Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Mix and match backbones for the formation of H-bonded duplexes†
Giulia
Iadevaia
a,
Alexander E.
Stross
a,
Anja
Neumann
b and
Christopher A.
Hunter
*a
aDepartment of Chemistry, University of Cambridge, Lensfield Road, Cambridge CB2 1EW, UK. E-mail: herchelsmith.orgchem@ch.cam.ac.uk
bDepartment of Chemistry, University of Sheffield, Sheffield S3 7HF, UK
First published on 7th January 2016
Abstract
The formation of well-defined supramolecular assemblies involves competition between intermolecular and intramolecular interactions, which is quantified by effective molarity. Formation of a duplex between two oligomers equipped with recognition sites displayed along a non-interacting backbone requires that once one intermolecular interaction has been formed, all subsequent interactions take place in an intramolecular sense. The efficiency of this process is governed by the geometric complementarity and conformational flexibility of the backbone linking the recognition sites. Here we report a series of phosphine oxide H-bond acceptor AA 2-mers and phenol H-bond donor DD 2-mers, where the two recognition sites are connected by isomeric backbone modules that vary in geometry and flexibility. All AA and DD combinations form stable AA·DD duplexes, where two cooperative H-bonds lead to an increase in stability of an order of magnitude compared with the corresponding A·D complexes that can only form one H-bond. For all six possible backbone combinations, the effective molarity for duplex formation is approximately constant (7–20 mM). Thus strict complementarity and high degrees of preorganisation are not required for efficient supramolecular assembly. Provided there is some flexibility, quite different backbone modules can be used interchangeably to construct stable H-bonded duplexes.
Introduction
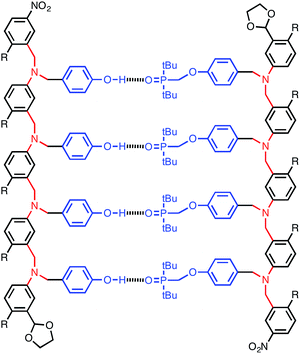
Linear polymers equipped with complementary recognition sites have the potential to reproduce the functional properties of nucleic acids: sequence selective duplex formation, templated synthesis, self-replication and forced evolution.1 Modified versions of DNA have been prepared, where the phosphate linker,2,3 the sugar,4 the backbone or the base pairing system5 have been replaced, and these systems all form stable duplexes.6 The success of these systems suggests that it might be possible to make completely different classes of synthetic information molecule that bear no resemblance to their biological counterparts.A number of synthetic systems that form duplexes via different non covalent interactions have been reported.7 Lehn described oligo(2,2′-bipyridine) ligands that self-assemble into length specific helical duplexes in the presence of a metal ion.7a Lehn also reported ligands containing sequences of bidentate (bipyridine) and tridentate (terpyridine) binding sites which form sequence specific helicates depending on the properties of the metal directing the assembly.7b Huc and Lehn described pyridinecarboxamide oligomers that form double helices due to aromatic stacking interactions.7c,d Anderson synthesized zinc porphyrin oligomers that assemble into ladders in the presence of 1,4-diazabicyclo[2.2.2]octane (DABCO). A linear increase in stability with the length of the ladder was observed, which indicates cooperative duplex formation, and when oligomers of different lengths were mixed in the presence of DABCO, only length complementary ladders were formed.7e,f deMendoza described guanidinium oligomers that form double helices held together by H-bonding interactions with the sulfate counterions.7g Hunter reported oligoamides that form duplexes via H-bonding and edge-to-face aromatic interactions. An increase in stability with increasing length was observed indicating cooperative assembly.7h,i Gong reported oligoamides containing different sequences of H-bond donor and H-bond acceptor sites that show sequence selective duplex formation.7j–o Chen reported self-complementary oligomeric hydrazide oligomers and amidourea oligomers both of which form H-bonded duplexes.7p–r Different length spacers between the H-bonding sites were used to obtain selective assembly of complementary oligomers. Krische reported aminotriazine and diaminopyridazine oligomers that form H-bonded duplexes.7s,t Yashima described oligomers that form helical duplexes through salt bridge interactions between amidinium and carboxylate sites.7u–w These systems show sequence selective and length selective duplex formation, and a template strand bearing two amidinium binding sites was used to direct the synthesis of a complementary strand bearing two carboxylate units.7x We recently reported a new class of linear oligomeric molecules that form stable duplexes via the formation of multiple cooperative H-bonding interactions (Fig. 1).8 Specifically, oligomers equipped with phenol H-bond donors formed 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexes with oligomers equipped with phosphine oxide H-bond acceptors. The stability of the duplex increases by of an order of magnitude for every additional H-bond formed.
1 complexes with oligomers equipped with phosphine oxide H-bond acceptors. The stability of the duplex increases by of an order of magnitude for every additional H-bond formed.
