were centrifugated at 200 × g for 5 min to obtain the cell pellets. Cell pellets were disrupted by sonication on ice for three times, with power setting at 200 W for 3 s, with 30 s intervals between the pulses. The cell suspension was centrifuged at 9000 × g for 20 min and the supernatant fraction (S9) was kept at −80 °C until use. The concentration of NMHN was 50 μM (near to Km value) and the final protein concentration was 0.5 mg mL−1. The hydrolysis activity of NMHN was determined in the S9 of human derived cell lines, including A549, HepG2, and Caco-2.
Acknowledgements
This work was supported by the National Basic Research Program of China (2013CB531800), NSF of China (81473181, 81302793, 21572029, 21421005), and the State Key Laboratory of Fine Chemicals (KF1304 and KF1408).
References
- S. Oda, T. Fukami, T. Yokoi and M. Nakajima, Drug Metab. Pharmacokinet., 2015, 30, 30–51 CrossRef PubMed.
- M. K. Ross and J. A. Crow, J. Biochem. Mol. Toxicol., 2007, 21, 187–196 CrossRef CAS PubMed.
- T. Imai, M. Taketani, M. Shii, M. Hosokawa and K. Chiba, Drug Metab. Dispos., 2006, 34, 1734–1741 CrossRef CAS PubMed.
- T. Imai, Drug Metab. Pharmacokinet., 2006, 21, 173–185 CrossRef CAS PubMed.
- Y. Sato, A. Miyashita, T. Iwatsubo and T. Usui, Drug Metab. Dispos., 2012, 40, 1389–1396 CrossRef CAS PubMed.
- K. Na, E. Y. Lee, H. J. Lee, K. Y. Kim, H. Lee, S. K. Jeong, A. S. Jeong, S. Y. Cho, S. A. Kim, S. Y. Song, K. S. Kim, S. W. Cho, H. Kim and Y. K. Paik, Proteomics, 2009, 9, 3989–3999 CrossRef CAS PubMed.
- M. R. Redinbo and P. M. Potter, Drug Discovery Today, 2005, 10, 313–325 CrossRef CAS PubMed.
- J. S. Xu, L. Y. Yin, Y. Xu, Y. Y. Li, M. Zalzala, G. Cheng and Y. Q. Zhang, PLoS One, 2014, 9, e109663 Search PubMed.
- D. Y. Hui and P. N. Howles, J. Lipid Res., 2002, 43, 2017–2030 CrossRef CAS.
- M. Friedrichsen, P. Poulsen, J. Wojtaszewski, P. R. Hansen, A. Vaag and H. B. Rasmussen, FASEB J., 2013, 8, e56861 CAS.
- E. K. Tarkiainen, J. T. Backman, M. Neuvonen, P. J. Neuvonen, M. Schwab and M. Niemi, Clin. Pharmacol. Ther. Ser., 2012, 92, 68–71 CrossRef CAS PubMed.
- E. V. Rudakova, G. F. Makhaeva, T. G. Galenko, A. Y. Aksinenko, V. B. Sokolov and I. V. Martynov, Dokl. Biochem. Biophys., 2013, 449, 87–89 CrossRef CAS PubMed.
- H. J. Zhu, D. I. Appel, Y. Jiang and J. S. Markowitz, Drug Metab. Dispos., 2009, 37, 1819–1825 CrossRef CAS PubMed.
- D. F. Yang, R. E. Pearce, X. L. Wang, R. Gaedigk, Y. J. Y. Wan and B. F. Yan, Biochem. Pharmacol., 2009, 77, 238–247 CrossRef CAS PubMed.
- M. X. Xie, D. F. Yang, L. Liu, B. Xue and B. F. Yan, Drug Metab. Dispos., 2002, 30, 541–547 CrossRef CAS.
- J. Lill, Mass Spectrom. Rev., 2003, 22, 182–194 CrossRef CAS PubMed.
