 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
‘Traceless’ tracing of proteins – high-affinity trans-splicing directed by a minimal interaction pair†‡
M.
Braner
,
A.
Kollmannsperger
,
R.
Wieneke
and
R.
Tampé
 *
*
Institute of Biochemistry, Biocenter, and Cluster of Excellence – Macromolecular Complexes, Goethe-University Frankfurt, Max-von-Laue-Str. 9, 60438 Frankfurt/M., Germany. E-mail: tampe@em.uni-frankfurt.de
First published on 21st December 2015
Abstract
Protein trans-splicing mediated by split inteins is a powerful technique for site-specific protein modification. Despite recent developments there is still an urgent need for ultra-small high-affinity intein tags for in vitro and in vivo approaches. To date, only very few in-cell applications of protein trans-splicing have been reported, all limited to C-terminal protein modifications. Here, we developed a strategy for covalent N-terminal intein-mediated protein labeling at (sub) nanomolar probe concentrations. Combined with a minimal synthetic lock-and-key element, the affinity between the intein fragments was increased more than 50-fold to 10 nM. Site-specific and efficient ‘traceless’ protein modification by high-affinity trans-splicing is demonstrated at nanomolar concentrations in living mammalian cells.
Introduction
An eminent challenge in life sciences remains the site-specific protein modification with minimal perturbation for in vitro as well as in vivo analyses. Although the use of fluorescent proteins facilitated fundamental insight into protein function and cellular processes, their applicability is inherently limited due to their low photostability and quantum yield, slow maturation time, as well as large size.1 Hence, alternative approaches have been developed to introduce synthetic fluorescent probes with improved photo-physical properties by self-labeling proteins, e.g. SNAP/CLIP/Halo tags,2–4 or by enzymatic modifications via phosphopantetheinyl transferases, sortases, or lipoic acid ligases.5–7 Yet, these enzymes need to be supplied at high concentrations (1–100 μM) and the large fusion domains (>20 kDa) have adverse effects on the function, interaction, and trafficking of the target protein.Besides these strategies, different chemoselective reactions and semi-synthetic techniques, such as native chemical ligation or intein-mediated protein splicing, have been established.8–10 Inteins are internal protein domains, which facilitate their own excision and thereby covalently fuse the flanking N- and C-terminal exteins, EN and EC, in a self-processive manner. A very promising strategy for protein semi-synthesis is protein trans-splicing (PTS), which has been applied for segmental isotope labeling,11 protein backbone cyclization,12 cyclic peptide generation,13–15 protein immobilization,9,16,17 as well as for N- and C-terminal protein labeling.18–21 In PTS, the autocatalytic domain is naturally or artificially split into two fragments, IN and IC, reconstituting the active intein complex.22
The excision process is virtually traceless, apart from a few flanking extein residues, required for efficient splicing. Whereas semi-synthetic PTS is widely applied in vitro, in-cell applications using split inteins are exclusively based on C-terminal protein modifications by trans-splicing.23–26 If the native C terminus is not accessible or essential for function, localization, or protein dynamics (e.g. lipid- and tail-anchored proteins,27 ubiquitin,28 and lamin A29), intracellular N-terminal protein modifications by PTS are indispensable. However, the large sizes of most split intein fragments with 100–130 amino acids (aa) for IN and 35–50 aa for IC, also including high-affinity inteins, e.g. Npu DnaE30 or Ssp DnaE,23,24,31 compromise their accessibility by solid-phase peptide synthesis (SPPS).32,33
Several attempts were made to create short intein fragments for these semi-synthetic approaches.20,34 Despite that, the affinities in the micromolar range of very small intein fragments18 impede in vivo applications.