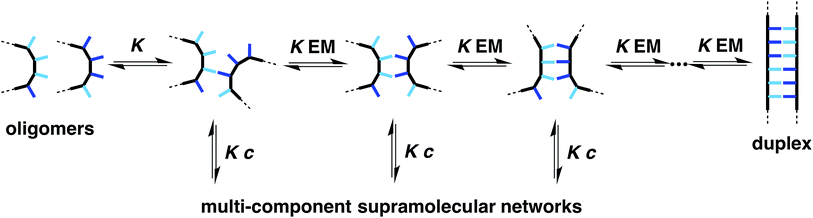
The efficiency of duplex formation is determined by the stepwise equilibria shown in Fig. 2.9 The first interaction in duplex assembly is an intermolecular H-bond which has an association constant K (≈300 M−1 for the phosphine oxide–phenol H-bond). The second H-bond is an intramolecular interaction with an equilibrium constant K![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EM, where EM is the effective molarity for the intramolecular process (≈10 mM for the duplex in Fig. 1). All subsequent H-bonds are intramolecular and were found to have similar effective molarities for the system in Fig. 1. Duplex formation requires that K
EM, where EM is the effective molarity for the intramolecular process (≈10 mM for the duplex in Fig. 1). All subsequent H-bonds are intramolecular and were found to have similar effective molarities for the system in Fig. 1. Duplex formation requires that K![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EM ≫ 1 to ensure that once the first intermolecular H-bond is made, all subsequent intramolecular H-bonds are highly favoured, and there is no competition from intermolecular interactions that would lead to uncontrolled aggregation (intermolecular channel in Fig. 2). For the system shown in Fig. 1, K
EM ≫ 1 to ensure that once the first intermolecular H-bond is made, all subsequent intramolecular H-bonds are highly favoured, and there is no competition from intermolecular interactions that would lead to uncontrolled aggregation (intermolecular channel in Fig. 2). For the system shown in Fig. 1, K![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EM ≈ 5, so duplex formation is reasonably efficient, but there are partially bound states present where the H-bonding interactions are broken some of the time. There are two strategies to improve duplex formation: increase K by changing the H-bonding groups, or increase EM by changing the supramolecular architecture. In this paper, we explore the effect of backbone architecture on the effective molarity for duplex formation.
EM ≈ 5, so duplex formation is reasonably efficient, but there are partially bound states present where the H-bonding interactions are broken some of the time. There are two strategies to improve duplex formation: increase K by changing the H-bonding groups, or increase EM by changing the supramolecular architecture. In this paper, we explore the effect of backbone architecture on the effective molarity for duplex formation.
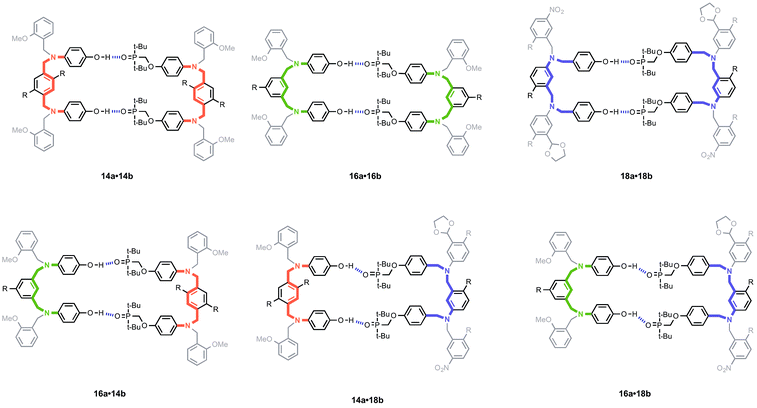
The design of the oligomer architecture in Fig. 1 is modular. Fig. 3 illustrates the basic blueprint. The recognition modules (blue), the chemistry used to synthesise oligomers (red) and the backbone (black) have been incorporated into the chemical structure in Fig. 1, such that different properties of the system can be independently varied, i.e. one module can be changed without affecting the other two. Fig. 3 illustrates how the backbone can be changed, but keeping the reductive amination chemistry that was used for synthesis of the compounds in Fig. 1 and the same phosphine oxide–phenol recognition module. Here we investigate changing the constitution of the backbone by measuring the effect of the three isomeric backbone modules in Fig. 3 on the EM for duplex formation for different combinations of AA and DD 2-mers. Each backbone module is each composed of an aromatic ring and two methylene groups, but differences in connectivity leads to a variation in the geometry and flexibility of the motif linking the recognition modules. The nomenclature N8/C8/N7 indicates the atom to which the recognition module is attached and the length of the linker. The C8 backbone has three flexible methylene groups connecting the recognition sites, while the N8 and N7 backbones each have two. Thus the N8 backbone is relatively rigid and extended, the N7 backbone is rigid and shorter, and the C8 backbone is flexible, so that it can explore both elongated and compact conformations.