- E. J. O'Reilly, P. J. Conroy, S. Hearty, T. E. Keyes, R. O'Kennedy, R. J. Forster and L. Dennany, RSC Adv., 2015, 5, 67874–67877 RSC.
- R. Zhang, K. C. Liu, Y. L. Cui, W. Zhang, L. S. He, S. Q. Guo, Y. Y. Chen, Q. X. Li, S. Z. Liu and B. M. Wang, RSC Adv., 2015, 5, 35874–35881 RSC.
- S. C. Laizure, R. B. Parker, V. L. Herring and Z. Y. Hu, Drug Metab. Dispos., 2014, 42, 201–206 CrossRef CAS PubMed.
- H. J. Zhu, X. W. Wang, B. E. Gawronski, B. J. Brinda, D. J. Angiolillo and J. S. Markowitz, J. Pharmacol. Exp. Ther., 2013, 344, 665–672 CrossRef CAS PubMed.
- R. B. Parker, Z. Y. Hu, B. Meibohm and S. C. Laizure, Clin. Pharmacokinet., 2015, 54, 627–638 CrossRef CAS PubMed.
- Z. M. Liu, L. Feng, G. B. Ge, X. Lv, J. Hou, Y. F. Cao, J. N. Cui and L. Yang, Biosens. Bioelectron., 2014, 57, 30–35 CrossRef CAS PubMed.
- D.-D. Wang, Q. Jin, J. Hou, L. Feng, N. Li, S.-Y. Li, Q. Zhou, L.-W. Zou, G.-B. Ge, J.-G. Wang and L. Yang, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2016, 1008, 212–218 CrossRef CAS PubMed.
- G. Zhou, G. K. Marathe, J. Hartiala, S. L. Hazen, H. Allayee, W. H. W. Tang and T. M. McIntyre, J. Biol. Chem., 2013, 288, 11940–11948 CrossRef CAS PubMed.
- T. S. Anirudhan and S. R. Rejeena, J. Colloid Interface Sci., 2012, 381, 125–136 CrossRef CAS PubMed.
- M. S. Benhabiles, N. Abdi, N. Drouiche, H. Lounici, A. Pauss, M. F. A. Goosen and N. Mameri, Mater. Sci. Eng., C, 2012, 32, 922–928 CrossRef CAS.
- V. Alterio, P. Pan, S. Parkkila, M. Buonanno, C. T. Supuran, S. M. Monti and G. De Simone, Biopolymers, 2014, 101, 769–778 CrossRef CAS PubMed.
- K. Stroobants, E. Moelants, H. G. T. Ly, P. Proost, K. Bartik and T. N. Parac-Vogt, Chem.–Eur. J., 2013, 19, 2848–2858 CrossRef CAS PubMed.
- S. Takusagawa, K. Yajima, A. Miyashita, S. Uehara, T. Iwatsubo and T. Usui, Xenobiotica, 2012, 42, 957–967 CrossRef CAS PubMed.
- R. Balti, F. Bougherra, A. Bougatef, B. Hayet, N. Nedjar-Arroume, P. Dhulster, D. Guillochon and M. Nasri, Food Chem., 2012, 130, 475–484 CrossRef CAS.
- A. Salvi, P. A. Carrupt, J. M. Mayer and B. Testa, Drug Metab. Dispos., 1997, 25, 395–398 CAS.
- W. Hakamata, S. Tamura, T. Hirano and T. Nishio, ACS Med. Chem. Lett., 2014, 5, 321–325 CrossRef CAS PubMed.
- D. I. Draganov, J. F. Teiber, A. Speelman, Y. Osawa, R. Sunahara and B. N. La Du, J. Lipid Res., 2005, 46, 1239–1247 CrossRef CAS PubMed.
- X. D. Zeng, X. L. Zhang, B. C. Zhu, H. Y. Jia, Y. M. Li and J. Xue, Analyst, 2011, 136, 4008–4012 RSC.
