A luminescence switch-on assay for the detection of specific gene deletion using G-quadruplex DNA and silver nanoclusters†
Modi
Wang‡
a,
Wanhe
Wang‡
a,
Chenfu
Liu
a,
Jinbiao
Liu
a,
Tian-Shu
Kang
b,
Chung-Hang
Leung
b and
Dik-Lung
Ma
*a
aDepartment of Chemistry, Hong Kong Baptist University, Kowloon Tong, Hong Kong, China. E-mail: edmondma@hkbu.edu.hk
bState Key Laboratory of Quality Research in Chinese Medicine, Institute of Chinese Medical Sciences, University of Macau, Macao, China
First published on 17th August 2016
Abstract
A switch-on gene deletion detection platform is reported based on the alteration of the effective distance between G-quadruplex DNA and silver nanoclusters (AgNCs). Moreover, the role of the G-quadruplex structure in interacting with AgNCs has also been demonstrated. The detection of the deletion mutants of LMP1 and CCR5 genes is performed as the proof-of-concept of our assay. Finally, the potential biological application of the assay was demonstrated in 10% v/v human serum.
Introduction
Gene deletion is a kind of DNA mutation that results in a part of the DNA being absent. Cytogenetically visible deletions occur in 1 in approximately every 7000 live births, and human disorders caused by chromosomal deletions include the cri du chat syndrome and the Prader–Willi syndrome.1 The phenotypic effects of deletions depend on the size of deletion, which can range from 5 bp to 2 bp, and the location of the deleted sequences on the genome. It has been reported that the resulting defect might be more drastic if more genes are involved with the larger gene deletion.2In consideration of the important role of gene deletion as a causative agent and as an indicator of diseases, it is of crucial importance to develop sensitive and efficient methods for gene deletion detection. Typical methods for the detection of gene deletions include capillary electrophoresis,3 polymerase chain reaction4 and rolling circle amplification.5 However these methods require time-consuming sample preparation and detection protocols as well as the use of expensive instruments.6,7 On the other hand, DNA oligonucleotides have been widely employed as recognition units targeting DNA, RNA, proteins and small molecules in various oligonucleotide-based detection platforms.
Silver nanoclusters (AgNCs) represent a new class of fluorophores that requires cytosine-rich nucleic acid templates for stabilization. AgNCs possess high quantum yields, good water solubility, high photostability, biocompatibility and low toxicity.8–12 Moreover, their emission can be fine-tuned by DNA sequence, size and the protecting layer of the NCs. Benefiting from these features, AgNCs have been widely utilized in optical sensing and biological imaging applications.13–16 In the literature, Willner's group has utilized AgNCs in conjunction with quantum dots and graphene oxide to develop a multiplexed analytical platform.17–19 Qu and co-workers have reported that molecular crowding could significantly facilitate the preparation of AgNCs20 thus widely employed in imaging.21–23 Furthermore, Martinez, Werner and co-workers reported that the fluorescence intensity of DNA/AgNCs can be increased when placed close to guanine-rich DNA sequences.24
The G-quadruplex is a DNA secondary structure formed from a guanine-rich DNA sequence. It consists of square-planar arrangements of guanine nucleobases stabilized by Hoogsteen hydrogen bonding and monovalent cations.25–29 Benefiting from the rich structural polymorphism, the G-quadruplex motif30 has stimulated the construction of numerous G-quadruplex-based sensing platforms for metal ions,31–35 DNA,36–39 small molecules,40,41 proteins42–47 and enzyme activity.36,48–52 While previous work has demonstrated that the emission of AgNCs is sensitive to the number of nearby guanine bases, we hypothesized that AgNCs may also respond to the formation of nearby G-quadruplex motifs. In this study, we investigated the effect of parameters such as the presence of the G-quadruplex and the distance between the guanine-rich sequence and AgNCs on the luminescence enhancement of AgNCs. As the number of nucleotide bases for gene deletion is closely related to the severity of the kind of disease, the detection of specific gene deletion is of great importance for medical diagnosis. By exploiting the sensitivity of AgNCs towards the distance between the AgNCs and guanine-rich DNA, we constructed a gene deletion detection platform to recognize deletion sequences of various lengths.