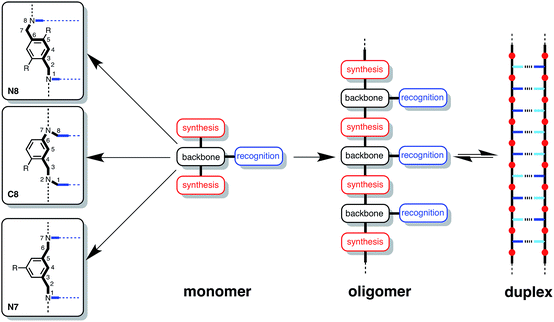 | ||
| Fig. 3 A blueprint for duplex forming molecules. There are three key design elements: one module which defines the chemistry for the synthesis of oligomers (red), the recognition module which controls intermolecular binding (blue) and the backbone module which links these components together. Three isomeric backbone modules are shown (R are sites for attachment of solubilising groups). Adapted from ref. 8. | ||
Results and discussion
Synthesis
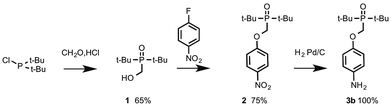
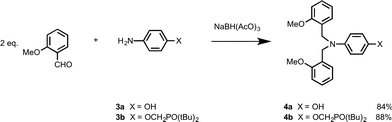
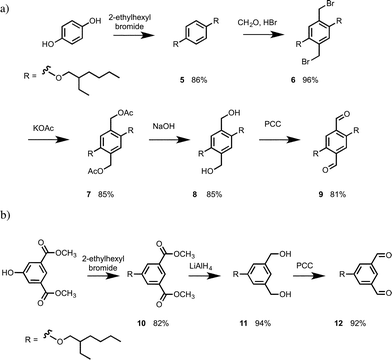
The N8 and N7 backbones were accessed by reductive amination of dialdehydes with anilines bearing the recognition groups. The H-bond donor aniline 4-aminophenol is commercially available. The H-bond acceptor aniline bearing a phosphine oxide group was synthesized as shown in Scheme 1. Di-t-butyl(chloro)phosphane was treated with formaldehyde to give alcohol 1, which was reacted with 4-fluoro-nitrobenzene in the presence of K2CO3 to give 2.10 Reduction of 2 gave aniline 3b.In order to determine effective molarities, H-bond acceptor and donor 1-mers are required to measure the intermolecular association constant K. The H-bond donor parameter of 3-dimethylaminophenol is somewhat lower than the value for phenol (α = 3.5 compared with 3.8), which suggests that there may be a substituent effect on the strengths of H-bonding interactions involving donors attached to the C8 backbone compared with donors attached to the N7 and N8 backbones.11 The 1-mers corresponding to the N7 and N8 backbones, 4a and 4b (A and D), were therefore synthesized from 3a or 3b by reductive amination with 2-methoxybenzaldehyde (Scheme 2).12
The 1,4-dialdehyde 9 required for the N8 backbone was synthesised using a sequence of bromomethylation–acetylation–reduction–oxidation reactions as shown in Scheme 3.13 The 1,3-dialdehdye 12 required for the N7 backbone was prepared from dimethyl 5-hydroxyisophthalate: alkylation of the phenol group with 2-ethylhexylbromide, followed by reduction of the esters with LiAlH4 gave diol 11, and oxidation of 11 with PCC gave 12 (Scheme 3).
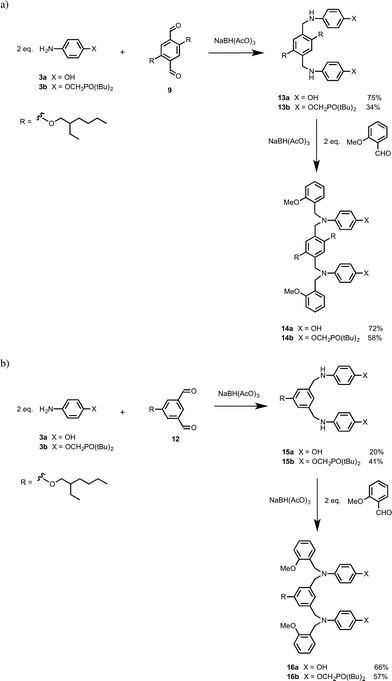
Compounds 13a and 13b were synthesized by reacting two equivalents of anilines 3a or 3b with dialdehyde 9 (Scheme 4a). A further reductive amination step was used to cap 13a and 13b with 2-methoxybenzaldehyde to give 14a and 14b, the N8 backbone DD and AA 2-mers. Similarly 15a was synthesized by reductive amination of dialdehyde 12 with aniline 3a (Scheme 4b). Compound 15b was obtained by reacting dialdehyde 12 and two equivalents of aniline 3b to obtain the diimine, which was then reduced with NaBH4 to give 15b. Compounds 15a and 15b were then capped by with 2-methoxybenzaldehyde under reductive amination conditions to obtain 16a and 16b, the N7 backbone DD and AA 2-mers (Scheme 4b).
The C8 backbone AA and DD 2-mers belong to the family of oligomers illustrated by the AAAA and DDDD 4-mers in Fig. 1. The synthesis of these compounds (18a and 18b) and the corresponding 1-mers (17a and 17b) used to determine effective molarities was reported previously (Fig. 4).8
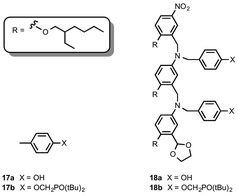 | ||
| Fig. 4 C8 backbone 1-mers (A and D) and 2-mers (AA and DD).8 | ||
Binding studies
Binding studies for pairwise combinations of the AA and DD 2-mers were carried out by means of 31P NMR titrations in toluene. The corresponding 1-mers, A and D, were used to measure the strength of a single intermolecular phenol–phosphine oxide H-bond. The H-bond acceptor phosphine oxides were used as the host, and a large increase in the 31P NMR chemical shift was observed upon guest addition in all cases, which is indicative of H-bond formation.14 The titration data were fit to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding isotherm to obtain the association constants and limiting complexation-induced changes in chemical shift (Table 1). The association constants for the two A·D complexes are similar, which indicates that substituent effects on the phenol–phosphine oxide H-bond are not significant in these systems. The association constants for the AA·DD complexes are significantly larger than the values for the corresponding A·D complexes in all cases, which implies that two H-bonds are formed in a cooperative manner in the 2-mer duplexes. The large limiting complexation-induced changes in 31P NMR chemical shift, Δδ, observed for the AA·DD complexes support this conclusion. Effective molarities for the formation of the second intramolecular H-bond in the AA·DD complexes were determined using eqn (1), and the values are reported in Table 1.