- C. M. Romero, L. M. Pera, F. Loto and M. D. Baigori, Catal. Lett., 2012, 142, 1361–1368 CrossRef CAS.
- S. J. Kim, H. W. Rhee, H. J. Park, H. Y. Kim, H. S. Kim and J. I. Hong, Bioorg. Med. Chem. Lett., 2013, 23, 2093–2097 CrossRef CAS PubMed.
- J. Cordova, J. D. Ryan, B. B. Boonyaratanakornkit and D. S. Clark, Enzyme Microb. Technol., 2008, 42, 278–283 CrossRef CAS.
- S. K. Quinney, S. P. Sanghani, W. I. Davis, T. D. Hurley, Z. Sun, D. J. Murry and W. F. Bosron, J. Pharmacol. Exp. Ther., 2005, 313, 1011–1016 CrossRef CAS PubMed.
- R. Thomsen, H. B. Rasmussen, K. Linnet and I. Consortium, Drug Metab. Dispos., 2014, 42, 126–133 CrossRef CAS PubMed.
- Z. M. Liu, L. Feng, J. Hou, X. Lv, J. Ning, G. B. Ge, K. W. Wang, J. N. Cui and L. Yang, Sens. Actuators, B, 2014, 205, 151–157 CrossRef CAS.
- L. Feng, Z. M. Liu, J. Hou, X. Lv, J. Ning, G. B. Ge, J. N. Cui and L. Yang, Biosens. Bioelectron., 2015, 65, 9–15 CrossRef CAS PubMed.
- G. B. Ge, J. Ning, L. H. Hu, Z. R. Dai, J. Hou, Y. F. Cao, Z. W. Yu, C. Z. Ai, J. K. Gu, X. C. Ma and L. Yang, Chem. Commun., 2013, 49, 9779–9781 RSC.
- L. Feng, Z. M. Liu, L. Xu, X. Lv, J. Ning, J. Hou, G. B. Ge, J. N. Cui and L. Yang, Chem. Commun., 2014, 50, 14519–14522 RSC.
- X. G. Tian, C. Wang, G. B. Ge, J. Ning, C. Z. Ai, J. Y. Hong, Y. X. Cai, X. K. Huo, J. Hou, K. X. Liu, H. Z. Sun and X. C. Ma, RSC Adv., 2015, 5, 5924–5927 RSC.
- D. S. Shi, J. Yang, D. F. Yang, E. L. LeCluyse, C. Black, L. You, F. Akhlaghi and B. F. Yan, J. Pharmacol. Exp. Ther., 2006, 319, 1477–1484 CrossRef CAS PubMed.
- E. T. Williams, H. Wang, S. A. Wrighton, Y. W. Qian and E. J. Perkins, Mol. Phylogenet. Evol., 2010, 57, 23–34 CrossRef CAS PubMed.
- T. Satoh, P. Taylor, W. F. Bosron, S. P. Sanghani, M. Hosokawa and B. N. La Du, Drug Metab. Dispos., 2002, 30, 488–493 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Materials and methods, results of correlation analysis, chemical inhibition and method validation, summary of the currently used probes of hCE1, NMR and HR-MS spectra of NMHN. See DOI: 10.1039/c5ra23614b |
|
| This journal is © The Royal Society of Chemistry 2016 |
Click here to see how this site uses Cookies. View our privacy policy here. 



![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g. The supernatant (10 μL) was taken and subjected to UFLC measurements.
000 × g. The supernatant (10 μL) was taken and subjected to UFLC measurements.

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution of a polyclonal anti-hCE1 antibody, followed by incubation with a 1
1000 dilution of a polyclonal anti-hCE1 antibody, followed by incubation with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3000 dilution of horseradish peroxidase-conjugated goat anti-rabbit secondary antibody for 2 h. The protein was detected by ECL chemiluminescence system.
3000 dilution of horseradish peroxidase-conjugated goat anti-rabbit secondary antibody for 2 h. The protein was detected by ECL chemiluminescence system.