Scheme 1 shows the proposed sensing principle of this assay. P1 and P2 oligonucleotides contain antisense regions (red) that are proposed to recognize DNA sequences flanking the deletion site of the target gene. The P1 oligonucleotide contains a guanine-rich sequence that could be converted into a G-quadruplex motif under physiological conditions, while the P2 oligonucleotide contains a cytosine-rich sequence that acts as a template for the formation of AgNCs. Upon the addition of wild-type DNA without any deletion, the two oligonucleotide overhangs are separated by a significant distance. Hence, the G-quadruplex structure and AgNCs are far away from each other, and thus the AgNCs display a weak luminescence signal. For mutant DNA with gene deletion, the G-quadruplex and AgNCs will be in close proximity because some bases have been deleted, promoting strong luminescence of the AgNCs.
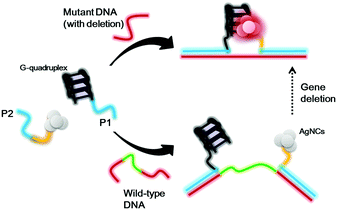 | ||
| Scheme 1 Schematic diagram of the detection strategy for gene deletion based on the distance between G-quadruplex DNA and silver nanoclusters. | ||
Results and discussion
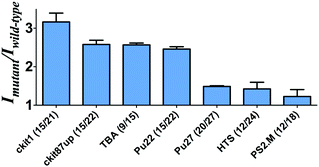
To investigate the feasibility of this assay, we first tested a 30-base deletion mutant of the latent membrane protein 1 (LMP1) gene (Fig. S1, ESI†). The system displayed a significant enhancement in the presence of a 30-base deletion mutant compared with wild-type DNA (DNA without deletion). As potassium ions affect the stabilization of the G-quadruplex, we anticipate that the performance of the assay may be affected by the concentration of potassium ions. To improve the performance of the assay, we investigated the effect of K+ concentration on the luminescence enhancement of the detection system. It was observed that the luminescence enhancement ratio between the mutant and wild-type DNA reached close to the maximum value at 50 mM K+ (Fig. S2, ESI†). As the control experiment, we utilized both the sequence with poly-C bases and the sequence without C bases to prepare AgNCs in parallel. Encouragingly, we found that the sample using the poly-C sequence as a template showed strong luminescence at λ = 630 nm, whereas a DNA template without C bases didn't show any luminescence (Fig. S3, ESI†). Therefore, it demonstrates that AgNCs could be formed with cytosine-rich nucleic acid templates.Next, we examined the relative luminescence enhancement of the system towards wild-type and deletion mutant DNA employing different kinds of G-quadruplex sequences, including c-kit87up, c-kit1, Pu27, Pu22, HTS, PS2.M and TBA (Fig. 1). The results showed that the mutant to wild-type luminescence enhancement ratio of the system was highly sensitive to the type of G-quadruplex sequence employed. The number of guanine bases in the G-quadruplex sequence was not necessarily desirable, as the Pu27 G-quadruplex containing the highest number of guanine bases (20) and total bases (27) showed a relatively low luminescence enhancement. The maximal ratio of ca. 3 was observed when P1 contained the c-kit1 sequence, which contains 15 guanine bases and 21 total bases. This result indicates that the G-quadruplex sequence plays an important role in the luminescence response of the AgNCs.
To further validate that the G-quadruplex structure plays a significant role in the luminescence enhancement of the AgNCs and investigate if there is any different luminescence enhancement of AgNCs with and without G-quadruplex formation, we prevented the formation of the G-quadruplex through the lack of the G-quadruplex stabilizing ion environment, as a control, to compare the luminescence enhancement of the G-quadruplex forming sequence with 50 mM K+ ions. A higher luminescence enhancement has been observed in the presence of 50 mM K+ ions (Fig. 2). It indicates that not only the number of guanine bases contribute to the luminescence enhancement but also the formation of the G-quadruplex.