1 binding isotherm to obtain the association constants and limiting complexation-induced changes in chemical shift (Table 1). The association constants for the two A·D complexes are similar, which indicates that substituent effects on the phenol–phosphine oxide H-bond are not significant in these systems. The association constants for the AA·DD complexes are significantly larger than the values for the corresponding A·D complexes in all cases, which implies that two H-bonds are formed in a cooperative manner in the 2-mer duplexes. The large limiting complexation-induced changes in 31P NMR chemical shift, Δδ, observed for the AA·DD complexes support this conclusion. Effective molarities for the formation of the second intramolecular H-bond in the AA·DD complexes were determined using eqn (1), and the values are reported in Table 1.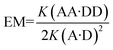 | (1) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding isotherm
1 binding isotherm
| Complexes | Backbone | K/M−1 | Δδ31P/ppm | EM/mM |
K![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EM EM |
|---|---|---|---|---|---|
| A·D | |||||
| 4a·4b | 250 ± 10 | 5.0 | |||
| 17a·17b 8 | 350 ± 20 | 4.9 | |||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| AA·DD | |||||
| 14a·14b | N8·N8 | 2500 ± 200 | 4.0 | 20 ± 2 | 5 ± 1 |
| 16a·16b | N7·N7 | 1200 ± 400 | 6.9 | 10 ± 3 | 2 ± 1 |
| 18a·18b 8 | C8·C8 | 1900 ± 600 | 5.3 | 8 ± 3 | 3 ± 1 |
| 16a·14b | N7·N8 | 900 ± 200 | 6.6 | 7 ± 2 | 2 ± 1 |
| 14a·18b | N8·C8 | 1400 ± 200 | 3.7 | 11 ± 2 | 3 ± 1 |
| 16a·18b | N7·C8 | 920 ± 60 | 6.0 | 7 ± 1 | 2 ± 1 |
The effective molarities are very similar for all backbone combinations, indicating that the ability to form a duplex does not depend strongly on the conformational properties of the backbone in these systems. The values of EM in Table 1 are consistent with the values found for other supramolecular systems which generally fall in the window 10–1000 mM.15,16Fig. 5 illustrates the six different backbone combinations investigated in this paper. The backbones units all have two methylene groups, and it seems that these linkages provide sufficient flexibility to allow the backbones to adapt to quite different geometrical requirements. The outcome is that any backbone combination leads to stable duplex formation, and the duplex architecture illustrated in Fig. 3 is truly modular with respect to the backbone component.
Table 1 also lists the values of K![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EM for the six different backbone combinations. The values are similar for all of the duplexes, and in all cases K
EM for the six different backbone combinations. The values are similar for all of the duplexes, and in all cases K![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EM is greater than one, indicating that intramolecular H-bonding is favoured. However, the values of K
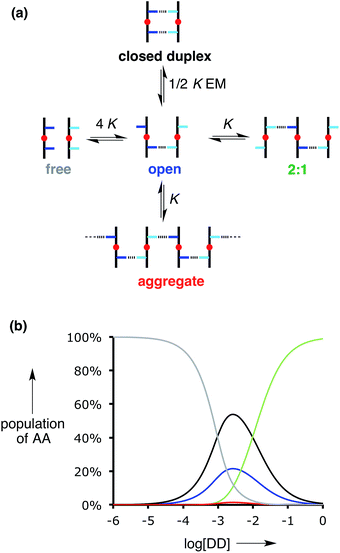
EM is greater than one, indicating that intramolecular H-bonding is favoured. However, the values of K![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EM are not very much greater than one, which implies that the partially bound open complex shown in Fig. 6a is populated to a significant extent. The open complex could also aggregate, and the other species that competes with formation of the closed doubly H-bonded 1
EM are not very much greater than one, which implies that the partially bound open complex shown in Fig. 6a is populated to a significant extent. The open complex could also aggregate, and the other species that competes with formation of the closed doubly H-bonded 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 duplex is formation of a 2
1 duplex is formation of a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex (Fig. 6a). Fig. 6b illustrates the speciation of AA for titration of DD into 1 mM AA for K
1 complex (Fig. 6a). Fig. 6b illustrates the speciation of AA for titration of DD into 1 mM AA for K![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EM = 5. Under these conditions, the population of polymeric aggregate (red) is negligible, and this will always be the case when the concentration of one of the oligomers is less than 1/K (≈3 mM). The duplex (black) is the major species present in a 1
EM = 5. Under these conditions, the population of polymeric aggregate (red) is negligible, and this will always be the case when the concentration of one of the oligomers is less than 1/K (≈3 mM). The duplex (black) is the major species present in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of AA and DD (≈50%). The partially bound open complex (blue) is also significantly populated (≈20%), and the 2
1 mixture of AA and DD (≈50%). The partially bound open complex (blue) is also significantly populated (≈20%), and the 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 AA·DD2 complex (green) dominates in the presence of excess DD. The titration data were also analysed using a 2
1 AA·DD2 complex (green) dominates in the presence of excess DD. The titration data were also analysed using a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding isotherm to allow for the formation of this species towards the end of the titration, but the association constants determined for the 1
1 binding isotherm to allow for the formation of this species towards the end of the titration, but the association constants determined for the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexes were not significantly affected.
