Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Kinetic and thermodynamic study of the interactions between human carbonic anhydrase variants and polystyrene nanoparticles of different size†
A. Assarsson§,
I. Nasir‡§
,
M. Lundqvist and
C. Cabaleiro-Lago*
and
C. Cabaleiro-Lago*
Lund University, Division of Biochemistry and Structural Biology, P O Box 124, SE 221 00, Lund, Sweden. E-mail: Celia.Cabaleiro-Lago@biochemistry.lu.se
First published on 12th April 2016
Abstract
The activity and adsorption of three variants of human carbonic anhydrase (HCA) with similar topology but variation in charge and stability were studied in the presence of carboxyl-modified polystyrene nanoparticles of different sizes ranging from 25 nm to 114 nm. The balance of forces driving the adsorption of carbonic anhydrase variants is affected by the physicochemical properties of the protein and the nanoparticle size. All enzymes are totally inhibited upon adsorption due to the transition towards a molten globule like state that lacks enzymatic activity. The size of the particle affects the adsorption of human carbonic anhydrase I and N-terminal truncated human carbonic anhydrase II. Investigations on pH effects indicate that the size of the particle modulates the lateral interactions at the protein layer for these particular variants whose adsorption is mainly driven by electrostatic forces. A third variant, human carbonic anhydrase II, instead shows no strong influence of nanoparticle size which supports an adsorption process mainly driven by the hydrophobic effect.
1. Introduction
Protein adsorption on solid surfaces is a relevant phenomenon to many applications in bionanotechnology. For example, the design of bionanocatalysts and bionanosensors benefits on the extensive immobilization of enzymes on the device due to the large surface–volume ratio of nanoparticles.1,2 Furthermore, surface functionalized nanoparticles are being designed to target a particular protein for biomedical applications.3,4 The adsorption of the protein at the particle surface is often a requisite for these type of applications. However, the adsorption of enzyme on solid surfaces can lead to loss or decrease in the enzymatic activity.5Protein adsorption on solid surfaces is driven by a combination of different contributions such as electrostatic forces, the hydrophobic effect, dehydration of surfaces or structural reorganization.6 The process is affected by several factors.6,7 The charge of the protein and the surface plays an important role on the adsorption process. As an example of their importance, it is often observed that maximum nanoparticle surface coverage by a protein is generally achieved when the pH of the solution matches the isoelectric point of the protein.6,8 In those conditions the net charge of the protein is zero and thus lateral repulsions between adsorbed proteins are minimized. Hydrophobicity is also a crucial factor that explains the common observation that proteins absorb on hydrophobic surfaces under unfavorable electrostatic conditions, i.e. when particle and protein bear the same charge sign.9 Protein conformational stability also affects the adsorption process. Proteins with low structural stability usually undergo significant structural changes upon adsorption that may lead to an increase of conformational entropy which favor the adsorption.9,10 The exact predominance of each factor depends on the particular properties of the particle and protein of interest.
Human carbonic anhydrases (HCA) belongs to a well characterized group of metalloenzymes that catalyzes the conversion of carbon dioxide and water into bicarbonate and proton. HCA variants play roles in several diseases and is therefore a target for therapeutic inhibition in treatment of glaucoma, epilepsy, mountain sickness, ulcers, osteoporosis, obesity and cancer as reviewed by McKenna and Supuran.11 There are currently 15 known human isoforms of carbonic anhydrase with different functions and distribution in the body. Development of more efficient and selective inhibitors for HCA is therefore interesting for the treatment of the named diseases.11 However due to the multiple variables that can affect protein adsorption, systematic studies are needed for the design of effective nanoparticle based inhibitors for HCA's.
Three variants of HCA are included in this study. Human carbonic anhydrase I (HCAI) and human carbonic anhydrase II (HCAII) are cytosolic enzymes with similar topology and 60% sequence homology.12 HCAI is 2 kcal mol−1 more stable than HCAII.13,14 The third variant, trHCAII, is a truncated version of HCAII lacking the 17 N-terminal amino acid residues. This mutant is active and folded but has a lower stability (5 kcal mol−1 relative to HCAII at pH 7.5).15
In this study the behavior of three HCA variants in presence of carboxyl-modified polystyrene nanoparticles of different sizes has been analyzed. We have studied how the nanoparticles affect the enzyme activity and complex formation to be able to investigate the importance of electrostatics forces and the hydrophobic effect in the inhibition of HCA variants by nanomaterials. Furthermore, the effect of particle size on the density of the adsorbed protein layer was also analyzed.