Here, we adopted the multivalent chelator tris-N-nitrilotriacetic acid (trisNTA),35–37 which allows for site-specific and reversible recognition of His6–10-tagged proteins in the nanomolar range (KD < 10 nM), even inside living cells.38,39 The trisNTA/His-tag system defines one of the smallest high-affinity recognition elements known to date. We developed a split intein system guided by this minimalistic interaction pair to promote N-terminal protein labeling at nanomolar concentrations by trans-splicing (Scheme 1). A related approach was realized with E. coli dihydrofolate reductase and its ligand trimethoprim.40 Nonetheless, this strategy suffers from a large fusion domain (25 kDa) and micromolar concentrations applied for labeling. Noteworthy, this affinity pair was utilized for extracellular labeling, whereas our high-affinity trisNTA/His-tag pair already demonstrated accurate labeling of proteins in living cells.38,39,41 Our approach combines this diminutive interaction pair with the smallest synthetically accessible IN (11 aa) and a recombinantly expressed IC (143 aa) of the artificially split DnaB M86 mini-intein from Synechocystis sp. PCC6803 (Ssp DnaB M86).42,43
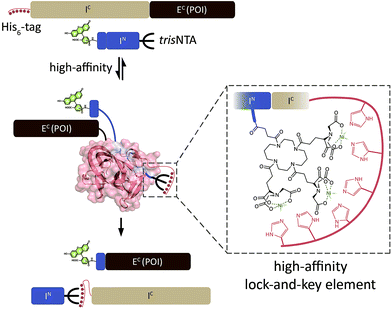 | ||
| Scheme 1 Protein trans-splicing guided by a minimalistic high-affinity interaction pair. Based on the high-affinity trisNTA/His-tag interaction, the intein fragments IN and IC react at nanomolar concentrations. PTS results in a ‘traceless’ N-terminal labeling of EC (POI, protein of interest) with e.g. a molecular probe. Ssp DnaB: 1MI8.pdb. | ||
Results and discussion
Protein trans-splicing directed by a high-affinity interaction pair
We generated a set of synthetic IN and recombinant IC fragments (Fig. 1a and b). IN was prepared by Fmoc-based SPPS with five native EN residues, KKESG, and either 5(6)-carboxyfluorescein or Cy5 as N-terminal fluorescent reporter. The multivalent chelator trisNTA was C-terminally introduced via diaminopropionic acid during SPPS (Fig. 1a and S1‡). Thioredoxin was chosen as a model protein and N-terminally fused with the IC fragment of Ssp DnaB M86. In addition, the N terminus of IC was equipped with a His6-tag and optionally a serine–glycine linker (SGGG, thereafter referred to as HICT or HLICT; Fig. 1b). For comparison, IC-Trx-His6 (ICTH) was used.42,43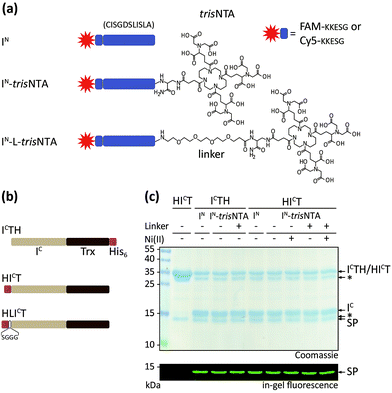 | ||
| Fig. 1 Semi-synthetic protein trans-splicing directed by a minimal high-affinity interaction pair. (a) Design of synthetic IN fragments with five EN residues (KKESG). TrisNTA was incorporated during Fmoc-based SPPS. 5(6)-Carboxyfluorescein (FAM) or Cy5 were used as fluorescent reporter. (b) Design of recombinant IC with thioredoxin (Trx). N-terminal His6-tagged HICT and HLICT were used for interaction analyses with IN-trisNTA and IN-L-trisNTA. ICTH with a C-terminal His6-tag served as control. (c) PTS of ICTH/HICT (10 μM) and FAM-labeled IN (40 μM) yielded fluorescently labeled Trx (1 h, 20 °C). SP, splice product; *, impurity/premature cleaved intein.20,44 | ||
We first investigated the trans-splicing reaction at micromolar concentrations typically used for in vitro experiments (Fig. 1c). After performing PTS, fluorescently labeled Trx and the excised IC fragment were detected by SDS-PAGE in-gel fluorescence and Coomassie staining. Densitometric analysis of IC conversion with His6-IC-Trx (HICT; 76 ± 8%) and His6-IC-linker-Trx (HLICT; 71 ± 10% for IN; Table S3‡) showed similar yields. These values are in accordance with data reported for ICTH and IN.42 >70% splice product (SP) formation was achieved for all constructs, with negligible formation of C-terminal by-products by premature cleavage of the intein. This phenomenon of self-induced C-terminal cleavage is known for the Ssp DnaB intein, but almost suppressed in the M86 mutant.42 SP formation was calculated as follows: [SP]/([SP] + [residual IC fragment] + [cleaved IC]).18,43 The amount of premature cleaved intein20,44 was not included in SP determination. Our emphasis was not to prevent premature cleavage, but rather to increase the affinities of the intein fragments.
At 37 °C, PTS showed a decreased efficiency with up to 5-fold higher C-terminal cleavage.42 Interestingly, the Ser–Gly linker between the His6-tag and IC (HLICT) caused ∼10% less IC conversion for IN-trisNTA and IN-L-trisNTA. The pseudo first-order rate of trans-splicing was kPTS = (2.0 ± 0.6) × 10−3 s−1 corresponding to a τ1/2 = 14 ± 3 min at 20 °C for HICT and IN. The rate is in perfect agreement with kPTS = (2.5 ± 0.1) × 10−3 s−1 for ICTH under comparable conditions.42 All other constructs reacted similarly (Table S3‡). In conclusion, the presence of Ni-trisNTA and N-terminal His-tag had no effect on the trans-splicing efficiency and reaction kinetics.