1 complexes were not significantly affected.
There are important implications of the results in Table 1 and the speciation diagram in Fig. 6 for the formation of longer duplexes from the building blocks described in this paper. As the lengths of the oligomers increase, the stabilities of the fully assembled duplexes will increase in proportion to (K![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EM)N, where N is the number of recognition modules. Although, the number of possible competing complexes will increase with N, the stabilities of these complexes will not increase proportionately, so off-pathway complexes will become increasingly less significant as N increases. Fig. 6b shows that even for N = 2, the values of K
EM)N, where N is the number of recognition modules. Although, the number of possible competing complexes will increase with N, the stabilities of these complexes will not increase proportionately, so off-pathway complexes will become increasingly less significant as N increases. Fig. 6b shows that even for N = 2, the values of K![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EM are sufficiently high that intermolecular processes leading to the formation of higher order complexes or polymeric aggregates do not compete with duplex formation at mM concentrations (<3% in total for a 1 mM mixture of AA and DD compared with 58% of the two 1
EM are sufficiently high that intermolecular processes leading to the formation of higher order complexes or polymeric aggregates do not compete with duplex formation at mM concentrations (<3% in total for a 1 mM mixture of AA and DD compared with 58% of the two 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexes). On the other hand, partially bound states of the duplex will be significantly populated for longer oligomers. Unless there is cooperative coupling of neighbouring H-bonding interactions along a duplex, the probability of breaking a H-bond will be independent of N and proportional to (K
1 complexes). On the other hand, partially bound states of the duplex will be significantly populated for longer oligomers. Unless there is cooperative coupling of neighbouring H-bonding interactions along a duplex, the probability of breaking a H-bond will be independent of N and proportional to (K![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EM)−1. Previous results on the C8 backbone oligomers confirm that the C8·C8 combination with K
EM)−1. Previous results on the C8 backbone oligomers confirm that the C8·C8 combination with K![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EM = 3 is capable of propagating the assembly longer duplexes (e.g. the AAAA·DDDD complex illustrated in Fig. 1) without competition from higher order aggregates. The results presented here suggest that any of the three different backbones can be used interchangeably to construct stable H-bonded duplexes.
EM = 3 is capable of propagating the assembly longer duplexes (e.g. the AAAA·DDDD complex illustrated in Fig. 1) without competition from higher order aggregates. The results presented here suggest that any of the three different backbones can be used interchangeably to construct stable H-bonded duplexes.
Conclusions
We previously reported a modular strategy for the construction of synthetic information molecules, where complementary H-bonding sites are displayed along a non-polar backbone. In this paper, we investigate how well different backbone modules are tolerated by investigating effect of isomeric linkers on the recognition properties of H-bond donor and acceptor 2-mers (DD and AA). Three different phosphine oxide AA 2-mers and three different phenol DD 2-mers were synthesised using linkers that vary in geometry and conformational flexibility. NMR titrations were used to characterise the AA·DD duplexes formed by all of the six possible backbone combinations. The association constants for formation of the AA·DD complexes (103 M−1) are all an order of magnitude higher than the association constants for formation of the corresponding A·D complexes (102 M−1), which can only form one H-bond. In addition, the complexation-induced changes in 31P NMR chemical shift are indicative of fully H-bonded complexes in all cases, indicating that all six backbone combinations lead to duplex formation with cooperative formation of two H-bonds. The values of EM measured for intramolecular H-bond formation leading to duplex formation are remarkably insensitive to the nature of the backbone (7–20 mM). Thus any of the backbone modules described in this paper could be used interchangeably to construct stable H-bonded duplexes of longer oligomers. There is no strong dependence of EM on geometric complementarity or conformational flexibility. It seems that provided the backbone has sufficient flexibility to allow the H-bonding sites to connect, the precise choice of linker is not critical.Acknowledgements
We thank the EPSRC and ERC for funding.Notes and references
- (a) S. Połowiński, Prog. Polym. Sci., 2002, 27, 537 CrossRef; (b) P. K. Lo and H. F. Sleiman, J. Am. Chem. Soc., 2009, 131, 4182 CrossRef CAS PubMed; (c) T. Terashima, T. Mes, T. F. A. de Greef, M. A. J. Gillissen, P. Besenius, A. R. A. Palmans and E. W. Meijer, J. Am. Chem. Soc., 2011, 133, 4742 CrossRef CAS PubMed; (d) C. R. South and M. Weck, Macromolecules, 2007, 40, 1386 CrossRef CAS; (e) M. Szwarc, J. Polym. Sci., 1954, 13, 317 CrossRef; (f) R. McHale, J. P. Patterson, P. B. Zetterlund and R. K. O'Reilly, Nat. Chem., 2012, 4, 491 CrossRef CAS PubMed; (g) Y. Kang, A. Lu, A. Ellington, M. C. Jewett and R. K. O'Reilly, ACS Macro Lett., 2013, 2, 581 CrossRef CAS; (h) J. Ferguson and S. A. O. Shah, Eur. Polym. J., 1968, 4, 343 CrossRef CAS; (i) J. Smid, Y. Y. Tan and G. Challa, Eur. Polym. J., 1983, 19, 853 CrossRef CAS; (j) J. Smid, Y. Y. Tan and G. Challa, Eur. Polym. J., 1984, 20, 887 CrossRef CAS; (k) J. Smid, Y. Y. Tan and G. Challa, Eur. Polym. J., 1984, 20, 1095 CrossRef CAS; (l) J. Smid, J. C. Speelman, Y. Y. Tan and G. Challa, Eur. Polym. J., 1985, 21, 141 CrossRef CAS; (m) J. Smid, Y. Y. Tan, G. Challa and W. R. Hagen, Eur. Polym. J., 1985, 21, 757 CrossRef CAS.