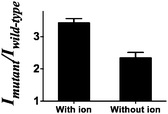 | ||
| Fig. 2 Mutant to wild-type intensity ratio of the system with/without the G-quadruplex stabilizing ions. | ||
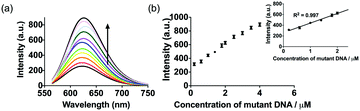
To investigate the linear range of the assay utilizing the ckit1 G-quadruplex, we carried out the emission titration experiment with increasing concentrations of mutant DNA. Encouragingly, we observed that the luminescence response of the system was increased with mutant DNA concentration (Fig. 3). The system showed a ca. 3.5-fold enhancement upon the addition of 4 µM of mutant DNA, with a detection limit of 53 nM using the three-sigma method. This result outlines that the AgNCs and G-quadruplex DNA become closer after hybridization with the mutant DNA.
To demonstrate the versatility of the gene deletion detection system, we also employed this platform to detect the mutant allele (CCR5-Δ32), which harbors a 32 bp deletion of the chemokine receptor gene (CCR5). Upon the addition of mutant DNA, a 2.4-fold greater enhancement was observed compared with wild-type DNA (Fig. S4, ESI†). Comparing with the DNA detection approach, the smaller fold change of the system may be due to the relatively short distance (30 nb) between P1 and P2 in the presence of wild type DNA in comparison with P1 and P2 as random coil in the DNA detection approach. Furthermore, we sought to investigate the effect of the number of deletion bases on the performance of the assay. Consistent with the proposed working principle of this assay, the increasing number of deleted bases in the LMP1 gene from 5 to 30 bases led to an improvement of the mutant to wild-type luminescence enhancement ratio of the system (Fig. 4). It is known that for some genetic diseases, there is a correlation between clinical severity and the size of deletion.53 For instance, the Cri du Chat syndrome may be caused by deletion sizes ranging from 5 to 40 Mb.54 We envision that the ability of the system to distinguish between long and short deletion mutants could potentially be used as a diagnostic tool to predict the clinical severity of a disease.
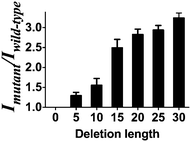 | ||
| Fig. 4 Mutant to wild type ratio of the system between mutant DNA of different deletion lengths (deletion length of 5, 10, 15, 20, 25, 30 bases) versus wild-type DNA (no deletion). | ||
Furthermore, the applicability of this detecting platform in a biological sample has also been evaluated. The stability of the luminescence of AgNCs is similar both in the presence and absence of G-rich sequences and 10% (v/v) human serum (Fig. S5, ESI†). Meanwhile, in a system containing 10% (v/v) human serum, the system experienced stronger luminecence in the presence of mutant DNA compared with wild-type DNA (Fig. 5). This result suggests that this assay could potentially be employed to biological samples even with the sample matrix.
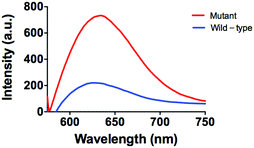 | ||
| Fig. 5 Emission spectra of the system (5 µM P1 and P2) in the presence of mutant DNA (5 µM) or wild-type DNA (5 µM) in 10% human serum. | ||
Experimental
Materials
Reagents, unless specified, were purchased from Sigma Aldrich (St. Louis, MO). All oligonucleotides were synthesized by Techdragon Inc. (Hong Kong, China).Synthesis of oligonucleotide-stabilized Ag nanoclusters
The oligonucleotide-stabilized Ag nanoclusters were synthesized according to previous reports. Briefly, a mixture of AgNO3 and DNA was prepared in a molar ratio of 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and the solution was then shaken vigorously for 30 s. After vigorous shaking, freshly prepared NaBH4 (10 µL) at the same concentration as the AgNO3 solution was added into this mixture for reduction followed by vigorous shaking of the mixture for 30 s. The reduced oligonucleotide–Ag solution was incubated at 4 °C and allowed to react for 1 h in the dark. Silver ions bound with cytosine were reduced to form nanoclusters.