2. Methods
2.1 Materials
Carboxyl-modified polystyrene nanoparticles (PSCOOH) were purchased from BangLabs and Polysciences and dialyzed against water before use. The sizes of the particles after dialysis were obtained by dynamic light scattering (DLS) and were in agreement with the manufacturer sizes. All chemicals were of analytical grade.2.2 Protein production
HCAI, purified from human blood, was provided by Professor Bengt-Harald Jonsson, Linköping University. HCAII (“pseudo wildtype” with C206S mutation) and trHCAII were expressed in E. coli BL21 (DE3) PlysS Star (plasmids were kindly provided by Professor Bengt-Harald Jonsson, Linköping University) and purified as previously described.16The concentration of the proteins was calculated by absorbance measurements at 280 nm using an Agilent 8453 spectrophotometer. The extinction coefficients are 54![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 M−1 cm−1, 44
800 M−1 cm−1, 44![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 M−1 cm−1 and 46
100 M−1 cm−1 and 46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 M−1 cm−1 for HCAII, trHCAII and HCAI respectively.17
800 M−1 cm−1 for HCAII, trHCAII and HCAI respectively.17
2.3 Isoelectric focusing
Isoelectric focusing experiments were performed in a Pharmacia PhastSystem High Speed Electrophoresis System using a Phast gel IEF 3-9 according to manufacturer protocol. The isoelectric points (pI) for the HCA variants were calculated from the gel using ImageQuant TL image analysis software.2.4 Particle size measurements
The hydrodynamic diameter of the particles upon titration of HCA variants were measured by DLS using a DynaPro DLS Plate Reader II from Wyatt Technology operating with a scattering angle of 158° at 30 °C, in a clear, flat bottom 96-well plate (Costar). Nanoparticle and protein solutions were prepared in 10 mM HEPES/NaOH buffer pH 7.4. The concentrations of different sized nanoparticles were selected to have equal surface area in solution (0.015 m2 mL−1 before the protein additions) except for the smallest particle. Due to detection issues, the concentration of 25 nm PSCOOH was doubled (0.030 m2 mL−1 before the protein addition). Aliquots of 3–5 μl stock solutions (10 or 24 μM) of the HCA variants were added to wells contained 200 μl stock solutions of 25, 41, 92, and 114 nm PSCOOH nanoparticles and measured after 1 minute incubation, collecting 10 acquisitions for each well. A general purpose analysis model (Cumulant for monomodal dispersions and Regularization for multimodal dispersions) was employed to determine the particle size upon addition of proteins, using Dynamics V7 Software.At the end of the kinetic and calorimetry measurements the size of the nanoparticle–HCA complexes as well as the size of the different HCA variants were measured and analysed as described above.
2.5 Kinetic measurements
The hydrolysis of p-nitrophenyl acetate (NPA) catalyzed by HCA variants18 was monitored by absorption UV-vis spectroscopy at 348 nm using a Fluostar Omega platereader (BMG Labtech, Offenburg, Germany). All kinetic runs were performed at 30 °C. The nanoparticles were incubated for 30–80 minutes with 0.35 μM HCA in 10 mM HEPES/NaOH buffer at pH 7.4 or 8.2 in a 96 well assay plate (Costar) before the initiation of the experiments. The substrate, NPA, was dissolved in acetonitrile in a 40 mM stock solution to avoid hydrolysis prior to the initiation of the experiments. The amount of acetonitrile in the assay reaction was always less than 2% (v/v). The reactions were initiated by addition of 5 μl NPA stock solution and shaking 10 s at 200 rpm to ensure good mixing. The increase in absorbance was followed during the initial linear range of the reaction (when less than 2% of the product is produced). The initial rate of autohydrolysis of NPA in water was subtracted from the initial rates of the enzymatically catalyzed reactions. Activity measurements were reproducible to within 8%.Assuming that the particles have n identical and independent protein binding sites and that the adsorbed proteins may have a residual enzymatic activity, a modified version of the Morrison equation19 for the fractional activity (Vi/V0) can be written (eqn (1)):
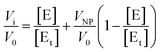 | (1) |
 | (2) |
By fitting these equations (in OriginPro, OriginLab corporation, Northampton, MA) to the experimental data the values for the stoichiometry of the complex, n, and the apparent inhibitor constant for each binding site, Kiapp, can be obtained.