High-affinity interaction determined by fluorescence anisotropy and microscale thermophoresis
We determined the affinity of the intein fragments by fluorescence anisotropy (FA; Fig. 2a and b) and by microscale thermophoresis (MST)45,46 using the splice-inactive IC(N154A, S+1A, H73A) mutant42,43 and 150 pM of Cy5-labeled IN (Fig. 2c–e and S6‡). The MST experiments represent the first use of MST for affinity measurements performed on inteins.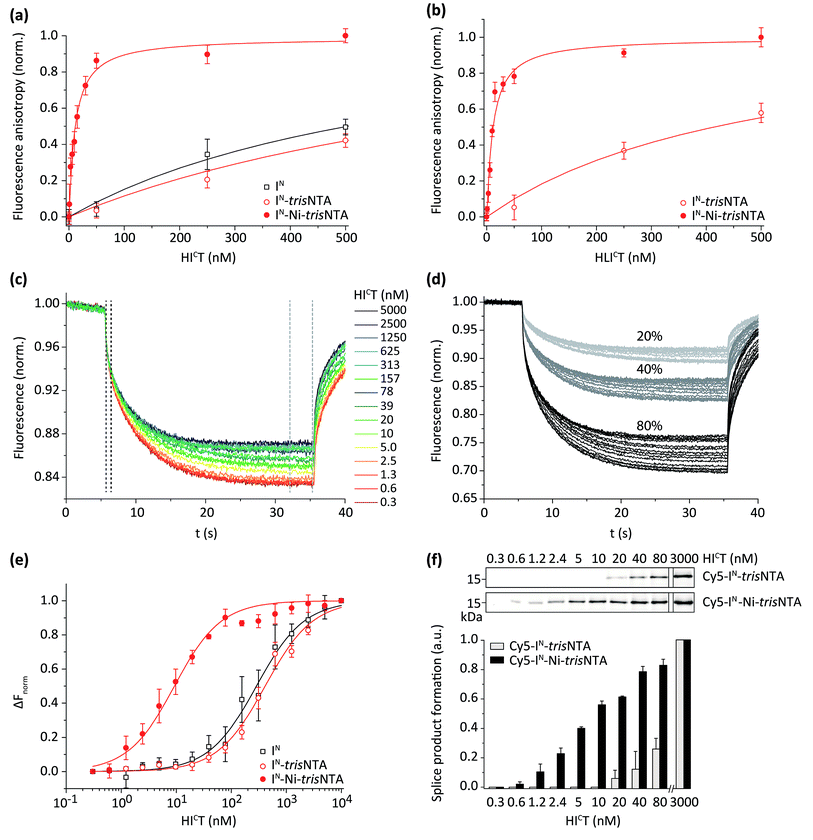 | ||
| Fig. 2 The trisNTA/His-tag pair triggers high-affinity interaction of intein fragments. (a) Interaction between FAM-IN or FAM-IN-trisNTA (100 nM each) with either HICT or HLICT (b) was analysed by fluorescence anisotropy. A dramatic increase in affinity was observed for FAM-IN-Ni-trisNTA. Fluorescence anisotropy measurements were performed with splice-inactive IC.42,43 (c) Representative MST traces of Cy5-IN (150 pM) with different concentrations of HICT as indicated. (d) Variable MST power (20–80%) was applied to optimize the trace resolution. (e) The interaction between HICT and Cy5-IN, Cy5-IN-trisNTA, or Cy5-IN-Ni-trisNTA (150 pM each) was determined by analysing changes in the MST signals. Interaction analysis was performed with splice-inactive IC.42,43 (f) Trans-splicing of Cy5-IN-trisNTA and Cy5-IN-Ni-trisNTA (1 nM) with increasing concentrations of HICT (1 h, 20 °C). PTS was followed by SDS-PAGE in-gel fluorescence analysis. Error bars: S.D. | ||
By FA, an equilibrium dissociation constant KD of 350 ± 60 nM was determined for the splice-inactive trans-splicing pair ICTH and IN. A similar KD was observed for HICT, whereas a negligible reduction of the affinity was detected for HLICT. In the case of IN-trisNTA and IN-L-trisNTA, we observed slight decreased affinities for HICT and HLICT (Table 1).