- (a) S. A. Benner and D. Hutter, Bioorg. Chem., 2002, 30, 62 CrossRef CAS PubMed; (b) Z. Huang, K. C. Schneider and S. A. Benner, J. Org. Chem., 1991, 56, 3869 CrossRef CAS; (c) Z. Huang and S. A. Benner, J. Org. Chem., 2002, 67, 3996 CrossRef CAS PubMed; (d) C. Richert, A. L. Roughton and S. A. Benner, J. Am. Chem. Soc., 1996, 118, 4518 CrossRef CAS; (e) B. R. Shaw, M. Dobrikov, X. Wang, J. Wan, K. He, J.-L. Lin, P. Li, V. Rait, Z. A. Sergueeva and D. Sergueev, Ann. N. Y. Acad. Sci., 2003, 1002, 12 CrossRef CAS PubMed; (f) P. Li, Z. A. Sergueeva, M. Dobrikov and B. R. Shaw, Chem. Rev., 2007, 107, 4746 CrossRef CAS PubMed; (g) H. Isobe, T. Fujino, N. Yamazaki, M. Guillot-Nieckowski and E. Nakamura, Org. Lett., 2008, 10, 3729 CrossRef CAS PubMed.
- (a) M. Eriksson and P. E. Q. Nielsen, Annu. Rev. Biophys. Biomol. Struct., 1996, 29, 369 CAS; (b) P. E. Nielsen, Chem. Biodiversity, 2010, 7, 786 CrossRef CAS PubMed; (c) P. E. Nielsen and M. Egholm, Curr. Issues Mol. Biol., 1999, 1, 89 CAS; (d) P. E. Nielsen and G. Haaima, Chem. Soc. Rev., 1997, 26, 73 RSC; (e) Y. Ura, J. M. Beierle, L. J. Leman, L. E. Orgel and M. R. Ghadiri, Science, 2009, 325, 73 CrossRef CAS PubMed.
- (a) K.-U. Schöning, P. Scholz, S. Guntha, X. Wu, R. Krishnamurthy and A. Eschenmoser, Science, 2000, 290, 1347 CrossRef; (b) A. Aerschot van, I. Verheggen, C. Hendrix and P. Herdewijn, Angew. Chem., Int. Ed. Engl., 1995, 34, 1338 CrossRef; (c) D. Renneberg and C. J. Leumann, J. Am. Chem. Soc., 2002, 124, 5993 CrossRef CAS PubMed; (d) D. A. Braasch and D. R. Corey, Chem. Biol., 2001, 8, 1 CrossRef CAS PubMed; (e) S. K. Singh, A. A. Koshkin and J. Wengel, Chem. Commun., 1998, 455 RSC; (f) H. V. Nguyen, Z.-Y. Zhao, A. Sallustrau, S. L. Horswell, L. Male, A. Mulas and J. H. R. Tucker, Chem. Commun., 2012, 48, 12165 RSC; (g) L. Zhang, A. Peritz and E. Meggers, J. Am. Chem. Soc., 2005, 127, 4174 CrossRef CAS PubMed; (h) M. K. Schlegel, A. E. Peritz, K. Kittigowittana, L. Zhang and E. Meggers, ChemBioChem, 2007, 8, 927 CrossRef CAS PubMed; (i) P. Karri, V. Punna, K. Kim and R. Krishnamurthy, Angew. Chem., Int. Ed., 2013, 52, 5840 CrossRef CAS PubMed.
- (a) J. A. Piccirilli, T. Krauch, S. E. Moroney and S. A. Benner, Nature, 1990, 343, 33 CrossRef CAS PubMed; (b) Z. Yang, D. Hutter, P. Sheng, A. M. Sismour and S. A. Benner, Nucleic Acids Res., 2006, 34, 6095 CrossRef CAS PubMed; (c) F. Wojciechowski and C. J. Leumann, Chem. Soc. Rev., 2011, 40, 5669 RSC; (d) S. A. Benner, Curr. Opin. Chem. Biol., 2012, 16, 581 CrossRef CAS PubMed; (e) H. Liu, J. Gao, S. R. Lynch, Y. D. Saito, L. Maynard and E. T. Kool, Science, 2003, 302, 868 CrossRef CAS PubMed; (f) E. T. Kool, Acc. Chem. Res., 2002, 35, 936 CrossRef CAS PubMed; (g) E. T. Kool, H. Lu, S. J. Kim, S. Tan, J. N. Wilson, J. Gao and H. Liu, Nucleic Acids Symp. Ser., 2006, 50, 15 CrossRef PubMed; (h) J. N. Wilson and E. T. Kool, Org. Biomol. Chem., 2006, 4, 4265 RSC.