2.6 Isothermal titration calorimetry (ITC)
ITC experiments were performed using a VP-ITC instrument from Microcal (Northhampton, MA). The carbonic anhydrase variants were titrated from a 30–40 μM stock. The concentration of 25 nm PSCOOH nanoparticle solution was 25 nM. All experiments were performed in 10 mM HEPES/NaOH buffer at 30 °C at pH 6.8, 7.4 or 8.2. The content of the cell was continuously stirred at 480 rpm. Control titrations for the heat of dilution of the proteins and nanoparticles into buffer were performed. The integrated heats for each experiment were calculated from the raw data and corrected with integrated heats from the control experiments. Data was analyzed using the Origin software (Microcal) assuming a “single set of identical sites” model or a “two sets of independent sites” model implemented in the fitting software. By an iterative fitting procedure the equilibrium dissociation constant Kd, n and ΔH can be calculated.3. Results
3.1 Size determination by DLS
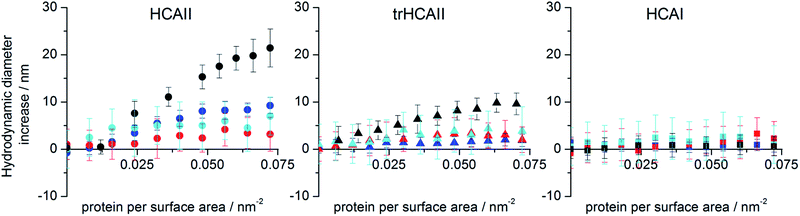
Particle colloidal stability ensures that the results on protein adsorption obtained by activity and ITC measurements are not compromised by significant particle aggregation. Hence, changes in particle size upon HCA titration were monitored by DLS in order to assess the stability of the colloidal dispersion before and after protein addition as well as analyzing the size of the protein–nanoparticle complex at pH 7.4 (Fig. S1†). All particles were stable in buffer in absence of protein. Fig. 1 shows that the hydrodynamic diameter of particles does not increase significantly in presence of HCAI whereas a slight increase of 3–4 nm is observed for trHCAII (except for 25 nm PSCOOH). On the other hand, the diameter for all particle sizes increases in presence of HCAII. For most of the particle sizes, a diameter increase upon titration between 6 and 10 nm is observed. If we take into account that a monolayer of natively folded HCA contributes to the particle size approximately around 6 to 8 nm (the hydrodynamic diameter obtained by DLS was 5.2 ± 1.8, 5.2 ± 3.8 and 5.0 ± 0.4 nm for HCAII, trHCAII and HCAI respectively), the size increase observed is compatible with the formation of a packed monolayer of adsorbed enzyme. However, in the case of the 25 nm particle, the diameter of the protein–particle complex shows an increase of about 20 nm for HCAII. This is not compatible with an ordered monolayer and may instead correspond to a disordered monolayer or multilayer. Alternatively, some degree of particle aggregation driven by protein adsorption may also contribute to an increase in the mean value of the size distribution. Indeed, the polydispersity of 25 nm PSCOOH increases as the concentration of HCAII increases. Yet, the colloidal dispersions of 25 nm PSCOOH after the addition of all HCA variants are stable for days and the population of big aggregates is lower than 3% in mass in all cases. The difference in size increase between HCAII and the other two variants may indicate that more HCAII is recruited at the PSCOOH nanoparticle surface than trHCAII and HCAI.3.2 Enzymatic activity in presence of nanoparticles
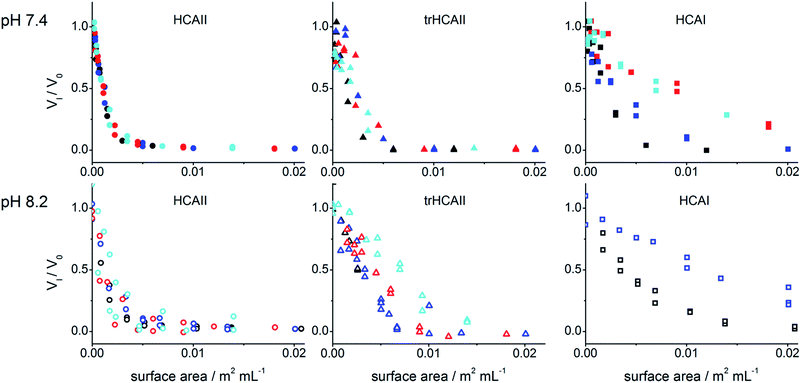