1, and the solution was then shaken vigorously for 30 s. After vigorous shaking, freshly prepared NaBH4 (10 µL) at the same concentration as the AgNO3 solution was added into this mixture for reduction followed by vigorous shaking of the mixture for 30 s. The reduced oligonucleotide–Ag solution was incubated at 4 °C and allowed to react for 1 h in the dark. Silver ions bound with cytosine were reduced to form nanoclusters.
Conclusions
To conclude, we report a luminescence switch-on detection platform for specific gene deletion utilizing G-quadruplex DNA and AgNCs. We demonstrated that the luminescence enhancement of the AgNCs was sensitive to the G-quadruplex sequence. And we demonstrated the proof-of-concept of our assay to detect the deletion mutants of the LMP1 and CCR5 genes. Finally, the potential biological application of the assay was demonstrated in 10% (v/v) human serum. We anticipate that the developed assay could potentially be used to monitor gene deletion mutants relevant to human diseases.Acknowledgements
This work was supported by Hong Kong Baptist University (FRG2/14-15/004 and FRG2/15-16/002), the Health and Medical Research Fund (HMRF/14130522), the Research Grants Council (HKBU/201811, HKBU/204612 and HKBU/201913), the French Agence Nationale de la Recherche/Research Grants Council Joint Research Scheme (AHKBU201/12; Oligoswitch ANR-12-IS07-0001), National Natural Science Foundation of China (21575121), Guangdong Province Natural Science Foundation (2015A030313816), Hong Kong Baptist University Century Club Sponsorship Scheme 2015, Interdisciplinary Research Matching Scheme (RC-IRMS/14-15/06), the Science and Technology Development Fund, Macao SAR (098/2014/A2), the University of Macau (MYRG091(Y3-L2)-ICMS12-LCH and MYRG2015-00137-ICMS-QRCM).Notes and references
- P. A. Jacobs, C. Browne, N. Gregson, C. Joyce and H. White, J. Med. Genet., 1992, 29, 103–108 CrossRef CAS PubMed.
- S. Clancy and K. M. Shaw, Nature Education, 2008, 1, 23 Search PubMed.
- K. Kleparnik and P. Bocek, Chem. Rev., 2007, 107, 5279–5317 CrossRef CAS PubMed.
- R. Saiki, D. Gelfand, S. Stoffel, S. Scharf, R. Higuchi, G. Horn, K. Mullis and H. Erlich, Science, 1988, 239, 487–491 CAS.
- D. Hourcade, D. Dressler and J. Wolfson, Proc. Natl. Acad. Sci. U. S. A., 1973, 70, 2926–2930 CrossRef CAS.
- D. Zhu, Y. Tang, D. Xing and W. R. Chen, Anal. Chem., 2008, 80, 3566–3571 CrossRef CAS PubMed.
- L. Yang, A. Pan, H. Zhang, J. Guo, C. Yin and D. Zhang, J. Agric. Food Chem., 2006, 54, 9735–9740 CrossRef CAS PubMed.
- S. H. Yau, N. Abeyasinghe, M. Orr, L. Upton, O. Varnavski, J. H. Werner, H.-C. Yeh, J. Sharma, A. P. Shreve, J. S. Martinez and T. Goodson Iii, Nanoscale, 2012, 4, 4247–4254 RSC.
- J. T. Petty, J. Zheng, N. V. Hud and R. M. Dickson, J. Am. Chem. Soc., 2004, 126, 5207–5212 CrossRef CAS PubMed.
- C. I. Richards, S. Choi, J.-C. Hsiang, Y. Antoku, T. Vosch, A. Bongiorno, Y.-L. Tzeng and R. M. Dickson, J. Am. Chem. Soc., 2008, 130, 5038–5039 CrossRef CAS PubMed.
- Y. Tao, M. Li, J. Ren and X. Qu, Chem. Soc. Rev., 2015, 44, 8636–8663 RSC.
- J. Sharma, R. C. Rocha, M. L. Phipps, H.-C. Yeh, K. A. Balatsky, D. M. Vu, A. P. Shreve, J. H. Werner and J. S. Martinez, Nanoscale, 2012, 4, 4107–4110 RSC.