- (a) A. Eschenmoser, Science, 1999, 284, 2118 CrossRef CAS PubMed; (b) S. A. Benner, Acc. Chem. Res., 2004, 37, 784 CrossRef CAS PubMed; (c) S. A. Benner, F. Chen and Z. Yang, Synthetic Biology, Tinkering Biology, and Artificial Biology: A Perspective from Chemistry, in Chemical Synthetic Biology, ed. P. L. Luisi and C. Chiarabelli, John Wiley & Sons, Ltd, Chichester, UK, 2011, pp. 69–106 Search PubMed; (d) S. A. Benner, Biol. Theory, 2013, 8, 357 CrossRef; (e) C. Wilson and A. D. Keefe, Curr. Opin. Chem. Biol., 2006, 10, 607 CrossRef CAS PubMed; (f) D. H. Appella, Curr. Opin. Chem. Biol., 2009, 13, 687 CrossRef CAS PubMed; (g) E. T. Kool, Curr. Opin. Chem. Biol., 2000, 4, 602 CrossRef CAS PubMed.
- (a) R. Kramer, J.-M. Lehn and A. Marquis-Rigault, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 5394 CrossRef CAS PubMed; (b) A. Marquis, V. Smith, J. Harrowfield, J.-M. Lehn, H. Herschbach, R. Sanvito, E. Leize-Wagner and A. van Dorsselaer, Chem.–Eur. J., 2006, 12, 5632 CrossRef CAS PubMed; (c) V. Berl, I. Huc, R. G. Khoury, M. J. Krische and J.-M. Lehn, Nature, 2000, 407, 720 CrossRef CAS PubMed; (d) V. Berl, I. Huc, R. G. Khoury and J.-M. Lehn, Chem.–Eur. J., 2001, 7, 2810 CAS; (e) H. L. Anderson, Inorg. Chem., 1994, 33, 972 CrossRef CAS; (f) P. N. Taylor and H. L. Anderson, J. Am. Chem. Soc., 1999, 121, 11538 CrossRef CAS; (g) J. Sánchez-Quesada, C. Seel, P. Prados, J. de Mendoza, I. Dalcol and E. Giralt, J. Am. Chem. Soc., 1996, 118, 277 CrossRef; (h) A. P. Bisson, F. J. Carver, D. S. Eggleston, R. C. Haltiwanger, C. A. Hunter, D. L. Livingstone, J. F. McCabe, C. Rotger and A. E. Rowan, J. Am. Chem. Soc., 2000, 122, 8856 CrossRef CAS; (i) A. P. Bisson and C. A. Hunter, Chem. Commun., 1996, 1723 RSC; (j) B. Gong, Y. Yan, H. Zeng, E. Skrzypczak-Jankunn, Y. W. Kim, J. Zhu and H. Ickes, J. Am. Chem. Soc., 1999, 121, 5607 CrossRef CAS; (k) B. Gong, Synlett, 2001, 582 CAS; (l) H. Zeng, R. S. Miller, R. A. Flowers and B. Gong, J. Am. Chem. Soc., 2000, 122, 2635 CrossRef CAS; (m) H. Zeng, H. Ickes, R. A. Flowers and B. Gong, J. Org. Chem., 2001, 66, 3574 CrossRef CAS PubMed; (n) B. Gong, Polym. Int., 2007, 56, 436 CrossRef CAS; (o) B. Gong, Acc. Chem. Res., 2012, 45, 2077 CrossRef CAS PubMed; (p) Y. Yang, Z.-Y. Yang, Y.-P. Yi, J.-F. Xiang, C.-F. Chen, L.-J. Wan and Z.-G. Shuai, J. Org. Chem., 2007, 72, 4936 CrossRef CAS PubMed; (q) W.-J. Chu, Y. Yang and C.-F. Chen, Org. Lett., 2010, 12, 3156 CrossRef CAS PubMed; (r) W.-J. Chu, J. Chen, C.-F. Chen, Y. Yang and Z. Shuai, J. Org. Chem., 2012, 77, 7815 CrossRef CAS PubMed; (s) E. A. Archer and M. J. Krische, J. Am. Chem. Soc., 2002, 124, 5074 CrossRef CAS PubMed; (t) H. Gong and M. J. Krische, J. Am. Chem. Soc., 2005, 127, 1719 CrossRef CAS PubMed; (u) Y. Tanaka, H. Katagiri, Y. Furusho and E. Yashima, Angew. Chem., 2005, 117, 3935 CrossRef; (v) H. Ito, Y. Furusho, T. Hasegawa and E. Yashima, J. Am. Chem. Soc., 2008, 130, 14008 CrossRef CAS PubMed; (w) Y. Furusho and E. Yashima, Macromol. Rapid Commun., 2011, 32, 136 CrossRef CAS PubMed; (x) H. Yamada, Y. Furusho, H. Ito and E. Yashima, Chem. Commun., 2010, 46, 3487 RSC; (y) C. A. Hunter, P. S. Jones, P. M. N. Tiger and S. Tomas, Chem. Commun., 2003, 1642 RSC; (z) G. K. Mittapalli, Y. M. Osornio, M. A. Guerrero, K. R. Reddy, R. Krishnamurthy and A. Eschenmoser, Angew. Chem., Int. Ed., 2007, 46, 2478 CrossRef CAS PubMed.
- A. E. Stross, G. Iadevaia and C. A. Hunter, Chem. Sci., 2016, 7, 94–101 RSC.
- C. A. Hunter and H. L. Anderson, Angew. Chem., Int. Ed., 2009, 48, 7488 CrossRef CAS PubMed.