The activity of the three HCA variants in presence of increasing concentrations of PSCOOH nanoparticles was monitored spectrophotometrically through the hydrolysis of NPA at two different pH values, pH 7.4 and pH 8.2 (Fig. S2†). The presence of PSCOOH nanoparticles decreases the activity for all three HCA variants down to a complete inhibition when the particle surface area is in excess. Due to concentration limitations (high background light scattering) no significant inactivation could be detected at pH 8.2 for HCAI in presence of 92 and 114 nm PSCOOH nanoparticles.For the same molar particle concentration the extent of inhibition increases as the size of the particle increases (Fig. S2†). This is in line with larger particles presenting larger surface area in solutions (at same molar concentration) and thus more interactions sites in the case the surface is homogeneous.
In order to further analyse the effect of nanoparticle size on the inactivation of HCA variants, the relative initial rates were plotted in Fig. 2 against the nanoparticle surface area in solution (m2 mL−1). HCAII consistently shows the same inactivation trend for surface area regardless the particle size. This indicates that the extent of inactivation is only governed by the accessible surface area. On the other hand, HCAI activity trend varies with the size of the particles at both studied pH. This result suggests that the curvature of the particle influences the amount of protein adsorbed for the same accessible surface area. The results for trHCAII at pH 7.4 are difficult to assess due to the higher dispersion of the experimental data (Fig. 2), but at pH 8.2 the effect of size on the trHCAII activity data clearly resembles the effect observed for HCAI.
To get a deeper understanding on the interaction between protein and nanoparticles, the data was analyzed using a model that assumes that the nanoparticles have n identical and individual sites to which proteins can bind (see Methods). Fig. S2† shows the best fits to the data. A global fit was performed for each HCA's kinetic data set assuming a common value for Kiapp for all nanoparticle sizes. Global Kiapp, n and VNP at two different pH were obtained (Table S1†). Due to the poor inhibition of HCAI by 92 and 114 nm PSCOOH within the working concentration range, n could not be extracted for these cases. The activity of the adsorbed enzymes is, in all cases, less than 1.5% of the free enzyme based on the Vnp value. This confirms total inactivation of the enzymes at the particle surface. For HCAI and HCAII, the Kiapp values are in a similar range (Table S1†). However, errors are larger for HCAI. For trHCA only a maximal Kiapp could be determined due to the experimental settings.
In order to compare the interactions between HCA variants and particles of different sizes, the protein surface coverage was estimated. The maximum number of binding sites for a fully packed monolayer (nmono) was calculated taking into account the HCA docking area and the mean surface area of the particles. These HCA variants can be described as prolate spheroids with the dimensions 3 × 4 nm and thus 10 nm2 side-on docking area. Protein coverage was calculated as the ratio between the experimental and the maximum number of binding sites (n/nmono × 100) (Table 1).
The obtained coverage data (Table 1) shows that HCAII has higher protein coverage than HCAI and trHCAII for all nanoparticle sizes. This difference in protein coverage increases as the pH increases from 7.4 to 8.2. At both pH 7.4 and pH 8.2, HCAII covers the particles to a similar degree regardless of the particle size. On the other hand, in the case of HCAI, a clear effect of the particle size on protein coverage is observed both at pH 7.4 and 8.2. HCAI coverage increases as the particle size decreases. This is also the case for trHCAII at pH 8.2.