| IC fragment | IN fragment | FA: KD (nM) | MST: KD (nM) |
|---|---|---|---|
| a n.d. not determined. | |||
| ICTH | IN | 350 ± 60 | n.d. |
| IN-trisNTA | 650 ± 60 | n.d. | |
| HICT | IN | 350 ± 90 | 285 ± 20 |
| IN-trisNTA | 560 ± 65 | 410 ± 30 | |
| IN-Ni-trisNTA | 11 ± 1 | 9 ± 1 | |
| IN-L-trisNTA | 590 ± 50 | 500 ± 50 | |
| IN-L-Ni-trisNTA | 13 ± 1 | 8 ± 2 | |
| HLICT | IN | 410 ± 90 | 480 ± 90 |
| IN-trisNTA | 510 ± 75 | 560 ± 40 | |
| IN-Ni-trisNTA | 10 ± 2 | 12 ± 3 | |
| IN-L-trisNTA | 610 ± 80 | 660 ± 70 | |
| IN-L-Ni-trisNTA | 9 ± 1 | 10 ± 4 | |
In the case of IN-Ni-trisNTA, an impressive increase in affinity between the intein fragments was observed (KD ∼ 10 nM). To further investigate high-affinity PTS, we employed MST. 150 pM of Cy5-IN were titrated with increasing concentrations of HICT (Fig. 2c). To better resolve the Cy5-fluorescence traces, diverse MST power was used (Fig. 2d). The fluorescence change ΔFnorm was deviated from Fhot/Fcold, whereat Fhot and Fcold are the mean fluorescence at the end of the measurement (Fig. 2c, light-grey dashed line) and 1 s after turning on the IR-laser (Fig. 2c, dark-grey dashed line).47 ΔFnorm of each MST trace is plotted against the respective IC concentration to preserve a dose–response curve (Fig. 2e), from which the affinity can be derived. Notably, the KD values, measured by two independent approaches, are very similar (Table 1). Importantly, the affinities between IN and IC were superimposed and promoted by the trisNTA/His-tag interaction.
We noted that the affinities of the trisNTA/His-tag modified intein pairs are in the same range as the naturally occurring Npu DnaE inteins for C-terminal modification,30 the rapamycin-triggered FKBP12-FRB,48,49 or the E. coli dihydrofolate reductase–trimethoprim interaction.40 However, in comparison, the ultra-small size of our interaction pair combined with the minimal synthetic IN fragment (11 aa) makes this system the smallest nanomolar affinity intein described until now.
Next, we compared the trans-splicing reaction in the micromolar and nanomolar range (Fig. 2f). In line with the drastic increased affinity promoted by the trisNTA/His-tag interaction, trans-splicing was observed down to the sub-nanomolar range (0.6 nM). In the absence of the small lock-and-key pair, e.g. nickel-deficient trisNTA, PTS was not observed at low probe concentrations (Fig. 2f). The trans-splicing efficiencies and kinetics are summarized in the ESI (Table S3, Fig. S7–S9‡).
Protein trans-splicing in the crowded cytosol of human cells
Encouraged by these results, we aimed at PTS in a crowded cellular environment. We analysed the high-affinity trans-splicing reaction in the cytosol of human cells (cell lysate). Using 10 μM of HICT or HICLT and 40 μM of the respective IN, a specific trans-splicing reaction and N-terminal modification of the target protein in cell lysate were detected (Fig. 3a). The trans-splicing efficiency was comparable to the in vitro results. A slightly lower SP efficiency was determined for IN-L-trisNTA, indicating that the Ser–Gly linker may affect the trans-splicing reaction. At nanomolar concentrations (1 nM), high-affinity PTS was exclusively observed with IN-Ni-trisNTA (Fig. 3b), whereas with IN or IN-trisNTA no SP formation was detectable.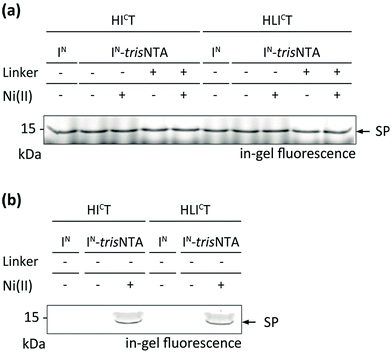 | ||
| Fig. 3 Protein trans-splicing in the cytosol of human cells promoted by the trisNTA/His-tag interaction. High-affinity protein labeling was performed with 10 μM of IC and 40 μM of IN fragments (a) as well as with 1 nM of IC and 4 nM of IN fragments (b) in HeLa cell lysate (20 mg mL−1). SP formation was analysed by SDS-PAGE in-gel fluorescence. Negative controls can be inferred from Fig. S10.‡ | ||
Protein trans-splicing inside mammalian cells