- X. Ran, Z. Wang, Z. Zhang, F. Pu, J. Ren and X. Qu, Chem. Commun., 2016, 52, 557–560 RSC.
- Z. Zhou, T. Li, H. Huang, Y. Chen, F. Liu, C. Huang and N. Li, Chem. Commun., 2014, 50, 13373–13376 RSC.
- H. Jiang, G. Xu, Y. Sun, W. Zheng, X. Zhu, B. Wang, X. Zhang and G. Wang, Chem. Commun., 2015, 51, 11810–11813 RSC.
- F. Cao, E. Ju, C. Liu, F. Pu, J. Ren and X. Qu, Chem. Commun., 2016, 52, 5167–5170 RSC.
- N. Enkin, F. Wang, E. Sharon, H. B. Albada and I. Willner, ACS Nano, 2014, 8, 11666–11673 CrossRef CAS PubMed.
- E. Sharon, N. Enkin, H. B. Albada and I. Willner, Chem. Commun., 2015, 51, 1100–1103 RSC.
- X. Liu, F. Wang, R. Aizen, O. Yehezkeli and I. Willner, J. Am. Chem. Soc., 2013, 135, 11832–11839 CrossRef CAS PubMed.
- Z. Huang, J. Ren, W. Yang and X. Qu, Chem. Commun., 2013, 49, 10856–10858 RSC.
- Y. Tao, Z. Li, E. Ju, J. Ren and X. Qu, Nanoscale, 2013, 5, 6154–6160 RSC.
- Y. Tao, Z. Li, E. Ju, J. Ren and X. Qu, Chem. Commun., 2013, 49, 6918–6920 RSC.
- Y. Teng, X. Jia, S. Zhang, J. Zhu and E. Wang, Chem. Commun., 2016, 52, 1721–1724 RSC.
- H.-C. Yeh, J. Sharma, J. J. Han, J. S. Martinez and J. H. Werner, Nano Lett., 2010, 10, 3106–3110 CrossRef CAS PubMed.
- J. Zhou, K. Murayama, S. Amrane, F. Rosu, H. Kashida, A. Bourdoncle, H. Asanuma and J.-L. Mergny, Chem. Sci., 2013, 4, 3693–3698 RSC.
- S. Neidle, Curr. Opin. Struct. Biol., 2009, 19, 239–250 CrossRef CAS PubMed.
- Y. Xue, Z.-y. Kan, Q. Wang, Y. Yao, J. Liu, Y.-h. Hao and Z. Tan, J. Am. Chem. Soc., 2007, 129, 11185–11191 CrossRef CAS PubMed.
- G. F. Salgado, C. Cazenave, A. Kerkour and J.-L. Mergny, Chem. Sci., 2015, 6, 3314–3320 RSC.
- H. Bertrand, S. Bombard, D. Monchaud, E. Talbot, A. Guedin, J.-L. Mergny, R. Grunert, P. J. Bednarski and M.-P. Teulade-Fichou, Org. Biomol. Chem., 2009, 7, 2864–2871 CAS.
- Y. Krishnan and F. C. Simmel, Angew. Chem., Int. Ed., 2011, 50, 3124–3156 CrossRef CAS PubMed.
- H. Xu, S. Gao, Q. Yang, D. Pan, L. Wang and C. Fan, ACS Appl. Mater. Interfaces, 2010, 2, 3211–3216 CAS.
- Z. Zhu, Y. Su, J. Li, D. Li, J. Zhang, S. Song, Y. Zhao, G. Li and C. Fan, Anal. Chem., 2009, 81, 7660–7666 CrossRef CAS PubMed.
- Y. Shi, H. Sun, J. Xiang, H. Chen, Q. Yang, A. Guan, Q. Li, L. Yu and Y. Tang, Chem. Commun., 2014, 50, 15385–15388 RSC.