3.3 Formation of protein–particle complex by ITC
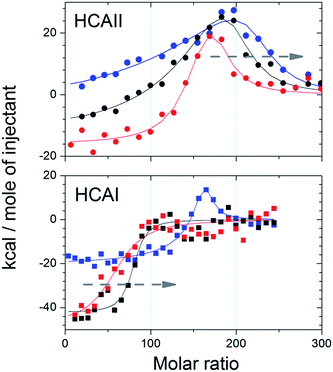
ITC experiments were performed in order to confirm the formation and to calculate the stoichiometry of the protein–particle complex for HCAII and HCAI at different pH. Fig. 3 shows the binding isotherms for the titration of the variants on 25 nm PSCOOH nanoparticles (for raw data see Fig. S3†). Fitting of an “one set of identical sites” or “two sets of independent sites” model allows us to obtain the number of binding sites per particle, n, (Table S2†) and, as explained above, the protein coverage with respect to the maximum theoretical coverage (n/nmono × 100) (Table 2). The data show that the protein coverage is lower for HCAI than HCAII for all pH studied. trHCAII has an intermediate coverage at pH 7.4 as shown by Fig. S3.† Moreover, the ITC data clearly show that the coverage for HCAI increases as the pH come closer to the pI of HCAI (pI = 6.6). The coverage at pH 6.8 reaches 80% of the maximum coverage most likely due to the reduction of the repulsive lateral forces between the adsorbed proteins. However HCAII does not show such pH dependence in the adsorption process. Even though an increase in coverage is observed as the protein gets more positively charged at pH 6.8 (HCAII pI = 7.6) the variation is not so noticeable (or falls within the errors of the measurements).| HCAII | HCAI | |
|---|---|---|
| pH 6.8 | 113 ± 40 | 81 ± 10 |
| pH 7.4 | 93 ± 40 | 43 ± 5 |
| pH 8.2 | 87 ± 25 | 37 ± 4 |
4. Discussion
Here, we have studied the effect of negatively charged hydrophobic nanoparticles of different size on the activity of adsorbed HCA variants at different pHs. The results show that all three variants interact with PSCOOH nanoparticle but at different extent. Regardless of the pH, the stability of the different variants does not correlate with the observed surface coverage. Moreover, the impact of nanoparticle size differs between the variants. HCAII coverage is not affected by the particle size. On the other hand the coverage of HCAI and trHCAII decreases as the particle size increases at pH 8.2. All these observations point towards a different balance of driving forces for the adsorption of these protein variants even though they have high structural similarity.4.1 Inactivation upon adsorption
Total inhibition of the variants is achieved at nanomolar concentration in all cases. Therefore, PSCOOH nanoparticles are strong inhibitors of the studied HCA's whilst free carboxyl groups were found to be specific but weak inhibitors of these enzymes.20 Carbonic anhydrases are known for having a folding intermediate between the native and the unfolded state known as the molten-globule state.21–23 This intermediate state has a compact core that retains most of the secondary structure of the native state while the tertiary structure is partially distorted. Furthermore, carbonic anhydrases molten-globule state is completely devoid of enzymatic activity.21,22HCAII and truncated versions can indeed adopt a molten-globule like state upon adsorption on silica nanoparticles with substantial loss of binding affinity towards substrate analogs24 and esterase activity.25 Here, total loss of esterase activity by PSCOOH is observed for all HCA variants. We have recently shown that trHCAII and HCAI experience structural rearrangements in the tertiary structure at the surface of PSCOOH nanoparticles.26 In addition, the increase of ANS signal, a fluorescent dye commonly used to detect molten globule states,27 suggests the formation of this intermediate upon adsorption on PSCOOH.26 Therefore, the loss of enzymatic activity observed here may be linked to a transition towards a molten globular like state upon adsorption of HCA's on PSCOOH nanoparticles. Alternatively, the access of the substrate to the active site might be partially hindered due to the orientation of the protein on the particle surface.