The successful PTS reaction in cell lysate incited us to perform trans-splicing in living mammalian cells. To date, only four in-cell approaches of PTS have been reported, all limited to C-terminal modifications.23–26 To establish the first in-cell application of semi-synthetic PTS at the N terminus, we used HIC-mEGFP as a model protein. To achieve a distinct cellular distribution for visual readout, mEGFP was additionally equipped with nuclear localization sequences (NLS, HIC-mEGFPNLS). For rapid intracellular delivery of Cy5-labeled IN fragments, two cell transduction methodologies were tested. The cell squeezing technique50 was applied for high-throughput delivery and subsequent ensemble measurements, whereas semi-permeabilization with streptolysin O (SLO) was additionally adapted to provide instantaneous excess to the cytosol for direct observation of intracellular processes by confocal laser-scanning microscopy (CLSM). Human cervical cancer (HeLa Kyoto) cells expressing HIC-mEGFPNLS were transduced in the presence of various concentrations of Cy5-labeled IN fragments by cell squeezing as well as SLO semi-permeabilization. In both cases, we first investigated the formation of covalently modified trans-splicing product (Cy5-mEGFPNLS) by SDS-PAGE in-gel-fluorescence analysis (Fig. 4a). At micromolar concentrations of Cy5-IN, we observed SP formation in squeezed as well as semi-permeabilized cells whereas nanomolar concentrations of Cy5-IN failed to yield the trans-splicing product Cy5-mEGFPNLS. In contrast, 100 nM of Cy5-IN-Ni-trisNTA still resulted in SP formation, demonstrating that the trans-splicing reaction is boosted by the minimal lock-and-key recognition element even in living cells (Fig. 4a and b and S13 and S14‡). Notably, no SP was detected in cells expressing mEGFPNLS lacking the HIC part. Interestingly, comparable SP yields were obtained for Cy5-IN and Cy5-IN-trisNTA at micromolar concentrations as well as for 100 nM of Cy5-IN-Ni-trisNTA, indicating that the chemical composition of the IN fragments does not bias the trans-splicing reaction. Moreover, the guidance by the minimal trisNTA/His-tag interaction is essential for SP formation at nanomolar concentrations, since no SP formation was observed for Cy5-IN-trisNTA lacking Ni(II). It is worth mentioning that no SP was detected in the external medium (Ext, Fig. 4a), confirming that high-affinity PTS occurred inside cells (Cell; Fig. 4a). Cytotoxic effects of Ni(II) ions can be excluded due to the high complex formation constant of 10−13 M of Ni-NTA. As a result, free nickel ions only exist in the femtomolar range. These results demonstrate that the high-affinity interaction pair does not only boost PTS in vitro and in cell lysates, but even inside living cells.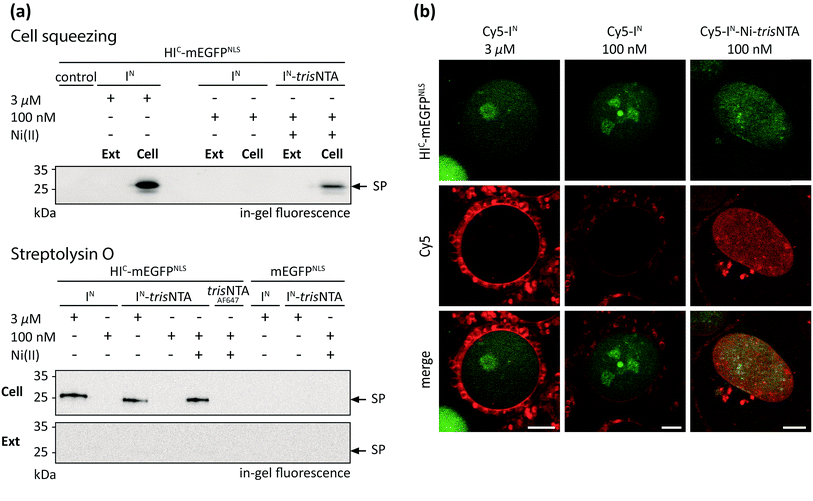 | ||
| Fig. 4 PTS in living mammalian cells. (a) Cy5-labeled IN fragments were delivered into the cytosol of HeLa Kyoto cells transfected with HIC-mEGFPNLS or mEGFPNLSvia cell squeezing or SLO semi-permeabilization. At nanomolar concentrations, in-cell PTS was only observed with Cy5-IN-Ni-trisNTA (100 nM), whereas similar concentrations of Cy5-IN or Cy5-IN-trisNTA showed no PTS reaction. In the absence of the high-affinity pair or without coordinated Ni(II), 3 μM of Cy5-IN or Cy5-IN-trisNTA was needed. For both permeabilization approaches, SP formation was detected only in cells (Cell) but not in the external medium (Ext). As a negative control for cell squeezing, 3 μM of Cy5-IN in mEGFPNLS transfected cells was used. Negative controls for SLO semi-permeabilization were performed with 3 μM of Cy5-IN and Cy5-IN-trisNTA, as well as 100 nM of Cy5-IN-Ni-trisNTA in mEGFPNLS transfected cells. (b) Cy5-labeled IN probes were delivered into the cytosol of SLO semi-permeabilized HeLa Kyoto cells and imaged using CLSM. At 100 nM of Cy5-IN-Ni-trisNTA, co-localization with mEGFPNLS was observed. In contrast, 100 nM of Cy5-IN did not lead to specific labeling. Using 3 μM of Cy5-IN, low signal-to-background labeling was detected at the nucleus (see also Fig. S16‡). Scale bar: 5 μm. | ||
The efficiency of trans-splicing in living cells was determined by immunoblotting analysis against GFP. The trans-splicing yield was determined to 2–8% for 3 μM of Cy5-IN and 2% for 100 nM of Cy5-IN-Ni-trisNTA (Fig. S15‡). In contrast to in vitro analyses and trans-splicing in cell lysates, the efficiency in living mammalian cells dropped 10- to 40-fold. Although the in vivo PTS efficiency decreased, the high-affinity trisNTA/His-tag interaction still enables SP formation in the nanomolar range, as demonstrated by SDS-PAGE in-gel fluorescence analysis of transduced cells. These observations again highlight the benefit of the minimal lock-and-key recognition element for PTS.