- (a) H. Salem, M. Schmitt, U. Herrlich, E. Kühnel, M. Brill, P. Nägele, A. L. Bogado, F. Rominger and P. Hofmann, Organometallics, 2013, 32, 29 CrossRef CAS; (b) S. Raeppel, F. Raeppel and J. Suffert, Synlett, 1998, 794 CrossRef CAS.
- (a) C. S. Calero, J. Farwer, E. J. Gardiner, C. A. Hunter, M. Mackey, S. Scuderi, S. Thompson and J. G. Vinter, Phys. Chem. Chem. Phys., 2013, 15, 18262 RSC; (b) C. A. Hunter, Angew. Chem., Int. Ed., 2004, 43, 5310 CrossRef CAS PubMed.
- (a) A. F. Abdel-Magid and S. J. Mehrman, Org. Process Res. Dev., 2006, 10, 971 CrossRef CAS; (b) A. F. Abdel-Magid, K. G. Carson, B. D. Harris, C. A. Maryanoff and R. D. Shah, J. Org. Chem., 1996, 61, 3849 CrossRef CAS PubMed.
- B. Wang and M. R. Wasielewski, J. Am. Chem. Soc., 1997, 119, 12–21 CrossRef CAS.
- (a) U. Mayer, V. Gutmann and W. Gerger, Monatsh. Chem., 1975, 106, 1235 CrossRef CAS; (b) U. Mayer, W. Gerger and V. Gutmann, Monatsh. Chem., 1977, 108, 489 CrossRef CAS; (c) V. Gutmann, Pure Appl. Chem., 1979, 51, 2197 CrossRef CAS.
- (a) C. A. Hunter, M. C. Misuraca and S. M. Turega, J. Am. Chem. Soc., 2011, 133, 582 CrossRef CAS PubMed; (b) C. A. Hunter, M. C. Misuraca and S. M. Turega, J. Am. Chem. Soc., 2011, 133, 20416 CrossRef CAS PubMed; (c) E. Chekmeneva, C. A. Hunter, M. C. Misuraca and S. M. Turega, Org. Biomol. Chem., 2012, 10, 6022 RSC; (d) C. A. Hunter, M. C. Misuraca and S. M. Turega, Chem. Sci., 2012, 3, 2462 RSC; (e) C. A. Hunter, M. C. Misuraca and S. M. Turega, Chem. Sci., 2012, 3, 589 RSC; (f) H. Adams, E. Chekmeneva, C. A. Hunter, M. C. Misuraca, C. Navarro and S. M. Turega, J. Am. Chem. Soc., 2013, 135, 1853 CrossRef CAS PubMed; (g) E. Chekmeneva, C. A. Hunter, M. J. Packer and S. M. Turega, J. Am. Chem. Soc., 2008, 130, 17718 CrossRef CAS PubMed; (h) M. A. Jinks, H. Sun and C. A. Hunter, Org. Biomol. Chem., 2014, 12, 1440 RSC; (i) H. Sun, C. A. Hunter and E. M. Llamas, Chem. Sci., 2015, 6, 1444 RSC; (j) H. Sun, C. Navarro and C. A. Hunter, Org. Biomol. Chem., 2015, 13, 4981 RSC.
- (a) V. M. Krishnamurthy, V. Semetey, P. J. Bracher, N. Shen and G. M. Whitesides, J. Am. Chem. Soc., 2007, 129, 1312 CrossRef CAS PubMed; (b) Z. Zhong, X. Li and Y. Zhao, J. Am. Chem. Soc., 2011, 133, 8862 CrossRef CAS PubMed; (c) M. Mammen, S.-K. Choi and G. M. Whitesides, Angew. Chem., Int. Ed., 1998, 37, 2754 CrossRef; (d) A. Mulder, J. Huskens and D. N. Reinhoudt, Org. Biomol. Chem., 2004, 2, 3409 RSC; (e) W. Jiang, K. Nowosinski, N. L. Löw, E. V. Dzyuba, F. Klautzsch, A. Schäfer, J. Huuskonen, K. Rissanen and C. A. Schalley, J. Am. Chem. Soc., 2012, 134, 1860 CrossRef CAS PubMed; (f) E. T. Mack, P. W. Snyder, R. Perez-Castillejos, B. Bilgiçer, D. T. Moustakas, M. J. Butte and G. M. Whitesides, J. Am. Chem. Soc., 2012, 134, 333 CrossRef CAS PubMed; (g) H. Sun, C. A. Hunter, C. Navarro and S. Turega, J. Am. Chem. Soc., 2013, 135, 13129 CrossRef CAS PubMed; (h) M. C. Misuraca, T. Grecu, Z. Freixa, V. Garavini, C. A. Hunter, P. van Leeuwen, M. D. Segarra-Maset and S. M. Turega, J. Org. Chem., 2011, 76, 2723 CrossRef CAS PubMed; (i) J.-M. Lehn, Angew. Chem., Int. Ed. Engl., 1988, 27, 89 CrossRef; (j) D. J. Cram, G. M. Lein, T. Kaneda, R. C. Helgeson, C. B. Knobler, E. Maverick and K. N. Trueblood, J. Am. Chem. Soc., 1981, 103, 6228 CrossRef CAS; (k) D. J. Cram, Angew. Chem., Int. Ed. Engl., 1986, 25, 1039 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available: Detailed experimental procedures with spectroscopic characterization data, 31P NMR titration spectra and binding isotherms. See DOI: 10.1039/c5sc04467g |
| This journal is © The Royal Society of Chemistry 2016 |