4.2 Balance between electrostatics forces and the hydrophobic effect
The net charge of the enzyme variants depends on the pH of the buffer and the pI of the proteins. The pI values obtained by isoelectric focusing were 7.6, 6.8 and 6.6 for HCAII, trHCAII and HCAI respectively and were consistent with previously reported values (Fig. S4†).28 Therefore, all three variants possess different net charge at any given pH. This difference may influence the adsorption process. HCAI is always negatively charged at all studied pH while HCAII goes from positively to negatively charge over the pH range used. The carboxylated nanoparticles are, on the other hand, always negatively charged at the selected experimental conditions.HCA adsorption takes place at the negatively charged hydrophobic nanoparticles surface even when the protein variant has a net negative charge. This means that long range electrostatic interactions are not the main driving force for the initial phase of the interaction. However, the non-uniform nature of the protein surface, consisting in a distribution of positively and negatively charged patches, might play a role in the initial docking by forming favorable local polar contacts with the surface. Another important factor for the adsorption is the hydrophobic effect. Indeed, proteins are known to adsorb on hydrophobic surfaces regardless unfavorable electrostatics interactions.6,29 Electrostatic repulsions between docked proteins may as well have an important role in the adsorption process and control the final protein packing at the surface.
Both of the methods used to calculate surface coverage, activity assays and ITC, report the same trends. HCAI and trHCAII coverage on PSCOOH nanoparticles is generally lower at high pH and increases when the pH decreases (Tables 1 and 2). However, HCAII adsorption is essentially invariable with respect to pH. This suggests that electrostatic forces are not the main driving force of the adsorption for this variant. The hydrophobic effect is rather one of the main contributors to the adsorption process of HCAII on PSCOOH nanoparticles as shown previously.16 In addition, HCAII can undergo considerable conformational changes upon adsorption.30 The subsequent conformational entropy gain will further contribute to the adsorption process. These observations are in agreement with the fact that proteins that easily undergo conformational changes on the surface are less sensitive to electrostatic interactions due to the nature of the forces that dominate their adsorption.9
On the other hand, HCAI shows a strong influence of pH on the adsorption. According to ITC experiments, coverage increases from 40% at high pH up to 80% surface coverage as the pH approaches the pI of the protein (Table 2). This indicates a strong role of electrostatic forces in the adsorption for this variant. Although the contribution from the hydrophobic effect may be similar for HCAII and HCAI based on sequence similarity, HCAI does not easily change conformation upon adsorption and behaves more as a hard sphere.17 We can therefore conclude that the contribution of electrostatic forces such as inter-protein interactions at the particle surface is still predominant and not overridden by the contribution from protein conformational rearrangements.
trHCAII adsorption is also affected by variation of pH. Therefore, electrostatic forces seem to have an important role in the process even though trHCAII has lower thermodynamic stability compared to HCAII. The difference between trHCAII and HCAII may rely on the nature of the N-terminal which is truncated in trHCAII. The N-terminal of HCAII contains a hydrophobic patch31 that may interact with the hydrophobic surface and significantly contribute to the adsorption process. Removal of the N-terminal may reduce the contribution of the hydrophobic effect to the adsorption and shifts the balance of forces in favor of electrostatics. In fact, a similar effect was observed on hydrophobic tag containing proteins where the length of the hydrophobic tag increased the adsorption of those proteins on hydrophobic surfaces.32,33
4.3 Effect of particle size on the balance of forces