Next, we aimed at visualization of SP formation in mammalian cells expressing HIC-mEGFPNLS. It is noteworthy that the fusion of the IC sequence (∼150 aa) did not affect the nuclear localization of HIC-mEGFPNLS (Fig. S12‡). Since cell squeezing dislocates mEGFPNLS and squeezed cells need time for reattachment to the surface, this technique is not appropriate in this case. Therefore, we delivered the IN fragments by SLO semi-permeabilization to provide instantaneous excess to the cytosol for direct observation of intracellular processes using confocal laser-scanning microscopy (CLSM) without a major time delay. Nuclear co-localization of the fluorescence signal for HIC-mEGFPNLS and Cy5-IN-Ni-trisNTA provided evidence of site-specific and high-affinity PTS at nanomolar probe concentration in living cells (Fig. 4a and b and S15‡). As demonstrated by SDS-PAGE in-gel fluorescence, this is indicative of PTS at nanomolar concentrations (Fig. 4a). Here, only 100 nM of Cy5-IN-Ni-trisNTA was offered to the cells, since this concentration proved to be sufficient for PTS (Fig. 4a). In contrast, applying the same concentrations of unmodified Cy5-IN, no specific Cy5-labeling of nuclear-targeted mEGFPNLS was detected. Although micromolar concentrations of Cy5-IN led to trans-splicing and nuclear Cy5-staining (Fig. 4a and b), this signal was outshone by the extensive background fluorescence (Fig. 4b and S16‡). Here, the laser intensities were adjusted to the high concentrations of unmodified Cy5-IN required for trans-splicing and hence suppress fluorescence signals emanating from nuclear localized Cy5-IN.
In summary, we demonstrated in-cell trans-splicing by using two different transduction techniques as well as two different readout methods. In all scenarios, the minimalistic lock-and-key element efficiently guided PTS in the nanomolar range, fully in line with the results achieved in vitro and in cell lysates.
Conclusions
We have developed a high-affinity split intein system for ‘traceless’ tracing of proteins. In our study, the affinity of the artificial split Ssp DnaB M86 intein was synergistically interconnected with the ultra-small trisNTA/His-tag recognition element. As a result, covalent N-terminal protein labeling in vitro and subsequently, for the first time, in living mammalian cells is demonstrated. Comprehensive in vitro characterization by SDS-PAGE in-gel fluorescence analysis, fluorescence anisotropy as well as microscale thermophoresis measurements demonstrated a more than 50-fold increase in affinity for the cognate intein fragments by the trisNTA/His-tag interaction pair. Despite this, the intrinsic intein properties were not affected in regard to yields and kinetics of the SP formation. Moreover, the minimalistic lock-and-key element even allowed performing PTS at (sub) nanomolar concentrations in vitro as well as ex vivo (human cell lysates) with comparable results. Hence, the trans-splicing reaction with minimal probe concentration is pivotal for advanced imaging techniques. As a first in vivo proof-of-concept, the developed high-affinity IN-trisNTA fragments were delivered into mammalian cells via microfluidic cell squeezing as well as semi-permeabilization. At nanomolar concentrations, SP formation in living cells was only detected in the presence of the high-affinity interaction pair and corroborated by SDS-PAGE in-gel fluorescence analysis. Thus, we established the first in-cell application of semi-synthetic protein trans-splicing at the N terminus. Although the trans-splicing efficiency was largely affected inside cells, the trans-splicing reaction is still guided by our minimal interaction pair and thereby promoted in vivo. To highlight opportunities for further development, particularly with respect to in-cell applications, FRET/quencher pairs to follow the trans-splicing reaction in real-time are currently under investigation. Taking advantage of the precision in labeling, we are exploring this high-affinity ‘traceless’ labeling approach for single-molecule tracking and localization analysis in live cells. This high-affinity N-terminal protein modification should advance our understanding of cellular networks.Acknowledgements
We thank Christine Le Gal for editing the manuscript. We gratefully acknowledge Prof. Dr. H. D. Mootz (University of Münster, Germany) for providing the plasmids coding for ICTH, Valentina Herbring for help with FX cloning, and Katrin Schanner for excellent technical assistance.References
- M. Chalfie, Y. Tu, G. Euskirchen, W. W. Ward and D. C. Prasher, Science, 1994, 263, 802–805 CAS.
- A. Keppler, S. Gendreizig, T. Gronemeyer, H. Pick, H. Vogel and K. Johnsson, Nat. Biotechnol., 2002, 21, 86–89 CrossRef PubMed.
- A. Gautier, A. Juillerat, C. Heinis, I. R. Correa Jr, M. Kindermann, F. Beaufils and K. Johnsson, Chem. Biol., 2008, 15, 128–136 CrossRef CAS PubMed.