- H. Sun, J. Xiang, W. Gai, Y. Liu, A. Guan, Q. Yang, Q. Li, Q. Shang, H. Su, Y. Tang and G. Xu, Chem. Commun., 2013, 49, 4510–4512 RSC.
- H. Chen, H. Sun, X. Zhang, X. Sun, Y. Shi and Y. Tang, RSC Adv., 2015, 5, 1730–1734 RSC.
- X. Su, C. Zhang, X. Zhu, S. Fang, R. Weng, X. Xiao and M. Zhao, Anal. Chem., 2013, 85, 9939–9946 CrossRef CAS PubMed.
- Y. Wen, Y. Xu, X. Mao, Y. Wei, H. Song, N. Chen, Q. Huang, C. Fan and D. Li, Anal. Chem., 2012, 84, 7664–7669 CrossRef CAS PubMed.
- H. Xu, Q. Yang, F. Li, L. Tang, S. Gao, B. Jiang, X. Zhao, L. Wang and C. Fan, Analyst, 2013, 138, 2678–2682 RSC.
- H. Xu, F. Geng, Y. Wang, M. Xu, X. Lai, P. Qu, Y. Zhang and B. Liu, Chem. Commun., 2015, 51, 8622–8625 RSC.
- Y. Peng, X. Wang, Y. Xiao, L. Feng, C. Zhao, J. Ren and X. Qu, J. Am. Chem. Soc., 2009, 131, 13813–13818 CrossRef CAS PubMed.
- C. Wang, J. Wu, C. Zong, H. Ju and F. Yan, Analyst, 2011, 136, 4295–4300 RSC.
- Y. Liu, Q. Zhou and A. Revzin, Analyst, 2013, 138, 4321–4326 RSC.
- M. Yin, Z. Li, Z. Liu, J. Ren, X. Yang and X. Qu, Chem. Commun., 2012, 48, 6556–6558 RSC.
- L. Cao, L. Cheng, Z. Zhang, Y. Wang, X. Zhang, H. Chen, B. Liu, S. Zhang and J. Kong, Lab Chip, 2012, 12, 4864–4869 RSC.
- T. Kwa, Q. Zhou, Y. Gao, A. Rahimian, L. Kwon, Y. Liu and A. Revzin, Lab Chip, 2014, 14, 1695–1704 RSC.
- M. Zou, Y. Chen, X. Xu, H. Huang, F. Liu and N. Li, Biosens. Bioelectron., 2012, 32, 148–154 CrossRef CAS PubMed.
- M. Wang, Z. Mao, T.-S. Kang, C.-Y. Wong, J.-L. Mergny, C.-H. Leung and D.-L. Ma, Chem. Sci., 2016, 7, 2516–2523 RSC.
- C.-H. Leung, D. S.-H. Chan, B. Y.-W. Man, C.-J. Wang, W. Lam, Y.-C. Cheng, W.-F. Fong, W. W.-L. Siu and D.-L. Ma, Anal. Chem., 2011, 83, 463–466 CrossRef CAS PubMed.
- L. Lu, H.-Z. He, D. S.-H. Chan, C.-H. Leung and D.-L. Ma, Chem. Sci., 2014, 5, 4561–4568 RSC.
- K.-H. Leung, B. He, C. Yang, C.-H. Leung, H.-M. D. Wang and D.-L. Ma, ACS Appl. Mater. Interfaces, 2015, 7, 24046–24052 CAS.
- X. Su, X. Zhu, C. Zhang, X. Xiao and M. Zhao, Anal. Chem., 2012, 84, 5059–5065 CrossRef CAS PubMed.
- Y. Lin, J. Ren and X. Qu, Acc. Chem. Res., 2014, 47, 1097–1105 CrossRef CAS PubMed.
- P. Cerruti Mainardi, Orphanet J. Rare Dis., 2006, 1, 1–9 CrossRef.
- A. D. Simmons, S. A. Goodart, T. D. Gallardo, J. Overhauser and M. Lovett, Hum. Mol. Genet., 1995, 4, 295–302 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6qm00068a |
| ‡ These authors contributed equally to this work. |
| This journal is © the Partner Organisations 2017 |