The size of the particles affects the surface coverage of trHCAII at pH 8.2 and HCAI at pH 7.4 and pH 8.2. The stoichiometry of the complex increases when the nanoparticle size decreases for those cases. The effect of nanoparticle size on protein adsorption has been previously related to the increase of contact area between nanoparticle and protein. Generally, an increase of contact area as the nanoparticle size increases leads to formation of more inter-molecular polar contacts that will in turn favor adsorption.17,34 However, this hypothesis does not explain the results observed here. In a previous work we have proposed a model to explain the effect of PSCOOH on the dynamics of adsorption of trHCAII and HCAI.26 According to that, lateral repulsive interactions between adsorbed proteins are geometrically minimized due to the increase of particle curvature. This reduction of repulsive forces helps the protein to pack more densely and therefore increase the coverage as shown in the case of HCAI and trHCAII. The findings in this paper support the previously proposed model. However, the modulating effect of particle size is only relevant depending on the balance of forces driving the interactions. For HCAI and trHCAII at high pH, lateral repulsion may strongly contribute to the final packing density at the surface. Therefore changes in particle size will reflect on the packing of the adsorbed protein at the surface due to geometrically minimized lateral repulsions. However, at lower pH the contribution of lateral repulsions decreases resulting in a smaller effect of the particle size on the overall adsorption process. For proteins where electrostatic forces are not the dominating driving force, like the case of HCAII, lateral repulsions are overcome by other stabilizing forces. Thus, no effect on particle curvature is observed for HCAII regardless the pH, i.e. the net charge of the protein.5. Concluding remarks
HCAII, trHCAII and HCAI are inhibited by carboxyl-modified polystyrene nanoparticles at experimental conditions where surface and protein are negatively charged. HCAII adsorption is mainly driven by non-electrostatic contributions while HCAI shows a strong contribution of electrostatic forces, especially lateral repulsion between adsorbed proteins. The removal of a hydrophobic patch in trHCAII shifts the balance of forces towards the HCAI case. This study also highlights the importance of particle size regarding the design of bionanomaterials since different sizes lead to the formation of complexes with different packing density of the adsorbed protein layer. Particle size affects protein adsorption by changing the nanoparticle–protein contact area but also by modulating the lateral interactions among the adsorbed proteins. In conclusion, even fairly similar proteins have a different set of dominant driving forces when adsorbing on surfaces which can be modulated by the relative size (respect to the protein) of the particle.Acknowledgements
This work was supported by Swedish Research Council (VR) and the Crafoord Foundation.Notes and references
- J. M. Slocik and R. R. Naik, Chem. Soc. Rev., 2010, 39, 3454–3463 RSC.
- N. T. K. Thanh and L. A. W. Green, Nano Today, 2010, 5, 213–230 CrossRef CAS.
- M. Stiti, A. Cecchi, M. Rami, M. Abdaoui, V. Barragan-Montero, A. Scozzafava, Y. Guari, J.-Y. Winum and C. T. Supuran, J. Am. Chem. Soc., 2008, 130, 16130–16131 CrossRef CAS PubMed.
- P. M. Tiwari, K. Vig, V. A. Dennis and S. R. Singh, Nanomaterials, 2011, 1, 31–63 CrossRef CAS.
- T. J. Maccormack, R. J. Clark, M. K. M. Dang, G. Ma, J. A. Kelly, J. G. C. Veinot and G. G. Goss, Nanotoxicology, 2012, 6, 514–525 CrossRef CAS PubMed.
- M. Malmsten, J. Colloid Interface Sci., 1998, 207, 186–199 CrossRef CAS PubMed.
- W. Norde, Macromol. Symp., 1996, 103, 5–18 CrossRef CAS.
- M. Rabe, D. Verdes and S. Seeger, Adv. Colloid Interface Sci., 2011, 162, 87–106 CrossRef CAS PubMed.
- C. A. Haynes and W. Norde, Colloids Surf., B, 1994, 2, 517–566 CrossRef CAS.
- C. A. Haynes and W. Norde, J. Colloid Interface Sci., 1995, 169, 313–328 CrossRef CAS.
- R. McKenna and C. T. Supuran, in Carbonic Anhydrase: Mechanism, Regulation, Links to Disease, and Industrial Applications, ed. S. C. Frost and R. McKenna, 2014, vol. 75, pp. 291–323 Search PubMed.
- L. Henderson, D. Henriksson and P. O. Nyman, Biochem. Biophys. Res. Commun., 1973, 52, 1388–1394 CrossRef CAS PubMed.
- P. O. Freskgard, U. Carlsson, L. G. Martensson and B. H. Jonsson, FEBS Lett., 1991, 289, 117–122 CrossRef CAS PubMed.