- G. V. Los, L. P. Encell, M. G. McDougall, D. D. Hartzell, N. Karassina, C. Zimprich, M. G. Wood, R. Learish, R. F. Ohana, M. Urh, D. Simpson, J. Mendez, K. Zimmerman, P. Otto, G. Vidugiris, J. Zhu, A. Darzins, D. H. Klaubert, R. F. Bulleit and K. V. Wood, ACS Chem. Biol., 2008, 3, 373–382 CrossRef CAS PubMed.
- Z. Zhou, P. Cironi, A. J. Lin, Y. Xu, S. Hrvatin, D. E. Golan, P. A. Silver, C. T. Walsh and J. Yin, ACS Chem. Biol., 2007, 2, 337–346 CrossRef CAS PubMed.
- C. S. Theile, M. D. Witte, A. E. Blom, L. Kundrat, H. L. Ploegh and C. P. Guimaraes, Nat. Protoc., 2013, 8, 1800–1807 CrossRef PubMed.
- M. Fernandez-Suarez, H. Baruah, L. Martinez-Hernandez, K. T. Xie, J. M. Baskin, C. R. Bertozzi and A. Y. Ting, Nat. Biotechnol., 2007, 25, 1483–1487 CrossRef CAS PubMed.
- G. Volkmann and H. D. Mootz, Cell. Mol. Life Sci., 2013, 70, 1185–1206 CrossRef CAS PubMed.
- T. W. Muir, Annu. Rev. Biochem., 2003, 72, 249–289 CrossRef CAS PubMed.
- M. Holt and T. Muir, Annu. Rev. Biochem., 2015, 84, 265–290 CrossRef CAS PubMed.
- A. E. Busche, A. S. Aranko, M. Talebzadeh-Farooji, F. Bernhard, V. Dötsch and H. Iwai, Angew. Chem., Int. Ed., 2009, 48, 6128–6131 CrossRef CAS PubMed.
- C. P. Scott, E. Abel-Santos, M. Wall, D. C. Wahnon and S. J. Benkovic, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 13638–13643 CrossRef CAS.
- J. K. Böcker, K. Friedel, J. C. Matern, A. L. Bachmann and H. D. Mootz, Angew. Chem., Int. Ed., 2015, 54, 2116–2120 CrossRef PubMed.
- K. Jagadish, R. Borra, V. Lacey, S. Majumder, A. Shekhtman, L. Wang and J. A. Camarero, Angew. Chem., Int. Ed., 2013, 52, 3126–3131 CrossRef CAS PubMed.
- K. Jagadish, A. Gould, R. Borra, S. Majumder, Z. Mushtaq, A. Shekhtman and J. A. Camarero, Angew. Chem., Int. Ed., 2015, 54, 8390–8394 CrossRef CAS PubMed.
- Y. Kwon, M. A. Coleman and J. A. Camarero, Angew. Chem., Int. Ed., 2006, 45, 1726–1729 CrossRef CAS PubMed.
- N. K. Chu, D. Olschewski, R. Seidel, K. F. Winklhofer, J. Tatzelt, M. Engelhard and C. F. Becker, J. Pept. Sci., 2010, 16, 582–588 CrossRef CAS PubMed.
- C. Ludwig, M. Pfeiff, U. Linne and H. D. Mootz, Angew. Chem., Int. Ed., 2006, 45, 5218–5221 CrossRef CAS PubMed.
- V. Schütz and H. D. Mootz, Angew. Chem., Int. Ed., 2014, 53, 4113–4117 CrossRef PubMed.
- J. H. Appleby, K. Zhou, G. Volkmann and X. Q. Liu, J. Biol. Chem., 2009, 284, 6194–6199 CrossRef CAS PubMed.
- A. S. Aranko, S. Zuger, E. Buchinger and H. Iwai, PLoS One, 2009, 4, e5185 Search PubMed.
- N. H. Shah and T. W. Muir, Chem. Sci., 2014, 5, 446 RSC.
- I. Giriat and T. W. Muir, J. Am. Chem. Soc., 2003, 125, 7180–7181 CrossRef CAS PubMed.
- R. Borra, D. Dong, A. Y. Elnagar, G. A. Woldemariam and J. A. Camarero, J. Am. Chem. Soc., 2012, 134, 6344–6353 CrossRef CAS PubMed.
- Y. David, M. Vila-Perello, S. Verma and T. W. Muir, Nat. Chem., 2015, 7, 394–402 CrossRef CAS PubMed.
- D. Jung, K. Sato, K. Min, A. Shigenaga, J. Jung, A. Otaka and Y. Kwon, Chem. Commun., 2015, 51, 9670–9673 RSC.
- W. Greenhalf, C. Stephan and B. Chaudhuri, FEBS Lett., 1996, 380, 169–175 CrossRef CAS PubMed.
- F. E. Reyes-Turcu, K. H. Ventii and K. D. Wilkinson, Annu. Rev. Biochem., 2009, 78, 363–397 CrossRef CAS PubMed.