- A. Kjellsson, I. Sethson and B. H. Jonsson, Biochemistry, 2003, 42, 363–374 CrossRef CAS PubMed.
- G. Aronsson, L. G. Martensson, U. Carlsson and B. H. Jonsson, Biochemistry, 1995, 34, 2153–2162 CrossRef CAS PubMed.
- A. Assarsson, I. Pastoriza-Santos and C. Cabaleiro-Lago, Langmuir, 2014, 30, 9448–9456 CrossRef CAS PubMed.
- M. Lundqvist, I. Sethson and B. H. Jonsson, Langmuir, 2004, 20, 10639–10647 CrossRef CAS PubMed.
- Y. Pocker and J. T. Stone, Biochemistry, 1967, 6, 668–678 CrossRef CAS PubMed.
- J. F. Morrison, Biochim. Biophys. Acta, 1969, 185, 269–286 CrossRef CAS.
- G. De Simone and C. T. Supuran, J. Inorg. Biochem., 2012, 111, 117–129 CrossRef CAS PubMed.
- D. A. Dolgikh, A. P. Kolomiets, I. A. Bolotina and O. B. Ptitsyn, FEBS Lett., 1984, 165, 88–92 CrossRef CAS PubMed.
- L. G. Maartensson, B. H. Jonsson, P. O. Freskgaard, A. Kihlgren, M. Svensson and U. Carlsson, Biochemistry, 1993, 32, 224–231 CrossRef CAS.
- D. Andersson, P. O. Freskgård, B. H. Jonsson and U. Carlsson, Biochemistry, 1997, 36, 4623–4630 CrossRef CAS PubMed.
- P. Billsten, P. O. Freskgard, U. Carlsson, B. H. Jonsson and H. Elwing, FEBS Lett., 1997, 402, 67–72 CrossRef CAS PubMed.
- M. Karlsson, L. G. Martensson, B. H. Jonsson and U. Carlsson, Langmuir, 2000, 16, 8470–8479 CrossRef CAS.
- I. Nasir, M. Lundqvist and C. Cabaleiro-Lago, Nanoscale, 2015, 7, 17504–17515 RSC.
- O. B. Ptitsyn, R. H. Pain, G. V. Semisotnov, E. Zerovnik and O. I. Razgulyaev, FEBS Lett., 1990, 262, 20–24 CrossRef CAS PubMed.
- S. Funakoshi and H. F. Deutsch, J. Biol. Chem., 1969, 244, 3438–3446 CAS.
- W. Norde, Adv. Colloid Interface Sci., 1986, 25, 267–340 CrossRef CAS PubMed.
- M. Lundqvist, C. Andresen, S. Christensson, S. Johansson, M. Karlsson, K. Broo and B. H. Jonsson, Langmuir, 2005, 21, 11903–11906 CrossRef CAS PubMed.
- A. E. Eriksson, T. A. Jones and A. Liljas, Proteins: Struct., Funct., Genet., 1988, 4, 274–282 CrossRef CAS PubMed.
- M. Malmsten and A. Veide, J. Colloid Interface Sci., 1996, 178, 160–167 CrossRef CAS.
- M. Malmsten, N. Burns and A. Veide, J. Colloid Interface Sci., 1998, 204, 104–111 CrossRef CAS PubMed.
- S. Lindman, I. Lynch, E. Thulin, H. Nilsson, K. A. Dawson and S. Linse, Nano Lett., 2007, 7, 914–920 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Table S1: number of binding sites n and apparent dissociation constant for the HCA variants at different pH extracted from activity measurements. Table S2: number of binding sites n for the HCA variants extracted from ITC experiments. Fig. S1: particle size obtained by DLS measurements. Fig. S2: activity of HCAII, trHCAII and HCAI in the presence of negatively charged nanoparticles at two different pH. Fig. S3: ITC data for the titration of CA variants to 25 nm PS nanoparticles at three different pH. Fig. S4: isoelectric focusing gel for the three carbonic anhydrase variants. See DOI: 10.1039/c6ra06175c |
| ‡ Current address: Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA USA 92037. |
| § These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2016 |