- J. L. Broers, B. M. Machiels, G. J. van Eys, H. J. Kuijpers, E. M. Manders, R. van Driel and F. C. Ramaekers, J. Cell Sci., 1999, 112, 3463–3475 CAS.
- N. H. Shah, M. Vila-Perello and T. W. Muir, Angew. Chem., Int. Ed., 2011, 50, 6511–6515 CrossRef CAS PubMed.
- H. Wu, Z. Hu and X.-Q. Liu, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 9226–9231 CrossRef CAS.
- H. D. Mootz, ChemBioChem, 2009, 10, 2579–2589 CrossRef CAS PubMed.
- V. Muralidharan and T. W. Muir, Nat. Methods, 2006, 3, 429–438 CrossRef CAS PubMed.
- I. V. Thiel, G. Volkmann, S. Pietrokovski and H. D. Mootz, Angew. Chem., Int. Ed., 2014, 53, 1306–1310 CrossRef CAS PubMed.
- C. Grunwald, K. Schulze, G. Giannone, L. Cognet, B. Lounis, D. Choquet and R. Tampé, J. Am. Chem. Soc., 2011, 133, 8090–8093 CrossRef CAS PubMed.
- I. T. Dorn, K. R. Neumaier and R. Tampé, J. Am. Chem. Soc., 1998, 120, 2753–2763 CrossRef CAS.
- S. Lata, A. Reichel, R. Brock, R. Tampé and J. Piehler, J. Am. Chem. Soc., 2005, 127, 10205–10215 CrossRef CAS PubMed.
- R. Wieneke, N. Laboria, M. Rajan, A. Kollmannsperger, F. Natale, M. C. Cardoso and R. Tampé, J. Am. Chem. Soc., 2014, 136, 13975–13978 CrossRef CAS PubMed.
- A. Kollmannsperger, A. Sharei, A. Raulf, M. Heilemann, R. Langer, K. F. Jensen, R. Wieneke and R. Tampé, Nat. Commun., 2015 DOI:10.1038/ncomms10372.
- T. Ando, S. Tsukiji, T. Tanaka and T. Nagamune, Chem. Commun., 2007, 4995–4997 RSC.
- R. Wieneke, A. Raulf, A. Kollmannsperger, M. Heilemann and R. Tampé, Angew. Chem., Int. Ed., 2015, 54, 10216–10219 CrossRef CAS PubMed.
- J. H. Appleby-Tagoe, I. V. Thiel, Y. Wang, Y. Wang, H. D. Mootz and X. Q. Liu, J. Biol. Chem., 2011, 286, 34440–34447 CrossRef CAS PubMed.
- A. Wasmuth, C. Ludwig and H. D. Mootz, Bioorg. Med. Chem., 2013, 21, 3495–3503 CrossRef CAS PubMed.
- A. S. Aranko, J. S. Oeemig, D. Zhou, T. Kajander, A. Wlodawer and H. Iwai, Mol. BioSyst., 2014, 10, 1023–1034 RSC.
- C. J. Wienken, P. Baaske, U. Rothbauer, D. Braun and S. Duhr, Nat. Commun., 2010, 1, 100 CrossRef PubMed.
- S. A. Seidel, P. M. Dijkman, W. A. Lea, G. van den Bogaart, M. Jerabek-Willemsen, A. Lazic, J. S. Joseph, P. Srinivasan, P. Baaske, A. Simeonov, I. Katritch, F. A. Melo, J. E. Ladbury, G. Schreiber, A. Watts, D. Braun and S. Duhr, Methods, 2013, 59, 301–315 CrossRef CAS PubMed.
- M. Jerabek-Willemsen, C. J. Wienken, D. Braun, P. Baaske and S. Duhr, Assay Drug Dev. Technol., 2011, 9, 342–353 CrossRef CAS PubMed.
- T. F. Massoud, R. Paulmurugan and S. S. Gambhir, Nat. Med., 2010, 16, 921–926 CrossRef CAS PubMed.
- R. Paulmurugan, Y. Umezawa and S. S. Gambhir, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 15608–15613 CrossRef CAS PubMed.
- A. Sharei, J. Zoldan, A. Adamo, W. Y. Sim, N. Cho, E. Jackson, S. Mao, S. Schneider, M. J. Han, A. Lytton-Jean, P. A. Basto, S. Jhunjhunwala, J. Lee, D. A. Heller, J. W. Kang, G. C. Hartoularos, K. S. Kim, D. G. Anderson, R. Langer and K. F. Jensen, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 2082–2087 CrossRef CAS PubMed.
Footnotes |
| † The German Research Foundation (EXC115 Cluster of Excellence – Macromolecular Complexes to R. W. and R. T., SPP 1623 to R. T.) supported the work. |
| ‡ Electronic supplementary information (ESI) available. See DOI: 10.1039/c5sc02936h |
| This journal is © The Royal Society of Chemistry 2016 |
